Significance
In 2014, Ebola virus became a household term. The ongoing outbreak in West Africa is the largest Ebola virus outbreak ever recorded, with over 20,000 cases and over 8,000 deaths to date. Very little is known about the human cellular immune response to Ebola virus infection, and this lack of knowledge has hindered development of effective therapies and vaccines. In this study, we characterize the human immune response to Ebola virus infection in four patients. We define the kinetics of T- and B-cell activation, and determine which viral proteins are targets of the Ebola virus-specific T-cell response in humans.
Keywords: Ebola infection, human immune response, immune activation, plasmablasts, T cells
Abstract
Four Ebola patients received care at Emory University Hospital, presenting a unique opportunity to examine the cellular immune responses during acute Ebola virus infection. We found striking activation of both B and T cells in all four patients. Plasmablast frequencies were 10–50% of B cells, compared with less than 1% in healthy individuals. Many of these proliferating plasmablasts were IgG-positive, and this finding coincided with the presence of Ebola virus-specific IgG in the serum. Activated CD4 T cells ranged from 5 to 30%, compared with 1–2% in healthy controls. The most pronounced responses were seen in CD8 T cells, with over 50% of the CD8 T cells expressing markers of activation and proliferation. Taken together, these results suggest that all four patients developed robust immune responses during the acute phase of Ebola virus infection, a finding that would not have been predicted based on our current assumptions about the highly immunosuppressive nature of Ebola virus. Also, quite surprisingly, we found sustained immune activation after the virus was cleared from the plasma, observed most strikingly in the persistence of activated CD8 T cells, even 1 mo after the patients’ discharge from the hospital. These results suggest continued antigen stimulation after resolution of the disease. From these convalescent time points, we identified CD4 and CD8 T-cell responses to several Ebola virus proteins, most notably the viral nucleoprotein. Knowledge of the viral proteins targeted by T cells during natural infection should be useful in designing vaccines against Ebola virus.
Ebola virus is a member of the Filoviridae family, which are filamentous, negative-stranded RNA viruses that are known to cause severe human disease (1). An ongoing outbreak of Ebola virus in West Africa has brought this virus and the disease it causes (Ebola virus disease; EVD) to the forefront. The World Health Organization has reported over 20,000 cases and 8,000 deaths in West Africa, with Sierra Leone, Guinea, and Liberia the most affected.
Our knowledge of the human immune response to Ebola virus has been severely limited due to the lack of infrastructure to perform such analyses in high containment levels (biosafety level 4; BSL-4). Minimal data exist regarding the human cellular immune response during acute Ebola virus infection, which indicate that aberrant cytokine responses (2–6), decreased CD4 and CD8 T cells, and increased CD95 expression on T cells are all associated with fatal outcomes (4). In vivo studies have revealed an association between apoptosis of lymphocytes and fatal outcome (3), and lymphocyte apoptosis has been seen both in vitro in infected human cells and in vivo in mouse and nonhuman primate models (7–9).
The natural serologic response to Ebola virus infection has been well-characterized, with specific IgM responses detected as early as 2 d after symptom onset but generally occurring 10–29 d after symptom onset in most patients. Ebola virus-specific IgG responses have been detected as early as 6 d post symptom onset, occurring ∼19 d after symptom onset in most individuals (10, 11). Serological responses to Ebola virus have been reported as absent or diminished in fatal cases; however, sample sizes have been very limited (3).
Data from in vitro studies have demonstrated that Ebola virus-infected dendritic cells are impaired in their ability to produce cytokines and activate autologous T cells (12), whereas infected macrophages exhibit impaired maturation (13). Ebola virus also encodes several proteins that can interfere with the innate immune response in infected cells (14). These in vitro studies, combined with the limited human data showing T-cell apoptosis, lymphopenia, and absent antibody responses in fatal cases, have led to the assumption that Ebola virus infection is immunosuppressive.
Here we examine the immune responses of four survivors of EVD who received care at Emory University Hospital. This first look, to our knowledge, at the human adaptive immune response during the acute phase of Ebola virus infection shows striking levels of T- and B-cell activation in all four patients.
Results
Analysis of Human Plasmablasts and Activated T Cells During Acute Ebola Virus Infection.
Between August and October of 2014, four patients with EVD received care at Emory University Hospital in the Serious Communicable Diseases Unit. We had the unique opportunity to evaluate the cellular and humoral immune responses during acute and convalescent disease phases in these patients. The clinical course of two of these cases has been described elsewhere (15). The four patients, EVD2, 5, 9, and 15, presented for care 12, 15, 5, and 2 d after self-reported onset of symptoms, respectively. EVD2 and 5 had moderate disease, EVD9 had severe disease, and EVD15 had mild disease.
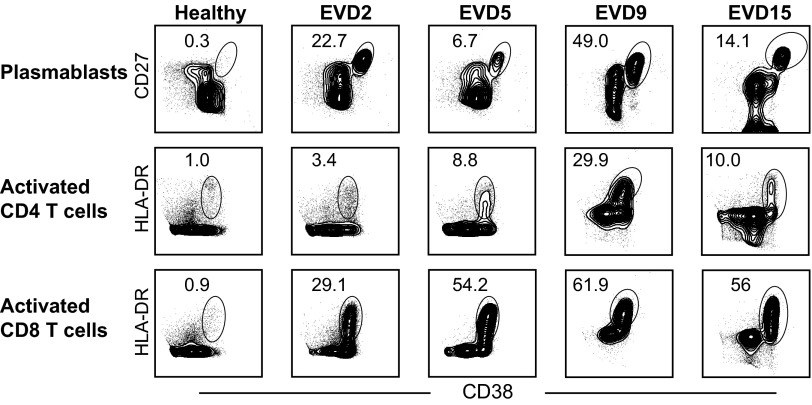
Initial studies focused on determining the frequency of activated T and B cells using phenotypic markers that have previously been defined in humans following infection or vaccination (16–19). CD4 and CD8 T cells were analyzed for their coexpression of the activation markers HLA-DR and CD38. Antibody-secreting cells (ASCs; plasmablasts) were defined by their expression of CD27 and CD38 on CD19+ cells. Representative flow plots for each cell type examined from each patient are depicted in Fig. 1. Compared with healthy controls, all four patients demonstrated increased numbers of plasmablasts and activated CD4 and CD8 T cells during infection. Striking frequencies of plasmablasts were seen in all patients, with up to 50% of CD19+ cells expressing CD27 and CD38. Activated CD4 and CD8 T cells were also seen in all patients, with 30–60% of the CD8 T cells expressing activation markers.
Fig. 1.
Representative flow cytometry plots of plasmablasts and activated CD4 and CD8 T-cell responses in Ebola patients. Samples from a healthy donor and from each Ebola virus-infected patient are depicted. EVD patient samples were obtained during the acute phase of disease: samples from EVD2 were obtained 16 d after symptom onset; from EVD5 at day 17; from EVD9 at day 17; and from EVD15 at day 3.
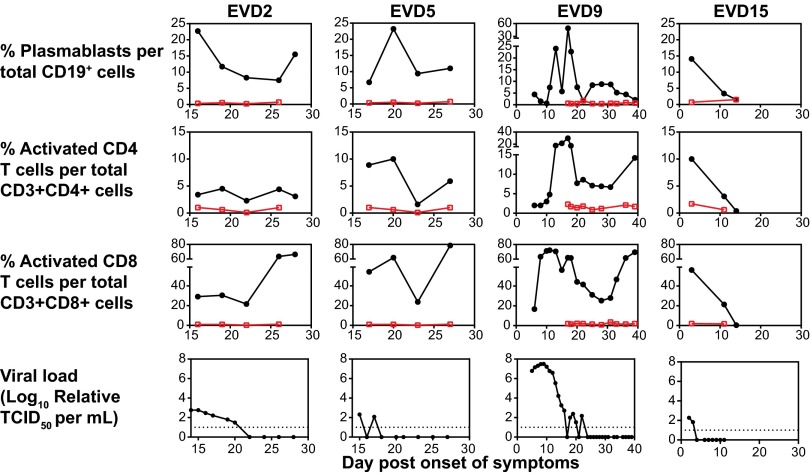
The kinetics of these activated responses were measured in all patients. Peak plasmablast frequencies were observed between 2 and 3 wk after onset of symptoms in EVD2, 5, and 9 (Fig. 2). EVD15, who had very mild disease that rapidly resolved, developed elevated plasmablast levels 3 d after onset of symptoms, but never to the same magnitude as the more severely ill patients (Fig. 2). Interestingly, activated CD8 T-cell levels remained elevated in EVD2, 5, and 9 even at the time of discharge from the hospital. In contrast, activated CD8 and CD4 T-cell levels in EVD15 were increased early on in the course of illness but declined to baseline as the illness rapidly resolved. Additionally, the moderate and severely affected patients also had an initial peak of activated CD4 and CD8 T cells followed by a second peak, likely representing the return of the tissue-based T cells to the peripheral blood after viral control in the affected tissues was achieved. Viral load in EVD2, 5, and 9 began to decline, or was noted to already be declining (in EVD2 and 5, whose care at the Emory University Hospital began during their second week of illness), coincidently with the presence of elevated levels of activated T and B cells (Fig. 2).
Fig. 2.
Kinetics of plasmablasts, activated CD4 and CD8 T-cell responses, and viral load in EVD patients. Percentages of plasmablasts (CD27+CD38+CD19+), activated CD4 T cells (HLA-DR+CD38+CD3+CD4+), activated CD8 T cells (HLA-DR+CD38+CD3+CD8+), and viral load are graphed as a function of time. Responses from a healthy control are shown in red. The dotted lines represent the limit of detection for the viral load assay. TCID50, 50% tissue culture infective dose.
Phenotyping Activated CD8 T Cells.
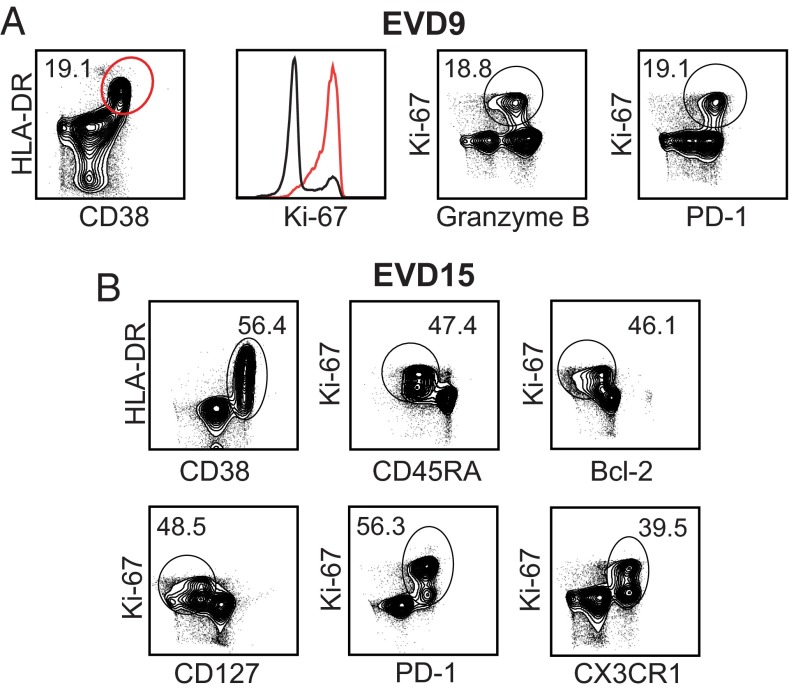
Our initial measurements defined CD8 T-cell populations that were phenotypically consistent with activation. To further define them as recently activated effector cells, we performed additional stains on select samples from EVD9 and 15. The majority of HLA-DR+/CD38+ CD8 T cells expressed Ki-67, identifying them as actively proliferating (Fig. 3A, red). Additionally, similar frequencies of HLA-DR+/CD38+ (19.1%), Ki-67+/granzyme B+ (18.8%), and Ki-67+/PD-1+ (19.1%) cells were seen in the CD8 T-cell compartment, consistent with an activated, proliferating effector phenotype (Fig. 3A). The expression of granzyme B in these cells defines them as cytotoxic, and the expression of the inhibitory receptor PD-1 is consistent with restraint of the magnitude of the CD8 T-cell response. The Ki-67+ CD8 T cells that were observed in these patient samples were CD45RAlow, Bcl-2low, CD127low, and PD-1high, all phenotypes consistent with antigen activation via the T-cell receptor as opposed to bystander activation (16) (Fig. 3B). Additionally, the activated CD8 T cells were also expressing CX3CR1, the fracktalkine receptor, indicating that these cells could home to sites of inflammation (Fig. 3B). These data all support the presence of a strong CD8 T-cell response in EVD patients that is characterized by proliferation and cytotoxic function.
Fig. 3.
Phenotyping activated CD8 T cells in EVD patients. The expression of various effector cell markers on CD3+CD8+ T cells from EVD9 on day 31 after onset of symptoms (A) and in EVD15 3–9 d after onset of symptoms (B). The Ki-67 histogram is derived from the population depicted in red in the first plot.
Sustained Activation of T-Cell Responses in EVD Patients.
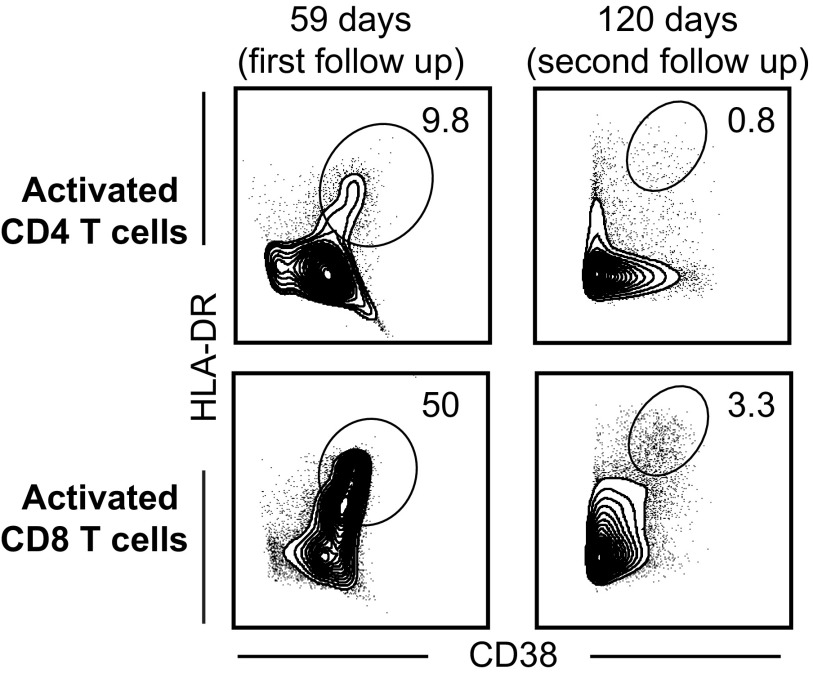
All four patients were seen in follow-up appointments after discharge from the hospital, and the same analyses were performed on blood samples obtained during the convalescent phase. Representative flow plots for EVD2, who was seen twice in follow-up, are depicted in Fig. 4. Significant numbers of activated CD4 and CD8 T cells were still present in EVD2’s peripheral blood at the first follow-up visit but not at the second follow-up. Similar levels were seen in EVD5 at the first follow-up appointment (Fig. S1). In both cases, the high levels of activated CD8 and CD4 T cells suggest ongoing antigen stimulation via the T-cell receptor. In contrast, activated T-cell levels of EVD9 and 15 were almost at baseline at their first follow-up visits (Fig. S1). However, for EVD9, the first follow-up appointment was 71 d after symptom onset, which is later in the disease course than the follow-up visits for the other patients. In contrast, EVD15, who had mild disease with rapid resolution of viremia, had low numbers of activated CD4 and CD8 T-cell populations even at discharge, and these levels remained low at the first follow-up visit.
Fig. 4.
CD4 and CD8 T-cell responses in convalescent EVD patients. Representative flow plots from EVD2 at two time points after discharge (first follow-up was 1 mo post discharge and the second follow-up was 3 mo post discharge) from the hospital demonstrating activated CD4 and CD8 T-cell populations. Days noted in all panels are days post symptom onset.
Development of Humoral Immunity in EVD Patients.
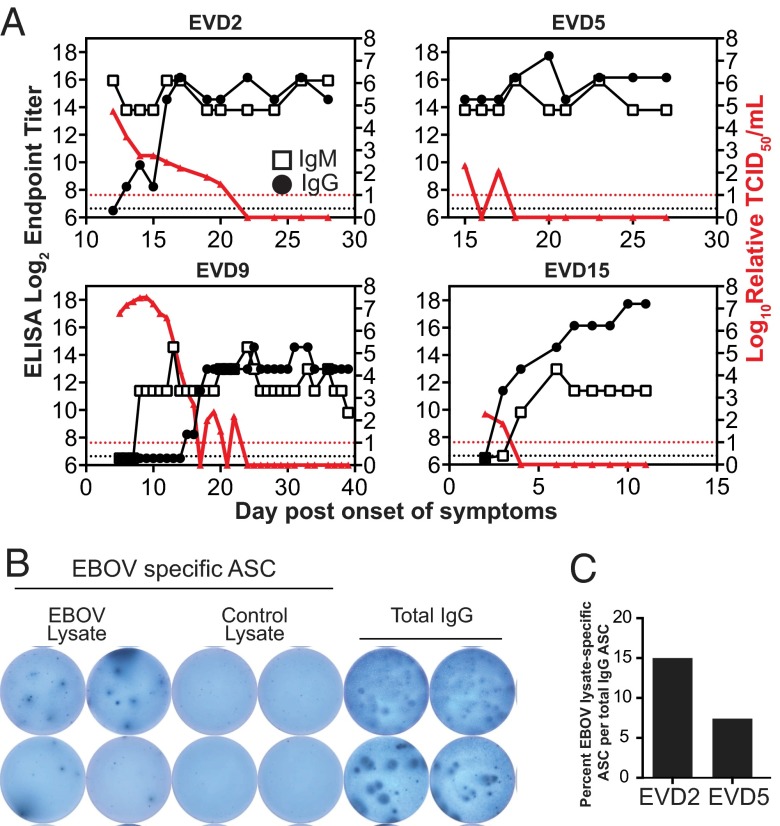
Serologic responses to the viral nucleoprotein were measured by ELISA. All patients developed peak IgG responses around 2–3 wk after symptom onset, consistent with their plasmablast responses (Fig. 5A). EVD5’s care at Emory began during the second week of illness; upon arrival, the patient had already developed both IgM and IgG responses. EVD2 and 9 demonstrated the classic kinetics of an IgM response before the IgG response. However, EVD15 had detectable IgM and IgG responses at about the same time. The frequencies of plasmablasts expressing IgA, IgM, and IgG were measured in select samples by intracellular staining of CD19+/CD27+/CD38+ cells. These samples demonstrated predominantly IgG-positive plasmablasts (data not shown).
Fig. 5.
B-cell responses during acute and convalescent EVD. (A) Nucleoprotein-specific antibody titers using ELISA and viral load are graphed as a function of time. Dotted lines represent the limit of detection for each assay. Open boxes represent IgM titers, filled black circles represent IgG titers, and filled red triangles indicate viral load. (B) A representative image of an ELISpot assay to detect Ebola virus (EBOV)-specific plasmablasts from a sample obtained from EVD2 at the first follow-up visit (59 d post onset of symptoms). (C) Graphical representation of antigen-specific ASCs, obtained from EVD2 (at day 59) and EVD5 (at day 60) at their first follow-up visit, as a percentage of total IgG-secreting cells. Whole-cell lysate of Ebola virus-infected Vero-E6 cells was used as antigen.
Plasmablast frequencies were very high in all patients during the acute phase of illness but had returned to baseline levels at the follow-up visits (Fig. S2). ELISpot assays were performed to quantitate Ebola virus-specific antibody-secreting cells at the first follow-up visits (approximately 2 mo post symptom onset). The data for EVD2 are shown in Fig. 5B, which shows plasmablasts clearly producing IgG that recognize the Ebola virus lysate but not a control lysate; total IgG-secreting cells are also depicted in this figure. Strikingly, both EVD2 and 5 still had 5–15% Ebola virus-specific plasmablasts at the time of their first follow-up visit (Fig. 5C). EVD2 and 5 both had plasmablasts that specifically secreted antibodies that bound to whole-cell lysate from Ebola virus-infected cells (Fig. 5C) and purified viral nucleoprotein (NP) (Fig. S3). The magnitude of the antigen-specific IgG responses was significantly higher when whole-cell lysate was used rather than NP alone, suggesting multiple viral antigen targets. Of note, whereas up to 15% of the plasmablasts were Ebola virus-specific, there was still a significant proportion of the IgG-producing plasmablasts that was not antigen-specific, suggesting that there may be polyclonal B-cell activation that is mediated by the inflammatory response. However, the time point at which these assays were performed was in convalescence (approximately 2 mo post symptom onset) and, at this time, only 3% of the CD19-positive cells are plasmablasts (Fig. S1). We suspect that during the acute phase, when up to 50% of the CD19+ cells were plasmablasts, the frequency of Ebola-specific plasmablasts would have been much higher.
Defining the Ebola Virus Proteins Targeted by Human T Cells.
To evaluate cytokine production and the breadth of the Ebola virus-specific T-cell response, we used a library of overlapping peptides encompassing all viral proteins except the L protein. The peptides were organized into eight pools, with a single pool for each of the viral proteins VP24, VP30, VP35, and VP40 and two pools each for NP and glycoprotein (GP). The peptide pools were used for in vitro stimulation of peripheral blood mononuclear cells (PBMCs) isolated from patients at the time of discharge (EVD2) or during postdischarge follow-up visits (EVD5 and 9). We then measured IFN-γ, TNF-α, and IL-2 production by CD8 and CD4 T cells using intracellular cytokine staining (Fig. 6).
Fig. 6.
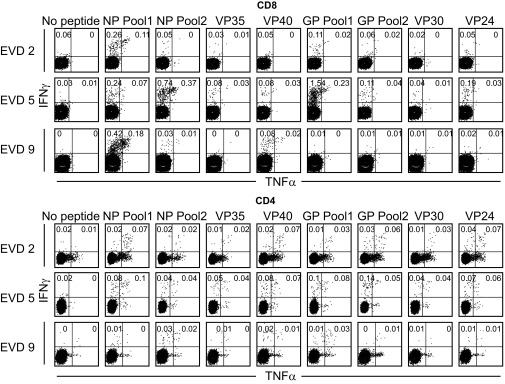
Antigen specificity of CD4 and CD8 T cells in EVD patients. Flow cytometry plots showing IFN-γ and TNF-α expression in response to stimulation with the indicated peptide pools from Ebola viral proteins. Assays were performed on PBMCs obtained from EVD2, EVD5, and EVD9 at 28, 144, and 71 d post onset of symptoms, respectively.
All three patients had readily detectable Ebola-specific CD8 T-cell responses. The response to NP peptides was dominant in all three patients. Epitopes were located in varying regions of the NP, as the response to the peptide pool spanning the N terminus was dominant in EVD2 and 9 (0.37% and 0.6% IFN-γ+ CD8, respectively), whereas the pool corresponding to the C terminus elicited a higher response in EVD5 (1.11% IFN-γ+ CD8). Reaction to GP peptides was more variable; the reaction was equally strong to GP as to NP in EVD5 (1.8% IFN-γ+ CD8), intermediate in EVD2, and absent in EVD9. VP40-specific CD8 T cells were also detected in all three patients, albeit at much lower frequencies (<0.2% IFN-γ+ CD8). The largest fraction of responding CD8 T cells were IFN-γ single producers (Fig. S4), whereas a smaller proportion was TNF-α/IFN-γ double producers (Fig. 6). Only a small frequency of the IFN-γ–producing CD8 T cells also produced IL-2 (Fig. S5).
CD4 T-cell responses were also detected in all of the patients tested. These responses were lower in magnitude compared with the CD8 T-cell responses but were much more diverse in terms of antigenic specificity. At least six of the eight peptide pools elicited measurable IFN-γ production in CD4 T cells from EVD2 and 5, whereas EVD9 had considerably lower frequencies of responding CD4 T cells overall. Among the CD4 T cells positive for IFN-γ, a substantial fraction was capable of also producing TNF-α (Fig. 6) and IL-2 (Fig. S5).
Together, these data support the notion that a strong antigen-specific T-cell response develops against Ebola virus during infection. The significance of this response as it relates to patient outcome remains to be determined; however, future T cell-based vaccine designs may benefit from including the NP antigen, because we have demonstrated that it is a major T-cell target during human infection.
Discussion
Caring for the EVD patients in the Emory University Hospital Serious Communicable Diseases Unit presented a unique opportunity to study the cellular immune responses to Ebola virus and to map the kinetics and specificity of these responses. The close proximity of the Centers for Disease Control and Prevention (CDC) and its BSL-4 facilities enabled us to perform these studies, which had previously been logistically impractical. We noted robust T- and B-cell activation during Ebola virus infection in these four patients. Ebola virus has been believed to be immunosuppressive, as inhibition of the IFN response (14, 20), inhibition of maturation and/or activation of infected antigen-presenting cells (13), and an overall lymphopenia and lymphocyte apoptosis (3, 21) had been noted previously in infected patients. Although three of the patients in our study did develop a marked lymphopenia, a significant proportion of their lymphocytes was activated, suggesting that these patients mounted a potent adaptive immune response to Ebola virus.
Phenotyping of CD8 T cells demonstrated that they were not only proliferating effector cells but also expressed surface markers consistent with activation via the T-cell receptor (HLA-DR+, CD38+, Bcl-2low, CD127low, granzyme Bhigh, and PD-1high) rather than a phenotype consistent with nonspecific bystander activation. The high level of PD-1 expression suggests that Ebola virus-specific CD8 T cells are being restrained by the PD-1 inhibitory pathway (22).
Plasmablast population peaks coincided with the generation of Ebola virus-specific IgG, demonstrating a robust humoral response in all patients. Phenotyping of the plasmablast population also showed that the majority of the plasmablasts were IgG+, consistent with the response expected and demonstrated in other acute viral infections (23). It is also worth noting that Ebola virus-specific plasmablasts were present in the blood even 2 mo after symptom onset.
In the patients studied here with EVD, high percentages of activated CD8 and CD4 T cells were observed up to 60 d after symptom onset. This finding was unexpected and is in contrast to the activated CD8 T-cell responses that have been observed in other acute viral infections, in which the percentages of activated T cells return to baseline much faster (16, 19). This implies the persistence of viral antigen that continually stimulated these responses. Viral antigen is most likely persisting in compartments other than the blood, because all patients had undetectable levels of viral RNA in their blood by 23 d after symptom onset. It is worth noting that an EVD patient who received care in Germany had detectable viral RNA in urine and sweat even after the plasma was negative for viral RNA (24), suggesting that Ebola virus antigen persistence is possible in at least these locations. The fact that prolonged activation was only seen in the most ill patients is consistent with much higher levels of viral antigen over a much longer time period. In marked contrast to our patients with moderate and severe disease, EVD15, who had mild disease, rapidly cleared viremia accompanied by an equally rapid contraction of activated T-cell populations.
Also of note is the fact that the mildly affected patient, EVD15, did not have two peaks of activated CD4 and CD8 T cells. This phenomenon was noted in the other three patients, and likely represents the return of the tissue-based T cells to the peripheral blood after control of the infection in the most affected tissues was achieved. Prolonged antigen expression in the affected tissues, most likely the liver, could have also contributed to the prolonged responses that were noted in the three more clinically affected patients.
As a first step toward defining the viral antigen targets of the human T-cell response to Ebola virus, we measured T-cell cytokine production in response to various pools of peptides derived from Ebola virus antigens. The strongest responses were CD8 T cell-mediated and directed against the Ebola virus NP, which is not surprising given that this is the most highly expressed protein in infected cells. All patients had low-level detectable CD8 T-cell responses against VP40, the viral matrix protein. Only two of the three EVD patients tested had detectable CD8 T-cell responses against the viral GP. Two Ebola virus vaccine candidates are being evaluated in phase 1 trials for human use (25), and both express only viral GP. One of these vaccine candidates, the cAd3-EBO vaccine, is aimed at generating T-cell responses, which correlate with protection induced by this vaccine in the nonhuman primate model (26). Our data could inform future vaccine design, and suggest that NP should be targeted in addition to GP to generate the most robust Ebola virus-specific human CD8 T-cell responses.
It is worth noting that these Ebola patients received experimental therapeutic interventions (Table S1). ZMapp (27) was given to EVD2 and 5, EVD9 received an siRNA product against Ebola virus (28), EVD15 received a DNA polymerase inhibitor, and convalescent serum was given to EVD9 and 15. It is unknown whether any of these experimental interventions impacted the course of disease in these patients. It is also not clear whether any of these treatments modified the immune response to Ebola virus. It remains plausible that infusion of ZMapp, a monoclonal antibody preparation directed against the Ebola GP, could have impacted the specificity of the responding B cells to the GP. Also, immune complexes generated by ZMapp antibodies could have influenced the T-cell response. However, it is important to note that all treatments were done after onset of symptoms and, in the case of ZMapp, was given 9 or 10 d after symptom onset. Thus, the initiation and activation of the adaptive immune response occurred under natural conditions without any interventions.
In summary, this study provides to our knowledge the first longitudinal kinetic data on the activation status of human B- and T-cell populations during acute Ebola virus infection. We have described the presence of robust B- and T-cell responses in four humans infected with Ebola virus. Not only was the magnitude of this response notable but the duration of these responses persisted into convalescence in some patients. Several viral proteins were targets of the Ebola virus-specific T-cell response, with the NP being the major viral target of CD8 T cells, suggesting the inclusion of this protein in future T cell-based vaccine designs.
Materials and Methods
Human Subjects and Biosafety.
Institutional review board approval (IRB) was obtained from Emory University (IRB00076700) and CDC (6643.0) prior to patient enrollment. Written consent was obtained from all patients. All work with infectious acute phase specimens was performed in the CDC BSL-4 laboratory space. Plasma samples were γ-irradiated with 5 × 106 rads prior to removal from BSL-4 for qRT-PCR and ELISA analyses. Throughout the paper, data are presented in days after onset of patient-reported symptoms.
Virus-Specific qRT-PCR.
RNA was isolated from plasma using the MagMax Total RNA Isolation Kit (Life Technologies), and qRT-PCR was performed using established primer/probe sets that target the nucleoprotein gene (29). A standard curve of known virus concentration, expressed as TCID50 per mL, was generated from a passage 3 stock of the virus isolated from EVD2 and used to convert raw Ct values to relative TCID50 per mL.
Antibodies, Flow Cytometry, ELISpot, and ELISA.
All staining reagents, including antibodies, lysis buffer, and cytofix/cytoperm kits, were obtained from BD, except for CX3CR1 (BioLegend) and Aqua live/dead stain (Life Technologies). All phenotype stains and intracellular cytokine stimulations were done as described previously (16). All samples were read using a BD Accuri C6 or LSRII and analyzed using Flowjo software (Treestar). Ebola virus NP (His-tagged; GenScript) was used to assay Ebola-specific IgG and IgM by ELISA and ASC by ELISPOT. Additionally, lysate from Ebola-infected or uninfected Vero E6 cells was used for the ASC ELISPOT. Further experimental details for the flow cytometry, PBMC peptide stimulations, ELISA, and ELISpot are presented in SI Materials and Methods.
Supplementary Material
Acknowledgments
We are most grateful to the patients for participating in the study, and thank the Emory Serious Communicable Diseases Unit team for their assistance. We thank Tim Uyeki for providing coordination between the clinical and research staff, and Tatyana Klimova for help with the manuscript. This project is supported by funding from the Defense Advanced Research Projects Agency (W31P4Q-14-1-0010) and NIH (UL1TR000454). A.K. McElroy is supported by a Pediatric Infectious Disease Society/St. Jude’s Fellowship Award, a Burroughs Wellcome Career Award, and NIH K12 HD072245.
Footnotes
The authors declare no conflict of interest.
See Commentary on page 4518.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1502619112/-/DCSupplemental.
References
- 1.Sanchez A, Geisbert T, Feldmann H. 2007. Filoviridae: Marburg and Ebola Viruses (Wolters Kluwer/Lippincott Williams & Wilkins, Philadelphia), 5th Ed.
- 2.Baize S, et al. Inflammatory responses in Ebola virus-infected patients. Clin Exp Immunol. 2002;128(1):163–168. doi: 10.1046/j.1365-2249.2002.01800.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Baize S, et al. Defective humoral responses and extensive intravascular apoptosis are associated with fatal outcome in Ebola virus-infected patients. Nat Med. 1999;5(4):423–426. doi: 10.1038/7422. [DOI] [PubMed] [Google Scholar]
- 4.Wauquier N, Becquart P, Padilla C, Baize S, Leroy EM. Human fatal Zaire Ebola virus infection is associated with an aberrant innate immunity and with massive lymphocyte apoptosis. PLoS Negl Trop Dis. 2010;4(10):e837. doi: 10.1371/journal.pntd.0000837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Villinger F, et al. Markedly elevated levels of interferon (IFN)-gamma, IFN-alpha, interleukin (IL)-2, IL-10, and tumor necrosis factor-alpha associated with fatal Ebola virus infection. J Infect Dis. 1999;179(Suppl 1):S188–S191. doi: 10.1086/514283. [DOI] [PubMed] [Google Scholar]
- 6.Ansari AA. Clinical features and pathobiology of Ebolavirus infection. J Autoimmun. 2014;55:1–9. doi: 10.1016/j.jaut.2014.09.001. [DOI] [PubMed] [Google Scholar]
- 7.Geisbert TW, et al. Apoptosis induced in vitro and in vivo during infection by Ebola and Marburg viruses. Lab Invest. 2000;80(2):171–186. doi: 10.1038/labinvest.3780021. [DOI] [PubMed] [Google Scholar]
- 8.Bradfute SB, et al. Mechanisms and consequences of Ebolavirus-induced lymphocyte apoptosis. J Immunol. 2010;184(1):327–335. doi: 10.4049/jimmunol.0901231. [DOI] [PubMed] [Google Scholar]
- 9.Gupta M, Spiropoulou C, Rollin PE. Ebola virus infection of human PBMCs causes massive death of macrophages, CD4 and CD8 T cell sub-populations in vitro. Virology. 2007;364(1):45–54. doi: 10.1016/j.virol.2007.02.017. [DOI] [PubMed] [Google Scholar]
- 10.Ksiazek TG, et al. Clinical virology of Ebola hemorrhagic fever (EHF): Virus, virus antigen, and IgG and IgM antibody findings among EHF patients in Kikwit, Democratic Republic of the Congo, 1995. J Infect Dis. 1999;179(Suppl 1):S177–S187. doi: 10.1086/514321. [DOI] [PubMed] [Google Scholar]
- 11.Rowe AK, et al. Clinical, virologic, and immunologic follow-up of convalescent Ebola hemorrhagic fever patients and their household contacts, Kikwit, Democratic Republic of the Congo. Commission de Lutte contre les Epidémies à Kikwit. J Infect Dis. 1999;179(Suppl 1):S28–S35. doi: 10.1086/514318. [DOI] [PubMed] [Google Scholar]
- 12.Mahanty S, et al. Cutting edge: Impairment of dendritic cells and adaptive immunity by Ebola and Lassa viruses. J Immunol. 2003;170(6):2797–2801. doi: 10.4049/jimmunol.170.6.2797. [DOI] [PubMed] [Google Scholar]
- 13.Bosio CM, et al. Ebola and Marburg viruses replicate in monocyte-derived dendritic cells without inducing the production of cytokines and full maturation. J Infect Dis. 2003;188(11):1630–1638. doi: 10.1086/379199. [DOI] [PubMed] [Google Scholar]
- 14.Basler CF, Amarasinghe GK. Evasion of interferon responses by Ebola and Marburg viruses. J Interferon Cytokine Res. 2009;29(9):511–520. doi: 10.1089/jir.2009.0076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lyon GM, et al. Emory Serious Communicable Diseases Unit Clinical care of two patients with Ebola virus disease in the United States. N Engl J Med. 2014;371(25):2402–2409. doi: 10.1056/NEJMoa1409838. [DOI] [PubMed] [Google Scholar]
- 16.Miller JD, et al. Human effector and memory CD8+ T cell responses to smallpox and yellow fever vaccines. Immunity. 2008;28(5):710–722. doi: 10.1016/j.immuni.2008.02.020. [DOI] [PubMed] [Google Scholar]
- 17.Precopio ML, et al. Immunization with vaccinia virus induces polyfunctional and phenotypically distinctive CD8(+) T cell responses. J Exp Med. 2007;204(6):1405–1416. doi: 10.1084/jem.20062363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lindgren T, et al. Longitudinal analysis of the human T cell response during acute hantavirus infection. J Virol. 2011;85(19):10252–10260. doi: 10.1128/JVI.05548-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Akondy RS, et al. The yellow fever virus vaccine induces a broad and polyfunctional human memory CD8+ T cell response. J Immunol. 2009;183(12):7919–7930. doi: 10.4049/jimmunol.0803903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Basler CF, et al. The Ebola virus VP35 protein functions as a type I IFN antagonist. Proc Natl Acad Sci USA. 2000;97(22):12289–12294. doi: 10.1073/pnas.220398297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sanchez A, et al. Analysis of human peripheral blood samples from fatal and nonfatal cases of Ebola (Sudan) hemorrhagic fever: Cellular responses, virus load, and nitric oxide levels. J Virol. 2004;78(19):10370–10377. doi: 10.1128/JVI.78.19.10370-10377.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Barber DL, et al. Restoring function in exhausted CD8 T cells during chronic viral infection. Nature. 2006;439(7077):682–687. doi: 10.1038/nature04444. [DOI] [PubMed] [Google Scholar]
- 23.Fink K. Origin and function of circulating plasmablasts during acute viral infections. Front Immunol. 2012;3:78. doi: 10.3389/fimmu.2012.00078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kreuels B, et al. A case of severe Ebola virus infection complicated by gram-negative septicemia. N Engl J Med. 2014;371(25):2394–2401. doi: 10.1056/NEJMoa1411677. [DOI] [PubMed] [Google Scholar]
- 25.Marzi A, Feldmann H. Ebola virus vaccines: An overview of current approaches. Expert Rev Vaccines. 2014;13(4):521–531. doi: 10.1586/14760584.2014.885841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Stanley DA, et al. Chimpanzee adenovirus vaccine generates acute and durable protective immunity against Ebolavirus challenge. Nat Med. 2014;20(10):1126–1129. doi: 10.1038/nm.3702. [DOI] [PubMed] [Google Scholar]
- 27.Qiu X, et al. Reversion of advanced Ebola virus disease in nonhuman primates with ZMapp. Nature. 2014;514(7520):47–53. doi: 10.1038/nature13777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Geisbert TW, et al. Postexposure protection of non-human primates against a lethal Ebola virus challenge with RNA interference: A proof-of-concept study. Lancet. 2010;375(9729):1896–1905. doi: 10.1016/S0140-6736(10)60357-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Towner JS, Sealy TK, Ksiazek TG, Nichol ST. High-throughput molecular detection of hemorrhagic fever virus threats with applications for outbreak settings. J Infect Dis. 2007;196(Suppl 2):S205–S212. doi: 10.1086/520601. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.