Significance
During meiosis, crossovers (COs) reshuffle homologous chromosomes, generating genetic diversity on which natural or human selection can act. However, CO numbers typically are very low, raising questions about the evolutionary forces that impose this constraint and limiting the efficiency of breeding programs. Here, we identified anti-CO factors in Arabidopsis and showed that several mechanisms actively antagonize CO formation in parallel. Disrupting these anti-CO factors provokes a large increase in CO frequency without affecting meiotic progression. These results suggest that COs are restrained not because a high number would impair chromosome segregation but because excessive recombination could break favorable genetic combinations built by past selection. These findings hold great promise for improving the efficiency of plant breeding programs.
Keywords: recombination, meiosis, crossover, Topoisomerase 3, RECQ4
Abstract
Meiotic crossovers (COs) have two important roles, shuffling genetic information and ensuring proper chromosome segregation. Despite their importance and a large excess of precursors (i.e., DNA double-strand breaks, DSBs), the number of COs is tightly regulated, typically one to three per chromosome pair. The mechanisms ensuring that most DSBs are repaired as non-COs and the evolutionary forces imposing this constraint are poorly understood. Here we identified Topoisomerase3α (TOP3α) and the RECQ4 helicases—the Arabidopsis slow growth suppressor 1 (Sgs1)/Bloom syndrome protein (BLM) homologs—as major barriers to meiotic CO formation. First, the characterization of a specific TOP3α mutant allele revealed that, in addition to its role in DNA repair, this topoisomerase antagonizes CO formation. Further, we found that RECQ4A and RECQ4B constitute the strongest meiotic anti-CO activity identified to date, their concomitant depletion leading to a sixfold increase in CO frequency. In both top3α and recq4ab mutants, DSB number is unaffected, and extra COs arise from a normally minor pathway. Finally, both TOP3α and RECQ4A/B act independently of the previously identified anti-CO Fanconi anemia of complementation group M (FANCM) helicase. This finding shows that several parallel pathways actively limit CO formation and suggests that the RECQA/B and FANCM helicases prevent COs by processing different substrates. Despite a ninefold increase in CO frequency, chromosome segregation was unaffected. This finding supports the idea that CO number is restricted not because of mechanical constraints but likely because of the long-term costs of recombination. Furthermore, this work demonstrates how manipulating a few genes holds great promise for increasing recombination frequency in plant-breeding programs.
Meiotic homologous recombination is initiated by the formation of DNA double-strand breaks (DSBs). DSBs are resected to form 3′ ssDNA overhangs which invade the intact homologous chromosome, producing DNA joint molecules (JMs). These JMs can be differentially processed to produce crossovers (COs) or non-COs (NCOs). In Arabidopsis thaliana, mammals, and budding yeast, two pathways of CO formation exist. The major pathway depends on the ZMM proteins (for Zip1-4, Msh4/5, and Mer3) in addition to MutL homolog 1 (MLH1) and MuL homolog 3 (MLH3) and produces interfering COs, so that one CO prevents the formation of another nearby (1). The second, pathway, producing noninterfering COs, depends on structure-specific endonucleases including MUS81 (1). These pro-CO pathways compete with anti-CO pathways, resulting in a minor portion of DSBs becoming COs; for instance, it is estimated that COs represent only 10% and 5% of DSBs in mouse and in Arabidopsis, respectively (2). Three helicases with meiotic anti-CO activities have been identified in different species: the Bloom syndrome (BLM) homolog, small growth suppressor 1 (Sgs1) in Saccharomyces cerevisiae (3, 4); regulator of telomere elongation helicase1 (RTEL-1) in Caenorhabditis elegans (5), and the Fanconi anemia of complementation group M (FANCM) helicase in Arabidopsis and Schizosaccharomyces pombe (6, 7). These helicases are thought to displace the invading strand, allowing its annealing with the other 3′ overhang end of the DSB, leading to NCO formation in a process called “synthesis-dependent strand annealing” (SDSA). Nevertheless, even when CO formation is increased threefold by the disruption of AtFANCM, DSBs still greatly outnumber COs (6), suggesting the existence of additional anti-CO pathways.
Results and Discussion
The meiotic anti-CO activity of FANCM was identified through a genetic screen because its mutation restores bivalent formation and fertility of zmm mutants (6). To identify additional meiotic anti-CO factors, we extended this screen and isolated one suppressor of human enhancer of invasion-10 (hei10), hei10(s)61, with increased fertility and bivalent number (Fig. 1A). The combination of genetic mapping, whole-genome sequencing, and functional complementation (24 independent transformants) identified the recessive causal mutation in Topoisomerase3α (TOP3α; At5g63920). In addition to hei10, this mutation was able to restore fertility and bivalent formation of mutS homolog 5 (msh5) (Fig. 1A), showing that TOP3α prevents CO formation in zmm mutants. The hei10(s)61 mutation changes Arg640 into a stop codon (top3α-R640X hereafter). Therefore, TOP3α-R640X has intact topoisomerase-primase (TOPRIM) and topoisomerase domains, but two predicted zinc-finger domains, which are conserved in TOP3α in plants and animals but not in yeasts, are truncated (Fig. S1). In Arabidopsis, two mutant alleles of TOP3α were described previously (8). The top3α-1 mutation leads to lethality early in development; top3α-2, a hypomorphic mutant, is viable but shows stunted growth and complete sterility, the latter being linked to aberrant bivalent-like structures followed by massive fragmentation at meiosis, suggesting an accumulation of unresolved JMs. The severe phenotypes associated with TOP3α disruption in Arabidopsis and various other species (9–12) highlight its essential role in the resolution of mitotic and meiotic DNA-repair intermediates. The phenotypes conferred by the top3α-R640X mutation contrast with the previously described alleles: the top3α-R640X plants did not show any somatic defect (Fig. S2), showed no fragmentation at meiosis (Fig. 1B), and were fertile (Table S1). The increased number of bivalents observed in hei10 top3α-R640X compared with hei10 (Fig. 1 A and B) suggests an elevated frequency of COs. Direct measurement of CO frequency using tetrad analysis (13) confirmed this increase (Fig. 1C and Dataset S1). Further, CO frequency increased 1.5-fold on average in the top3α-R640X single mutant compared with wild type (Fig. 1D). Furthermore, in plants carrying one allele of TOP3α-R640X and one allele of either top3α-1 or top3α-2, early lethality and growth defects were complemented (Fig. S2), and no fragmentation was observed at meiosis, suggesting that one dose of TOP3α-R640X is enough to process JMs. In this context, CO frequency was increased further (2.5-fold compared with wild type) (Fig. 1D), revealing that TOP3α-R640X has retained some anti-CO activity and that TOP3α is an important barrier to CO formation in wild-type plants.
Fig. 1.
Meiotic recombination is increased in the top3α-R640X and recq4a recq4b mutants. (A) Average number of bivalents per male meiocyte. Light blue bars represent rod bivalents, which have a rod shape with enlarged ends, indicating that one arm has at least one CO, whereas the other arm has no CO. Dark blue bars indicate ring bivalents, which have a lozenge shape, indicating that they have at least one CO on both arms. Red bars indicate pairs of univalents. The number of cells analyzed is indicated in parentheses. N.S., P > 0.01; **P < 0.01; ***P < 0.001, CHI2 test. (B and E) Chromosome spreads at metaphase I and anaphase I. Ring bivalents (marked as “a” in B), rod bivalents (marked as “b” in B), and univalents (marked as “u” in B) are indicated. (Scale bars, 10 µM.) (C, D, and F) Genetic distances measured using fluorescent-tagged lines. N.S., P > 0.05; **P < 0.01; ***P < 0.001, Z test.
TOP3α is a member of the human BLM-TOP3α-RMI1-RMI2 (or S. cerevisiae Sgs1-Top3-Rmi1) complex, which is essential for DNA repair in somatic cells (1). Using a tandem affinity purification (TAP) tagging strategy in an Arabidopsis somatic cell culture, we showed that TOP3α forms a complex in vivo with RecQ-mediated genome instabiliy 1 (RMI1) and RecQ-mediated genome instabiliy 2 (RMI2), and the RECQ4A proteins (Table S2 and Dataset S2). RECQ4A is one of the seven Arabidopsis family members related to the Sgs1/BLM helicase (14, 15), confirming that this complex is conserved in plants. Also, in another zmm suppressor screen (msh4 in a Landsberg erectra strain of Arabidopsis thaliana), we isolated two allelic suppressors, msh4(s)84 and msh4(s)101 (Fig. S3), that contained mutations in the RECQ4A gene (Fig. S1). Mutation of RECQ4A in a different strain, Columbia-0, previously was shown not to restore fertility and bivalent formation in msh4 (16). RECQ4A has a close paralog, RECQ4B, that arose from a duplication specific to the Brassicaceae lineage (Fig. S4). The RECQ4B sequence in the two strains diverged (17), including a premature stop codon upstream of the helicase domain (Q427 > STOP) (Fig. S1) in Lansdberg, strongly suggesting that RECQ4B is not functional in this strain. We hypothesized that the recq4a mutation in Columbia failed to suppress the msh4 defect because of redundancy with RECQ4B during meiosis. We combined the msh4, recq4a, and recq4b mutations in the Columbia strain. As previously shown (16), the double mutants msh4 recq4a and msh4 recq4b were quasi-sterile, like msh4 (Table S1), and at metaphase I had bivalent numbers similar to those of msh4 (Fig. 1 A and E). In contrast, the triple mutant msh4 recq4a-4 recq4b-2 was fertile, and the bivalent number was restored to wild-type levels (Fig. 1 A and E and Table S1). Similarly, the recq4a recq4b double mutation was able to restore the fertility and bivalent formation of another zmm, shortage in chiasmata 1 (shoc1)/Atzip2 (Fig. 1A). Therefore, in Columbia, RECQ4A and its closest paralogue, RECQ4B, redundantly prevent bivalent formation in zmm mutants (Fig. 1 A and E). We then measured the effects of the recq4a and recq4b mutations on CO frequency through tetrad analysis (Fig. 1F) (13). In the single recq4a and recq4b mutants, genetic distances were not significantly different from wild-type (P > 0.1), but the distances increased greatly in the double mutant, by 6.2-fold on average (P < 10−9). This large increase in recombination did not impair chromosome segregation (Fig. 1E) and fertility (Table S1). These results show that RECQ4A/B has the strongest meiotic anti-CO activity identified to date in any species. The RECQ4A/B depletion has a much more pronounced effect on increasing CO than the mutation of their single homolog in S. cerevisiae, Sgs1. In sgs1 mutants, multichromatid JMs accumulate but eventually are resolved with a modest, if any, eventual increase in CO (3, 4, 18–21). This difference may be caused by a much higher excess of DSBs than COs in Arabidopsis (DSBs/COs ratio ∼25) as compared with yeast (∼2) (2), allowing a greater increase in the number of COs in Arabidopsis.
We then investigated the origin of the extra COs. In top3α-R640X and recq4a recq4b, COs required SPO11-induced DSBs (Fig. 1A). The number of DNA meiotic recombinase 1 (DMC1) foci was unchanged (Fig. S5), suggesting that the CO increase is not associated with increased DSBs. In both top3α-R640X and recq4a recq4b, the number of MLH1 foci was unaffected, suggesting that the number of class I COs is unchanged (Fig. 2 A and B). Further, CO interference was no longer detected (Fig. 2C), which is compatible with an increase in noninterfering COs. In addition, abnormal bivalents at metaphase I and disastrous anaphase I, with chromatin bridges and chromosome fragmentation, were observed in top3α-R640X mus81 (Fig. 2D), showing that MUS81 becomes important for complete resolution of JMs in top3α-R640X. Somatic growth also was affected in top3α-R640X mus81 (Fig. 2E). The recq4a mus81 double mutant is not viable (22 and this study), nor is the recq4a recq4b mus81 triple mutant (this study). MUS81 thus is essential for somatic DNA repair in the recq4a and recq4a recq4b backgrounds, and we speculate that it also is essential at meiosis. Taken together, these findings suggest that TOP3α and RECQ4A/B, similar to FANCM (6), limit class II CO formation, apparently without affecting the number of class I COs.
Fig. 2.
Extra COs are class II COs. (A) MLH1 foci counts per cell at diakinesis. (B) MLH1 immunolocalization at diakinesis (green). DNA is stained with DAPI (red). (Scale bars, 10 µM.) (C) Mean interference ratio (IR). ***P(IR) = 1 < 0.001. (D) Chromosome spreads at metaphase I and anaphase I. (E) Four-week-old plants. The top3α-R640X mus81-1 double mutant shows a synthetic growth defect.
Two activities of the BLM/Sgs1-TOP3 proteins have been described in vitro. First, BLM alone can disassemble D-loops (23), but this activity is enhanced by TOP3α (24). Second, BLM-TOP3α-RMI1 and Sgs1-Top3-Rmi1 can promote NCO formation through double Holliday junction dissolution (25–27). In vivo, these proteins appear to function both in complex and independently of each other, as suggested by the mutants’ shared and unique defects in DNA repair (1). In Arabidopsis, the recq4a recq4b mutant is viable and proficient in meiotic DSB repair, whereas the top3α-1–null mutant is inviable. The somatic lethality of top3α-1 is partially suppressed by the recq4a mutation (8), recapitulating the ability of sgs1 to rescue top3 slow growth in yeasts (9, 28). This observation led to the proposal that RECQ4A/Sgs1 produces molecules that require TOP3α for repair (25). The recq4b mutation neither rescued the top3α-1 lethality (14 and this study) nor further improved the top3α-1 recq4a growth defect (this study, Fig. S6), suggesting that, in somatic cells, RECQ4B does not act redundantly with RECQ4A to produce substrates that TOP3α can process. Strong meiotic defects with abnormal bivalents and catastrophic anaphase I were observed in top3α-1 recq4a and top3α-1 recq4a recq4b (Fig. 3A), showing that the absence of both RECQ4 paralogs does not alleviate the requirement for TOP3α in processing toxic recombination intermediates at meiosis. Thus, TOP3α appears to be important for complete resolution of meiotic JMs independently of RECQ4A/B.
Fig. 3.
Genetic interactions between the top3α-R640X and recq4a recq4b mutations. (A and B) Chromosome spreads at metaphase I and anaphase I. (Scale bars, 10 μm.) (C) Genetic distances measured using fluorescent-tagged lines.
Next we addressed whether RECQ4A/B and TOP3α act together to limit meiotic CO formation. Although no chromosome fragmentation was observed in single or double mutants, the top3α-R640X recq4a recq4b triple mutant showed meiotic catastrophe associated with complete sterility. Bivalents with aberrant shapes were observed, followed by chromosome fragmentation and chromatin bridges at anaphase I (Fig. 3B). This result suggests that TOP3α-R640X is not fully able to resolve JMs produced in the absence of RECQ4A/B. Sterility of top3α-R640X recq4a recq4b prevented the measurement of CO frequency in this background. Nevertheless tetrad analysis in the fertile genotypes revealed that CO frequency was higher in recq4a top3α-R640X than in top3α-R640X, whereas recq4b top3α-R640X was similar to top3α-R640X (Fig. 3C). Thus, we observed a gradient in CO increase compared with wild type, from no effect in recq4a and recq4b single mutants, an ∼1.5-fold increase in top3α-R640X and top3α-R640X recq4b, an approximately threefold increase in top3α-R640X/top3α-1, and an approximately sixfold increase in top3α-R640X recq4a and recq4a recq4b double mutants. An attractive model explaining these results is that RECQ4A/B helicases promote NCO formation via D-loop displacement and SDSA, as proposed for its ortholog Sgs1 (3, 4), and that TOP3α is a cofactor of this activity. This model is supported by the findings that human TOP3α promotes the DNA-unwinding activity of BLM in vitro (24). However, our data also are compatible with an alternative, but not mutually exclusive, model in which TOP3α prevents CO formation independently of RECQ4A/B-dependent SDSA, possibly through dissolution of JMs.
While this article was under review, three independent studies also addressed the function of Top3 in DSB repair and meiosis (29–31). They revealed that in S. Cerevisiae Top3-Rmi1 acts with Sgs1 to channel JMs into CO and NCO pathways and prevent aberrant recombination intermediate accumulation. In addition, Top3-Rmi1 has a Sgs1-independent role that ensures complete recombination intermediate resolution and chromosome segregation. This suggests that BLM-TOP3α-RMI1 (BTR) has a conserved prevalent role to ensure accurate completion of meiotic recombination.
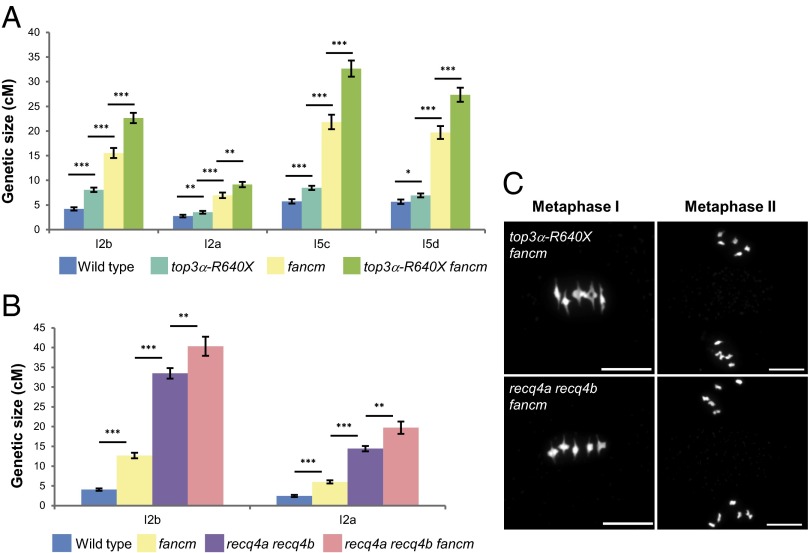
FANCM was the first meiotic anti-CO gene described in Arabidopsis (6). The effects of the top3α-R640X and recq4a recq4b mutations on CO formation were cumulative with fancm. Compared with wild-type plants, CO frequency was increased by 4.8-fold, on average, in top3α-R640X fancm and by ninefold in the recq4a recq4b fancm triple mutant (Fig. 4 A and B). This result demonstrates that at least two pathways prevent CO formation in parallel. Although previously it was thought that different organisms used a different helicase to ensure the same activity (i.e., RTEL-1, FANCM, Sgs1) (1), our data show that FANCM and RECQ4A/B, at least, have specific activities that independently prevent COs, possibly by unwinding different JM substrates (e.g., nascent versus extended D-loop).
Fig. 4.
TOP3α and RECQ4A/B limit meiotic COs in parallel with FANCM. (A and B) Genetic distances measured by using fluorescent-tagged lines. P < 0.05, **P < 0.01, ***P < 0.001, Z test. (C) Chromosome spreads at metaphase I and metaphase II. (Scale bars, 10 µM.)
In wild-type Arabidopsis, it is estimated that ∼5% (11/250) of the DSBs become COs. In recq4a recq4b fancm, we observed a ninefold increase in CO frequency, which, extrapolated genomewide, would mean that about half the DSBs become COs. This finding is reminiscent of observations in the yeast sgs1 mutant in which all DSBs form stable JMs that are resolved by nucleases (including MUS81) in an unbiased manner to form both COs and NCOs in equal numbers (3, 4, 20). If JM resolution also is unbiased in the absence of RECQ4A/B and FANCM, the level of COs would have reached almost the theoretical maximum of ∼125 CO per meiosis, suggesting that these helicases together are responsible for most of the SDSA events in Arabidopsis.
Despite an unprecedented increase in recombination, the top3α-R640X fancm and the recq4a recq4b fancm triple mutants grew normally and were fully fertile (Table S1). Further, we closely examined meiotic chromosome behavior in recq4a recq4b fancm, in which the highest level of recombination was observed. Among >450 meiocytes, including 135 post-anaphase I pictures, no defect was detected (Fig. 4C). Notably, no missegregation, lagging chromosomes, precocious sister chromatid separation, or fragmentation was detected. Thus, greatly increasing the CO frequency does not seem to affect meiosis completion. However, we cannot completely exclude the existence of a weak effect that would disturb less than a few percent of the meiotic divisions.
Although CO frequency varies between one (obligatory CO) and three per chromosome in most eukaryotes, we showed that COs can be unleashed without immediate negative effects on meiosis and fertility. Translation of these findings to crops may have a significant impact on plant breeding. In addition, this finding argues against the hypothesis that a high number of COs would impair chromosome segregation and thus supports the idea that natural selection constrains COs below their possible physical maximum because of the long-term costs of recombination (i.e., “recombination load,” the breaking of favorable genetic combinations built by past selection) (32, 33).
Materials and Methods
Genetic Resources.
The lines used in this study were hei10-2 (N514624) (34), msh5-2 (N526553) (35), spo11-1-3 (N646172) (36), top3α-2 (N445612) (8), top3α-1(N639357) (8), msh4 (cshl_GT14269) (37), msh4 (N636296) (38), recq4a-4 (N419423) (15), recq4b-2 (N511130) (15), shoc1-1 (N557589) (39), mus81-2 (N607515) (40), and fancm-1 (6). Tetrad analysis lines were I2ab (FTL1506/FTL1524/FTL965/qrt1-2), I1bc (FTL567/FTL1262/FTL992/ qrt1-2), and I5cd (FTL1143U/FTL1963U/FTL2450L/ qrt1-2) from Gregory Copenhaver, University of North Carolina at Chapel Hill, Chapel Hill, NC (13). Suppressors hei10(s)61, msh4(s)84, and msh4(s)101 were sequenced using Illumina technology (The Genome Analysis Centre). Mutations were identified through the MutDetect pipeline (41). The causal mutations were C-to-T substitutions at positions TAIR10 chr5:25579403 for hei10(s)61, TAIR10 chr1:3652474 for msh4(s)84, and TAIR10 chr1:3650343 for msh4(s)101.
Cytology Techniques.
Meiotic chromosome spreads were performed as described previously (42). Immunolocalizations of MLH1 were performed as described in ref. 43. Immunolocalizations of DMC1 and ZYP1 were performed as described in ref. 44. Observations were made using a Zeiss Axio Observer microscope. Scatter plots were made using the GraphPad software Prism6 (www.graphpad.com).
Cloning of a TOP3α Genomic Fragment and Plant Transformation.
A 10-kb genomic fragment containing a TOP3α locus was amplified with DNA primers bearing AttB1 and AttB2 and was cloned into Gateway vector pDONR207 using BP recombination (Invitrogen), producing the entry vector, which then was sequenced. Primers used for PCR amplification on Columbia genomic DNA were GGGGACAAGTTTGTACAAAAAAGCAGGCTAGAGCTCATCAAGCAACGAATCTG and GGGGACCACTTTGTACAAGAAAGCTGGGTGATGGCCGACATGGCATGAGAACTTG. An LR reaction between the entry vector and the destination vector pGWB1 (45) produced the final vector. The vector was introduced in Agrobacterium tumefaciens strain C58C1 (pMP90) by electroporation. Plants were transformed as described by Clough and Bent (46). Complementation analyses were carried out on T1 transformant plants previously selected on 30 µg/mL hygromycin.
Fluorescent Tagged Lines Analysis.
For each fluorescent tagged line (FTL) experiment, all genotypes, including wild-type controls, were siblings segregating for the tested mutations. Tetrad slides were prepared as in ref. 13, and counting was performed through an automated detection of tetrads using a pipeline developed on the Metafer Slide Scanning Platform (www.metasystems-international.com/metafer). For each tetrad, attribution to a specific class (A to L) was confirmed by hand (13). The genetic size of each interval was calculated using the Perkins equation (47):
(See www.molbio.uoregon.edu/∼fstahl for details.)
The interference ratio (IR) was calculated as in ref. 13, with pooled data from all experiments containing the relevant genotypes. For two adjacent intervals, I1 and I2, two populations of tetrads are considered: those with at least one CO in I2 and those without any CO in I2. The genetic size of I1 then is calculated for these two populations using the Perkins equation (above), namely D1 (I1 with CO in I2) and D2 (I1 without a CO in I2). The IR is thus defined as IR = D1/D2. If the genetic size of I1 is lowered by the presence of a CO in I2, IR < 1, and interference is detected. If not, IR is close to 1, and no interference is detected. A χ2 test is performed to test the null hypothesis (H0: D1 = D2.). The average of the two reciprocal IRs is shown on the graphs (Fig. 2C).
TAP.
Cloning of transgenes encoding the GSrhino tag (48), fusions under control of the constitutive cauliflower tobacco mosaic virus 35S promoter, and transformation of Arabidopsis cell-suspension cultures (PSB-D) with direct selection in liquid medium were carried out as previously described (49). TAP experiments were performed with 100 mg of total protein extract as input as described in ref. 48. Protein interactors were identified by mass spectrometry using an LTQ Orbitrap Velos mass spectrometer. Proteins with at least two matched high-confidence peptides were retained. Background proteins were filtered out based on the frequency of the occurrence of the copurified proteins in a large dataset containing 543 TAP experiments using 115 different baits (48).
Supplementary Material
Acknowledgments
We thank Gregory Copenhaver for providing the FTL lines; Bertrand Dubreucq for help in developing the FTL counting module; and Mathilde Grelon, Christine Mézard, and Eric Jenczewski for helpful discussions. This work was funded by European Research Council Grant ERC 2011 StG 281659 (MeioSight) (to R.M.). Research in the R.M. laboratory is supported by the Fondation Schlumberger pour l’Enseignement et la Recherce (FRM-FSER, 2014).
Footnotes
Conflict of interest statement: A provisional patent based on the work has been filed by the National Institute for Agricultural Research.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1423107112/-/DCSupplemental.
References
- 1.Kohl KP, Sekelsky J. Meiotic and mitotic recombination in meiosis. Genetics. 2013;194(2):327–334. doi: 10.1534/genetics.113.150581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.de Massy B. Initiation of meiotic recombination: How and where? Conservation and specificities among eukaryotes. Annu Rev Genet. 2013;47:563–599. doi: 10.1146/annurev-genet-110711-155423. [DOI] [PubMed] [Google Scholar]
- 3.De Muyt A, et al. BLM helicase ortholog Sgs1 is a central regulator of meiotic recombination intermediate metabolism. Mol Cell. 2012;46(1):43–53. doi: 10.1016/j.molcel.2012.02.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zakharyevich K, Tang S, Ma Y, Hunter N. Delineation of joint molecule resolution pathways in meiosis identifies a crossover-specific resolvase. Cell. 2012;149(2):334–347. doi: 10.1016/j.cell.2012.03.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Youds JL, et al. RTEL-1 enforces meiotic crossover interference and homeostasis. Science. 2010;327(5970):1254–1258. doi: 10.1126/science.1183112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Crismani W, et al. FANCM limits meiotic crossovers. Science. 2012;336(6088):1588–1590. doi: 10.1126/science.1220381. [DOI] [PubMed] [Google Scholar]
- 7.Lorenz A, et al. The fission yeast FANCM ortholog directs non-crossover recombination during meiosis. Science. 2012;336(6088):1585–1588. doi: 10.1126/science.1220111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hartung F, Suer S, Knoll A, Wurz-Wildersinn R, Puchta H. Topoisomerase 3alpha and RMI1 suppress somatic crossovers and are essential for resolution of meiotic recombination intermediates in Arabidopsis thaliana. PLoS Genet. 2008;4(12):e1000285. doi: 10.1371/journal.pgen.1000285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Goodwin A, Wang SW, Toda T, Norbury C, Hickson ID. Topoisomerase III is essential for accurate nuclear division in Schizosaccharomyces pombe. Nucleic Acids Res. 1999;27(20):4050–4058. doi: 10.1093/nar/27.20.4050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Plank JL, Chu SH, Pohlhaus JR, Wilson-Sali T, Hsieh T-S. Drosophila melanogaster topoisomerase IIIalpha preferentially relaxes a positively or negatively supercoiled bubble substrate and is essential during development. J Biol Chem. 2005;280(5):3564–3573. doi: 10.1074/jbc.M411337200. [DOI] [PubMed] [Google Scholar]
- 11.Li W, Wang JC. Mammalian DNA topoisomerase IIIalpha is essential in early embryogenesis. Proc Natl Acad Sci USA. 1998;95(3):1010–1013. doi: 10.1073/pnas.95.3.1010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kim YC, Lee J, Koo HS. Functional characterization of Caenorhabditis elegans DNA topoisomerase IIIalpha. Nucleic Acids Res. 2000;28(9):2012–2017. doi: 10.1093/nar/28.9.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Berchowitz LE, Copenhaver GP. Fluorescent Arabidopsis tetrads: A visual assay for quickly developing large crossover and crossover interference data sets. Nat Protoc. 2008;3(1):41–50. doi: 10.1038/nprot.2007.491. [DOI] [PubMed] [Google Scholar]
- 14.Hartung F, Puchta H. The RecQ gene family in plants. J Plant Physiol. 2006;163(3):287–296. doi: 10.1016/j.jplph.2005.10.013. [DOI] [PubMed] [Google Scholar]
- 15.Hartung F, Suer S, Puchta H. Two closely related RecQ helicases have antagonistic roles in homologous recombination and DNA repair in Arabidopsis thaliana. Proc Natl Acad Sci USA. 2007;104(47):18836–18841. doi: 10.1073/pnas.0705998104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Higgins JD, Ferdous M, Osman K, Franklin FCH. The RecQ helicase AtRECQ4A is required to remove inter-chromosomal telomeric connections that arise during meiotic recombination in Arabidopsis. Plant J. 2011;65(3):492–502. doi: 10.1111/j.1365-313X.2010.04438.x. [DOI] [PubMed] [Google Scholar]
- 17.Gan X, et al. Multiple reference genomes and transcriptomes for Arabidopsis thaliana. Nature. 2011;477(7365):419–423. doi: 10.1038/nature10414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jessop L, Rockmill B, Roeder GS, Lichten M. Meiotic chromosome synapsis-promoting proteins antagonize the anti-crossover activity of sgs1. PLoS Genet. 2006;2(9):e155. doi: 10.1371/journal.pgen.0020155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jessop L, Lichten M. Mus81/Mms4 endonuclease and Sgs1 helicase collaborate to ensure proper recombination intermediate metabolism during meiosis. Mol Cell. 2008;31(3):313–323. doi: 10.1016/j.molcel.2008.05.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Oh SD, et al. BLM ortholog, Sgs1, prevents aberrant crossing-over by suppressing formation of multichromatid joint molecules. Cell. 2007;130(2):259–272. doi: 10.1016/j.cell.2007.05.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Oh SD, Lao JP, Taylor AF, Smith GR, Hunter N. RecQ helicase, Sgs1, and XPF family endonuclease, Mus81-Mms4, resolve aberrant joint molecules during meiotic recombination. Mol Cell. 2008;31(3):324–336. doi: 10.1016/j.molcel.2008.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hartung F, Suer S, Bergmann T, Puchta H. The role of AtMUS81 in DNA repair and its genetic interaction with the helicase AtRecQ4A. Nucleic Acids Res. 2006;34(16):4438–4448. doi: 10.1093/nar/gkl576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bachrati CZ, Borts RH, Hickson ID. Mobile D-loops are a preferred substrate for the Bloom’s syndrome helicase. Nucleic Acids Res. 2006;34(8):2269–2279. doi: 10.1093/nar/gkl258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Daley JM, Chiba T, Xue X, Niu H, Sung P. Multifaceted role of the Topo IIIα-RMI1-RMI2 complex and DNA2 in the BLM-dependent pathway of DNA break end resection. Nucleic Acids Res. 2014;42(17):11083–11091. doi: 10.1093/nar/gku803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cejka P, Plank JL, Bachrati CZ, Hickson ID, Kowalczykowski SC. Rmi1 stimulates decatenation of double Holliday junctions during dissolution by Sgs1-Top3. Nat Struct Mol Biol. 2010;17(11):1377–1382. doi: 10.1038/nsmb.1919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wu L, Hickson ID. The Bloom’s syndrome helicase suppresses crossing over during homologous recombination. Nature. 2003;426(6968):870–874. doi: 10.1038/nature02253. [DOI] [PubMed] [Google Scholar]
- 27.Wu L, et al. BLAP75/RMI1 promotes the BLM-dependent dissolution of homologous recombination intermediates. Proc Natl Acad Sci USA. 2006;103(11):4068–4073. doi: 10.1073/pnas.0508295103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gangloff S, McDonald JP, Bendixen C, Arthur L, Rothstein R. The yeast type I topoisomerase Top3 interacts with Sgs1, a DNA helicase homolog: A potential eukaryotic reverse gyrase. Mol Cell Biol. 1994;14(12):8391–8398. doi: 10.1128/mcb.14.12.8391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tang S, Wu MK, Zhang R, Hunter N. Pervasive and essential roles of the Top3-rmi1 decatenase orchestrate recombination and facilitate chromosome segregation in meiosis. Mol Cell. 2015;57(4):607–621. doi: 10.1016/j.molcel.2015.01.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kaur H, De Muyt A, Lichten M. Top3-rmi1 DNA single-strand decatenase is integral to the formation and resolution of meiotic recombination intermediates. Mol Cell. 2015;57(4):583–594. doi: 10.1016/j.molcel.2015.01.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Fasching CL, Cejka P, Kowalczykowski SC, Heyer WD. Top3-rmi1 dissolve rad51-mediated D loops by a topoisomerase-based mechanism. Mol Cell. 2015;57(4):595–606. doi: 10.1016/j.molcel.2015.01.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hadany L, Comeron JM. Why are sex and recombination so common? Ann N Y Acad Sci. 2008;1133:26–43. doi: 10.1196/annals.1438.011. [DOI] [PubMed] [Google Scholar]
- 33.Otto SP. The evolutionary enigma of sex. Am Nat. 2009;174(Suppl 1):S1–S14. doi: 10.1086/599084. [DOI] [PubMed] [Google Scholar]
- 34.Chelysheva L, et al. The Arabidopsis HEI10 is a new ZMM protein related to Zip3. PLoS Genet. 2012;8(7):e1002799. doi: 10.1371/journal.pgen.1002799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Higgins JD, et al. AtMSH5 partners AtMSH4 in the class I meiotic crossover pathway in Arabidopsis thaliana, but is not required for synapsis. Plant J. 2008;55(1):28–39. doi: 10.1111/j.1365-313X.2008.03470.x. [DOI] [PubMed] [Google Scholar]
- 36.Stacey NJ, et al. Arabidopsis SPO11-2 functions with SPO11-1 in meiotic recombination. Plant J. 2006;48(2):206–216. doi: 10.1111/j.1365-313X.2006.02867.x. [DOI] [PubMed] [Google Scholar]
- 37.Drouaud J, et al. Contrasted patterns of crossover and non-crossover at Arabidopsis thaliana meiotic recombination hotspots. PLoS Genet. 2013;9(11):e1003922. doi: 10.1371/journal.pgen.1003922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Higgins JD, Armstrong SJ, Franklin FCH, Jones GH. The Arabidopsis MutS homolog AtMSH4 functions at an early step in recombination: Evidence for two classes of recombination in Arabidopsis. Genes Dev. 2004;18(20):2557–2570. doi: 10.1101/gad.317504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Macaisne N, et al. SHOC1, an XPF endonuclease-related protein, is essential for the formation of class I meiotic crossovers. Curr Biol. 2008;18(18):1432–1437. doi: 10.1016/j.cub.2008.08.041. [DOI] [PubMed] [Google Scholar]
- 40.Berchowitz LE, Francis KE, Bey AL, Copenhaver GP. The role of AtMUS81 in interference-insensitive crossovers in A. thaliana. PLoS Genet. 2007;3(8):e132. doi: 10.1371/journal.pgen.0030132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Girard C, et al. FANCM-associated proteins MHF1 and MHF2, but not the other Fanconi anemia factors, limit meiotic crossovers. Nucleic Acids Res. 2014;42(14):9087–9095. doi: 10.1093/nar/gku614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ross KJ, Fransz P, Jones GH. A light microscopic atlas of meiosis in Arabidopsis thaliana. Chromosome Res. 1996;4(7):507–516. doi: 10.1007/BF02261778. [DOI] [PubMed] [Google Scholar]
- 43.Chelysheva L, et al. An easy protocol for studying chromatin and recombination protein dynamics during Arabidopsis thaliana meiosis: Immunodetection of cohesins, histones and MLH1. Cytogenet Genome Res. 2010;129(1-3):143–153. doi: 10.1159/000314096. [DOI] [PubMed] [Google Scholar]
- 44.Armstrong SJ, Caryl APP, Jones GH, Franklin FCH. Asy1, a protein required for meiotic chromosome synapsis, localizes to axis-associated chromatin in Arabidopsis and Brassica. J Cell Sci. 2002;115(Pt 18):3645–3655. doi: 10.1242/jcs.00048. [DOI] [PubMed] [Google Scholar]
- 45.Nakagawa T, et al. Development of series of gateway binary vectors, pGWBs, for realizing efficient construction of fusion genes for plant transformation. J Biosci Bioeng. 2007;104(1):34–41. doi: 10.1263/jbb.104.34. [DOI] [PubMed] [Google Scholar]
- 46.Clough SJ, Bent AF. Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 1998;16(6):735–743. doi: 10.1046/j.1365-313x.1998.00343.x. [DOI] [PubMed] [Google Scholar]
- 47.Perkins DD. Biochemical Mutants in the Smut Fungus Ustilago Maydis. Genetics. 1949;34(5):607–626. doi: 10.1093/genetics/34.5.607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Van Leene J, et al. An improved toolbox to unravel the plant cellular machinery by tandem affinity purification of Arabidopsis protein complexes. Nat Protoc. 2015;10(1):169–187. doi: 10.1038/nprot.2014.199. [DOI] [PubMed] [Google Scholar]
- 49.Van Leene J, et al. Isolation of transcription factor complexes from Arabidopsis cell suspension cultures by tandem affinity purification. Methods Mol Biol. 2011;754:195–218. doi: 10.1007/978-1-61779-154-3_11. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.