Significance
Large offspring syndrome (LOS) is a fetal overgrowth condition that mimics the human syndrome Beckwith–Wiedemann. These conditions have been observed with higher incidence in offspring conceived with the use of assisted reproductive technologies and are believed to be the result of misregulation of a set of genes that are expressed only from the maternally or paternally inherited chromosomes. These genes are known as imprinted genes. In our study, we demonstrate that the kidney, brain, muscle, and liver of LOS fetuses show misregulation of multiple imprinted genes when compared with controls. Furthermore, we show that the magnitude of overgrowth in LOS fetuses correlates with the number of misregulated imprinted genes. Our results may help create diagnostics for these fetal syndromes.
Keywords: genomic imprinting, assisted reproductive technologies, large offspring syndrome, Beckwith–Wiedemann syndrome, loss of imprinting
Abstract
Embryos generated with the use of assisted reproductive technologies (ART) can develop overgrowth syndromes. In ruminants, the condition is referred to as large offspring syndrome (LOS) and exhibits variable phenotypic abnormalities including overgrowth, enlarged tongue, and abdominal wall defects. These characteristics recapitulate those observed in the human loss-of-imprinting (LOI) overgrowth syndrome Beckwith–Wiedemann (BWS). We have recently shown LOI at the KCNQ1 locus in LOS, the most common epimutation in BWS. Although the first case of ART-induced LOS was reported in 1995, studies have not yet determined the extent of LOI in this condition. Here, we determined allele-specific expression of imprinted genes previously identified in human and/or mouse in day ∼105 Bos taurus indicus × Bos taurus taurus F1 hybrid control and LOS fetuses using RNAseq. Our analysis allowed us to determine the monoallelic expression of 20 genes in tissues of control fetuses. LOS fetuses displayed variable LOI compared with controls. Biallelic expression of imprinted genes in LOS was associated with tissue-specific hypomethylation of the normally methylated parental allele. In addition, a positive correlation was observed between body weight and the number of biallelically expressed imprinted genes in LOS fetuses. Furthermore, not only was there loss of allele-specific expression of imprinted genes in LOS, but also differential transcript amounts of these genes between control and overgrown fetuses. In summary, we characterized previously unidentified imprinted genes in bovines and identified misregulation of imprinting at multiple loci in LOS. We concluded that LOS is a multilocus LOI syndrome, as is BWS.
Genomic imprinting is a series of precisely regulated epigenetic processes that lead to parental allele-specific expression of a subset of genes in mammals (1). Proper allelic expression of imprinted genes plays an important role in embryonic and neonatal growth, placental function, and postnatal behavior (2). Allele-specific DNA methylation at discrete regions established during gametogenesis defines the functional asymmetry of parental alleles (1). These regions, termed differentially methylated regions (DMRs), are required to regulate the imprinted expression of these genes. DNA methylation of DMRs is erased in primordial germ cells, re-established during gametogenesis, and maintained when the global DNA demethylation occurs during preimplantation development (1). In addition to DNA methylation, other epigenetic modifications and mechanisms such as noncoding RNAs (ncRNAs) and histone posttranslational modifications may contribute to parental allele-specific expression of these genes (1).
Because of the dynamic epigenetic reprogramming that occurs during oocyte growth and preimplantation development (3), environmental perturbations during this time period, such as the use of assisted reproductive technologies (ART), can affect imprint establishment and maintenance (4). Numerous prospective studies in animals (5–8) and retrospective studies in humans (9) have shown that ART can induce improper regulation of genomic imprinting. ART is commonly used in clinics to treat subfertility and infertility, and each year as high as 5.9% of infants born in developed countries are conceived by the use of these technologies (10, 11). In agriculture, ART is also widely used to increase the number of offspring produced from genetically superior individuals in a shortened period (12). Multiple reports have indicated that ART-conceived offspring are more likely to develop imprinting disorders such as Beckwith–Wiedemann syndrome (BWS) in human (9, 13, 14) and large offspring syndrome (LOS) in ruminants (15–19).
BWS is the most common pediatric overgrowth syndrome, which is characterized by complex and variable symptoms such as prenatal and postnatal overgrowth, ear creases, macroglossia, umbilical hernia, and predisposition to develop childhood tumors (20). BWS has an estimated worldwide frequency of 1 in 13,700 live births (20) with no noted sex bias (21) and a weighted relative risk of 5.2 in children conceived with the use of ART (9). Most BWS cases are sporadic and are associated with epimutations in human chromosome 11p15.5, a region that harbors KCNQ1 and H19/IGF2 imprinted loci (20). Approximately 50% of BWS cases are associated with the loss of methylation at the KvDMR1 (i.e., KCNQ1 locus) and 2–7% are associated with the gain of methylation of the DMR at the H19/IGF2 locus (20). Recent studies have shown that a subset of BWS individuals with hypomethylation at the KCNQ1 locus also exhibited aberrant DNA methylation at other imprinted loci (13, 22–28).
LOS in ruminants exhibits a variable combination of anomalies that recapitulate the phenotypes commonly observed in BWS (16). These anomalies can have detrimental effects on both the dam and offspring, including difficult delivery due to the oversized nature of the fetus and the inability of the newborn to suckle and breathe. We have recently reported that phenotypic and epigenetic similarities exist between LOS and BWS, such as macroglossia, macrosomia, and ear malformation, as well as loss of methylation at the KvDMR1 on the maternal allele (29). These parallels make LOS an appropriate animal model for the study of BWS and the understanding of the etiology of these overgrowth syndromes (29).
The incidence of LOS complications is variable and little is known about the molecular cause(s) of this complex phenotype. Given the variable phenotypes of BWS and LOS and the fact that only 50% of individuals exhibit LOI at the KCNQ1 locus (20, 29), we hypothesize that these overgrowth syndromes exhibit LOI at loci beyond those primarily used for diagnosis of BWS, namely KCNQ1 and H19/IGF2 DMRs.
In this study, we assessed the allelic expression of imprinted genes previously identified in human and/or mouse in somatic tissues of day ∼105 (d105) bovine control and LOS fetuses (term of ∼280 d; SI Appendix, SI Methods). We identified 20 genes exhibiting monoallelic expression in control fetal tissues and found that approximately half were biallelically expressed in at least one tissue in LOS. Furthermore, we observed that biallelic expression of imprinted genes was associated with loss of DNA methylation at DMRs in a tissue-specific manner. Finally, our data show that misregulation of imprinted genes goes beyond loss of allele-specific expression and provide insights into the variable gene expression observed in this condition.
Results
Control and LOS Bovine Fetal Tissues Preparation and RNA Sequencing Reads Processing.
In the current study, we used high-throughput RNA sequencing (from here on referred to as “RNAseq”) to determine allelic expression of imprinted genes previously identified in human and/or mouse in somatic tissues of d105 control and LOS bovine female fetuses (Fig. 1A). Fetuses used in this study were Bos taurus indicus (B. t. indicus) × Bos taurus taurus (B. t. taurus) F1 hybrids (29). Only females were used in this study to avoid any potential sex-specific effects on gene expression (30). On average, control fetuses weighed 405 g and LOS fetuses weighed 592 g (P = 0.008; SI Appendix, Fig. S1 and Table S1). Kidney, brain, skeletal muscle (from here on referred to as “muscle”), and liver from each fetus were used for RNAseq analysis. These tissues were selected as they are representative of the primary germ-cell lineages: ectoderm (brain), mesoderm (kidney and muscle), and endoderm (liver). Moreover, kidney and liver are organs that have been documented to be susceptible to tumors in children affected by BWS (20).
Fig. 1.
Loss of imprinting in LOS. (A) Flowchart of methodology. (B) Allele-specific expression analysis of imprinted genes identified by RNAseq in day ∼105 B. t. indicus x B. t. taurus fetal kidney. The parental origin of imprinted gene expression (IGE) is defined according to the known imprinting status in human and/or mouse. Samples are arranged from left to right in ascending order by weight and by treatment group. A sample with at least 15% expression from the repressed allele was considered biallelically expressed. Genes that showed biallelic expression in LOS but not in control fetuses are indicated by red boxes. Missing data for RTL1 indicate lack of discriminating SNPs between parental alleles in the reads. We obtained the allelic expression of the long ncRNA KCNQ1OT1 (indicated by asterisk) in a previous study (29). (C) Heat map illustrating the degree of loss of imprinting for the biallelically expressed genes. Numbers represent the percentage of transcripts expressed from the normally repressed allele. A “0” represents <15% expression from the repressed allele (actual percentages may be found in SI Appendix, Table S3).
We first aligned the reads to the bovine whole-genome assembly B. t. taurus UMD3.1 (SI Appendix, Table S2). The reads that aligned to the known imprinted genes were then compared among all four tissues of each fetus, and genes with at least one consistent variant nucleotide in at least two tissues were used for subsequent analysis. Of the 105 known imprinted genes detected by RNAseq in our study, 72 genes (SI Appendix, Table S4) had at least one single nucleotide polymorphism (SNP) with sufficient read depth (≥10) to assess allelic expression. A gene was considered to be monoallelically expressed if the expression of the allele with the fewer reads accounted for less than 15% of the total reads (SI Appendix, SI Methods). The other 33 genes were not used for further analysis as the heterozygous nature of the bovine makes it impossible for us to determine with any certainty if the reads originated from one or both alleles. For example, if all reads have a base that differs from the reference genome, two scenarios are possible: (i) the gene is monoallelically expressed or (ii) the gene is biallelically expressed and both alleles contain the same base.
Allelic Determination of Imprinted Genes in d105 Control Fetal Tissues.
Of the 72 genes assessable for allelic expression, 52 (SI Appendix, Table S4) were biallelically expressed in d105 fetal kidney, brain, muscle, and liver. The remaining 20 genes were observed to be imprinted in at least one tissue from any fetus. Of these, 18 were expressed monoallelically in the kidney and 14, 17, and 14 in brain, muscle, and liver, respectively (Fig. 1B and SI Appendix, Figs. S2–S4 and Table S3). Thirteen genes (i.e., DIRAS3, DLK1, GNAS, GTL2, H19, MAGEL2, NAP1L5, NNAT, PEG3, PEG10, PLAGL1, RTL1, and SNRPN) exhibited monoallelic expression in all of the tissues analyzed where the genes were expressed, whereas 7 genes (i.e., BEGAIN, CDKN1C, IGF2, IGF2R, INPP5F, PHLDA2, and SGCE) showed tissue-specific imprinting (Fig. 1B and SI Appendix, Figs. S2–S4 and Table S3).
Allelic Expression of Imprinted Genes in LOS.
Allelic expression of the 20 genes identified to be imprinted in control tissues was then determined in tissues of LOS fetuses. For a gene to be described as experiencing loss-of-imprinted gene expression (as defined by our 15% cutoff), that gene must be monoallelically expressed in the same tissue in all control fetuses. We observed loss-of-imprinted gene expression in nine, eight, eight, and six genes in kidney, brain, muscle, and liver, respectively (Fig. 1 B and C and SI Appendix, Figs. S2–S4). Of these, IGF2R showed universal LOI in LOS kidney, muscle, and liver whereas INPP5F was biallelically expressed in the brain of all LOS fetuses. In other tissues, however, these genes had variable allelic expression in control and/or LOS fetuses (SI Appendix, Table S3). Fetuses LOS #3 and #4 displayed LOI of NNAT in all of the tissues analyzed whereas LOS #4 had loss-of-imprinted expression of PLAGL1 in brain, kidney, and muscle. Notably, there is a positive correlation between the number of imprinted genes showing LOI in each LOS fetus and their body weights (SI Appendix, Fig. S5).
Confirmation of SNPs Identified by RNAseq.
We first performed Sanger sequencing of genomic DNA from eight B. t. taurus animals, the B. t. indicus sire, and several of the B. t. indicus × B. t. taurus F1 hybrids to verify 37 of the SNPs identified by RNAseq. For 35 SNPs, both the chromosome position and allele variants matched between RNAseq and Sanger sequencing (SI Appendix, Table S6). The other two SNPs were not confirmed by Sanger sequencing, possibly due to RNAseq alignment errors. The confirmed SNPs indicated that SGCE, PEG10, NAP1L5, SNRPN, PEG3, and MAGEL2 were paternally expressed in the d105 bovine fetal tissues analyzed, showing conservation of maternal-allele silencing of these genes in bovine, human, and mouse. We were not able to determine parental allele-specific expression of NNAT because the five SNPs within the exons are heterozygous in both B. t. indicus and B. t. taurus (Fig. 2B and SI Appendix, Table S6).
Fig. 2.
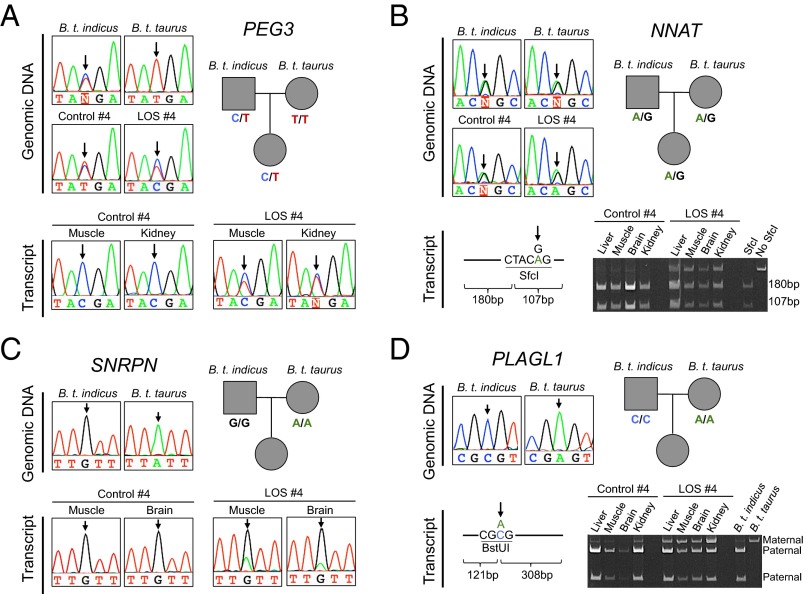
Verification of the allelic expression of PEG3, NNAT, SNRPN, and PLAGL1 in control and LOS fetuses. (A–D, Top Left) Genotyping results obtained by Sanger sequencing of B. t. indicus, B. t. Taurus, and/or F1 hybrid fetuses for the SNPs identified by RNAseq analysis. SNP positions are indicated by arrows. Pedigree diagrams indicate the SNPs identified by Sanger sequencing for B. t. indicus (square, paternalallele in F1), B. t. taurus (top circle, maternal allele in the F1), and female F1 d105 fetus (bottom circle). (A and C, Bottom) Sanger sequencing results of RT-PCR amplicons of PEG3 and SNRPN, respectively. Double peaks demonstrate biallelic expression. (B and D, Bottom) Allele-specific restriction of RT-PCR amplicons followed by acrylamide gel electrophoresis for NNAT and PLAGL1, respectively. Restriction enzyme (i.e., SfcI for NNAT and BstUI for PLAGL1), restriction site, and fragment length are illustrated on the Bottom Left of each panel.
Confirmation of Loss-of-Imprinted Expression Identified by RNAseq.
To exclude potential false-positive imprinted expression caused by RNAseq systemic errors (31), we verified allele-specific expression of PEG3, NNAT, SNRPN, and PLAGL1 by conventional methods (Fig. 2). Paternal expression was validated for PEG3, SNRPN, and PLAGL1 in control fetuses. In addition, imprinted expression was also confirmed for NNAT although parental origin of the transcript was not assessable due to the lack of subspecies-specific polymorphisms (Fig. 2B). In contrast to controls, PEG3, SNRPN, NNAT, and PLAGL1 were biallelically expressed in LOS fetuses (Fig. 2 and SI Appendix, Fig. S6).
Association Between Allelic Expression and Transcript Abundance of Imprinted Genes.
To assess if the biallelic expression of imprinted genes was associated with the increased amount of transcripts, we normalized the read counts aligned to each gene locus to determine the transcript abundance. We found that loss of imprinted gene expression in LOS tissues did not always correlate with the amount of transcript (SI Appendix, Fig. S7). For example, the increased number of transcripts of SNRPN, NNAT, and PLAGL1 was associated with biallelic expression in kidney and muscle (P < 0.01), but not in brain. Furthermore, IGF2R had a lower total level of expression in both liver and muscle of all LOS fetuses, although IGF2R was biallelically expressed in both tissues.
DNA Methylation Analysis of PLAGL1, SNRPN, and NNAT DMRs.
As described above, SNRPN, NNAT, and PLAGL1 exhibited LOI in several of the LOS fetuses. These genes have also been demonstrated to be associated with BWS (22–24, 28). Thus, we assessed allele-specific DNA methylation of the three DMRs.
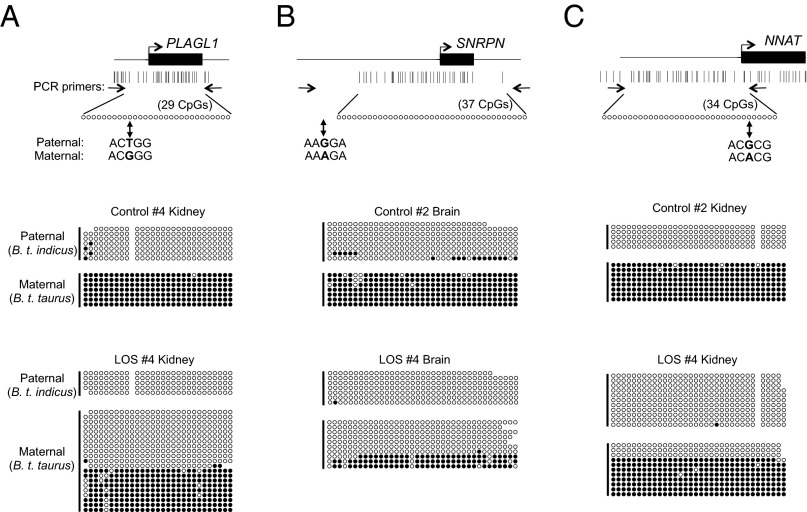
DNA methylation of PLAGL1 was examined in a region within the DMR (32) that contained 28 CpGs on the paternal (B. t. indicus) and 29 CpGs on the maternal (B. t. taurus) allele (Fig. 3A and SI Appendix, Table S7). PLAGL1 is maternally repressed and acquires maternal methylation during oocyte growth in bovine (32). As expected, we observed methylated maternal alleles and unmethylated paternal alleles in kidney, brain, muscle, and liver of the control fetus analyzed (Fig. 3A and SI Appendix, Fig. S8A). Methylation of CpGs within this region was substantially reduced on the maternal allele in the kidney of LOS #4 (Fig. 3A) coincident with the biallelic expression of this gene in this tissue (Fig. 1B). However, biallelic expression of PLAGL1 did not coincide with a reduction in DNA methylation in brain and muscle (SI Appendix, Fig. S8A).
Fig. 3.
DNA methylation of the PLAGL1, SNRPN, and NNAT DMRs in control and LOS fetuses. (A–C, Top) Schematics of the regions analyzed. Bent arrows and black boxes represent transcription start sites and first exons. Vertical lines illustrate the CpGs distributed across the region (drawn to scale). The positions of bisulfite-converted DNA-specific PCR primers are shown as arrows facing each other. Single bisulfite maps represent the number of CpGs contained within each amplicon. Circles depict the CpGs examined within the region of interest (the unmethylated maternal allele is shown as an example). SNPs (in boldface) used to determine parental origins are indicated by vertical arrows. (A–C, Bottom) Allele-specific bisulfite sequencing of PLAGL1, SNRPN, and NNAT DMRs, respectively. Each row = one strand of DNA; open circles, unmethylated CpG sites; filled circle, methylated CpG sites; missing circles are due to SNPs or undetermined bases because of sequencing issues.
We next examined the methylation at the DMR (32) of the maternally imprinted gene SNRPN (Fig. 3B). The SNRPN DMR displayed differential methylation in the brain of the control fetus analyzed (Fig. 3B and SI Appendix, Table S7) but showed a substantial loss of methylation in the brain of LOS #4 fetus. Loss of methylation of this DMR in this tissue (Fig. 3B) is associated with the gain of maternal expression of this gene (SI Appendix, Fig. S2).
Finally, we analyzed DNA methylation at NNAT in control and LOS fetuses. Because the NNAT DMR has not been previously reported in bovine, we chose to analyze a CpG island at the NNAT promoter region (Fig. 3C) that was syntenic to the human NNAT DMR (33). We examined a region within the CpG island that contained 33 CpGs on the paternal and 34 CpGs on the maternal allele (Fig. 3C and SI Appendix, Table S7). Control fetus #2 displayed an unmethylated paternal allele and a methylated maternal allele in both kidney and brain (Fig. 3C and SI Appendix, Fig. S8B), which strongly suggests that NNAT is maternally imprinted in bovine as in human and mouse (34, 35). LOS fetus #4 showed reduced DNA methylation on the maternal allele in kidney but not in brain (Fig. 3C and SI Appendix, Fig. S8B).
Determination of the Transcript Level of Imprinted Genes Previously Identified in Human and/or Mouse in Bovine d105 Fetuses.
Given the diverse phenotype and various loss-of-imprinted gene expressions in LOS, we compared imprinted gene transcript abundance of each LOS fetus to the average of four controls to identify differentially expressed imprinted genes for each LOS individual using edgeR (36, 37). With the threshold of false discovery rate set at < 0.05, we identified 53, 21, 47, and 35 imprinted genes as being differentially expressed in kidney, brain, muscle, and liver, respectively, in at least one LOS fetus (SI Appendix, Fig. S9). The fetuses differed in the type of tissue that displayed the highest number of misregulated genes with the kidney of LOS #3 exhibiting inappropriate levels of expression in 50 of the 53 genes and the muscle of LOS #4 showing altered expression in 40 of the 47 genes (SI Appendix, Fig. S9).
Discussion
Beckwith–Wiedemann syndrome and large offspring syndrome are similar fetal overgrowth conditions (29, 38). Phenotypically, these are heterogeneous syndromes characterized by variable developmental anomalies including macrosomia, macroglossia, abdominal wall defect, and ear malformations. These syndromes also share LOI at the KCNQ1 locus (29), the most common epimutation in BWS, observed in 50% of patients (20). Even though reports describing BWS date back to the early 1960s (39) and the first cases of ART-induced LOS were reported in 1995 (15, 18), no molecular signature has been identified that can consistently and reliably predict and diagnose these syndromes and/or their varied phenotypes. To date, only a small set of imprinted genes have been queried in LOS (29, 40, 41). With the current study, we advance the field by describing the imprinted signature of d105 unaffected bovine fetuses and by identifying misregulated imprinted loci in LOS.
Approximately 200 imprinted genes have been identified in human and mouse (igc.otago.ac.nz/search.html and www.mousebook.org/imprinting-gene-list, respectively), and little is known about the allele and tissue specificity of these genes in bovine. In our study we detected expression of 105 of these genes with 52 being biallelically expressed and 20 having monoallelic expression in the fetal tissues analyzed. The remaining 33 genes were not assessable in our system. RNAseq analysis confirmed the maternal expression of CDKN1C, H19, and PHLDA2 and the paternal expression of IGF2 and PLAGL1 (29, 42). In addition, we demonstrated paternal expression of NNAT, NAP1L5, MAGEL2, PEG3, PEG10, SGCE, and SNRPN, which is similar to what has been reported for human and mouse (43). Furthermore, we also show that BEGAIN, DIRAS3, DLK1, GNAS, GTL2, IGF2R, INPP5F, and RTL1 are monoallelically expressed, although our analysis precludes us from ascribing the allelic origin of the transcripts. Because monoallelic expression of imprinted genes is tissue- and stage-specific (1), it is possible that the biallelically expressed genes in the present study are imprinted in other tissues and/or developmental stages. An example is TSSC4, which has been previously reported to be imprinted in bovine placenta (44); however, this gene exhibited biallelic expression in fetal d105 kidney, brain, muscle, and liver.
We demonstrate that half of the imprinted genes in controls displayed biallelic expression in LOS, making this a multilocus LOI syndrome, as is BWS (13, 22–28). The profile of LOI was dependent on the tissue, fetus, and degree of macrosomia and may provide insights into the understanding of the etiology of the anomalies observed in LOS and BWS. For example, Wilm’s tumor of the kidney, a high-risk childhood cancer in BWS, displayed hypomethylation at the NNAT promoter that coincided with up-regulation of the NNAT transcript (33). In our study we observed a similar situation where biallelic expression and an increased transcript amount of NNAT was associated with hypomethylation of its DMR in LOS kidney. Furthermore, we detected biallelic expression of the paternal gene PLAGL1, which corresponded with the increased transcript amount of this gene in the muscle and kidney of the largest LOS fetus. PLAGL1 is a known regulator of embryonic growth, and mice lacking the paternal allele of this gene exhibit growth retardation (45). Interestingly, PLAGL1 has been documented to bind the DMR of the KCNQ1 locus in a methylation-dependent manner, suggesting the involvement of this molecule in the misregulation of this locus, as has been suggested by others (46).
In the current study, we also determined the transcript amount of imprinted genes to determine if their misregulation in LOS transcends parental allele-specific expression. Similar to what others have reported in the mouse, biallelic expression of imprinted genes does not always correlate with increased transcript level in bovine (7, 47). For example, even though IGF2R was biallelically expressed in liver and muscle of LOS fetuses, the total transcript amount was lower than in the controls where monoallelic expression of this gene was observed. Lower IGF2R levels have been previously observed in LOS (40).
Furthermore, differentially expressed imprinted genes in each LOS fetus were analyzed in the GeneCard database (www.genecards.org) (Table 1) to assess the attributes of these genes (e.g., gene ontology, disorders, phenotypes, expression patterns). The analysis showed that these genes were most significantly enriched with the descriptors “Beckwith–Wiedemann syndrome,” “growth size and body phenotype,” “cellular phenotypes,” and “tumors,” indicating similarities in the types of imprinted genes misregulated in LOS and BWS. In addition, differentially expressed genes were implicated in tumor and/or body size control, which suggests that misregulation of these genes may contribute to the overgrowth phenotype of LOS. The analysis allowed us not only to confirm the imprinted genes previously known to be associated with LOS such as IGF2R (40) and CDKN1C (29), but also to identify previously unidentified candidates that can be used to predict or diagnose LOS at the molecular level. A gene of interest, for example, is FBXO40, which was down-regulated by at least twofold in the muscle of all LOS fetuses. Recently, Shi and coworkers demonstrated that FBXO40 is a negative regulator of IGF1 signaling in muscle differentiation and observed increased body weight and muscle mass in FBXO40 null mice at 6 wk of age (48). Other genes with a potential link to the overgrowth seen in LOS are MKRN3 and NNAT, which were up-regulated in the muscle of the two largest LOS fetuses by >3.5- and 2.37-fold, respectively, compared with controls. MKRN3 and NNAT are protein-coding genes with currently undefined function in muscle.
Table 1.
Shared descriptors of differentially expressed imprinted genes in LOS fetuses
| Rank | Descriptor | No. of genes | Associated imprinted genes | P value |
| 1 | Beckwith–Wiedemann syndrome | 13 | CDKN1C, H19, IGF2, IGF2R, KCNQ1, NAP1L4, PEG3, PHLDA2, PLAGL1, SLC22A18, SNRPN, TSPAN32, TSSC4 | 1.00 × 10−16 |
| 2 | Growth/size/body phenotype | 34 | ANO1, CD81, CDKN1C, COMMD1, DCN, DHCR7, DIO3, DLK1, DNMT1, GATM, GLIS3, GNAS, H19, IGF2, IGF2R, KCNQ1, LIN28B, MAGEL2, MEST, PDE10A, PDE4D, PEG10, PEG3, PHLDA2, PLAGL1, PON3, RASGRF1, RB1, RTL1, SLC22A3, SNRPN, TCEB3, TP73, UBE3A | 1.00 × 10−16 |
| 3 | Cellular phenotype | 30 | AMPD3, AXL, CDKN1C, DCN, DIO3, DLK1, DNMT1, GNAS, H19, IGF2, IGF2R, L3MBTL1, MAGEL2, MEST, NDN, PDE4D, PEG10, PEG3, PHLDA2, PLAGL1, PON2, PON3, RASGRF1, RB1, SGCE, SLC22A3, SNRPN, TCEB3, TP73, UBE3A | 2.11 × 10−15 |
| 4 | Tumors | 32 | AMPD3, ANO1, AXL, BLCAP, CALCR, CD81, CDKN1C, DCN, DIO3, DLK1, DNMT1, GNAS, H19, IGF2, IGF2R, L3MBTL1, NAP1L4, NDN, NNAT, NTM, PEG3, PLAGL1, QPCT, RB1, SLC22A18, SLC22A3, TCEB3, TFPI2, TP73, TSPAN32, UBE3A, WIF1 | 1.02 × 10−14 |
In summary, our study characterizes previously unknown expression of imprinted genes in unaffected and LOS bovine fetuses. We conclude that LOS is a multilocus LOI syndrome, as is BWS. Future studies will assess if genes identified in this study are similarly misregulated in BWS.
Materials and Methods
An expanded and detailed version of our methodology may be found in SI Appendix, SI Methods.
Fetal Tissue Collection and Illumina RNAseq.
Day 105 B. t. indicus × B. t. taurus F1 hybrid control and LOS fetuses were used to analyze allele-specific expression and DNA methylation of imprinted genes. Fetuses used in this study were produced by us as part of a previous study (29). All animal procedures were performed at TransOva Genetics by veterinarians, and all procedures were approved by their animal care and use committee. Serum was purposely used in that study to supplement the culture media to increase the incidence of the overgrowth phenotype. This method enables us to have a reliable system to study the etiology and progression of LOS. In our previous study, we generated nine control fetuses (five females and four males) and seven LOS fetuses (four females and three males). In the current study, all four females in the LOS group were used. For the control group, we randomly chose four of the five females to analyze the same number of samples in each treatment group.
Total RNA was isolated from kidney, brain, muscle, and liver of four control and four LOS females using TRIzol Reagent (Invitrogen) according to the manufacturer’s instructions. RNA quality was assessed using spectrometry and agarose gel electrophoresis. RNAseq libraries were prepared using standard Illumina protocol and sequenced on an Illumina HiSeq. 2000 platform as single-end reads. The raw FASTQ files are publically available at Gene Expression Omnibus (accession no. GSE63509).
Bioinformatics Analysis.
After quality trimming, RNAseq reads were aligned to the bovine genome reference (UMD3.1) using TopHat2 (49). Only reads that had >95% identity with the reference genome were used for the analyses. Uniquely aligned RNAseq reads were normalized to the library size to compute counts per million. Known imprinted genes were selected based on the human and mouse imprinted gene databases (igc.otago.ac.nz/search.html; www.geneimprint.com/site/genes-by-species; www.mousebook.org/mousebook-catalogs/imprinting-resource). Imprinted genes annotated by RefSeq or Ensembl were analyzed.
SNPs between B. t. indicus and B. t. taurus were identified by the following criteria: (i) at least 10-read coverage at a given chromosome position; (ii) presence of only one variant allele (different from the reference allele); and (iii) at least three reads for the lower expressed allele. To further reduce the false-positive SNPs, only the SNPs that exist in at least two of the four tissues (e.g., liver and muscle) from a single individual were kept for further analysis. Only genes that had one or more SNPs and at least 10 SNP-containing read counts were analyzed. Only genes with at least 15% of reads from the repressed allele were considered biallelically expressed.
SNP Validation by Sanger Sequencing.
DNA was isolated from B. t. taurus, B. t. indicus, and B. t. indicus × B. t. taurus F1 hybrid tissues by phenol–chloroform extraction. PCR amplifications were performed using GoTaq Hot Start polymerase (Promega), and amplicons were sequenced at the University of Missouri DNA core using the 96-capillary Applied Biosystems 3730 DNA analyzer with Big Dye Terminator.
Analysis of Allelic Expression.
Allele-specific expression of PEG3 and SNRPN was validated using RT-PCR followed by Sanger sequencing. The PCR reactions were prepared and sequenced as described above. Allelic expression of NNAT and PLAGL1 were determined using RT-PCR followed by restriction fragment length polymorphism and PAGE.
DNA Methylation Analysis.
Genomic DNA was mutagenized with sodium bisulfite using the Imprint DNA Modification Kit (Sigma) according to the manufacturer’s instructions. Amplified PCR products were ligated with pCC1 vector and cloned using a CopyControl PCR cloning kit according to the manufacturer’s instructions, except all of the cloning incubation procedures were performed at room temperature. Colonies were usually visible within 2 d. Positive colonies were sequenced as described above.
Supplementary Material
Acknowledgments
We thank Dr. Christopher Childers, Colin Diesh, and Dr. Shu Tao for assistance with the RNAseq analysis and Ms. Kira Marshall for assistance with the manuscript. This work was supported by the National Institutes of Health (Grant 5R21HD062920), the Food for the 21st Century Program at the University of Missouri, and by Agriculture and Food Research Initiative Competitive Grant 2010-65205-20647 from the US Department of Agriculture National Institute of Food and Agriculture.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1422088112/-/DCSupplemental.
This article is a PNAS Direct Submission.
Data deposition: The RNAseq data reported in this paper have been deposited in the Gene Expression Omnibus (GEO) database, www.ncbi.nlm.nih.gov/geo (accession no. GSE63509).
References
- 1.Bartolomei MS, Ferguson-Smith AC. Mammalian genomic imprinting. Cold Spring Harb Perspect Biol. 2011;3(7) doi: 10.1101/cshperspect.a002592. pii: a002592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Miyoshi N, Barton SC, Kaneda M, Hajkova P, Surani MA. The continuing quest to comprehend genomic imprinting. Cytogenet Genome Res. 2006;113(1-4):6–11. doi: 10.1159/000090808. [DOI] [PubMed] [Google Scholar]
- 3.Morgan HD, Santos F, Green K, Dean W, Reik W. Epigenetic reprogramming in mammals. Hum Mol Genet. 2005;14(Spec No 1):R47–R58. doi: 10.1093/hmg/ddi114. [DOI] [PubMed] [Google Scholar]
- 4.Denomme MM, Mann MR. Genomic imprints as a model for the analysis of epigenetic stability during assisted reproductive technologies. Reproduction. 2012;144(4):393–409. doi: 10.1530/REP-12-0237. [DOI] [PubMed] [Google Scholar]
- 5.Rivera RM, et al. Manipulations of mouse embryos prior to implantation result in aberrant expression of imprinted genes on day 9.5 of development. Hum Mol Genet. 2008;17(1):1–14. doi: 10.1093/hmg/ddm280. [DOI] [PubMed] [Google Scholar]
- 6.Market-Velker BA, Zhang L, Magri LS, Bonvissuto AC, Mann MR. Dual effects of superovulation: Loss of maternal and paternal imprinted methylation in a dose-dependent manner. Hum Mol Genet. 2010;19(1):36–51. doi: 10.1093/hmg/ddp465. [DOI] [PubMed] [Google Scholar]
- 7.de Waal E, et al. Primary epimutations introduced during intracytoplasmic sperm injection (ICSI) are corrected by germline-specific epigenetic reprogramming. Proc Natl Acad Sci USA. 2012;109(11):4163–4168. doi: 10.1073/pnas.1201990109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.de Waal E, et al. Gonadotropin stimulation contributes to an increased incidence of epimutations in ICSI-derived mice. Hum Mol Genet. 2012;21(20):4460–4472. doi: 10.1093/hmg/dds287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Vermeiden JP, Bernardus RE. Are imprinting disorders more prevalent after human in vitro fertilization or intracytoplasmic sperm injection? Fertil Steril. 2013;99(3):642–651. doi: 10.1016/j.fertnstert.2013.01.125. [DOI] [PubMed] [Google Scholar]
- 10. Centers for Disease Control and Prevention, American Society for Reproductive Medicine, Society for Assisted Reproductive Technology (2013) 2011 Assisted Reproductive Technology National Summary Report (US Department of Health and Human Services, Atlanta)
- 11.Kupka MS, et al. European IVF-Monitoring Consortium for the European Society of Human Reproduction and Embryology Assisted reproductive technology in Europe, 2010: Results generated from European registers by ESHRE. Hum Reprod. 2014;29(10):2099–2113. doi: 10.1093/humrep/deu175. [DOI] [PubMed] [Google Scholar]
- 12.IETS 2012 Statistics of Embryo Collection and Transfer in Domestic Farm Animals. 2013 (International Embryo Transfer Society, Champaign, IL). Available at www.iets.org/pdf/comm_data/December2013.pdf. Accessed March 11, 2015.
- 13.Lim D, et al. Clinical and molecular genetic features of Beckwith-Wiedemann syndrome associated with assisted reproductive technologies. Hum Reprod. 2009;24(3):741–747. doi: 10.1093/humrep/den406. [DOI] [PubMed] [Google Scholar]
- 14.Halliday J, Oke K, Breheny S, Algar E, J Amor D. Beckwith-Wiedemann syndrome and IVF: A case-control study. Am J Hum Genet. 2004;75(3):526–528. doi: 10.1086/423902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Farin PW, Farin CE. Transfer of bovine embryos produced in vivo or in vitro: Survival and fetal development. Biol Reprod. 1995;52(3):676–682. doi: 10.1095/biolreprod52.3.676. [DOI] [PubMed] [Google Scholar]
- 16.Young LE, Sinclair KD, Wilmut I. Large offspring syndrome in cattle and sheep. Rev Reprod. 1998;3(3):155–163. doi: 10.1530/ror.0.0030155. [DOI] [PubMed] [Google Scholar]
- 17.Bertolini M, et al. Morphology and morphometry of in vivo- and in vitro-produced bovine concepti from early pregnancy to term and association with high birth weights. Theriogenology. 2002;58(5):973–994. doi: 10.1016/s0093-691x(02)00935-4. [DOI] [PubMed] [Google Scholar]
- 18.Behboodi E, et al. Birth of large calves that developed from in vitro-derived bovine embryos. Theriogenology. 1995;44(2):227–232. doi: 10.1016/0093-691x(95)00172-5. [DOI] [PubMed] [Google Scholar]
- 19.Farin PW, Piedrahita JA, Farin CE. Errors in development of fetuses and placentas from in vitro-produced bovine embryos. Theriogenology. 2006;65(1):178–191. doi: 10.1016/j.theriogenology.2005.09.022. [DOI] [PubMed] [Google Scholar]
- 20.Weksberg R, Shuman C, Beckwith JB. Beckwith-Wiedemann syndrome. Eur J Hum Genet. 2010;18(1):8–14. doi: 10.1038/ejhg.2009.106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pettenati MJ, et al. Wiedemann-Beckwith syndrome: Presentation of clinical and cytogenetic data on 22 new cases and review of the literature. Hum Genet. 1986;74(2):143–154. doi: 10.1007/BF00282078. [DOI] [PubMed] [Google Scholar]
- 22.Azzi S, et al. Multilocus methylation analysis in a large cohort of 11p15-related foetal growth disorders (Russell Silver and Beckwith Wiedemann syndromes) reveals simultaneous loss of methylation at paternal and maternal imprinted loci. Hum Mol Genet. 2009;18(24):4724–4733. doi: 10.1093/hmg/ddp435. [DOI] [PubMed] [Google Scholar]
- 23.Bliek J, et al. Hypomethylation at multiple maternally methylated imprinted regions including PLAGL1 and GNAS loci in Beckwith-Wiedemann syndrome. Eur J Hum Genet. 2009;17(5):611–619. doi: 10.1038/ejhg.2008.233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Court F, et al. Genome-wide allelic methylation analysis reveals disease-specific susceptibility to multiple methylation defects in imprinting syndromes. Hum Mutat. 2013;34(4):595–602. doi: 10.1002/humu.22276. [DOI] [PubMed] [Google Scholar]
- 25.Eggermann T, et al. Clinical utility gene card for: Beckwith-Wiedemann Syndrome. Eur J Hum Genet. 2014 doi: 10.1038/ejhg.2013.132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Maeda T, et al. 2014. Comprehensive and quantitative multilocus methylation analysis reveals the susceptibility of specific imprinted differentially methylated regions to aberrant methylation in Beckwith-Wiedemann syndrome with epimutations. Genet Med 16(12):903–912.
- 27.Poole RL, et al. International Clinical Imprinting Consortium Targeted methylation testing of a patient cohort broadens the epigenetic and clinical description of imprinting disorders. Am J Med Genet A. 2013;161A(9):2174–2182. doi: 10.1002/ajmg.a.36049. [DOI] [PubMed] [Google Scholar]
- 28.Tee L, et al. 2013. Epimutation profiling in Beckwith-Wiedemann syndrome: Relationship with assisted reproductive technology. Clin Epigenetics 5(1):23.
- 29.Chen Z, Robbins KM, Wells KD, Rivera RM. 2013. Large offspring syndrome: A bovine model for the human loss-of-imprinting overgrowth syndrome Beckwith-Wiedemann. Epigenetics 8(6):591–601.
- 30.Ingleby FC, Flis I, Morrow EH. Sex-biased gene expression and sexual conflict throughout development. Cold Spring Harb Perspect Biol. 2014;7(1):a017632. doi: 10.1101/cshperspect.a017632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.DeVeale B, van der Kooy D, Babak T. Critical evaluation of imprinted gene expression by RNA-Seq: A new perspective. PLoS Genet. 2012;8(3):e1002600. doi: 10.1371/journal.pgen.1002600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.O’Doherty AM, O’Shea LC, Fair T. Bovine DNA methylation imprints are established in an oocyte size-specific manner, which are coordinated with the expression of the DNMT3 family proteins. Biol Reprod. 2012;86(3):67. doi: 10.1095/biolreprod.111.094946. [DOI] [PubMed] [Google Scholar]
- 33.Hubertus J, et al. Selective methylation of CpGs at regulatory binding sites controls NNAT expression in Wilms tumors. PLoS ONE. 2013;8(6):e67605. doi: 10.1371/journal.pone.0067605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Evans HK, Wylie AA, Murphy SK, Jirtle RL. The neuronatin gene resides in a “micro-imprinted” domain on human chromosome 20q11.2. Genomics. 2001;77(1-2):99–104. doi: 10.1006/geno.2001.6612. [DOI] [PubMed] [Google Scholar]
- 35.Kagitani F, et al. Peg5/Neuronatin is an imprinted gene located on sub-distal chromosome 2 in the mouse. Nucleic Acids Res. 1997;25(17):3428–3432. doi: 10.1093/nar/25.17.3428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Robinson MD, McCarthy DJ, Smyth GK. edgeR: A Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26(1):139–140. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.McCarthy DJ, Chen Y, Smyth GK. Differential expression analysis of multifactor RNA-Seq experiments with respect to biological variation. Nucleic Acids Res. 2012;40(10):4288–4297. doi: 10.1093/nar/gks042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kalish JM, Jiang C, Bartolomei MS. Epigenetics and imprinting in human disease. Int J Dev Biol. 2014;58(2-4):291–298. doi: 10.1387/ijdb.140077mb. [DOI] [PubMed] [Google Scholar]
- 39.Weksberg R, Shuman C, Smith AC. Beckwith-Wiedemann syndrome. Am J Med Genet C Semin Med Genet. 2005;137C(1):12–23. doi: 10.1002/ajmg.c.30058. [DOI] [PubMed] [Google Scholar]
- 40.Young LE, et al. Epigenetic change in IGF2R is associated with fetal overgrowth after sheep embryo culture. Nat Genet. 2001;27(2):153–154. doi: 10.1038/84769. [DOI] [PubMed] [Google Scholar]
- 41.Hori N, et al. Aberrant CpG methylation of the imprinting control region KvDMR1 detected in assisted reproductive technology-produced calves and pathogenesis of large offspring syndrome. Anim Reprod Sci. 2010;122(3-4):303–312. doi: 10.1016/j.anireprosci.2010.09.008. [DOI] [PubMed] [Google Scholar]
- 42.Robbins KM, Chen Z, Wells KD, Rivera RM. Expression of KCNQ1OT1, CDKN1C, H19, and PLAGL1 and the methylation patterns at the KvDMR1 and H19/IGF2 imprinting control regions is conserved between human and bovine. J Biomed Sci. 2012;19:95. doi: 10.1186/1423-0127-19-95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Morison IM, Ramsay JP, Spencer HG. A census of mammalian imprinting. Trends Genet. 2005;21(8):457–465. doi: 10.1016/j.tig.2005.06.008. [DOI] [PubMed] [Google Scholar]
- 44.Zaitoun I, Khatib H. Comparative genomic imprinting and expression analysis of six cattle genes. J Anim Sci. 2008;86(1):25–32. doi: 10.2527/jas.2007-0150. [DOI] [PubMed] [Google Scholar]
- 45.Varrault A, et al. Zac1 regulates an imprinted gene network critically involved in the control of embryonic growth. Dev Cell. 2006;11(5):711–722. doi: 10.1016/j.devcel.2006.09.003. [DOI] [PubMed] [Google Scholar]
- 46.Arima T, et al. ZAC, LIT1 (KCNQ1OT1) and p57KIP2 (CDKN1C) are in an imprinted gene network that may play a role in Beckwith-Wiedemann syndrome. Nucleic Acids Res. 2005;33(8):2650–2660. doi: 10.1093/nar/gki555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Susiarjo M, Sasson I, Mesaros C, Bartolomei MS. Bisphenol a exposure disrupts genomic imprinting in the mouse. PLoS Genet. 2013;9(4):e1003401. doi: 10.1371/journal.pgen.1003401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Shi J, Luo L, Eash J, Ibebunjo C, Glass DJ. The SCF-Fbxo40 complex induces IRS1 ubiquitination in skeletal muscle, limiting IGF1 signaling. Dev Cell. 2011;21(5):835–847. doi: 10.1016/j.devcel.2011.09.011. [DOI] [PubMed] [Google Scholar]
- 49.Kim D, et al. TopHat2: Accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol. 2013;14(4):R36. doi: 10.1186/gb-2013-14-4-r36. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.