Significance
MDMX protein is a critical regulator of p53 and a novel drug target. The current generation of MDM2 inhibitors does not inhibit MDMX. Therefore, their therapeutic efficacy will be influenced by poorly characterized MDMX functional status in tumors. Efforts to develop MDMX inhibitors have been largely unsuccessful, indicating gaps in our understanding of the structure and regulation of MDMX. This study provides evidence that MDMX-p53 binding is regulated by an autoinhibitory mechanism that involves intramolecular interaction in MDMX through p53 mimicry. The results suggest a mechanism by which DNA damage signaling inhibits MDMX and activates p53.
Keywords: MDMX, p53, intramolecular, acidic domain, protease
Abstract
The p53 inhibitor MDMX is controlled by multiple stress signaling pathways. Using a proteolytic fragment release (PFR) assay, we detected an intramolecular interaction in MDMX that mechanistically mimics the interaction with p53, resulting in autoinhibition of MDMX. This mimicry is mediated by a hydrophobic peptide located in a long disordered central segment of MDMX that has sequence similarity to the p53 transactivation domain. NMR spectroscopy was used to show this hydrophobic peptide interacts with the N-terminal domain of MDMX in a structurally analogous manner to p53. Mutation of two critical tryptophan residues in the hydrophobic peptide disrupted the intramolecular interaction and increased p53 binding, providing further evidence for mechanistic mimicry. The PFR assay also revealed a second intramolecular interaction between the RING domain and central region that regulates MDMX nuclear import. These results establish the importance of intramolecular interactions in MDMX regulation, and validate a new assay for the study of intramolecular interactions in multidomain proteins with intrinsically disordered regions.
The p53 tumor suppressor is activated by numerous cellular and environmental signals and induces the expression of genes that regulate metabolism, cell growth, cell cycle, apoptosis, and senescence (1). MDM2 and MDMX are key regulators that control the cellular level and transcriptional activity of p53 through direct binding. Mouse knockout experiments showed that both MDM2 and MDMX are essential for controlling p53 activity during embryogenesis. Somatic knockout experiments showed that MDM2 is indispensable for regulating p53 in adult tissues, whereas MDMX deletion does not lead to cell death (2). MDM2 is a well-established p53 transcriptional target that forms a negative feedback loop by binding to the N-terminal transcriptional activation domain of p53 and, subsequently, ubiquitinating the C-terminal regulatory domain, which leads to degradation of p53 by the proteasome. p53 binding sites are also found in intron 1 of human MDMX, and p53 activation leads to moderate induction of MDMX transcription (3). Therefore, MDMX is a p53 target gene that may also provide dynamic feedback in response to p53 activation.
MDMX alone does not have E3 ligase activity, but it is important for regulating p53 transcriptional function. MDMX expression and phosphorylation by the ATM/Chk2 pathway is important for the p53-mediated DNA damage response in mice (4, 5). MDMX levels are controlled by MDM2-mediated ubiquitination in a stress-dependent fashion (6, 7). Significant degradation of MDMX occurs after DNA damage through phosphorylation at several C-terminal sites (S342 and S367 by Chk2, S403 by ATM) (8). Furthermore, ribosomal stress promotes MDMX degradation through L11–MDM2 interaction (9), and oncogenic stress promotes MDMX degradation through ARF expression (10). Therefore, key signaling mechanisms that block p53 degradation simultaneously enhance MDMX degradation by MDM2. These observations underscore the coordinated control of MDM2 and MDMX that regulate the cellular dynamics of p53 in response to DNA damage.
MDMX knockout in mice leads to p53 activation without significant stabilization (11). Inhibiting MDMX-p53 binding leads to p53 activation, suggesting that MDMX-p53 binding is critical for the regulation of p53. Both MDM2 and MDMX bind to p53 through a hydrophobic pocket at the N-terminal domain, but the interactions appear to be regulated differently. A previous study showed that CK1α kinase stably binds to MDMX (but not MDM2) and stimulates MDMX-p53 binding (12). CK1α interacts with the central region of MDMX, including the partially disordered acidic region and zinc finger, and promotes phosphorylation of S289 (12). CK1α appears to inhibit a putative intramolecular interaction between the p53 binding domain and central domain of MDMX, suggesting a mechanism by which CK1α stimulates MDMX-p53 binding (13). DNA damage inhibits MDMX–CK1α binding that, in turn, leads to decreased MDMX-p53 binding (13).
Intramolecular interactions in multidomain proteins often have important functions (14). Analysis of intramolecular interactions by X-ray crystallography is often difficult, because flexible regions interfere with crystallization. Most studies rely on GST pull down, coprecipitation, and yeast two-hybrid assays. These approaches may create overexpression artifacts or false negative results due to abnormal folding or low affinity of separated domains. We established an assay for analyzing protein intramolecular interactions, which we referred to as proteolytic fragment release assay (PFR). Using the PFR assay, we detected multiple intramolecular interactions by using full-length MDMX produced in human cells. We identified an autoinhibitory sequence in the MDMX central domain that binds to the N-terminal pocket in a manner similar to p53. We also detected changes in MDMX intramolecular binding induced by mutations or interaction with CK1α. These results provide new insight on the mechanism of MDMX regulation and validate an approach for analyzing protein intramolecular interactions.
Results
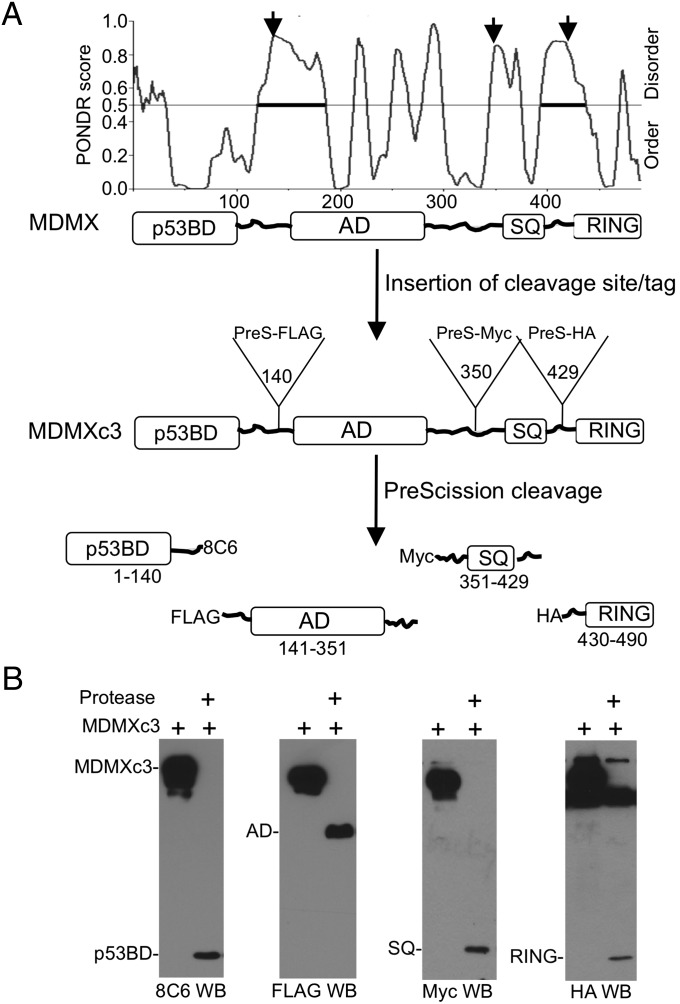
Design of a Protease Cleavable MDMX Construct.
To analyze the intramolecular interactions between MDMX domains, PreScission protease cleavage site followed by an epitope tag was inserted into three disordered regions of MDMX selected by using the PONDR predictor of natural disordered regions (15), generating MDMXc3 (Fig. 1A). Cleavage with PreScission produced four fragments each with a unique epitope: the p53 binding domain (p53BD, recognized by the antibody 8C6 with an epitope located within residues 101–140), the central acidic region (AD, FLAG tag), C-terminal ATM/Chk2 phosphorylation region (SQ, Myc tag), and the RING domain (RING, HA tag). It was expected that after cleavage of the linkers, domains that interact in the full-length protein would dissociate slowly compared with noninteracting domains. The binding between fragment pairs was detected by immobilizing MDMXc3 using different epitope-specific antibodies, on-bead digestion with PreScission, and analyzing the retention/release ratio of each domain by Western blot. The accessibility of all three cleavage sites were confirmed by the ability of PreScission protease to cleave MDMXc3 in H1299 lysate in <10 min, producing the epitope-tagged fragments (Fig. 1B). Functional analyses showed that MDMXc3 retained p53/CK1α/MDM2 binding and degradation by MDM2 (SI Appendix, Fig. S1).
Fig. 1.
Design of protease cleavable MDMXc3. (A) Identification of MDMX disordered regions for the insertion of cleavage sites. PreScission cleavage site and epitope tags were inserted after residues 140, 350, and 429 to create MDMXc3. Cleavage by PreScission protease produces four fragments each containing a unique epitope. (B) Lysate of H1299 transfected with MDMXc3 was digested with 0.1 µg/µL PreScission for 10 min at 4 °C and analyzed by Western blot to detect the production of individual fragments.
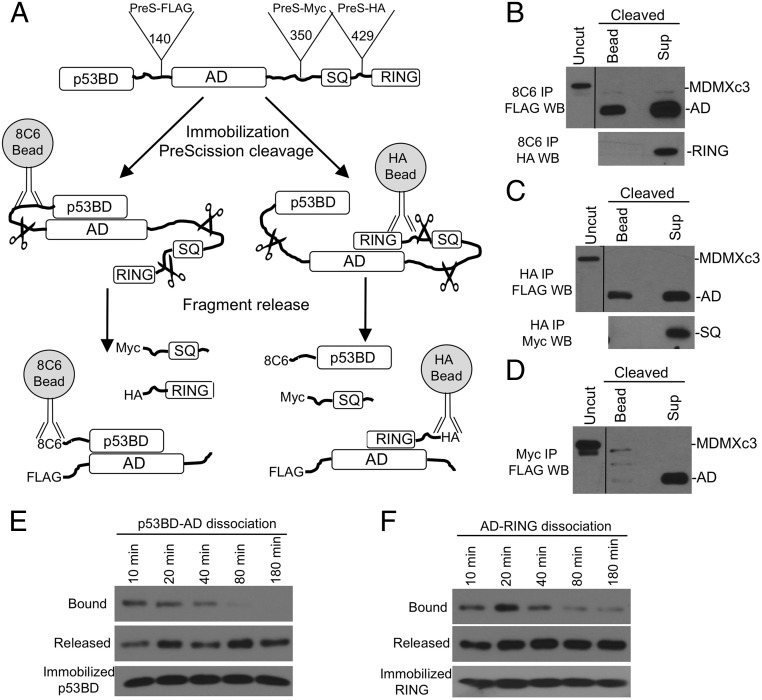
Detection of MDMX Intramolecular Interactions Using PFR Assay.
When MDMXc3 expressed in H1299 cells was immobilized with N-terminal antibody 8C6 and cleaved with PreScission, ∼25% of the AD remained bound to the beads (Fig. 2B), indicating an intramolecular binding between the p53BD and AD. Immobilization using HA antibody resulted in ∼25–50% of the AD remained bound to the beads, suggesting that the AD also interacts with the RING (Fig. 2C). In contrast, when MDMXc3 was immobilized by using Myc antibody, the AD was completely released after cleavage, suggesting a lack of SQ-AD binding (Fig. 2D). Similar analyses detected no p53BD-RING binding (Fig. 2B) and no SQ-RING binding (Fig. 2C). These negative results provided internal controls for specificity. Overall, the assay revealed that the MDMX AD forms simultaneous or competing internal complexes with the p53BD and the RING domain (Fig. 2A). The result likely provides a conservative measure of the MDMX population present in a closed conformation. Hereon we refer to such experiments as proteolytic fragment release, or PFR assay.
Fig. 2.
Proteolytic fragment release (PFR) assay. (A) Diagram of PFR assay for MDMX intramolecular interactions. MDMXc3 was immobilized on beads by using different antibodies and cleaved by PreScission for <40 min. The beads and supernatant were analyzed by Western blot to detect fragment release from the beads. (B) MDMXc3 expressed in H1299 was immobilized on 8C6 beads (captures MDMXc3 through the p53BD fragment) and incubated with PreScission for 40 min. The beads and supernatant were analyzed by FLAG and HA Western blot. The presence of AD fragment in the bead fraction indicates slow dissociation from the immobilized p53BD. Absence of RING fragment in the bead fraction indicates lack of binding to the p53BD. (C) Immobilization of MDMXc3 through the RING domain revealed RING binding to AD but not SQ fragment. (D) Immobilization of MDMX through the SQ fragment revealed lack of binding to the AD. (E) MDMXc3 captured on 8C6 beads was cleaved with PreScission in <5 min. Beads and supernatants were separated at indicated time points and analyzed by FLAG Western blot to detect the dissociation of AD from p53BD. (F) MDMXc3 immobilized using HA antibody was analyzed for the dissociation of AD from RING after cleavage at indicated time points.
Time course analysis showed that the p53BD–AD complex dissociated in 1–2 h after cleavage of the linker (Fig. 2E), whereas AD–RING dissociation was slow (Fig. 2F). Both AD–p53BD and AD–RING interactions were disrupted in high detergent or high salt buffers, suggesting that hydrophobic and electrostatic interactions were both important for the internal binding (SI Appendix, Fig. S2A). We also performed cleavage of MDMXc3 in cell extract, followed by IP-Western blot to detect the coprecipitation of different domains. Robust coprecipitation was detected between the AD and RING fragment by using HA IP (SI Appendix, Fig. S2C). However, p53BD–AD coprecipitation was very weak by using 8C6 IP (SI Appendix, Fig. S2C, arrow), suggesting that the p53BD–AD complex dissociated completely during the 16 h IP. AD-SQ coprecipitation was not detectable by using Myc IP.
To compare the fragment release assay to GST pull down assay, GST fusion containing MDMX p53BD, MDMX AD, MDMX RING, MDM2 RING, and Praja RING control was incubated with PreScission-digested MDMXc3 and the pull down products were blotted for MDMX fragments. The results showed that this pull down assay specifically detected RING-RING dimerization and RING–MDMXc3 (full-length) interactions (SI Appendix, Fig. S2B, Top and Top Middle), but p53BD–AD and AD–RING interactions were not detectable under these conditions (SI Appendix, Fig. S2B, Bottom and Bottom Middle). These results suggest that the PFR assay was more sensitive in detecting weak or transient intramolecular interactions compared with GST pull down and co-IP assays.
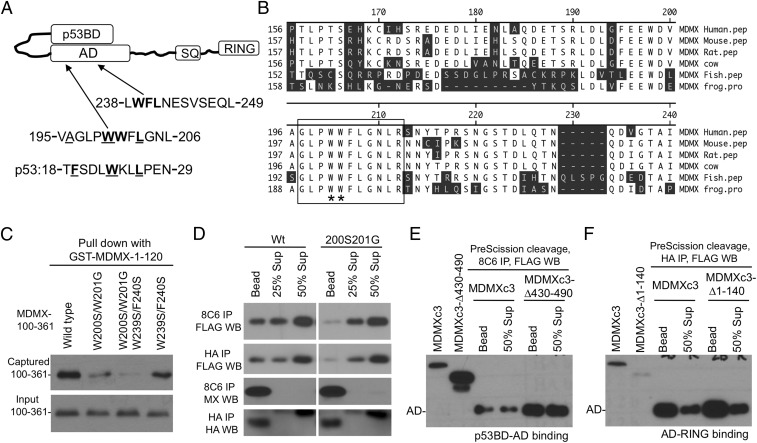
Identification of an Autoinhibitory Sequence in MDMX.
Our recent study suggested that the MDMX AD regulates the N-terminal domain (13). Inspection of the MDMX sequence identified two central regions with hydrophobic residues that may serve as p53 mimetics (Fig. 3A). The sequence around residue 200 (referred to as WW) contains highly conserved hydrophobic residues (Fig. 3B). Point mutation analysis showed that W200S/W201G double substitution (referred to as SG) disrupted the p53BD–AD binding in a GST pull-down assay (Fig. 3C) (13). Next, the SG mutation was introduced into MDMXc3 and analyzed by PFR assay. The results showed that the SG mutation reduced both p53BD–AD and AD–RING intramolecular interactions (Fig. 3D). In contrast, W239S/F240S substitution only had minor effects. These results suggest that the WW sequence was critical for AD intramolecular binding to both N- and C-terminal domains.
Fig. 3.
Identification of an internal p53 mimetic sequence in MDMX. (A) Proposed model of p53BD–AD intramolecular interaction and the presence of two potential pocket-binding sequences in the AD. (B) Alignment of MDMX WW sequence from various species. (C) Indicated substitutions were introduced into MDMX-100–361 fragment and expressed in H1299 cells. The cell lysate was incubated with beads loaded with GST-MDMX-1-120, the captured 100–361 fragment was detected by 8C6 Western blot. (D) The SG mutations were introduced into MDMXc3, expressed in H1299 cells, and analyzed for p53BD–AD and AD–RING interactions using the PFR assay. (E) MDMXc3 was subcloned to delete the RING domain and analyzed for p53BD–AD binding using PFR assay. (F) MDMXc3 was subcloned to delete the p53BD and analyzed for AD–RING binding by using PFR assay.
To test whether the N and C terminus compete for internal binding to AD, the p53BD or RING domains were deleted from the MDMXc3 construct. The remaining internal binding was analyzed by using PFR assay. The result showed that deleting the p53BD (MDMXc3-∆1-140) did not enhance AD–RING binding (Fig. 3E), and deleting the RING (MDMXc3-∆430-490) had no effect on p53BD–AD binding (Fig. 3F). Therefore, both the N and C terminus may interact with the AD simultaneously through nonoverlapping binding sites.
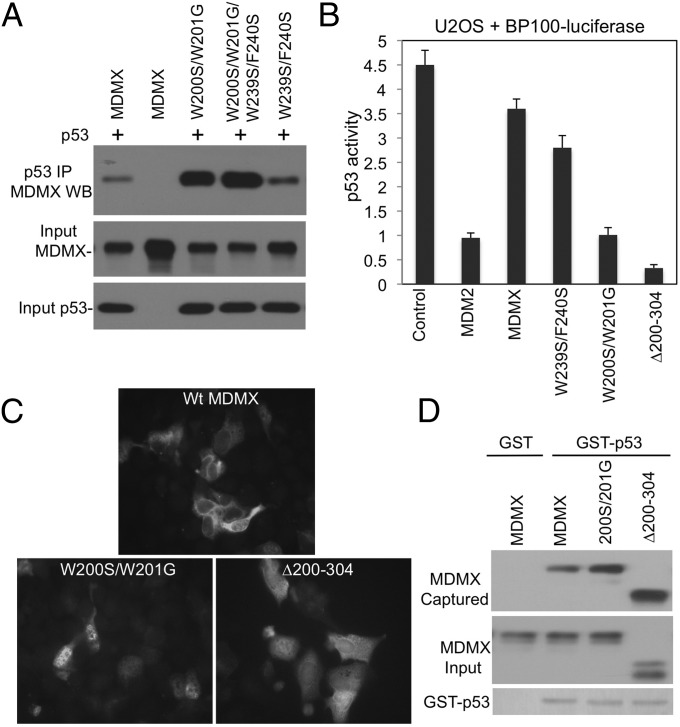
The WW Sequence Regulates p53 Binding and MDMX Localization.
When introduced into full-length MDMX, the SG mutation strongly enhanced p53 binding (Fig. 4A). Unlike MDM2, MDMX is a poor inhibitor of p53 transcriptional activity in the reporter gene assay. However, the SG mutant strongly inhibited p53 activity, similar to AD internal deletion mutant ∆200–304 (Fig. 4B). When the MDMX constructs were transiently expressed in U2OS cells, the SG mutant and ∆200–304 also blocked p53 induction of p21 more efficiently than wild-type MDMX (SI Appendix, Fig. S3A).
Fig. 4.
Regulation of p53 binding and localization by the p53 mimetic sequence. (A) Mutations of the putative p53 mimetic sequences were introduced into full-length MDMX and coexpressed with p53 in H1299 cells. MDMX-p53 binding was analyzed by IP-Western blot. (B) U2OS was cotransfected with p53-response reporter BP100-luciferase and MDMX mutants. Endogenous p53 activity was measured by luciferase assay and normalized to cotransfected CMV-lacZ. (C) U2OS cells were transiently transfected with MDMX mutants and stained for MDMX localization by using 8C6 immunofluorescence. (D) Beads loaded with GST-p53 were incubated with H1299 lysate expressing MDMX mutants. p53 binding to MDMX mutants was determined by Western blot.
MDMX was predominantly located in the cytoplasm (Fig. 4C), which was mediated by a cryptic NLS in the RING domain (16). The SG mutation caused MDMX to accumulate in the nucleus; deletion of the AD also increased MDMX nuclear localization (Fig. 4C). This result suggests that the AD–RING interaction is also involved in regulating MDMX nuclear import by masking the NLS. However, this observation also raised a concern that the increased p53 binding by SG (Fig. 4A) was due to colocalization with p53 in the nucleus. To demonstrate that the SG mutant has increased p53-binding affinity, glutathione beads loaded with GST-p53 were incubated with SG in vitro, confirming the increased binding (Fig. 4D). The MDMX-∆200-304 mutant also showed increased p53 binding in vitro (Fig. 4D), as expected from the loss of the WW sequence. When the 100–361 fragment was coexpressed in U2OS cells, the ability of ∆200–304 to inhibit p53 was partially blocked (SI Appendix, Fig. S3B). This result is consistent with the weak binding of the central domain to the N terminus when acting in trans. Overall, these results indicate that the WW region is important for internal binding to the N and C terminus, autoinhibition of p53 binding, and inhibition of MDMX nuclear import.
Interaction of the WW Sequence with the Hydrophobic Pocket.
To test whether the WW sequence is directly involved in binding to the p53BD, the interaction was analyzed by isothermal titration calorimetry (ITC). The ITC analysis using MDMX-191-209 peptide (EEWDVAGLPWWFLGNLRSN) and p53BD (MDMX-23-111) suggested multiple interactions with KD of 8 µM (presumably from pocket binding) and 340 µM (nonspecific binding) respectively (SI Appendix, Fig. S4 A and C). This binding affinity was ∼16-fold weaker compared with p53-14-28 peptide (SQETFSDLWKLLPE) (KD ∼ 0.5 µM; SI Appendix, Fig. S4 B and C), which was expected for an intramolecular inhibitor that can respond to dynamic regulation.
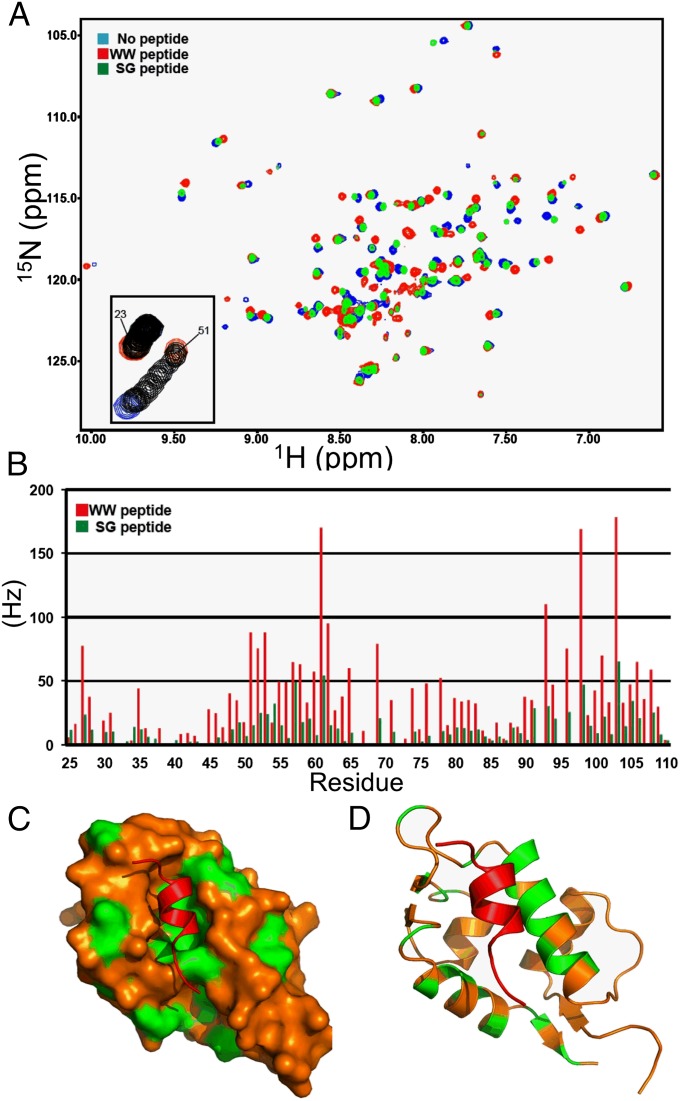
To further determine whether the WW sequence blocks the N-terminal p53-binding pocket, 15N-labeled MDMX-23-111 was incubated with the unlabeled MDMX-191–209 peptide. Analysis of the HSQC NMR spectra (Fig. 5 A and B) by using published assignments showed that the MDMX-191-209 peptide induced chemical shifts on residues localized to the p53-binding pocket (Fig. 5 C and D) (17). The behavior of the resonances during the titration indicates binding of the WW peptide is in the fast exchange regime, with a KD in the range of 10–50 µM, which is close to the KD measured using ITC. As expected, the chemical shifts induced by the SG peptide were significantly reduced but still affected similar residues (Fig. 5B). The NMR results provide direct evidence that the WW peptide interacts with the p53-binding pocket of MDMX, supporting our mutagenesis mapping and the intramolecular p53 mimetic model.
Fig. 5.
Interaction of MDMX WW and SG peptides with the p53-binding pocket. (A) Overlay of 1H-15N HSQC spectra for 15N-MDMX (blue), 15N-MDMX+WW peptide (red), and 15N-MDMX+SG peptide (green). (B) Chemical shift changes for MDMX p53BD residues binding to the WW peptide (red bars) or the SG peptide (green bars). (C) Surface image of the MDMX p53BD structure. The residues that have combined chemical shifts close to or greater than 50 Hz (upon binding the WW peptide) are highlighted in green. (D) Cartoon showing the chemical shifts on the back side of two helices obscured on the surface image.
CK1α Binding Alters MDMX Intramolecular Interactions.
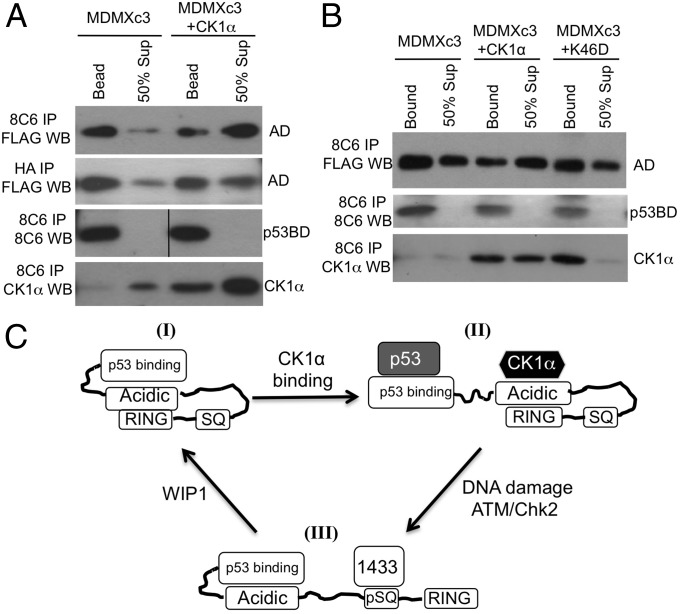
Our previous study suggested that CK1α binds to the MDMX central region and disrupts p53BD–AD interaction to expose the p53-binding pocket. Using the PFR assay, we were able to demonstrate that CK1α expression inhibited p53BD–AD intramolecular binding in full-length MDMX (Fig. 6A). Furthermore, the CK1α that coprecipitated with MDMXc3 was preferentially released into the supernatant after protease cleavage (Fig. 6A), consistent with the interpretation that it’s binding to the AD fragment weakens p53BD–AD binding. In contrast, CK1α only had minor effect on AD–RING interaction (Fig. 6A). The kinase-inactive CK1α-K46D mutant did not inhibit p53BD–AD binding, suggesting that phosphorylation of MDMX by CK1α is required for disrupting p53BD–AD interaction (Fig. 6B). The SG mutation disrupted MDMX binding to CK1α (SI Appendix, Fig. S1B), suggesting that the WW sequence also directly or indirectly participates in binding to CK1α. CK1a binding may conceal the WW sequence, thus preventing its interaction with the N-terminal pocket to enhance the p53-binding affinity. Overall, the results demonstrated the ability of the PFR assay to detect changes in intramolecular interactions by using proteins under physiological regulation.
Fig. 6.
Regulation of MDMX intramolecular interactions by CK1α. (A and B) MDMXc3 was cotransfected with CK1α and CK1α-K46D kinase-dead mutant into U2OS cells, and the changes in intramolecular interactions were analyzed by using PFR assay. (C) A model of MDMX intramolecular interactions. MDMX can assume multiple conformational states. I, a closed state of weak p53 binding and cytoplasmic localization due to intramolecular interactions. II, interaction with CK1α opens the N terminus for p53 binding. III, DNA damage recruits 14-3-3 and disrupts binding to CK1α, inhibits the N terminus, and exposes the RING to mediate nuclear import and heterodimerization with MDM2. ATM/Chk2 and WIP1-mediated phosphorylation and dephosphorylation alter the balance between the conformational states.
Discussion
Although the detailed mechanism of p53 inhibition by MDMX is still unclear, previous studies using p53-mimetic peptides and small molecule disruptors showed that MDMX-p53 binding is a critical step for p53 inactivation (18, 19). MDMX-p53 binding is stimulated by CK1α, which is a major binding partner of MDMX. DNA damage inhibits MDMX-p53 binding through a mechanism that involves the disruption of MDMX–CK1α interaction (13), further enhancing p53 activity. Our previous findings and the results from this report together reveal that MDMX autoinhibition is an important mechanism for linking p53 binding to DNA damage response. The model suggests that the MDMX central AD normally interacts with the N terminus to inhibit p53 binding (Fig. 6C). CK1α disrupts this intramolecular interaction, stimulating N-terminal binding to p53. DNA damage inhibits the MDMX–CK1α interaction (13), thus enhancing the MDMX internal binding to conceal the p53-binding pocket.
Given the low affinity between the MDMX N terminus and the AD, it was unclear from the previous study how frequently MDMX adopts the closed conformation. In this report, we unambiguously detected the preexisting intramolecular interactions in MDMX isolated from mammalian cells. By cleaving full-length MDMX into multiple domains and observing the slow dissociation of fragments over the time scale of minutes to hours, the PFR assay revealed that >25–50% of the MDMX N terminus is associated with the AD, explaining the strong autoinhibitory effect on p53 binding. We identified a p53-mimetic sequence critical for the intramolecular binding and autoinhibition. NMR and ITC analyses confirmed that the WW peptide binds to the N-terminal hydrophobic pocket in a manner similar to p53, but with ∼16-fold lower affinity. Its strong functional impact is likely due to a combination of direct binding between the p53BD and the AD as well as a significant contribution from the intramolecular linkage that increases avidity. Although the affinity between the N terminus and AD is low (Kd ∼ 8 µM), the slow dissociation of fragments after cleavage of MDMX suggests that conformational changes (induced fit) may have occurred during the intramolecular interaction.
Our analysis also detected a previously unidentified AD–RING internal binding in MDMX. We recently showed that the AD–RING internal interaction in MDM2 activates the ubiquitin E3 ligase activity of the RING domain (20). Here we found that in MDMX, the AD–RING binding is involved in suppressing nuclear translocation, presumably by concealing the NLS in the RING domain (16). Whether AD–RING interaction has other functions in MDMX remains to be determined. It is possible that by simultaneously interacting with the AD, the N- and C-terminal domains allosterically communicate to coordinate p53 regulation and stress response. MDMX has been shown to stimulate MDM2 E3 activity by forming a heterodimer. It remains to be determined whether MDMX AD–RING interaction also stimulates the E3 activity of the heterodimer. RING–AD internal interaction appears to be a common feature of MDM2 and MDMX, whereas p53BD–AD interaction and the WW motif are unique to MDMX.
A publication by Bista et al. also discovered the autoinhibitory WW sequence by comparing the chemical shift of MDMX N-terminal domain and full-length MDMX in NMR analysis (21). In their study, the significant change of N-terminal conformation in the presence of the AD suggests that a large fraction of MDMX engages in intramolecular binding. This result is consistent with the significant internal binding observed by using the PFR assay. Therefore, both studies reveal that MDMX predominantly adopts an inactive conformation for p53 binding. Our results also shed light on a puzzling observation that full-length MDMX is a poor inhibitor of p53 in reporter gene assays, whereas the MDMX-S alternative splice product without the central and C-terminal domains is a strong inhibitor of p53 and its expression in cancer correlates with poor prognosis (22).
Currently, therapeutic targeting of MDM2 and MDMX mainly focuses on disrupting binding to p53 (23). Given that MDMX contains an intrinsic inhibitory domain, identifying molecules that strengthen the autoinhibitory interaction may also inhibit MDMX binding to p53 and provide new leads for drug development. For unknown reasons, targeting of MDMX–p53 interaction using small molecules has been largely unsuccessful, whereas several potent MDM2 inhibitors have entered clinical trials. The discovery of MDMX autoinhibition may provide clues to this conundrum, suggesting that full-length MDMX rather than the p53-binding domain should be used as a target in drug screening.
Atomic structures have been obtained for the MDMX N-terminal domain and RING domain in isolation (24, 25). Our results suggest that in the context of the full-length protein, these domains are often present in complex with the central domain. Therefore, structural analysis of MDMX using individual domains does not capture important conformational states of the full-length protein. Dynamic intramolecular interaction is important for the function and regulation of many proteins. Certain long-range allosteric effects may also involve dynamic intramolecular interactions to cause distant conformational change without a rigid structural path (26). The PFR assay described here may facilitate the study of such multidomain proteins and unstructured regions.
Materials and Methods
Plasmids and Cell Lines.
Point mutants of human MDMX were generated by site-directed mutagenesis using the QuikChange kit (Stratagene). The cDNA-encoding MDMXc3 was produced by gene synthesis (Genscript) and cloned into pcDNA3 vector. MDMXc3 contains LEVLFQGPDYKDDDDK, LEVLFQGPEEQKLISEEDL, and LEVLFQGPYPYDVPDYA inserted after MDMX residues 140, 350, and 429, respectively. GST-PreScission protease fusion was purified from Escherichia coli by using glutathione agarose column.
Proteolytic Fragment Release Assay.
H1299 or U2OS cells were transiently or stably transfected with MDMXc3 by using calcium phosphate precipitation protocol. Cells were lysed by using IP buffer [150 mM NaCl, 50 mM Tris⋅HCl pH 8.0, 0.5% NP50, 2 mM NaF, 0.5 mM DTT, 10% (vol/vol) glycerol]. Cell lysate (1 mL) from ∼2 × 106 cells were immunoprecipitated by using 20 µL of packed protein A beads with chemically cross-linked 8C6, FLAG, Myc, or HA mouse monoclonal antibodies for 2 h at 4 °C. The beads were washed two times with PreScission buffer (150 mM NaCl, 10 mM Hepes pH 7.5, 0.5 mM DTT, 10% glycerol) and suspended in 100 µL of PreScission buffer. PreScission protease was added to 0.2 µg/µL, and the beads were incubated at 4 °C with shaking for 5–20 min. The protease digestion mixture was centrifuged for 10 s, and the beads (bound material) and supernatant (released material) were separated. The beads were washed once with PreScission buffer. The beads and supernatant were boiled in Laemmli buffer, and analyzed by SDS/PAGE and Western blot by using affinity-purified rabbit anti-FLAG, Myc, or HA antibodies to determine the bound/released ratio of each fragment. Monoclonal antibodies 3G9, 4B2, and 4B11 were also used for detection of different MDM2 fragments (27).
GST Pull Down, ITC, Protein Expression, and NMR Analysis.
Experimental procedures for GST pull down, ITC, and NMR are described in SI Appendix, SI Materials and Methods.
Supplementary Material
Acknowledgments
This work has been supported in part by the Chemical Biology Core Facility at the H. Lee Moffitt Cancer Center & Research Institute, a National Cancer Institute-designated Comprehensive Cancer Center (P30-CA076292). This work is also supported by National Institutes of Health Grants CA141244 and CA109636 and Florida Department of Health Grant 4BB14 (to J.C.) and a National Science Foundation Grant MCB-0939014 (to G.W.D.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1420833112/-/DCSupplemental.
References
- 1.Vousden KH, Lane DP. p53 in health and disease. Nat Rev Mol Cell Biol. 2007;8(4):275–283. doi: 10.1038/nrm2147. [DOI] [PubMed] [Google Scholar]
- 2.Grier JD, Xiong S, Elizondo-Fraire AC, Parant JM, Lozano G. Tissue-specific differences of p53 inhibition by Mdm2 and Mdm4. Mol Cell Biol. 2006;26(1):192–198. doi: 10.1128/MCB.26.1.192-198.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Li B, Cheng Q, Li Z, Chen J. p53 inactivation by MDM2 and MDMX negative feedback loops in testicular germ cell tumors. Cell Cycle. 2010;9(7):1411–1420. doi: 10.4161/cc.9.7.11255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Terzian T, et al. Haploinsufficiency of Mdm2 and Mdm4 in tumorigenesis and development. Mol Cell Biol. 2007;27(15):5479–5485. doi: 10.1128/MCB.00555-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wang YV, Leblanc M, Wade M, Jochemsen AG, Wahl GM. Increased radioresistance and accelerated B cell lymphomas in mice with Mdmx mutations that prevent modifications by DNA-damage-activated kinases. Cancer Cell. 2009;16(1):33–43. doi: 10.1016/j.ccr.2009.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kawai H, et al. DNA damage-induced MDMX degradation is mediated by MDM2. J Biol Chem. 2003;278(46):45946–45953. doi: 10.1074/jbc.M308295200. [DOI] [PubMed] [Google Scholar]
- 7.Pan Y, Chen J. MDM2 promotes ubiquitination and degradation of MDMX. Mol Cell Biol. 2003;23(15):5113–5121. doi: 10.1128/MCB.23.15.5113-5121.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chen L, Gilkes DM, Pan Y, Lane WS, Chen J. ATM and Chk2-dependent phosphorylation of MDMX contribute to p53 activation after DNA damage. EMBO J. 2005;24(19):3411–3422. doi: 10.1038/sj.emboj.7600812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gilkes DM, Chen L, Chen J. MDMX regulation of p53 response to ribosomal stress. EMBO J. 2006;25(23):5614–5625. doi: 10.1038/sj.emboj.7601424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Li X, et al. Abnormal MDMX degradation in tumor cells due to ARF deficiency. Oncogene. 2012;31(32):3721–3732. doi: 10.1038/onc.2011.534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Francoz S, et al. Mdm4 and Mdm2 cooperate to inhibit p53 activity in proliferating and quiescent cells in vivo. Proc Natl Acad Sci USA. 2006;103(9):3232–3237. doi: 10.1073/pnas.0508476103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chen L, Li C, Pan Y, Chen J. Regulation of p53-MDMX interaction by casein kinase 1 alpha. Mol Cell Biol. 2005;25(15):6509–6520. doi: 10.1128/MCB.25.15.6509-6520.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wu S, Chen L, Becker A, Schonbrunn E, Chen J. Casein kinase 1α regulates an MDMX intramolecular interaction to stimulate p53 binding. Mol Cell Biol. 2012;32(23):4821–4832. doi: 10.1128/MCB.00851-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pufall MA, Graves BJ. Autoinhibitory domains: Modular effectors of cellular regulation. Annu Rev Cell Dev Biol. 2002;18:421–462. doi: 10.1146/annurev.cellbio.18.031502.133614. [DOI] [PubMed] [Google Scholar]
- 15.Li X, Romero P, Rani M, Dunker AK, Obradovic Z. Predicting protein disorder for N-, C-, and internal regions. Genome Inform Ser Workshop Genome Inform. 1999;10:30–40. [PubMed] [Google Scholar]
- 16.LeBron C, Chen L, Gilkes DM, Chen J. Regulation of MDMX nuclear import and degradation by Chk2 and 14-3-3. EMBO J. 2006;25(6):1196–1206. doi: 10.1038/sj.emboj.7601032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sanchez MC, Renshaw JG, Davies G, Barlow PN, Vogtherr M. MDM4 binds ligands via a mechanism in which disordered regions become structured. FEBS Lett. 2010;584(14):3035–3041. doi: 10.1016/j.febslet.2010.05.058. [DOI] [PubMed] [Google Scholar]
- 18.Graves B, et al. Activation of the p53 pathway by small-molecule-induced MDM2 and MDMX dimerization. Proc Natl Acad Sci USA. 2012;109(29):11788–11793. doi: 10.1073/pnas.1203789109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chang YS, et al. Stapled α-helical peptide drug development: A potent dual inhibitor of MDM2 and MDMX for p53-dependent cancer therapy. Proc Natl Acad Sci USA. 2013;110(36):E3445–E3454. doi: 10.1073/pnas.1303002110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cheng Q, Song T, Chen L, Chen J. Autoactivation of the MDM2 E3 ligase by intramolecular interaction. Mol Cell Biol. 2014;34(15):2800–2810. doi: 10.1128/MCB.00246-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bista M, Petrovich M, Fersht AR. MDMX contains an autoinhibitory sequence element. Proc Natl Acad Sci USA. 2013;110(44):17814–17819. doi: 10.1073/pnas.1317398110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lenos K, et al. Alternate splicing of the p53 inhibitor HDMX offers a superior prognostic biomarker than p53 mutation in human cancer. Cancer Res. 2012;72(16):4074–4084. doi: 10.1158/0008-5472.CAN-12-0215. [DOI] [PubMed] [Google Scholar]
- 23.Wade M, Li YC, Wahl GM. MDM2, MDMX and p53 in oncogenesis and cancer therapy. Nat Rev Cancer. 2013;13(2):83–96. doi: 10.1038/nrc3430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Linke K, et al. Structure of the MDM2/MDMX RING domain heterodimer reveals dimerization is required for their ubiquitylation in trans. Cell Death Differ. 2008;15(5):841–848. doi: 10.1038/sj.cdd.4402309. [DOI] [PubMed] [Google Scholar]
- 25.Phan J, et al. Structure-based design of high affinity peptides inhibiting the interaction of p53 with MDM2 and MDMX. J Biol Chem. 2010;285(3):2174–2183. doi: 10.1074/jbc.M109.073056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hilser VJ, Wrabl JO, Motlagh HN. Structural and energetic basis of allostery. Ann Rev Biophys. 2012;41:585–609. doi: 10.1146/annurev-biophys-050511-102319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chen J, Marechal V, Levine AJ. Mapping of the p53 and mdm-2 interaction domains. Mol Cell Biol. 1993;13(7):4107–4114. doi: 10.1128/mcb.13.7.4107. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.