Significance
Mutations in the JAK pseudokinase domain are bona fide oncogenic drivers that underlie many myeloproliferative and autoimmune diseases in humans. The JAK2 V617F mutation is responsible for ∼95% of polycythemia vera and ∼60% of primary myelofibrosis and essential thrombocytosis cases. Currently, developed JAK2 tyrosine kinase inhibitors have not been able to eradicate disease caused by mutated JAK2. The data presented here show that alteration of the ATP binding site of the pseudokinase domain has the potential to suppress JAK hyperactivation caused by pathogenic mutations, with minimal effects on wild-type JAK, thus establishing the ATP binding site of the pseudokinase domain as a potential pharmacological target.
Keywords: JAK, pseudokinase domain, nucleotide binding, cytokine, myeloid neoplasia
Abstract
Pseudokinases lack conserved motifs typically required for kinase activity. Nearly half of pseudokinases bind ATP, but only few retain phosphotransfer activity, leaving the functional role of nucleotide binding in most cases unknown. Janus kinases (JAKs) are nonreceptor tyrosine kinases with a tandem pseudokinase–kinase domain configuration, where the pseudokinase domain (JAK homology 2, JH2) has important regulatory functions and harbors mutations underlying hematological and immunological diseases. JH2 of JAK1, JAK2, and TYK2 all bind ATP, but the significance of this is unclear. We characterize the role of nucleotide binding in normal and pathogenic JAK signaling using comprehensive structure-based mutagenesis. Disruption of JH2 ATP binding in wild-type JAK2 has only minor effects, and in the presence of type I cytokine receptors, the mutations do not affect JAK2 activation. However, JH2 mutants devoid of ATP binding ameliorate the hyperactivation of JAK2 V617F. Disrupting ATP binding in JH2 also inhibits the hyperactivity of other pathogenic JAK2 mutants, as well as of JAK1 V658F, and prevents induction of erythrocytosis in a JAK2 V617F myeloproliferative neoplasm mouse model. Molecular dynamic simulations and thermal-shift analysis indicate that ATP binding stabilizes JH2, with a pronounced effect on the C helix region, which plays a critical role in pathogenic activation of JAK2. Taken together, our results suggest that ATP binding to JH2 serves a structural role in JAKs, which is required for aberrant activity of pathogenic JAK mutants. The inhibitory effect of abrogating JH2 ATP binding in pathogenic JAK mutants may warrant novel therapeutic approaches.
The Janus kinases (JAK1–3, TYK2) are a family of nonreceptor tyrosine kinases with essential functions in the regulation of hematopoiesis, the immune system, and cellular metabolism. JAKs interact specifically with various cytokine receptors and couple cytokine binding to cytoplasmic signaling cascades, including the signal transducers and activators of transcription (STAT) pathway. JAKs consist of an N-terminal FERM domain, an SH2-like (Src homology 2) domain, a pseudokinase domain (JAK homology 2, JH2), and the C-terminal tyrosine kinase domain (JH1). JH2 mediates critical regulatory functions in JAKs and primarily serves to inhibit basal JH1 activity. Experimental deletion of JH2 increases JH1 activity in full-length JAK in the absence of stimulation (1–3), and in recombinant systems addition of JH2 suppresses JH1 activity (4–6). JH2 is, however, also required for ligand-induced activation of full-length JAKs in cell (1–3, 6, 7). The regulatory functions of JH2 are corroborated by the multitude of human disease mutations identified in the domain. The most common JAK2 mutation, V617F, leads to cytokine-independent signaling through the exclusively JAK2-dependent homotypic receptors for erythropoietin (EPO), granulocyte colony stimulating factor (G-CSF), and thrombopoietin (8). The V617F mutation is found in ∼95% of patients with polycythemia vera (PV) (9–12) as well as in ∼60% of patients with essential thrombocythemia (ET) and primary myelofibrosis (PMF). After identification of the V617F mutation, a multitude of other mutations in JAK2, JAK1, and JAK3 have been found that are linked to myeloid and lymphoid malignancies and to immunological diseases as well as to some solid cancers (13, 14). The mutations cluster mainly in exon 12 in the SH2–JH2 linker (numbering for human JAK2), exon 14 near Val617, and exon 16 (13). Although most JAK JH2 mutations are gain-of-function, some JH2 mutations in JAK3 suppress JH1 activity, leading to severe combined immunodeficiency (2, 15).
The mechanism by which JH2 regulates JAK activity has long been enigmatic, but recent studies have provided previously unidentified insights. The crystal structure of JAK2 JH2 (16) revealed a prototypical kinase-domain fold that binds ATP (17), but with a noncanonical binding mode. Additionally, JAK2 JH2 was found to possess weak kinase activity in vitro and autophosphorylate two regulatory sites: Ser523 in the SH2–JH2 linker and Tyr570 in JH2 itself (17). The structure of JAK2 JH2 V617F is highly similar to wild-type JH2 but shows a rigidified C helix (αC) in the kinase N lobe and a slightly altered ATP binding cleft (16). These structural differences, however, do not provide an obvious explanation for the mechanism of pathogenic activation. A recent simulation-based model of the JAK2 tandem kinase domains (JH2–JH1) (18) and a crystal structure of TYK2 JH2–JH1 (5) show an extensive interaction interface between JH2 and the backside of JH1, providing a rationale for the autoinhibitory interaction mediated by JH2. Importantly, practically all known disease-causing JH1 and JH2 mutations localize in or near the JH2–JH1 interface and are expected to destabilize the interaction (13, 18). The structures of JAK1 and TYK2 JH2 are highly similar to JAK2 JH2 (5, 19), and all three JH2s bind ATP (20). The regulatory residues Ser523 and Tyr570 in JAK2 are not conserved in other JAK family members, and the catalytic function of JH2 appears to be a unique characteristic of JAK2, which is also the only JAK to function as homodimers on type I cytokine receptors. The conserved function of nucleotide binding in all JAK JH2s, however, is currently unknown.
Research on JH2 also ties into the field of pseudokinases in general. Pseudokinases are kinase-like proteins that lack one or more conserved catalytic residues and constitute almost 10% of the human kinome (21). Many pseudokinases have retained the ability to bind nucleotides, yet the physiological function of this binding has remained unknown in most cases. Deciphering the function of these nucleotide binding sites is of practical importance, as the ATP binding pocket is a well-validated pharmacological target (22). Intrigued by these questions, we set out to investigate the functional role of ATP binding in JAK JH2, focusing on its role in the regulation of JAK2 signaling in wild-type and pathogenic contexts.
Results
Establishing JAK2 JH2 ATP Binding Site Mutations.
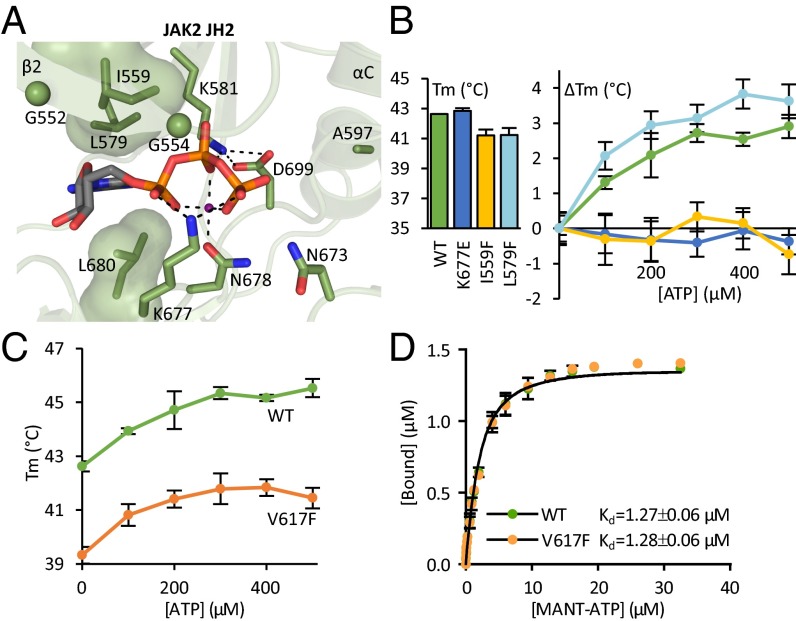
JAK2 JH2 has an ATP binding pocket with unusual characteristics (Fig. 1A) (16). To study the role of nucleotide binding, we used a systematic, structure-based mutagenesis approach designed to distinguish the effect of nucleotide binding from possible structural effects caused by the mutation, as has been observed with, for example, β-strand 3 (β3) lysine mutations (23, 24). We thus mutated not only the β3 lysine (K581A) and its interaction partner (D699A) but also the glycine-rich loop (G552A G554A) residues on the catalytic loop interacting directly with ATP (K677E) or the divalent cation (N678A), as well as residues lining the purine binding pocket (I559F, L579F). To compare the ATP binding site mutations with a structurally destabilizing mutation, we mutated Phe739 to arginine in the hydrophobic core of the C lobe (16) (Table 1).
Fig. 1.
Characterizing the ATP binding pocket of JAK2 JH2. (A) The ATP binding pocket of JAK2 JH2 (16) [Protein Data Bank (PDB) ID code 4FVQ] highlighting the noncanonical mode of nucleotide binding. JAK2 JH2 contains a bulky leucine (Leu579) on the N-lobe side of the purine pocket substituting for the canonical alanine in the VAIK motif. The glycine-rich loop consists of two glycines (shown as spheres) rather than three. The phosphates of bound ATP (shown as sticks) are coordinated by only one divalent cation (shown in magenta) instead of two in typical kinases. Furthermore, Asp699 of the DPG motif (consensus DFG) forms a salt bridge to the β3 lysine (Lys581), and Lys677 from the catalytic loop binds directly to the α and γ phosphates of ATP. Dotted lines highlight hydrogen bonds and salt bridges participating in the binding of ATP. The hydrophobic amino acids lining the purine base and sugar moiety binding site are shown as volume-filling models. (B) Fluorometric TSA of recombinant JAK2 JH2. Tms of JAK2 JH2 ATP binding site mutants are shown in the bar graph. TSA shows no thermal stabilization for JAK2 JH2 I559F or JAK2 JH2 K677E upon addition of ATP (ΔTm, line graph). (C) Thermal stability of recombinant JAK2 JH2 wild type and V617F upon addition of ATP to wild-type and V617F JH2 shows similar ATP responses yet overall reduced thermal stability in V617F. The data for wild type are the same as shown in B. (D) MANT-ATP binding assay on recombinant JAK2 JH2 reveals identical MANT-ATP binding affinities for wild type and V617F. All experiments were done in the presence of Mg2+. All error bars are standard deviations (SD) from triplicate experiments.
Table 1.
Summary of JAK2 and JAK1 point mutations used to study the function of the ATP binding site of JH2
| Mutation | Rationale/effect | Producible JH2 |
| JAK2 | ||
| S523A | Removes Ser523 phosphorylation | — |
| K539L | Hyperactivating MPN mutation in JH2–JH1 interface (5, 18) | — |
| G552A G554A | Removes flexible glycines usually needed for ATP binding | No |
| I559F | β2; designed to sterically inhibit ATP binding; verifiably inhibits ATP binding (Fig. 1B) | Yes |
| I559E | β2; designed to electrostatically inhibit ATP binding | No |
| Y570F | Removes Tyr570 phosphorylation | — |
| K581A | Removes β3 lysine needed for ATP binding | No |
| L579F | β3; designed to sterically inhibit ATP binding; does not inhibit ATP binding (Fig. 1B) | Yes |
| V617F | Hyperactivating MPN mutation | Yes |
| K677E | Exchanges catalytic loop lysine interacting with α and γ phosphates of ATP with oppositely charged residue; verifiably inhibits ATP binding (Fig. 1B) | Yes |
| N678A | Removes catalytic loop asparagine coordinating binding of cation needed for ATP binding | No |
| R683S | Hyperactivating MPN mutation directly in JH2–JH1 interface (5, 18) | — |
| D699A | DFG (DPG in JAK2) motif; disrupts Lys581-Asp699 salt bridge | No |
| F739R | Designed to disrupt JH2 by introducing charged residue into hydrophobic core of C lobe (16) | No |
| JAK1 | ||
| G590A G592A | Analogous to JAK2(G552A G554A) | — |
| K622A | Analogous to JAK2(K581A) | — |
| V658F | Analogous to JAK2(V617F) | — |
Constructs not tested for recombinant production are marked with a “—.”
To verify the effects of the mutations on ATP binding, we produced recombinant JAK2 JH2. Fluorometric thermal-shift analysis (TSA) showed that the mutations K677E, I559F, and L579F have only a marginal effect on the melting temperature (Tm) of apo JH2 (Fig. 1B, bar graph). ATP binding-induced stabilization, however, was completely abrogated in K677E and I559F but not in L579F (Fig. 1B, line graph). These results indicate that I559F and K677E do not affect the proper folding of the domain but that they effectively block ATP binding. L579F evidently did not prevent ATP binding. The effect of the other mutations described above could not be explicitly analyzed due to lack of recombinant expression (Table 1).
ATP Binding in JAK2 JH2 V617F.
The crystal structure of JAK2 JH2 V617F is highly similar to the wild-type structure (16), with small changes in αC and the β3–αC linker and a slight change in the topography of the ATP binding pocket due to alignment of Phe617, Phe595, and Phe594 (the latter two in αC) in V617F (Fig. S1). To gauge whether these differences have any effect on the stability of JH2 and its affinity for ATP, we used TSA and a FRET-based 2’/3′-(N-methyl-anthraniloyl)–ATP (MANT-ATP) binding assay on recombinant JAK2 JH2. TSA showed that the V617F mutation lowers the overall thermal stability of the domain (Tm = 39.3 ± 0.3 °C compared with 42.6 ± 0.2 °C for wild type). Addition of Mg-ATP caused similar stabilization for both domains, with Tm shifts of up to 3 °C, which was, however, still not enough to bring the Tm of V617F to the level of wild type (Fig. 1C). Quantification of Mg–MANT-ATP binding affinity showed no difference between JAK2 JH2 wild type and V617F, with dissociation constants of 1.3 ± 0.1 µM for both (Fig. 1D). Thus, V617F lowers the thermal stability of JH2 but does not substantially affect ATP binding.
Analysis of JAK2 JH2 ATP Binding Site Mutations in the Absence of JAK2-Associated Type I Receptors.
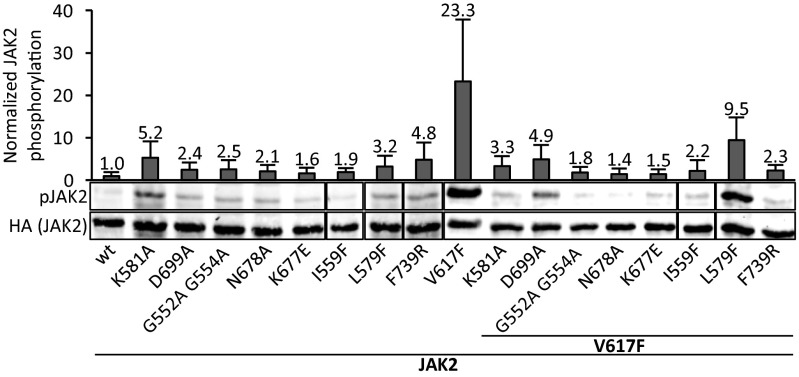
The ATP-coordinating residues were mutated in the context of full-length JAK2, and activation was analyzed in transfected JAK2-deficient γ2A cells by measuring JH1 activation-loop phosphorylation (pY1007–pY1008) (Fig. 2). γ2A is a fibroblast cell line and thus lacks expression of JAK2-associated homotypic type I myeloid cytokine receptors (EPOR, MPL, and G-CSF-R). In the context of wild-type JAK2, the JH2 mutations resulted generally in small increases in basal JAK2 activity. Specifically, the verified ATP binding-deficient mutants K677E and I559F showed 1.6- and 1.9-fold increases over wild-type JAK2, whereas the less conservative change K581A caused a larger ∼fivefold increase in pY1007–pY1008 levels, as did the structurally disruptive F739R mutation (Fig. 2).
Fig. 2.
Mutation of the JAK2 JH2 ATP binding site removes V617F-mediated hyperactivation. Shown is the whole-cell lysate immunoblot from γ2A cells transfected with full-length human JAK2-HA without exogenous receptors. JAK2 expression levels are shown using anti-HA staining from the same blots. Bar graph shows pJAK2(Y1007–Y1008) quantification normalized to HA levels from immunoblots like the one shown, as averages from three independent experiments. Error bars are SDs. See Table 1 for explanation of the mutants.
Next, the different ATP binding site mutations were introduced into full-length JAK2 V617F, and the role of JH2 nucleotide binding on cytokine-independent activation was analyzed. Expression of V617F resulted in >20-fold hyperphosphorylation compared with wild type. Strikingly, almost all ATP binding site mutations reverted the high basal activity of V617F to near wild-type levels (Fig. 2). The inhibition was most prominent in the glycine-rich loop mutant (G552A G554A) and both catalytic loop mutants (N678A, K677E), with pY1007–pY1008 levels nearly wild type (1.4–1.8-fold of wild type). L579F, which did not abrogate ATP binding (Fig. 1B), gave the smallest reduction in hyperphosphorylation (Fig. 2). Furthermore, mutation of JH2 autophosphorylation sites (Ser523 and Tyr570) (17) did not lower V617F hyperactivity (Fig. S2), demonstrating that the suppression of aberrant activation by the JH2 ATP binding mutations is not due to loss of JH2 catalytic activity.
Removal of ATP Binding Is Distinct From Structural Disruption of JH2 and Does Not Affect Type I or Type II Cytokine Receptor Signaling.
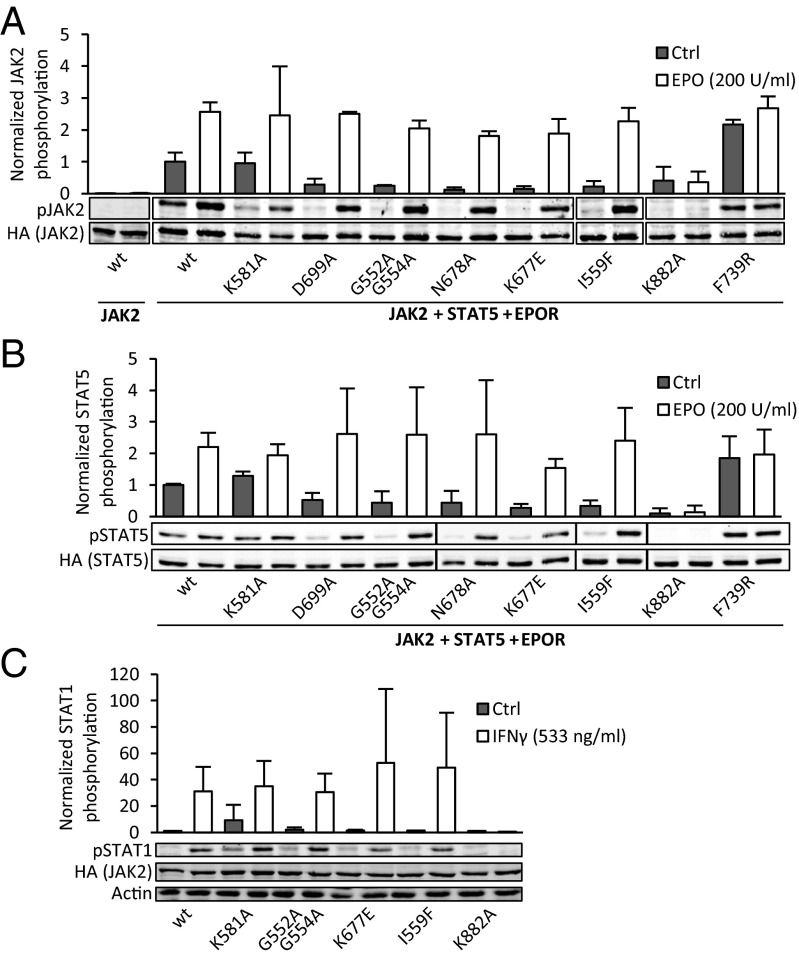
To assess the effect of JH2 ATP binding site mutations on the cytokine inducibility of JAK2, we coexpressed type I cytokine receptor EPOR, STAT5A, and the JAK2 mutants in γ2A cells. A kinase-inactivating mutation in JH1 (K882A) was included as a control. In the presence of EPOR, the ATP binding site mutations did not increase basal JAK2 activation compared with wild-type JAK2, whereas the destabilizing mutation F739R still caused a marked increase (Fig. 3A). This result was also evident in downstream signaling, with anti-pSTAT5A blotting showing low basal STAT5A phosphorylation for the ATP binding site mutants yet increased phosphorylation for F739R (Fig. 3B). EPO-induced JAK2 activation and signaling remained essentially unchanged in all ATP site mutants. In contrast, the destabilizing JH2 mutation (F739R) was refractory to EPO stimulation (Fig. 3 A and B), resembling the effect of JH2 deletion (1, 6, 7).
Fig. 3.
Disruption of the JAK2 JH2 ATP binding site is distinct from structural disruption. (A) JAK2(Y1007–Y1008) phosphorylation of JAK2 mutants in the presence of type I cytokine receptor (EPOR) in γ2A cells. Basal JAK2 phosphorylation in the absence of EPOR expression is shown on the left. (B) STAT5A(Y694) phosphorylation from the same samples as pJAK2 in A. Phosphorylation was measured from whole-cell lysates of transfected γ2A cells using immunoblotting and normalized to JAK2-HA and STAT5-HA expression levels, respectively. Expression levels of EPOR were analyzed by immunoblotting with anti-HA and found to be equal. (C) STAT1(Y701) phosphorylation of endogenous STAT1 in γ2A cells. Bar graph shows quantification of pSTAT1 from immunoblots normalized to basal pSTAT1 levels in cells transfected with wild-type JAK2 (leftmost sample). Actin is shown as a loading control. All error bars are SDs from three independent experiments.
In addition to homodimeric type I cytokine receptors, JAK2 functions on type II cytokine receptors where signaling relies on trans-activation of two different JAKs. We thus analyzed activation of STAT1 through the interferon γ (IFNγ) receptor in γ2A cells. Congruent with the effect observed on pJAK2, basal pSTAT1 was increased in K581A, whereas the less invasive G552A G554A, K677E, and I559F did not affect basal STAT1 phosphorylation (Fig. 3C). Upon IFNγ stimulation, all four tested JH2 mutants showed induction of STAT1 phosphorylation (Fig. 3C).
Taken together, these results indicate that an intact ATP binding site in JAK2 JH2 is required for full JH2-mediated autoinhibition of JH1 activity in the absence of type I cytokine receptors. However, the ATP binding ability of JAK2 JH2 is not essential for ligand-induced signaling via type I or type II cytokine receptors. Furthermore, these results clearly indicate that loss of JH2 ATP binding is distinct from structural disruption of JH2.
Loss of ATP Binding in JAK2 JH2 Suppresses the V617F Phenotype.
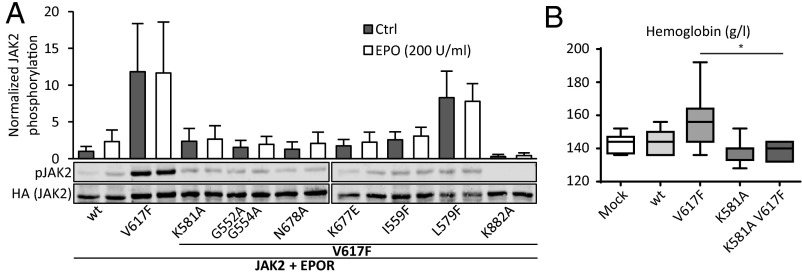
Cytokine-independent signaling of JAK2 V617F has previously been shown to be reliant on expression of type I cytokine receptors, when JAK2 is expressed at physiological levels (25, 26). Even when coexpressed with EPOR, the JH2 ATP binding site mutants were found to suppress V617F-induced hyperactivity (Fig. 4A). In accordance with the results in the absence of type I receptor expression, activity of V617F L579F also remained high in the presence of EPOR (Fig. 4A). Inhibition of JH2 ATP binding also suppressed cytokine inducibility of JAK2 V617F (Fig. 4A and Fig. S3).
Fig. 4.
Loss of ATP binding in JH2 suppresses the V617F phenotype. (A) JAK2(Y1007–Y1008) phosphorylation in γ2A cells normalized as explained for Fig. 2. Error bars are SDs from three independent experiments. (B) Hemoglobin levels from mice transplanted with retrovirally transduced bone marrow. Mice were analyzed 12 wk posttransplantation. Results show mean ± SEM. n = 8 for each group. *P < 0.05.
To assess the effects of JH2 ATP binding-deficient mutations in vivo, the K581A mutation was introduced into human JAK2, wild type and V617F, in a pMSCV–IRES GFP vector, and its effect on development of myeloid lineage cells was analyzed in a mouse bone marrow transplantation model (27). Hematological analysis showed that mice expressing JAK2 V617F developed erythrocytosis within 3 mo, typical for V617F-induced cytokine-independent JAK2 signaling, with increased hemoglobin (Fig. 4B), mean corpuscular volume (MCV), hematocrit, and reticulocyte numbers (Fig. S4) (27). Mature red blood cell, platelet, and neutrophil counts did not differ between control and any of the JAK2 constructs (Fig. S4). Mice expressing JAK2 K581A V617F did not develop erythrocytosis and showed blood counts indistinguishable from wild-type JAK2-transplanted mice (Fig. 4B and Fig. S4). In line with the cell culture data with coexpressed EPOR, K581A in otherwise wild-type JAK2 did not affect the myeloid phenotype (Fig. 4B and Fig. S4).
An Intact ATP Binding Pocket of JH2 Is Required for Hyperactivity of JAK2 Exon 12 and 16 Mutations and JAK1 V658F.
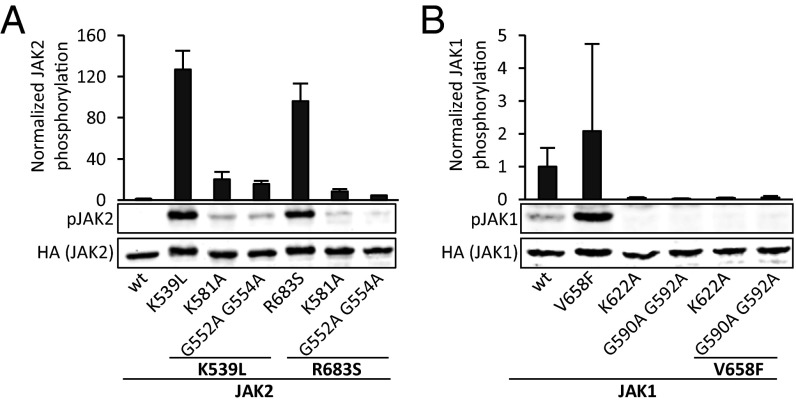
To analyze whether other hyperactivating JAK2 mutations are also dependent on ATP binding to JH2, K581A and G552A G554A were inserted to exon 12 (K539L, mutated in PV) and exon 16 JAK2 mutants (R683S, mutated in acute B lymphoblastic leukemia) (13). K581A and G552A G554A significantly lowered hyperphosphorylation of both mutants (Fig. 5A).
Fig. 5.
Analysis of ATP binding site mutants in other JAK2 disease mutations and JAK1 JH2. (A) JAK2(Y1007–Y1008) phosphorylation in γ2A cells transfected with JAK2 mutants. K539L and R683S are disease mutations located in JAK2 JH2. (B) JAK1(Y1022–Y1023) phosphorylation from COS7 cells transfected with full-length JAK1-HA. JAK1 V658F is analogous to JAK2 V617F. Bar graph shows quantification of phosphorylation from immunoblots, as described for Fig. 2. Error bars are SDs from three independent experiments.
To test the generality and functional conservation of ATP binding in JH2 within the JAK family, studies were performed with JAK1. Binding of MANT-ATP to JAK1 JH2 revealed tight binding in the presence of both Mg2+ and Mn2+, with a Kd of 3.1 ± 0.2 µM (Fig. S5), which is comparable to that of JAK2 JH2 (1.3 µM). Analysis of JAK1 V658F (analogous to JAK2 V617F) showed that mutating the ATP binding site of JAK1 JH2 with K622A or G590A G592A (corresponding to K581A and G552A G554A in JAK2, respectively) resulted in abrogation of V658F-induced JAK1 hyperphosphorylation (Fig. 5B). In contrast to the results for JAK2, basal phosphorylation levels of JAK1 were reduced to nearly undetectable levels in K622A and G590A G592A in the context of both wild type and V658F.
Molecular Dynamic Simulations Imply a Stabilizing Effect for ATP Binding on the Structure of JH2.
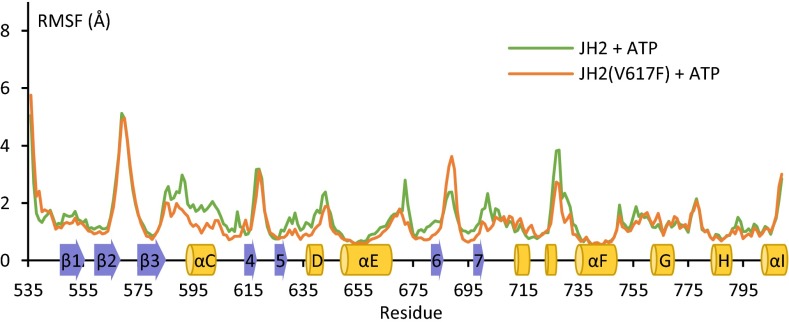
Molecular dynamic simulations were performed to explore the structural consequences of JH2 ATP binding. JAK2 JH2 wild type and V617F were simulated for 3.5 µs in the presence of ATP, and the results were compared with simulations of the domains without ATP (16). An overall reduction in flexibility of JH2 was observed upon ATP binding, with reductions in root-mean-square fluctuation and deviation (Fig. S6 A–C). The largest stabilization was seen in loop regions in both the N and C lobes that participate in the autoinhibitory JH2–JH1 interaction (5, 18), such as the β2–β3 loop (containing Tyr570) and the β6–β7 loop (containing Arg683), the latter of which showed large reduction in mobility only in wild-type JH2 (Fig. S6A). Interestingly, the dynamics of ATP-bound wild-type and V617F JH2 were highly similar, with the exception of the region encompassing αC, which was more stable in V617F (Fig. 6). Secondary structure analysis of αC showed that the time αC residues (587–602) were in an α-helical conformation increased upon ATP binding from 41% to 65% in wild type and from 73% to 84% in V617F (Fig. S6D) (16). Taken together, these data imply a stabilizing effect for ATP binding on JH2 and a difference in the αC region of ATP-bound JAK2 JH2 V617F compared with wild type.
Fig. 6.
Comparison of ATP-bound JAK2 JH2 wild type and V617F in molecular dynamic simulations. Root-mean-square fluctuation of each residue in JAK2 JH2 wild type and V617F with ATP bound over the course of the simulation (3.5 µs). The secondary structure of JAK2 JH2 is shown schematically on the x axis.
Discussion
A recent study evaluating nucleotide binding in 30 pseudokinase domains indicated that almost half of pseudokinases retain nucleotide binding, which in most cases is not associated with phosphotransfer activity (20). These and similar observations (23, 28–31) have brought up the question about the functional role of ATP binding in pseudokinases. Here we have addressed this question in the JAK2 pseudokinase domain, focusing on its pathogenic mutants. The most striking finding of our study is that JH2 nucleotide binding plays a critical role in pathogenic activation of JAKs. Specifically, our in vitro and in vivo results show that ATP binding-deficient JH2 mutants suppress the hyperactivation of pathogenic JAK2, whereas the same alterations do not significantly affect the activation characteristics of wild-type JAK2.
Our results are consistent with an earlier study, in which mutation of the JH2 β3 lysine (K581R) was found to decrease JAK2 V617F hyperphosphorylation (32). Our characterization of the nucleotide binding and thermal stability of JH2 ATP binding site mutants (Fig. 1B) distinguishes the effect of ATP binding from potential structural destabilization caused by the mutations. This leads us to conclude that the inhibition of V617F hyperactivity is due to changes in JH2 caused by loss of nucleotide binding (Figs. 2 and 4A). The loss of pathogenic activation was observed with three pathogenic mutations (V617F, R683S, and K539L) (Figs. 2 and 5), all of which disrupt the autoinhibitory JH2–JH1 interdomain interaction (18). The mutations are located in different regions of JH2, indicating that the underlying mechanism for reduction of hyperactivity is not mutation specific but rather is a common effect on aberrantly activated JAK2.
The effects of JH2 ATP binding site mutations on otherwise wild-type JAK2 were small: normal cytokine stimulation but a slightly increased basal activity in the absence of type I cytokine receptors (Fig. 2), which was abolished by expression of EPOR (Fig. 3A). This is distinctly different from structurally disrupted JH2, as demonstrated by F739R, which effectively removes both aspects of JH2 function––namely, inhibition of basal activity and response to stimulation (Fig. 3 A and B)––thus mimicking JAK JH2 deletion (1, 6, 7). Interestingly, mutating the ATP binding site of JAK1 JH2 showed reduced basal activity (Fig. 5B), which we speculate is due to different modes of regulation of basal JAK activity in homo- (JAK2) and heterodimeric (all JAKs) receptor configurations.
The JH2 domain likely arose early in evolution through duplication of JH1, and the two domains have since taken a different evolutionary path with distinct regions conserved in each of them (33), indicative of their respective separate functional roles. As JH1 maintained the substrate phosphorylation function, the catalytic function in JH2 became redundant and has consequently been lost, with the exception of regulatory autophosphorylation in JAK2. Interestingly, JH2 ATP binding ability is conserved in JAK1 and TYK2 (JAK3 unknown), even though a precise function in wild-type JAK1/TYK2 cannot be ascertained from current data. Nevertheless, the relatively high binding affinity (Fig. 1D, Fig. S5, and ref. 17), combined with low or absent hydrolysis and high cellular ATP concentrations, likely results in constitutively bound ATP in JH2, thus making ATP essentially a structural component of the JAK pseudokinase domain.
The effects of nucleotide binding to the structure of JH2 are subtle: mainly rigidification of αC in the ATP-bound form (16). Why is it then that the loss of nucleotide binding has a pronounced effect in V617F but not in wild type? TSA shows that the V617F mutation significantly reduces the Tm of JAK2 JH2 compared with wild type (Fig. 1C), yet ATP binding to both wild type and V617F causes equal thermal stabilization. These data suggest that the V617F mutation destabilizes JAK2 JH2 and renders it thus more sensitive to the loss of the stabilizing effect of ATP, as even ATP-bound JH2 V617F does not reach the Tm of wild type (Fig. 1C). Whether this sensitivity is due to overall destabilization of JH2 V617F or a specific structural alteration cannot be definitively determined from current data, but molecular dynamic simulations hint at a critical role for the αC region.
The main structural effects due to ATP binding in JH2 are observed in αC, and mutations in αC (e.g., F594A, F595A) have been shown to reverse hyperactivation of several mutations scattered throughout the JAK2 JH2–JH1 interface (18, 34, 35). Although in the case of V617F, which is proximal to αC, F595A might reconstitute a disrupted autoinhibitory JH2–JH1 interface, a more likely explanation for the suppressive effects of ATP binding mutations (and of F594A, F595A) is that pathogenic hyperactivation is dependent on a yet-to-be-characterized positive regulatory interaction mediated by JH2, which probably involves αC and is therefore sensitive to its conformation.
The modulatory nature of nucleotide binding pocket occupation has previously been documented in the kinase and pseudokinase literature. Protein kinase C, for example, has been shown to be regulated by noncatalytic nucleotide binding (30). Also, in the pseudokinases STRADα (28, 36) and integrin-linked kinase (ILK) (23, 31), ATP binding is required to enable critical protein–protein interactions. Interestingly, ATP binding is necessary for ILK function, even though no major ATP binding-induced structural changes could be detected using multiple biophysical methods (23). Also, the allosteric regulatory function of the HER3 pseudokinase has been shown to be sensitive to modulation by an ATP mimetic inhibitor (29). Furthermore, some pseudokinases that are incapable of binding ATP, like Vaccinia-related kinase 3, effectively mimic the ATP-bound conformation through bulky and acidic amino acid substitutions in the active site (37). Because almost half of pseudokinases studied so far possess some form of nucleotide binding activity (20), it seems likely that future studies will find even more evidence for a functional role of ATP binding in this group of proteins.
The results presented here reveal that the ATP binding site of JAK JH2 has characteristics to serve as a potential target site for modulators and/or mutant-selective inhibitors of JAK activity. Loss of JH2 ATP binding abrogates hyperactivation of mutant JAK2 in cells and in vivo while leaving wild-type JAK2 largely unaffected. Current pharmacological interventions at JAKs target JH1, and although these inhibitors have brought important advances in the treatment of PMF and PV patients, they are unable to eradicate the disease and they also affect wild-type JAKs (38). Targeting JH2 with conformation-specific ATP binding site inhibitors may give rise to novel pharmacological compounds able to allosterically inhibit pathogenic JAK activity.
Materials and Methods
Cell Culture, Transfection, and Immunoblotting.
JAK2-deficient γ2A fibrosarcoma cells and COS7 cells were cultured using standard cell culture methods and transfected with full-length human JAK2-HA, human STAT5A-HA, and human EPOR-HA using FuGENE6 (Promega) or Xtreme-GENE9 (Roche) according to the manufacturers’ instructions. After 10 h cells were starved in serum-free medium overnight and stimulated for 30 min with human EPO or human IFNγ. After stimulation, cells were lysed into Triton-X cell lysis buffer and centrifuged, and the supernatant was used directly for SDS/PAGE and immunoblotting. Blots were double-stained with phosphospecific antibodies and anti-HA and detected with a mix of IRDye-labeled secondaries. Blots were read and quantified using a LI-COR Odyssey CLx. A minimum of three independent experiments were performed for each condition.
Retroviral Transduction and Bone Marrow Transplantation.
For details on retroviral transduction and bone marrow transplantation, see SI Materials and Methods. All experiments were performed in strict adherence to Swiss laws for animal welfare and approved by the Swiss Cantonal Veterinary Office of Basel-Stadt.
Molecular Dynamic Simulations.
Simulations were carried out as described previously (16). Trajectories were analyzed using VMD (visual molecular dynamics) (39).
In Vitro Biochemical Assays on Recombinant Proteins.
Recombinant proteins were expressed in Sf9 cells as detailed earlier (17). After cell collection and Ni-NTA purification, protein was either used as such [JAK2(536–812-6xHis) for TSA] or subjected to anion exchange purification as described earlier (17) [JAK2(513–827-6xHis) and JAK1(553–856-6xHis) for MANT-ATP binding assays]. TSA experiments were carried out essentially as described in ref. 20. The MANT-ATP binding assay is described in ref. 40.
Supplementary Material
Acknowledgments
We thank Yibing Shan for kindly providing the JAK2 JH2 molecular dynamic simulation data, Heidi Peussa, Anna U. Laitinen, and Ellin-Kristina Hillert for excellent technical assistance, and Kaury Kucera for valuable comments on the manuscript. This work was supported in part by the Medical Research Council of the Academy of Finland, Sigrid Juselius Foundation, Medical Research Fund of Tampere University Hospital, Finnish Cancer Foundation, Novo Nordisk Foundation, and Tampere Tuberculosis Foundation (to O.S.); National Institutes of Health Grant R21 AI095808 (to S.R.H.); and Swiss National Science Foundation Grants 310000-120724/1 and 32003BB_135712/1 and Swiss Cancer League Grant KLS-2950-02-2012 (to R.C.S.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1423201112/-/DCSupplemental.
References
- 1.Saharinen P, Silvennoinen O. The pseudokinase domain is required for suppression of basal activity of Jak2 and Jak3 tyrosine kinases and for cytokine-inducible activation of signal transduction. J Biol Chem. 2002;277(49):47954–47963. doi: 10.1074/jbc.M205156200. [DOI] [PubMed] [Google Scholar]
- 2.Chen M, et al. Complex effects of naturally occurring mutations in the JAK3 pseudokinase domain: Evidence for interactions between the kinase and pseudokinase domains. Mol Cell Biol. 2000;20(3):947–956. doi: 10.1128/mcb.20.3.947-956.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Yeh TC, Dondi E, Uzé G, Pellegrini S. A dual role for the kinase-like domain of the tyrosine kinase Tyk2 in interferon-α signaling. Proc Natl Acad Sci USA. 2000;97(16):8991–8996. doi: 10.1073/pnas.160130297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sanz Sanz A, et al. The JH2 domain and SH2-JH2 linker regulate JAK2 activity: A detailed kinetic analysis of wild type and V617F mutant kinase domains. Biochim Biophys Acta. 2014;1844(10):1835–1841. doi: 10.1016/j.bbapap.2014.07.003. [DOI] [PubMed] [Google Scholar]
- 5.Lupardus PJ, et al. Structure of the pseudokinase-kinase domains from protein kinase TYK2 reveals a mechanism for Janus kinase (JAK) autoinhibition. Proc Natl Acad Sci USA. 2014;111(22):8025–8030. doi: 10.1073/pnas.1401180111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Saharinen P, Vihinen M, Silvennoinen O. Autoinhibition of Jak2 tyrosine kinase is dependent on specific regions in its pseudokinase domain. Mol Biol Cell. 2003;14(4):1448–1459. doi: 10.1091/mbc.E02-06-0342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Saharinen P, Takaluoma K, Silvennoinen O. Regulation of the Jak2 tyrosine kinase by its pseudokinase domain. Mol Cell Biol. 2000;20(10):3387–3395. doi: 10.1128/mcb.20.10.3387-3395.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Vainchenker W, Constantinescu SN. JAK/STAT signaling in hematological malignancies. Oncogene. 2013;32(21):2601–2613. doi: 10.1038/onc.2012.347. [DOI] [PubMed] [Google Scholar]
- 9.Baxter EJ, et al. Cancer Genome Project Acquired mutation of the tyrosine kinase JAK2 in human myeloproliferative disorders. Lancet. 2005;365(9464):1054–1061. doi: 10.1016/S0140-6736(05)71142-9. [DOI] [PubMed] [Google Scholar]
- 10.James C, et al. A unique clonal JAK2 mutation leading to constitutive signalling causes polycythaemia vera. Nature. 2005;434(7037):1144–1148. doi: 10.1038/nature03546. [DOI] [PubMed] [Google Scholar]
- 11.Kralovics R, et al. A gain-of-function mutation of JAK2 in myeloproliferative disorders. N Engl J Med. 2005;352(17):1779–1790. doi: 10.1056/NEJMoa051113. [DOI] [PubMed] [Google Scholar]
- 12.Levine RL, et al. Activating mutation in the tyrosine kinase JAK2 in polycythemia vera, essential thrombocythemia, and myeloid metaplasia with myelofibrosis. Cancer Cell. 2005;7(4):387–397. doi: 10.1016/j.ccr.2005.03.023. [DOI] [PubMed] [Google Scholar]
- 13.Haan C, Behrmann I, Haan S. Perspectives for the use of structural information and chemical genetics to develop inhibitors of Janus kinases. J Cell Mol Med. 2010;14(3):504–527. doi: 10.1111/j.1582-4934.2010.01018.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lipson D, et al. Identification of new ALK and RET gene fusions from colorectal and lung cancer biopsies. Nat Med. 2012;18(3):382–384. doi: 10.1038/nm.2673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Candotti F, et al. Structural and functional basis for JAK3-deficient severe combined immunodeficiency. Blood. 1997;90(10):3996–4003. [PubMed] [Google Scholar]
- 16.Bandaranayake RM, et al. Crystal structures of the JAK2 pseudokinase domain and the pathogenic mutant V617F. Nat Struct Mol Biol. 2012;19(8):754–759. doi: 10.1038/nsmb.2348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ungureanu D, et al. The pseudokinase domain of JAK2 is a dual-specificity protein kinase that negatively regulates cytokine signaling. Nat Struct Mol Biol. 2011;18(9):971–976. doi: 10.1038/nsmb.2099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Shan Y, et al. Molecular basis for pseudokinase-dependent autoinhibition of JAK2 tyrosine kinase. Nat Struct Mol Biol. 2014;21(7):579–584. doi: 10.1038/nsmb.2849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Toms AV, et al. Structure of a pseudokinase-domain switch that controls oncogenic activation of Jak kinases. Nat Struct Mol Biol. 2013;20(10):1221–1223. doi: 10.1038/nsmb.2673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Murphy JM, et al. A robust methodology to subclassify pseudokinases based on their nucleotide-binding properties. Biochem J. 2014;457(2):323–334. doi: 10.1042/BJ20131174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Manning G, Whyte DB, Martinez R, Hunter T, Sudarsanam S. The protein kinase complement of the human genome. Science. 2002;298(5600):1912–1934. doi: 10.1126/science.1075762. [DOI] [PubMed] [Google Scholar]
- 22.Knapp S, Sundström M. Recently targeted kinases and their inhibitors—The path to clinical trials. Curr Opin Pharmacol. 2014;17:58–63. doi: 10.1016/j.coph.2014.07.015. [DOI] [PubMed] [Google Scholar]
- 23.Fukuda K, Knight JD, Piszczek G, Kothary R, Qin J. Biochemical, proteomic, structural, and thermodynamic characterizations of integrin-linked kinase (ILK): Cross-validation of the pseudokinase. J Biol Chem. 2011;286(24):21886–21895. doi: 10.1074/jbc.M111.240093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Iyer GH, Garrod S, Woods VL, Jr, Taylor SS. Catalytic independent functions of a protein kinase as revealed by a kinase-dead mutant: Study of the Lys72His mutant of cAMP-dependent kinase. J Mol Biol. 2005;351(5):1110–1122. doi: 10.1016/j.jmb.2005.06.011. [DOI] [PubMed] [Google Scholar]
- 25.Lu X, et al. Expression of a homodimeric type I cytokine receptor is required for JAK2V617F-mediated transformation. Proc Natl Acad Sci USA. 2005;102(52):18962–18967. doi: 10.1073/pnas.0509714102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lu X, Huang LJS, Lodish HF. Dimerization by a cytokine receptor is necessary for constitutive activation of JAK2V617F. J Biol Chem. 2008;283(9):5258–5266. doi: 10.1074/jbc.M707125200. [DOI] [PubMed] [Google Scholar]
- 27.Tiedt R, et al. Ratio of mutant JAK2-V617F to wild-type Jak2 determines the MPD phenotypes in transgenic mice. Blood. 2008;111(8):3931–3940. doi: 10.1182/blood-2007-08-107748. [DOI] [PubMed] [Google Scholar]
- 28.Zeqiraj E, et al. ATP and MO25α regulate the conformational state of the STRADalpha pseudokinase and activation of the LKB1 tumour suppressor. PLoS Biol. 2009;7(6):e1000126. doi: 10.1371/journal.pbio.1000126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Littlefield P, Moasser MM, Jura N. An ATP-competitive inhibitor modulates the allosteric function of the HER3 pseudokinase. Chem Biol. 2014;21(4):453–458. doi: 10.1016/j.chembiol.2014.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 30.Cameron AJ, Escribano C, Saurin AT, Kostelecky B, Parker PJ. PKC maturation is promoted by nucleotide pocket occupation independently of intrinsic kinase activity. Nat Struct Mol Biol. 2009;16(6):624–630. doi: 10.1038/nsmb.1606. [DOI] [PubMed] [Google Scholar]
- 31.Lange A, et al. Integrin-linked kinase is an adaptor with essential functions during mouse development. Nature. 2009;461(7266):1002–1006. doi: 10.1038/nature08468. [DOI] [PubMed] [Google Scholar]
- 32.Andraos R, et al. Modulation of activation-loop phosphorylation by JAK inhibitors is binding mode dependent. Cancer Discov. 2012;2(6):512–523. doi: 10.1158/2159-8290.CD-11-0324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gu J, Wang Y, Gu X. Evolutionary analysis for functional divergence of Jak protein kinase domains and tissue-specific genes. J Mol Evol. 2002;54(6):725–733. doi: 10.1007/s00239-001-0072-3. [DOI] [PubMed] [Google Scholar]
- 34.Dusa A, Mouton C, Pecquet C, Herman M, Constantinescu SN. JAK2 V617F constitutive activation requires JH2 residue F595: A pseudokinase domain target for specific inhibitors. PLoS ONE. 2010;5(6):e11157. doi: 10.1371/journal.pone.0011157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gnanasambandan K, Magis A, Sayeski PP. The constitutive activation of Jak2-V617F is mediated by a π stacking mechanism involving phenylalanines 595 and 617. Biochemistry. 2010;49(46):9972–9984. doi: 10.1021/bi1014858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zeqiraj E, Filippi BM, Deak M, Alessi DR, van Aalten DM. Structure of the LKB1-STRAD-MO25 complex reveals an allosteric mechanism of kinase activation. Science. 2009;326(5960):1707–1711. doi: 10.1126/science.1178377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Scheeff ED, Eswaran J, Bunkoczi G, Knapp S, Manning G. Structure of the pseudokinase VRK3 reveals a degraded catalytic site, a highly conserved kinase fold, and a putative regulatory binding site. Structure. 2009;17(1):128–138. doi: 10.1016/j.str.2008.10.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sonbol MB, et al. Comprehensive review of JAK inhibitors in myeloproliferative neoplasms. Ther Adv Hematol. 2013;4(1):15–35. doi: 10.1177/2040620712461047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Humphrey W, Dalke A, Schulten K. VMD: Visual molecular dynamics. J Mol Graph. 1996;14(1):33–38, 27–28. doi: 10.1016/0263-7855(96)00018-5. [DOI] [PubMed] [Google Scholar]
- 40.Niranjan Y, et al. Analysis of steady-state Förster resonance energy transfer data by avoiding pitfalls: Interaction of JAK2 tyrosine kinase with N-methylanthraniloyl nucleotides. Anal Biochem. 2013;442(2):213–222. doi: 10.1016/j.ab.2013.07.020. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.