SUMMARY
Gibberellins (GAs) play a critical role in fruit-set and fruit growth. Gibberellin is perceived by its nuclear receptors GA INSENSITIVE DWARF1s (GID1s), which then trigger degradation of downstream repressors DELLAs. To understand the role of the three GA receptor genes (GID1A, GID1B and GID1C) in Arabidopsis during fruit initiation, we have examined their temporal and spatial localization, in combination with analysis of mutant phenotypes. Distinct expression patterns are revealed for each GID1: GID1A is expressed throughout the whole pistil, while GID1B is expressed in ovules, and GID1C is expressed in valves. Functional study of gid1 mutant combinations confirms that GID1A plays a major role during fruit-set and growth, whereas GID1B and GID1C have specific roles in seed development and pod elongation, respectively. Therefore, in ovules, GA perception is mediated by GID1A and GID1B, while GID1A and GID1C are involved in GA perception in valves. To identify tissue-specific interactions between GID1s and DELLAs, we analyzed spatial expression patterns of four DELLA genes that have a role in fruit initiation (GAI, RGA, RGL1 and RGL2). Our data suggest that GID1A can interact with RGA and GAI in all tissues, whereas GID1C–RGL1 and GID1B–RGL2 interactions only occur in valves and ovules, respectively. These results uncover specific functions of each GID1–DELLA in the different GA-dependent processes that occur upon fruit-set. In addition, the distribution of GA receptors in valves along with lack of expression of GA biosynthesis genes in this tissue, strongly suggests transport of GAs from the developing seeds to promote fruit growth.
Keywords: fruit-set, gibberellin, GA INSENSITIVE DWARF1, DELLA, Arabidopsis
INTRODUCTION
Fruit-set is the activation of the developmental program that transforms a pistil into a developing fruit. It is initiated by the fertilization of ovules, which promotes synthesis and signaling of hormones, mainly auxin and gibberellin (GA; Dorcey et al., 2009). These hormones form the initial instructive signals that promote the fruit development program under physiological conditions (Alabadi et al., 2009).
Gibberellins are plant tetracyclic diterpenoids that control a wide range of processes throughout plant development, including seed germination, leaf expansion, stem and root elongation, floral induction, and flower development (Fleet and Sun, 2005; Swain and Singh, 2005; Sun, 2011). Studies also indicate that GAs are key factors for fruit-set and development. GA treatment of unpollinated pistils promotes fruit initiation, probably by mimicking GA production upon ovule fertilization (Vivian-Smith and Koltunow, 1999; Dorcey et al., 2009). In fact, upon pollination, GA biosynthesis genes are up-regulated, and bioactive GA1 and its precursor GA20 levels increase (Ben-Cheikh et al., 1997). The last steps of GA biosynthesis are catalyzed by GA20-oxidases (GA20ox) and GA 3-oxidases (GA3ox), which convert inactive GA precursors into bioactive GAs (Hedden and Thomas, 2012). Significant progress has been made in defining the roles of both GA20ox and GA3ox genes during reproductive growth in Arabidopsis (Hu et al., 2008; Rieu et al., 2008; Plackett et al., 2012). Expression of most of the GA20ox and GA3ox genes is induced at fruit initiation, and each gene shows a specific temporal expression pattern; most are up-regulated in the ovules after fertilization (Dorcey et al., 2009). GA20ox1 and GA20ox2 play a major role in fertility and fruit growth (Rieu et al., 2008). Reduced fruit length in the double ga20ox1 ga20ox2 mutant has a maternal origin, pointing out to a defect in GA-dependent fruit elongation in the absence of these GA20ox activities. In a similar way, GA3ox3, GA3ox4, along with GA3ox1, have major roles in providing the bioactive GAs during flower development and fruit-set (Hu et al., 2008). GA3ox4 is involved in promoting seed-dependent fruit elongation. Similar up-regulation of GA biosynthesis genes upon fruit-set has also been reported in other species (Garcia-Martinez et al., 1997; Serrani et al., 2007).
Gibberellin signaling relies on the perception of the hormone by its receptor GA INSENSITIVE DWARF1 (GID1). GID1 was first described in rice as a nuclear localized protein similar to hormone-sensitive lipase family without hydrolase activity (Ueguchi-Tanaka et al., 2005). In Arabidopsis, there are three GID1 orthologs (GID1A, GID1B, and GID1C; Griffiths et al., 2006; Nakajima et al., 2006). The active GA binds to GID1 and promotes a conformational change in its N-terminus region (Murase et al., 2008; Shimada et al., 2008). The GA-GID1 complex can then bind to the GA-signaling repressor DELLA proteins, promoting a conformational modification, which allows their recognition by GID2 or SLEEPY1 (SLY1) F-box proteins in rice and Arabidopsis, respectively (McGinnis et al., 2003; Sasaki et al., 2003; Griffiths et al., 2006; Willige et al., 2007; Murase et al., 2008; Hirano et al., 2010). The DELLA proteins are then polyubiquitinated and degraded through the 26S-proteasome pathway (Sasaki et al., 2003; Dill et al., 2004; Fu et al., 2004). Studies using the sly1 and gid2 mutants further show that GA-GID1 binding to DELLA can also inhibit DELLA activity without DELLA degradation (Ariizumi et al., 2008, 2013; Ueguchi-Tanaka et al., 2008). Both proteolysis-dependent and -independent mechanisms of GA signaling call for a close interaction between GID1s and DELLAs. This interaction may occur only if these proteins are temporally and spatially co-expressed. In addition, GID1s and DELLAs showed differential binding affinity in vitro (Nakajima et al., 2006 and Suzuki et al., 2009), which may contribute to establishing proper GA-signaling responses in specific tissues during plant development.
The presence of three GID1s in Arabidopsis suggests that each GA receptor may have distinct roles during plant development. In contrast, gid1 mutant analysis points to partial redundancy. While the single mutants did not show any obvious phenotype, the doubles show partially reduced GA-response phenotypes: gid1a-1 gid1c-1 is a semi-dwarf and has compromised germination, and gid1a-1 gid1b-1 shows stamen filament shortening that results in reduced fertility (Griffiths et al., 2006; Nakajima et al., 2006; Iuchi et al., 2007; Voegele et al., 2011). The null mutation in all three genes leads to an extremely dwarf and GA-insensitive plant (Ueguchi-Tanaka et al., 2005; Griffiths et al., 2006; Iuchi et al., 2007). Therefore, GID1A and GID1C play a major role during seed germination (Voegele et al., 2011) as well as in stem elongation (Suzuki et al., 2009). In addition, the role of GID1A and GID1C is consistent with their spatial expression in stems and seeds.
Fruit-set and subsequent development are controlled by GAs. This study aims to understand the role of GA receptors in Arabidopsis upon fertilization by a combination of expression and mutant phenotype analyses. We show that GID1A plays a major role during fruit-set, whereas GID1B and GID1C have partially redundant function with GID1A in seed development and pod elongation, respectively. In addition, the GA-dependent degradation of the endocarp during fruit senescence and maturation, necessary to pod shattering, is controlled mainly by GID1A. We also identified distinct co-expression patterns of GID1s and DELLAs, which uncover specific roles of several GID1-DELLA combinations during seed-set and fruit growth.
RESULTS
GID1 genes are differentially expressed in pistils and fruits
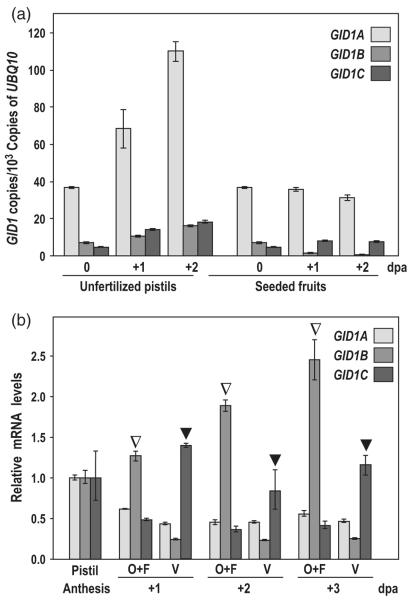
To investigate the role of each of the GID1 GA receptors during fruit-set and early fruit development, we first quantified their expression in pistils and fruits, by quantitative RT-PCR (qRT-PCR). mRNAs for the three GID1 genes were detected in pistils and fruits, with GID1A being expressed at higher levels than GID1B and GID1C (Figures 1a and S1a). The expression of the three GID1s increased after anthesis in the unpollinated pistils. However, the expression levels of GID1A and GID1C remained unaltered, but GID1B was decreased upon pollination. The increased GID1 expression in unfertilized pistils may be due to the feedback mechanism caused by low levels of GAs in this tissue.
Figure 1.
GID1 genes are differentially expressed in pistils and fruits.
(a) Time-course of GID1 expression during unfertilized pistil and early fruit development. Expression is represented as the copies of cDNA per 103 copies of UBQ10.
(b) GID1 expression in ovules + funiculi (O + F) and valve (V) of hand-dissected unfertilized pistils at 1, 2, and 3 dpa. Expression was normalized to UBQ10 and to the expression in the whole pistil at anthesis. Open and closed triangles indicate preferential expression of GID1B in ovules and GID1C in valves, respectively. Each experiment was repeated twice using independent samples with similar results (Figure S1). Data are the mean ± standard deviation (SD) of a single representative experiment. dpa, days post-anthesis.
To determine whether GID1s are expressed in specific tissues in the pistil, we performed qRT-PCR analysis using hand-dissected ovules + funiculi and valves from unfertilized pistils. Data in Figures 1(b) and S1(b) indicated that GID1B and GID1C mRNAs were preferentially expressed in ovules + funiculi and valves, respectively, while GID1A was expressed throughout the pistil.
GID1 proteins show specific spatial expression patterns
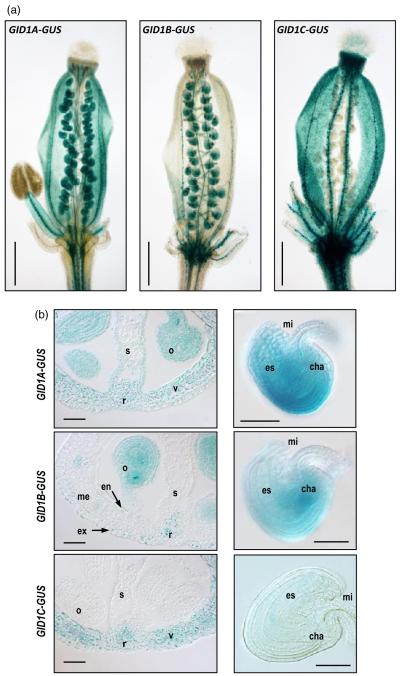
To finely study the localization of GID1 proteins in pistils and fruits, we analyzed their expression patterns using transgenic Arabidopsis lines carrying GID1 promoter:GID1-GUS translational fusions (Suzuki et al., 2009). GUS staining in whole pistil at anthesis clearly showed that GID1 proteins are differentially expressed (Figure 2a). GID1A was localized in the ovary and style, but was absent in the stigma. In the ovary, it was expressed in nearly all tissues, with the exception of the central area of the septum, the transmitting track (Figure 2b). In contrast to the broader expression of GID1A, GID1B and GID1C showed restricted expression. GID1B was mainly localized in ovules and funiculi, and it was also expressed at much lower levels in the exocarp and endocarp a. GID1B was absent from the mesocarp, stigma or style (Figure 2a,b). Expression of GID1C was localized specifically in the valve and style, but was completely absent in the ovules and funiculi (Figure 2a,b). GID1C is the only GA receptor that showed some expression in stigma (Figure 2a). All three receptors are expressed in the medial and lateral vascular tissues of the pistil.
Figure 2.
GID1 expression is differentially localized in tissues of pistils and fruits.
GID1 expression was analyzed using translational fusion lines pGID1:GID1-GUS (Suzuki et al., 2009).
(a) Expression in whole pistils at anthesis. Pistils were slightly squeezed between slide and coverslip to differentiate expression in valves and ovules. Scale bar represents 500 μm.
(b) Expression in cross-sections of pistils (left panels) and ovules (right panels) at anthesis. o, ovule; r, replum; s, septum-transmitting track; v, valve, ex, exocarp, me, mesocarp, en, endocarp. mi, micropyle; es, embryo sac; cha, chalaza. Scale bar represents 50 μm.
Expression of GID1A and GID1B in ovules showed slight variations in localization (Figure 2b); while GID1A was localized in all tissues with stronger expression surrounding the embryo sac, GID1B was localized mainly in the chalaza, at the base of the embryo sac. GID1C expression was undetectable in ovules/seeds. In summary, GID1A and GID1C are expressed in valves, while GID1A and GID1B are expressed in ovules. The different spatial and temporal expression of GID1s may reflect their involvement in different GA-mediated processes during fruit-set and development.
RGL1 and RGL2 are differentially expressed in pistils and fruits
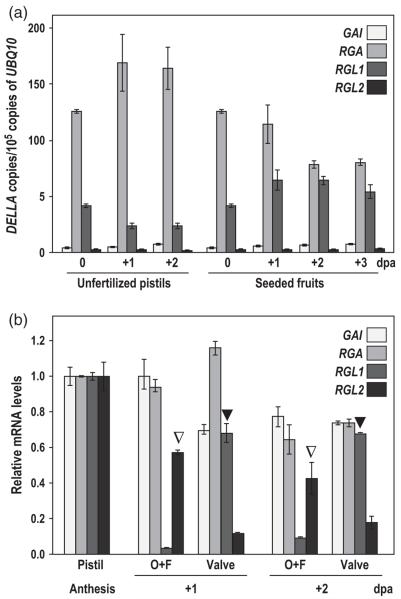
DELLAs are involved in the control of fruit-set in Arabidopsis. The elimination of four of the five DELLA genes (GAI, RGA, RGL1 and RGL2) is sufficient to promote facultative parthenocarpy (Dorcey et al., 2009). To study further the role of each DELLA in fruit-set, we analyzed their mRNA levels in pistils and fruits by qRT-PCR. RGA and RGL1 were expressed at higher levels, while GAI and RGL2 were expressed at much lower levels (Figures 3a and S2a). Upon fertilization, the expression of GAI and RGL1 slightly increased, the expression of RGA decreased, while no changes in expression were observed for RGL2. We also examined the expression of the four DELLAs in hand-dissected pistils to uncover their spatial expression within the pistil. GAI and RGA were expressed similarly in ovules + funiculi and valves (Figures 3b and S2b), while RGL1 and RGL2 were preferentially expressed in valves and ovules + funiculi, respectively (Figure 3b), suggesting their tissue-specific functions in GA-mediated signaling.
Figure 3.
DELLA genes are differentially expressed in pistils and fruits.
(a) Time-course of DELLA expression during unfertilized pistil and early fruit development. Expression is represented as the copies of cDNA per 105 copies of UBQ10.
(b) DELLA expression in ovules + funiculi (O + F) and valve (V) of hand-dissected unfertilized pistils at 1 and 2 dpa. Expression was normalized to UBQ10 and to the expression in the whole pistil at anthesis. Open and closed triangles indicate preferential expression of RGL2 in ovules and RGL1 in valves, respectively. Each experiment was repeated twice using independent samples with similar results (Figure S2). Data are the mean ± standard deviation (SD) of a single representative experiment. dpa, day post-anthesis.
DELLAs co-localize with GID1s in ovules and developing seeds
Gibberellin receptors interact, in a GA-dependent manner, with the DELLA proteins, to alleviate DELLA repression on GA signaling. Although the kinetics of GID1-DELLA interaction have been the focus of several reports by in vitro assays (Nakajima et al., 2006; Iuchi et al., 2007 and Suzuki et al., 2009), the formation of such protein complexes is only possible by co-localization of GID1s and DELLAs both in time and space. Therefore, to determine which GID1-DELLA complex is mediating GA-dependent fruit-set events, it is important to analyze the tissue-specific expression patterns of DELLAs in pistils, and compare them with those for the GID1s. Toward this goal, we generated promoter-GUS transcriptional fusion transgenic lines for GAI, RGA and RGL1 (see Experimental procedures for details). For the study of the spatial expression of RGL2, the line rgl2-5 was used (a translational GUS fusion, Lee et al., 2002; Figure 4).
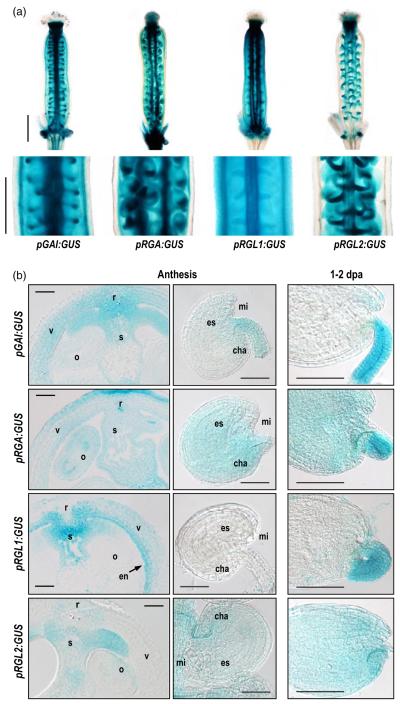
Figure 4.
DELLA expression is differentially localized in tissues of pistils and fruits.
DELLA expression was analyzed using transcriptional fusion lines pGAI:GUS, pRGA:GUS, and pRGL1:GUS (see Experimental procedures), and mutant line rgl2-5 for RGL2.
(a) DELLA expression in whole pistils at anthesis. Scale bar represents 500 μm.
(b) Expression in cross-sections of pistils and ovules at anthesis (left and middle panels), and seeds at 1 or 2 dpa (right panels). o, ovule; r, replum; s, septum-transmitting track; v, valve, en, endocarp; mi, micropyle; es, embryo sac; cha, chalaza. Scale bar represents 50 μm.
While RGA and GAI were expressed throughout the pistil, RGLl and RGL2 showed more restricted expression patterns (Figure 4). RGA was detected at high levels in all tissues, but GAI expression showed high levels in valves, especially in mesocarp and endocarp cell layers, and in funiculi; weaker GAI expression was detected in ovules at anthesis. Interestingly, the expression patterns for GAI and RGA resembled those for GID1A. On the other hand, RGL1 was expressed mainly in the endocarp, and at lower levels in mesocarp and exocarp (Figure 4), as well as in funiculi at 2 dpa. It was not detected in ovules or seeds. In contrast to RGL1, RGL2 expression was detected in both ovules at anthesis and seeds, including funiculi, but it was totally absent in valves. RGL1 and RGL2 showed strikingly similar expression patterns to GID1C and GID1B, respectively.
Overall, the GID1 and DELLA expression patterns using GUS assays are consistent with the results of qRT-PCR analysis. The comparison of DELLA and GID1 expression patterns in different tissues allowed us to speculate spatial-specific GID1-DELLA interactions that mediate GA signaling (Figures 2 and 4). In ovules, GID1A and GID1B would potentially interact only with GAI, RGA and RGL2. In contrast, in valves, GID1A and GID1C would interact with GAI, RGA, and RGL1.
Mutation of GID1 genes results in maternal defects in fertility
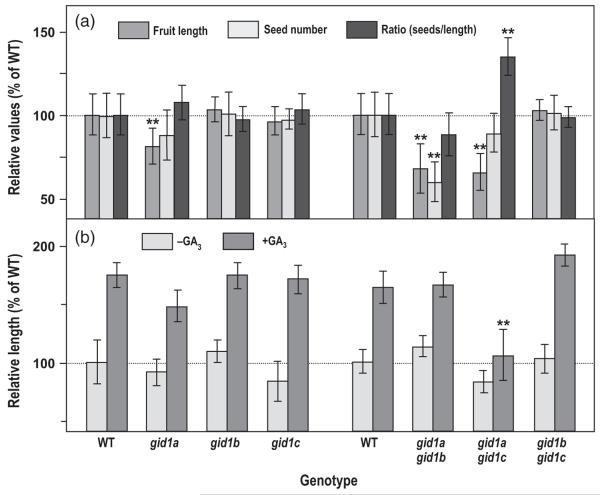
In this study, we showed that GID1A and GID1C are expressed in valves, while GID1A and GID1B are expressed in ovules. These expression patterns suggest that GID1A and GID1C mediate GA-induced fruit growth, whereas GID1A and GID1B promote seed development. Consistent with GID1 expression patterns, the single gid1a and the double gid1a gid1b and gid1a gid1c mutants display compromised fertility (Griffiths et al., 2006). gid1a had some decrease in seed number and a slight reduction in fruit length. The double mutant gid1a gid1c showed a strong reduction in silique length with a little reduction of seed number. In contrast, the double mutant gid1a gid1b had much reduced seed number and silique length; this phenotype was attributed to the shortening of the stamen filament that reduced pollination (Griffiths et al., 2006; Iuchi et al., 2007; Plackett et al., 2011). Fruit growth and fertility is the result of the combinatory effect of male (pollen) and female (ovule and ovary) factors. Considering that GID1A and GID1B are expressed in ovules, the reduced seed number and silique length phenotype of the double gid1a gid1b could also result from reduced seed-set due to defects in the ovule. To test this possibility, we carried out an experiment in which pistils of emasculated WT (Col-0) and gid1 mutant flowers were hand pollinated with WT pollen. In this way, reduced fertility would only be a consequence of pistil defect. Fruits were harvested at maturity, just before pod opening, and pod length, seed number and the ratio between seed number and pod length for each individual fruit were determined (Figure 5a). Among the single mutants, only gid1a showed a slight reduction in silique length and seed content, which resulted in the seed/silique length ratio similar to WT. Double mutants showed different phenotypes (Figure 5a). Similarly to gid1a, gid1a gid1b had a proportional but stronger reduction in both silique length and seed number that resulted in a ratio similar to WT. In contrast, gid1a gid1c showed the same seed number reduction as in gid1a but a strong reduction in silique length, which resulted in a significant increase of seed/length ratio, i.e., a higher seed density or increased packing phenotype in these fruits. gid1b gid1c did not show any defects. Almost identical results were obtained with self-pollinated pistils for the three gid1 double mutants (Figure S3), indicating that the phenotypes of gid1a gid1b and gid1a gid1c are mainly caused by maternal defects.
Figure 5.
GID1s control female fertility and pod elongation.
(a) Maternal effect of the gid1 null mutations. Flowers of gid1 mutants and wild-type (WT) were emasculated 1 day before anthesis and hand pollinated at anthesis with WT pollen. Mature fruits were individually harvested, and fruit length and seed number were measured. Ratio (seed number versus length) was determined. Values were normalized to the WT.
(b) Gibberellin (GA) response of unfertilized pistils of the gid1 null mutants as measured by relative pistil length in comparison to untreated WT pistils. Flowers of GID1 mutants and WT were emasculated 1 day before anthesis; half of the pistils were treated at anthesis with 300 μm GA3 (+GA3), and the other half were treated with mock solution (−GA3). Fruit or pistil length was measured at 10 dpa. Mean and standard deviation (SD) were calculated from at least 50 pistils/fruits per treatment. The experiment was repeated three times with similar results. Significant differences (Student’s t-test analysis) between the WT and mutants are marked with asterisks (**P-value < 0.001).
Elimination of two out of the three GA receptors in each double mutant combination may cause up-regulated expression of the remaining GID1 gene, which may alleviate reduced GA-response phenotype. Indeed, expression of GID1A in the double gid1b gid1c is enhanced in seedlings and pistils (Figure S4), which contributes to normal growth even in the absence of GID1B and GID1C activity. In contrast, no alteration of the expression of GID1B or GID1C was observed in either seedling or pistils of gid1a gid1c and gid1a gid1b, respectively.
There is a close correlation between the final fruit size and the seed number (Cox and Swain, 2006; Dorcey et al., 2009). Therefore, a reduction in seed number with no alteration of the slope of the best fit line of the data (a measure of seed-dependent fruit growth) may reveal defects of seed-set, while a decrease in the slope may reflect impairment of pod elongation, which results in increased packing of seeds in the fruit. To distinguish between defects in seed-set and fruit elongation, we analyzed siliques from the double gid1 mutants manually pollinated with different amounts of WT pollen, and recorded final fruit length and seed number (Figure S5). The gid1a gid1b double mutant showed, as expected, a reduced seed number with no significant alteration of the seed number/fruit length ratio, indicating that the smaller fruit size is due to decreased seed-set. In contrast, gid1a gid1c showed a significant decrease in the slope, which suggested that this mutant has defects in pod elongation even in the presence of a significant number of seeds, possibly due to a reduced sensitivity to GAs. Again, gid1b gid1c behaved as the WT plant. Finally, without pollination, all three gid1 double mutants showed similar pistil length as WT (approximately 4 mm).
Consistent with the expression patterns of GID1s in the fruit, the phenotypes observed for gid1a gid1b and gid1a gid1c reflect GA-insensitivity in ovules and valves that causes reduced seed-set and pod elongation, respectively. To determine if the gid1a gid1c pistil is indeed insensitive to GA treatment, the response of unfertilized pistils to GAs in double null mutants was recorded (Figure 5b). gid1a gid1b and gid1b gid1c showed normal GA response; in contrast, the gid1a gid1c pistils displayed little or no GA response. Therefore, the reduced fruit growth in gid1a gid1c is due to limited GA-mediated pod elongation.
In summary, our data suggest that the three GID1 GA receptors act partially redundantly in pistil development. The defects observed in the gid1 double mutants are due to GA perception defects in maternal tissues. The contribution of filament shortening to fertility in gid1a gid1b seems to be minor.
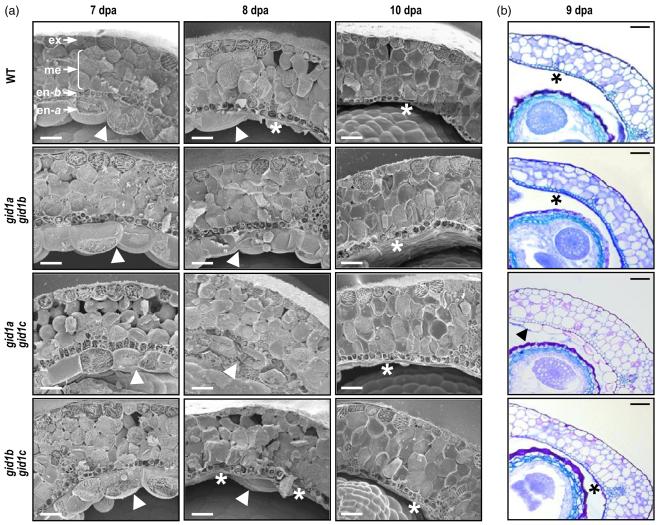
Mutation of GID1 genes promotes alterations at the endocarp
The inner cell layers of the Arabidopsis silique, the endocarp, differentiate during fruit development to facilitate pod shattering and seed dispersal. During fruit development, the endocarp a, the adaxial cell layer facing the carpel space, is degraded, along with the lignification of the support cell layer, the endocarp b. We previously showed that both processes are mediated by GAs (Dorcey et al., 2009). Early degradation and lignification was observed in GA-treated unfertilized pistils, as well as in the DELLA quadruple mutant (gai-t6 rga-24 rgl1-1 rgl2-1). To determine which GA receptors participate in this process, we studied pod structure during fruit development in double gid1 mutants (Figure 6). Morphological analysis suggested that GID1A has a major role in the regulation of the differentiation of the inner cell layers of the fruit pod, followed by GID1C (Figure 6). Although the single gid1a mutant did not show significant alteration, double mutants gid1a gid1b and gid1a gid1c showed 1 and 2 days delay, respectively, in the degradation of endocarp a and the lignification of endocarp b. This correlates with the intensity and distribution of their expression: GID1A is highly expressed in pods, followed by GID1C (Figure 2b). Expression of GID1B is also detected, to a much lesser extent, in the endocarp b, as well as in the adaxial layer, the exocarp. Even though both GID1B and GID1C are expressed in the endocarp, degradation and lignification in gid1b gid1c was identical to that of the WT plants (Figure 6). The enhanced expression of GID1A in all tissues of this mutant may prevent detectable alterations of the process.
Figure 6.
The gid1 null mutants show morphological alterations in fruit structure.
(a) Delayed degradation of endocarp a in double gid1a gid1b and gid1a gid1c mutants. Transverse cryosections of fruits at 7, 8 and 10 dpa of wild-type (WT) and double mutants are shown. In the left top image, the different tissues are labeled: ex, exocarp; me, mesocarp; en-b, endocarp b; en-a, endocarp a. Presence or degradation of en-a are indicated by an arrowhead or asterisk, respectively. Scale bar represents 50 μm.
(b) Delayed lignification of en-b and degradation of en-a in double gid1a gid1c mutant. Transversal sections of fruits at 9 dpa of WT and the double gid1 mutants are shown. Presence of end-a and delayed lignification of end-b in gid1a gid1c mutant is indicated by an arrow head; degradation of en-a and strong lignification of en-b in WT and in gid1a gid1b and gid1b gid1c mutant is indicated by an asterisk. Scale bar represents 50 μm.
Expression of GA biosynthesis genes in ovules and seeds co-localize with GID1s
Differential spatial localization of GID1 proteins suggests that each GID1 may perceive GAs synthesized in, or transported to, different tissues within the pistil/fruit. Previously, we showed that most of the GA biosynthesis genes have a coordinated temporal expression patterns in ovules upon fertilization (Dorcey et al., 2009). In addition, GA20ox1 and GA20ox2, as well as GA3ox1, GA3ox3, and GA3ox4, have been shown by mutant analyses to have crucial roles in fruit-set and fertility (Hu et al., 2008; Rieu et al., 2008; Plackett et al., 2012). All four GA3ox genes are expressed in young developing seeds (Mitchum et al., 2006; Hu et al., 2008). In addition, GA3ox1 contributes to bioactive GA synthesis in maternal tissues, including replum, funiculus and receptacle (Hu et al., 2008; Arnaud et al., 2010). However, spatial and temporal expression of GA20oxs in ovules and developing seeds has not been reported previously. Therefore, we have studied the expression of GA20ox1 and GA20ox2 in ovules and seeds with translational fusion GUS lines (Figure 7). GA20ox1-GUS (Desgagne-Penix et al., 2005) was expressed only in the pollen and pollen tube (Figure 7a). Expression could also be detected in proximal site of the embryo sac just after fertilization. Interestingly, reciprocal crosses revealed that the GA20ox1-GUS activity came from the sperm cells rather than a de novo expression in the egg cell. Figure S6(a) shows that GA20ox1-GUS activity could be detected in both self-pollinated GUS reporter line and in WT plants pollinated with pollen from the GUS line, but GUS expression was absent when the GUS line was pollinated with WT pollen or in unfertilized pistils. Therefore, GUS activity in the GA20ox1-GUS was dependent on the male tissues; GA20ox1 expression was only detected when pollen was from the GA20ox1-GUS line.
Figure 7.
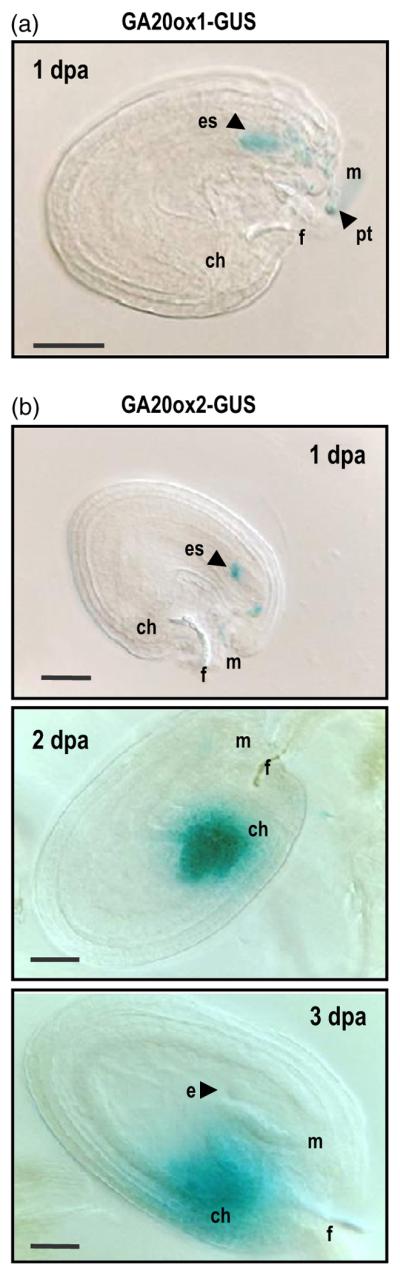
Expression of gibberellin (GA) biosynthesis genes is located in seeds shortly upon fertilization.
(a) Expression of GA20ox1-GUS (Desgagne-Penix et al., 2005) in seeds at 1 dpa.
(b) Expression of GA20ox2-GUS (Frigerio et al., 2006) in seeds at 1, 2 or 3 dpa. ch, chalaza; e, embryo; es, embryo sac; f, funiculus; m, micropyle; pt, pollen tube. Scale bar represents 40 μm.
GA20ox2-GUS (Frigerio et al., 2006) was expressed in both pollen tubes and seeds (Figure 7b). At 1 dpa, GUS activity was detected in embryo sac, but at 2–3 dpa its expression shifted to the chalazal pole of the developing seed. This change in spatial expression may correlate with its differential expression in male and female tissues at different stages after fertilization. Indeed, as it was observed for the GA20ox1-GUS line, GA20ox2-GUS expression 1d after fertilization in the embryo sac corresponded to male tissue (pollen tube discharge in embryo upon plasmogamy; Figure S6b). On the other hand, expression in the chalaza at 2–3 dpa corresponded to the sporophytic female tissue, as this expression was detected only in the GA20ox2-GUS line, regardless the precedence of the pollen. In contrast, neither GA20ox1 nor GA20ox2 showed any expression in valve (Figure S6).
In summary, GUS assays confirm previous qRT-PCR data indicating that the GAs are mostly synthesized in the developing seeds by GA20ox and GA3ox. With the exception of GA3ox1 expression in receptacle, replum and funiculus, none of the GA3ox or GA20ox genes were expressed in the valves. The strong expression of GID1A and GID1C in valves and their function in GA-induced pod elongation imply that GAs perceived in valves are transported mainly from the seed.
DISCUSSION
Differential expression of GID1s in ovules and valves
By qRT-PCR analysis, we observed that all three GID1s are expressed in pistils, with GID1A expressed at higher levels, followed by GID1B and GID1C. The predominant expression for GID1A reveals its major role in GA perception through the plant, including pistils and fruits. Within pistils, GID1s show differential spatial distribution as indicated by our results of qRT-PCR analysis and expression study using GID1-GUS transgenic lines. In valves, GID1A and GID1C participate in GA perception, while in the ovules GA perception is carried out by GID1A and GID1B, but not GID1C. Therefore, distinct GID1 combinations perceive GAs synthesized in, or transported to, specific tissues of the pistil to regulate growth and development of the seeds and the surrounding pod. Interestingly, most of the GA20ox and GA3ox genes that control the final steps of GA biosynthesis are only expressed in the developing seeds, but not in the fruit (Hu et al., 2008; Dorcey et al., 2009; this study). The only exception is GA3ox1, which is not expressed in young seeds, but is expressed in the replum, funiculus and receptacle of the fruit (Hu et al., 2008). None of the GA20ox or GA3ox genes are expressed in the valves (Hu et al., 2008; Arnaud et al., 2010; Plackett et al., 2012). Therefore, the bioactive GAs in the valve for promoting fruit elongation are likely transported from the developing seeds and/or from other surrounding tissues (replum, funiculus and receptacle). How bioactive GAs are transported to the valve remains an open question.
GID1s and DELLAs are co-expressed in pistils during fruit-set
Gibberellin signaling occurs through the GA-dependent interaction of GID1 and DELLA proteins (Sun, 2011). Our spatial expression analysis of DELLAs and GID1s suggests that certain GID1 and DELLA combinations interact in vivo to trigger the GA response in pistils. GID1A along with RGA, and GAI to a lesser extent, are expressed throughout the pistil. We propose that GID1A–GAI and GID1A–RGA interactions account for most of the GA-mediated processes in both valve and ovules. In addition, GID1B–RGL2 interaction can also occur in ovules, and GID1C–RGL1 interaction may take place in valves. The latter two interactions may account for tissue-specific GA-mediated processes during fruit-set and development, such as fertility and seed development for GID1B–RGL2 and fruit growth for GID1C–RGL1.
In addition to these co-expression patterns, differential affinity of GID1s towards the DELLAs may also modulate their interactions (Nakajima et al., 2006; Suzuki et al., 2009). For example, in vitro binding assays have revealed a strong affinity of RGA–GID1B, RGL2–GID1A, and GAI–GID1B proteins (Suzuki et al., 2009), each pair being co-expressed in ovules/seeds (this study). On the other hand, in addition to RGA–GID1B interaction, RGL1 can strongly interact with GID1A and GID1C, all of these combinations being expressed in the valves. The same in vitro assay revealed that RGL1 and RGL2 interact with very low affinity to GID1B and GID1C, respectively (Suzuki et al., 2009). Interestingly, RGL1–GID1B and RGL2–GID1C are not co-expressed in pistils, which may reflect co-evolution of GID1 and DELLA that results in distinct expression patterns and binding affinity. Figure 8 represents the possible GID1 and DELLA interactions in pistils, based on spatial gene expression and mutant phenotype analysis. Based on this model, we analyzed the silique phenotypes (seed number and pod length) of the triple mutants rga gai rgl2 and rga gai rgl1, but did not find significant differences comparing to the WT plants. This could be explained by the presence of GA and GID1s in these mutants. To test the specific roles of different DELLAs in ovule development and pod elongation, expression of gain-of-function rga gai rgl2 versus gain-of-function rga gai rgl1 using their endogenous promoters will be necessary.
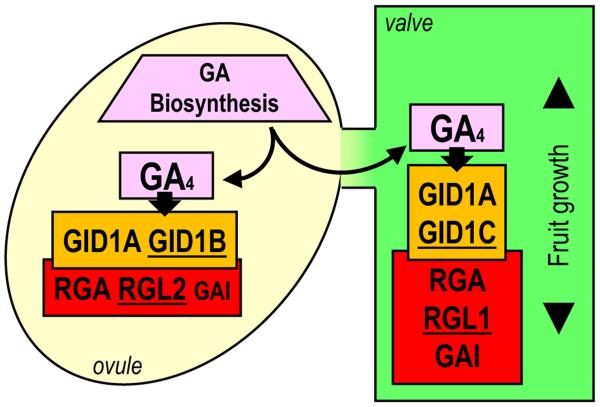
Figure 8.
Model of gibberellin (GA) biosynthesis, perception and signaling in seeds and valves of pistils/fruits.
Bioactive GAs, mainly GA4, are synthesized in developing seeds by GA20ox and GA3ox activities. In ovules/seeds GAs are perceived by GID1A and GID1B, which interact with RGA, GAI, and RGL2 to promote seed-specific GA signaling. GAs are also transported to the valve and perceived by GID1A and GID1C, which interact with RGA, GAI, and RGL1, to promote GA-specific valve responses (mainly pod growth and degradation of endocarp a). Proteins with tissue-specific expression are underlined.
GID1s have partial redundant function in fertility and fruit growth
Our study showed that the expression patterns of GID1s in developing fruit correlate well with the phenotypes of gid1 mutants. Lacking functional GID1A, which is expressed in nearly all pistil tissues, caused a slight reduction in fertility. Removing both GID1A and GID1B that are expressed in ovules resulted in reduced fertility. In contrast, mutations in both GID1A and GID1C that are expressed in the valves led to reduced fruit length.
The reduced silique length and low seed yield were previously correlated with shortening of the stamen filament, which decreased pollination (Griffiths et al., 2006; Iuchi et al., 2007). This is in agreement with the similar expression levels of both GID1A and GID1B in filaments (Suzuki et al., 2009). However, we found that the double mutant gid1a gid1b, which does not have GID1 activity in ovules, showed reduced seed-set even when fertilized by WT pollen. These results indicate that the reduced seed-set of the gid1a gid1b mutant is due to defects in maternal tissues, likely because of the absence of the main GA receptors in the ovule (GID1A and GID1B). GID1C expression is not detected in either ovules or funiculi, and it is not induced in the double mutant gid1a gid1b. It is intriguing that despite the lack of functional GA receptors in seeds, the double mutant gid1a gid1b still produces a significant number of seeds, suggesting the presence of a GA or GID1-independent pathway in promoting seed-set. However, this hypothesis is difficult to be verified because neither the GA-deficient mutant ga1 nor the triple gid1a gid1b gid1c mutant produces fertile flowers.
In contrast to gid1a gid1b, gid1a gid1c produced shorter silique with little reduction in seed number, even when pollinated with WT pollen, indicating that this phenotype is also due to maternal defects. The reduced fruit length in this gid1 double mutant is caused by limited GA perception in the silique pod. The double gid1b gid1c mutant did not show any fruit defects, indicating that GID1A activity is sufficient for GA perception in the pistil. In addition, GID1A expression in gid1b gid1c was slightly enhanced in seedlings and pistils (Figure S4), which further contributes to normal growth. Interestingly, expression of GID1B or GID1C was not induced in either seedling or pistils of gid1a gid1c and gid1a gid1b, respectively. Previous study showed that expression of GID1A and GID1B (but not GID1C) is controlled by the DELLA-dependent feedback mechanism (Griffiths et al., 2006). The up-regulation of GID1A in gid1b gid1c may be consequence of this feedback mechanism. However, GID1B was not up-regulated in seedlings or pistils of gid1a gid1c. Interestingly, up-regulation of GID1B in gid1a gid1c was described in floral buds, although none of the double mutants showed up-regulation of any of the GID1s in the inflorescence stem (Suzuki et al., 2009). All together, these data suggest that the feedback mechanism may be affected by developmental processes.
In summary, our study revealed that GID1A in combination with GID1B and GID1C controls seed-set and fruit growth, respectively. The distribution of GA receptors in valves, along with the lack of expression of late-stage GA biosynthesis genes, strongly suggests GA transportation from the seeds to promote fruit growth. Finally, the co-expression of GID1s and DELLAs in different tissues of the pistil suggests specific roles of each GID1–DELLA combination in the GA-dependent fruit-set and development. Detailed analysis of this interaction and the study of their specific roles in each tissue would be necessary to dissect and understand the GA-mediated molecular mechanisms taking place in pistils and fruits.
EXPERIMENTAL PROCEDURES
Plant material and fruit-set assays
Arabidopsis thaliana plants are in the Col-0 background. gid1a-1 (SALK_142767), gid1b-1 (SM_3_30227), and gid1c-1 (SALK_023529) were described by Griffiths et al. (2006). These lines will be mentioned in this report as gid1a, gid1b, and, gid1c, respectively. Double mutants gid1a-1 gid1b-1, gid1a-1 gid1c-1, and gid1b-1 gid1c-1 were generated by genetic cross and confirmed by genotyping, using the oligos listed in Table S1.
Seeds were surface-sterilized in EtOH and plated onto ½MS (Murashige and Skoog, 1962) media, kept at 4°C for 4 days, and transferred to a growth chamber at 22°C in long day photoperiod (16 h/8 h) for 10 days. Seedlings were then transferred to soil (a mix of peat moss, vermiculite and perlite, 2:1:1) and grown to maturity in a growth chamber at 22°C in long day photoperiod (16 h/8 h).
Fertility was scored using two different approaches: auto-pollinated fruits or fruits from emasculated flowers fully pollinated with WT pollen. Fruits were collected at maturity (12 days post-anthesis, dpa), seed number was counted and silique length was measured with a digital caliper. Ratio (seed number versus length) was determined. Parthenocarpy was assayed by application of GA3 to unfertilized pistils. Flowers were emasculated 1 day before anthesis and treated the next day with 330 μm GA3 (Fluka, http://www.sigmaaldrich.com/) in 0.01% (v/v) Tween-80, pH 7. Fruit and pistils were harvested 10 days after treatment, and scanned to measure final length with ImageJ software (Abramoff et al., 2004). Experiments were repeated three times with similar results.
Generation of transcriptional DELLA–GUS gene fusions and plant transformation
Transcriptional DELLA–GUS gene fusions were constructed for RGA, GAI and RGL1 genes. RGA gene fusions were made in binary vector pOCA28. pRGA:GUS contains approximately 7.7 kb of the RGA promoter region upstream of the ATG start site fused to the GUS reporter gene. GAI and RGL1 gene fusions were generated using vector pBI101.1. pGAI:GUS contains approximately 4 kb of the GAI promoter region upstream of the translational start site fused to the GUS reporter gene. pRGL1:GUS contains approximately 4 kb of the RGL1 promoter region upstream of the translational start site fused to the GUS reporter gene. For the pRGL2: GUS fusion, the rgl2-5 allele (Lee et al., 2002) was used. Detailed procedures for making the DELLA–GUS gene fusion constructs are described in Methods S1, using oligos listed in the Table S2. Plant transformations using DELLA–GUS plasmids and isolation of homozygous transgenic lines that contain a single insertion site were conducted as previously described (Hu et al., 2008).
Gene expression analysis by qRT-PCR
Pistils and fruits were harvested at different time points. For the dissection assay, ovules and valves of 1, 2, and 3 dpa unfertilized pistils (from emasculated flowers) were hand-dissected using acupuncture needles under a stereomicroscope. In addition, whole unfertilized pistils of 0 dpa were also harvested as reference. Total RNA was extracted using the RNeasy Plant Mini Kit (Qiagen, http://www.qiagen.com/). Genomic DNA was eliminated with 50 units of DNase I (Qiagen) for 15 min at room temperature. cDNA was synthesized using the SuperScript First-Strand Synthesis System for RT-PCR (Invitrogen, http://www.invitrogen.com/). qRT-PCR was carried out using the SYBR® GREEN PCR Master Mix (Applied Biosystems, http://www.lifetechnologies.com/) in an ABI PRISM 7000 Sequence Detection System (Applied Biosystems), essentially as described in Dorcey et al. (2009).
For the quantification of gene expression, cDNAs for each gene were cloned in pGEM-T Easy vector (Promega, http://www.promega.com/), using oligos listed in the Table S3. Absolute expression values were calculated basically as described in Whelan et al. (2003). Standard curves were generated using serial dilutions of purified plasmid DNA for each gene, ranging from 10−7 to 10−10. The copy number of plasmid in each dilution was calculated based on the molecular weight and the initial concentration of each construct (Whelan et al., 2003). Log10 of copy number of each transcript were calculated from the standard curve and the value of the threshold cycle (Ct) from the qRT-PCR data. The number of copies of each gene was normalized using the number of copies of ubiquitin10 (UBQ10, At4 g05320; Czechowski et al., 2005) in each sample. Primers (Table S1) were designed with the Primer Express™ v2.0 software (Applied Biosystems) and tested for efficiency.
Scanning electron microscopy (SEM)
Samples were harvested, mounted on the specimen holder of a CT-1000C cryo-transfer system (Oxford Instruments, www.oxford-instruments.com), interfaced with a JEOL JSM-5410 scanning electron microscope, and frozen in liquid N2. Samples were fractured, and sublimated at −85°C. Finally, samples were observed at incident electron energy of 10 kV with ×10 to ×100 magnification.
β-Glucuronidase (GUS) histochemical assay and histological procedures
The pGA20ox1:GA20ox1-GUS (Hay et al., 2002) and pGA20ox2:GA20ox2-GUS (Frigerio et al., 2006) transgenic Arabidopsis lines were provided by Dr Hedden (Rothamsted Research Center). The pGID1A:GID1A-GUS, pGID1B:GID1B-GUS and pGID1C:GID1C-GUS transgenic lines (Suzuki et al., 2009) were provided by Dr Nakajima (University of Tokyo). GUS assay and histological procedures were basically as previously described (Carbonell-Bejerano et al., 2010). K3Fe(CN)6 and K4Fe(CN)6, concentrations were adjusted for each line to obtain optimal signal (5 mm for pGID1A:GID1AGUS and pGID1B:GID1B-GUS, 10 mm for pGID1C:GID1C-GUS, 0.2 mm for pGA20ox:GA20ox-GUS, 5 mm for pGAI:GUS and pRGA:GUS; 2 mm for pRGL1:GUS, and 4 mm for rgl2-5). For the detection of GUS activity in thin resin sections, after staining with X-GlcA, samples were dehydrated in a series of 20, 35, and 50% (v/v) ethanol, post-fixed for 30 min in FAE (50% [v/v] ethanol, 5% [v/v] formaldehyde, 10% [v/v] acetic acid), and further dehydrated to 100% (v/v) EtOH and embedded in Technovit 7100 resin.
Supplementary Material
ACKNOWLEDGEMENTS
We wish to thank Dr Masatoshi Nakajima (University of Tokyo, Japan) for providing the pGID1:GID1-GUS lines, and Dr Peter Hedden (Rothamsted Research, UK) for the pGA20ox:GA20ox-GUS lines. We also thank Ms C. Fuster and M.A. Argomániz for technical assistance. This work has been supported by grants BIO2008-01039 and BIO2011-26302 from the Spanish Ministry of Science and Innovation and ACOMP/2010/079 and ACOMP/2011/287 from the Generalitat Valenciana for M.A.P.-A. and USDA grants 2010-65116-20460 and 2014-67013-21548 for T.P.S. C.G.-G. received a JAE PhD fellowship from the Spanish Council for Scientific Research (CSIC).
Footnotes
SUPPORTING INFORMATION Additional Supporting Information may be found in the online version of this article.
REFERENCES
- Abramoff MD, Magelhaes PJ, Ram SJ. Image processing with ImageJ. Biophotonics Int. 2004;11:36–42. [Google Scholar]
- Alabadi D, Blazquez MA, Carbonell J, Ferrandiz C, Perez-Amador MA. Instructive roles for hormones in plant development. Int. J. Dev. Biol. 2009;53:1597–1608. doi: 10.1387/ijdb.072423da. [DOI] [PubMed] [Google Scholar]
- Ariizumi T, Murase K, Sun TP, Steber CM. Proteolysis-independent downregulation of DELLA repression in Arabidopsis by the gibberellin receptor GIBBERELLIN INSENSITIVE DWARF1. Plant Cell. 2008;20:2447–2459. doi: 10.1105/tpc.108.058487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ariizumi T, Hauvermale AL, Nelson SK, Hanada A, Yamaguchi S, Steber CM. Lifting DELLA repression of Arabidopsis seed germination by nonproteolytic gibberellin signaling. Plant Physiol. 2013;162:2125–2139. doi: 10.1104/pp.113.219451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnaud N, Girin T, Sorefan K, Fuentes S, Wood TA, Lawrenson T, Sablowski R, Østergaard L. Gibberellins control fruit patterning in Arabidopsis thaliana. Genes Dev. 2010;24:2127–2132. doi: 10.1101/gad.593410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ben-Cheikh W, Perez-Botella J, Tadeo FR, Talon M, Primo-Millo E. Pollination increases gibberellin levels in developing ovaries of seeded varieties of citrus. Plant Physiol. 1997;114:557–564. doi: 10.1104/pp.114.2.557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carbonell-Bejerano P, Urbez C, Carbonell J, Granell A, Perez-Amador MA. A fertilization-independent developmental program triggers partial fruit development and senescence processes in pistils of Arabidopsis. Plant Physiol. 2010;154:163–172. doi: 10.1104/pp.110.160044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox CM, Swain SM. Localised and non-localised promotion of fruit development by seeds in Arabidopsis. Funct. Plant Biol. 2006;33:1–8. doi: 10.1071/FP05136. [DOI] [PubMed] [Google Scholar]
- Czechowski T, Stitt M, Altmann T, Udvardi MK, Scheible WR. Genome-wide identification and testing of superior reference genes for transcript normalization in Arabidopsis. Plant Physiol. 2005;139:5–17. doi: 10.1104/pp.105.063743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desgagne-Penix I, Eakanunkul S, Coles JP, Phillips AL, Hedden P, Sponsel VM. The auxin transport inhibitor response 3 (tir3) allele of BIG and auxin transport inhibitors affect the gibberellin status of Arabidopsis. Plant J. 2005;41:231–242. doi: 10.1111/j.1365-313X.2004.02287.x. [DOI] [PubMed] [Google Scholar]
- Dill A, Thomas SG, Hu J, Steber CM, Sun TP. The Arabidopsis F-box protein SLEEPY1 targets gibberellin signaling repressors for gibberellin-induced degradation. Plant Cell. 2004;16:1392–1405. doi: 10.1105/tpc.020958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorcey E, Urbez C, Blazquez MA, Carbonell J, Perez-Amador MA. Fertilization-dependent auxin response in ovules triggers fruit development through the modulation of gibberellin metabolism in Arabidopsis. Plant J. 2009;58:318–332. doi: 10.1111/j.1365-313X.2008.03781.x. [DOI] [PubMed] [Google Scholar]
- Fleet CM, Sun TP. A DELLAcate balance: the role of gibberellin in plant morphogenesis. Curr. Opin. Plant Biol. 2005;8:77–85. doi: 10.1016/j.pbi.2004.11.015. [DOI] [PubMed] [Google Scholar]
- Frigerio M, Alabadí D, Perez-Gomez J, Garcia-Carcel L, Phillips AL, Hedden P, Blazquez MA. Transcriptional regulation of gibberellin metabolism genes by auxin signaling in Arabidopsis. Plant Physiol. 2006;142:553–563. doi: 10.1104/pp.106.084871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu X, Richards DE, Fleck B, Xie D, Burton N, Harberd NP. The Arabidopsis mutant sleepy1gar2-1 protein promotes plant growth by increasing the affinity of the SCFSLY1 E3 ubiquitin ligase for DELLA protein substrates. Plant Cell. 2004;16:1406–1418. doi: 10.1105/tpc.021386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Martinez JL, Lopez-Diaz I, Sanchez-Beltran MJ, Phillips AL, Ward DA, Gaskin P, Hedden P. Isolation and transcript analysis of gibberellin 20-oxidase genes in pea and bean in relation to fruit development. Plant Mol. Biol. 1997;33:1073–1084. doi: 10.1023/a:1005715722193. [DOI] [PubMed] [Google Scholar]
- Griffiths J, Murase K, Rieu I, et al. Genetic characterization and functional analysis of the GID1 gibberellin receptors in Arabidopsis. Plant Cell. 2006;18:3399–3414. doi: 10.1105/tpc.106.047415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hay A, Kaur H, Phillips A, Hedden P, Hake S, Tsiantis M. The gibberellin pathway mediates KNOTTED1-type homeobox function in plants with different body plans. Curr. Biol. 2002;12:1557–1565. doi: 10.1016/s0960-9822(02)01125-9. [DOI] [PubMed] [Google Scholar]
- Hedden P, Thomas SG. Gibberellin biosynthesis and its regulation. Biochem. J. 2012;444:11–25. doi: 10.1042/BJ20120245. [DOI] [PubMed] [Google Scholar]
- Hirano K, Asano K, Tsuji H, Kawamura M, Mori H, Kitano H, Ueguchi-Tanaka M, Matsuoka M. Characterization of the molecular mechanism underlying gibberellin perception complex formation in rice. Plant Cell. 2010;22:2680–2696. doi: 10.1105/tpc.110.075549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu J, Mitchum MG, Barnaby N, et al. Potential sites of bioactive gibberellin production during reproductive growth in Arabidopsis. Plant Cell. 2008;20:320–336. doi: 10.1105/tpc.107.057752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iuchi S, Suzuki H, Kim YC, et al. Multiple loss-of-function of Arabidopsis gibberellin receptor AtGID1s completely shuts down a gibberellin signal. Plant J. 2007;50:958–966. doi: 10.1111/j.1365-313X.2007.03098.x. [DOI] [PubMed] [Google Scholar]
- Lee S, Cheng H, King KE, Wang W, He Y, Hussain A, Lo J, Harberd NP, Peng J. Gibberellin regulates Arabidopsis seed germination via RGL2, a GAI/RGA-like gene whose expression is up-regulated following imbibition. Genes Dev. 2002;16:646–658. doi: 10.1101/gad.969002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGinnis KM, Thomas SG, Soule JD, Strader LC, Zale JM, Sun TP, Steber CM. The Arabidopsis SLEEPY1 Gene Encodes a Putative F-Box Subunit of an SCF E3 Ubiquitin Ligase. Plant Cell. 2003;15:1120–1130. doi: 10.1105/tpc.010827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchum MG, Yamaguchi S, Hanada A, Kuwahara A, Yoshioka Y, Kato T, Tabata S, Kamiya Y, Sun TP. Distinct and overlapping roles of two gibberellin 3-oxidases in Arabidopsis development. Plant J. 2006;45:804–818. doi: 10.1111/j.1365-313X.2005.02642.x. [DOI] [PubMed] [Google Scholar]
- Murase K, Hirano Y, Sun TP, Hakoshima T. Gibberellin-induced DELLA recognition by the gibberellin receptor GID1. Nature. 2008;456:459–463. doi: 10.1038/nature07519. [DOI] [PubMed] [Google Scholar]
- Murashige T, Skoog F. A revised medium for rapid growth and bioassays with tobacco tissue culture. Physiol. Plant. 1962;15:437–497. [Google Scholar]
- Nakajima M, Shimada A, Takashi Y, et al. Identification and characterization of Arabidopsis gibberellin receptors. Plant J. 2006;46:880–889. doi: 10.1111/j.1365-313X.2006.02748.x. [DOI] [PubMed] [Google Scholar]
- Plackett AR, Thomas SG, Wilson ZA, Hedden P. Gibberellin control of stamen development: a fertile field. Trends Plant Sci. 2011;16:568–578. doi: 10.1016/j.tplants.2011.06.007. [DOI] [PubMed] [Google Scholar]
- Plackett AR, Powers SJ, Fernandez-Garcia N, et al. Analysis of the developmental roles of the Arabidopsis gibberellin 20-oxidases demonstrates that GA20ox1, -2, and -3 are the dominant paralogs. Plant Cell. 2012;24:941–960. doi: 10.1105/tpc.111.095109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rieu I, Ruiz-Rivero O, Fernandez-Garcia N, et al. The gibberellin biosynthetic genes AtGA20ox1 and AtGA20ox2 act, partially redundantly, to promote growth and development throughout the Arabidopsis life cycle. Plant J. 2008;53:488–504. doi: 10.1111/j.1365-313X.2007.03356.x. [DOI] [PubMed] [Google Scholar]
- Sasaki A, Itoh H, Gomi K, et al. Accumulation of phosphorylated repressor for gibberellin signaling in an F-box mutant. Science. 2003;299:1896–1898. doi: 10.1126/science.1081077. [DOI] [PubMed] [Google Scholar]
- Serrani JC, Sanjuan R, Ruiz-Rivero O, Fos M, Garcia-Martinez JL. Gibberellin regulation of fruit set and growth in tomato. Plant Physiol. 2007;145:246–257. doi: 10.1104/pp.107.098335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimada A, Ueguchi-Tanaka M, Nakatsu T, Nakajima M, Naoe Y, Ohmiya H, Kato H, Matsuoka M. Structural basis for gibberellin recognition by its receptor GID1. Nature. 2008;456:520–523. doi: 10.1038/nature07546. [DOI] [PubMed] [Google Scholar]
- Sun TP. The molecular mechanism and evolution of the GA-GID--DELLA signaling module in plants. Curr. Biol. 2011;21:338–345. doi: 10.1016/j.cub.2011.02.036. [DOI] [PubMed] [Google Scholar]
- Suzuki H, Park SH, Okubo K, et al. Differential expression and affinities of Arabidopsis gibberellin receptors can explain variation in phenotypes of multiple knock-out mutants. Plant J. 2009;60:48–55. doi: 10.1111/j.1365-313X.2009.03936.x. [DOI] [PubMed] [Google Scholar]
- Swain SM, Singh DP. Tall tales from sly dwarves: novel functions of gibberellins in plant development. Trends Plant Sci. 2005;10:123–129. doi: 10.1016/j.tplants.2005.01.007. [DOI] [PubMed] [Google Scholar]
- Ueguchi-Tanaka M, Ashikari M, Nakajima M, et al. GIBBERELLIN INSENSITIVE DWARF1 encodes a soluble receptor for gibberellin. Nature. 2005;437:693–698. doi: 10.1038/nature04028. [DOI] [PubMed] [Google Scholar]
- Ueguchi-Tanaka M, Hirano K, Hasegawa Y, Kitano H, Matsuoka M. Release of the repressive activity of rice DELLA protein SLR1 by gibberellin does not require SLR1 degradation in the gid2 mutant. Plant Cell. 2008;20:2437–2446. doi: 10.1105/tpc.108.061648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vivian-Smith A, Koltunow AM. Genetic analysis of growth-regulator-induced parthenocarpy in Arabidopsis. Plant Physiol. 1999;121:437–451. doi: 10.1104/pp.121.2.437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voegele A, Linkies A, Müller K, Leubner-Metzger G. Members of the gibberellin receptor gene family GID1 (GIBBERELLIN INSENSITIVE DWARF1) play distinct roles during Lepidium sativum and Arabidopsis thaliana seed germination. J. Exp. Bot. 2011;62:5131–5147. doi: 10.1093/jxb/err214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whelan JA, Russell NB, Whelan MA. A method for the absolute quantification of cDNA using real-time PCR. J. Immunol. Methods. 2003;278:261–269. doi: 10.1016/s0022-1759(03)00223-0. [DOI] [PubMed] [Google Scholar]
- Willige BC, Ghosh S, Nill C, Zourelidou M, Dohmann EMN, Maier A, Schwechheimer C. The DELLA domain of GA INSENSITIVE mediates the interaction with the GA INSENSITIVE DWARF1A gibberellin receptor of Arabidopsis. Plant Cell. 2007;19:1209–1220. doi: 10.1105/tpc.107.051441. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.