Significance
An important challenge for improving cell-based approaches for Parkinson’s disease is the development of techniques that facilitate greater standardization of the donor material. This report describes the enrichment of transplantable progenitors for dopamine neurons from the ventral mesencephalon based on targeting of transmembrane proteins. It is an important step toward the development of clinically relevant techniques that allow for greater standardization of cell preparations used in transplantation and potentially, more predictable clinical outcomes. The findings are highly relevant for current efforts to develop stem cell-based therapies for Parkinson’s disease, where current techniques yield mixed cell populations that may contain unwanted cell types and thus, would benefit from a cell selection step prior to grafting.
Keywords: Parkinson’s disease, cell sorting, Alcam, microarray, transplantation
Abstract
An important challenge for the continued development of cell therapy for Parkinson’s disease (PD) is the establishment of procedures that better standardize cell preparations for use in transplantation. Although cell sorting has been an anticipated strategy, its application has been limited by lack of knowledge regarding transmembrane proteins that can be used to target and isolate progenitors for midbrain dopamine (mDA) neurons. We used a “FACS-array” approach to identify 18 genes for transmembrane proteins with high expression in mDA progenitors and describe the utility of four of these targets (Alcam, Chl1, Gfra1, and Igsf8) for isolating mDA progenitors from rat primary ventral mesencephalon through flow cytometry. Alcam and Chl1 facilitated a significant enrichment of mDA neurons following transplantation, while targeting of Gfra1 allowed for robust separation of dopamine and serotonin neurons. Importantly, we also show that mDA progenitors isolated on the basis of transmembrane proteins are capable of extensive, functional innervation of the host striatum and correction of motor impairment in a unilateral model of PD. These results are highly relevant for current efforts to establish safe and effective stem cell-based procedures for PD, where clinical translation will almost certainly require safety and standardization measures in order to deliver well-characterized cell preparations.
Although cell replacement therapy for Parkinson’s disease (PD) has been successful for some patients, the high variability in clinical outcome has limited its continued development as a conventional treatment option (1). The main drawback has been the reliance on human fetal tissue as a source of donor cells for the implantation of new midbrain dopamine (mDA) neurons. Not only does this represent an unsustainable resource, with multiple fetal donors required per patient, it is impossible to standardize with respect to the number and type of cells in the resulting graft. Key challenges for further progression of cell therapy for PD are the establishment of sustainable cell sources (e.g., stem cells) and also, the development of procedures that allow for greater standardization of the donor cell preparation.
Identification and preselection of mDA progenitors from mixed cell populations are part of a promising avenue for standardizing the donor cell preparation in order to achieve greater consistency in graft composition and clinical outcome. Recent studies have used genetic labeling approaches to demonstrate that such an approach is feasible using fetal ventral mesencephalon (VM) tissue from transgenic mice (2, 3) as well as differentiated embryonic stem cell reporter lines (4–6). Fluorescent protein expression driven by regulatory elements for genes involved in the embryonic development of mDA neurons, including neurogenin2 (Ngn2), Nr4a2 (Nurr1), and Pitx3, allows for isolation of cells within these gene linages through fluorescent activated cell sorting (FACS) and a significant enrichment of mDA neurons in the resulting grafts.
While these proof-of-principal studies support the feasibility and potential of a cell-sorting approach, the genetic labeling complicates clinical translation. An alternative strategy with a record of clinical success in the hematopoietic field is the targeting of cell-specific surface antigens as a means to separate defined cell fractions from mixed populations. Central to the development of the technique was the identification of transmembrane proteins that selectively define a target cell population. For example, isolation of CD34+ and Thy1+ cells facilitates purification of hematopoietic stem cells that can reconstitute the hematopoietic system as part of treatment for cancer patients (7–10). A current obstacle for the development of similar techniques for the purification of specific neural progenitor phenotypes, including mDA progenitors, is a lack of knowledge regarding the identity of suitable transmembrane targets.
To address this issue, we profiled gene expression in genetically labeled cell fractions enriched for transplantable mDA progenitors through microarray analysis of RNA extracted from the GFP-positive (GFPpos) cell fraction from the VM of Ngn2-GFP mice at embryonic day 12.5 (E12.5). Previous FACS transplantation studies have shown that the GFPPos fraction in the VM of Ngn2-GFP mice represents <30% of all cells but contains virtually all transplantable mDA neurons at E12.5 (2, 3). Here, we report the gene expression profile for this cell population, which includes genes for a number of transmembrane proteins that show discrete expression patterns in the developing VM. Importantly, transplantation experiments showed that FACS isolation of VM cells based on these transmembrane proteins produced functional grafts that provided extensive innervation of the host striatum and were significantly enriched for mDA neurons compared with conventional grafts of unsorted VM tissue.
Results
Genetic Profiling of Transplantable mDA Progenitors.
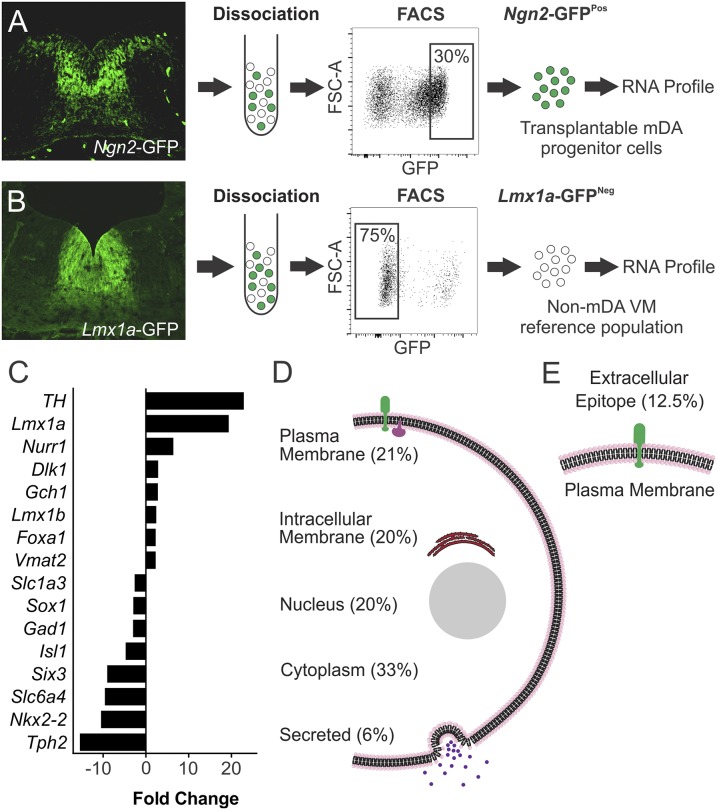
Expression of GFP in the VM of embryonic Ngn2-GFP mice identifies a distinct subpopulation of cells within the Ngn2 lineage (Fig. 1A). In dissected pieces of VM used for transplantation procedures, selective isolation of this GFPPos fraction yields ∼30% of all viable cells (Fig. 1A) but contains virtually all of the mDA progenitors, thus allowing for a significant enrichment of this population (2, 3). Here, we sought to exploit this to identify genes with some degree of selective expression in mDA progenitors through microarray analysis of mRNA prepared from FACS-isolated GFPPos cells from Ngn2-GFP E12.5 VM. Notably, the Ngn2-GFPPos population extends lateral to the mDA germinal zone and identifies progenitors for other non-mDA neuronal subtypes (Fig. 1A). To reduce the likelihood of identifying gene transcripts specific to this lateral, non-mDA Ngn2-GFPPos population, we performed a subtractive analysis using GFP-negative (GFPNeg) cells from Lmx1a-GFP VM (i.e., a definitively non-mDA VM fraction) as a reference cell population (Fig. 1B).
Fig. 1.
Gene expression profiling of transplantable mDA progenitors. (A) Immunohistochemistry for GFP in coronal sections of E12.5 mouse VM shows expression throughout the DA neurogenic region, most prominently in the intermediate zone, and also, in a lateral population of cells outside the DA germinal region, whereas (B) expression in Lmx1a-GFP mice is exclusively in the DA germinal zone and throughout all stages of differentiation. The GFPPos cell fraction was isolated by FACS from Ngn2-GFP VM, and mRNA expression in this population was compared with definitively nondopaminergic VM cells represented by the GFPNeg fraction isolated from Lmx1a-GFP VM. (C) Fold change in mRNA expression between Ngn2-GFPPos and Lmx1a-GFPNeg cells showed greater expression of genes closely associated with mDA phenotype in the Ngn2-GFPPos cell fractions, whereas genes associated with other neurotransmitter phenotypes or regional identities were underrepresented. (D) Gene ontology browsing and literature mining were used to classify genes up-regulated in the Ngn2-GFPPos fraction based on localization to different cellular compartments. (E) The plasma membrane classification is further divided into genes encoding for proteins expressed in the plasma membrane and predicted to have an extracellular epitope available for antibody binding to allow FACS isolation of mDA progenitors. FSC-A, forward scatter area.
Comparison of mRNA levels across triplicate samples for each group revealed 144 genes significantly enriched in the GFPPos fraction from Ngn2-GFP VM based on highly conservative inclusion criteria consisting of fold increase in expression of >2, P < 0.05 and seven of a possible nine scored as increased expression from direct pairwise comparisons of all samples (Table S1). Genes with the greatest fold-change up-regulation in the Ngn2-GFPPos group included many established mDA-specific genes [e.g., Th, Nurr1, Lmx1a, Lmx1b, Dlk1, Gch1, Foxa1, and Slc18a2 (Vmat2)] (Fig. 1C). Genes with significantly decreased expression in the Ngn2-GFPPos fraction (i.e., highly expressed in the Lmx1a-GFPNeg group) included those associated with other neurotransmitter phenotypes, including cholinergic (Isl1), serotonergic (Slc6a4 and Tph2), or glutamatergic (Gad1 and Slc1a3) neurons, as well as transcriptional determinants of regional identity outside the mDA germinal zone (Nkx2-2, Six3, and Sox1) (Fig. 1C and Table S1).
Classification of Ngn2-GFPPos up-regulated genes by ontology was consistent with developing neural tissue and showed that the majority could be grouped as genes associated with neuronal growth, migration, and differentiation. Classification based on cellular localization showed that 59 genes (41%) encoded for proteins that were membrane-associated, 30 (21%) were definitively plasma membrane (as opposed to internal vesicles), and 18 (12.5%) were verified or predicted to feature an extracellular sequence (Fig. 1 D and E and Table S2). These 18 extracellular targets consisted largely of receptors and cell adhesion molecules and included a number of genes encoding for proteins previously identified in association with mDA phenotype, such as Corin, Ret, Gfra1, and Dlk1, but importantly, also, a number of proteins not previously described in the context of mDA neuronal development (Table 1). We screened commercially available antibodies against six of these transmembrane targets (Table 1, asterisk) to assess expression in the developing VM relative to the location of the transplantable mDA progenitor population.
Table 1.
Ngn2-GFPPos up-regulated genes encoding for transmembrane proteins containing known or predicted extracellular sequences and ordered by fold change relative to the Lmx1a-GFPNeg reference population
| UGRepAcc | Symbol | Name | Fold change | P value |
| NM_001122756 | Corin | Corin | 9.12 | 0.00063 |
| NM_009655* | Alcam | Activated leukocyte cell adhesion molecule | 6.65 | 0.00001 |
| NM_001159317 | Il1rap | IL-1 receptor accessory protein | 5.83 | 0.00004 |
| NM_001039347* | Kcnd3 | Potassium voltage-gated channel, Shal-related family, member 3 | 5.36 | 0.00128 |
| NM_001080780 | Ret | Ret proto-oncogene | 5.16 | 0.00000 |
| NM_001079847 | Gpr64 | G protein-coupled receptor 64 | 5.12 | 0.01411 |
| NM_010279* | Gfra1 | Glial cell line-derived neurotrophic factor family receptor α1 | 2.92 | 0.00002 |
| NM_001110843 | Cacna2d1 | Calcium channel, voltage-dependent, α2/δ subunit 1 | 2.89 | 0.00069 |
| NM_007697* | Chl1 | Cell adhesion molecule with homology to L1CAM | 2.87 | 0.00025 |
| NM_001190703 | Dlk1 | δ-Like 1 homolog (Drosophila) | 2.60 | 0.00487 |
| NM_080419* | Igsf8 | Ig superfamily, member 8 | 2.52 | 0.00391 |
| NM_001077403 | Nrp2 | Neuropilin 2 | 2.39 | 0.00006 |
| NM_001122758 | Pcdh7 | Protocadherin 7 | 2.33 | 0.00345 |
| NM_001083917* | Scn3b | Sodium channel, voltage-gated, type III, β | 2.30 | 0.00501 |
| NM_053144 | Pcdhb19 | Protocadherin β-19 | 2.19 | 0.03557 |
| NM_001163348 | Ntng1 | Netrin G1 | 2.09 | 0.00020 |
| NM_001198811 | Frem1 | Fras1-related extracellular matrix protein 1 | 2.03 | 0.00394 |
| NM_001042607 | Ryk | Receptor-like tyrosine kinase | 2.03 | 0.01915 |
The inclusion criteria for up-regulated Ngn2-GFPPos genes included a present call in two of three arrays for at least one of two groups, average pairwise comparisons fold change greater than two, seven of nine direct pairwise comparisons in the same direction, and a P value < 0.05. Additional refinement included elimination of candidates known to be expressed in domains unlikely to support cell-sorting applications (e.g., Gria3 and Chrna3). Fold change, GCOS (Gene Chip Operating Software) fold change; Name, UniGene Name; Symbol, UniGene Symbol; UGRepACC, UniGene representative accession number.
Indicate transmembrane targets investigated in this study.
Expression of Transmembrane Proteins in the Developing Ventral Midbrain.
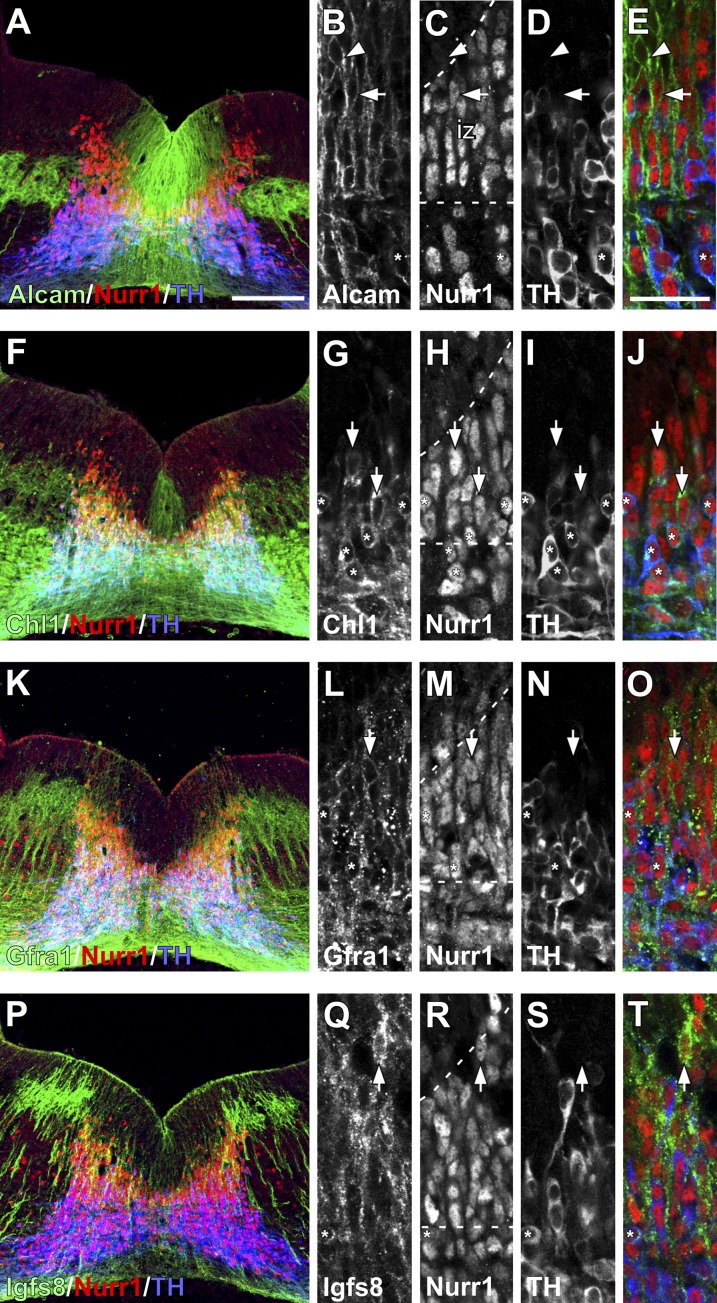
Antibodies targeting extracellular motifs were identified for 4 of 18 candidates—Alcam, Chl1, Gfra1, and Igsf8. An additional two antibodies were found to target internal sequences on ion channel subunits (Kcnd3 and Scn3b) and therefore, were informative for expression studies but not cell-sorting procedures. Immunohistochemical analysis of E12.5 mouse VM showed that all six proteins were expressed in the mDA germinal zone with cellular localizations consistent with transmembrane proteins (Fig. 2 and Figs. S1 and S2).
Fig. 2.
Expression of transmembrane proteins in the mouse VM at E12.5. Immunohistochemistry shows expression of (A–E) Alcam, (F–J) Chl1, (K–O) Gfra1, and (P–T) Igsf8 relative to Nurr1 and TH. Low magnification images illustrate the pattern of expression throughout the VM, whereas the adjacent higher magnification images show the overlap between each transmembrane protein and VM progenitors at different stages of maturation. Nurr1 and TH expression approximately demarcate boundaries (dashed lines) for the proliferative ventricular zone (Nurr1−/TH−; arrowheads), newly postmitotic neuroblasts in the intermediate zone (Nurr1+/TH−; arrows), and immature DA neurons in the mantle zone (Nurr1+/TH+; asterisks). Alcam was widely expressed throughout the VM, including (A) in a discrete population of cells lateral to the DA neurogenic zone and (B–E) at all stages of maturation. Chl1 was expressed (F) weakly in a subset of ventricular zone cells around the midline and (F–J) more prominently in intermediate Nurr1+/TH− neuroblasts and Nurr1+/TH+ neurons. (K–O) Gfra1 had a punctate expression pattern, particularly on dendrites, and was not expressed in the early ventricular zone cells but was prominent within more mature cells in the intermediate and mantle zones. (P–T) Igsf8 also has a punctate expression pattern and distribution on more mature Nurr1+ cells, with little expression in the ventricular zone. iz, Intermediate zone. (Scale bar: A, F, K, and P, 100 µm; B–E, G–J, L–O, and Q–T, 20 µm.)
ALCAM was expressed widely throughout the Nurr1+ mDA domain, including ventricular zone progenitors, intermediate zone Nurr1+ neuroblasts, and immature tyrosine hydroxylase+ (TH+) mDA neurons in the mantle zone (Fig. 2 A–E and Fig. S1). Expression was absent from the lateral, non-mDA Ngn2-GFPPos population; however, ALCAM+ cells could be seen immediately ventral to this lateral Ngn2-GFPPos cell group.
Chl1 was also expressed throughout different stages of mDA maturation, but it was more restricted to the midline within the mDA domain (Fig. 2 F–J). Expression was weaker in the ventricular zone and more prominent in the intermediate zone neuroblasts and early TH+ neurons. Intensely stained Chl1 fibers were found throughout the mantle zone both within and lateral to the mDA domain.
Gfra1 expression was relatively widespread throughout the ventral midbrain, including the medial Nurr1+ mDA domain (Fig. 2 K–O) and also, extending lateral to this area. Cellular distribution showed radial processes that were densely labeled with a punctate pattern of Gfra1 protein, whereas there was relatively little Gfra1 immunoreactivity on the cell soma (Fig. 2 L–O). This expression pattern made it difficult to unambiguously characterize expression in distinct states of mDA progenitor maturity, although the pattern is indicative of a low level of expression within ventricular zone progenitors and relatively higher expression in Nurr1+ progenitors in the intermediate zone, with maintained expression in immature TH+ cells (Fig. 2 L–O).
Igsf8 expression was seen primarily in the intermediate zone throughout the VM including the medial Nurr1+ mDA progenitors (Fig. 2 P–T) but also, the lateral area outside of the Nurr1+ domain. Igsf8 was absent from the earliest ventricular zone progenitors but clearly expressed by Nurr1+ neuroblasts in the intermediate zone, and it persisted in the TH+ mantle zone neurons (Fig. 2 P–T).
The ion channel subunits Kcnd3 and Scn3b were both expressed at the protein level in the developing VM, with prominent expression in intermediate zone Ngn2-GFPPos progenitors persisting in TH+ neurons (Fig. S2 A–F). Both subunits were also expressed in cells lateral to the mDA germinal zone. Interestingly, analysis of expression in the adult midbrain showed that both proteins were more strongly expressed in A9 mDA neurons, with less frequent overlap and lower labeling levels within the A10 population (Fig. S2 G–J).
Immunohistochemical labeling of Alcam, Chl1, Igsf8, and Gfra1 in sagittal sections showed that expression of all four proteins extended on the anterior–posterior axis beyond the midbrain (Fig. S3). Labeling for serotonin (5HT) showed transmembrane protein expression throughout the medial hindbrain, including the developing raphe nucleus, but not overlapping with early 5HT+ neurons. The pattern of expression for all six proteins was found to be virtually identical in rat VM of corresponding developmental age (E14.5) using the same antibodies.
Isolation of Ventral Midbrain Progenitors Based on Transmembrane Proteins.
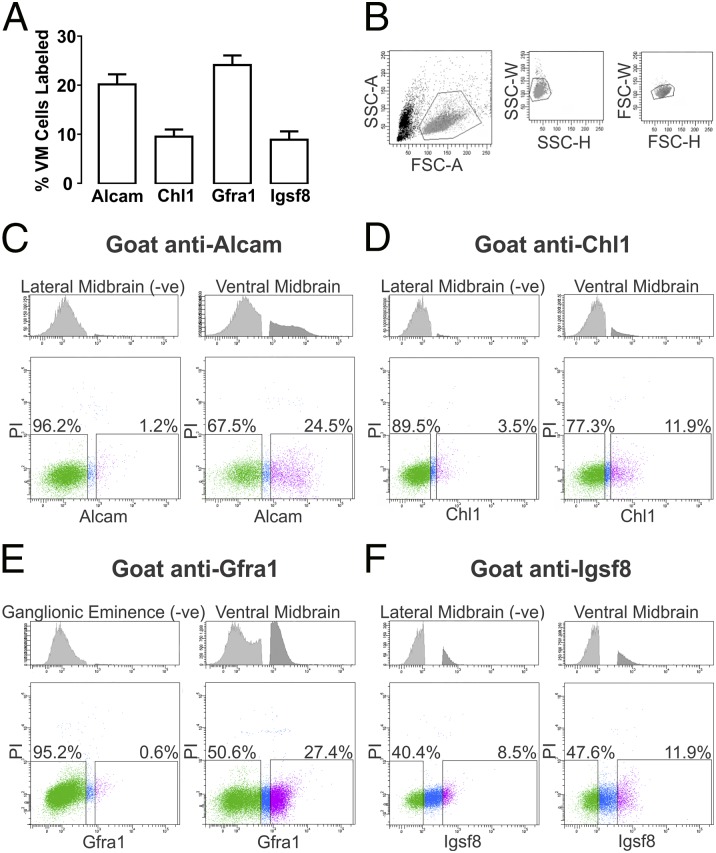
Commercial antibodies for Alcam, Chl1, Gfra1, and Igsf8 identify extracellular motifs on each of these proteins and thus, allowed for FACS-based isolation of the corresponding cell populations through attachment of an antibody–fluorophore complex. Using single-cell preparations of dissected pieces of E14.5 rat VM, cells were sorted into positive and negative fractions based on the expression of each transmembrane protein. Complete biological replicates using separate VM preparations were used to assess the proportion of cells expressing each transmembrane protein and showed 20.2% ± 2.0% Alcam (n = 4), 9.5% ± 1.5% Chl1 (n = 3), 24.1% ± 1.9% Gfra1 (n = 3), and 8.9% ± 1.7% Igsf8 (n = 3) as the average fraction of the viable (propidium iodide excluding) cell pool (Fig. 3A).
Fig. 3.
FACS analysis of single-cell preparations of E14.5 rat VM. (A) The average percentage of viable cells expressing Alcam, Chl1, Gfra1, or Igsf8 in E14.5 rat VM preparations was estimated by FACS analysis of at least three biological replicates. (B) Initial FACS gatings for each preparation were set to eliminate cell debris and doublets based on forward and side scatter profiles (representative example shown). Antibodies targeting extracellular regions of (C) Alcam, (D) Chl1, (E) Gfra1, or (F) Igsf8 were used to assess the fraction of cells in VM preparations expressing those proteins. The threshold for positive detection of antibody-labeled protein was determined for each antibody using control E14.5 rat cell preparations verified by immunohistochemistry to have no or little expression of each protein. Control preparations included lateral midbrain for Alcam, Chl1, and Igsf8 and ganglionic eminence for Gfra1—Upper in C–F shows representative frequency of cells unlabeled (light gray) and labeled (dark gray) in (Left) control and (Right) VM tissue. Representative FACS plots show the distribution of viable, antibody-labeled cells for each protein based on fluorescence intensity (x axis) against the fluorescent signal for propidium iodide (y axis; Lower in C–F). x And y scales are in arbitrary log units for FACS plots. The y scale for histograms represents frequency of events. FSC-A/H/W, forward scatter area/height/width; PI, propidium iodide; s-A/H/W, side scatter area/height/width.
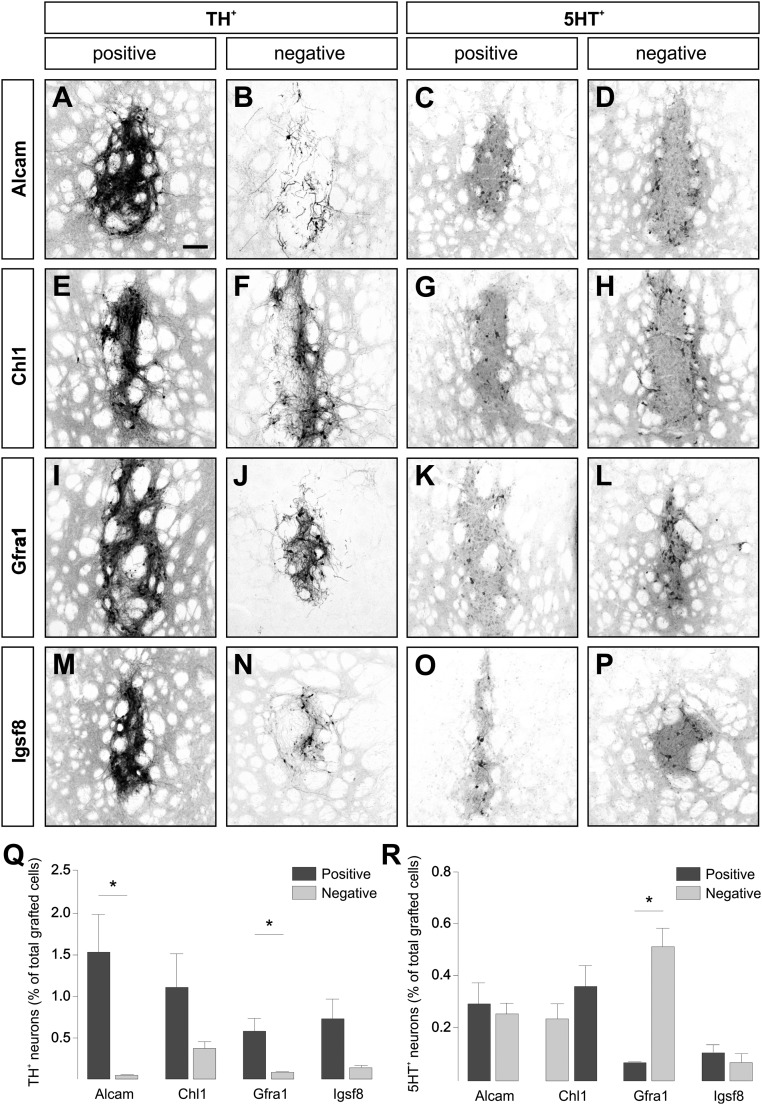
To determine the distribution of transplantable mDA progenitors relative to cells expressing specific transmembrane proteins, separate preparations from the positive and negative fractions for each protein were transplanted into the striatum of 6-hydroxydopamine (6-OHDA) –lesioned rats (Fig. 3 B–F). Histological analysis 4 wk later showed varying degrees of enrichment for dopamine (DA) neurons in the resulting grafts derived from the positive fractions of FACS-isolated cells (Fig. 4). Sorting against Alcam provided the greatest separation of grafted DA neurons between positive and negative fractions (Fig. 4 A and B), with ∼30-fold more mDA neurons in grafts from the positive fraction, and also, the greatest yield as a percentage of total cells grafted (1.5% ± 0.49%) (Fig. 4Q). Gfra1 sorting also provided a significant separation of DA neurons (Fig. 4 I and J), with sevenfold greater numbers in the positive grafts, although a lower total yield of 0.58% ± 0.08% of grafted cells (Fig. 4Q). Both Chl1 and Igsf8 sorting resulted in less distinct separation of DA neurons (Fig. 4 E, F, M, and N): three- and fivefold greater numbers in the positive fractions, respectively, and yields of 1.1% ± 0.44% and 0.73% ± 0.27%, respectively, of grafted cells (Fig. 4Q). We considered that DA neurons distributed across both positive and negative fractions may represent different mDA neuronal subtypes; however, quantification of mDA neurons expressing subtype-specific proteins Girk2 and Calbindin showed a mixed distribution in grafts from all groups (Fig. S4 B–F).
Fig. 4.
Distribution of DA and serotonin neurons after FACS and transplantation of E14.5 rat VM using antibodies against transmembrane proteins. Representative images of immunohistochemistry for (A, B, E, F, I, J, M, and N) TH and (C, D, G, H, K, L, O, and P) 5HT illustrate the presence of DA and serotonin neurons, respectively, 4 wk after grafting of positive or negative cell fractions isolated by FACS based on expression of (A–D) Alcam, (E–H) Chl1, (I–L) Gfra1, and (M–P) Igsf8. The mean (± SEM) number of grafted (Q) TH+ and (R) 5HT+ neurons in each transplantation group is shown as a percentage of the total number of cells grafted (positive fraction is in dark gray and negative fraction is in light gray). Paired t tests show significant differences in the average number of (Q) TH+ (Alcam, *P = 0.02; Gfra1, *P = 0.04) and (R) 5HT+ (Gfra1, *P < 0.0001) neurons in grafts generated from positive and negative fractions. Group numbers for Q and R: AlcamPos and AlcamNeg (n = 4); Chl1Pos (n = 3), Chl1Neg (n = 4); Gfra1Pos (n = 6), Gfra1Neg (n = 5); and Igsf8Pos and Igsf8Neg (n = 3). (Scale bar: A–P, 150 µm.)
Immunohistochemistry for 5HT showed that serotonin neurons were not distinctly segregated between grafts from positive or negative fractions after sorting based on Alcam, Chl1, or Igsf8 (Fig. 4 C, D, G, H, K, L, O, and P). Grafts generated from Gfra1-sorted fractions, however, showed a distinct separation of serotonin neurons, with a ninefold enrichment in the negative cell fraction (Fig. 4R). Fig. S4G shows the total number of cells grafted and the average TH+ and 5HT+ cell counts for all groups.
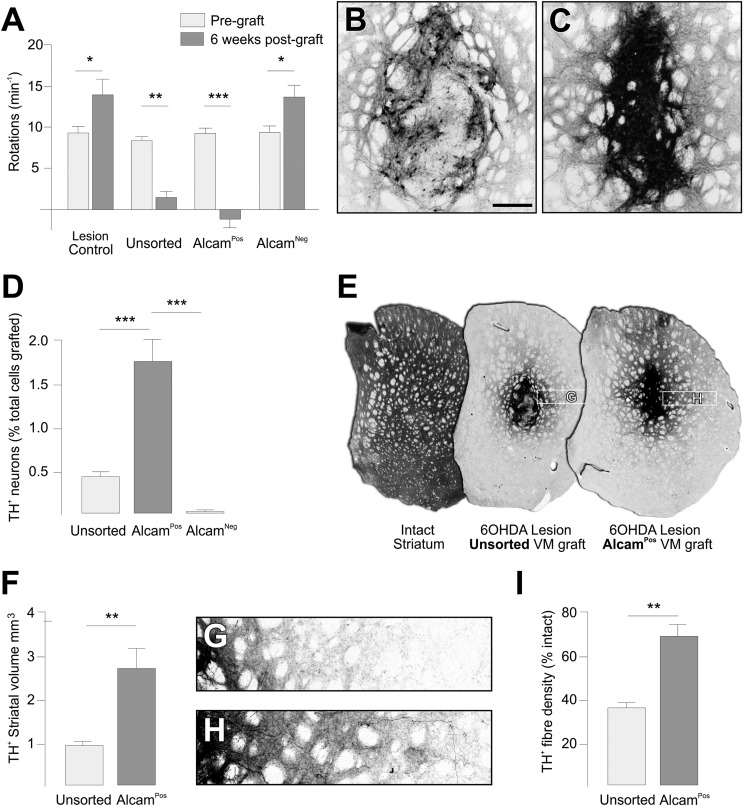
The high yield of DA neurons in the AlcamPos group motivated a second round of transplantation experiments to assess the functional and anatomical properties of grafts enriched for DA neurons by AlcamPos selection relative to conventional, unsorted grafts of fetal VM. At 6 wk, both unsorted VM grafts and grafts of AlcamPos VM cells completely ameliorated amphetamine-induced rotational asymmetry in rats with unilateral 6-OHDA lesions, whereas animals grafted with AlcamNeg cells or ungrafted controls showed no improvement (Fig. 5A). At the histological level, a striking feature of the AlcamPos grafts was the density of TH+ cells within the graft (9.17 × 10−2 cells/µm3), which was significantly greater than for unsorted grafts (3.68 × 10−2 cells/µm3; P < 0.05) (Fig. 5 B and C). In fact, preselection of AlcamPos cells from VM preparations resulted in a significant fourfold enrichment of grafted mDA neurons, with a yield of 1.7% ± 0.25% of all cells transplanted compared with 0.42% ± 0.11% from unsorted cells from the same FACS-processed VM preparation (Fig. 5D). Although the average size of the grafts themselves was not significantly different, the volume of host striatal territory innervated by TH+ fibers was significantly greater in grafts arising from AlcamPos cells (2.6 ± 0.48 mm3) compared with unsorted cells (0.9 ± 0.1 mm3) (Fig. 5 E and F). The density of innervation was also significantly greater in AlcamPos grafts compared with unsorted grafts (Fig. 5 G–I).
Fig. 5.
Integration and function of intrastriatal grafts generated from FACS selection of AlcamPos cells from E14.5 rat VM. (A) Animals with unilateral 6-OHDA lesion of the mDA system showed a robust rotational response to d-amphetamine (3.5 mg/kg i.p.) 4 wk after lesioning (light gray bars). Rotational asymmetry was significantly corrected in animals grafted with AlcamPos (n = 5) or unsorted VM cells (n = 5) but not in ungrafted animals (n = 5) or animals receiving AlcamNeg cells (n = 5). In both ungrafted and AlcamNeg animals, the rotational response was, in fact, increased 6 wk after transplantation (dark gray bars). *P < 0.05, **P < 0.01, and ***P < 0.005 for paired t tests for pregraft and 6 wk postgraft time points. Immunohistochemistry for TH 6 wk after grafting shows (B) the relatively sparse distribution of DA neurons within unsorted grafts compared with (C) the densely packed arrangement in grafts of AlcamPos cells. (D) The survival of DA neurons in the AlcamPos group was significantly greater than in the unsorted and AlcamNeg groups (mean TH+ cells ± SEM as a percentage of total cells grafted). ***P < 0.005 for one-way ANOVA with Tukey’s posthoc test. (E) TH immunohistochemistry shows the normal level of TH+ fiber innervation in the intact striatum and territory of reinnervated striatum in the lesioned host after grafting of unsorted or AlcamPos E14.5 rat VM cells. (F) The total volume of TH+ fiber staining (mean ± SEM) was estimated from quantification of TH+ area in coronal sections and showed a significantly greater average volume in the AlcamPos group (n = 5) compared with the unsorted group (n = 5) based on Student’s t test. **P < 0.01. This result is illustrated in G and H (locations indicated in E), showing a greater density and distance of TH+ fiber outgrowth emanating from (G) AlcamPos compared with (H) unsorted grafts. (I) The average fiber density (± SEM) measured directly lateral to the graft by OD was significantly greater in the AlcamPos (n = 5) compared with the unsorted group (n = 5) based on Student’s t test. **P < 0.01. (Scale bar: B and C, 300 µm.)
Discussion
These results show that transplantable progenitors for mDA neurons can be identified and isolated from heterogeneous cell populations by targeting of transmembrane proteins. This finding supports and extends previous work in this area, in which fluorescent transgenes have been used to isolate mDA progenitors that can form functional mDA neurons after transplantation (2, 3, 5, 6). Although the concept of targeting surface antigens is not in itself new, its practical development has been limited by a lack of knowledge of transmembrane proteins with some degree of selectivity for mDA neurons. So-called “FACS-array” studies provide a powerful means to examine gene expression in predefined cell populations isolated by flow cytometry. Some early examples of the approach in the neural field have made substantial contributions to our understanding of gene expression patterns in cell types that are otherwise difficult to isolate or distinguish between in heterogeneous systems, including neural crest stem cells and developing Schwann cells (11) as well as striatopalladial and striatonigral projection neurons (12). More recently (13), gene expression was successfully profiled from fluorescently labeled photoreceptor progenitors specifically to identify surface antigens that could be used to isolate and transplant those cells as a treatment for retinal pathologies.
Here, we report a list of promising transmembrane targets generated from transcriptome analysis of VM cells isolated from the Ngn2-GFP reporter mouse. Previous transplantation studies have shown that the GFPPos fraction in these mice represents ∼30% of cells in typical VM dissections and yet contains virtually all transplantable mDA progenitors (2, 3), thus allowing for a significant enrichment of the progenitor pool. Transcriptional profiling of this cell fraction identified a range of genes encoding for transcription factors and other transcriptional regulators, components of signaling pathways, and secreted molecules. Several of these genes have been identified and verified in previous gene expression studies (Dmrta1 and Dlk1) (14, 15) or have well-established roles in mDA neurogenesis and differentiation (TH, Lmx1a, Nurr1, En1, and Foxa2) (reviewed in refs. 16 and 17). Many of the genes, however, have not been previously identified in the context of mDA development and may play roles in key processes underlying mDA development and differentiation. Importantly for the purposes of this study, 12.5% of the genes encoded for transmembrane proteins that are either known or predicted to contain extracellular regions.
We investigated the VM expression of six of these proteins using commercially available antibodies. Two antibodies detected intracellular sequences on protein subunits for ion channels, including the potassium channel Kcnd3 and the calcium channel Scn3b, and thus, were informative for histological analysis but not cell-sorting procedures. Kcnd3 has previously been shown to regulate the intrinsic pacemaker activity of A9 mDA neurons (18), whereas to our knowledge, Scn3b has not previously been described in the context of mDA biology. Immunohistochemical analysis showed that, like Kcnd3, Scn3b is most prominently expressed in the A9 subset of mDA neurons in the adult midbrain. Its functional role is an area that may be worth additional exploration given the selective vulnerability of A9 neurons in PD (19, 20).
The main focus of the study was on the four transmembrane proteins that could be targeted with antibodies recognizing extracellular epitopes. A first round of FACS transplantation experiments using E14.5 rat VM showed a variable degree of enrichment of mDA neurons in grafts generated from positive fractions based on selection of Alcam-, Chl1-, Gfra1-, and Igsf8-expressing cells compared to the corresponding negative fractions or unsorted control grafts. The number of TH+ mDA neurons in unsorted grafts from E14.5 rat VM was around 0.5% as a fraction of the total number of cells grafted, which is similar to the ∼0.4% yield that we have reported previously from FACS experiments using mouse VM at a similar developmental stage (2). These figures are substantially lower than yields for primary rodent VM grafts in our hands (3–4%) (21) and highlight that the tissue handling associated with the FACS procedures has a detrimental impact on mDA neuron survival. Preselecting for AlcamPos or Chl1Pos cells significantly enriched the yield of grafted mDA neurons by 30- (1.5% of total grafted) and 3-fold (1.1% of total grafted), respectively, compared with unsorted grafts. This result compares favorably with previous studies showing enrichment based on FACS selection of GFPPos cells from E12.5 VM from the Ngn2-GFP mouse (approximately twofold) (2, 3). Selection based on Gfra1 or Igsf8 showed that, although the transplantable progenitor pool was largely derived from the positive fractions, the overall yield was similar to unsorted control grafts, suggesting more limited selectivity and/or survival when using these antibodies. Nonetheless, a notable feature of Gfra1 selection was the significant segregation of serotonin neurons to the negative fraction. It is an interesting finding given recent reports that inclusion of high numbers of serotonin neurons in primary VM grafts may contribute to graft-induced dyskinesia (22; reviewed in ref. 23) and highlights the potential of cell-sorting strategies for eliminating unwanted cell types before grafting.
A number of variables will likely contribute to the overall capacity of different transmembrane targets to select for mDA progenitors from VM preparations, including expression pattern within the progenitor domain as well as the binding properties of the antibody and any associated functional consequences for the cells. During mDA neurogenesis, the mDA germinal zone will contain a mixture of mDA progenitors in different states of maturity. We have previously reported that the transplantable mDA progenitor pool largely resides within the early, dividing mDA progenitors in the ventricular zone as well as newly postmitotic neuroblasts in the intermediate zone (2, 24; reviewed in ref. 25). The more mature TH+ mDA neurons in the mantle zone have a poor capacity to survive the FACS transplantation procedure. These previous observations may partly explain the improved yield of grafted mDA neurons selected on the basis of the cell adhesion molecules Alcam and Chl1, which were both expressed in the ventricular and intermediate zone progenitors, relative to Gfra1, which was expressed in some Nurr1+ intermediate zone progenitors but more prominently in the young TH+ neurons. It should also be considered that functional antagonism of signaling mediated by the targeted protein after antibody binding may also impact on cell survival. This possibility may be particularly relevant for trophic factor receptors, such as Gfra1. Furthermore, the binding properties of the antibody will also determine efficiency in cell-sorting procedures. The short incubation times required when working with live cells means that antibodies with low affinity or high nonspecific binding properties will have limited efficacy for cell-sorting applications. This explanation may underlie the relatively poor yield resulting from selection using the Igsf8 antibody. The high nonspecific binding profile necessitated a conservative separation of FACS gatings in order to reasonably define the positive and negative fractions, and thus, the positive fraction likely underestimates the actual Igsf8-expresssing population. Nonetheless, the expression pattern of Igsf8 in the developing VM, including prominent expression on Nurr1+ intermediate zone progenitors, suggests that it is a promising target for isolation of transplantable mDA progenitors. Development of Igsf8 antibodies with greater specificity may improve efficacy in FACS transplantation procedures and thus, the value of Igsf8 as an mDA selection target.
An important consideration for FACS-based approaches to enrich mDA progenitors is that they do not impinge on the integration and function of the resulting grafts. We performed a second round of VM transplantation experiments to assess the integration and function of grafts generated from positive and negative cell fractions defined on the basis of Alcam expression compared with unsorted VM control grafts. The results showed that grafts generated from AlcamPos cells were just as effective as the unsorted VM grafts at reversing rotational bias in rats with unilateral lesions of the mDA system. This finding is in line with observations from our earlier studies showing that significant change to the composition of VM grafts [for example, the complete exclusion of glial cells when sorting on the basis of Ngn2-GFP (2)] does not impact the survival or function of grafted DA neurons. Sorting on the basis of Alcam significantly increased not only the fraction of mDA neurons per unit of grafted tissue but also, the volume of host striatal territory innervated by the grafted mDA neurons. Both animal studies (26–30) and clinical observations (31–33) strongly suggest that the degree of recovery of motor function after grafting is closely related to the level of dopaminergic reinnervation of the striatum.
These results compare favorably with previous attempts to sort and graft VM progenitors by targeting of the transmembrane protein Corin (2), a marker for floor plate cells in the developing VM (34). These earlier experiments provided the important proof of principle that mDA progenitors could be identified and selected through targeting of transmembrane proteins, however, the overall efficiency of selection was low and insufficient for generating functional grafts, even when pooling a very large (>100) number of VM pieces. This previous observation likely reflects the limited expression of Corin in the developing VM, where it is only transiently expressed in the early ventricular zone progenitors and restricted to a narrow pattern of expression in the most medial part of the mDA germinal zone (Fig. S1). Nonetheless, recent studies using mouse (35) or human (36) pluripotent stem cells show that, when the initial cell pool is sufficiently large (i.e., as afforded by highly expandable stem cell populations), Corin can be used to isolate enough DA progenitors to generate functional grafts in rodent models of PD. This finding is an important advance for clinical application of pluripotent stem cells in neural grafting procedures, where some form of cell selection will almost certainly be required to eliminate potentially dangerous cell types, for example those capable of uncontrolled growth after transplantation. It extends on previous work in this area using fluorescent transgenes (4–6) or targeting of neurodevelopmental stage-specific transmembrane proteins (37, 38) to eliminate tumorigenic cells before transplantation.
These results suggest that Alcam, Chl1, Gfra1, and Igsf8 are promising new targets for selection of mDA progenitors from preparations of partially differentiated stem cells. All four proteins show robust expression on the transplantable mDA pool in the developing VM and substantially improved efficiency for selection of mDA progenitors relative to our previous work using Corin. In fact, the results from Alcam and Chl1 sorting are the first, to our knowledge, to demonstrate that utilization of specific transmembrane proteins can significantly enrich the fraction of mDA neurons in VM grafts relative to conventional unsorted grafts. Notably, sorting for Gfra1 also provided a unique means to separate progenitors for mDA neurons from those for serotonin neurons and highlights the value of the approach as a means to eliminate unwanted cell types and standardize cell preparations. Further development of an optimal cell selection strategy for isolating mDA progenitors may well come from development of specific antibodies targeting other transmembrane targets identified in the transcriptome analysis. New antibodies may also facilitate combinatorial cell-sorting protocols, where multiple transmembrane proteins are targeted to further refine the selection process (for example, where the transplantable mDA progenitor pool may be more precisely defined by multiple targets).
In summary, we report here a series of transmembrane proteins expressed by progenitors for mDA neurons in the developing VM and demonstrate the utility of targeting these proteins for selecting mDA progenitors from mixed cell populations before grafting. It forms an important step in the context of current efforts to develop strategies to better define and standardize cell preparations for use in transplantation procedures for PD. Although the limited availability and survival of human VM tissue in current grafting procedures limit the feasibility of flow cytometry, continued development of methods to expand primary VM preparations and improve the yield of grafted DA neurons (39, 40) may allow for the introduction of cell sorting in the longer term. The more likely benefit will be as a means to introduce safety and standardization measures to facilitate clinical translation of stem cell-based cell replacement approaches for PD.
Methods
Ethical Approval and Animal Housing.
The use of animals in this study conformed to the Australian National Health and Medical Research Council’s published Code of Practice for the Use of Animals in Research and the guidelines of the Ethical Committee for the Use of Laboratory Animals at Lund University. Transplantation experiments were approved by the Florey Neuroscience Institute Animal Ethics Committee. All animals were housed under a 12-h light/dark cycle with ad libitum access to food and water.
FACS of Embryonic Midbrain Progenitors.
GFP-labeled cell preparations for RNA collection and microarray.
Isolation of the GFPPos or GFPNeg cell fractions from the VM of transgenic reporter mice was performed as previously described (2, 3). Briefly, time-mated litters were generated by breeding female WT NMRI mice with male Ngn2-GFP (gift from Francois Guillemot, London, United Kingdom) or Lmx1a-GFP (gift from Johan Ericsson and Thomas Perlmann, Stockholm, Sweden) (41) heterozygous knockin mice. The VM was dissected from GFP+ embryos in cold L15 medium (Gibco) at E12.5 as previously described (42). Individual pieces were pooled from six to eight litters (∼35–50 GFP+ VM pieces) from each of the Ngn2 and Lmx1a reporter lines and prepared as separate single-cell suspensions (3 × 106 cells/mL) through incubation in HBSSCa2+Mg2+ with 0.1% trypsin (Invitrogen) and 0.05% DNase (Invitrogen) for 20 min at 37 °C followed by washing and mechanical dissociation in HBSSCa2+Mg2+ with 0.05% DNase. The cell preparation was filtered using a 70-µm cell strainer and resuspended at 3 × 106 cells/mL in HBSSCa2+Mg2+ containing 1% BSA, 0.05% DNase, and 1 mM EDTA. The GFPPos and GFPNeg cell fractions were separated using a FACS Diva Flow Cytometer (Becton Dickinson) after initial filtering for cell debris and doublets as well as the dead cell (7-aminoactionomycin-D–labeled) fraction. The detection threshold for GFP was established using cell preparations of WT VM tissue. Purity in the GFPPos and GFPNeg populations was found to be >98% based on reanalysis of the separated fractions. The cells were collected in HBSSCa2+Mg2+ containing 1% FBS, pelleted by centrifugation, immediately resuspended in RLT Lysis Buffer (Qiagen), and stored at −80 °C. These procedures were repeated in triplicate for each of the Ngn2-GFP and Lmx1a-GFP preparations to obtain three biological replicate RNA samples for microarray analysis.
Antibody-labeled cell preparations for transplantation.
These procedures were based on those described above and in detail elsewhere (2). Dissected VM pieces were pooled from E14.5 Sprague–Dawley rats and prepared as single-cell suspensions by incubation in HBSSCa2+Mg2+ with 0.05% DNase with either collagenase-dispase (1 mg/mL; Sigma) for Alcam and Chl1 or Accutase (StemPro) for Gfra1 and Igsf8 (30 min at 37 °C) followed by mechanical dissociation (note that we found optimal dissociation using different proteases for the different transmembrane targets based on initial FACS screening). The dissociated cells were collected by centrifugation (500 × g for 5 min) and incubated with primary antibody [goat anti-Alcam (1:400; R&D Systems), goat anti-Chl1 (1:400; R&D Systems), goat anti-Gfra1 (1:100; R&D Systems), or goat anti-Igsf8 (1:200; R&D Systems)] in HBSSCa2+Mg2+ containing 10% FBS and 1 mM EDTA for 20 min at 4 °C. After washing (resuspension in HBSSCa2+Mg2+ with 0.05% DNase and 1 mM EDTA after centrifugation), cells were blocked for 5 min in 5% (vol/vol) donkey serum before incubation for 15 min at 4 °C with secondary antibody (donkey anti-goat Dylight 649; 1:400; Jackson Labs) in 5% donkey serum and 1 mM EDTA in HBSSCa2+Mg2+ followed by final washing and preparation for FACS through addition of propidium iodide and filtering through a 70-µm cell strainer. The detection threshold for antibody fluorophore-labeled cells was defined using the same procedure on tissue preparations where the target proteins are not widely expressed (lateral midbrain for Alcam, Chl1, and Igsf8 and ganglionic eminence for Gfra1). Unsorted cells were subject to the same procedures, passed through the FACS, and collected without sorting. Cells were prepared for transplantation by centrifugation (500 × g for 5 min) and resuspension in HBSSCa2+Mg2+ with 1% FBS and 1 mM EDTA at ∼0.5–1 × 105 cells/µL based on numbers indicated by the flow cytometer. The final density of viable cells for each preparation used for transplantation was calculated manually from FACS-separated cell fractions.
Microarray and Bioinformatic Analyses.
Whole RNA was extracted from all samples using the RNeasy Micro Kit (Qiagen), and the yield and integrity of each sample assessed using a Bioanalyser (Agilent) before microarray processing. To determine mRNA expression levels in each sample, RNA was labeled using the High-Yield RNA Transcript Labeling Kit (Qiagen) and hybridized to Mouse Genome 430 2.0 Arrays (Affymetrix). The data were normalized using the MAS5 algorithm (Affymetrix Microarray Suite, version 5.0), and statistical analysis was carried out with the limma package (43) using the R (version 2.14.2) statistical computing environment.
To identify gene expression patterns enriched in the DA neural progenitor domain, expression levels in the GFPPos fractions (n = 3) from Ngn2-GFP mice were compared with the GFPNeg fractions (n = 3) from Lmx1a-GFP mice (i.e., a domain that is definitively outside the mDA germinal zone) (Fig. 1B). The criteria for differentially expressed genes between Ngn2-GFPPos and Lmx1a-GFPNeg groups were a present call in two of three arrays for at least one of two groups, average pairwise comparisons fold change greater than two, seven of nine direct pairwise comparisons in the same direction, and a P value < 0.05. The resulting list of genes shown to be up-regulated in the Ngn2-GFPPos fractions were classified by cellular component using the Swiss-Prot database to identify and distinguish between those encoding for proteins containing transmembrane domains. This list was further refined by ontology browsing (44) and literature mining using conservative criteria to classify proteins with a strong likelihood of providing targets for FACS isolation of mDA progenitors. The classification criteria included genes encoding for transmembrane proteins expressed on the plasma membrane, containing an extracellular sequence available for antibody binding, and localized on the cell body [for example, elimination of candidates known to be expressed in domains unlikely to support cell-sorting applications (e.g., Gria3 and Chrna3, which are expressed at the synaptic cleft)]. All microarray data are available through the Gene Expression Omnibus database (accession no. GSE65094).
Surgical Procedures and Rotational Behavior.
For transplantation experiments, rats were used in favor of mice because of the greater reliability in tests of motor function. Anesthesia for all surgical procedures was established and maintained by inhaled isoflurane (2% in air). Adult (250 g) female Sprague–Dawley rats received complete, unilateral lesions of the mDA projection system through stereotaxic delivery of 6-OHDA into the medial forebrain bundle as previously described (45). Briefly, 2 µL 6-OHDA (3.5 µg/µL free base dissolved in a solution of 0.2 mg/mL l-ascorbic acid in 0.9% wt/vol NaCl) was injected into the medial forebrain bundle (4.4 mm anterior and 1.2 mm lateral to Bregma and 7.8 mm below the dural surface) using a 10-mL Hamilton syringe fitted with a glass capillary as previously described in detail (46).
A first cohort of rats (n = 32) received intrastriatal (0.5 mm anterior and 3.0 mm lateral to Bregma and 4.0 mm below the dural surface) grafts of cells prepared from positive or negative E14.5 VM cell fractions following Alcam/Chl1/Gfra1/Igsf8 FACS procedures in a total volume of 2 µL as previously described (47) and were perfused for histological analysis 4 wk later. The total number of cells implanted for each group is shown in Fig. S4A. A second group (n = 30) was tested for rotational behavior in response to administration of d-amphetamine sulfate (3.5 mg/kg i.p.) 3 wk after 6-OHDA based on procedures originally described in ref. 48. One week later, preparations of E14.5 VM AlcamPos, AlcamNeg, or unsorted FACS-processed control cells were grafted into the striatum, and all animals were again tested for rotational behavior at 6 wk and subsequently, perfused for histology.
Tissue Processing and Immunohistochemistry.
Procedures were as previously described in detail (45). Mouse embryos were immersion-fixed overnight in 4% (wt/vol) paraformaldehyde in 0.1 m PBS and cryoprotected in 20% (wt/vol) sucrose in 0.1 m phosphate buffer before freezing and sectioning at 20 µm in six series onto glass slides. Adult rats received a terminal dose (100 mg/kg i.p.) of sodium pentobarbitone (Virbac; Peakhurst), were transcardially perfused with saline (50 mL) followed by paraformaldehyde (250 mL) and postfixed for another 2 h followed by cryoprotection in 30% sucrose before sectioning at 30 µm in 12 series on a freezing microtome.
Immunohistochemistry was performed on slide-mounted sections from embryonic mice or free-floating sections from adult rats. The tissue was incubated overnight with primary antibodies diluted in 0.1 m PBS containing 5% normal serum and 0.25% Triton X-100 (Amresco) as follows: goat anti-Alcam (1:800; R&D Systems), goat anti-Chl1 (1:800; R&D Systems), rabbit anti-Corin (1:200; custom generated; Abbomax), goat anti-Gfra1 (1:400; R&D Systems), goat anti-Igsf8 (1:400; R&D Systems), goat anti-Scn3b (1:200; Santa Cruz), Kcnd3 (1:500; Alomone Labs), rabbit anti-GFP (1:20,000; Abcam), chicken anti-GFP (1:1,000; Abcam), rabbit anti-Nurr1 (1:200; Abcam), rabbit anti-TH (1:800; PelFreez), sheep anti-TH (1:600; PelFreez), chicken anti-TH (1:400; Abcam), mouse anti-Calbindin (1:1,000; Sigma), rabbit anti-Girk2 (1:400; Alomone Labs), and mouse anti-5HT (1:10,000; Immunostar). Detection of the primary–secondary antibody complexes was through peroxidase-driven precipitation of diaminobenzidine (DAB) or conjugation of a fluorophore. Secondary antibodies generated in donkey were applied for 2 h at room temperature at a dilution of 1:400 for fluorescent detection using 488-, 549-, or 649-conjugated anti-mouse, anti-chicken, anti-rabbit, anti-sheep, or anti-goat (Jackson ImmunoResearch). Chromogenic detection of antibody–DAB complex was carried out using biotin-conjugated donkey anti-rabbit (1:500; 2 h; Jackson ImmunoResearch) followed by peroxidase conjugated streptavidin (1 h; Vectastain ABC Kit; Vector Laboratories) and incubation with DAB (0.5 mg/mL for 5 min), which was precipitated by the addition of 1% wt/vol H2O2. Fluorescently labeled sections were cover-slipped with fluorescent mounting media (Dako), and chromogenic-labeled sections were dehydrated in alcohol and xylene and cover-slipped with DePeX Mounting Media (BDH Chemicals).
Imaging.
Fluorescent images were captured using a Zeiss Pascal Confocal Microscope System, and chromogenic images were captured on a Leica DM6000 Microscope equipped with a motor-driven stage.
Quantification and Statistics.
The total number of grafted TH+ neurons or 5HT+ neurons in each animal was estimated by extrapolation of the number of cells counted in every sixth serial coronal section, and double-counting error was corrected according to the method used in ref. 49. The total volume of TH+ striatal fiber innervation was also estimated through quantification in every sixth serial section using LAS Image Analysis Software (Leica) and extrapolation according to the principle by Cavalieri (50). The boundary of innervation was defined by homogenous patterns of dense TH+ fiber staining and did not include territory where only isolated fibers were found. The level of TH+ fiber density was compared between groups through measurement of OD in three consecutive 40× fields of view from the edge of the graft toward the lateral part of the striatum (near the corpus callosum). Measurements were also obtained from the contralateral intact striatum for comparison, and the OD was quantified using ImageJ.
All quantitative data are represented as means ± SEMs unless otherwise stated. Comparison of values for statistical significance between groups was performed using one-way ANOVA with Tukey’s (Fig. 5D) multiple comparisons test or Student’s t test for comparison of two data points (Figs. 4 Q and R and 5 A, F, and I). GraphPad Prism 6 software was used for statistical analysis.
Supplementary Material
Acknowledgments
The authors thank Mong Tien for expert technical assistance in the tissue preparation. We acknowledge Dr. Francois Guillemot (MRC National Institute for Medical Research) for provision of the Ngn2-GFP mice and Dr. Thomas Perlmann (Ludwig Institute for Cancer Research) and Dr. Johan Ericson (Karolinska Institutet) for provision of the Lmx1a-GFP mice. The Florey Institute of Neuroscience and Mental Health acknowledges the support of the Victorian Government and in particular, funding from the Operational Infrastructure Support Grant. C.R.B. is supported by an NHMRC (National Health and Medical Research Council) Peter Doherty Training Fellowship, C.L.P. is a Viertel Senior Research Fellow, and L.H.T. is supported by an NHMRC Career Development Fellowship. This work was supported by NHMRC Project Grant 1042584.
Footnotes
The authors declare no conflict of interest.
Data deposition: The microarray data reported in this paper have been deposited in the Gene Expression Omnibus (GEO) database, www.ncbi.nlm.nih.gov/geo (accession no. GSE65094).
See Commentary on page 4512.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1501989112/-/DCSupplemental.
References
- 1.Winkler C, Kirik D, Björklund A. Cell transplantation in Parkinson’s disease: How can we make it work? Trends Neurosci. 2005;28(2):86–92. doi: 10.1016/j.tins.2004.12.006. [DOI] [PubMed] [Google Scholar]
- 2.Jönsson ME, Ono Y, Björklund A, Thompson LH. Identification of transplantable dopamine neuron precursors at different stages of midbrain neurogenesis. Exp Neurol. 2009;219(1):341–354. doi: 10.1016/j.expneurol.2009.06.006. [DOI] [PubMed] [Google Scholar]
- 3.Thompson LH, et al. Neurogenin2 identifies a transplantable dopamine neuron precursor in the developing ventral mesencephalon. Exp Neurol. 2006;198(1):183–198. doi: 10.1016/j.expneurol.2005.11.025. [DOI] [PubMed] [Google Scholar]
- 4.Fukuda H, et al. Fluorescence-activated cell sorting-based purification of embryonic stem cell-derived neural precursors averts tumor formation after transplantation. Stem Cells. 2006;24(3):763–771. doi: 10.1634/stemcells.2005-0137. [DOI] [PubMed] [Google Scholar]
- 5.Ganat YM, et al. Identification of embryonic stem cell-derived midbrain dopaminergic neurons for engraftment. J Clin Invest. 2012;122(8):2928–2939. doi: 10.1172/JCI58767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hedlund E, et al. Embryonic stem cell-derived Pitx3-enhanced green fluorescent protein midbrain dopamine neurons survive enrichment by fluorescence-activated cell sorting and function in an animal model of Parkinson’s disease. Stem Cells. 2008;26(6):1526–1536. doi: 10.1634/stemcells.2007-0996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tricot G, et al. Collection, tumor contamination, and engraftment kinetics of highly purified hematopoietic progenitor cells to support high dose therapy in multiple myeloma. Blood. 1998;91(12):4489–4495. [PubMed] [Google Scholar]
- 8.Bomberger C, et al. Lymphoid reconstitution after autologous PBSC transplantation with FACS-sorted CD34+ hematopoietic progenitors. Blood. 1998;91(7):2588–2600. [PubMed] [Google Scholar]
- 9.Quyyumi AA, et al. CD34(+) cell infusion after ST elevation myocardial infarction is associated with improved perfusion and is dose dependent. Am Heart J. 2011;161(1):98–105. doi: 10.1016/j.ahj.2010.09.025. [DOI] [PubMed] [Google Scholar]
- 10.Negrin RS, et al. Transplantation of highly purified CD34+Thy-1+ hematopoietic stem cells in patients with metastatic breast cancer. Biol Blood Marrow Transplant. 2000;6(3):262–271. doi: 10.1016/s1083-8791(00)70008-5. [DOI] [PubMed] [Google Scholar]
- 11.Buchstaller J, et al. Efficient isolation and gene expression profiling of small numbers of neural crest stem cells and developing Schwann cells. J Neurosci. 2004;24(10):2357–2365. doi: 10.1523/JNEUROSCI.4083-03.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lobo MK, Karsten SL, Gray M, Geschwind DH, Yang XW. FACS-array profiling of striatal projection neuron subtypes in juvenile and adult mouse brains. Nat Neurosci. 2006;9(3):443–452. doi: 10.1038/nn1654. [DOI] [PubMed] [Google Scholar]
- 13.Lakowski J, et al. Effective transplantation of photoreceptor precursor cells selected via cell surface antigen expression. Stem Cells. 2011;29(9):1391–1404. doi: 10.1002/stem.694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gennet N, et al. Doublesex and mab-3-related transcription factor 5 promotes midbrain dopaminergic identity in pluripotent stem cells by enforcing a ventral-medial progenitor fate. Proc Natl Acad Sci USA. 2011;108(22):9131–9136. doi: 10.1073/pnas.1016679108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Surmacz B, et al. DLK1 promotes neurogenesis of human and mouse pluripotent stem cell-derived neural progenitors via modulating Notch and BMP signalling. Stem Cell Rev. 2012;8(2):459–471. doi: 10.1007/s12015-011-9298-7. [DOI] [PubMed] [Google Scholar]
- 16.Andressoo JO, Saarma M. Signalling mechanisms underlying development and maintenance of dopamine neurons. Curr Opin Neurobiol. 2008;18(3):297–306. doi: 10.1016/j.conb.2008.07.005. [DOI] [PubMed] [Google Scholar]
- 17.Pruszak J, Isacson O. Molecular and cellular determinants for generating ES-cell derived dopamine neurons for cell therapy. Adv Exp Med Biol. 2009;651:112–123. doi: 10.1007/978-1-4419-0322-8_11. [DOI] [PubMed] [Google Scholar]
- 18.Liss B, et al. Tuning pacemaker frequency of individual dopaminergic neurons by Kv4.3L and KChip3.1 transcription. EMBO J. 2001;20(20):5715–5724. doi: 10.1093/emboj/20.20.5715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Damier P, Hirsch EC, Agid Y, Graybiel AM. The substantia nigra of the human brain. II. Patterns of loss of dopamine-containing neurons in Parkinson’s disease. Brain. 1999;122(Pt 8):1437–1448. doi: 10.1093/brain/122.8.1437. [DOI] [PubMed] [Google Scholar]
- 20.Mendez I, et al. Cell type analysis of functional fetal dopamine cell suspension transplants in the striatum and substantia nigra of patients with Parkinson’s disease. Brain. 2005;128(Pt 7):1498–1510. doi: 10.1093/brain/awh510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sui Y, Vermeulen R, Hökfelt T, Horne MK, Stanić D. Female mice lacking cholecystokinin 1 receptors have compromised neurogenesis, and fewer dopaminergic cells in the olfactory bulb. Front Cell Neurosci. 2013;7:13. doi: 10.3389/fncel.2013.00013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Politis M, et al. Graft-induced dyskinesias in Parkinson's disease: High striatal serotonin/dopamine transporter ratio. Mov Disord. 2011;26(11):1997–2003. doi: 10.1002/mds.23743. [DOI] [PubMed] [Google Scholar]
- 23.Lane EL, Björklund A, Dunnett SB, Winkler C. Neural grafting in Parkinson’s disease unraveling the mechanisms underlying graft-induced dyskinesia. Prog Brain Res. 2010;184:295–309. doi: 10.1016/S0079-6123(10)84015-4. [DOI] [PubMed] [Google Scholar]
- 24.Bye CR, Thompson LH, Parish CL. Birth dating of midbrain dopamine neurons identifies A9 enriched tissue for transplantation into parkinsonian mice. Exp Neurol. 2012;236(1):58–68. doi: 10.1016/j.expneurol.2012.04.002. [DOI] [PubMed] [Google Scholar]
- 25.Villaescusa JC, Arenas E. Transplantable midbrain dopamine neurons: A moving target. Exp Neurol. 2010;222(2):173–178. doi: 10.1016/j.expneurol.2009.12.028. [DOI] [PubMed] [Google Scholar]
- 26.Dunnett SB, Björklund A, Schmidt RH, Stenevi U, Iversen SD. Intracerebral grafting of neuronal cell suspensions. IV. Behavioural recovery in rats with unilateral 6-OHDA lesions following implantation of nigral cell suspensions in different forebrain sites. Acta Physiol Scand Suppl. 1983;522:29–37. [PubMed] [Google Scholar]
- 27.Dunnett SB, Hernandez TD, Summerfield A, Jones GH, Arbuthnott G. Graft-derived recovery from 6-OHDA lesions: Specificity of ventral mesencephalic graft tissues. Exp Brain Res. 1988;71(2):411–424. doi: 10.1007/BF00247501. [DOI] [PubMed] [Google Scholar]
- 28.Grealish S, et al. The A9 dopamine neuron component in grafts of ventral mesencephalon is an important determinant for recovery of motor function in a rat model of Parkinson’s disease. Brain. 2010;133(Pt 2):482–495. doi: 10.1093/brain/awp328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nakao N, et al. Overexpressing Cu/Zn superoxide dismutase enhances survival of transplanted neurons in a rat model of Parkinson’s disease. Nat Med. 1995;1(3):226–231. doi: 10.1038/nm0395-226. [DOI] [PubMed] [Google Scholar]
- 30.Redmond DE, Jr, Vinuela A, Kordower JH, Isacson O. Influence of cell preparation and target location on the behavioral recovery after striatal transplantation of fetal dopaminergic neurons in a primate model of Parkinson’s disease. Neurobiol Dis. 2008;29(1):103–116. doi: 10.1016/j.nbd.2007.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kordower JH, et al. Fetal nigral grafts survive and mediate clinical benefit in a patient with Parkinson’s disease. Mov Disord. 1998;13(3):383–393. doi: 10.1002/mds.870130303. [DOI] [PubMed] [Google Scholar]
- 32.Kordower JH, et al. Functional fetal nigral grafts in a patient with Parkinson’s disease: Chemoanatomic, ultrastructural, and metabolic studies. J Comp Neurol. 1996;370(2):203–230. doi: 10.1002/(SICI)1096-9861(19960624)370:2<203::AID-CNE6>3.0.CO;2-6. [DOI] [PubMed] [Google Scholar]
- 33.Piccini P, et al. Factors affecting the clinical outcome after neural transplantation in Parkinson’s disease. Brain. 2005;128(Pt 12):2977–2986. doi: 10.1093/brain/awh649. [DOI] [PubMed] [Google Scholar]
- 34.Ono Y, et al. Differences in neurogenic potential in floor plate cells along an anteroposterior location: Midbrain dopaminergic neurons originate from mesencephalic floor plate cells. Development. 2007;134(17):3213–3225. doi: 10.1242/dev.02879. [DOI] [PubMed] [Google Scholar]
- 35.Chung S, et al. ES cell-derived renewable and functional midbrain dopaminergic progenitors. Proc Natl Acad Sci USA. 2011;108(23):9703–9708. doi: 10.1073/pnas.1016443108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Doi D, et al. Isolation of human induced pluripotent stem cell-derived dopaminergic progenitors by cell sorting for successful transplantation. Stem Cell Rev. 2014;2(3):337–350. doi: 10.1016/j.stemcr.2014.01.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pruszak J, Sonntag KC, Aung MH, Sanchez-Pernaute R, Isacson O. Markers and methods for cell sorting of human embryonic stem cell-derived neural cell populations. Stem Cells. 2007;25(9):2257–2268. doi: 10.1634/stemcells.2006-0744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sundberg M, et al. Improved cell therapy protocols for Parkinson’s disease based on differentiation efficiency and safety of hESC-, hiPSC-, and non-human primate iPSC-derived dopaminergic neurons. Stem Cells. 2013;31(8):1548–1562. doi: 10.1002/stem.1415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Parish CL, et al. Wnt5a-treated midbrain neural stem cells improve dopamine cell replacement therapy in parkinsonian mice. J Clin Invest. 2008;118(1):149–160. doi: 10.1172/JCI32273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ribeiro D, et al. Efficient expansion and dopaminergic differentiation of human fetal ventral midbrain neural stem cells by midbrain morphogens. Neurobiol Dis. 2013;49:118–127. doi: 10.1016/j.nbd.2012.08.006. [DOI] [PubMed] [Google Scholar]
- 41.Deng Q, et al. Specific and integrated roles of Lmx1a, Lmx1b and Phox2a in ventral midbrain development. Development. 2011;138(16):3399–3408. doi: 10.1242/dev.065482. [DOI] [PubMed] [Google Scholar]
- 42.Dunnett SB, Björklund A. Dissecting embryonic neural tissues for transplantation. In: Dunnett SB, Boulton AA, Baker GB, editors. Neuromethods: Cell and Tissue Transplantation in the CNS. Humana Press; Totowa, NJ: 2000. pp. 3–25. [Google Scholar]
- 43.Smyth GK. Linear models and empirical bayes methods for assessing differential expression in microarray experiments. Stat Appl Genet Mol Biol. 2004;3(2004):3. doi: 10.2202/1544-6115.1027. [DOI] [PubMed] [Google Scholar]
- 44.Huang W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4(1):44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 45.Thompson L, Barraud P, Andersson E, Kirik D, Björklund A. Identification of dopaminergic neurons of nigral and ventral tegmental area subtypes in grafts of fetal ventral mesencephalon based on cell morphology, protein expression, and efferent projections. J Neurosci. 2005;25(27):6467–6477. doi: 10.1523/JNEUROSCI.1676-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Thompson LH, Parish CL. Transplantation of fetal midbrain dopamine progenitors into a rodent model of Parkinson’s disease. Methods Mol Biol. 2013;1059:169–180. doi: 10.1007/978-1-62703-574-3_15. [DOI] [PubMed] [Google Scholar]
- 47.Nikkhah G, Winkler C, Rödter A, Samii M. Microtransplantation of nigral dopamine neurons: A “step by step” recipe. In: Dunnett SB, Boulton AA, Baker GB, editors. Neuromethods: Cell and Tissue Transplantation in the CNS. Humana Press; Totowa, NJ: 2000. pp. 207–231. [Google Scholar]
- 48.Ungerstedt U, Arbuthnott GW. Quantitative recording of rotational behavior in rats after 6-hydroxy-dopamine lesions of the nigrostriatal dopamine system. Brain Res. 1970;24(3):485–493. doi: 10.1016/0006-8993(70)90187-3. [DOI] [PubMed] [Google Scholar]
- 49.Abercrombie M. Estimation of nuclear population from microtome sections. Anat Rec. 1946;94:239–247. doi: 10.1002/ar.1090940210. [DOI] [PubMed] [Google Scholar]
- 50.Cavalieri B. Geometric Degl: Indivisible. Unione Tipografico, Editrice; Turin Italy: 1966. pp. 1–543. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.