Significance
The development of an embryo from a single fertilized cell is orchestrated by a large set of key regulatory genes whose activities need to be precisely controlled. Proteins from the Polycomb group (PcG) family maintain the inactive state of these genes by modifying the surrounding histone H3 tails. Here we report that these inactive genes contact other PcG-rich regions within otherwise active environments in the cell nucleus, suggesting the presence of repressive microenvironments. PcG positive genes interact independently from neighboring genes, depending on their linear distance and more global chromosome folding characteristics, with only moderate changes during embryonic development.
Keywords: Hox genes, Polycomb, long-range chromatin contacts, nuclear organization
Abstract
Embryogenesis requires the precise activation and repression of many transcriptional regulators. The Polycomb group proteins and the associated H3K27me3 histone mark are essential to maintain the inactive state of many of these genes. Mammalian Hox genes are targets of Polycomb proteins and form local 3D clusters centered on the H3K27me3 mark. More distal contacts have also been described, yet their selectivity, dynamics, and relation to other layers of chromatin organization remained elusive. We report that repressed Hox genes form mutual intra- and interchromosomal interactions with other genes located in strong domains labeled by H3K27me3. These interactions occur in a central and active nuclear environment that consists of the HiC compartment A, away from peripheral lamina-associated domains. Interactions are independent of nearby H3K27me3-marked loci and determined by chromosomal distance and cell-type–specific scaling factors, thus inducing a moderate reorganization during embryogenesis. These results provide a simplified view of nuclear organization whereby Polycomb proteins may have evolved to repress genes located in gene-dense regions whose position is restricted to central, active, nuclear environments.
Embryonic development relies on a precise implementation of transcriptional programs in space and time. These programs are controlled by a balance between the presence and absence of many transcriptional regulators, which will determine future cell-type or tissue specificities (1). Proteins from the Polycomb group (PcG) family and their associated H3K27me3 histone mark maintain the inactive state of these transcriptional regulators (2, 3). In particular, PcG repression is initially observed at Hox gene clusters, which need to be silenced early on to avoid inappropriate temporal and spatial activation (4, 5). Mammalian Hox genes are found at four genomic loci and their embryonic regulation involves a progressive reorganization in time of two local 3D microcompartments, labeled by the opposing H3K4me3 and H3K27me3 histone marks (6–8).
Previously, we reported that the H3K27me3-marked part of the HoxD cluster interacts with the Dlx1/Dlx2 gene cluster located 3 Mb upstream and also labeled with H3K27me3 (7). In Drosophila, similar PcG-driven interactions were detected between the Hox gene subclusters (ANTP-C and BX-C) and other Polycomb targets (9, 10). In mouse ES cells, a similar tendency for H3K27me3-marked Hox clusters to interact with each other was detected at lower resolution, as well as with a few other H3K27me3 targets (11). However, the dynamics, selectivity, and potential functional relevance of PcG target interactions remained elusive.
Here we show that H3K27me3-marked loci, including Hox clusters, form a network of intra- and interchromosomal interactions. These interactions occur in an active nuclear environment [the active HiC compartment A (12), located away from peripheral lamina-associated domains (LADs) (13)]. Interactions are established independently of nearby-located H3K27me3-marked loci, based on the linear distance between loci, the strength of the involved H3K27me3 domains, and the cell-type–specific chromatin folding patterns. Accordingly, we detect only moderate reorganization of these interactions during cellular differentiation.
Results
Hox Clusters Are Part of a Network of H3K27me3-Marked Long-Range Interactions, Extending Beyond Topologically Associating Domains.
To further document the fact that inactive Hoxd genes interact with the H3K27me3-marked Dlx1/2 cluster (7), we assessed their interactions in different cellular contexts. Both Hox and Dlx loci are inactive in ES cells, in embryonic day 10.5 (E10.5) forebrain cells, as well as in axial “junction” cells from the distal part of the future mandibule (SI Appendix, Fig. S1 A and B). Circularized chromosome conformation capture sequencing (4C-seq) using Hoxd13 as a bait showed strong interactions with the inactive Dlx1/2 cluster in all three cell types (Fig. 1A). Moreover, a robust interaction was scored with the H3K27me3-marked Sp9 gene located in between. Reciprocal experiments with the Dlx1 and Sp9 genes as baits confirmed the interactions (SI Appendix, Fig. S1C). Analyses at both HoxB and HoxA clusters also identified interactions with H3K27me3-marked loci in the neighboring 4 Mb, whereas such interactions were not detected at HoxC (SI Appendix, Fig. S2 and below). These PcG-mediated interactions were not restricted to topologically associating domains (TADs) and thus likely extend beyond common enhancer–promoter contacts (14, 15) (SI Appendix, Figs. S1C and S2).
Fig. 1.
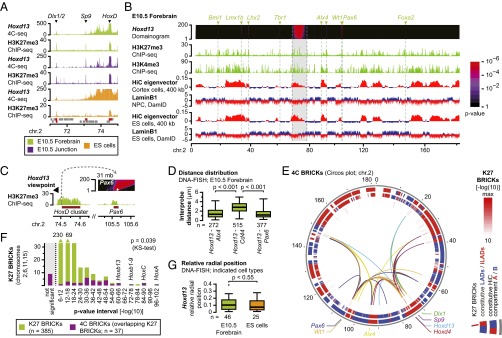
The inactive HoxD cluster is involved in a network of intrachromosomal H3K27me3-marked interactions. (A) Patterns of 4C signal Hoxd13 as a bait within a 4-Mb interval identifies interactions with the H3K27me3-marked Dlx1/2 and Sp9 genes in multiple cell types. Genes are indicated below the patterns, with H3K27me3 peaks in red. (B) Domainogram with intrachromosomal interactions of the Hoxd13 gene. Significant interacting regions are H3K27me3 positive (arrowheads with H3K27me3-marked genes on Top) and are primarily located in the active HiC compartment A and in iLADs (red signal). The 10-Mb saturated region around the viewpoint is indicated in gray. (C) Magnification into the domainogram at the Pax6 locus. The “stem” of the interaction overlaps the H3K27me3 signal (white arrowhead). (D) Distance between Hoxd13 and three genes in single cells. The H3K27me3-marked Alx4 and Pax6 genes are on average closer to Hoxd13 than the interspersed, non–H3K27me3-marked Cd44 gene. (E) Circos plot with intrachromosomal interactions for seven H3K27me3-marked viewpoints showing mutual interactions between significant H3K27me3-marked loci (inner ring). Interactions occur primarily in the parts of the genome that have an active signature (middle and outer rings; legend on the Right). (F) Intrachromosomal interactions on four chromosomes are significantly enriched for H3K27me3-rich regions. K27 BRICKs that are too close to viewpoints to be analyzed are indicated in gray. (G) Relative radial position of the HoxD cluster in single cells. The Hoxd13 gene is mostly located near the nuclear periphery but does not overlap the LaminB1 signal (see also SI Appendix, Fig. S6). Relative radial position 0 = nuclear periphery and 0.5 = nuclear center.
To assess the generality of these observations, we systematically analyzed the long-range intrachromosomal interactions of H3K27me3-marked loci. A domainogram approach was used to identify interactions at high resolution. Using this approach, significantly interacting blocks of regulators in chromosomal context (BRICKs) can be identified based on the clustering of positive signal (16–18). Using the Hoxd13 dataset in E10.5 forebrain cells, we detected nine significant intrachromosomal interactions outside the saturated 10 Mb around HoxD (Fig. 1B and SI Appendix, Fig. S3A). To correlate these interactions with H3K27me3 marks, we subjected the ChIP-seq data to a domainogram analysis using the same parameters as for the 4C analysis (SI Appendix). This approach identifies H3K27me3-marked regions (K27 BRICKs) and assigns them a significance score. Eight of the nine 4C BRICKs overlapped with highly significant K27 BRICKs (P < 10−18, SI Appendix, Fig. S3A). Significant 4C BRICKs, up to 70 Mb away from the Hoxd13 viewpoint, precisely stemmed from the H3K27me3-marked domains, confirming the link between the PcG coverage and the interactions (Fig. 1C, arrowhead and SI Appendix, Fig. S3B). In individual cells the Hoxd13 gene was on average more proximal to the interacting Alx4 and Pax6 genes than to the interspersed, non–H3K27me3-marked Cd44 gene locus, supporting the observations (Fig. 1D and SI Appendix, Fig. S3C).
We expanded the analysis by adding six H3K27me3-marked viewpoints on chromosome 2 and found that most interactions were shared with Hoxd13 (Fig. 1E and SI Appendix, File S1). A linear visualization showed the influence of distance, with more centromeric genes contacting fewer telomeric loci, and vice versa (SI Appendix, Fig. S3A). The analysis of two non–H3K27me3-marked loci did not identify these interactions, confirming that they were not common to all loci on chromosome 2 (SI Appendix, Fig. S3D). We confirmed that networks of H3K27me3-marked interactions also existed on other chromosomes by reexploring previous data for other Hox clusters in E10.5 forebrain cells (7). All Hox clusters, which are the most significant K27 BRICK on their respective chromosomes, also interacted with other K27 BRICKs (SI Appendix, Fig. S3E). To explore the possibility of interchromosomal interactions, we also generated genome-wide domainograms. By using the same H3K27me3-marked viewpoints on chromosome 2 and in other Hox clusters, we identified 68 significant interchromosomal 4C BRICKs. Of these BRICKs, 40 overlapped with highly significant K27 BRICKs (P < 10−18) and many were contacted by viewpoints located on different chromosomes (SI Appendix, Fig. S4A). However, the 4C signals of these interchromosomal interactions were considerably weaker than for intrachromosomal interactions (SI Appendix, Fig. S4B).
Long-Range Interactions Involve Strong H3K27me3 Targets.
Even though long-range interactions were directly associated with H3K27me3 marks, we did not detect significant interactions among all K27 BRICKs, suggesting some selectivity. We scored the significance of the K27 BRICKs, which combines both signal level and size of the domain (16), and found a direct link with the detection of interactions (Fig. 1F). Of the 37 unique intrachromosomal 4C BRICKs obtained from the 16 H3K27me3-marked viewpoints located on four chromosomes, 28 overlapped with a significant K27 BRICK (P values 10−6 to 10−102). Whereas most low significant K27 BRICKs were not contacted (less than 1% for the P value range from 10−6 to 10−12), highly significant BRICKs were contacted more often (Kolmogorov–Smirnov test). Interchromosomal interactions behaved similarly, with the 40 H3K27me3-anchored 4C BRICKs being clearly enriched among the most significant K27 BRICKs (SI Appendix, Fig. S4C). Moreover, the number of viewpoints that picked up the interaction directly correlated with the significance of the K27 BRICKs (SI Appendix, Fig. S4 A and D). These data overlap and expand a previous set of interchromosomal interactions between H3K27me3-marked loci in ES cells (11).
PcG-Dependent Interactions Occur in the Active, Non-LAD nuclear domain.
We next assessed how PcG-mediated interactions relate to the global organization of repressed domains in the nucleus. HiC studies in human cells have reported that H3K27me3 marks are primarily found within the “HiC compartment A,” a global gene-dense chromatin domain that is enriched for active genes (12). A DamID study, on the other hand, reported that H3K27me3 marks are enriched at the borders of so-called LADs, which are regions that interact with the repressive LaminB1 protein (13). To integrate our data in this context, we reanalyzed HiC and LaminB1 DamID data from multiple mouse cell types (Fig. 1 B and E and SI Appendix, Fig. S3) (14, 15, 19, 20), by using the published strategy (12) at resolutions of 1 Mb, 400 kb, and 80 kb. With the latter resolution, a reliable output was obtained for only nine chromosomes (SI Appendix, Fig. S5 A–C).
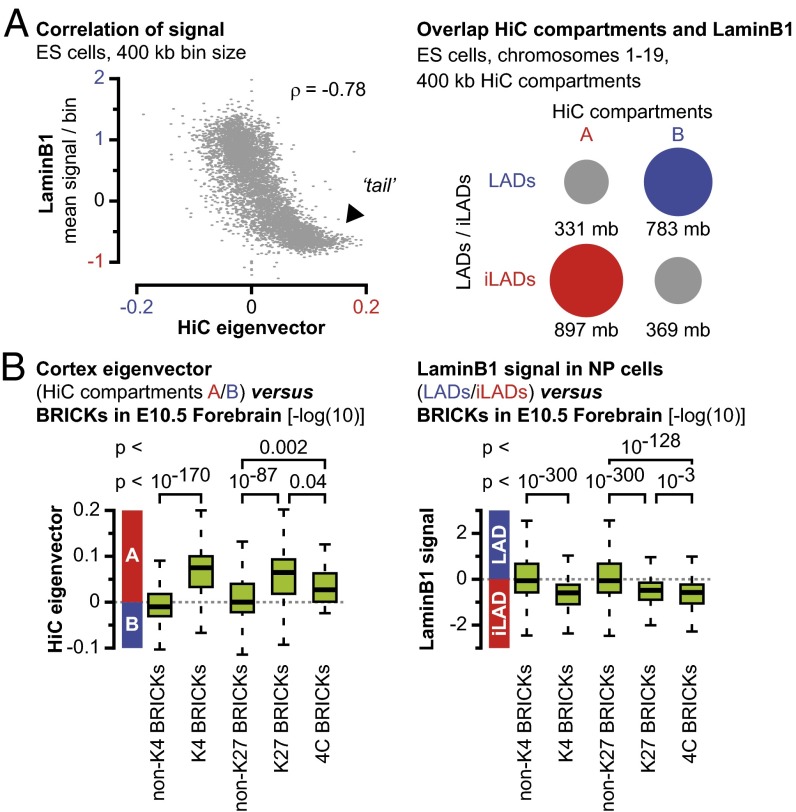
We observed a high degree of similarity between the HiC compartments on the one hand and the LaminB1 signal on the other hand (Fig. 1 B and E and SI Appendix, Figs. S3 and S5A). Although a certain degree of similarity had previously been noticed (21), we more systematically analyzed this potential correlation between the HiC data and LaminB1 DamID data and observed an almost complete correlation in ES cells over a more than 10-fold range in HiC resolution (80 kb–1 Mb, Fig. 2A, Left and SI Appendix, Fig. S5D). Importantly, HiC and LaminB1 DamID data are obtained via technically unrelated techniques and with different resolutions (12, 13). Particularly prominent in the analysis was the large tail in the graphs at all resolutions, showing that the regions most associated with the active HiC compartment A are those largely depleted for LaminB1 interactions (Fig. 2A, arrowhead). The intersection between HiC compartments (as extracted from ES and cortex cell data) and domains enriched or depleted for the LaminB1 protein [LADs and inter-LADs (iLADs)] (20) further revealed a high degree of cell-type–independent overlap (Fig. 2A, Right and SI Appendix, Fig. S5E). Moreover, this overlap increased with a decreasing resolution of the HiC data, suggesting that the remaining difference may be technical rather than biological. Accordingly, we conclude that at distances of over several megabases, HiC compartments are a proxy for nuclear lamina interactions. At a global scale, the nucleus may thus be divided into two mutually exclusive environments: a transcriptionally competent environment that consists of the combined HiC compartment A and iLADs, and a transcriptionally inactive environment that includes the combined HiC compartment B and LADs.
Fig. 2.
HiC compartments are a proxy for interactions with the LaminB1 protein. (A, Left) The HiC eigenvector at 400-kb resolution in ES cells anticorrelates with the LaminB1 signal. (Right) In ES cells, HiC compartment A overlaps mainly with iLADs, whereas compartment B overlaps with LADs. (B) Similar to active regions marked with H3K4me3, significant H3K27me3-marked regions [K27 BRICKs with –log(10) P value > 6] and intrachromosomal 4C BRICKs are enriched in the active HiC compartment A and in iLADs.
We compared the position of the HoxD cluster versus these domains and found a consistent association with a positive HiC eigenvector (i.e., HiC compartment A) and a depletion for LaminB1 interactions (i.e., iLADs), both features associated with the active nuclear environment (Fig. 1 B and E and SI Appendix, Fig. S3A). The other Hox clusters were also positioned within HiC compartment A/iLADs (SI Appendix, Fig. S3E). DNA-immuno-FISH experiments confirmed this location in forebrain and ES cells, with Hoxd13 located at a short distance from the nuclear periphery but not overlapping with the LaminB1 signal (Fig. 1G and SI Appendix, Fig. S6 A and B). This rather peripheral position may come from the upstream neighboring Lunapark (Lnp) gene that is enriched for LaminB1 interactions in ES and differentiated brain cells (SI Appendix, Fig. S6C).
Upon visual inspection, we noticed that most long-range interaction partners were also located in the HiC compartment A and iLADs (Fig. 1 B and E and SI Appendix, Fig. S3). We therefore decided to correlate H3K27me3-mediated interactions (4C BRICKs) and, more generally, H3K27me3-marked loci (K27 BRICKs with P value < 10−6), with both HiC compartments and LaminB1-associated domains and compared the outcome to active genomic regions as determined by the presence of H3K4me3 marks (K4 BRICKs with P value < 10−6). HiC bins containing the most significant active regions in the genome were indeed enriched in HiC compartment A (average significantly more positive eigenvector) compared with bins that do not contain such H3K4me3 BRICKs. This enrichment further increased with decreasing HiC bin size (Fig. 2B, Left and SI Appendix, Fig. S7). Likewise, significant H3K27me3-marked regions (K27 BRICKs > 6) and H3K27me3-marked regions involved in intrachromosomal interactions (4C BRICKs) were enriched in the active HiC compartment A (Fig. 2B, Left and SI Appendix, Figs. S7 and S8). In contrast to previous work on human cells (12), we saw this correlation persisting at resolutions below 100 kb, when either mouse ES or cortex cell HiC data were correlated with K27 BRICKs (SI Appendix, Fig. S7). This difference may be due to the BRICK-based approach, which scores for the significance of H3K27me3-marked regions rather than for a H3K27me3 signal. These results established that significant H3K27me3 domains reside in the active HiC compartment A, even at sub-100 kb resolutions. Similarly, although using an inversed approach due to the higher resolution of the DamID data, the H3K27me3 domains and long-range interactions were also scored versus LaminB1 interactions (LADs and iLADs). Significant K4 BRICKs (active domains), significant H3K27me3 domains (K27 BRICKs > 6), and intrachromosomal 4C BRICKs were consistently depleted for LaminB1 signal, reflecting their localization within active iLADs (Fig. 2B, Right and SI Appendix, Figs. S8 and S9). A comparable enrichment for HiC compartment A and for iLADs was scored for interchromosomal 4C BRICKs (SI Appendix, Fig. S4E).
Long-Range Interactions Are Moderately Reorganized During Embryonic Development.
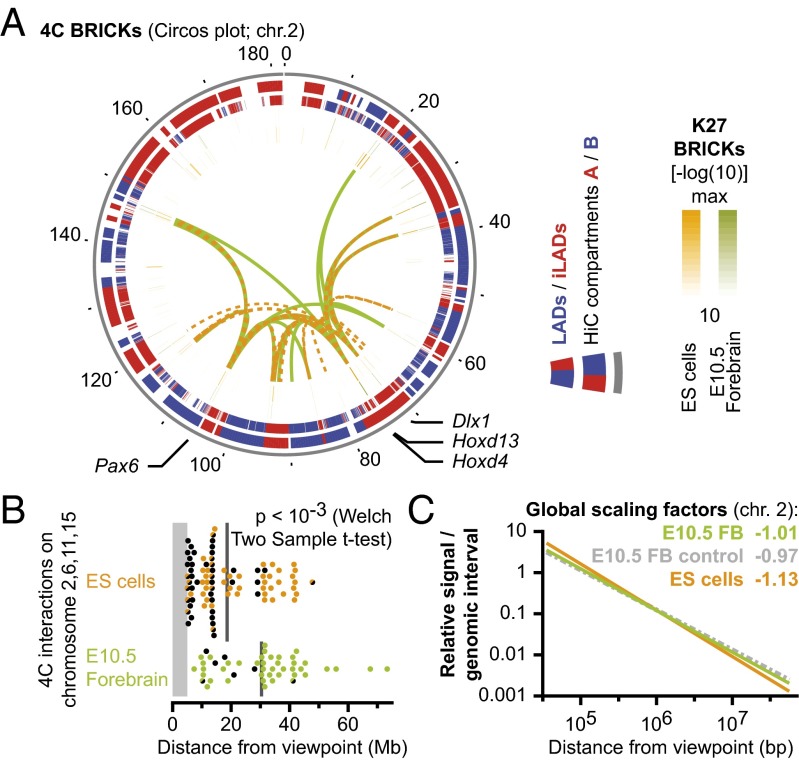
We next asked whether H3K27me3-marked long-range interactions were reorganized during cellular differentiation. The 4C-seq data from chromosome 2 were compared between ES and E10.5 forebrain cells. In both cell types, the H3K27me3-marked viewpoints interacted with the most significant H3K27me3-marked domains (Fig. 3A and SI Appendix, Fig. S10A). Moreover, these interactions were similarly enriched in the active nuclear compartment (HiC compartment A and iLADs, Figs. 1G and 3A and SI Appendix, Figs. S7–S10A). Our pinpointed intrachromosomal interactions in ES cells, using Hoxd13 and Hoxd4 as viewpoints, were contained within the larger regions identified in a previous study of HoxD at lower resolution (11), confirming the consistency of our approach (SI Appendix, Fig. S10B). Likewise, viewpoints in other Hox clusters identified H3K27me3-marked interactions in ES cells (SI Appendix, Fig. S10C). Whereas many highly significant interactions were shared between ES and forebrain cells, the same viewpoints had a significantly different distance distribution in their range of contacts (Fig. 3B and SI Appendix, Fig. S10D). In ES cells the average significant interaction was 20 Mb away, but in forebrain cells this had increased to 30 Mb. Furthermore, a substantial fraction of nearby interactions scored in ES cells overlapped with low and nonsignificant K27 BRICKs (P value > 10−18). This phenomenon was particularly prominent for the HoxC cluster, which had many interactions within the 12-Mb distant Brd1 to Rab12 locus, a gene-dense iLAD that contains housekeeping genes interspersed with low significance K27 BRICKs (P value > 10−18, SI Appendix, Fig. S10E). In forebrain cells, the Brd1 to Rab12 locus contained such low significant K27 BRICKs as well, yet the region was not significantly contacted.
Fig. 3.
H3K27me3-dependent contacts in ES cells are less distal due to overall chromosome folding patterns. (A) Circos plot with interactions for four H3K27me3-marked viewpoints in ES (orange) or in E10.5 forebrain (green) cells. Interactions occur between highly significant H3K27me3-marked loci (two central rings). In both cell types, interacting loci are mainly located within genomic regions with an active signature (red blocks in outer rings; legend at Right). (B) Interactions are significantly more proximal in ES cells than in forebrain cells. Colored dots represent 4C BRICKs that overlap significant K27 BRICKs (P value < 10−18). Half dots indicate low significant K27 BRICKs (P < 10−6) and black dots indicate nonsignificant K27 BRICKs. Gray lines indicate mean distance. (C) The overall folding pattern of chromosome 2, as determined by 4C-seq, follows a different scaling between ES and E10.5 forebrain cells, but is independent from the H3K27me3 mark.
Because Hox clusters seem to interact with the surrounding TADs slightly more robustly in ES cells than in forebrain cells (8), we wondered if this may reflect a more global phenomenon that may account for the different PcG-mediated interactions in these cell types. Indeed considerable differences in global scaling factors have been reported between cell types (22). To evaluate whether chromosome-folding patterns may affect the selectivity of long-range interactions, we subjected our 4C data to a similar analysis. As for HiC data, 4C interactions followed a power law, with a typical pattern of steep decline at distances between 500 kb and 5 Mb, bordered by more gradual slopes (12) (SI Appendix, Fig. S11). The scaling factors for H3K27me3-marked 4C viewpoints on different chromosomes had a consistently steeper decay of interactions in ES cells than in forebrain (Fig. 3C, orange versus green line and SI Appendix, Fig. S11). Interestingly, this pattern was independent of the H3K27me3 mark at the viewpoint (Fig. 3C, green versus gray line). Even though the logarithmic visualization makes these differences appear moderate, the signal is enriched about twofold in forebrain cells, at distances over 10 Mb. These global scaling patterns thus allow the detection of significant interactions up to larger distances in forebrain, which may explain the moderate reorganization of H3K27me3-mediated interactions during cellular differentiation.
Hoxd Genes Independently Form Long-Range H3K27me3-Marked Interactions.
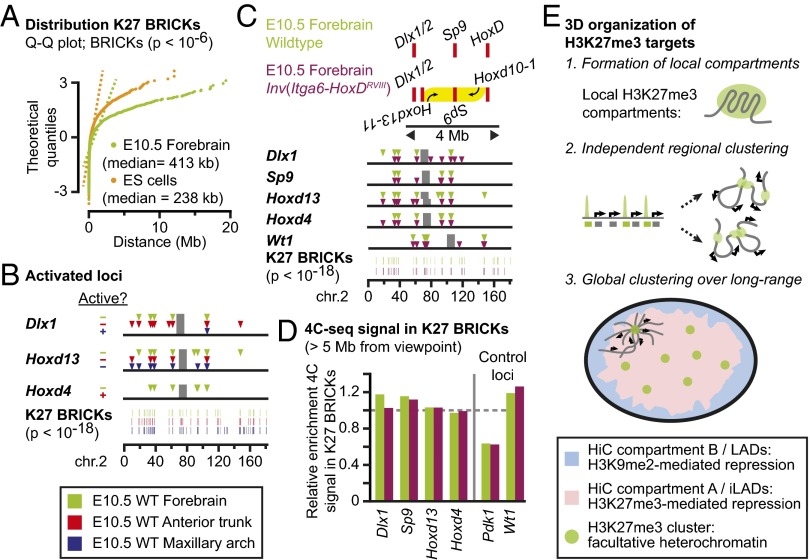
Finally, we assessed whether the presence of closely located H3K27me3 domains may promote and/or enhance the establishment of long-range interactions. Such an effect, if associated with a functional outcome, could have participated in the maintenance of paralogous syntenic relationships, such as the conserved clustering of Hox, Dlx, and Sp genes, which is found on multiple chromosomes and thus predates the 2R genome duplications that occurred at the root of the vertebrate lineage (23, 24). In both ES and forebrain cells, significant K27 BRICKs are unequally distributed, with clusters of multiple K27 BRICKs being separated by large intervals of several megabases (Fig. 4A and SI Appendix, Fig. S12A).
Fig. 4.
Lack of cooperation between interactions among H3K27me3-marked loci. (A) Significant H3K27me3 BRICKs in E10.5 forebrain and ES cells are clustered rather than normally distributed (dashed lines indicate normal distributions). (B) The active Dlx1/2 (blue) and Hoxd4 (red) genes lose intrachromosomal contacts, yet nearby H3K27me3-marked viewpoints maintain their interactions. (C) A 3-Mb inversion that brings the Hoxd11–13 genes near the Dlx1/2 cluster (Top) does not drastically change long-range contacts of viewpoints. (D) Positioning of the Hoxd11–Hoxd13 genes near the Dlx1 gene and away from Hoxd4 does not visibly modulate the contact efficiency of each viewpoint with other K27 BRICKs on chromosome 2. (E) A model for clustering of H3K27me3 targets in the cell nucleus. (Top) Individual H3K27me3-marked loci form 3D compartments. (Middle) The 3D compartments interact independently with nearby H3K27me3 targets. (Bottom) H3K27me3 targets, which tend to be embedded in active, gene-rich regions, form long-range clusters in the active nuclear environment that is depleted for interactions with the peripheral nuclear lamina.
We first looked at the cooperativity of interactions in cells where the size or number of H3K27me3 domains was reduced at HoxD or surrounding genes, due to transcriptional activation of parts of the genes. In the anterior trunk (SI Appendix, Fig. S1 A and B), four of nine Hoxd genes are active, which is paralleled by an approximately 50% reduction in H3K27me3 domain size (SI Appendix, Fig. S12B). Likewise, in the maxillary part of the first pharyngeal arch, the Dlx1 gene is active in most cells and thus carries only residual traces of H3K27me3 (SI Appendix, Figs. S1 A and B and S12B). Similar to previous observations (7, 9), the active Hoxd4 or Dlx1 viewpoints displayed virtually no interactions with H3K27me3-marked loci (Fig. 4B and SI Appendix, Fig. S12C). In contrast, whereas interactions were scored when the neighboring H3K27me3-marked viewpoints were used, they remained unaffected compared with ES cells. In anterior trunk, the inactive Dlx1 and Hoxd13 viewpoints maintained their mutual interactions, their interactions with the Sp9 gene, and with more distant K27 BRICKs. Similarly, in the maxillary arch, the inactive Hoxd13 gene maintained its interaction with the Sp9 gene and other distant K27 BRICKs (Fig. 4B and SI Appendix, Fig. S12C). Therefore, a reduction in the number or size of H3K27me3 domains did not drastically alter the occurrence of long-range contacts of nearby H3K27me3 domains.
We also analyzed whether the clustering of H3K27me3 domains could promote the establishment of interactions by using a large engineered chromosomal inversion. In this Inv(Itga6-HoxDRVIII) allele (25), the respective locations of the H3K27me3 domains were reorganized. Animals carrying this 3-Mb large inversion had Hoxd11–Hoxd13 repositioned at only 300 kb from the Dlx1/2 cluster, via a split of the cluster into two pieces (Fig. 4C, Top). After inversion, the combined H3K27me3 signal at HoxD remains virtually untouched, even though it was split into separate domains (6). ChIP-seq experiments on mutant and control forebrain cells also showed no drastic differences in the 4 Mb surrounding the inversion (SI Appendix, Fig. S13). Similarly, interactions among the H3K27me3-marked genes within this 4-Mb region remained as expected, although the inverted Hoxd13 viewpoint now strongly contacted the nearby Dlx1/2 cluster instead of the noninverted Hoxd10–Hoxd1 genes (SI Appendix, Fig. S13). However, the interactions with the Sp9 gene and the noninverted part of the HoxD cluster remained at the expected levels for distances of 1–3 Mb (SI Appendix, Fig. S13). Domainograms of distant intrachromosomal 4C BRICKs in control and inverted cells did not reveal any consistent differences (Fig. 4D). Neither did the Dlx1 viewpoint contact more distant regions when the H3K27me3-carrying Hoxd11–Hoxd13 genes were placed nearby, nor did the Hoxd4 viewpoint reduce its interactions when it was separated from the latter Hox genes (Fig. 4D).
Because a domainogram approach may not be sufficient to detect subtle modifications induced by the reorganization of H3K27me3 domains, we also scored for the general enrichment of intrachromosomal 4C-seq signals covering K27 BRICKs in control and mutant cells (SI Appendix, Fig. S4D). Whereas 4C signal in K27 BRICKs was generally enriched about twofold in comparison with the active Pdk1 locus, the enrichment for Dlx1 did not increase when paired with the Hoxd11–Hoxd13 genes, nor did the enrichment for Hoxd4 decrease when separated from these genes (Fig. 4D). These quantitative approaches did not provide any evidence that a “local” clustering of H3K27me3-marked genes may either promote or reinforce long-range clustering. Rather, it suggests that H3K27me3 targets establish long-range interactions rather independently from locus to locus.
Discussion
Here we report that mammalian genes associated with H3K27me3 modifications, including Hox genes, are partners in a network of mutual intra- and interchromosomal interactions. Interactions are established autonomously, with a frequency based on both the linear genomic distance and the importance of the H3K27me3 domains and cell-type–specific chromosome folding patterns. Transcriptional activation of these genes and the concurrent loss of H3K27me3 marks correlate with a loss of these long-range interactions. Finally, we report that interactions between H3K27me3 targets occur within an active nuclear environment, as defined by the overlap between iLADs (13, 20) and the HiC compartments A (12).
Our data suggest a model whereby facultative repression by the H3K27me3 marks occurs in a spatial domain distinct from H3K9me2-marked repressed chromatin. Loci decorated with H3K27me3 marks form local compartments that cluster in a noncooperative manner with other nearby and distant H3K27me3 targets (Fig. 4E). In contrast, H3K9me2-marked loci associate with the peripheral nuclear lamina (LADs) for exerting their repression (13, 26). Whereas LADs are gene poor, H3K27me3 marks are enriched in gene-dense regions of the genome (13, 27). Accordingly, we suggest that both mechanisms evolved along with distinct spatial modalities, to fulfill different repressive requirements. Whereas the G9A/H3K9me2 system can relocate genes in otherwise gene-poor regions to the peripheral LAD compartment, the Polycomb/H3K27me3 system forms repressive microenvironments in the more active central nuclear environment. Genes silenced following the latter mechanism may require robust activities in other cell types, making their presence in active gene-dense regions necessary to allow for more versatility.
Our data however did not support a role for genomic clustering of nearby H3K27me3-marked loci to help cooperatively form clusters with distant loci. Long-range interactions may therefore reflect the affinity of H3K27me3-marked domains to aggregate whenever located at each other’s vicinity, rather than being an active mechanism to silence distant gene loci in a collaborative manner. Such a probabilistic mechanism is further supported by the fact that chromosome folding is the primary determinant of cell-type–specific differences (besides the presence of the H3K27me3 mark). Our experiments using a targeted inversion located within a single HiC compartment A do not allow us to state whether the biological relevance of clustering may be higher at the scales of a few megabases or, alternatively, whether the efficiency of clustering depends on the co-occupancy within the same HiC compartment without a need for precisely defined positions. Moreover, our current analytical precision may not allow ruling out a role for these 3D clusters in more precise fine tuning of gene repression.
Likewise, these results do not support a role for H3K27me3-mediated clustering in the evolutionary conservation of paralogous syntenies (23), as determined here for the Hox, Dlx, and Sp loci. Similarly, we found no evidence that the transcriptional activation of these specific genes relies upon contacts among each other, or with shared genomic elements. Rather, both in the inactive and active states, the Hoxd, Dlx1/2, and Sp9 genes maintained the bulk of their interactions within their individual TADs. These results appear at odds with reports that active genes engage in widespread contacts among each other (28, 29). The requirement for highly significant interactions in our domainogram analysis, combined with the limited set of active genes may not be able to reliably score for global promoter–promoter contacts. In contrast, this work suggests that Polycomb-mediated contacts constitute a set of intra- and interchromosomal interactions that may be an important component of the chassis that stabilizes overall genome organization as well (see ref. 30 for other components).
Finally, our finding that HiC data at distances of over several megabases are a proxy for LaminB1 interactions provides a simplified view of nuclear organization, with (nonrepetitive) chromatin globally residing either in an active, central nuclear domain (iLAD, HiC compartment A) or in a more peripheral, inactive domain (LAD, HiC compartment B). This refinement extends beyond nuclear genome organization, as both HiC compartments and LADs are implicated in diverse cellular processes. TAD borders can overlap with LAD/iLAD or HiC compartment borders (14), suggesting that bordering TADs may be separated by placement in opposing nuclear domains. In addition, early and late replicating regions in the human genome correlate with HiC compartments (31), suggesting that replication timing may be linked to the presence in LADs and iLADs.
Materials and Methods
All experiments were performed in agreement with the Swiss law on animal protection (LPA), with the appropriate legal authorizations to D.D. The 4C-seq and ChIP-seq experiments were performed as described (7). The 4C-seq and ChIP-seq BRICKs (16) were calculated as described (17, 18), with ChIP-seq data translated to mean signal per NlaIII fragment to allow the comparison with 4C-seq data. FISH experiments were performed as described (32). All data are according to Mouse genome assembly NCBIM37 (mm9). An extended description of the materials, methods, and data analysis is provided in SI Appendix.
Supplementary Material
Acknowledgments
We thank the members of the D.D. laboratories for discussions; Elisabeth Joye for ES cell culture; Shelagh Boyle and Wendy Bickmore (Medical Research Council Human Genetics Unit) for help with setting up FISH procedures; Bas van Steensel (Dutch Cancer Institute) for useful comments; Sandra Gitto and Thi Hanh Nguyen Huynh (University of Geneva) for animal care; Mylène Docquier, Christelle Barraclough, Céline Delucinge, and Natacha Civic (Geneva Genomics Platform) for generating sequencing data; Olivier Burri and Romain Guiet [École Polytechnique Fédérale de Lausanne (EPFL) Bioimaging and Optics Platform] for help with image analysis; and Jacques Rougemont (EPFL Bioinformatics and Biostatistics Core Facility, BBCF) for assistance in the data analysis. Bioinformatic computations were performed at the Vital-IT Center for high-performance computing (www.vital-it.ch) of the Swiss Institute of Bioinformatics using tools developed by the BBCF (bbcf.epfl.ch). This work was supported by funds from the EPFL, the University of Geneva, the Swiss National Research Fund, a European Research Council grant SystemsHox.ch, and the Claraz Foundation (D.D.).
Footnotes
The authors declare no conflict of interest.
Data deposition: The data reported in this paper have been deposited in the Gene Expression Omnibus (GEO) database, www.ncbi.nlm.nih.gov/geo (accession no. GSE61372).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1504783112/-/DCSupplemental.
References
- 1.Spitz F, Furlong EE. Transcription factors: From enhancer binding to developmental control. Nat Rev Genet. 2012;13(9):613–626. doi: 10.1038/nrg3207. [DOI] [PubMed] [Google Scholar]
- 2.Boyer LA, et al. Polycomb complexes repress developmental regulators in murine embryonic stem cells. Nature. 2006;441(7091):349–353. doi: 10.1038/nature04733. [DOI] [PubMed] [Google Scholar]
- 3.Bracken AP, Dietrich N, Pasini D, Hansen KH, Helin K. Genome-wide mapping of Polycomb target genes unravels their roles in cell fate transitions. Genes Dev. 2006;20(9):1123–1136. doi: 10.1101/gad.381706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bernstein BE, et al. A bivalent chromatin structure marks key developmental genes in embryonic stem cells. Cell. 2006;125(2):315–326. doi: 10.1016/j.cell.2006.02.041. [DOI] [PubMed] [Google Scholar]
- 5.Tschopp P, Tarchini B, Spitz F, Zakany J, Duboule D. Uncoupling time and space in the collinear regulation of Hox genes. PLoS Genet. 2009;5(3):e1000398. doi: 10.1371/journal.pgen.1000398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Soshnikova N, Duboule D. Epigenetic temporal control of mouse Hox genes in vivo. Science. 2009;324(5932):1320–1323. doi: 10.1126/science.1171468. [DOI] [PubMed] [Google Scholar]
- 7.Noordermeer D, et al. The dynamic architecture of Hox gene clusters. Science. 2011;334(6053):222–225. doi: 10.1126/science.1207194. [DOI] [PubMed] [Google Scholar]
- 8.Noordermeer D, et al. Temporal dynamics and developmental memory of 3D chromatin architecture at Hox gene loci. eLife. 2014;3:e02557. doi: 10.7554/eLife.02557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bantignies F, et al. Polycomb-dependent regulatory contacts between distant Hox loci in Drosophila. Cell. 2011;144(2):214–226. doi: 10.1016/j.cell.2010.12.026. [DOI] [PubMed] [Google Scholar]
- 10.Tolhuis B, et al. Interactions among Polycomb domains are guided by chromosome architecture. PLoS Genet. 2011;7(3):e1001343. doi: 10.1371/journal.pgen.1001343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Denholtz M, et al. Long-range chromatin contacts in embryonic stem cells reveal a role for pluripotency factors and polycomb proteins in genome organization. Cell Stem Cell. 2013;13(5):602–616. doi: 10.1016/j.stem.2013.08.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lieberman-Aiden E, et al. Comprehensive mapping of long-range interactions reveals folding principles of the human genome. Science. 2009;326(5950):289–293. doi: 10.1126/science.1181369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Guelen L, et al. Domain organization of human chromosomes revealed by mapping of nuclear lamina interactions. Nature. 2008;453(7197):948–951. doi: 10.1038/nature06947. [DOI] [PubMed] [Google Scholar]
- 14.Dixon JR, et al. Topological domains in mammalian genomes identified by analysis of chromatin interactions. Nature. 2012;485(7398):376–380. doi: 10.1038/nature11082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shen Y, et al. A map of the cis-regulatory sequences in the mouse genome. Nature. 2012;488(7409):116–120. doi: 10.1038/nature11243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.de Wit E, Braunschweig U, Greil F, Bussemaker HJ, van Steensel B. Global chromatin domain organization of the Drosophila genome. PLoS Genet. 2008;4(3):e1000045. doi: 10.1371/journal.pgen.1000045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Noordermeer D, et al. Variegated gene expression caused by cell-specific long-range DNA interactions. Nat Cell Biol. 2011;13(8):944–951. doi: 10.1038/ncb2278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Splinter E, et al. The inactive X chromosome adopts a unique three-dimensional conformation that is dependent on Xist RNA. Genes Dev. 2011;25(13):1371–1383. doi: 10.1101/gad.633311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Peric-Hupkes D, et al. Molecular maps of the reorganization of genome-nuclear lamina interactions during differentiation. Mol Cell. 2010;38(4):603–613. doi: 10.1016/j.molcel.2010.03.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Meuleman W, et al. Constitutive nuclear lamina-genome interactions are highly conserved and associated with A/T-rich sequence. Genome Res. 2013;23(2):270–280. doi: 10.1101/gr.141028.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhu J, et al. Genome-wide chromatin state transitions associated with developmental and environmental cues. Cell. 2013;152(3):642–654. doi: 10.1016/j.cell.2012.12.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Barbieri M, et al. Complexity of chromatin folding is captured by the strings and binders switch model. Proc Natl Acad Sci USA. 2012;109(40):16173–16178. doi: 10.1073/pnas.1204799109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lundin LG, Larhammar D, Hallböök F. Numerous groups of chromosomal regional paralogies strongly indicate two genome doublings at the root of the vertebrates. J Struct Funct Genomics. 2003;3(1-4):53–63. [PubMed] [Google Scholar]
- 24.Sundström G, Larsson TA, Larhammar D. Phylogenetic and chromosomal analyses of multiple gene families syntenic with vertebrate Hox clusters. BMC Evol Biol. 2008;8:254. doi: 10.1186/1471-2148-8-254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Spitz F, Herkenne C, Morris MA, Duboule D. Inversion-induced disruption of the Hoxd cluster leads to the partition of regulatory landscapes. Nat Genet. 2005;37(8):889–893. doi: 10.1038/ng1597. [DOI] [PubMed] [Google Scholar]
- 26.Kind J, et al. Single-cell dynamics of genome-nuclear lamina interactions. Cell. 2013;153(1):178–192. doi: 10.1016/j.cell.2013.02.028. [DOI] [PubMed] [Google Scholar]
- 27.Pauler FM, et al. H3K27me3 forms BLOCs over silent genes and intergenic regions and specifies a histone banding pattern on a mouse autosomal chromosome. Genome Res. 2009;19(2):221–233. doi: 10.1101/gr.080861.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Schoenfelder S, et al. Preferential associations between co-regulated genes reveal a transcriptional interactome in erythroid cells. Nat Genet. 2010;42(1):53–61. doi: 10.1038/ng.496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Li G, et al. Extensive promoter-centered chromatin interactions provide a topological basis for transcription regulation. Cell. 2012;148(1-2):84–98. doi: 10.1016/j.cell.2011.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Phillips-Cremins JE. Unraveling architecture of the pluripotent genome. Curr Opin Cell Biol. 2014;28:96–104. doi: 10.1016/j.ceb.2014.04.006. [DOI] [PubMed] [Google Scholar]
- 31.Ryba T, et al. Evolutionarily conserved replication timing profiles predict long-range chromatin interactions and distinguish closely related cell types. Genome Res. 2010;20(6):761–770. doi: 10.1101/gr.099655.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Morey C, Da Silva NR, Perry P, Bickmore WA. Nuclear reorganisation and chromatin decondensation are conserved, but distinct, mechanisms linked to Hox gene activation. Development. 2007;134(5):909–919. doi: 10.1242/dev.02779. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.