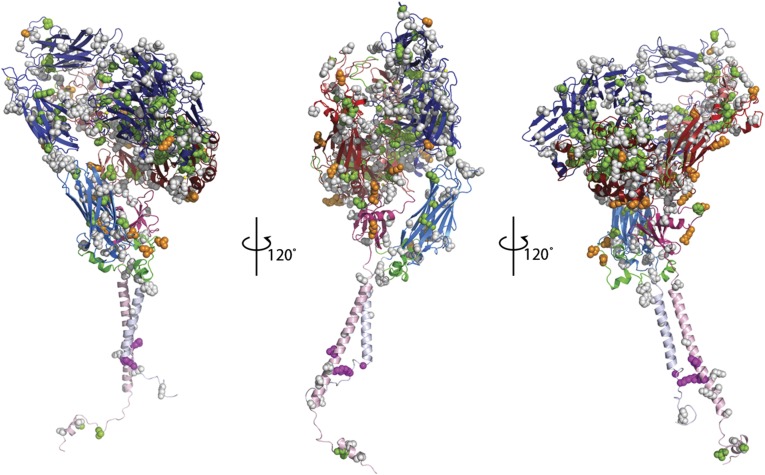Significance
Next-generation sequencing is identifying millions of novel gene variants, presenting challenges to researchers and clinicians. Variations in the genes ITGA2B and ITGB3 affect integrin αIIbβ3, leading to the bleeding disorder Glanzmann thrombasthenia. We analyzed novel missense variants on ∼32,000 alleles of ITGA2B and ITGB3 and found missense variants affecting ∼10% of the amino acids in each protein in ∼1.3% of the population. Almost all variants are rare, indicating recent entry into the population. Two novel variants we predicted would be deleterious profoundly affected recombinant protein expression. At cut-off values that correctly predicted at least 69% of the known Glanzmann thrombasthenia mutations as deleterious, three variant prediction algorithms predicted that at least 27% of the novel variants are deleterious.
Keywords: Glanzmann, integrin, single-nucleotide variants, next-generation sequencing, molecular modeling
Abstract
Next-generation sequencing is transforming our understanding of human genetic variation but assessing the functional impact of novel variants presents challenges. We analyzed missense variants in the integrin αIIbβ3 receptor subunit genes ITGA2B and ITGB3 identified by whole-exome or -genome sequencing in the ThromboGenomics project, comprising ∼32,000 alleles from 16,108 individuals. We analyzed the results in comparison with 111 missense variants in these genes previously reported as being associated with Glanzmann thrombasthenia (GT), 20 associated with alloimmune thrombocytopenia, and 5 associated with aniso/macrothrombocytopenia. We identified 114 novel missense variants in ITGA2B (affecting ∼11% of the amino acids) and 68 novel missense variants in ITGB3 (affecting ∼9% of the amino acids). Of the variants, 96% had minor allele frequencies (MAF) < 0.1%, indicating their rarity. Based on sequence conservation, MAF, and location on a complete model of αIIbβ3, we selected three novel variants that affect amino acids previously associated with GT for expression in HEK293 cells. αIIb P176H and β3 C547G severely reduced αIIbβ3 expression, whereas αIIb P943A partially reduced αIIbβ3 expression and had no effect on fibrinogen binding. We used receiver operating characteristic curves of combined annotation-dependent depletion, Polyphen 2-HDIV, and sorting intolerant from tolerant to estimate the percentage of novel variants likely to be deleterious. At optimal cut-off values, which had 69–98% sensitivity in detecting GT mutations, between 27% and 71% of the novel αIIb or β3 missense variants were predicted to be deleterious. Our data have implications for understanding the evolutionary pressure on αIIbβ3 and highlight the challenges in predicting the clinical significance of novel missense variants.
Next-generation sequencing is transforming our understanding of human genetic variation (1) and providing profound insights into the impact of both inherited and de novo variants on human health (2, 3). At the same time, the data from these studies present serious challenges in providing information to individuals who are found to have variant forms of different proteins. To highlight these challenges, in this report we describe our experience in analyzing missense variants of the platelet αIIbβ3 integrin receptor from The Human Genome Mutation Database (HGMD), the 1000 Genomes project (1000G), the United Kingdom 10K Whole Exome Sequencing project (U.K.10KWES), the United Kingdom 10K Whole Genome Sequencing project (U.K.10KWGS), and The National Heart, Lung and Blood Institute Exome Sequencing Project (ESP); the latter four sources encompass ∼32,000 alleles derived from 16,108 individuals.
The αIIbβ3 receptor has a number of virtues as a model system. First, it is required for hemostasis because platelet aggregation requires cross-linking of the activated form of αIIbβ3 by macromolecular ligands (4). Thus, defects in its biogenesis, activation, or ligand binding lead to the rare bleeding diathesis Glanzmann thrombasthenia (GT), an autosomal recessive disorder (5, 6). Patients with GT come to medical attention because of their hemorrhagic symptoms, and thus have been carefully analyzed clinically and with tests of platelet function for nearly 50 y (5, 7). The biochemical and molecular abnormalities in GT have been studied for nearly 40 y (4, 6, 8–10). In the past 10 y, high-resolution crystallography, electron microscopy, and computational studies of the αIIbβ3 receptor have provided atomic-level information on the correlation between receptor structure and function (11–21). In addition, ethnic groups with relatively high prevalence of GT have been defined that share the same genetic abnormality based on founder mutations, and the dates that some of the mutations entered the population have been estimated (22–28). An on-line registry of GT abnormalities, including patient phenotypes was developed in 1997 (29) and currently contains 51 αIIb and 43 β3 missense variants linked to the disorder (sinaicentral.mssm.edu/intranet/research/glanzmann). The frequency of GT in the general population has not been established but it has a world-wide distribution, and based on data from hematologic practices, it is rare except in areas with a high rate of consanguineous mating (30).
Second, alloimmune disorders, including neonatal thrombocytopenia and posttransfusion purpura, due to amino acid substitutions in either αIIb or β3, have been characterized at the molecular biological level and correlated with mechanisms of immunologic recognition (31).
Third, inherited macrothrombocytopenia and anisothrombocytopenia have been associated with heterozygous missense variants or deletions in αIIb or β3. All of these appear to induce constitutive activation of the receptor and impair proplatelet formation (32–38).
Fourth, αIIbβ3 contributes to pathological platelet thrombus formation in human ischemic cardiovascular disease and αIIbβ3 is a validated target for antithrombotic therapy (39–41).
Fifth, αIIbβ3 is a member of the large integrin family of receptors, which includes 24 receptors derived from 18 α- and 8 β-subunits (41, 42). These receptors are involved in important biologic processes, including development, cell migration, homing, cell survival, and adaptive immunity (41–43). More is known about the structure–function relationships of αIIbβ3 than the other members of the group, and so it serves as the paradigmatic integrin receptor (44, 45).
Sixth, 3D molecular models have been built based on crystallographic and NMR data to analyze the effects of novel amino acid substitutions on receptor structure and function and the generation of alloantigens (15, 46–53). The data from these models and assessments of the severity of the amino acid change in the variants have the potential to aid in predicting whether a novel variant is likely to affect receptor function and immunogenicity (54–59).
Results
Glanzmann Thrombasthenia.
The ThromboGenomics αIIbβ3 compendium developed from the five databases (HGMD, 1000G, U.K.10KWES, U.K.10KWGS, and ESP) contains 319 missense variants in either αIIb or β3, and these are reported in Table S1 A and B. Putative mutations producing GT were considered to be confirmed as disease-causing if they were associated with a GT phenotype and the negative impact of the mutation on αIIbβ3 expression, activation, and/or ligand binding was established in a recombinant system based on a review of the publications describing each of the mutations. Mutations reported in association with a GT phenotype that were not studied in a recombinant system were considered presumptive GT mutations. Because some patients with GT have at least one allele with no exonic variant, it is possible that a nondeleterious (silent) variant that just happened to be on such an allele will be mistaken for a GT-causing variant. This possibility is highlighted by the identification of a silent variant with a relatively high frequency (1%) on an allele in a patient with GT that had no other exonic variants (60). This variant might have been misidentified as disease-causing if its functional consequences were not analyzed in a recombinant expression system.
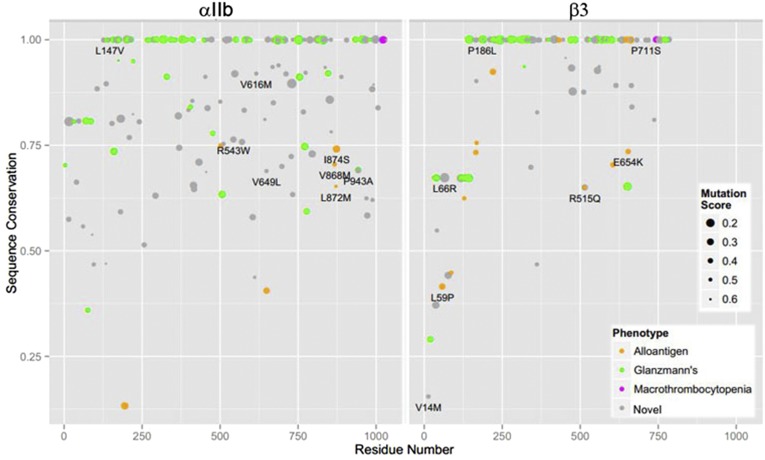
A total of 111 GT missense mutations were identified (56 in αIIb and 55 in β3), of which 52 were considered confirmed and 59 presumptive. The locations of these mutants on the complete αIIbβ3 molecular model are indicated by green spheres in Fig. 1 and Movie S1. For each of these mutations, we analyzed the degree of conservation of the substituted amino acid across species (based on a scale of 0–1, with 1 being the most conserved). A mutation score was then assigned to each missense mutation as an indication of the severity of the amino acid change (based on a scale from 0 to 1, with 0 being the most severe), that is, taking into account the likely amino acid substitutions during evolution. The results are shown in Fig. 2, with green dots indicating the GT mutants, and the different dot sizes indicating the severity of the variant. The majority of disease-causing GT missense mutations involve highly conserved amino acids (scores at or near 1.0) and severe amino acid changes that are less tolerated evolutionarily (55% of variants responsible for GT have mutation scores <0.4).
Fig. 1.
Model of αIIbβ3 with Glanzmann thrombasthenia mutations (green spheres), macro/anisothrombocytopenia mutations (magenta spheres), alloantigens (orange spheres), and novel missense variants (gray spheres).
Fig. 2.
Analysis of the degree of conservation of the substituted amino acids across species (based on a scale of 0–1, with 1 being the most conserved). A mutation score is also assigned to each missense variant as an indication of the severity of the amino acid change (based on a scale from 0 to 1, with 0 and the larger dot size indicating the most severe variant), that is, taking into account likely amino acid substitutions during evolution. Circles identified as 0.2 represent mutations scores between 0–0.2, those identified as 0.3 represent mutation scores between 0.2–0.3, and so forth.
It is notable that not one of the established GT mutations was identified on any of the up to ∼32,000 alleles analyzed in the four WES and WGS studies represented in the databases, suggesting that all of these GT mutations have allele frequencies less than 0.01% in the studied populations.
Alloantigens.
The recognized alloantigens on αIIb and β3 are listed in Table S2 and shown as orange spheres in Fig. 1 and on the complete αIIbβ3 molecular model in Movie S1. The αIIb HPA-3b/Baka/b (I874S) alloantigen is by far the most common polymorphism, being found in 37% of the alleles and identified in individuals in all five databases. Three of the other reported αIIb alloantigens were identified in at least one of the four WES and WGS studies in addition to HGMD, and they had minor allele frequencies (MAFs) of 0.4% (HPA-27b/Cab3,Ak-L872M), 0.2% (HPA-9b/Max-V868M), and 0.03% (HPA-20b/Kno-T650M). Two of the reported αIIb alloantigens (HPA-24b/Cab2,In-S503N and HPA-22b/She,Sey-K195T) were not identified in any of the four studies and thus presumably have MAFs < 0.01% in the studied populations. HPA-1b/PlA2-L59P was the most common β3 alloantigen with an MAF of 14%. The other β3 alloantigens had MAFs of 0.9% or less and only HPA-6b/Ca/Tua-R515Q was identified in all four WES and WGS studies. When analyzed on the basis of amino acid conservation, compared with GT missense mutations, the variants responsible for the alloantigens (orange dots in Fig. 2) were less likely to affect highly conserved amino acids or to exhibit severe amino acid changes (40% of variants responsible for alloantigens have a mutation scores <0.4; differences in conservation score values between GT and alloantigens: P = 9 × 10−5; difference in location = 0.25; Wilcoxon rank sum test).
Macro/Anisothrombocytopenia.
Five heterozygous αIIbβ3 missense mutations affecting four different amino acids (αIIb G1022C, R1026Q, and R1026W; β3 L744P and D749H), along with three different deletions, have been associated with macrothrombocytopenia or platelet anisocytosis and variable abnormalities in platelet function and clinical hemorrhage (32–38). The corresponding positions on the complete αIIbβ3 molecular model are shown as magenta spheres in Fig. 1 and Movie S1. All of the mutations appear to result in constitutive activation of the receptor, which in turn impairs proplatelet formation. None of these mutations was identified in the four WES and WGS databases, suggesting that they, like the mutations causing GT, are likely rare (MAF < 0.01%). The conservation and mutation scores for these amino acids are shown in Fig. 2.
Novel Variants.
A total of 114 novel missense variants in αIIb and 68 novel missense variants in β3 were identified. These are shown as gray spheres in Fig. 1 and on the complete αIIbβ3 molecular model in Movie S1. In total, 11.2% of the 1,025 amino acids in mature αIIb and 8.7% of the 784 amino acids in mature β3 were mutated. Only 3.8% of the variants had MAFs > 0.1%, indicating their relative rarity, consistent with the expectation under the Fisher-Wright model (61) that most variants are rare. Six αIIb amino acids were mutated twice (40 T/I and T/A, 369 H/Q and H/D, 433 R/C and R/H, 543 R/W and R/Q, 728 A/T and A/V, and 988 R/W and R/Q) as were two β3 amino acids (14 V/L and V/M and 186 P/S and P/L) among the novel variants and the previously described incidental nondisease-causing variants. The severity of the amino acid change and the conservation across species of the reference amino acid of each variant are shown in Fig. 2 (gray dots). Compared with the GT missense mutations, there was greater representation of variants at less conserved amino acids and, in general, the variants produced less severe amino acid changes (30% of the novel variants have mutation scores ≤ 0.4).
Assessment of the Effect of Three Novel Variants on αIIbβ3 Expression and Function.
Five of the novel variants in our study affected an amino acid in αIIb previously reported to be associated with GT (P176H, L214F, H813L, L830F, and P943A) and two affected an amino acid in β3 previously reported to be associated with GT (R131W and C547G). The allele frequency for P943A was 0.46%, whereas it was 0.01% or less for the other variants. It is notable that αIIb P176 appears to be a mutational hotspot because four patients from three ethnically distinct kindreds (Mennonite, Dutch, and Italian) have been reported to have P176A mutations and a Chinese GT patient has been reported with a P176L mutation (62, 63).
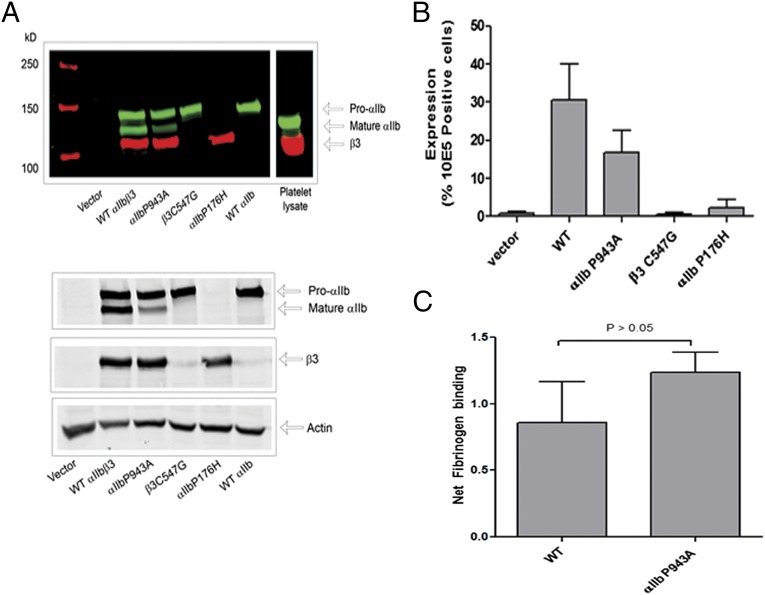
We assessed the effect of the three variants with the most severe variant scores, αIIb P176H and P943A, and β3 C547G, by expressing them in HEK293 cells and analyzing both receptor expression and the ability of the receptor to bind fibrinogen when activated with the reducing agent DTT (53). Fig. 3A shows the immunoblot data from the cell lysates. The WT αIIbβ3 bands correspond to proαIIb, mature αIIb (which aligns with platelet αIIb), and β3 (which migrates slightly slower than platelet β3 because it contains a V5 tag). The αIIb P943A variant expressed similar amounts of proαIIb, but reduced amounts of mature αIIb, suggesting a partial defect in αIIbβ3 maturation. In contrast, the αIIb P176H variant showed a dramatic reduction in proαIIb expression and virtually no mature αIIb, and the β3 C547G variant resulted in a profound decrease in β3 expression. Flow cytometry using an antibody specific for the αIIbβ3 complex (mAb 10E5) confirmed that the P943A variant produced a partial reduction in αIIbβ3 expression and that the other two variants produced profound reductions in surface expression of the αIIbβ3 complex (Fig. 3B). The cells expressing the αIIb P943A variant were then further tested for their ability to bind fibrinogen in the presence of DTT. When the amount of fibrinogen bound was normalized for αIIbβ3 expression, the αIIb P943A variant was found to bind as much fibrinogen as normal αIIbβ3 (Fig. 3C), suggesting that this variant partially affects expression, but does not alter receptor function.
Fig. 3.
(A) HEK293 cells (105 cells/mL) were transfected with cDNA harboring the indicated variant in αIIb or β3. Cell lysates were subject to immunoblotting with antibodies to αIIb and β3. Immunoblot shown is representative of five experiments. (Upper) Color image showing detection with Odyssey system. (Lower) Black and white image of the examined proteins. (B) Transfected HEK293 cells were analyzed by flow cytometry for αIIbβ3 expression. Cells were incubated with Alexa488-conjugated mAb 10E5 (10 µg/mL) or (as a control for nonspecific binding) 10 µg/mL Alexa488-conjugated mAb 10E5 in the presence of excess (125 µg/mL) unlabeled 10E5. The graph depicts the mean ± SEM; n = 5. (C) To assess αIIbβ3 function, DTT (10 mM) was added to the cells and Alexa647-conjugated fibrinogen binding was assessed via flow cytometry. Binding is expressed as net fibrinogen binding. Data are expressed as mean ± SEM; n = 5.
Novel Variants Affecting Amino Acids Mutated in Reported Alloantigens.
We identified two novel variants affecting the same amino acid responsible for an alloantigen, β3 R515W affecting the same amino acid as HPA-6b (R515Q) and β3 R662H affecting the same amino acid as HPA-11b (R662C). Both novel variants were identified in only one of the studies. R515W has an allele frequency of 0.1% among the 32,216 alleles analyzed, whereas the alloantigen R515Q has an allele frequency of 0.01% based on analysis of 13,306 alleles. R662H has the same allele frequency as the alloantigen variant R662C (0.02%).
Predicting the Consequences of Individual Variants.
A number of algorithms have been developed to predict whether a given amino acid substitution is likely to be deleterious—that is, whether it will alter expression or function. We used three different commonly used algorithms: combined annotation-dependent depletion (CADD) (57), Polyphen 2-HDIV (58), and sorting intolerant from tolerant (SIFT) (59), and analyzed the missense variants based on whether they were confirmed GT mutants, presumptive GT mutants, alloantigens, or novel variants. We anticipated that the confirmed and presumptive GT mutants would be judged to have the highest likelihood of being deleterious, the alloantigens would have the lowest likelihood, and the novel variants would have the greatest range of likelihoods. The results varied with the prediction tool, but in general were consistent with our expectations (Fig. 4A). Moreover, each of the algorithms identified a cluster of three presumptive GT variants with a low probability of being deleterious, and it may be valuable to study these in more detail [αIIb P76A and β3 M321L (64), and H652L (65)]. The concordance between prediction tools for individual variants is displayed as a heat map in Fig. S1 and demonstrates variable degrees of discordance among the tools.
Fig. 4.
(A) A prediction algorithms to assess the likelihood that a variant will be deleterious. The results of the analysis with the algorithms CADD, SIFT, and Polyphen 2-HDIV are presented for the alloantigens, the presumptive and confirmed GT variants, and the ThromboGenomics and Exome Aggregation Consortium (ExAC) novel variants. Black arrows on ordinate denote optimal rank score cut-off values. (B) ROC curves of data based on considering confirmed GT and macro/anisothrombocytopenia as true positives and 20 alloantigens as true negatives. The optimal discriminator points are identified by dots and the corresponding rank scores are shown next to the dots. (C) Concordance of algorithms in predicting whether novel variants are likely to be deleterious using the optimal cut-offs determined by the ROC curves. D, deleterious; ND, nondeleterious.
To obtain an estimate of the sensitivity and specificity of each of the prediction tools in judging missense variants, we assessed their sensitivity in identifying the confirmed GT and macro/anisothrombocytopenia mutations as deleterious (positive controls) and their specificity in identifying the 20 alloantigens as not being deleterious (negative controls). We then used the results to create receiver operating characteristic (ROC) curves for each of the prediction tools (Fig. 4B). All three gave similar patterns, with areas under the curves of 0.91 [median; 95% confidence intervals (CI) 0.83, 0.99], 0.90 (CI 0.81, 0.99), and 0.88 (CI 0.80, 0.97) for CADD, Polyphen 2-HDIV, and SIFT, respectively (Table 1) (P = not significant for all pair-wise comparisons). To estimate the percentage of the novel variants we identified that are likely to be deleterious, we applied the cut-off value of each prediction tool that best differentiated the positive controls from the negative controls according to the ROC curve and then applied this value to the novel variants. These optimal cut-off values, achieved median sensitivities of 98%, 94%, and 69% and specificities of 75%, 80%, and 95% for CADD, Polyphen 2-HDIV, and SIFT, respectively. The percentages of novel αIIb missense variants predicted to be deleterious were 43.0%, 45.6%, and 27.2% for CADD, Polyphen 2-HDIV, and SIFT, respectively, and the comparable values for β3 missense variants were 70.6%, 54.4%, and 32.4%, respectively. Because it is possible that some of the low-frequency alloantigens are deleterious, we also created ROC curves using only the 11 of 20 alloantigens that were judged by all three algorithms to have a low probability of being deleterious (rank scores < 0.5) as negative controls (Table S1A). As expected, the areas under the curves were higher (1.00, 0.99, and 0.99, respectively). The optimal cut-off values for each algorithm yielded sensitivities of 98%, 94%, and 96%, respectively and 100% specificity for all three (Table 1). The percentages of novel αIIb missense variants predicted to be deleterious based on these optimal cut-off points were 49.1%, 47.4%, and 59.7%, respectively; the comparable values for β3 were 73.5%, 59.9%, 67.7%, respectively. Thus, only SIFT showed a major change in the prediction of deleteriousness, and this reflects the more step-wise nature of the prediction rank scores with this algorithm in comparison with the more continuous rank scores with the other algorithms (Fig. 4A).
Table 1.
ROC analysis of αIIbβ3 variants
| Algorithm | AUC | AUC Confidence Interval | Sensitivity | Specificity | Cut-off | Percentage of novel variants predicted to be deleterious | |
| ITGA2B | ITGB3 | ||||||
| Specificity based on all 20 alloantigens | |||||||
| CADD | 0.91 | 0.83–0.99 | 0.98 | 0.75 | 0.51 | 43.0 | 70.6 |
| SIFT | 0.88 | 0.80–0.88 | 0.69 | 0.95 | 0.80 | 27.2 | 32.4 |
| Polyphen-2 | 0.90 | 0.81–0.90 | 0.94 | 0.80 | 0.58 | 45.6 | 54.4 |
| Specificity based on 11 alloantigens with low probability of being deleterious | |||||||
| CADD | 1.00 | 0.99–1.00 | 0.98 | 1 | 0.46 | 49.1 | 73.5 |
| SIFT | 0.99 | 0.98–0.99 | 0.96 | 1 | 0.44 | 59.7 | 67.7 |
| Polyphen-2 | 0.99 | 0.98–0.99 | 0.94 | 1 | 0.55 | 47.4 | 55.9 |
AUC, area under the ROC curve.
Concordance in Predictions Among the Three Algorithms.
The concordance among the three different algorithms in classifying the novel variants is shown in Fig. 4C. All three algorithms classified 22.8% of the ITGA2B and 26.5% of the ITGB3 variants as deleterious and 43.0% of the ITGA2B and 29.4%% of the ITGB3 variants as nondeleterious. Thus, in total, there was concordance among all three in classifying 65.8% of the ITGA2B variants and 55.9% of the ITGB3 variants. The greatest discordance was found in the classification of ITGB3 variants as being deleterious by both CADD and Polyphen 2-HDIV, but nondeleterious by SIFT, which accounted for 25.0% of the novel ITGB3 variants.
It is of particular note that Polyphen 2-HDIV and SIFT identified the αIIb P176H and β3 C547G variants that we found to have profound defects in surface expression as nearly certain to be deleterious, with rank scores much higher than the optimal cut-off values. In contrast, CADD gave both lower rankings (0.66 and 0.61, respectively), but still ones above the optimal cut-off value of 0.51. As a result, all three algorithms classified these variants as deleterious. In contrast, the rank scores given to the αIIb P943A variant were more broadly distributed, with SIFT giving it the lowest ranking (0.31), Polyphen 2-HDIV an intermediate ranking (0.50), and CADD the highest ranking (0.80). Based on each of their optimal cut-off values, this would result in SIFT and Polyphen2-HDIV classifying this mutation as nondeleterious and CADD ranking it as deleterious. These results are particularly interesting because we found that the variant partially decreased αIIbβ3 expression.
Analysis of the Exome Aggregation Consortium Database.
After our studies were completed, the Exome Aggregation Consortium made available the results of 60,706 unrelated individuals whose DNA was sequenced as part of various disease-specific and population genetic studies. We analyzed the 319 novel variants (that is, those remaining after removing the GT and macro/anisothrombocytopenia mutations and the alloantigens) in αIIb (72 of which are also reported in the ThromboGenomics dataset) and the 216 novel variants in β3 (49 of which are also reported in the ThromboGenomics dataset) using the optimal cut-off values for each algorithm based on the ROC curves. CADD, Polyphen 2-HDIV, and SIFT predicted that 40.8%, 35.7%, and 23.5%, respectively, of the αIIb and 60.6%, 46.3%, and 29.6%, respectively, of the β3 variants would be deleterious (Fig. 4A), and all three predicted that 17.6% of αIIb and 22.7% of the β3 variants would be deleterious, which are values similar to those predicted using the ThromboGenomics dataset.
Discussion
Our study highlights the opportunity and challenges in interpreting the potential impact of novel variants of αIIbβ3 on receptor function, with broader implications for interpreting novel variants in other genes and gaining insights into the evolutionary pressures that have affected αIIbβ3. We found that three different prediction algorithms were able to differentiate confirmed GT and macro/anisothrombocytopenia mutations from alloantigens with reasonably high sensitivity and specificity, but there was some overlap between the groups and the three algorithms were only about 60% concordant in classifying novel variants. Thus, it is important to augment the results of these analyses with additional data. Table S3 summarizes some of the considerations that a clinician can bring to bear on deciding whether a novel variant in αIIb or β3 is likely to result in a medically meaningful phenotype. If a propositus is available for study, αIIbβ3 can be quantified by flow cytometry and its function assessed directly by platelet aggregation studies using ADP or other agonists that rely on the αIIbβ3 receptor. Individuals who are heterozygous for mutations that affect expression of the receptor usually have about 50–60% of the normal level of platelet αIIbβ3 expression, but there is some overlap between the values of heterozygotes and normals (66). Homozygotes have markedly decreased platelet aggregation, but heterozygotes have little or no decrease in platelet aggregation. If the propositus is not available, as might occur if a fetus is tested, data from the prediction tools can be combined with additional data, including whether the variant affects an amino acid previously associated with causing GT or macro/anisothrombocytopenia, or whether the amino acid is highly conserved or is in a region known to have very stringent structural requirements (for example, the blades of the β-propeller in αIIb) or to affect ligand binding. If molecular modeling or molecular dynamics simulations are available, they can provide more detailed information on the likely impact of the variant. Additional information can come from producing a recombinant version of the variant in a cell line and testing expression and ligand binding, but this requires considerable time and research expertise, and there are regulatory issues in providing information for diagnostic purposes from assays that are not performed in certified laboratories. The MAF, if known, can also provide valuable information, because one would expect a deleterious variant with a high MAF to have already come to medical attention. By combining all of this information, the clinician will be able to make a reasoned judgment, but in many cases there will still be an irreducible level of uncertainty that needs to be carefully conveyed to the individual requesting the information.
The results from recent analyses of human exomes and genomes in large populations provides context for interpreting our data. The explosive growth in the human population over the past 2,500 y, estimated to be from ∼50 million to ∼7 billion people, implies departure from population genetic equilibrium and predicts a large numbers of relatively new variants that have had little time to undergo selective evolutionary pressure (2, 3, 67, 68). Because they entered the gene pool recently, these variants are expected to have a low allele frequency, and vice versa, unless the variant confers a strong negative effect on fertility in the heterozygous state, one can surmise that variants with a low MAF entered the gene pool recently and thus are relatively new. Variants with an MAF < 0.01% probably originated within the past 250 y and those with an MAF of ∼0.5% probably originated ∼2,500 y ago (68). By definition, one must sequence large numbers of individuals with high precision to robustly establish a low MAF, and so such data have only become available in the past few years (3). Racial differences also need to be considered, because the African population is the oldest and thus most diverse, and the European population, which is presumed to have undergone two population bottlenecks, probably has had the most rapid recent population growth (2, 3). Establishing that rare variants are not the result of sequencing errors using current methodology requires careful attention, and new methods to minimize the risk of mistaking sequencing errors for variants are being developed (69).
It is important to recognize the limitations of our study. In particular, the number of variants identified may be underestimated if the sequencing does not include all of the exons, as was the case for several exons in the β3 subunit gene in the ESP study, representing ∼40% of the exonic bases (Fig. S2), or if the filters are stringent in the face of relatively low coverage. On the other hand, if there is low sequencing coverage of a region, as was the case for one complete exon and a portion of another exon in the αIIb subunit gene in the ESP study, and in the low-coverage U.K.10KWGS study (70), it is possible that a sequencing error may be misreported as a variant. To minimize this risk, a series of filters are applied to the raw sequencing data before making a final determination of whether a variant is present. In the case of the U.K.10KWGS study, we found it reassuring that the percentage of individuals with missense mutations (1.27%) was similar to the combined value of 1.39% for the other three studies. Moreover, 37 of the 48 missense variants identified in the U.K.10KWGS were also identified by at least one other study. Overestimation of variant frequency may also occur if one uses cultured immortalized lymphoblastoid cells as the source of DNA because they acquire variants during culture that are not present in the germ line genome (71). Lymphoblastoid cells were used as a source of DNA only in the 1000G database. Thus, with the above caveats, we believe our dataset provides a reasonable reflection of the missense variants in the populations studied. We did not include variants involving DNA base pair deletions or insertions because these usually are much less common than missense variants, and next-generation sequencing is less accurate in detecting them.
Because not one of the reported GT missense mutants reported in the literature (HGMD) (Table S1B) was identified in the alleles included in this analysis, they all presumably have very low MAFs, implying that they entered the population recently. Independent support for the recent origin of GT missense mutations comes from studies of two populations with relatively high rates of GT because of intragroup marriage, namely Israeli and Palestinian Arabs and French Manouche gypsies, in which the founder mutations were estimated to be only ∼300–600 and ∼300–400 y old, respectively (23, 24).
Against this background, our data provide new insights into the frequency and potential impact of novel variants of αIIbβ3. In the ∼16,500 individuals studied, we found 114 novel missense variants affecting ∼10% of the amino acids in αIIb and 68 novel missense variants affecting ∼9% of the amino acids in β3. Thus, with the limitations noted above, we found that ∼1.3% of the population studied harbors a variant in either αIIb or β3.
αIIbβ3 is expressed at ∼100,000 copies per platelet, which is approximately twice the density required for normal platelet aggregation (72, 73). As a result, individuals who are heterozygous for GT variants are usually asymptomatic or have relatively mild symptoms (66, 74–76), and thus are unlikely to be subjected to selective pressure. Because hemorrhage can result in death before reaching sexual maturity or during or after child-birth, it is possible that there would be selective pressure against αIIb or β3 variants that decreased receptor expression or function in mammals if they had a lower density of αIIbβ3 receptors than modern humans or had a high percentage of intragroup matings. The lack of GT mutations with high MAFs is consistent with selective pressure against deleterious variants in the distant past. Platelets are known to be present in all mammals, including monotremes (which lay eggs) and marsupials (which do not have placental births), and so the hemorrhagic risk associated with placental births cannot explain the appearance of platelets in mammals (77). To our knowledge however, the densities of αIIbβ3 receptors in monotreme and marsupial platelets have not been reported, and so it remains possible that placental birth selected for high receptor density. For comparison, the density of αIIbβ3 receptors on chicken thrombocytes has been reported to be only ∼5% of that of human platelets (78). Because humans increasingly mate with individuals outside their kinship, heterozygous deleterious variants are likely to continue to accumulate. Such variants may become more consequential in the future as they spread through the population. Additional variants are likely to enter the population as de novo variants. Individuals are estimated to have an average of ∼74 de novo single-nucleotide variants (79); with a world population of ∼7 billion people, one can expect more than 500 billion variants per generation in the “humanome,” enough on average, to affect each of the 3 billion genome bases more than 100-times over. Thus, with time these variants will increase the likelihood of GT because of double heterozygosity.
Our studies of αIIbβ3 serve as a paradigm for the opportunities and challenges one can anticipate as we enter the era in which many individuals (and fetuses) undergo whole-exome and whole-genome sequencing and want to know whether a novel variant is deleterious. We applied three algorithms that are currently used to estimate the percentage of novel variants that are deleterious and found that using their optimal ROC rank cut-off values, they predicted between 27.2% and 45.6% of the novel αIIb variants and between 32.4% and 70.6% of the novel β3 variants are deleterious; all three predicted that 22.8% of αIIb variants and 26.5% of β3 variants would be deleterious. These percentages can be compared with the data from the ESP study (2), where 74% of nonsynonymous variants (98% of which were missense variants) were judged deleterious by at least one of seven algorithms, but only 27.9% met the criterion of being predicted deleterious by at least four of the seven algorithms. We would emphasize, however, that the optimal ROC cut-off value is based on discriminating between two groups, with sensitivity and specificity given equal weight. Clinically, however, one may wish to set a minimum sensitivity or specificity to address a specific goal, for example, choosing a high-sensitivity value so as to avoid a false-negative diagnosis. This can be achieved by identifying the rank cut-off value corresponding to the desired sensitivity/specificity point on the ROC curve.
Two of the novel variants we identified (αIIb P176H and β3 C547G) were predicted by all three algorithms to be deleterious and we found that both profoundly affected αIIbβ3 expression. We anticipated that the αIIb P176H variant would decrease expression because of its strategic location and the major change in the side-chain structure, and that the β3 C547G variant would decrease receptor expression because of its impact on disulfide bonding. The αIIb P943A variant was predicted by two of the three algorithms to be deleterious and we found that it partially decreased αIIbβ3 expression and did not impair fibrinogen binding induced by DTT. We suspected that this latter variant would not cause a severe defect in αIIbβ3 expression or function because, with its relatively high MAF (0.46%), one would have expected it to have already come to medical attention in homozygous or double-heterozygous GT patients. It is possible that hypomorphic variants like P943A, which partially reduce αIIbβ3 expression, account for some of the wide range of αIIbβ3 expression observed among apparently healthy individuals (66). Thus, variants like P943A highlight that even with the availability of multiple prediction algorithms and an advanced understanding of a protein’s structure and function, it may be challenging to predict the functional consequences of any given variant.
Similar considerations bear on the ability to predict whether a given variant will result in the generation of an alloantigen. Despite advances in molecular modeling of platelet alloantigens (47) and predicting the immunogenicity of HLA variants (80) (www.hlamatchmaker.net/), and the ability of peptides to bind to specific major histocompatibility molecules (81), it is challenging to predict the likelihood that a mismatch between parents in their sequences of αIIb or β3 will result in medically meaningful fetal and neonatal alloimmune thrombocytopenia because many other immunologic factors may influence the outcome (82). Whether β3 variants induce antibodies that affect αVβ3 function, potentially leading to intrauterine growth restriction or vascular abnormalities contributing to hemorrhage, or whether de novo αIIb or β3 variants can result in constitutively active receptors that may lead to thrombus formation in the placenta (82), are additional questions of import. It is possible that the latter abnormalities have not come to medical attention to date because they result in fetal loss.
In conclusion, next-generation sequencing, coupled with advanced bioinformatics algorithms, is making it possible to both identify novel missense variants and in many cases make a reasoned estimate of the likelihood that the variant will affect protein expression and/or function. Current algorithms are not definitive, however, in a substantial percentage of cases, and so these tools need to be used judiciously and coupled with additional clinical, laboratory, and population-based data.
Methods
Variant Analysis.
This study is part of the ThromboGenomics working group (see SI Methods for description). Data for the present study on αIIb and β3 variants were obtained from the whole-exome and whole-genome sequencing data from five different databases: (i) HGMD (www.hgmd.cf.ac.uk/ac/index.php), which collates published gene lesions responsible for human inherited disease; (ii) The 1000 Genomes project (1,000GP; www.1000genomes.org), representing 1,092 individuals of East Asian, South Asian, African, European, and American ancestry who underwent whole exome or low pass whole genome sequencing (83); (iii) The U.K.10KWES (www.uk10k.org/), representing up to 4,732 members of the British population (77); (iv) The U.K.10KWGS, representing 3,781 members of the British population; and (v) The National Heart, Lung and Blood Institute ESP, representing up to 6,503 Americans of European and African ancestry (84). We restricted our analyses to single-nucleotide variants that lead to amino acid substitutions (missense variants). The variants were expressed in a variant call format, starting with the first based in the human genome reference sequence, which includes the signal peptide. As a result, the numbers are 31 amino acids higher than those for mature αIIb and 26 higher than those for mature β3. The effects of variants were predicted on the standard ITGA2B reference transcript ENST00000262407 (NM_000419.3, LRG_479t1) and the ITGB3 transcript ENST0000055948 (NM_000212.2, LRG_481t1) using the variant effect predictor.
Data on coverage of the exons in ITGA2B and ITGB3 are available from the ESP study and the results shown in Fig. S2. All of the exons in ITG2A had more than 15× coverage, except for the 5′ end of exon 2 and exon 27. Exons 1–10 in ITGB3 had greater than 15× coverage, but exons 11–15 were not analyzed. In the U.K.10KWGS study, sequencing was performed to a median depth of 7.6 reads per sample at single-nucleotide variant sites (70). Each study defined a series of criteria by which they validated the variants they identified (83, 84) (www.uk10k.org/data.html#datasets).
Catalogue of GT Missense Mutants.
We combined data from the online registry of GT variants and the five databases identified above into Table S1, where part A of the table includes data from HGMD only and part B of the table includes combined data from 1000G, U.K.10KWES, U.K.10KWGS, and ESP. All reported variants where reviewed and those in which the proposed GT mutant was shown to lead to a defect in receptor expression and function in a recombinant system were categorized as confirmed GT mutations. All other variants where considered presumptive GT mutations. Defects in GT patients were also characterized as either homozygous or heterozygous if the information was available. The MAF of each missense variant is reported based on the combined data from the different databases. Finally, the database in which the variant was reported is identified.
Analysis of Missense Variants.
A complete αIIbβ3 3D model was built using a combination of X-ray crystallographic data of the αIIbβ3 ectodomain [PDB ID code 3FCS (18)] and NMR data of the transmembrane and cytoplasmic domains [PDB ID codes 2K9J (85), 2KNC (86), and 2KV9 (87)]. Missing segments of the αIIb calf-2 domain (amino acid residues 763–775 and 839–874), the β3 hybrid domain (amino acids residues 74–79), and the link between the EGF-1 and EGF-2 domains (amino acids residues 476–483) were built ab initio using Rosetta 2.2 (88) (Movie S1).
Missense variants were analyzed in terms of: (i) the level of conservation of the residue position in the alignment of the wild-type sequence of αIIb or β3 with sequences from different species (the ordinate of the graph in Fig. 2), and (ii) the severity of the specific amino acid change (the size of each circle in Fig. 2).
Both the level of conservation across different species and the severity of the missense variant were assessed based on a mutation score calculated as the ”sum-of-pairs” score with a modified PET91 substitution matrix (89) by Valdar and Thornton (see ref. 54 and equation 37 in ref. 55) using the Scorecons server at www.ebi.ac.uk/thornton-srv/databases/cgi-bin/valdar/scorecons_server.pl. Severe mutations are less tolerated during evolution and are therefore characterized by low scores in the substitution matrix, as opposed to mutations to amino acids with similar properties. Large circle sizes in Fig. 1 (variant score = 1 − “sum-of-pairs” score) were chosen to represent severe variants.
In the calculation of sequence conservation, the mutation score consists of the average of the pair scores calculated for each amino acid in an aligned column, weighted using the dissimilarity between the sequences comprising each pair to normalize against sequence redundancy in the alignment. Sequence alignments of all human and vertebrate αIIb or β3 sequences were obtained using the multiple sequence alignment Hidden Markov Model/clustal W method (56).
Mutagenesis, Cell Transfection, and αIIbβ3 Analysis.
For mutagenesis, cell transfection, and αIIbβ3 analysis, see SI Methods.
Prediction of Variant Impact.
We curated the variants using the dbNSFP v2.4 database (database for nonsynonymous SNP’s functional predictions; https://sites.google.com/site/jpopgen/dbNSFP). This database integrates prediction scores from several different algorithms, including CADD (57), Polyphen 2-HDIV (58), and SIFT (59). We then plotted the ranked prediction of CADD, Polyphen-HDIV, and SIFT for the variants in ITG2A and ITGB3. Variants were grouped as alloantigens, confirmed GT mutations, presumptive GT mutations, and novel variants. The ROC analysis was done through R package of pROC v1.7.3 (cran.r-project.org/web/packages/pROC/index.html) (90).
To examine the concordance between the different algorithms, the ranked scores from the different methods were first displayed as a heat map. We also displayed the ranked scores for all of the variants by group for each algorithm.
Statistical Analysis.
Each experiment was repeated at least four times. Results are expressed as mean ± SEM with number of observations n. Data were analyzed using GraphPad Prism 5 software. Significant differences were determined using Student t test and considered significant at P ≤ 0.05.
The CI of ROC analysis is calculated by CI function of pROC package (with default parameters (95% CI through 2,000 stratified bootstrap replicates) (90).
Supplementary Material
Acknowledgments
We thank Dr. Davide Provasi for technical assistance, and Drs. Paul Bray, Peter Newman, Jean-Laurent Casanova, and Yuval Itan for reviewing the manuscript and helpful suggestions. This study was supported, in part, by Grant HL19278 from the National Heart, Lung and Blood Institute, Grant UL1 TR000043 from the National Center for Advancing Translational Sciences, National Institutes of Health Clinical and Translational Science Award program, and funds from Stony Brook University.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
3A complete list of the ThromboGenomics Consortium can be found in the SI Methods.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1422238112/-/DCSupplemental.
References
- 1.Koboldt DC, Steinberg KM, Larson DE, Wilson RK, Mardis ER. The next-generation sequencing revolution and its impact on genomics. Cell. 2013;155(1):27–38. doi: 10.1016/j.cell.2013.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Tennessen JA, et al. Broad GO Seattle GO NHLBI Exome Sequencing Project Evolution and functional impact of rare coding variation from deep sequencing of human exomes. Science. 2012;337(6090):64–69. doi: 10.1126/science.1219240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Nelson MR, et al. An abundance of rare functional variants in 202 drug target genes sequenced in 14,002 people. Science. 2012;337(6090):100–104. doi: 10.1126/science.1217876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Coller BS, Shattil SJ. The GPIIb/IIIa (integrin alphaIIbbeta3) odyssey: A technology-driven saga of a receptor with twists, turns, and even a bend. Blood. 2008;112(8):3011–3025. doi: 10.1182/blood-2008-06-077891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Coller BS, Seligsohn U, Peretz H, Newman PJ. Glanzmann thrombasthenia: New insights from an historical perspective. Semin Hematol. 1994;31(4):301–311. [PubMed] [Google Scholar]
- 6.Nurden AT, Fiore M, Nurden P, Pillois X. Glanzmann thrombasthenia: A review of ITGA2B and ITGB3 defects with emphasis on variants, phenotypic variability, and mouse models. Blood. 2011;118(23):5996–6005. doi: 10.1182/blood-2011-07-365635. [DOI] [PubMed] [Google Scholar]
- 7.Caen JP, et al. Congenital bleeding disorders with long bleeding time and normal platelet count. I. Glanzmanns thrombasthenia (report of 15 patients) Am J Med. 1966;41(1):4–26. doi: 10.1016/0002-9343(68)90070-3. [DOI] [PubMed] [Google Scholar]
- 8.Nurden AT, Caen JP. An abnormal platelet glycoprotein pattern in three cases of Glanzmann’s thrombasthenia. Br J Haematol. 1974;28(2):253–260. doi: 10.1111/j.1365-2141.1974.tb06660.x. [DOI] [PubMed] [Google Scholar]
- 9.Phillips DR, Agin PP. Platelet membrane defects in Glanzmann’s thrombasthenia. Evidence for decreased amounts of two major glycoproteins. J Clin Invest. 1977;60(3):535–545. doi: 10.1172/JCI108805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.French DL, Seligsohn U. Platelet glycoprotein IIb/IIIa receptors and Glanzmann’s thrombasthenia. Arterioscler Thromb Vasc Biol. 2000;20(3):607–610. doi: 10.1161/01.atv.20.3.607. [DOI] [PubMed] [Google Scholar]
- 11.Negri A, Li J, Naini S, Coller BS, Filizola M. Structure-based virtual screening of small-molecule antagonists of platelet integrin αIIbβ3 that do not prime the receptor to bind ligand. J Comput Aided Mol Des. 2012;26(9):1005–1015. doi: 10.1007/s10822-012-9594-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Provasi D, Negri A, Coller BS, Filizola M. Talin-driven inside-out activation mechanism of platelet αIIbβ3 integrin probed by multimicrosecond, all-atom molecular dynamics simulations. Proteins. 2014;82(12):3231–3240. doi: 10.1002/prot.24540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhu J, et al. Structure-guided design of a high-affinity platelet integrin alphaIIbbeta3 receptor antagonist that disrupts Mg2+ binding to the MIDAS. Sci Transl Med. 2012;4(125):125ra132. doi: 10.1126/scitranslmed.3003576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhu J, et al. Closed headpiece of integrin αIIbβ3 and its complex with an αIIbβ3-specific antagonist that does not induce opening. Blood. 2010;116(23):5050–5059. doi: 10.1182/blood-2010-04-281154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Murcia M, Jirouskova M, Li J, Coller BS, Filizola M. Functional and computational studies of the ligand-associated metal binding site of beta3 integrins. Proteins. 2008;71(4):1779–1791. doi: 10.1002/prot.21859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Filizola M, Hassan SA, Artoni A, Coller BS, Weinstein H. Mechanistic insights from a refined three-dimensional model of integrin alphaIIbbeta3. J Biol Chem. 2004;279(23):24624–24630. doi: 10.1074/jbc.M400243200. [DOI] [PubMed] [Google Scholar]
- 17.Choi WS, Rice WJ, Stokes DL, Coller BS. Three-dimensional reconstruction of intact human integrin αIIbβ3: New implications for activation-dependent ligand binding. Blood. 2013;122(26):4165–4171. doi: 10.1182/blood-2013-04-499194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhu J, et al. Structure of a complete integrin ectodomain in a physiologic resting state and activation and deactivation by applied forces. Mol Cell. 2008;32(6):849–861. doi: 10.1016/j.molcel.2008.11.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Eng ET, Smagghe BJ, Walz T, Springer TA. Intact alphaIIbbeta3 integrin is extended after activation as measured by solution X-ray scattering and electron microscopy. J Biol Chem. 2011;286(40):35218–35226. doi: 10.1074/jbc.M111.275107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Xiao T, Takagi J, Coller BS, Wang JH, Springer TA. Structural basis for allostery in integrins and binding to fibrinogen-mimetic therapeutics. Nature. 2004;432(7013):59–67. doi: 10.1038/nature02976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Provasi D, Murcia M, Coller BS, Filizola M. Targeted molecular dynamics reveals overall common conformational changes upon hybrid domain swing-out in beta3 integrins. Proteins. 2009;77(2):477–489. doi: 10.1002/prot.22463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rosenberg N, et al. Glanzmann thrombasthenia caused by an 11.2-kb deletion in the glycoprotein IIIa (beta3) is a second mutation in Iraqi Jews that stemmed from a distinct founder. Blood. 1997;89(10):3654–3662. [PubMed] [Google Scholar]
- 23.Rosenberg N, et al. A 13-bp deletion in alpha(IIb) gene is a founder mutation that predominates in Palestinian-Arab patients with Glanzmann thrombasthenia. J Thromb Haemost. 2005;3(12):2764–2772. doi: 10.1111/j.1538-7836.2005.01618.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fiore M, Pillois X, Nurden P, Nurden AT, Austerlitz F. Founder effect and estimation of the age of the French Gypsy mutation associated with Glanzmann thrombasthenia in Manouche families. Eur J Hum Genet. 2011;19(9):981–987. doi: 10.1038/ejhg.2011.61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Seligsohn U, Rososhansky S. A Glanzmann’s thrombasthenia cluster among Iraqi Jews in Israel. Thromb Haemost. 1984;52(3):230–231. [PubMed] [Google Scholar]
- 26.Khanduri U, et al. Glanzmann’s thrombasthenia. A review and report of 42 cases from South India. Thromb Haemost. 1981;46(4):717–721. [PubMed] [Google Scholar]
- 27.Schlegel N, et al. The molecular genetic basis of Glanzmann’s thrombasthenia in a gypsy population in France: Identification of a new mutation on the alpha IIb gene. Blood. 1995;86(3):977–982. [PubMed] [Google Scholar]
- 28.Awidi AS. Rare inherited bleeding disorders secondary to coagulation factors in Jordan: A nine-year study. Acta Haematol. 1992;88(1):11–13. doi: 10.1159/000204587. [DOI] [PubMed] [Google Scholar]
- 29.French DL, Coller BS. Hematologically important mutations: Glanzmann thrombasthenia. Blood Cells Mol Dis. 1997;23(1):39–51. doi: 10.1006/bcmd.1997.0117. [DOI] [PubMed] [Google Scholar]
- 30.Nurden AT, Pillois X, Nurden P. Understanding the genetic basis of Glanzmann thrombasthenia: Implications for treatment. Expert Rev Hematol. 2012;5(5):487–503. doi: 10.1586/ehm.12.46. [DOI] [PubMed] [Google Scholar]
- 31.Peterson JA, McFarland JG, Curtis BR, Aster RH. Neonatal alloimmune thrombocytopenia: Pathogenesis, diagnosis and management. Br J Haematol. 2013;161(1):3–14. doi: 10.1111/bjh.12235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kashiwagi H, et al. Demonstration of novel gain-of-function mutations of αIIbβ3: Association with macrothrombocytopenia and Glanzmann thrombasthenia-like phenotype. Mol Genet Genomic Med. 2013;1(2):77–86. doi: 10.1002/mgg3.9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kunishima S, et al. Heterozygous ITGA2B R995W mutation inducing constitutive activation of the αIIbβ3 receptor affects proplatelet formation and causes congenital macrothrombocytopenia. Blood. 2011;117(20):5479–5484. doi: 10.1182/blood-2010-12-323691. [DOI] [PubMed] [Google Scholar]
- 34.Ghevaert C, et al. A nonsynonymous SNP in the ITGB3 gene disrupts the conserved membrane-proximal cytoplasmic salt bridge in the alphaIIbbeta3 integrin and cosegregates dominantly with abnormal proplatelet formation and macrothrombocytopenia. Blood. 2008;111(7):3407–3414. doi: 10.1182/blood-2007-09-112615. [DOI] [PubMed] [Google Scholar]
- 35.Nurden AT, Pillois X, Fiore M, Heilig R, Nurden P. Glanzmann thrombasthenia-like syndromes associated with macrothrombocytopenias and mutations in the genes encoding the αIIbβ3 integrin. Semin Thromb Hemost. 2011;37(6):698–706. doi: 10.1055/s-0031-1291380. [DOI] [PubMed] [Google Scholar]
- 36.Peyruchaud O, et al. R to Q amino acid substitution in the GFFKR sequence of the cytoplasmic domain of the integrin IIb subunit in a patient with a Glanzmann’s thrombasthenia-like syndrome. Blood. 1998;92(11):4178–4187. [PubMed] [Google Scholar]
- 37.Schaffner-Reckinger E, et al. Overexpression of the partially activated alpha(IIb)beta3D723H integrin salt bridge mutant downregulates RhoA activity and induces microtubule-dependent proplatelet-like extensions in Chinese hamster ovary cells. J Thromb Haemost. 2009;7(7):1207–1217. doi: 10.1111/j.1538-7836.2009.03494.x. [DOI] [PubMed] [Google Scholar]
- 38.Jayo A, et al. L718P mutation in the membrane-proximal cytoplasmic tail of beta 3 promotes abnormal alpha IIb beta 3 clustering and lipid microdomain coalescence, and associates with a thrombasthenia-like phenotype. Haematologica. 2010;95(7):1158–1166. doi: 10.3324/haematol.2009.018572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bledzka K, Smyth SS, Plow EF. Integrin αIIbβ3: From discovery to efficacious therapeutic target. Circ Res. 2013;112(8):1189–1200. doi: 10.1161/CIRCRESAHA.112.300570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bosch X, Marrugat J, Sanchis J. Platelet glycoprotein IIb/IIIa blockers during percutaneous coronary intervention and as the initial medical treatment of non-ST segment elevation acute coronary syndromes. Cochrane Database Syst Rev. 2013;11:CD002130. doi: 10.1002/14651858.CD002130.pub4. [DOI] [PubMed] [Google Scholar]
- 41.Cox D, Brennan M, Moran N. Integrins as therapeutic targets: Lessons and opportunities. Nat Rev Drug Discov. 2010;9(10):804–820. doi: 10.1038/nrd3266. [DOI] [PubMed] [Google Scholar]
- 42.Hynes RO. Integrins: Bidirectional, allosteric signaling machines. Cell. 2002;110(6):673–687. doi: 10.1016/s0092-8674(02)00971-6. [DOI] [PubMed] [Google Scholar]
- 43.Avraamides CJ, Garmy-Susini B, Varner JA. Integrins in angiogenesis and lymphangiogenesis. Nat Rev Cancer. 2008;8(8):604–617. doi: 10.1038/nrc2353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bennett JS, Berger BW, Billings PC. The structure and function of platelet integrins. J Thromb Haemost. 2009;7(Suppl 1):200–205. doi: 10.1111/j.1538-7836.2009.03378.x. [DOI] [PubMed] [Google Scholar]
- 45.Du X, Ginsberg MH. Integrin alpha IIb beta 3 and platelet function. Thromb Haemost. 1997;78(1):96–100. [PubMed] [Google Scholar]
- 46.Mor-Cohen R, et al. Unique disulfide bonds in epidermal growth factor (EGF) domains of β3 affect structure and function of αIIbβ3 and αvβ3 integrins in different manner. J Biol Chem. 2012;287(12):8879–8891. doi: 10.1074/jbc.M111.311043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Jallu V, Poulain P, Fuchs PF, Kaplan C, de Brevern AG. Modeling and molecular dynamics of HPA-1a and -1b polymorphisms: Effects on the structure of the β3 subunit of the αIIbβ3 integrin. PLoS ONE. 2012;7(11):e47304. doi: 10.1371/journal.pone.0047304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Mehrbod M, Trisno S, Mofrad MR. On the activation of integrin αIIbβ3: Outside-in and inside-out pathways. Biophys J. 2013;105(6):1304–1315. doi: 10.1016/j.bpj.2013.07.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Laguerre M, et al. Molecular dynamics analysis of a novel β3 Pro189Ser mutation in a patient with Glanzmann thrombasthenia differentially affecting αIIbβ3 and αvβ3 expression. PLoS ONE. 2013;8(11):e78683. doi: 10.1371/journal.pone.0078683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Blue R, et al. Effects of limiting extension at the alphaIIb genu on ligand binding to integrin alphaIIbbeta3. J Biol Chem. 2010;285(23):17604–17613. doi: 10.1074/jbc.M110.107763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Vomund AN, Stuhlsatz-Krouper S, Dimitry J, Song Y, Frazier WA. A naturally occurring extracellular alpha-beta clasp contributes to stabilization of beta3 integrins in a bent, resting conformation. Biochemistry. 2008;47(44):11616–11624. doi: 10.1021/bi8015108. [DOI] [PubMed] [Google Scholar]
- 52.Levin L, et al. A single disulfide bond disruption in the β3 integrin subunit promotes thiol/disulfide exchange, a molecular dynamics study. PLoS ONE. 2013;8(3):e59175. doi: 10.1371/journal.pone.0059175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kamata T, et al. Structural requirements for activation in alphaIIb beta3 integrin. J Biol Chem. 2010;285(49):38428–38437. doi: 10.1074/jbc.M110.139667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Valdar WS, Thornton JM. Protein-protein interfaces: Analysis of amino acid conservation in homodimers. Proteins. 2001;42(1):108–124. [PubMed] [Google Scholar]
- 55.Valdar WS. Scoring residue conservation. Proteins. 2002;48(2):227–241. doi: 10.1002/prot.10146. [DOI] [PubMed] [Google Scholar]
- 56.Sievers F, et al. Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol Syst Biol. 2011;7:539. doi: 10.1038/msb.2011.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kircher M, et al. A general framework for estimating the relative pathogenicity of human genetic variants. Nat Genet. 2014;46(3):310–315. doi: 10.1038/ng.2892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Adzhubei IA, et al. A method and server for predicting damaging missense mutations. Nat Methods. 2010;7(4):248–249. doi: 10.1038/nmeth0410-248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Ng PC, Henikoff S. SIFT: Predicting amino acid changes that affect protein function. Nucleic Acids Res. 2003;31(13):3812–3814. doi: 10.1093/nar/gkg509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Losonczy G, et al. Three novel mutations in the glycoprotein IIb gene in a patient with type II Glanzmann thrombasthenia. Haematologica. 2007;92(5):698–701. doi: 10.3324/haematol.10847. [DOI] [PubMed] [Google Scholar]
- 61.Wright S. The distribution of gene frequencies in populations. Proc Natl Acad Sci USA. 1937;23(6):307–320. doi: 10.1073/pnas.23.6.307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Basani RB, et al. A naturally occurring mutation near the amino terminus of alphaIIb defines a new region involved in ligand binding to alphaIIbbeta3. Blood. 2000;95(1):180–188. [PubMed] [Google Scholar]
- 63.D’Andrea G, et al. GLAnzmann’s Thrombasthenia Italian Team (GLATIT) Glanzmann’s thrombasthenia: Identification of 19 new mutations in 30 patients. Thromb Haemost. 2002;87(6):1034–1042. [PubMed] [Google Scholar]
- 64.Nair S, Ghosh K, Shetty S, Mohanty D. Mutations in GPIIIa molecule as a cause for Glanzmann thrombasthenia in Indian patients. J Thromb Haemost. 2005;3(3):482–488. doi: 10.1111/j.1538-7836.2005.01159.x. [DOI] [PubMed] [Google Scholar]
- 65.Vinciguerra C, et al. Description of 10 new mutations in platelet glycoprotein IIb (alphaIIb) and glycoprotein IIIa (beta3) genes. Platelets. 2001;12(8):486–495. doi: 10.1080/095371001317126383. [DOI] [PubMed] [Google Scholar]
- 66.Coller BS, et al. Immunologic and biochemical characterization of homozygous and heterozygous Glanzmann thrombasthenia in the Iraqi-Jewish and Arab populations of Israel: Comparison of techniques for carrier detection. Br J Haematol. 1986;62(4):723–735. doi: 10.1111/j.1365-2141.1986.tb04096.x. [DOI] [PubMed] [Google Scholar]
- 67.Keinan A, Clark AG. Recent explosive human population growth has resulted in an excess of rare genetic variants. Science. 2012;336(6082):740–743. doi: 10.1126/science.1217283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Coventry A, et al. Deep resequencing reveals excess rare recent variants consistent with explosive population growth. Nat Commun. 2010;1:131. doi: 10.1038/ncomms1130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Schmitt MW, et al. Detection of ultra-rare mutations by next-generation sequencing. Proc Natl Acad Sci USA. 2012;109(36):14508–14513. doi: 10.1073/pnas.1208715109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Browning BL, Browning SR. Detecting identity by descent and estimating genotype error rates in sequence data. Am J Hum Genet. 2013;93(5):840–851. doi: 10.1016/j.ajhg.2013.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Schafer CM, et al. Whole exome sequencing reveals minimal differences between cell line and whole blood derived DNA. Genomics. 2013;102(4):270–277. doi: 10.1016/j.ygeno.2013.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Coller BS, Folts JD, Smith SR, Scudder LE, Jordan R. Abolition of in vivo platelet thrombus formation in primates with monoclonal antibodies to the platelet GPIIb/IIIa receptor. Correlation with bleeding time, platelet aggregation, and blockade of GPIIb/IIIa receptors. Circulation. 1989;80(6):1766–1774. doi: 10.1161/01.cir.80.6.1766. [DOI] [PubMed] [Google Scholar]
- 73.Wagner CL, et al. Analysis of GPIIb/IIIa receptor number by quantification of 7E3 binding to human platelets. Blood. 1996;88(3):907–914. [PubMed] [Google Scholar]
- 74.Reichert N, Seligsohn U, Ramot B. Clinical and genetic aspects of Glanzmann’s thrombasthenia in Israel: Report of 22 cases. Thromb Diath Haemorrh. 1975;34(3):806–820. [PubMed] [Google Scholar]
- 75.Cronberg S, Nilsson IM, Zetterqvist E. Investigation of a family with members with both severe and mild degree of thrombasthenia. Acta Paediatr Scand. 1967;56(2):189–197. doi: 10.1111/j.1651-2227.1967.tb15363.x. [DOI] [PubMed] [Google Scholar]
- 76.Stormorken H, Gogstad GO, Solum NO, Pande H. Diagnosis of heterozygotes in Glanzmann’s thrombasthenia. Thromb Haemost. 1982;48(2):217–221. [PubMed] [Google Scholar]
- 77.Levin J. The evolution of mammalian platelets. In: Michelson AD, editor. Platelets. 3rd Ed. Academic; London: 2013. pp. 3–26. [Google Scholar]
- 78.Schmaier AA, et al. Occlusive thrombi arise in mammals but not birds in response to arterial injury: Evolutionary insight into human cardiovascular disease. Blood. 2011;118(13):3661–3669. doi: 10.1182/blood-2011-02-338244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Veltman JA, Brunner HG. De novo mutations in human genetic disease. Nat Rev Genet. 2012;13(8):565–575. doi: 10.1038/nrg3241. [DOI] [PubMed] [Google Scholar]
- 80.Duquesnoy RJ. The antibody response to an HLA mismatch: A model for nonself-self discrimination in relation to HLA epitope immunogenicity. Int J Immunogenet. 2012;39(1):1–9. doi: 10.1111/j.1744-313X.2011.01042.x. [DOI] [PubMed] [Google Scholar]
- 81.Carrasco Pro S, Zimic M, Nielsen M. Improved pan-specific MHC class I peptide-binding predictions using a novel representation of the MHC-binding cleft environment. Tissue Antigens. 2014;83(2):94–100. doi: 10.1111/tan.12292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Kaplan CNH, Freedman J. Alloimmune thrombocytopenia. In: Michelson AD, editor. Platelets. 3rd Ed. Academic; London: 2013. pp. 953–970. [Google Scholar]
- 83.Abecasis GR, et al. 1000 Genomes Project Consortium An integrated map of genetic variation from 1,092 human genomes. Nature. 2012;491(7422):56–65. doi: 10.1038/nature11632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Fu W, et al. NHLBI Exome Sequencing Project Analysis of 6,515 exomes reveals the recent origin of most human protein-coding variants. Nature. 2013;493(7431):216–220. doi: 10.1038/nature11690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Lau TL, Kim C, Ginsberg MH, Ulmer TS. The structure of the integrin alphaIIbbeta3 transmembrane complex explains integrin transmembrane signalling. EMBO J. 2009;28(9):1351–1361. doi: 10.1038/emboj.2009.63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Yang J, et al. Structure of an integrin alphaIIb beta3 transmembrane-cytoplasmic heterocomplex provides insight into integrin activation. Proc Natl Acad Sci USA. 2009;106(42):17729–17734. doi: 10.1073/pnas.0909589106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Metcalf DG, et al. NMR analysis of the alphaIIb beta3 cytoplasmic interaction suggests a mechanism for integrin regulation. Proc Natl Acad Sci USA. 2010;107(52):22481–22486. doi: 10.1073/pnas.1015545107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Wang C, Bradley P, Baker D. Protein-protein docking with backbone flexibility. J Mol Biol. 2007;373(2):503–519. doi: 10.1016/j.jmb.2007.07.050. [DOI] [PubMed] [Google Scholar]
- 89.Jones DT, Taylor WR, Thornton JM. The rapid generation of mutation data matrices from protein sequences. Comput Appl Biosci. 1992;8(3):275–282. doi: 10.1093/bioinformatics/8.3.275. [DOI] [PubMed] [Google Scholar]
- 90.Robin X, et al. pROC: An open-source package for R and S+ to analyze and compare ROC curves. BMC Bioinformatics. 2011;12:77. doi: 10.1186/1471-2105-12-77. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.