Significance
Phagocytes are key to innate immunity, as they ingest, kill, and degrade foreign particles, such as microbes. Particle uptake results in the formation of phagosomes that eventually fuse with lysosomes, the main degradative compartments in cells. Phagosome–lysosome fusion (PLF) is crucial for the degradation of phagosome contents and antigen presentation. Most pathogenic microbes that multiply within phagocytes and cause severe diseases (e.g. tuberculosis), prevent PLF. Deciphering the mechanisms that govern PLF will help to understand how phagocytic cells clear microbial infections and how pathogens evade being delivered to microbicidal phagolysosomes. Here, we identify specific phosphoinositide lipids [i.e. PI(4)P and PI(3)P] and the enzymes that produce them as regulators of PLF, which explains why these lipids are manipulated by some pathogens.
Keywords: phosphoinositides, cell-free membrane fusion, phagosome, lysosome, Legionella pneumophila
Abstract
Professional phagocytic cells ingest microbial intruders by engulfing them into phagosomes, which subsequently mature into microbicidal phagolysosomes. Phagosome maturation requires sequential fusion of the phagosome with early endosomes, late endosomes, and lysosomes. Although various phosphoinositides (PIPs) have been detected on phagosomes, it remained unclear which PIPs actually govern phagosome maturation. Here, we analyzed the involvement of PIPs in fusion of phagosomes with various endocytic compartments and identified phosphatidylinositol 4-phosphate [PI(4)P], phosphatidylinositol 3-phosphate [PI(3)P], and the lipid kinases that generate these PIPs, as mediators of phagosome–lysosome fusion. Phagosome–early endosome fusion required PI(3)P, yet did not depend on PI(4)P. Thus, PI(3)P regulates phagosome maturation at early and late stages, whereas PI(4)P is selectively required late in the pathway.
Phagosomes are formed when phagocytes engulf particles of more than 0.4 µm in diameter and mature into degradative phagolysosomes by sequential fusion with early endosomes, late endosomes, and lysosomes (1, 2). Protein and lipid compositions of the phagosome membrane change dramatically during phagosome maturation (3).
Phosphoinositides (PIPs) are mono, bis-, or tris-phosphorylated derivatives of the glycerophospholipid phosphatidylinositol (PI). The distinct subcellular localization of PIP phosphatases and PIP kinases (PIKs) results in the accumulation of PIPs in specific compartments (4). Various PIPs have been implicated in phagosome maturation (5) and some microbial “intracellular” pathogens, which evade being delivered to and killed within phagolysosomes, manipulate the PIP composition of the phagosomes in which they reside (6). However, the impact of these lipids on late stages of phagosome maturation, in particular phagosome–lysosome fusion (PLF), is ill-defined.
Here, we tested directly whether PLF requires PIPs, using an assay which reconstitutes fusion between phagosomes and endocytic compartments in a cell-free system (2).
Results
PLF Requires Phosphatidylinositol 4-Phosphate and Phosphatidylinositol 3-Phosphate.
Involvement of PIPs in PLF is difficult to investigate because of the experimental complications that arise from the vectorial organization of the phagosome maturation subreactions in macrophages: phagosomes will not fuse with lysosomes, if any of the preceding maturation steps is inhibited. Thus, if a certain PIP were required both early and late, degradation or sequestration of this PIP would block phagosome maturation early and, hence, obscure the effects of such treatment on later stages. We circumvented this problem using a cell-free assay that specifically reconstitutes PLF (2).
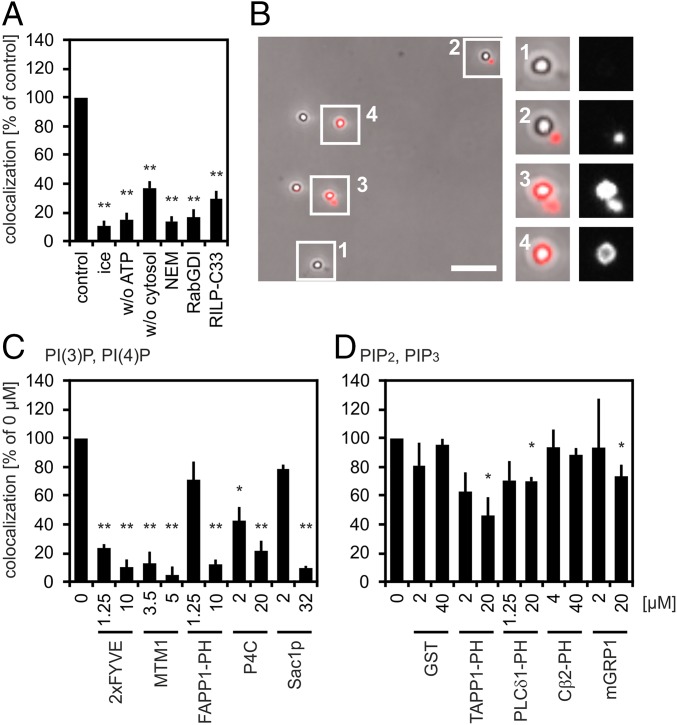
Cell-free PLF was performed as described previously (2) and presented all features observed earlier; that is, dependence on a physiological temperature, ATP, and cytosol and sensitivity to N-ethylmaleimide and Rab guanosine nucleotide dissociation inhibitor (RabGDI) (Fig. 1 A and B). PLF, moreover, was inhibited by Rab7-interacting lysosomal protein-C33, a truncated Rab effector that binds to Rab7(GTP) (7) and by Rab34(GTP) (8), both of which have been implicated in PLF (9, 10). Phagosomes used in our fusion studies were generated by feeding 1-µm latex beads to macrophages and were purified after 30 min of uptake (pulse) and 60 min of maturation (chase). These latex bead phagosomes (LBPs) were enriched in cathepsin D, lysosome-associated membrane protein 1 (LAMP1), and a fluorescent tracer preloaded into lysosomes, but lacked early endocytic transferrin receptor (TfR) and Rab5b (Fig. S1 A and B). Paramagnetically labeled lysosomes were purified as described previously (2). Thirty minute/60 min (pulse/chase) LBPs fused more frequently with purified lysosomes than early (10 min/0 min) or late (10 min/20 min) LBPs (Fig. S1C), which is in line with the vectorial organization of the phagosome maturation subreactions (2, 3, 11).
Fig. 1.
PLF requires PI(4)P and PI(3)P. (A) Phagosomes containing 1-µm latex beads (LBPs) and lysosomes containing BSA-rho-bio were mixed under fusion-promoting conditions. LBPs were isolated from reaction mixtures, spun onto coverslips, and assayed for colocalization with the lysosomal fluor. Colocalization under standard conditions (control) was set as 100% (4–10% absolute colocalization). Reactions were incubated for 60 min on ice or at 37 °C in the absence of ATP or cytosol or in the presence of 4 mM N-ethylmaleimide (NEM), 10 µM RabGDI, or 6 µM Rab7-interacting lysosomal protein-C33 (RILP-C33). (B) Micrograph showing LBPs from a cell-free fusion reaction prepared for fluorescence microscopy. Fusion of LBPs with lysosomes results in colocalization between LBPs and lysosomal BSA-rho-bio. Enlargements show LBPs colocalizing (nos. 3 and 4) or not colocalizing (nos. 1 and 2) with the lysosomal fluor. (Scale bar, 5 µm.) Enlargements (2.5-fold) show LBPs colocalizing (3 and 4) or not colocalizing. (C and D) Recombinant proteins were added to standard fusion reactions as indicated. Colocalization in the absence of purified proteins (0 µM) was defined as 100% (4–20% of phagosomes containing the lysosomal tracer). Data are means ± SEM from at least three independent experiments. Proteins added to fusion reactions were: (C) PI(3)P binder 2xFYVE, PI(3)P phosphatase MTM1, PI(4)P binders FAPP1 PH domain and SidC P4C, or PI(4)P phosphatase Sac1p; (D) binders to poly-phosphorylated PIPs (PI(3,4)P2, TAPP1 PH domain; PI(4,5)P2, PLCδ1 PH domain; PI(3,5)P2, Centaurinß2 PH domain (Cβ2-PH); PI(3,4,5)P3, mGRP1) or GST. Final protein concentrations are indicated. *P < 0.05, **P < 0.01 compared with untreated controls (= control, = 0 µM).
To test the impact of PIPs on PLF, specific PIP-binding proteins were added to cell-free fusion reactions and they were expected to inhibit fusion, if the lipid they sequestered was relevant (12). Phosphatidylinositol 4-phosphate [PI(4)P] binders [FAPP1 PH domain (13) and SidC P4C from the pathogen Legionella pneumophila (14)], phosphatidylinositol 3-phosphate [PI(3)P]-binding Hrs 2xFYVE domain (15), and enzymes which dephosphorylate PI(4)P [Sac1p (16)] or PI(3)P [MTM1 (17)] blocked fusion (Fig. 1C), suggesting that PLF requires PI(4)P and PI(3)P. Binders of the other PIPs were less, if at all, inhibitory (Fig. 1D). GST, the purification tag of the PIP binders, did not block PLF (Fig. 1D).
Phagolysosomes and Lysosomes Contain PI(4)P and PI(3)P.
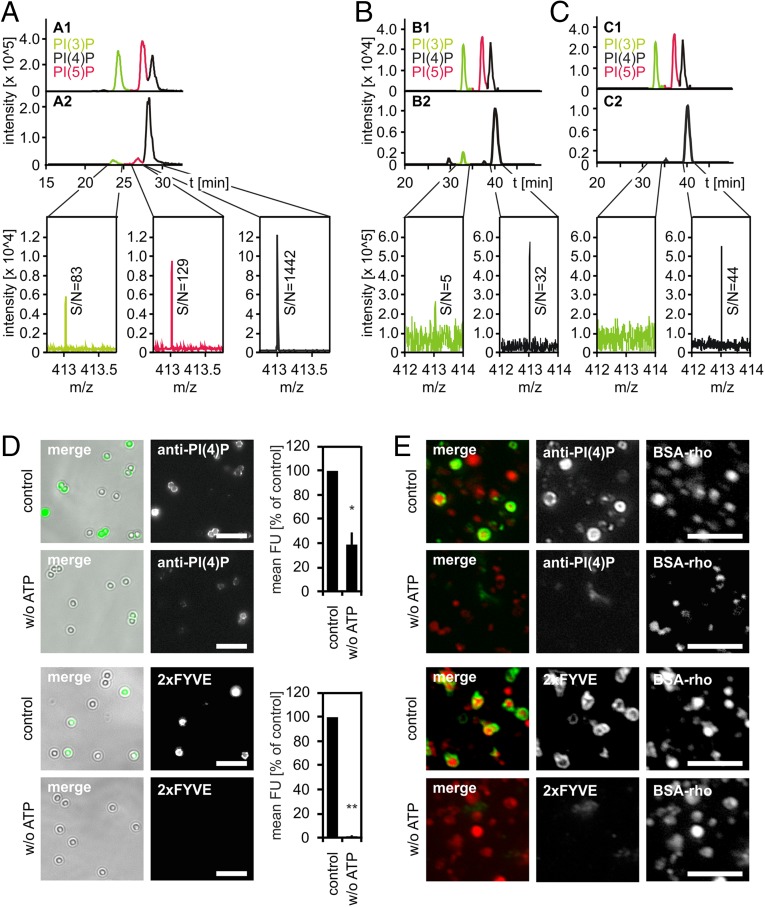
PI(4)P is enriched in the Golgi complex and endoplasmic reticulum (18) and PI(3)P in early endosomes, yet both lipids have occasionally been observed in late endosomes (19, 20) and phagosomes (21, 22). Because PLF required free PI(4)P and PI(3)P, we tested using a reverse-phase high performance liquid chromatography-mass spectrometry (RP-HPLC-MS) approach (23) whether our LBP and lysosome preparations do contain these PIPs: LBPs or lysosomes were incubated under fusion assay conditions and lipids were extracted, deacylated, and analyzed for monophosphorylated PIPs. LBP and lysosome preparations contained both PI(4)P and PI(3)P, with PI(4)P being markedly enriched relative to PI(3)P (Fig. 2 A and B). Lipid extracts from lysosomes contained little PI(3)P (Fig. 2B), yet, as would be expected, we did not detect PI(3)P when the phosphatidylinositol 3-kinase (PI3K) inhibitor wortmannin (WM) was included in the incubations (Fig. 2C).
Fig. 2.
Phagolysosomes and lysosomes contain PI(4)P and PI(3)P. LBPs or lysosomes were purified from J774E cells and incubated under fusion assay conditions for 60 min. Compartments were harvested from the reaction mixture by centrifugation, lipids were extracted, deacylated, and analyzed by RP-HPLC-MS. (A) Analysis of deacylated lipids extracted from LBPs. (A1) Extracted-ion chromatogram (EIC) of separation of PIP standards [i.e., PI(3)P, PI(5)P, or PI(4)P)]. (A2) EIC of separation of lipids prepared from purified LBPs and mass spectra of the elution time range of PI(3)P, PI(4)P, or PI(5)P. Signal-to-noise ratios (S/N) are indicated. (B and C) EICs showing separation of PIP standards (B1, C1) or of PIPs prepared from purified lysosomes upon incubation under fusion conditions in the absence (B2) or presence (C2) of WM. (B2 and C2) Mass spectra of the elution time range of PI(3)P or PI(4)P. S/N-ratios are given. (D) LBPs or (E) lysosomes were incubated with PI(3)P-binding 2xFYVE domain or a PI(4)P-binding antibody under fusion conditions in the presence or absence of ATP. Compartments were harvested from reaction mixtures and stained for bound lipid probes. Representative micrographs of LBPs (D) or lysosomes (E). (Scale bars, 5 µm.) (D) Quantification of the amount of lipid probes on individual LBPs. Mean fluorescence intensities (mean FU) from three independent experiments ± SEM, *P < 0.05, **P < 0.01.
In line with the above observations, a PI(4)P-specific antibody and PI(3)P-binding Hrs 2xFYVE domain bound to LBPs and lysosomes under fusion conditions, as revealed by immunofluorescence microscopy (Fig. 2 D and E). Intriguingly, much less of these lipid probes bound to LBPs and lysosomes when ATP was absent (Fig. 2 D and E), indicating that during the in vitro incubation both PI(4)P and PI(3)P were generated in an ATP-dependent fashion, likely by phosphorylation of PI.
Class II Phosphatidylinositol 4-Kinases Regulate PLF.
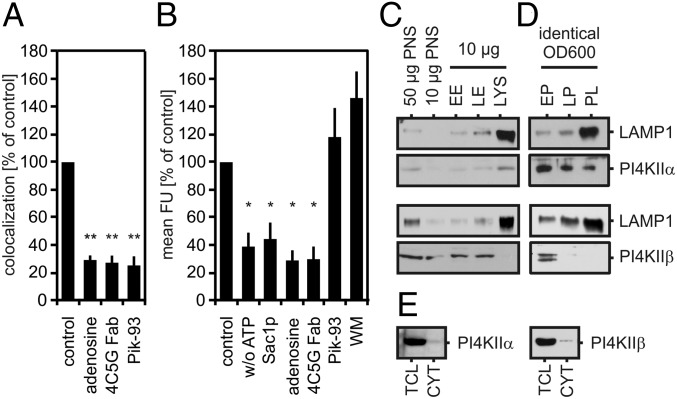
Mammalian cells possess four different phosphatidylinositol 4-kinases (PI4Ks) (i.e., PI4KIIα, PI4KIIβ, PI4KIIIα, and PI4KIIIβ), all of which catalyze the formation of PI(4)P from PI. PLF was inhibited by the general PI4K inhibitors, Pik-93, WM, and adenosine, and by Fab fragments of 4C5G, a monoclonal antibody that specifically inhibits class II PI4K activity (24) (Figs. 3A and 4A). To test whether these inhibitors blocked PI(4)P formation in phagosomes, we incubated purified LBPs with the above PI4K inhibitors under fusion conditions and visualized PI(4)P using a PI(4)P-binding antibody. Incubation in the presence of class II PI4K inhibitors (i.e., adenosine or 4C5G) or of PI(4)P-dephosphorylating Sac1p strongly decreased LBP contents in PI(4)P, as did omission of ATP from reaction mixtures (Fig. 3B). Class III PI4K inhibitors (i.e., Pik-93 or 1 µM WM), by contrast, did not affect phagosome PI(4)P levels (Fig. 3B). Thus, PLF-relevant PI(4)P is generated by class II PI4Ks. Notably, treatments which decreased phagosomal PI(4)P levels also reduced phosphatidylinositol 4,5-bisphosphate [PI(4,5)P2] contents of LBPs (Fig. S2), showing that, under fusion conditions, phagosomal PI(4)P is partially consumed by PI(4)P 5-kinases (PI4,5Ks) during formation of PI(4,5)P2.
Fig. 3.
PI4K activities are involved in PLF. (A) Cell-free fusion between LBPs and lysosomes in the presence of 8 mM adenosine, 12.5 µg/mL Fab fragments of 4C5G antibody (4C5G Fab), or 4 µM Pik-93. (B) Purified LBPs were incubated under fusion conditions in the absence of ATP (w/o ATP) or in the presence of ATP and 32 µM Sac1p, 8 mM adenosine, 12.5 µg/mL 4C5G Fab, 1 µM WM, or 250 nM Pik-93. LBPs were analyzed for PI(4)P contents using an anti-PI(4)P antibody as in Fig. 2D. Data are means ± SEM from at least three independent experiments. *P < 0.05, **P < 0.01 compared with control. (C) Early endosomes (EE), late endosomes (LE), and lysosomes (LYS) or (D) early phagosomes (EP), late phagosomes (LP), and phagolysosomes (PL) were purified, adjusted to identical protein content (C) or identical OD600 (D), and analyzed for LAMP1 and PI4KIIα or PI4KIIβ by immunoblotting. (E) Total cell lysates (TCL) or cytosol (CYT) were prepared from identical numbers of J774E cells. Twenty micrograms of the cytosol preparation and the corresponding volume of the respective TCL were analyzed for PI4KIIα or PI4KIIβ by immunoblotting.
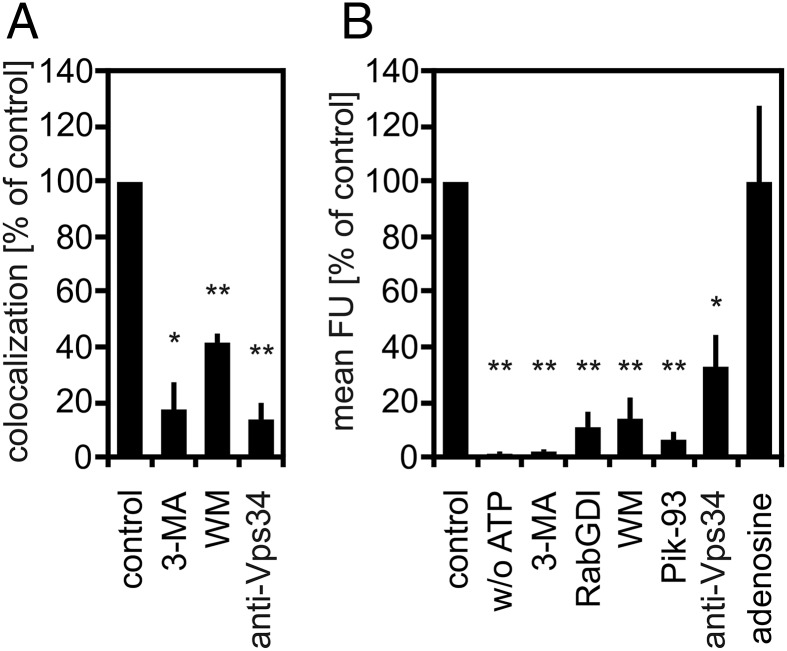
Fig. 4.
PI3K inhibitors block PLF and reduce PI(3)P levels in phagosome membranes. (A) PLF in the presence of 4 mM 3-MA, 40 nM WM, or 50 µg/mL of Vps34-inhibiting antibody. (B) Purified LBPs were incubated under fusion conditions with 2 µM of PI(3)P-binding 2xFYVE domain and the above PI3K-inhibiting reagents, 10 µM RabGDI, 4 µM Pik-93, or 8 mM adenosine. The amount of LBP-associated 2xFYVE domain was quantified by fluorescence microscopy as in Fig. 2D. Mean fluorescence intensity of LBPs under standard conditions was defined as 100%. Data are means ± SEM from at least three independent experiments. *P < 0.05, **P < 0.01 compared with control.
Purified lysosomes and phagolysosomes contained PI4KIIα but lacked PI4KIIβ (Fig. 3 C and D). Moreover, cytosol preparations contained neither kinase (Fig. 3E). Hence, reconstituted PLF depended on PI4KIIα rather than PI4KIIβ.
Class III PI3K Vps34 Regulates PLF.
PLF requires PI3K activity, as reconstituted PLF was blocked by the PI3K inhibitors 3-methyladenine (3-MA), WM (Fig. 4A), or Pik-93 (Fig. 3A). In mammalian cells, D3-phosphorylated PIPs can be generated by three different classes of PI3Ks. At the concentrations applied, WM, 3-MA, and Pik-93 inhibit all classes of PI3K and potentially interfere with synthesis of any D3-PIP. Because PI(3)P had the strongest impact on PLF (Fig. 1C), we asked whether PI3K inhibitors block the formation of PI(3)P in LBP membranes. To this end, purified LBPs were incubated under fusion conditions with the PI(3)P-binding 2xFYVE domain and the above PI3K inhibitors. The amount of LBP-associated 2xFYVE domain, indicative of PI(3)P in phagosome membranes, was then quantified by fluorescence microscopy.
PI3K inhibitors, 3-MA, WM, and Pik-93 led to decreased steady-state levels of PI(3)P in phagosomes (Fig. 4B) and lysosomes (Fig. S3), whereas the PI4K inhibitor adenosine did not (Fig. 4B). All classes of PI3K catalyze the phosphorylation of PI to PI(3)P in vitro (25). However, in vivo, PI(3)P is thought to be mainly generated by the class III PI3K Vps34 (26). Here, an antibody that specifically inhibits Vps34 activity (5, 27–29), blocked PLF (Fig. 4A) and decreased LBP contents in PI(3)P to the same degree as the PI3K inhibitors WM, 3-MA, or Pik-93 (Fig. 4B). Hence, PLF in vitro required formation of PI(3)P by class III PI3K Vps34 (Fig. 4). RabGDI, which extracts Rab(GDP) proteins from membranes, also decreased phagosome (Fig. 4B) and lysosome contents in PI(3)P (Fig. S3), which is in line with the described role of Vps34 as a Rab protein effector.
To test whether progression of late phagosomes to phagolysosomes requires PI3Ks also in intact cells, we applied WM to cells 20 min after addition of latex beads, a time when LBPs have passed the PI(3)P-dependent early maturation step and contained late endocytic marker proteins (Fig. S1 A and B). Then, cells were incubated for an additional 60 min. WM completely inhibited late phagosome-lysosome fusion and decreased the levels of PI(3)P in LBPs (Fig. S4). Completion of phagolysosome biogenesis in intact macrophages thus requires PI3K activity both upstream (30) and, intriguingly, downstream of early-to-late phagosome transition.
PI(4)P Regulates Late Phagosome Maturation Only, Whereas PI(3)P Is Required Both Early and Late in the Pathway.
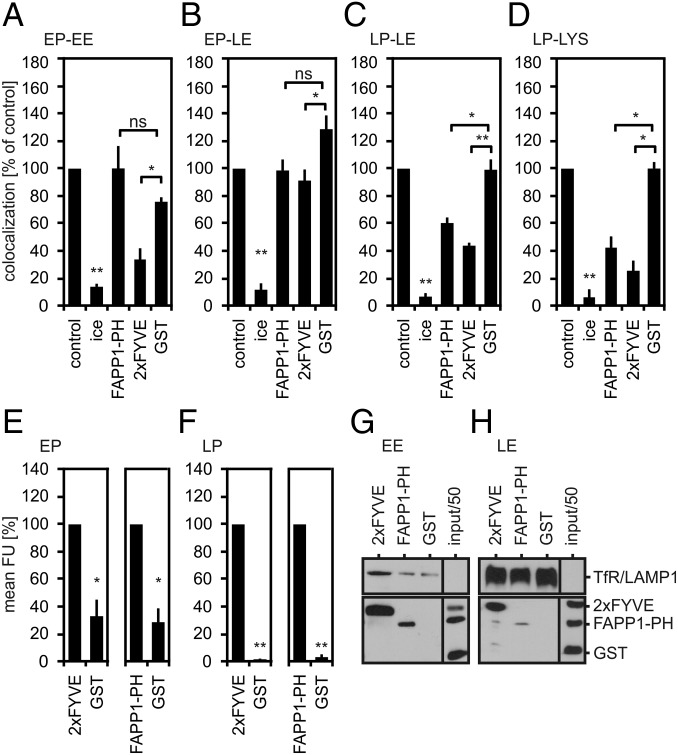
Given these results, we addressed the question whether the requirement for PI(4)P and PI(3)P was limited to fusion between phagolysosomes and lysosomes. To this end, we reconstituted fusion between phagosomes and endosomes at earlier stages of maturation and tested whether these fusion events were sensitive to the PI(4)P-binding FAPP1 PH domain or PI(3)P-binding 2xFYVE domain (Fig. 5 A–D and Fig. S5). The FAPP1 PH domain inhibited neither early phagosome–early endosome (EP–EE) fusion, nor early phagosome–late endosome fusion (EP–LE) (Fig. 5 A and B). However, it blocked late phagosome–late endosome fusion (LP–LE) (Fig. 5C) and late phagosome–lysosome fusion (LP– LYS) (Fig. 5D). Notably, sensitivity of phagosome–endosome fusion to sequestration of PI(4)P increased with increasing maturation of the compartments (Fig. 5 C and D).
Fig. 5.
PI(4)P and PI(3)P regulate phagosome maturation. Purified early phagosomes or late phagosomes were assayed for fusion with early endosomes, late endosomes, or lysosomes, as indicated (A–D). Fusion reactions were incubated on ice or at 37 °C and in the absence (= control) or presence of 20 µM FAPP1-PH, 2xFYVE, or GST. Purified early phagosomes (E) or late phagosomes (F) were incubated under fusion assay conditions in the presence of 2 µM 2xFYVE, FAPP1-PH, or GST. The amount of phagosome-associated lipid-binding probes was quantified as in Fig. 2D. Data are means ± SEM from three independent experiments. *P < 0.05, **P < 0.01, ns: not significant. Purified early endosomes (G) or late endosomes (H) were incubated under fusion assay conditions in the presence of 2 µM 2xFYVE, FAPP1-PH, or GST. Compartments were isolated from reaction mixtures, adjusted to identical protein content, and analyzed for bound lipid probes or GST by immunoblotting. Staining of TfR (G) or LAMP1 (H) was used as loading control.
As expected, PI(3)P-binding 2xFYVE domain efficiently blocked EP–EE fusion (Fig. 5A) (31). Moreover, it inhibited LP–LE fusion (Fig. 5C) and LP–LYS fusion (Fig. 5D). In contrast, EP–LE fusion was almost resistant to the 2xFYVE domain (Fig. 5B). Despite the differential sensitivity of the various fusion events to 2xFYVE and FAPP1-PH, these lipid probes bound to early phagosomes/endosomes (Fig. 5 E and G) and late phagosomes/endosomes (Fig. 5 F and H), indicating that these compartments do contain the corresponding lipids, similar to phagolysosomes and lysosomes (Fig. 2). In sum, phagosome maturation requires PI(3)P at early and late stages, whereas PI(4)P is selectively required late in the pathway.
Discussion
Previous studies have analyzed PIP dynamics on nascent and maturing phagosomes in intact cells using expression of fluorescent protein-tagged lipid-binding protein domains. These studies revealed accumulation of PI(4,5)P2 and phosphatidylinositol 3,4,5-trisphosphate [PI(3,4,5)P3] at sites of phagocytosis (32, 33) and of PI(3)P on early phagosomes (29, 34), yet no specific PIP had been assigned to phagolysosomes. To address the function rather than the localization of these lipids, previous reports used pharmacological inhibitors of PIKs, such as PI3K inhibitors WM (29), LY94002 (35), and PI-103 (5) or PikFYVE inhibitor YM201636 (36); researchers used siRNA-mediated knockdown of PIK expression (22) or microinjection of PIK-inhibiting antibodies (29). In all these experiments, phagocytic cells had to be manipulated before being infected. Hence, if several steps of phagosome maturation were affected by the treatment, analysis of later steps was hampered by early inhibition. Using cell-free, stage-specific fusion reactions, we show here the functional impact of PIPs on defined subreactions of phagosome maturation, especially PLF. PIP involvement was assessed by sequestering or degrading defined PIP species, in an approach similar as used with yeast vacuole fusion (12, 37) and yeast COPII vesicle–Golgi fusion (38). We observed that both PI(4)P and PI(3)P are absolutely required for PLF.
Because of the localization of PIKs and PIP phosphatases within cells, PI(3)P is strongly enriched on early endosomes and PI(4)P on the Golgi complex (18), but both lipids have also been observed at other subcellular sites, including late endocytic compartments (19–22, 39, 40). Importantly and surprisingly, we detected here both PI(4)P and PI(3)P in purified phagolysosomes and lysosomes using the corresponding lipid-binding domains as probes. Because detection of these lipids via lipid-binding domains is indirect and excludes protein-bound PIPs, we further analyzed LBPs or lysosomes directly for monophosphoinositides by RP-HPLC-MS. Both, PI(4)P and PI(3)P were detected in LBP and lysosome preparations, complementing our microscopic data. Notably, to our knowledge this is the first time that defined subcellular compartments have been analyzed for their content in PIPs using a direct, nonradioactive assay that can distinguish between the different monophosphoinositides (23).
A central result of this study is the, to our knowledge, first-time definition of PI(4)P as a central regulator of late phagosome maturation. Conversely, early subreactions of phagosome maturation (i.e., fusion of early phagosomes with early endosomes or late endosomes) did not require PI(4)P (this study). Both, PI(4)P and PI(4,5)P2 have occasionally been detected in phagolysosomes (21) and lysosomes (19, 20), yet specific functions have only been attributed to PI(4,5)P2. Even in other cellular processes, PI(4)P was long thought to be merely the precursor of PI(4,5)P2 (41) and here, too, phagosomal PI(4)P was converted to PI(4,5)P2 under fusion conditions. However, in contrast to PI(4)P, PI(4,5)P2 was not required for PLF, in line with a direct effector function for PI(4)P. This does, of course, not exclude a role of PI(4,5)P2 in late phagosome maturation in vivo. For example, conversion of PI(4)P to PI(4,5)P2 on phagosomes is required for polymerization of F-actin (21), which facilitates their fusion with lysosomes (42). This possible in vivo role of PI(4,5)P2 would not be reconstituted in our cell-free PLF assay, which does not require actin polymerization (2).
Our study shows that PLF-relevant PI(4)P is produced by a type II PI4K, as fusion of phagosomes with lysosomes and PI(4)P production were blocked by an inhibitory antibody specific to type II PI4Ks and by adenosine. Early phagosomes contained both type II PI4Ks, which is in line with earlier reports (5, 43). In contrast, phagolysosomes and lysosomes contained PI4KIIα (our data, and refs. 44 and 45), but lacked PI4KIIβ. Moreover, our cytosol preparations were almost devoid of both type II PI4Ks. Hence, PLF in our system depends on PI4KIIα, because PI4KIIβ is absent. Our data nicely support previous observations made in intact cells: PI4KIIα colocalizes with lysosomal proteins LAMP1, CD63 (44), and LIMP2 (46), and depletion of PI4KIIα causes enlarged LAMP1-positive late endosomes (45, 46), which fail to fuse with lysosomes (44).
We observed that PLF requires formation of PI(3)P by class III PI3K Vps34. Vps34 is an effector of Rab5 (28) and Rab7 (47), both of which are key regulator GTPases of endosome (48) and phagosome (31) maturation. Vps34 promotes homotypic early endosome fusion (28) and has been proposed to also regulate late endosome (49) and phagosome (29, 30, 39, 40) maturation. We now show that Vps34 and its lipid product PI(3)P are absolutely required for PLF. Because we observed that formation of PI(3)P in phagosomes or lysosomes and fusion of these compartments was inhibited by RabGDI, Vps34 recruitment to late phagosomes and endosomes depends on Rab proteins, likely Rab7 (47).
The in vivo relevance of these observations is manifested in the finding that WM completely blocked PLF and decreased levels of PI(3)P in phagosome membranes, even if applied 30 min postphagocytosis; that is, after completion of the early phagosome stage (present study). Because WM inhibits activity of all PI3K isoforms, inhibition of phagosome maturation downstream of early-to-late transition could still be explained by a role for PI3Ks other than Vps34 in PLF. Although a recent study did indeed report that class I PI3K p110α and PI(3,4,5)P3 are required for phagosome maturation (22), we observed that PLF per se was independent of PI(3,4,5)P3 and, hence, of class I PI3Ks. Thus, p110α might regulate subreactions of phagosome maturation other than PLF (e.g., LAMP- and microtubule-dependent phagosome transport) (22, 50).
In addition to the herein described roles in late phagosome maturation, we confirmed PI(3)P as a regulator of fusion between early phagosomes and early endosomes (31). Taken together, these data support previous reports on the biphasic requirement for PI(3)P of both phagosome (39, 40) and endosome maturation (51).
In sum, our data reveal new and direct roles of PI(4)P, class II PI4Ks, PI(3)P, and Vps34 in PLF. Although, we did not unravel the mechanism by which PI(4)P and PI(3)P govern PLF, both lipids will likely act by recruiting and activating “tethering” or “docking” effector proteins (12, 52), such as the HOPS complex (53, 54) or SNAREs (2, 55, 56). Another important aspect of this study is the demonstration that the effector protein SidC from pathogenic Legionella pneumophila inhibited PLF. Although it is not clear whether this phenomenon contributes to the inhibition of PLF in Legionella-infected cells (6), it shows the power of a biochemical approach to understand the reprogramming of host cell membrane trafficking by pathogens.
Materials and Methods
Chemicals.
Chemicals, antibodies, plasmids, and recombinant proteins are described in SI Text.
Purification of LBPs or Endosomes from J774E Macrophages.
LBPs and endosomes were purified from J774E cells as in ref. 2, with modifications. In brief, cells were washed once in PBS and a suspension of 1-µm NeutrAvidin-coated latex beads (SI Text) in 37 °C DMEM (6 × 108 particles/mL) was added (pulse). Cells were rinsed thrice in PBS, new DMEM/FCS [DMEM (Gibco), 10% (vol/vol) FCS (FCS; PAA), 1% (wt/vol) glutamax (Gibco)] was added, and cells were incubated at 37 °C (chase) to isolate early phagosomes (10 min/0 min, pulse/chase), late phagosomes (10 min/20 min), or phagolysosomes (30 min/60 min). DMEM/FCS was discarded, PBS was added, and cells were harvested using a plastic cell scraper. Cells were washed sequentially in PBS/5 mM EDTA and homogenization buffer [HB: 20 mM Hepes/KOH (pH 7.2), 8.6% (wt/vol) sucrose, 0.5 mM EGTA] (160 × g, 4 °C, 7 min), resuspendend in HB containing 1× protease inhibitor mixture, and homogenized in a Dounce homogenizer. A postnuclear supernatant was prepared (800 × g, 4 °C, 5 min), adjusted to 35% (wt/vol) sucrose by addition of HB/62% [HB containing 62% (wt/vol) sucrose], overlayed with 5 mL of HB/25% [HB containing 25% (wt/vol) sucrose] and 3 mL of HB. After centrifugation (42,000 × g, 4 °C, 30 min), LBPs were harvested from the HB/25%/HB interface. Endocytic compartments were labeled with ferrofluid or fluorescent tracers (SI Text) and purified as in ref. 2. Immunoblot analysis was performed as detailed in SI Text.
In Vitro Fusion of LBPs with Endocytic Compartments.
Cell-free fusion of LBPs with endocytic compartments was performed as in ref. 2 with modifications. LBPs contained NeutrAvidin-coated 1-µm latex beads and endosomes were fluid-phase labeled by either calcein or BSA rhodamine biotin (BSA-rho-bio) (SI Text).
A cell-free fusion reaction (total volume: 30 µL) contained 0.625 OD600/mL of purified LBPs, 0.4 mg protein/mL endosomes, 0.12 mg/mL biotin-coupled BSA (BSA-bio) (SI Text), 2 mg/mL J774E cytosol (2), 1× ATP-regenerating system (2), 1× salt mixture (2), and 1 mM DTT. Cell-free fusion between early LBPs and calcein-loaded early endosomes was performed in the absence of BSA-bio. Inhibitors to be tested were incubated for 10 min at 4 °C with purified compartments before addition of the remaining components and incubation at 37 °C for 60 min. Reactions were set on ice and incubated in the presence of 0.2 mg/mL proteinase K (Qiagen) for 15 min. Then, 1.75 mg/mL PMSF were added and reactions were adjusted to 200 µL by addition of HB. Reaction mixtures were layered on top of 1 mL HB/25% cushions and centrifuged (1,800 × g, 30 min, 4 °C). LBPs were collected from the HB/25%/HB interface, spun onto glass coverslips (690 × g, 15 min, 4 °C), and fixed for 16 h at 4 °C in HB containing 2.5% (vol/vol) glutardialdehyde and 2% (wt/vol) formaldehyde (FA). Samples were quenched with 0.1 mg/mL NaBH4 in HB for 30 min at ambient temperature, rinsed once in PBS, mounted in Mowiol, and analyzed by fluorescence microscopy. Colocalization of LBPs with endosomal BSA-rho-bio or calcein, indicative of LBP-endosome fusion, was quantified from at least 600 LBPs.
Detection of Lipid Species on Purified LBPs or Endosomes.
Purified (vol/vol) LBPs (1.25 OD600/mL) or endosomes (0.4 mg protein/mL) were incubated under fusion assay conditions in the presence of 2 µM GST-tagged lipid probe or a PI(4)P-specific antibody (1:200 dilution) for 60 min. The analysis of endosome lipids is detailed in SI Text. For analysis of phagosome lipids, reactions were adjusted to 35% (wt/vol) sucrose by addition of HB/62%, overlayed with 1 mL of HB/25% and 200 µL of HB, and gradients were centrifuged (30 min, 1,800 × g, 4 °C). LBPs were harvested from the HB/25%/HB interface, adjusted to 500 µL HB and 2 mg/mL BSA, spun onto glass coverslips (690 × g, 15 min, 4 °C), and fixed in 4% (wt/vol) FA in HB. Fixative was quenched for 30 min with HB/50 mM NH4Cl for 30 min and PBS/4% (wt/vol) BSA (IF blocking buffer) was added for 30 min. GST-tagged lipid probes were visualized with an anti-GST antibody (Santa Cruz Biotechnology, No. sc-459,1:200 in IF blocking buffer) and an Alexa488-conjugated secondary antibody (goat anti-rabbit, 1:200 in IF blocking buffer). PI(4)P-specific antibodies were visualized with an Alexa488-coupled secondary antibody (goat anti-mouse, 1:200 in IF blocking buffer). Samples were mounted in Mowiol and the fluorescence intensities around phagosomes in random fields were analyzed using a Zeiss Axio Observer.Z1 epifluorescence microscope and ImageJ software (SI Text). Analysis of phagosome or endosome lipids by RP-HPLC-MS is detailed in SI Text and Table S1.
Statistical Analysis.
Means and SEMs were calculated from independent experiments. Data were analyzed by the two-tailed unpaired Student’s t test with significance assumed at P < 0.05 (*) and high significance at P < 0.01 (**).
Supplementary Material
Acknowledgments
We thank all colleagues who generously contributed plasmids and antibodies. This study was supported by Deutsche Forschungsgemeinschaft Grants SFB 645 (to A.H.) and SPP 1580 (to A.H., B.L., and H.H.) and by the intramural research program of the Eunice Kennedy Shriver National Institute of Child Health and Human Development of the National Institutes of Health, Bethesda, MD (to M.J. and T.B.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1423456112/-/DCSupplemental.
References
- 1.Haas A. The phagosome: Compartment with a license to kill. Traffic. 2007;8(4):311–330. doi: 10.1111/j.1600-0854.2006.00531.x. [DOI] [PubMed] [Google Scholar]
- 2.Becken U, Jeschke A, Veltman K, Haas A. Cell-free fusion of bacteria-containing phagosomes with endocytic compartments. Proc Natl Acad Sci USA. 2010;107(48):20726–20731. doi: 10.1073/pnas.1007295107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Desjardins M, Huber LA, Parton RG, Griffiths G. Biogenesis of phagolysosomes proceeds through a sequential series of interactions with the endocytic apparatus. J Cell Biol. 1994;124(5):677–688. doi: 10.1083/jcb.124.5.677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kutateladze TG. Translation of the phosphoinositide code by PI effectors. Nat Chem Biol. 2010;6(7):507–513. doi: 10.1038/nchembio.390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bohdanowicz M, Cosío G, Backer JM, Grinstein S. Class I and class III phosphoinositide 3-kinases are required for actin polymerization that propels phagosomes. J Cell Biol. 2010;191(5):999–1012. doi: 10.1083/jcb.201004005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Weber SS, Ragaz C, Hilbi H. Pathogen trafficking pathways and host phosphoinositide metabolism. Mol Microbiol. 2009;71(6):1341–1352. doi: 10.1111/j.1365-2958.2009.06608.x. [DOI] [PubMed] [Google Scholar]
- 7.Cantalupo G, Alifano P, Roberti V, Bruni CB, Bucci C. Rab-interacting lysosomal protein (RILP): The Rab7 effector required for transport to lysosomes. EMBO J. 2001;20(4):683–693. doi: 10.1093/emboj/20.4.683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wang T, Hong W. Interorganellar regulation of lysosome positioning by the Golgi apparatus through Rab34 interaction with Rab-interacting lysosomal protein. Mol Biol Cell. 2002;13(12):4317–4332. doi: 10.1091/mbc.E02-05-0280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Harrison RE, Bucci C, Vieira OV, Schroer TA, Grinstein S. Phagosomes fuse with late endosomes and/or lysosomes by extension of membrane protrusions along microtubules: Role of Rab7 and RILP. Mol Cell Biol. 2003;23(18):6494–6506. doi: 10.1128/MCB.23.18.6494-6506.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kasmapour B, Gronow A, Bleck CK, Hong W, Gutierrez MG. Size-dependent mechanism of cargo sorting during lysosome-phagosome fusion is controlled by Rab34. Proc Natl Acad Sci USA. 2012;109(50):20485–20490. doi: 10.1073/pnas.1206811109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jahraus A, et al. In vitro fusion of phagosomes with different endocytic organelles from J774 macrophages. J Biol Chem. 1998;273(46):30379–30390. doi: 10.1074/jbc.273.46.30379. [DOI] [PubMed] [Google Scholar]
- 12.Fratti RA, Jun Y, Merz AJ, Margolis N, Wickner W. Interdependent assembly of specific regulatory lipids and membrane fusion proteins into the vertex ring domain of docked vacuoles. J Cell Biol. 2004;167(6):1087–1098. doi: 10.1083/jcb.200409068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dowler S, et al. Identification of pleckstrin-homology-domain-containing proteins with novel phosphoinositide-binding specificities. Biochem J. 2000;351(Pt 1):19–31. doi: 10.1042/0264-6021:3510019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ragaz C, et al. The Legionella pneumophila phosphatidylinositol-4 phosphate-binding type IV substrate SidC recruits endoplasmic reticulum vesicles to a replication-permissive vacuole. Cell Microbiol. 2008;10(12):2416–2433. doi: 10.1111/j.1462-5822.2008.01219.x. [DOI] [PubMed] [Google Scholar]
- 15.Gillooly DJ, et al. Localization of phosphatidylinositol 3-phosphate in yeast and mammalian cells. EMBO J. 2000;19(17):4577–4588. doi: 10.1093/emboj/19.17.4577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Maehama T, Dixon JE. [PTEN tumor suppressor: Functions as a lipid phosphatase] Tanpakushitsu Kakusan Koso. 2000;45(14):2405–2414. Japanese. [PubMed] [Google Scholar]
- 17.Taylor GS, Maehama T, Dixon JE. Myotubularin, a protein tyrosine phosphatase mutated in myotubular myopathy, dephosphorylates the lipid second messenger, phosphatidylinositol 3-phosphate. Proc Natl Acad Sci USA. 2000;97(16):8910–8915. doi: 10.1073/pnas.160255697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Vicinanza M, D’Angelo G, Di Campli A, De Matteis MA. Function and dysfunction of the PI system in membrane trafficking. EMBO J. 2008;27(19):2457–2470. doi: 10.1038/emboj.2008.169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hammond GRV, Machner MP, Balla T. A novel probe for phosphatidylinositol 4-phosphate reveals multiple pools beyond the Golgi. J Cell Biol. 2014;205(1):113–126. doi: 10.1083/jcb.201312072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Arneson LS, Kunz J, Anderson RA, Traub LM. Coupled inositide phosphorylation and phospholipase D activation initiates clathrin-coat assembly on lysosomes. J Biol Chem. 1999;274(25):17794–17805. doi: 10.1074/jbc.274.25.17794. [DOI] [PubMed] [Google Scholar]
- 21.Defacque H, et al. Phosphoinositides regulate membrane-dependent actin assembly by latex bead phagosomes. Mol Biol Cell. 2002;13(4):1190–1202. doi: 10.1091/mbc.01-06-0314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Thi EP, Lambertz U, Reiner NE. Class IA phosphatidylinositol 3-kinase p110α regulates phagosome maturation. PLoS ONE. 2012;7(8):e43668. doi: 10.1371/journal.pone.0043668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kiefer S, et al. Separation and detection of all phosphoinositide isomers by ESI-MS. J Pharm Biomed Anal. 2010;53(3):552–558. doi: 10.1016/j.jpba.2010.03.029. [DOI] [PubMed] [Google Scholar]
- 24.Endemann GC, Graziani A, Cantley LC. A monoclonal antibody distinguishes two types of phosphatidylinositol 4-kinase. Biochem J. 1991;273(Pt 1):63–66. doi: 10.1042/bj2730063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Vanhaesebroeck B, Guillermet-Guibert J, Graupera M, Bilanges B. The emerging mechanisms of isoform-specific PI3K signalling. Nat Rev Mol Cell Biol. 2010;11(5):329–341. doi: 10.1038/nrm2882. [DOI] [PubMed] [Google Scholar]
- 26.Di Paolo G, De Camilli P. Phosphoinositides in cell regulation and membrane dynamics. Nature. 2006;443(7112):651–657. doi: 10.1038/nature05185. [DOI] [PubMed] [Google Scholar]
- 27.Siddhanta U, McIlroy J, Shah A, Zhang Y, Backer JM. Distinct roles for the p110alpha and hVPS34 phosphatidylinositol 3′-kinases in vesicular trafficking, regulation of the actin cytoskeleton, and mitogenesis. J Cell Biol. 1998;143(6):1647–1659. doi: 10.1083/jcb.143.6.1647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Christoforidis S, et al. Phosphatidylinositol-3-OH kinases are Rab5 effectors. Nat Cell Biol. 1999;1(4):249–252. doi: 10.1038/12075. [DOI] [PubMed] [Google Scholar]
- 29.Vieira OV, et al. Distinct roles of class I and class III phosphatidylinositol 3-kinases in phagosome formation and maturation. J Cell Biol. 2001;155(1):19–25. doi: 10.1083/jcb.200107069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Vieira OV, et al. Modulation of Rab5 and Rab7 recruitment to phagosomes by phosphatidylinositol 3-kinase. Mol Cell Biol. 2003;23(7):2501–2514. doi: 10.1128/MCB.23.7.2501-2514.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Vieira OV, Botelho RJ, Grinstein S. Phagosome maturation: Aging gracefully. Biochem J. 2002;366(Pt 3):689–704. doi: 10.1042/BJ20020691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Botelho RJ, et al. Localized biphasic changes in phosphatidylinositol-4,5-bisphosphate at sites of phagocytosis. J Cell Biol. 2000;151(7):1353–1368. doi: 10.1083/jcb.151.7.1353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Marshall JG, et al. Restricted accumulation of phosphatidylinositol 3-kinase products in a plasmalemmal subdomain during Fc gamma receptor-mediated phagocytosis. J Cell Biol. 2001;153(7):1369–1380. doi: 10.1083/jcb.153.7.1369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ellson CD, et al. Phosphatidylinositol 3-phosphate is generated in phagosomal membranes. Curr Biol. 2001;11(20):1631–1635. doi: 10.1016/s0960-9822(01)00447-x. [DOI] [PubMed] [Google Scholar]
- 35.Fratti RA, Backer JM, Gruenberg J, Corvera S, Deretic V. Role of phosphatidylinositol 3-kinase and Rab5 effectors in phagosomal biogenesis and mycobacterial phagosome maturation arrest. J Cell Biol. 2001;154(3):631–644. doi: 10.1083/jcb.200106049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kerr MC, et al. Inhibition of the PtdIns(5) kinase PIKfyve disrupts intracellular replication of Salmonella. EMBO J. 2010;29(8):1331–1347. doi: 10.1038/emboj.2010.28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mayer A, et al. Phosphatidylinositol 4,5-bisphosphate regulates two steps of homotypic vacuole fusion. Mol Biol Cell. 2000;11(3):807–817. doi: 10.1091/mbc.11.3.807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lorente-Rodríguez A, Barlowe C. Requirement for Golgi-localized PI(4)P in fusion of COPII vesicles with Golgi compartments. Mol Biol Cell. 2011;22(2):216–229. doi: 10.1091/mbc.E10-04-0317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chua J, Deretic V. Mycobacterium tuberculosis reprograms waves of phosphatidylinositol 3-phosphate on phagosomal organelles. J Biol Chem. 2004;279(35):36982–36992. doi: 10.1074/jbc.M405082200. [DOI] [PubMed] [Google Scholar]
- 40.Lu N, et al. Two PI 3-kinases and one PI 3-phosphatase together establish the cyclic waves of phagosomal PtdIns(3)P critical for the degradation of apoptotic cells. PLoS Biol. 2012;10(1):e1001245. doi: 10.1371/journal.pbio.1001245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.D’Angelo G, Vicinanza M, Di Campli A, De Matteis MA. The multiple roles of PtdIns(4)P—Not just the precursor of PtdIns(4,5)P2. J Cell Sci. 2008;121(Pt 12):1955–1963. doi: 10.1242/jcs.023630. [DOI] [PubMed] [Google Scholar]
- 42.Jahraus A, et al. ATP-dependent membrane assembly of F-actin facilitates membrane fusion. Mol Biol Cell. 2001;12(1):155–170. doi: 10.1091/mbc.12.1.155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Pizarro-Cerdá J, et al. Type II phosphatidylinositol 4-kinases promote Listeria monocytogenes entry into target cells. Cell Microbiol. 2007;9(10):2381–2390. doi: 10.1111/j.1462-5822.2007.00967.x. [DOI] [PubMed] [Google Scholar]
- 44.Minogue S, et al. Phosphatidylinositol 4-kinase is required for endosomal trafficking and degradation of the EGF receptor. J Cell Sci. 2006;119(Pt 3):571–581. doi: 10.1242/jcs.02752. [DOI] [PubMed] [Google Scholar]
- 45.Craige B, Salazar G, Faundez V. Phosphatidylinositol-4-kinase type II alpha contains an AP-3-sorting motif and a kinase domain that are both required for endosome traffic. Mol Biol Cell. 2008;19(4):1415–1426. doi: 10.1091/mbc.E07-12-1239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Jović M, et al. Two phosphatidylinositol 4-kinases control lysosomal delivery of the Gaucher disease enzyme, β-glucocerebrosidase. Mol Biol Cell. 2012;23(8):1533–1545. doi: 10.1091/mbc.E11-06-0553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Stein MP, Feng Y, Cooper KL, Welford AM, Wandinger-Ness A. Human VPS34 and p150 are Rab7 interacting partners. Traffic. 2003;4(11):754–771. doi: 10.1034/j.1600-0854.2003.00133.x. [DOI] [PubMed] [Google Scholar]
- 48.Huotari J, Helenius A. Endosome maturation. EMBO J. 2011;30(17):3481–3500. doi: 10.1038/emboj.2011.286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Johnson EE, Overmeyer JH, Gunning WT, Maltese WA. Gene silencing reveals a specific function of hVps34 phosphatidylinositol 3-kinase in late versus early endosomes. J Cell Sci. 2006;119(Pt 7):1219–1232. doi: 10.1242/jcs.02833. [DOI] [PubMed] [Google Scholar]
- 50.Huynh KK, et al. LAMP proteins are required for fusion of lysosomes with phagosomes. EMBO J. 2007;26(2):313–324. doi: 10.1038/sj.emboj.7601511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Cao C, Backer JM, Laporte J, Bedrick EJ, Wandinger-Ness A. Sequential actions of myotubularin lipid phosphatases regulate endosomal PI(3)P and growth factor receptor trafficking. Mol Biol Cell. 2008;19(8):3334–3346. doi: 10.1091/mbc.E08-04-0367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Poccia D, Larijani B. Phosphatidylinositol metabolism and membrane fusion. Biochem J. 2009;418(2):233–246. doi: 10.1042/BJ20082105. [DOI] [PubMed] [Google Scholar]
- 53.Stroupe C, Collins KM, Fratti RA, Wickner W. Purification of active HOPS complex reveals its affinities for phosphoinositides and the SNARE Vam7p. EMBO J. 2006;25(8):1579–1589. doi: 10.1038/sj.emboj.7601051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Pols MS, ten Brink C, Gosavi P, Oorschot V, Klumperman J. The HOPS proteins hVps41 and hVps39 are required for homotypic and heterotypic late endosome fusion. Traffic. 2013;14(2):219–232. doi: 10.1111/tra.12027. [DOI] [PubMed] [Google Scholar]
- 55.Dai S, Zhang Y, Weimbs T, Yaffe MB, Zhou D. Bacteria-generated PtdIns(3)P recruits VAMP8 to facilitate phagocytosis. Traffic. 2007;8(10):1365–1374. doi: 10.1111/j.1600-0854.2007.00613.x. [DOI] [PubMed] [Google Scholar]
- 56.Pryor PR, et al. Combinatorial SNARE complexes with VAMP7 or VAMP8 define different late endocytic fusion events. EMBO Rep. 2004;5(6):590–595. doi: 10.1038/sj.embor.7400150. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.