Significance
Despite extensive studies of protein trafficking across length scales of many microns, how proteins correctly localize within the smaller length scales of bacterial cells is still poorly understood. Recently, we proposed that slight membrane curvature, defined by the surface geometry of a bacterium, can drive the localization of certain shape-sensing proteins. Here, we developed an assay to quantify membrane curvature recognition by the small bacterial protein SpoVM and used NMR to determine the structural basis of curvature recognition. NMR and molecular dynamics simulations suggested a model wherein unusually deep membrane insertion allows the protein to sense subtle acyl chain packing differences between differently curved membranes, a distinct curvature-sensing mechanism from those used by proteins that sense high membrane curvature.
Keywords: ArfGAP1, SpoIVA, DivIVA, curvature sensing, lipid packing
Abstract
In bacteria, certain shape-sensing proteins localize to differently curved membranes. During sporulation in Bacillus subtilis, the only convex (positively curved) surface in the cell is the forespore, an approximately spherical internal organelle. Previously, we demonstrated that SpoVM localizes to the forespore by preferentially adsorbing onto slightly convex membranes. Here, we used NMR and molecular dynamics simulations of SpoVM and a localization mutant (SpoVMP9A) to reveal that SpoVM’s atypical amphipathic α-helix inserts deeply into the membrane and interacts extensively with acyl chains to sense packing differences in differently curved membranes. Based on binding to spherical supported lipid bilayers and Monte Carlo simulations, we hypothesize that SpoVM’s membrane insertion, along with potential cooperative interactions with other SpoVM molecules in the lipid bilayer, drives its preferential localization onto slightly convex membranes. Such a mechanism, which is distinct from that used by high curvature-sensing proteins, may be widely conserved for the localization of proteins onto the surface of cellular organelles.
Subcellular protein localization studies typically focus on the sorting of proteins across tens of microns, such as vesicle-mediated transport of cargo destined for a particular cellular organelle (1). Mechanisms by which proteins correctly localize within shorter length scales of a few microns or less, such as organization within the confines of bacterial cells, are less well understood (2). In bacteria, many proteins that will undergo localization are synthesized in the cytosol and then diffuse to the correct destination (3), where they are captured by a prelocalized landmark, typically a protein or a lipid (4). In this “diffusion and capture” model, it remains largely unresolved how the landmark itself recognizes its own correct location, although several mechanisms have been proposed (reviewed in ref. 5). For example, proteins that are recruited to the cell division site may be left behind after cytokinesis to form a scar that identifies one pole of a rod-shaped bacterium (6, 7). Proteins can also use clustering interactions to define the longest axis of the cell; in rod-shaped bacteria, this self-assembly mechanism can be used to mark and recruit proteins to the poles (8, 9). Moreover, the architecture of the bacterial cell itself, such as convex (positively curved) and concave (negatively curved) membrane surfaces, may provide geometric cues for the localization of landmark proteins (10, 11).
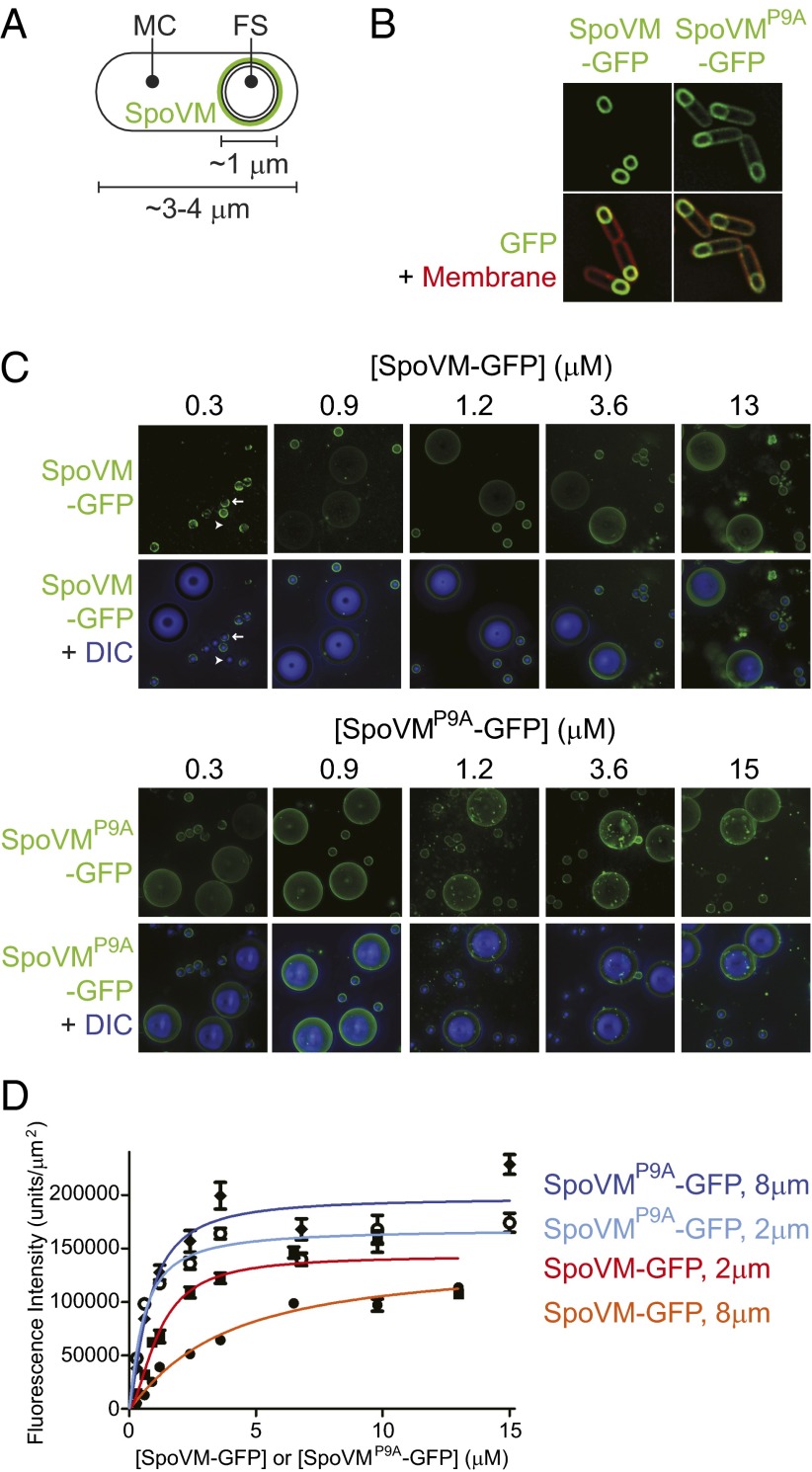
During spore formation in the bacterium Bacillus subtilis, the rod-shaped cell elaborates a spherical internal organelle, the “forespore,” which eventually becomes a dormant spore (Fig. 1A) (12). A tough proteinaceous shell (the “coat”) composed of proteins made in the outer “mother cell” is built around the forespore and protects the genetic material in the dormant spore from environmental insults (13). Assembly of the coat begins with a basement layer (14, 15), the structural protein of which is an unusual cytoskeletal protein that polymerizes into a static structure via a mechanism requiring both ATP binding and ATP hydrolysis (14, 16–19). This protein is anchored onto the surface of the forespore (20) by the small (26 aa) amphipathic α-helical protein SpoVM (21–23). Interestingly, substitution of a Pro residue at position 9 of SpoVM resulted in the promiscuous mislocalization of SpoVMP9A in vivo, suggesting that this residue is critical for the recognition of a forespore landmark that mediates SpoVM localization (Fig. 1B) (23).
Fig. 1.
SpoVM, but not SpoVMP9A, preferentially localizes to convex membranes. (A) Schematic of a sporulating B. subtilis cell, in which the rod-shaped mother cell (MC) elaborates a spherical internal organelle termed the forespore (FS). SpoVM (green) is produced exclusively in the MC and localizes to the FS surface. Membranes are depicted as black lines. (B) Fluorescence micrographs of sporulating B. subtilis cells. In vivo localization of SpoVM-GFP (Left) or SpoVMP9A-GFP (Right). Overlay of GFP signal (green) and membranes (red) visualized using the fluorescent dye FM4-64. (C) Fluorescence micrographs of SSLBs 2 or 8 μm in diameter that were incubated with increasing concentrations of purified SpoVM-GFP (Upper) or SpoVMP9A-GFP (Lower). Aggregate GFP fluorescence across several z-stacks (top of each data set) is depicted in green; the bottom of each data set shows the overlay of GFP fluorescence (green) and SSLBs visualized by differential interference contrast (DIC) microscopy (blue). Arrow: bead displaying a nonuniform pattern of fluorescence; arrowhead: bead displaying little or no fluorescence (see Fig. S1 for magnified view). (D) Quantification of SpoVM-GFP fluorescence on 2-μm (red) or 8-μm (orange) SSLBs or SpoVMP9A-GFP on 2-μm (light blue) or 8-μm (blue) SSLBs per square micron, as a function of protein concentration. Each data point represents the mean intensity of 30–52 SSLBs; errors bars are SEM; data were fit with an allosteric sigmoidal model.
Previously, we demonstrated that the landmark recognized by SpoVM is the slightly convex membrane surface of the forespore, the only convex surface in the mother cell cytosol (24). Membrane curvature recognition depends on a complex interplay of protein–lipid, protein–protein, and lipid–lipid interactions. Recent studies have suggested two major mechanisms for the sensing of membrane curvature. Some sensor proteins may directly recognize specific membrane geometries through a scaffolding mechanism in which the structure of the sensing protein closely matches the curvature of a membrane surface (25). Other curvature-sensing molecules shallowly insert hydrophobic regions, such as an amphipathic helix, into one leaflet of the bilayer to detect stress due to lipid packing defects (26, 27). A high density of packing defects in the lipid headgroup region of the outer leaflet of ∼50-nm-diameter vesicles has been implicated in the recognition of highly curved membranes (28). However, this mechanism does not address how slightly curved membranes, such as the outer surface of the forespore, are recognized.
Here, we develop an in vitro assay based on spherical supported lipid bilayers (SSLBs) to quantify SpoVM adsorption; we solve the NMR structures of SpoVM and the SpoVMP9A variant; and we perform long-timescale molecular dynamics simulations to probe their dynamic interactions with membranes. Using Monte Carlo simulations, we show that, unlike proteins that sense highly curved surfaces, SpoVM likely relies on a combination of small increases in binding affinity and cooperativity to localize to slightly convex membranes with curvature similar to that of the forespore. NMR studies and molecular dynamics simulations reveal that the SpoVM structure represents an atypical amphipathic α-helix deeply embedded in the membrane, unlike many molecules sensitive to highly curved membranes that shallowly insert into the membrane. Our data indicate that SpoVM exploits a novel mechanism for recognition of slightly curved membranes, appropriate for acting as a landmark of the forespore geometry.
Results
A Previously Unidentified Mechanism for Curvature-Mediated Adsorption of SpoVM.
To determine the biochemical basis for the preferential adsorption of SpoVM onto slightly convex membrane surfaces, we first sought to generate a saturation-binding curve by incubating membrane surfaces of a given curvature with a range of concentrations of purified SpoVM-GFP. Previously, we demonstrated that purified SpoVM-GFP selectively bound to lipid vesicles similar in size to the forespore in a mixture of giant unilamellar vesicles of various sizes (24). In the current investigation, we eliminated limitations associated with giant unilamellar vesicles (variability in vesicle size and associated variability in membrane stiffness) by using SSLBs, in which a single phospholipid bilayer is assembled on the surface of silica beads of defined size (29, 30). In addition to providing a more strictly defined membrane radius of curvature (determined by the diameter of the silica bead), the use of a supported bilayer system eliminates osmotic-dependent variability in membrane tension across vesicle curvatures (31).
We generated 2- and 8-μm SSLBs (Materials and Methods) and incubated them with increasing concentrations of purified SpoVM-GFP or SpoVMP9A-GFP. At the lowest concentration of SpoVM-GFP tested (0.3 μM), the protein bound to the surface of the 2-μm beads, but was largely absent from the surface of the 8-μm beads (Fig. 1C, Upper), suggesting preferential binding to more positively curved surfaces. At higher concentrations of SpoVM-GFP, fluorescence intensity increased on the surface of both types of beads, but more rapidly on the 2-μm beads; at 13 μM SpoVM-GFP, the protein bound approximately equally well to both 2- and 8-μm beads (Fig. 1C, Upper). In contrast, purified SpoVMP9A-GFP adsorbed similarly to both the 2- and 8-μm beads at all protein concentrations we tested (Fig. 1C, Lower), suggesting that SpoVMP9A-GFP does not discriminate between differently curved surfaces in vitro.
At low concentrations of SpoVM-GFP, but not SpoVMP9A-GFP, we noticed that some 2-μm beads displayed obvious qualitative signs of binding, but other 2-μm beads were almost completely devoid of fluorescence signal (Fig. 1C, arrowhead; magnified view and distribution of quantified fluorescence signals in Fig. S1). We hypothesized that this bimodal distribution of fluorescence was due to a clustering interaction between SpoVM-GFP molecules in which subsequent binding events were positively influenced by SpoVM molecules that had already adsorbed. Consistent with this hypothesis, we observed that the spatial pattern of SpoVM-GFP fluorescence on smaller beads at lower concentrations was often nonuniform (Fig. 1C, arrow; Fig. S1A), whereas the adsorbance of SpoVMP9A-GFP was more uniform (Fig. 1C, Lower).
To quantify the binding of SpoVM and SpoVMP9A on differently curved surfaces, we integrated deconvolved fluorescence intensities across multiple z-stacks for many beads and measured the mean fluorescence intensity per square micron of surface area as a function of SpoVM-GFP or SpoVMP9A-GFP concentration (Fig. 1D). Fitting these saturation-binding curves to an allosteric sigmoidal model revealed differences in the parameters that described the adsorption of SpoVM-GFP onto either 2- or 8-μm SSLBs. The fitted Bmax values were not appreciably different (Table 1), suggesting that the number of SpoVM-GFP binding sites per square micron was similar on the two surfaces and therefore did not contribute to the preferential binding of SpoVM-GFP onto the smaller beads. Thus, our results strongly indicate that SpoVM discriminates between surfaces with slight differences in curvature via a different mechanism than the one used by other amphipathic α-helices that recognize extremely highly curved surfaces (28).
Table 1.
Parameters for the allosteric sigmoidal fit for SpoVM-GFP and SpoVMP9A-GFP adsorption onto differently curved surfaces
| Protein (bead diameter) | ||||
| SpoVM-GFP (2 μm) | SpoVM-GFP (8 μm) | SpoVMP9A-GFP (2 μm) | SpoVMP9A-GFP (8 μm) | |
| Bmax, units/μm2 | 143,125 (± 4,487) | 139,618 (± 12,491) | 167,977 (± 6,014) | 197,454 (± 7,520) |
| H | 1.7 (± 0.2) | 1.1 (± 0.1) | 1.2 (± 0.2) | 1.4 (± 0.2) |
| K1/2, μM | 1.18 (± 0.31) | 3.64 (± 0.52) | 0.56 (± 0.14) | 0.77 (± 0.23) |
Bmax, maximal binding value; h, Hill coefficient; K1/2, concentration of protein producing half maximal binding. Errors are SEM (n = 30–52).
The Hill coefficient for SpoVM-GFP binding onto 8-μm beads was 1.1 ± 0.1 (Table 1), suggesting little to no cooperativity, whereas the Hill coefficient for 2-μm beads was 1.7 ± 0.2 (Table 1), indicating a slight increase in binding cooperativity (32). The binding curves also revealed a difference in the half-maximal concentration of binding between the two bead geometries, with K1/2 = 1.18 ± 0.31 μM for 2-μm beads and 3.64 ± 0.52 μM for 8-μm beads (Table 1), indicating an approximately threefold higher affinity of SpoVM-GFP for the more convex surfaces. In contrast, SpoVMP9A-GFP did not display noticeable differences between its adsorption onto 2- or 8-μm beads (Table 1), consistent with its promiscuous localization in vivo. Interestingly, SpoVMP9A-GFP bound with almost twofold higher affinity than wild-type SpoVM-GFP did to 2-μm beads, and also had a slightly higher Bmax (Table 1), indicating that more binding sites were available for SpoVMP9A-GFP. Finally, the GFP fusion did not influence the ability of SpoVM to preferentially bind to more convex surfaces, because SpoVM labeled with an organic fluorescent dye behaved similarly to SpoVM-GFP (Fig. S2).
Taken together, these data indicate that SpoVM preferentially binds onto membranes similar in curvature to the surface of the forespore compared with flatter surfaces via a new mechanism involving a small degree of cooperativity and a modest increase in intrinsic affinity, rather than any increase in the density of binding sites (28).
Curvature-Dependent Adsorption Through Slight Increases in Binding Cooperativity and Affinity.
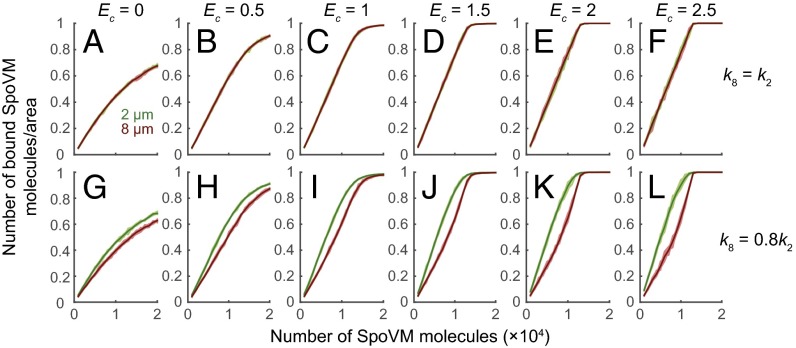
To better understand how small increases in binding cooperativity and affinity result in preferential absorption, we conducted Monte Carlo simulations to mimic the binding of SpoVM molecules onto the surfaces of 2- and 8-μm beads (Fig. 2). In these simulations, we varied two parameters, k8_on/k2_on, the ratio of on rates in the absence of cooperativity between the two bead sizes to reflect an affinity difference, and Ec, the cooperativity energy, such that all nearest-neighbor sites of a bound SpoVM undergo an increase in binding rate by the factor exp(Ec/kBT). For simplicity, we represented the beads as periodic 2D square lattices, such that each binding site has n = 4 nearest neighbors. As a positive control, we first simulated SpoVM adsorption when the on rate was equal for both bead sizes, in the absence of cooperativity (Fig. 2A). As expected, in this condition SpoVM molecules did not discriminate between 2- and 8-μm beads, binding equally well to both surfaces as a function of protein concentration. Next, we increased cooperativity while maintaining an equal on rate (k8_on/k2_on = 1; Fig. 2 B–F). Under these conditions, SpoVM molecules bound strongly to both 2- and 8-μm beads, with no curvature preference.
Fig. 2.
Slight increases in cooperativity and affinity are sufficient for preferential localization of SpoVM onto convex membranes. Monte Carlo simulations (SI Materials and Methods) of SpoVM molecules binding onto 2-μm (green) or 8-μm (red) beads as a function of increasing number of SpoVM molecules as the cooperative energy (Ec, in units of kBT) was increased from 0 to 2.5. (A–F) On rate for adsorption onto 2- and 8 μm- diameter beads is equal (k8_on = k2_on). (G–L) On rate for adsorption onto 8-μm-diameter beads is 20% less than that onto 2-μm diameter beads (k8_on = 0.8k2_on).
We then introduced a small preference for SpoVM adsorption onto the 2-μm beads (k8_on = 0.8 k2_on) while incrementally increasing cooperativity (Fig. 2 G–L). With only a 20% difference between k8_on and k2_on, binding to the 2-μm beads was accelerated by the nonzero Ec, because each bound SpoVM molecule increased the affinity of its nearby binding sites for free SpoVM molecules, resulting in the net preferential adsorption of SpoVM onto the more convex surface (Fig. 2 G–L). In these simulations, clustering was evident on both bead types, particularly on the 2-μm beads due to the accelerated binding (Fig. S3), consistent with the heterogeneity in fluorescence we observed experimentally (Fig. 1C; Fig. S1). The difference in binding between 2- and 8-μm beads initially increased when cooperativity was introduced, but saturated at higher cooperativity. Our Monte Carlo simulations therefore indicate that geometric cue-mediated binding of SpoVM requires only a small increase in affinity for more convex surfaces in combination with modest cooperativity.
SpoVM and SpoVMP9A Structures in Bilayer-Like Bicelles.
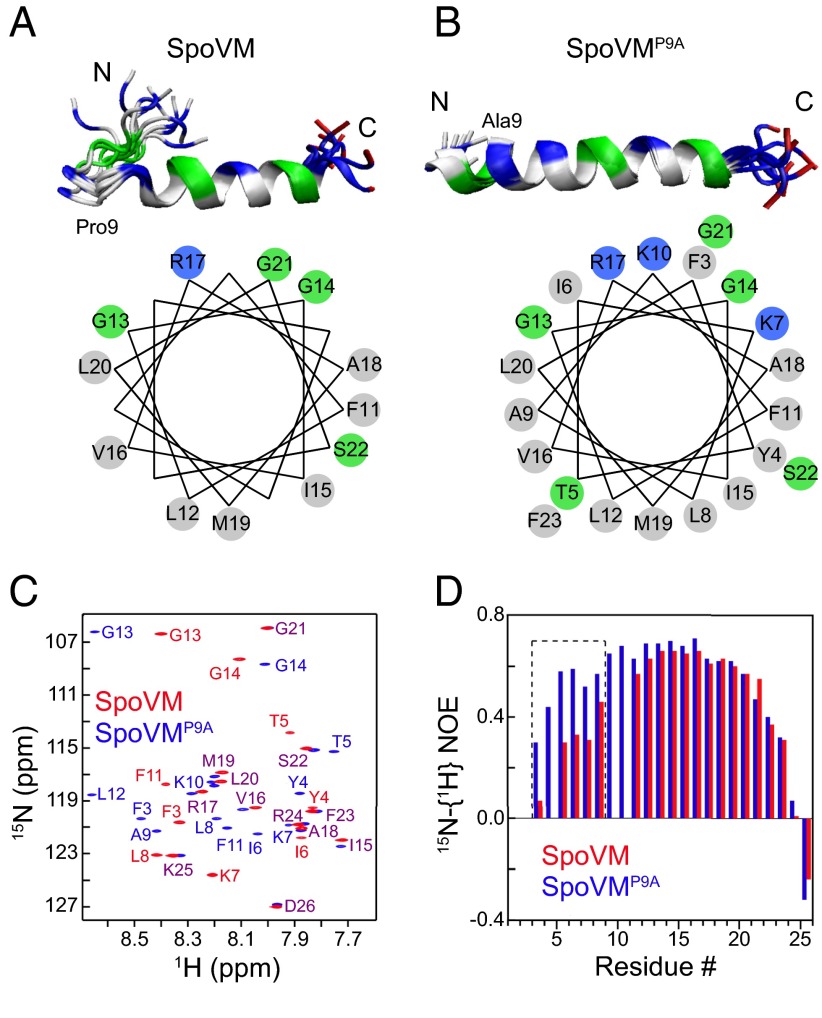
To determine whether a particular structural feature of SpoVM was altered upon substitution of Pro9 with Ala, which in turn abolished geometric cue recognition by SpoVM, we determined the structures of SpoVM and SpoVMP9A in isotropic bicelles using solution NMR (Table S1). Compared with the highly curved surfaces of micelles, the planar surface of disk-like bicelles better mimics the very slightly curved (almost planar) outer surface of the forespore. The solution structures of SpoVM (Fig. 3A) revealed two interesting features. First, instead of forming a long amphipathic α-helix, the actual helical region of SpoVM is short and extends only from residues 11 to 23 (Fig. 3A). Moreover, this helix is not a typical amphipathic α-helix, because roughly two-thirds of its face consists of hydrophobic residues and its hydrophilic face contains only one positively charged residue (R17) and three Gly residues (G13, 14, and 21). Second, residues 5–8 form a loop (Fig. 3A). NMR relaxation measurements indicated that this loop is flexible (Fig. 3D; Fig. S4A) and may transiently interact with some of the helical residues because replacement of Ile6 in the loop with Ala perturbed resonance from nearby residues 3–8 and helical residues 11–14 (Fig. S5).
Fig. 3.
NMR reveals the structures and flexibilities of SpoVM and SpoVMP9A in bilayer-like bicelles. (A and B, Upper) Overlay of 10 lowest-energy structures of SpoVM (A) and SpoVMP9A (B) (PDB ID codes 2MVH and 2MVJ, respectively). Gray, nonpolar residues; green, Gly and polar residues; blue, positively charged residues; red, negatively charged residues. (Lower) Helical wheel plots of SpoVM (A) and SpoVMP9A (B) α-helical regions. (C) Overlay of 2D 15N-1H TROSY spectra with resonance assignments. Extensive and large shifts in resonance positions for residues F3 to G14 indicate that the N termini of SpoVM and SpoVMP9A adopt different structures. Residues that displayed substantial differences are labeled in red for SpoVM and in blue for SpoVMP9A. Residues that displayed no or small shifts are labeled in purple. (D) Heteronuclear 15N-{1H} NOEs. The substantial NOE differences for residues of F3 to L8 (dashed box) show that the N terminus of SpoVM is more flexible than that of SpoVMP9A.
In contrast, the structure of SpoVMP9A (Fig. 3B) revealed that substitution of Pro, a known helix-breaker, at position 9 with an Ala resulted in a straight amphipathic α-helix. Rearrangement of the N-terminal loop in SpoVM to a helix in SpoVMP9A was consistent with extensive and large chemical shift perturbations in their TROSY spectra (Fig. 3C) and with increased backbone steady-state heteronuclear 15N-{1H} NOEs for residues 3–8 (Fig. 3D; Fig. S4). In addition, the circular dichroism spectra of SpoVMP9A displayed a somewhat higher helical content than those of SpoVM (Fig. S6). As a result of this structural change, the Lys residues of SpoVMP9A at positions 7 and 10 align with the polar face of the helix. This change, along with the presence of R17, increases the number of positively charged residues on the polar face from just one in SpoVM to three in SpoVMP9A (Fig. 3 A and B, Lower), suggesting the possibility of increased electrostatic interactions between SpoVMP9A and phospholipid head groups. Taken together, our NMR data provide the structural basis for the critical role of the Pro9 residue for curvature sensitivity.
Membrane Topology of SpoVM.
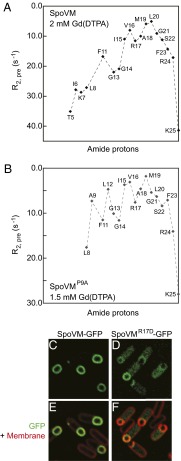
Previous computational studies proposed a link between the topology of a peripheral membrane protein and its ability to detect highly curved membranes (33–35). To determine the insertion depth and membrane orientation of SpoVM in a lipid bilayer, we used NMR paramagnetic relaxation enhancements (PREs) using a water-soluble probe gadopentetic acid [Gd(DTPA)] and a membrane-soluble probe 16-doxylstearic acid (16-DSA) (36, 37). In this technique, amino acid residues accessible by solvent molecules will experience large PRE effects from Gd(DTPA) and small effects from 16-DSA, whereas residues embedded in the membrane will have small PREs from Gd(DTPA) and large effects from 16-DSA. SpoVM residues at the N and C termini exhibited large PREs from Gd(DTPA) (Fig. 4A; Fig. S7A), suggesting that these residues were solvent accessible. In contrast, helical residues 15–22 displayed relatively small PREs from Gd(DTPA) (Fig. 4A; Fig. S7A), and large effects from 16-DSA (Fig. S7C), indicating that the helical region of SpoVM is inserted deeply into the membrane, in contrast to the reported surface association of amphipathic helices that sense highly curved membranes (38). In addition, the observation that PREs from SpoVM helical residues have similar values indicated that the helical axis is oriented approximately parallel to the plane of the membrane, consistent with previous genetic studies suggesting that the hydrophobic face of SpoVM is buried within the phospholipid bilayer (20). Assuming that residue K25 is located close to the bilayer surface and that Gd(DTPA) is inert toward the protein and freely diffusing in solution, the estimated insertion depth for residues M19 and L20 can be more than 10 Å into the membrane (39); the side chains of these residues would penetrate deeply into the hydrocarbon region of the lipid bilayer. For SpoVMP9A, the C terminus of its helix displayed similar small PRE values to those of the SpoVM helix (Fig. 4B; Fig. S7B), suggesting a similar C-terminal orientation. However, its N-terminal residues experienced substantial PRE effects at a low concentration of 0.5 mM Gd(DTPA) (Fig. S7B), indicating that the N-terminal part of the SpoVMP9A helix was solvent accessible. At 1.5 mM Gd(DTPA) (Fig. 4B), the N-terminal residues experienced large line broadening, preventing reliable measurements of their PREs and a quantitative comparison of membrane insertion depth between SpoVM and SpoVMP9A; this is likely due to nonspecific interaction between positively charged residues K2 and K7 and the Gd(DTPA) probe, which is negatively charged.
Fig. 4.
Orientation and insertion depth of SpoVM and SpoVMP9A in the membrane underlie differential interactions with lipid bilayers. (A) PRE effects of Gd(DTPA) on SpoVM. The fact that the effects are small and similar for SpoVM residues 15–22 indicate that the helix is deeply inserted into the membrane and with its axis oriented approximately parallel to the membrane plane, respectively. (B) PRE effects of Gd(DTPA) on SpoVMP9A. Whereas the C terminus of SpoVMP9A experiences similar PRE effects as those of SpoVM, its N terminus PRE effects differ. (C and D) Fluorescence micrographs of in vivo localization of SpoVM-GFP (C) or SpoVMR17D-GFP (D) in sporulating B. subtilis cells. (E and F) Overlay of GFP signals in C and D and membranes visualized with FM4-64 (red).
Given this depth, we wondered whether the positive charge of the R17 side chain could extend, or “snorkel” (40), to interact with the negatively charged phospholipid head groups and thereby help stabilize SpoVM in the membrane. To test this idea, we substituted R17 with a negatively charged Asp and monitored the localization of SpoVMR17D-GFP in vivo. Whereas SpoVM-GFP almost exclusively localized to the forespore surface, substantial SpoVMR17D-GFP fluorescence was detected in the mother cell cytosol (Fig. 4 C–F), suggesting that substitution of the positive charge at position 17 with a negative charge affected the adsorption of SpoVM onto the membrane. Interestingly, substitution of R17 with the hydrophobic Ala did not disrupt localization of SpoVMR17A-GFP (23). Thus, the different in vivo localization of SpoVMR17D-GFP relative to SpoVM-GFP, as well as the NMR PRE measurements, was consistent with a topology of SpoVM in the membrane in which both the backbone and side chains penetrate deeply into the lipid bilayer.
All-Atom Molecular Dynamics Simulations Support Deep Embedding of SpoVM and SpoVMP9A in Membranes.
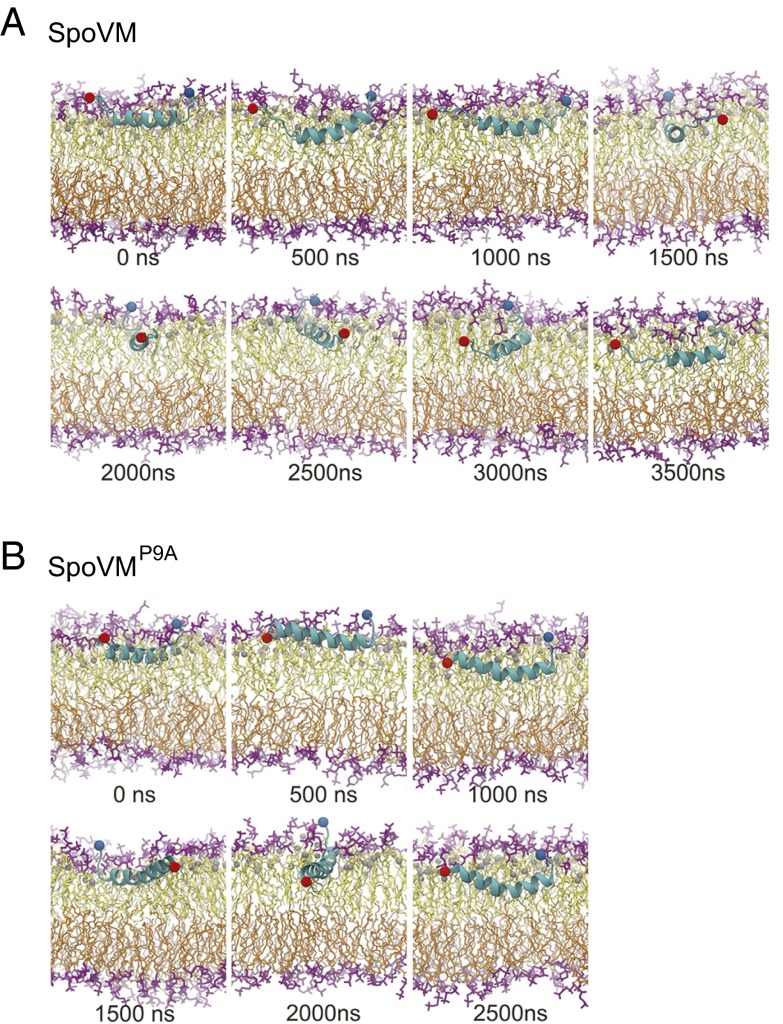
To probe the dynamic interactions between SpoVM or SpoVMP9A and its membrane environment, we conducted microsecond-timescale molecular dynamics simulations on two protein–membrane systems. These simulations were initiated using the NMR structures obtained in this study, but no additional constraints were imposed on the system. In these two simulations, the SpoVM and SpoVMP9A molecules not only remained associated with the membrane, but also were submerged below the head groups of the top bilayer (Fig. 5 A and B and Movies S1 and S2), consistent with our PRE data (Fig. 4). For SpoVM, the central helical region was very stable, whereas the terminal ends were flexible (Fig. 5A; Fig. S8).
Fig. 5.
Molecular dynamics simulations reveal conformations of single molecules of SpoVM and SpoVMP9A in a lipid bilayer. In A and B, snapshots of the membrane–protein systems are shown after every 500-ns interval of the trajectories. Protein is shown in a secondary structure representation in light blue, and the Cα atoms at residue 1 and 26 are shown as red and blue spheres, respectively. Lipid head groups are shown in purple, and tails are shown in yellow and orange for the top and bottom bilayers, respectively. The C2 atoms of the lipid acyl chains used to define the membrane plane and the protein immersion depth are shown as gray spheres. Full trajectories are available as Movies S1 and S2.
We then tracked the average immersion depth of each amino acid in wild-type SpoVM (defined by the distance of each amide proton below the plane of the C2 atoms of the lipid acyl chains in the top bilayer), and found that, with the exception of the C terminus, most amino acids were buried in the membrane (Fig. S9A). However, by examining the SD of the immersion depth, we noted that fluctuations in immersion depth were much higher at the terminal ends (Fig. S9A, Inset). This phenomenon was recapitulated when we measured the average number of water molecules in contact with each amide proton (within 4 Å), and found that the N terminus was solvent accessible (Fig. S9B). This solvent accessibility may be important for the recruitment of cytoplasmic sporulation proteins or for establishing contacts with other SpoVM molecules. Previous genetic and biochemical studies revealed that I6 of SpoVM directly contacts a C-terminal residue of the cytoplasmic coat protein SpoIVA (20); our simulation indicates that such a contact would be physically possible despite the deep insertion of SpoVM into the membrane. As indicated by our NMR data (Fig. 3D; Fig. S3), the N terminus of SpoVM is structurally flexible; in our molecular dynamics simulation, we observed that the motion of the N terminus extensively scanned the direction normal to the membrane plane as well as the direction along the membrane plane (Fig. 5A; Fig. S8C), potentially facilitating the search for interacting partners.
In contrast to SpoVM, SpoVMP9A maintained its mostly helical structure throughout the simulation, and flexibility was only prominent at the C terminus (Fig. 5B, Fig. S8A, and Movie S2). Nonetheless, both SpoVM and SpoVMP9A persistently positioned their helical regions parallel to the membrane plane. Our molecular dynamics simulations support a scenario in which SpoVM and SpoVMP9A have a stable orientation parallel to the plane of the membrane, and indicate that both proteins are capable of inserting deep into the membrane bilayer without displacing the phospholipid head groups. Furthermore, despite the insertion depth of SpoVM, the flexibility of the N terminus of SpoVM makes it physically possible for N-terminal residues to become solvent accessible to potentially contact other SpoVM molecules or to contact cytoplasmic proteins in the mother cell during sporulation.
Discussion
Despite the general lack of organelles and vesicle-mediated protein-sorting machinery in bacterial cells, they are capable of localizing certain proteins to distinct subcellular locations. However, the mechanisms by which landmark proteins initially achieve their own proper localization are frequently unclear. Previously, we proposed that during sporulation in B. subtilis, the slightly convex surface of the developing forespore provides a geometric cue for the initial localization of the shape-sensing amphipathic α-helical protein SpoVM (24). In this report, we describe a previously unidentified structure-motivated mechanism by which SpoVM discriminates between differently curved membrane surfaces.
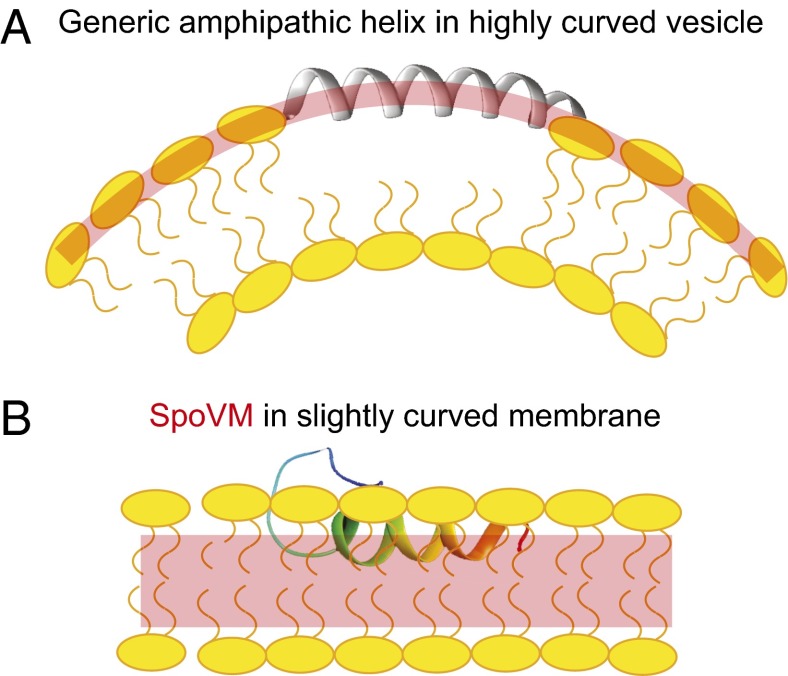
Current understanding of membrane curvature sensing mainly comes from the study of the recognition of highly curved membranes, such as those formed by ∼50 nm-diameter vesicles (28, 38, 41–43). In those scenarios, the prevailing model for curvature recognition is based on the premise that highly curved surfaces provide more binding sites for curvature-recognizing proteins, presumably because acute membrane curvature drives phospholipid head groups in the outer leaflet apart, and increases the occurrences of packing defects that are manifested near the surface of the membrane (26, 28) (Fig. 6A). Shallow insertion of an amphipathic helix into the membrane is thought to alleviate these stresses (44). Consistent with this model, biochemical studies have shown that curvature-sensing proteins bind to small lipid vesicles (more curved surfaces) with a higher Bmax, but with similar binding affinity and cooperativity (28).
Fig. 6.
Model for the preferential adsorption of SpoVM onto slightly curved membranes. (A) Depiction of a standard amphipathic α-helix (gray) inserted into a highly curved membrane, such as that found on ∼50-nm-diameter budding vesicles. Lipid-packing defects (curved red bar) at this scale are manifested near the head groups (yellow) of the outer leaflet; these defects provide a higher density of binding sites for amphipathic α-helices. (B) Depiction of SpoVM (multicolored) inserted into a slightly curved membrane, such as that found on ∼1,000-nm-diameter forespores. At this scale, packing defects are likely not detectable near the lipid head groups. We propose that the deep insertion of SpoVM allows for the detection of differences in packing near the acyl chains (highlighted in red) in the hydrocarbon region of the lipid bilayer between differently curved membranes.
Compared with the highly curved membranes of small (∼50 nm) vesicles, the substrate recognized by SpoVM is much less curved (the forespore is ∼1,000 nm in diameter), and any asymmetry between leaflets of these bilayers would likely be negligible to a single molecule of SpoVM (45) (Fig. 6B). Consistent with this notion, here we found that SpoVM does not use a Bmax-driven mechanism for its preferential adsorption to the surface of the forespore, but rather uses increased affinity for positively curved membranes (decreased K1/2) combined with a modest cooperativity (increased h; Fig. 1 C and D; Table 1). In contrast, the functionally null SpoVMP9A, which mislocalizes in vivo, showed similar binding affinity and cooperativity to 2- and 8-μm SSLBs in vitro (Fig. 1 C and D; Table 1). Monte Carlo simulations indicated that small increases in affinity and cooperative interactions are sufficient for producing SpoVM membrane curvature selectivity (Fig. 2). The unusually deep insertion of SpoVM into the membrane likely precludes any significant interaction with the phospholipid head groups, and instead implicates more significant interactions with the acyl chains, which we predict must be differentially packed between membranes that sit atop 2- and 8-μm SSLBs (Fig. 6B). Thus, the preferential adsorbance of SpoVM to substrates with slight positive curvature is enabled through a fundamentally distinct membrane curvature-recognition mechanism from those used by high curvature-sensing proteins. SpoVM curvature recognition is driven by affinity and cooperativity, rather than enhanced maximal binding produced by increased numbers of membrane defects, and SpoVM interacts extensively with the acyl chains and therefore likely detects lipid packing differences deep in the membrane, not at the surface (Fig. 6) (43).
Beyond classic roles as membrane anchors and membrane-destabilizing agents, amphipathic helices have increasingly been implicated in sensing and modulating membrane properties. Our study highlights how an amphipathic helix can be tuned for membrane curvature sensing despite its structural simplicity. Compared with the SpoVMP9A structure, which was α-helical throughout its entire length and harbored multiple positive charges on its polar face, the wild-type protein contained a relatively short helical segment with a flexible N terminus and a single positive charge on its polar face (Fig. 3). Which of these structural features is critical for curvature sensing by SpoVM? The recognition of highly convex membranes relies on a delicate balance between electrostatic and hydrophobic interactions (46). In the amphipathic lipid packing sensor motif, the lack of positively charged residues in the polar face (e.g., ArfGAP1) or the absence of bulky hydrophobic residues at nonpolar faces (e.g., α-synuclein) weakens the protein’s interactions with the membrane. As a result, the association with membranes requires packing defects, a hallmark of highly curved membranes, and the proteins become sensitive to membrane curvature by preferentially embedding into surfaces that display this feature (Fig. 6A). It is conceivable that the increase in the number of positive charges on the polar face increases the strength of electrostatic interactions between SpoVMP9A and lipids, thereby disrupting the balance between electrostatic and hydrophobic interactions and leading to the mutant’s loss of sensitivity to membrane curvature. Second, increased flexibility of an α-helix due to a kink from a Pro residue has been linked to the abilities of the melittin peptide (47–49) and antimicrobial peptides (27, 50) to multimerize in the membrane. In SpoVM, Pro9 disrupted the helix and resulted in a flexible and solvent-accessible N terminus, as seen by both molecular dynamics simulations and NMR. At the very least, this accessibility would permit SpoVM to interact with and anchor a cytosolic sporulation protein, as proposed previously (20). Furthermore, we speculate that the flexibility of the N terminus may permit the interaction of SpoVM molecules with one another (absent in SpoVMP9A), thereby potentially providing a molecular mechanism by which cooperativity may be directly achieved within a lipid bilayer. Thus, the highly conserved Pro9 of SpoVM (23) provides a flexible N terminus and an altered helical charge profile, and is required for recognition of slightly curved membranes such as the forespore.
The orientation and insertion depth of a membrane protein in the bilayer directly affects protein–lipid interactions and is a critical determinant of the functions of some proteins (41, 44). In this study, we found that SpoVM is deeply embedded in the membrane (Fig. 4A), as opposed to a presumed shallow insertion for other curvature-sensing molecules. This scenario is likely a direct result of the chemistry of SpoVM’s atypical amphipathic structure, which has a large nonpolar surface and only one positively charged residue (R17) at the polar surface. The long side-chain of R17 can extend its guanidinium group toward the membrane surface to interact with the negatively charged phospholipid head groups, whereas the hydrophobic side chains of SpoVM remain inside the nonpolar region of the membrane (Fig. 4A). Consistent with this notion, replacement of Arg17 with a negatively charged Asp residue resulted in mislocalization of the protein in vivo (Fig. 4 C–F), presumably due to a perturbation in its membrane insertion depth by removing this snorkeling effect. Therefore, unlike amphipathic α-helical motifs that induce membrane curvature by inserting shallowly into the membrane and pushing apart phospholipid head groups to create asymmetry in the lipid bilayer (51), SpoVM primarily interacts with the acyl chains of the membrane (Fig. 6B). Recent computational studies hypothesized that deep membrane insertion of a hydrophobic moiety increases the stability and rate of protein self-assembly (34, 35, 52), whereas shallow insertion effectively produces or stabilizes highly curved membranes (33, 44). We postulate that deep membrane insertion may promote direct SpoVM–SpoVM intermolecular interactions to form higher-order structures, or promote clustering of SpoVM molecules within the lipid bilayer indirectly through membrane–protein interactions, to produce the observed modest binding cooperativity.
What, then, is the physical difference sensed by SpoVM molecules that allows them to distinguish between a membrane that sits atop a 2-μm bead from one on an 8-μm bead? Given that SpoVM differs in adsorption behavior, α-helical structure, and membrane insertion depth from amphipathic α-helices that recognize highly curved membranes, we hypothesize that there may be subtle differences in the way that acyl chains are packed between membrane bilayers harboring these different radii of curvature. The acyl chain environment curved around 2-μm lipid vesicles may offer SpoVM a slightly more stable interaction than one that is curved around 8-μm vesicles. This lipid environment may also provide favorable driving forces for interactions between SpoVM molecules, perhaps mediated directly by the protein’s flexible N terminus and indirectly by the membrane (53). Together, the slight increases in affinity and binding cooperativity ultimately lead to the preferential adsorption of SpoVM onto convex membranes that we observed experimentally (Fig. 1 B–D).
The unique structural properties of SpoVM provide a template for predicting the existence of other proteins that preferentially bind to slightly curved membrane surfaces, such as those that encapsulate organelles. For example, short helical regions that are amphipathic, display a large hydrophobic face, and are combined with a flexible terminus or loop region may be ideal candidates for proteins that recognize geometric localization cues. Our assay using SSLBs may also enable the biochemical isolation of proteins in other systems that preferentially bind to slightly curved membranes. Given the relatively ancient evolutionary origins of bacterial endospores, perhaps the recognition of geometric localization cues represents a widely exploited strategy for subcellular protein localization.
Materials and Methods
B. subtilis strains used in this study are derivatives of PY79 (54). SpoVM-GFP-His6 or SpoVMP9A-GFP-His6 purification (24), SSLB preparation (29, 30), and microscopy (7) were performed as described previously. All NMR data were acquired at 37 °C on Bruker 600- or 850-MHz spectrometers equipped with cryoprobes. The data were processed using NMRPipe and analyzed using NMRView. Molecular dynamics simulations were carried out on the special-purpose Anton supercomputer (55) and analyzed with VMD (56). Details of strain construction and growth, assays, structure determination, and simulations are described in SI Materials and Methods.
Supplementary Material
Acknowledgments
We thank S. Lecuyer and J. Flanagan for discussion; S. Gottesman, R. Losick, A. Hitchcock-Camp, and members of the K.C.H., F.T., and K.S.R. laboratories for comments on the manuscript; Cordelia Weiss for construction of strain CW245; and A. Benesi and E. Hatzakis at the NMR facility of Pennsylvania State University for assistance with the 850-MHz instrument. Long time-scale MD simulations were carried out on the Anton supercomputer at the Pittsburgh Supercomputing Center, supported by National Institute of General Medical Sciences (NIGMS) National Center for Multiscale Modeling of Biological Systems Grant P41GM103712-S1. Access to the Anton supercomputer was granted through Proposal PSCA13052P (to K.C.H. and J.H.). Auxiliary simulations were conducted with computer time provided by the Extreme Science and Engineering Discovery Environment, supported by National Science Foundation Grant OCI-1053575, Allocation TG-MCB110056 (to K.C.H. and J.H.). This work was funded by NIH NIGMS Grant R01GM105963 (to F.T. and K.C.H.), NIH Ruth L. Kirschstein National Research Service Award 1F32GM100677 (to J.H.), and the Intramural Research Program of the NIH, National Cancer Institute, Center for Cancer Research (K.S.R.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The atomic coordinates have been deposited in the Protein Data Bank, www.pdb.org (PDB ID codes 2MVH and 2MVJ).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1423868112/-/DCSupplemental.
References
- 1.Zanetti G, Pahuja KB, Studer S, Shim S, Schekman R. COPII and the regulation of protein sorting in mammals. Nat Cell Biol. 2012;14(1):20–28. doi: 10.1038/ncb2390. [DOI] [PubMed] [Google Scholar]
- 2.Shapiro L, McAdams HH, Losick R. Why and how bacteria localize proteins. Science. 2009;326(5957):1225–1228. doi: 10.1126/science.1175685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Deich J, Judd EM, McAdams HH, Moerner WE. Visualization of the movement of single histidine kinase molecules in live Caulobacter cells. Proc Natl Acad Sci USA. 2004;101(45):15921–15926. doi: 10.1073/pnas.0404200101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rudner DZ, Pan Q, Losick RM. Evidence that subcellular localization of a bacterial membrane protein is achieved by diffusion and capture. Proc Natl Acad Sci USA. 2002;99(13):8701–8706. doi: 10.1073/pnas.132235899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rudner DZ, Losick R. Protein subcellular localization in bacteria. Cold Spring Harb Perspect Biol. 2010;2(4):a000307. doi: 10.1101/cshperspect.a000307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lam H, Schofield WB, Jacobs-Wagner C. A landmark protein essential for establishing and perpetuating the polarity of a bacterial cell. Cell. 2006;124(5):1011–1023. doi: 10.1016/j.cell.2005.12.040. [DOI] [PubMed] [Google Scholar]
- 7.Eswaramoorthy P, et al. Asymmetric division and differential gene expression during a bacterial developmental program requires DivIVA. PLoS Genet. 2014;10(8):e1004526. doi: 10.1371/journal.pgen.1004526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Greenfield D, et al. Self-organization of the Escherichia coli chemotaxis network imaged with super-resolution light microscopy. PLoS Biol. 2009;7(6):e1000137. doi: 10.1371/journal.pbio.1000137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mukhopadhyay R, Huang KC, Wingreen NS. Lipid localization in bacterial cells through curvature-mediated microphase separation. Biophys J. 2008;95(3):1034–1049. doi: 10.1529/biophysj.107.126920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ramamurthi KS. Protein localization by recognition of membrane curvature. Curr Opin Microbiol. 2010;13(6):753–757. doi: 10.1016/j.mib.2010.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Strahl H, Hamoen LW. Finding the corners in a cell. Curr Opin Microbiol. 2012;15(6):731–736. doi: 10.1016/j.mib.2012.10.006. [DOI] [PubMed] [Google Scholar]
- 12.Higgins D, Dworkin J. Recent progress in Bacillus subtilis sporulation. FEMS Microbiol Rev. 2012;36(1):131–148. doi: 10.1111/j.1574-6976.2011.00310.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.McKenney PT, Driks A, Eichenberger P. The Bacillus subtilis endospore: Assembly and functions of the multilayered coat. Nat Rev Microbiol. 2013;11(1):33–44. doi: 10.1038/nrmicro2921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Driks A, Roels S, Beall B, Moran CP, Jr, Losick R. Subcellular localization of proteins involved in the assembly of the spore coat of Bacillus subtilis. Genes Dev. 1994;8(2):234–244. doi: 10.1101/gad.8.2.234. [DOI] [PubMed] [Google Scholar]
- 15.McKenney PT, et al. A distance-weighted interaction map reveals a previously uncharacterized layer of the Bacillus subtilis spore coat. Curr Biol. 2010;20(10):934–938. doi: 10.1016/j.cub.2010.03.060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Roels S, Driks A, Losick R. Characterization of spoIVA, a sporulation gene involved in coat morphogenesis in Bacillus subtilis. J Bacteriol. 1992;174(2):575–585. doi: 10.1128/jb.174.2.575-585.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ramamurthi KS, Losick R. ATP-driven self-assembly of a morphogenetic protein in Bacillus subtilis. Mol Cell. 2008;31(3):406–414. doi: 10.1016/j.molcel.2008.05.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Castaing JP, Nagy A, Anantharaman V, Aravind L, Ramamurthi KS. ATP hydrolysis by a domain related to translation factor GTPases drives polymerization of a static bacterial morphogenetic protein. Proc Natl Acad Sci USA. 2013;110(2):E151–E160. doi: 10.1073/pnas.1210554110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Castaing JP, et al. An autoinhibitory conformation of the Bacillus subtilis spore coat protein SpoIVA prevents its premature ATP-independent aggregation. FEMS Microbiol Lett. 2014;358(2):145–153. doi: 10.1111/1574-6968.12452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ramamurthi KS, Clapham KR, Losick R. Peptide anchoring spore coat assembly to the outer forespore membrane in Bacillus subtilis. Mol Microbiol. 2006;62(6):1547–1557. doi: 10.1111/j.1365-2958.2006.05468.x. [DOI] [PubMed] [Google Scholar]
- 21.Prajapati RS, Ogura T, Cutting SM. Structural and functional studies on an FtsH inhibitor from Bacillus subtilis. Biochim Biophys Acta. 2000;1475(3):353–359. doi: 10.1016/s0304-4165(00)00089-1. [DOI] [PubMed] [Google Scholar]
- 22.Levin PA, et al. An unusually small gene required for sporulation by Bacillus subtilis. Mol Microbiol. 1993;9(4):761–771. doi: 10.1111/j.1365-2958.1993.tb01736.x. [DOI] [PubMed] [Google Scholar]
- 23.van Ooij C, Losick R. Subcellular localization of a small sporulation protein in Bacillus subtilis. J Bacteriol. 2003;185(4):1391–1398. doi: 10.1128/JB.185.4.1391-1398.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ramamurthi KS, Lecuyer S, Stone HA, Losick R. Geometric cue for protein localization in a bacterium. Science. 2009;323(5919):1354–1357. doi: 10.1126/science.1169218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Daumke O, Roux A, Haucke V. BAR domain scaffolds in dynamin-mediated membrane fission. Cell. 2014;156(5):882–892. doi: 10.1016/j.cell.2014.02.017. [DOI] [PubMed] [Google Scholar]
- 26.Bigay J, Antonny B. Curvature, lipid packing, and electrostatics of membrane organelles: Defining cellular territories in determining specificity. Dev Cell. 2012;23(5):886–895. doi: 10.1016/j.devcel.2012.10.009. [DOI] [PubMed] [Google Scholar]
- 27.Yang ST, et al. Contribution of a central proline in model amphipathic alpha-helical peptides to self-association, interaction with phospholipids, and antimicrobial mode of action. FEBS J. 2006;273(17):4040–4054. doi: 10.1111/j.1742-4658.2006.05407.x. [DOI] [PubMed] [Google Scholar]
- 28.Hatzakis NS, et al. How curved membranes recruit amphipathic helices and protein anchoring motifs. Nat Chem Biol. 2009;5(11):835–841. doi: 10.1038/nchembio.213. [DOI] [PubMed] [Google Scholar]
- 29.Bayerl TM, Bloom M. Physical properties of single phospholipid bilayers adsorbed to micro glass beads. A new vesicular model system studied by 2H-nuclear magnetic resonance. Biophys J. 1990;58(2):357–362. doi: 10.1016/S0006-3495(90)82382-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gopalakrishnan G, Rouiller I, Colman DR, Lennox RB. Supported bilayers formed from different phospholipids on spherical silica substrates. Langmuir. 2009;25(10):5455–5458. doi: 10.1021/la9006982. [DOI] [PubMed] [Google Scholar]
- 31.Wasnik V, Wingreen NS, Mukhopadhyay R. Modeling curvature-dependent subcellular localization of the small sporulation protein SpoVM in Bacillus subtilis. PLoS ONE. 2015;10(1):e0111971. doi: 10.1371/journal.pone.0111971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bhatia VK, Hatzakis NS, Stamou D. A unifying mechanism accounts for sensing of membrane curvature by BAR domains, amphipathic helices and membrane-anchored proteins. Semin Cell Dev Biol. 2010;21(4):381–390. doi: 10.1016/j.semcdb.2009.12.004. [DOI] [PubMed] [Google Scholar]
- 33.Campelo F, McMahon HT, Kozlov MM. The hydrophobic insertion mechanism of membrane curvature generation by proteins. Biophys J. 2008;95(5):2325–2339. doi: 10.1529/biophysj.108.133173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Li S, Zhang X, Wang W. Cluster formation of anchored proteins induced by membrane-mediated interaction. Biophys J. 2010;98(11):2554–2563. doi: 10.1016/j.bpj.2010.02.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yue TT, Li SY, Zhang XR, Wang WC. The relationship between membrane curvature generation and clustering of anchored proteins: A computer simulation study. Soft Matter. 2010;6(24):6109–6118. [Google Scholar]
- 36.Clore GM, Iwahara J. Theory, practice, and applications of paramagnetic relaxation enhancement for the characterization of transient low-population states of biological macromolecules and their complexes. Chem Rev. 2009;109(9):4108–4139. doi: 10.1021/cr900033p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Beel AJ, et al. Structural studies of the transmembrane C-terminal domain of the amyloid precursor protein (APP): Does APP function as a cholesterol sensor? Biochemistry. 2008;47(36):9428–9446. doi: 10.1021/bi800993c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bigay J, Casella JF, Drin G, Mesmin B, Antonny B. ArfGAP1 responds to membrane curvature through the folding of a lipid packing sensor motif. EMBO J. 2005;24(13):2244–2253. doi: 10.1038/sj.emboj.7600714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Respondek M, Madl T, Göbl C, Golser R, Zangger K. Mapping the orientation of helices in micelle-bound peptides by paramagnetic relaxation waves. J Am Chem Soc. 2007;129(16):5228–5234. doi: 10.1021/ja069004f. [DOI] [PubMed] [Google Scholar]
- 40.Strandberg E, Killian JA. Snorkeling of lysine side chains in transmembrane helices: How easy can it get? FEBS Lett. 2003;544(1-3):69–73. doi: 10.1016/s0014-5793(03)00475-7. [DOI] [PubMed] [Google Scholar]
- 41.Qiang W, Sun Y, Weliky DP. A strong correlation between fusogenicity and membrane insertion depth of the HIV fusion peptide. Proc Natl Acad Sci USA. 2009;106(36):15314–15319. doi: 10.1073/pnas.0907360106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bigay J, Gounon P, Robineau S, Antonny B. Lipid packing sensed by ArfGAP1 couples COPI coat disassembly to membrane bilayer curvature. Nature. 2003;426(6966):563–566. doi: 10.1038/nature02108. [DOI] [PubMed] [Google Scholar]
- 43.Peter BJ, et al. BAR domains as sensors of membrane curvature: The amphiphysin BAR structure. Science. 2004;303(5657):495–499. doi: 10.1126/science.1092586. [DOI] [PubMed] [Google Scholar]
- 44.Ambroso MR, Hegde BG, Langen R. Endophilin A1 induces different membrane shapes using a conformational switch that is regulated by phosphorylation. Proc Natl Acad Sci USA. 2014;111(19):6982–6987. doi: 10.1073/pnas.1402233111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Huang KC, Ramamurthi KS. Macromolecules that prefer their membranes curvy. Mol Microbiol. 2010;76(4):822–832. doi: 10.1111/j.1365-2958.2010.07168.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Drin G, et al. A general amphipathic alpha-helical motif for sensing membrane curvature. Nat Struct Mol Biol. 2007;14(2):138–146. doi: 10.1038/nsmb1194. [DOI] [PubMed] [Google Scholar]
- 47.Terwilliger TC, Eisenberg D. The structure of melittin. II. Interpretation of the structure. J Biol Chem. 1982;257(11):6016–6022. [PubMed] [Google Scholar]
- 48.Dempsey CE, et al. Contribution of proline-14 to the structure and actions of melittin. FEBS Lett. 1991;281(1-2):240–244. doi: 10.1016/0014-5793(91)80402-o. [DOI] [PubMed] [Google Scholar]
- 49.Raghuraman H, Chattopadhyay A. Melittin: A membrane-active peptide with diverse functions. Biosci Rep. 2007;27(4-5):189–223. doi: 10.1007/s10540-006-9030-z. [DOI] [PubMed] [Google Scholar]
- 50.Bobone S, et al. The importance of being kinked: Role of Pro residues in the selectivity of the helical antimicrobial peptide P5. J Pept Sci. 2013;19(12):758–769. doi: 10.1002/psc.2574. [DOI] [PubMed] [Google Scholar]
- 51.Kozlov MM, et al. Mechanisms shaping cell membranes. Curr Opin Cell Biol. 2014;29:53–60. doi: 10.1016/j.ceb.2014.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Morozova D, Guigas G, Weiss M. Dynamic structure formation of peripheral membrane proteins. PLOS Comput Biol. 2011;7(6):e1002067. doi: 10.1371/journal.pcbi.1002067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Phillips R, Ursell T, Wiggins P, Sens P. Emerging roles for lipids in shaping membrane-protein function. Nature. 2009;459(7245):379–385. doi: 10.1038/nature08147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Youngman P, Perkins JB, Losick R. Construction of a cloning site near one end of Tn917 into which foreign DNA may be inserted without affecting transposition in Bacillus subtilis or expression of the transposon-borne erm gene. Plasmid. 1984;12(1):1–9. doi: 10.1016/0147-619x(84)90061-1. [DOI] [PubMed] [Google Scholar]
- 55.Shaw DE, et al. Anton, a special-purpose machine for molecular dynamics simulation. Commun ACM. 2008;51(7):91–97. [Google Scholar]
- 56.Humphrey W, Dalke A, Schulten K. VMD: Visual molecular dynamics. J Mol Graph. 1996;14(1):33–38, 27–28. doi: 10.1016/0263-7855(96)00018-5. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.