ABSTRACT
In this study, we examined the requirement for host dynein adapter proteins such as dynein light chain 1 (DYNLL1), dynein light chain Tctex-type 1 (DYNLT1), and p150Glued in early steps of human immunodeficiency virus type 1 (HIV-1) replication. We found that the knockdown (KD) of DYNLL1, but not DYNLT1 or p150Glued, resulted in significantly lower levels of HIV-1 reverse transcription in cells. Following an attempt to determine how DYNLL1 could impact HIV-1 reverse transcription, we detected the DYNLL1 interaction with HIV-1 integrase (IN) but not with capsid (CA), matrix (MA), or reverse transcriptase (RT) protein. Furthermore, by mutational analysis of putative DYNLL1 interaction motifs in IN, we identified the motifs 52GQVD and 250VIQD in IN as essential for DYNLL1 interaction. The DYNLL1 interaction-defective IN mutant HIV-1 (HIV-1INQ53A/Q252A) exhibited impaired reverse transcription. Through further investigations, we have also detected relatively smaller amounts of particulate CA in DYNLL1-KD cells or in infections with HIV-1INQ53A/Q252A mutant virus. Overall, our study demonstrates the novel interaction between HIV-1 IN and cellular DYNLL1 proteins and suggests the requirement of this virus-cell interaction for proper uncoating and efficient reverse transcription of HIV-1.
IMPORTANCE Host cellular DYNLL1, DYNLT1, and p150Glued proteins have been implicated in the replication of several viruses. However, their roles in HIV-1 replication have not been investigated. For the first time, we demonstrated that during viral infection, HIV-1 IN interacts with DYNLL1, and their interaction was found to have a role in proper uncoating and efficient reverse transcription of HIV-1. Thus, interaction of IN and DYNLL1 may be a potential target for future anti-HIV therapy. Moreover, while our study has evaluated the involvement of IN in HIV-1 uncoating and reverse transcription, it also predicts a possible mechanism by which IN contributes to these early viral replication steps.
INTRODUCTION
The steps of early stage human immunodeficiency virus type 1 (HIV-1) replication include virus entry, uncoating, reverse transcription, intracytoplasmic retrograde transportation (i.e., the migration of HIV from the cytoplasmic periphery to the perinuclear space), nuclear import, and genomic integration (reviewed in reference 1). Following HIV-1 entry into the cell, viral genomic RNA and associated proteins are released into the cytoplasm as a ribonucleoprotein complex referred as the reverse transcription complex (RTC). Within the RTC, HIV-1 genomic RNA is reverse transcribed into a cDNA, which then forms a high-molecular-weight preintegration complex (PIC). HIV-1 cDNA enters the nucleus as a part of PIC by active nuclear import and subsequently integrates into the host cell genome (reviewed in reference 2).
HIV-1 utilizes various cellular proteins for replication mostly by interacting with its viral proteins. Genome-wide small interfering RNA (siRNA)/short hairpin RNA (shRNA) screening as well as other functional studies have uncovered a large number of host proteins with putative roles in HIV-1 replication (reviewed in references 3, 4, and 5). Additionally, functional studies from our laboratory, as well as from other groups, have uncovered key viral and cellular protein interactions that promote successful HIV-1 nuclear import and integration (reviewed in references 2 and 6). However, molecular events associated with HIV-1 reverse transcription, uncoating, or retrograde transport in the cytoplasm are not well understood. To date, evidence suggests that gem-associated protein 2 (Gemin2) interacts with HIV-1 integrase (IN) in target cells and contributes to reverse transcription by an unknown mechanism(s) (7, 8). Similarly, accumulated evidence suggests that cyclophilin A (CypA) and peptidyl-prolyl cis-trans isomerase NIMA-interacting 1 (prolyl isomerase Pin1) proteins interact with HIV-1 capsid (CA) protein in target cells and facilitate the proper uncoating of HIV-1 (9, 10). In addition, some other cellular factors with putative roles in HIV-1 reverse transcription and uncoating have been described in recent studies (11–14). Although the exact mechanism(s) by which these cellular factors contribute to HIV-1 reverse transcription and/or uncoating is not very clear, the accumulated evidence so far clearly suggests a key role for cellular cofactors in HIV-1 uncoating and reverse transcription.
Dynein adapter proteins such as dynein light chain 1 (DYNLL1, LC8, DLC1), dynein light chain Tctex-type 1 (DYNLT1), and p150Glued have been implicated in cargo recruitment to the dynein complex during retrograde transport (15–18). The dynein complex is a microtubule (MT)-associated protein complex that mediates retrograde transport of macromolecules in the cytoplasm (19). Interestingly, recent studies have indicated that the functional dynein complex or intact MT network is essential for efficient HIV-1 uncoating (20, 21), retrograde transport (20, 22), and restriction by rhesus tripartite motif protein 5 isoform alpha (TRIM5α) (23). It is also important to note that the dynein adapter proteins are involved in various cellular functions outside the dynein complex (reviewed in references 24 and 25). In the past, studies have implicated the DYNLL1 and DYNLT1 proteins in different aspects of viruses replication (reviewed in reference 24). For example, DYNLL1 interacts with specific rabies virus phosphoprotein and contributes to the viral gene expression (26–28). Also, DYNLL1 interacts with the CA protein of bovine immunodeficiency virus (BIV) and contributes to retrograde transport of BIV (18). DYNLT1 interacts with the CA protein of human papillomavirus type 16 (HPV-16) and contributes to HPV-16 replication at an unknown replication step(s) (29). Unlike the case for DYNLL1 and DYNLT1, potential involvement of p150Glued in the replication of viruses is currently not very clear. Nevertheless, the dynactin structure of the dynein complex, with which the p150Glued is associated (30), has been implicated in the replication and/or retrograde transport of viruses (15, 31, 32). To date, potential contributions of the dynein adapter proteins in early steps of HIV-1 replication or interactions between HIV-1 and the dynein adapter proteins have not been investigated.
In this study, we investigated the role of dynein adapter proteins in different early steps of HIV-1 replication and DYNLL1 interaction with HIV-1 IN. We detected significantly lower levels of total HIV-1 DNA synthesis in DYNLL1 knockdown (KD) cells but not in p150Glued- or DYNLT1-KD cells. Our data also showed that DYNLL1 was specifically coprecipitated with the HIV-1 IN protein in an in vitro interaction assay and a cell-based coimmunoprecipitation (Co-IP) assay performed using 293T cells or HIV-1-infected C8166 CD4+ T cells. By mutational analysis, the 52GQVD and 250VIQD motifs in IN were shown to be essential for DYNLL1 interaction. DYNLL1 interaction-defective IN mutant HIV-1 (HIV-1INQ53A/Q252A) demonstrated impaired HIV-1 replication and reverse transcription activity. Further investigations demonstrated relatively lower levels of particulate CA (i.e., CA associated with RTC/PIC) in DYNLL1-KD cells as well as in infections involving the HIV-1INQ53A/Q252A mutant virus. From these data, we suggest that HIV-1 utilizes the cellular protein DYNLL1 for efficient reverse transcription and proper uncoating of HIV-1 through the interaction with its IN protein.
MATERIALS AND METHODS
Antibodies and chemicals.
The antibodies (Abs) used for Western blot (WB), immunoprecipitation, and fluorescence-activated cell sorter (FACS) analyses were as follows: a rabbit anti-green fluorescent protein (anti-GFP) polyclonal antibody (Ab) (Molecular Probes), a mouse anti-T7 monoclonal Ab (Novagen), a rabbit anti-DYNLL1 monoclonal Ab (Abcam), a mouse anti-dynein intermediate chain 1 (DIC1) Ab (Abcam), an anti-β-tubulin Ab (Sigma), a horseradish peroxidase (HRP)-conjugated anti-GFP Ab (Miltenyi Biotec), an HRP-conjugated donkey anti-rabbit IgG (Amersham Biosciences), a sheep anti-mouse IgG (Amersham Biosciences), and a goat anti-glutathione S-transferase (anti-GST) polyclonal Ab (Amersham Biosciences). The mouse anti-HIV-1 p24Gag Ab was described previously (33). A rabbit anti-IN Ab (catalog no. 757) was obtained from the NIH AIDS Reagent Program. Anti-CD4-allophycocyanin (APC) Ab was purchased from eBioscience, USA. The chemicals and reagents used in this study are as follows. Purified recombinant GST-IN and GST-MA proteins were produced in Escherichia coli using the pGEX4T3-GST-IN and pGEX4T3-GST-matrix (MA) expression vectors, respectively, as previously described (34). The purified recombinant DYNLL1 protein was purchased from ProSpec-Tany TechnoGene Ltd., Israel. The ECL WB detection kit was purchased from Perkin-Elmer Life Science (Boston, MA, USA). Puromycin and NP-40 Alternative were obtained from Calbiochem. Phorbol 12-myristate 13-acetate (PMA) was purchased from EMD Millipore, Canada. Pronase was purchased from Sigma-Aldrich, USA. Phytohemagglutinin (PHA) was purchased from Sigma-Aldrich, USA, and human recombinant interleukin-2 (hIL-2) was purchased from Roche, Germany.
Construction of expression plasmids and HIV-1 proviruses.
The construction of GFP-IN and MA-yellow fluorescent protein (YFP) mammalian expression vectors has been described previously (34). A GFP-CA mammalian expression vector was constructed by cloning cDNA for the CA gene into a cytomegalovirus (CMV)-AcGFPc plasmid (Clontech) at BamHI and XbaI restriction enzyme sites, in frame with the GFP. The cDNA for the CA gene was PCR amplified using HIV-1Bru as a template. The primers 5′-GCCAGGTCGGATCCCCTATAGTGCAG-3′ (forward) and 5′-TTGTTACGCGGCCGCTCTAGATTACAAAACTCTTGC-3′ (reverse) were used for the CA gene amplification. A GFP-IN (Moloney murine leukemia virus [MMLV]) mammalian expression vector was constructed by cloning cDNA for the MMLV IN gene into the CMV-AcGFPc plasmid at BamHI and XbaI restriction sites, in frame with the GFP. The cDNA for the MMLV IN gene was PCR amplified from pVPack-GP (MMLV gag-pol expression vector) (Stratagene). The primers 5′-CTCGGATCCGAGAATTCATCACCCTA-3′ (forward) and 5′-GCAGCTAGCTTAGGGAGCTTCGCGGGTTAACCT-3′ (reverse) were used for MMLV IN gene amplification. A GFP-reverse transcriptase (RT) fusion protein expression vector was generated by cloning the cDNA for the RT gene into the CMV-AcGFPc plasmid (Clontech) at the BamHI restriction enzyme site, in frame with the GFP. The coding region for the RT gene was obtained by digesting a CMV-T7-RT expression vector with BamHI and BglII restriction enzymes. The CMV-T7-RT expression vector has been described earlier (35). A ProLabel (PL)-DYNLL1 expression vector was constructed by subcloning the DYNLL1 gene into a CMV-PL-Ku70 expression vector at BamHI and NotI restriction enzyme sites, by replacing the Ku70 gene. The coding region for the DYNLL1 gene was obtained by digesting a pCDNA3-T7-DYNLL1 expression vector with BamHI and NotI restriction enzymes. The pCDNA3-T7-DYNLL1 was obtained from Addgene, USA. CMV-PL-Ku70 was constructed by cloning cDNA for the Ku70 gene into a CMV-PL plasmid (Clontech) at BamHI and NotI restriction enzyme sites, in frame with the PL tag. The construction of wild-type CMV-GFP-IN (CMV-GFP-INWt) and deletion mutants (amino acids [aa] 1 to 212, 50 to 288, 117 to 288, and 1 to 230) has been previously described (36). A CMV GFP-IN(180–230) deletion mutant expression vector was constructed by cloning the coding cDNA for the IN180–230 amino acids into the CMV-AcGFPc at the HindIII and BamHI restriction enzyme sites, in frame with the GFP. CMV-PL-INWt has been previously described (37). To construct GFP-INQ53A, GFP-INQ252A, GFP-INQ209A, GFP-INQ53A/Q252A, and GFP-INK186A/R187A expression vectors, IN cDNAs with the desired mutations were generated by a two-step PCR method (38) by using the forward primer 5′-GCGCTCGAGAAGCTTGGCTTTTTAGATGGAATAG-3′ containing a HindIII restriction enzyme site, the reverse primer 5′-CTAAACGGATCCATGTTCTAA-3′ containing a BamHI restriction enzyme site, and complementary primers with the intended mutations. Subsequently, cDNAs were cloned into CMV-AcGFPc at HindIII and BamHI restriction enzyme sites. The INK186A/R187A mutant was found to be defective for multimerization (39). A SvCMVin-Vpr-RT-INQ53A/Q252A mutant fusion protein expression vector was generated by introducing specific mutations into the SvCMVin-Vpr-RT-IN using a two-step PCR method, as previously described (40). For the construction of SvCMVin-Vpr-RT-IN mutants, a forward primer targeting the RT gene at a natural NheI restriction enzyme site (5′-GCAGCTAGCAGGGAGACTAA-3′), the reverse primer with a PstI restriction enzyme site targeting the 3′ end of IN (5′-CTGTTCCTGCAGCTAATCCTCATCCTG-3′), and complementary primers with the intended mutations were used. The construction of SvCMVin-Vpr-RT-(stop)IN has been previously described (41). pGEX4T3-GST-IN and pGEX4T3-GST-MA were constructed by cloning the cDNAs for the IN and MA genes, respectively, into a pGEX Vector using the EcoRI and NotI restriction enzyme sites. The pGEX4T3 plasmid was obtained from AMRAD Corporation Ltd. pLET-Lai, an X4 trophic HIV-1 envelope expression vector, was a gift from Vicente Planelles at the University of Utah. HIV-1pNL4.3/R−/E−/Luc+ and HIV-1pNL4.3/R−/E−/GFP+ provirus DNAs (36), HxBruR−/ΔRI/E+ (42), and HIV-1HxBru-IN-HA (34) were previously described.
Cell culture, transfection, gene knockdown, cell proliferation assay, and FACS assay.
Human 293T cells and MMLV-packaging Phoenix cells were maintained in complete Dulbecco modified Eagle medium (DMEM) (i.e., supplemented with 1% penicillin-streptomycin and 10% fetal calf serum). ACH-2 and CD4+ C8166T lymphocyte cell lines were maintained in complete RPMI 1640 medium. Peripheral blood mononuclear cells (PBMCs) were isolated from the blood of healthy adult human volunteers through sedimentation on a Ficoll (Lymphoprep; Axis-Shield) gradient. Primary human CD4+ T lymphocytes were purified from PBMCs using the EasySep human CD4+ T cell enrichment kit (Stemcell technologies) according to the manufacturer's instructions. CD4+ T cells were stimulated with PHA (3 μg/ml) and hIL-2 (10 IU/ml) for 4 days. DNA transfection experiments were performed in 293T cells using a standard calcium phosphate DNA precipitation method. For DYNLL1-, DYNLT1-, or p150Glued-KD, different shRNAs for each gene were tested, and the one best shRNA for each gene was selected. For DYNLL1, three different clones of pLKO.1 lentiviral vector for DYNLL1 shRNA (i.e., clones 1, 2, and 3) were obtained from Open Biosystems (belonging to GE Dharmacon, USA). The sense oligonucleotide sequences of the shRNA in clone 1, 2, and 3 are AGAAGGACATTGCGGCTCATA, GCGCTGGAGAAATACAACATA, and ACATGAAACCAAACACTTCAT, respectively. Vesicular stomatitis virus glycoprotein G (VSV-G) envelope-pseudotyped lentiviral particles (LVPs) harboring different shRNAs were produced in 293T cells, as described earlier (36). A pLKO.1 vector plasmid having no shRNA, which was a gift from Sam Kung at the University of Manitoba, was used to produce control vector particles. The control or different LVPs for DYNLL1 (400 ng of p24Gag/2 × 106 cells) were transduced into C8166T cells. After 48 h postransduction, the cells were selected using 0.5 μg/ml of puromycin. On the third day of LVP transduction, the KD of DYNLL1 was determined by detecting DYNLL1 protein expression by WB using an anti-DYNLL1 antibody. For DYNLL1-KD in ACH-2 cells, ACH-2 cells were transduced with control or DYNLL1 (clone 2) LVPs, and DYNLL1-KD analysis was carried out as described above. The LVPs expressing different shRNAs for DYNLT1- or p150Glued-KD were similarly produced. On day 3 of LVP transduction, the KD of DYNLT1 or p150Glued was determined by detecting protein expression via WB using the corresponding Abs. The sense oligonucleotide sequences of the best shRNAs used in this study for DYNLT1 and p150Glued were 5′-CCACAAATGTAGTAGAACAAA-3′ and 5′-GCTGGAGACATTGAATCAATT-3′, respectively. A WST-1 cell proliferation assay kit (Roche, Germany) was used to measure the proliferation of C8166T cells transduced with different LVPs. Briefly, control or DYNLL1 LVP-transduced C8166T cells were cultured at a density of 2 × 104 cells/well in a 96-well format and maintained at 37°C. On different days, WST-1 cell proliferation reagent was added to the cultures at 10 μl/well. Subsequently, the cultures were incubated at 37°C for 4 h, and the absorbance was measured at 450 nm using a microplate reader. To examine the expression of the CD4 receptor on the surfaces of C8166T cells transduced with different LVPs, the cells were stained with anti-CD4-APC or the respective isotype control Ab in FACS buffer (0.5% bovine serum albumin [BSA] in phosphate-buffered saline [PBS]) (eBioscience, USA) on ice for 20 min. Subsequently, the cells were washed in FACS buffer and analyzed by flow cytometry. Sample data were collected using a BD LSR II flow cytometer (BD Biosciences, USA), and the data were analyzed using FCS Express software (De Novo Software, USA).
Cell-based Co-IP and in vitro protein interaction assay.
To detect the interaction of HIV-1 IN, MA, CA, or RT with DYNLL1, 293T cells were transfected with the GFP-IN, MA-YFP, GFP-CA, or GFP-RT expression vector. At 48 h posttransfection, 9/10 of the cells were lysed in 0.12% NP-40 lysis buffer (RPMI medium containing 0.12% NP-40 Alternative and protease inhibitor cocktail [Roche]) on ice for 30 min. The lysates were subjected to immunoprecipitation using an anti-GFP Ab. The immunoprecipitates were resolved by 12% SDS-PAGE, and coprecipitation of endogenous DYNLL1 was analyzed by WB using an anti-DYNLL1 Ab. The immunoprecipitation of GFP-IN, MA-YFP, GFP-CA, or GFP-RT was analyzed by WB using an HRP-conjugated anti-GFP Ab. Endogenous DYNLL1 expression in 293T cells was detected by WB using an anti-DYNLL1 Ab. To detect the interaction of INWt or various IN deletion or substitution mutants with PL-DYNLL1, GFP, GFP-INWt, or GFP-INMt (deletion or substitution mutants), the expression vector was cotransfected with the PL-DYNLL1 vector into 293T cells. At 48 h posttransfection, the cells were lysed, and the lysates were subjected to chemiluminescent coimmunoprecipitation (Co-IP) using an anti-GFP Ab. The coprecipitation of PL-DYNLL1 with GFPWt/Mt in the immunoprecipitates was detected by measuring PL activity using the ProLabel detection kit II (Clontech, USA) as described in the manufacturer's instructions. Similarly, DIC1 interaction with GFP-INWt/Mt or GFP-CA was analyzed through a Co-IP using an anti-GFP Ab followed by detection of DIC1 by WB using an anti-DIC1 Ab. The interaction of IN with endogenous DYNLL1 from HIV-1-infected C8166T cells was detected by Co-IP. Briefly, 15 × 106 C8166T cells were infected with equal amounts of the HxBru or HxBru-IN-HA virus (50 ng of HIV-1 p24Gag). After 72 h of infection, the cells were lysed in 0.12% NP-40 lysis buffer for 30 min on ice. The lysates were immunoprecipitated using an anti-HA Ab. The immunoprecipitates were resolved by 12% SDS-PAGE, and DYNLL1 and IN-HA were detected by WB using anti-DYNLL1 and anti-HA Abs, respectively. The p24Gag in the total cell lysates was detected by WB using an anti-p24Gag Ab. To determine the in vitro interaction of DYNLL1 with GST-IN or GST-MA, 0.2 μg of recombinant DYNLL1 was incubated with an equal amount of recombinant GST, GST-IN, or GST-MA protein. The mixtures were incubated with GST beads at 4°C for 4 h, and the eluates were resolved by 12% SDS-PAGE. The coprecipitation of DYNLL1 was examined by WB using an anti-DYNLL1 Ab. The presence of GST, GST-IN, or GST-MA in immunoprecipitates was detected by WB using an anti-GST Ab. The DYNLL1 in supernatants was detected by WB using an anti-DYNLL1 Ab.
Virus production and infection.
A single-cycle replicating luciferase (Luc) reporter HIV-1pNL4.3/R−/Luc+ (HIV-Luc) was produced in 293T cells by cotransfecting HIV-1pNL4.3/R−/E−/Luc+ and pLET-Lai, as described earlier (36). Single-cycle-replicating HIV-1Wt or HIV-1INQ53A/Q252A mutant virus was produced in 293T cells by cotransfecting HxBruR−/ΔRI/E+ with the CMV-Vpr-RT-INWt or -INQ53A/Q252A expression vector, respectively, as previously described (40). Similarly, single-cycle-replicating HIV-1ΔIN virus was produced in 293T cells by cotransfecting HxBruR−/ΔRI/E+ with CMV-Vpr-RT-(stop)IN. The above-mentioned viruses consist of autologous HIV-1 envelope. The p24Gag titers for viruses were determined by using an HIV-1p24Gag enzyme-linked immunosorbent assay (ELISA). Control or DYNLL1-KD C8166T cells (0.5 × 106) were infected with different doses of HIV-Luc (i.e., 30, 10, and 3.3 ng of p24Gag). At 24 h postinfection (p.i.), the Luc activity was measured as previously described (34). C8166T cells (0.5 × 106) were infected with equal amounts of HIV-1Wt, HIV-1INQ53A/Q252A, or HIV-1ΔIN (at 10 ng of virus-associated p24Gag). At 72 h p.i., the p24Gag content in the supernatants was determined using an HIV-1p24Gag ELISA. The production and infection of Luc reporter MMLV vector particles (MMLV-Luc) were done as previously described (36).
Virus composition analysis.
HIV-1Wt or various HIV-1IN deletion/substitution mutant virus stocks were treated with subtilisin, as previously described (43). Subsequently, virus stocks were purified on a 20% sucrose cushion for 1.5 h at 125,000 × g and 4°C. Equal amounts of sucrose-purified virus stocks were subsequently lysed in 4× Laemmli buffer, and the lysates were resolved by 10% SDS-PAGE. Virus-incorporated RT, IN, and p24Gag proteins were detected by WB using the corresponding Abs.
HIV-1 entry analysis.
The cellular entry of HIV-1 was analyzed using a previously published cell fractionation protocol with minor modifications (44–46). Briefly, 3 × 106 control or DYNLL1-KD C8166T cells were infected with HIV-Luc (at 50 ng of p24Gag) on ice for 20 min to allow virus attachment, followed by transfer to a 37°C cell culture chamber for 2 h. Control cells (3 × 106) were infected with HIV-Luc (at 50 ng of p24Gag) on ice for 20 min and similarly processed as a negative control. Following harvest, the cells were treated for 3 min with pronase (1 mg/ml) to remove viruses adsorbed at the cell surface. The cells were washed three times using RPMI supplemented with 10% fetal calf serum to remove pronase. Subsequently, the cells were incubated with ice-cold digitonin buffer (10 mM Tris [pH 7.5], 10 mM NaCl, 0.15 mM spermine, 0.5 mM spermidine, 1 mM EDTA, and 100 μg/ml digitonin) for 10 min at 4°C to selectively permeabilize the plasma membrane. The cytoplasmic fraction was separated by centrifugation at 3,000 rpm for 4 min at 4°C. The virus entry was examined by quantifying HIV-1 genomic RNA from the cytoplasmic fractions using reverse transcription-quantitative PCR (qPCR) for total HIV-1 DNA analysis.
Real-time qPCR analysis.
The qPCR analysis was performed as described previously (36). Briefly, 1.5 × 106 control and DYNLL1-KD cells were infected with HIV-Luc (at 10 ng of virus-associated p24Gag), which was treated with 340 IU/ml of DNase (Roche) for 1 h at 37°C to remove residual plasmid DNA. At 12 and 24 h p.i., total HIV-1 DNA and 2-long-terminal-repeat (2-LTR) circle DNA were quantified from the total cellular DNA samples by using the Mx3000P real-time PCR system (Stratagene, USA). The primer and probe sets used for total HIV-1 and 2-LTR circle DNA quantification were previously described (40). Total HIV-1 DNA and 2-LTR circle DNA were expressed as copy numbers per cell, and the DNA templates were normalized using β-globin gene amplification (40). To examine the IN-dependent loss of reverse transcription in DYNLL1-KD cells, 1.5 × 106 control and DYNLL1-KD C8166T cells were infected with the DNase-treated HIV-1Wt or ΔIN viruses (at 50 ng of virus-associated p24Gag), and the total HIV-1 DNA synthesis was quantified at 12 h p.i. by using qPCR, as described above. To examine the cDNA synthesis of the DYNLL1 interaction-defective IN mutant HIV-1, 1.5 × 106 control and DYNLL1-KD C8166T cells were infected with the DNase treated HIV-1Wt, HIV-1INQ53A/252A, or HIV-1ΔIN virus (at 50 ng of virus-associated p24Gag). At 12 h p.i., the total HIV-1 DNA was quantified using qPCR, as described above. A heat-inactivated virus (INA) (70°C for 30 min) was treated with DNase and included in parallel as a control for monitoring the carryover plasmid DNA contamination during qPCR analyses.
Fate-of-capsid assay.
The fate-of-capsid assay was performed as previously described (47). Briefly, control or DYNLL1-KD C8166T cells were infected with HIV-1Wt, or C8166T cells were infected with equal amounts of the HIV-1Wt, HIV-1INQ53A/Q252A, or HIV-1ΔIN virus. Cells were incubated at 4°C for 20 min to allow virus attachment and transferred to a 37°C cell culture chamber. At 4 h of incubation, the cells were treated for 3 min with pronase (1 mg/ml) and washed three times using RPMI supplemented with 10% fetal calf serum. A negative control was obtained by incubating a panel of cells with HIV-1Wt on ice for 20 min and harvesting after treatment with pronase followed by three washes with complete RPMI medium. One-tenth of cells were lysed in 0.5% NP-40 lysis buffer, and HIV-1 p24Gag in total cell lysates was evaluated by ELISA to determine the virus entry. The cells were cultured at 37°C until the time of harvest. At 4, 8, and 12 h p.i., the cells were harvested and subjected to the fate-of-capsid assay. Briefly, the cells were incubated with hypotonic buffer (10 mM Tris-HCl [pH 8.0], 10 mM KCl, and 1 mM EDTA) for 15 min on ice. The cell pellets were then mixed with glass beads and vortexed for three times for 10 s each to lyse the cells. The cell lysis was confirmed through staining of cell lysates with trypan blue dye and observing under a microscope. The lysates were clarified by centrifugation for 3 min at 2,000 × g. Supernatants were overlaid onto 7 ml of a 50% sucrose cushion prepared in 1× PBS and centrifuged for 2 h at 125,000 × g at 4°C using a Beckman SW41 rotor. After centrifugation, pellet fractions were collected in 100 μl of hypotonic lysis buffer. The concentration of HIV-1 p24Gag (i.e., particulate CA) in pellet fractions was determined by using an HIV-1 p24Gag ELISA.
RESULTS
DYNLL1, but not p150Glued or DYNLT1, is essential for HIV-1 reverse transcription.
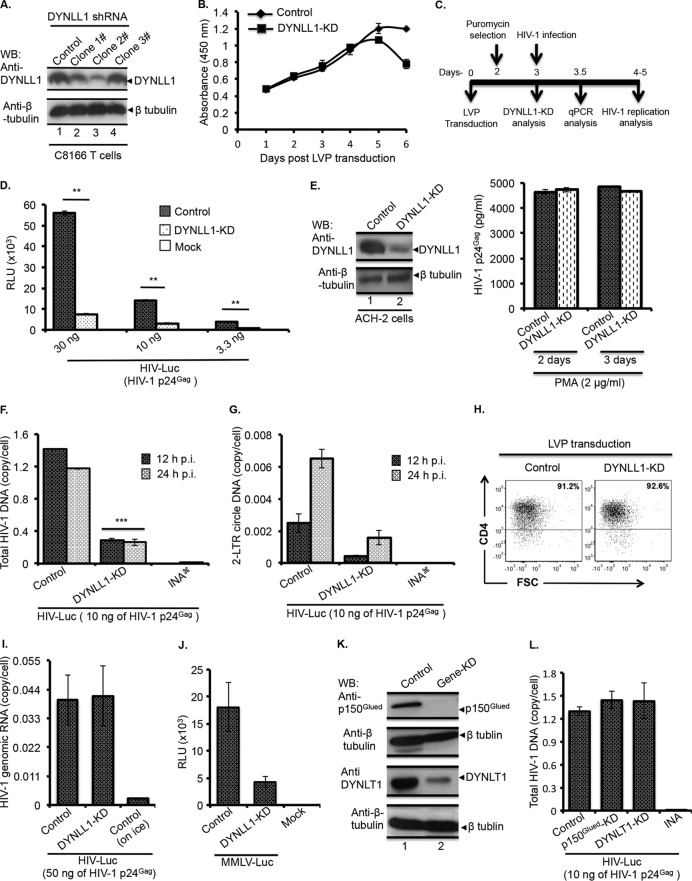
In order to explore the contributions of dynein adapter proteins in the early stage of HIV-1 replication, we examined the requirement of DYNLL1, DYNLT1, or p150Glued in different early steps of HIV-1 replication by employing a gene KD approach. For DYNLL1-KD, C8166T cells were transduced with LVPs expressing different clones of shRNAs for DYNLL1. After 3 days of transduction, the extent of DYNLL1-KD was examined by WB using an anti-DYNLL1 antibody. DYNLL1-KD was most efficient (i.e., approximately 70% KD) in cells that were transduced with LVPs for DYNLL1 shRNA clone 2 (Fig. 1A, top panel, lane 3), and this clone was used in our DYNLL1-KD experiments. First, we examined the viability of the DYNLL1-KD cells using a WST-1 cell proliferation assay. DYNLL1 and control LVP-transduced C8166T cells proliferated to a similar extent until 5 days after transduction; however, the DYNLL1-KD cell proliferation was moderately lower after 5 days of transduction (Fig. 1B). Therefore, we carried out all our experiments within 5 days of transduction, as depicted in Fig. 1C. First, to know the involvement of DYNLL1 in early steps of HIV-1 replication, we infected the control or DYNLL1-KD C8166T cells with single-cycle replication-competent Luc reporter HIV-1 (HIV-Luc), as described in Materials and Methods. At 24 h p.i., we detected 5- to 7-fold-lower Luc activity in DYNLL1-KD cells at all investigated infection doses (Fig. 1D). In order to test whether DYNLL1 is involved in postintegration steps (i.e., the late stage) of HIV-1 replication, DYNLL1 was knocked down in ACH-2 cells. (Fig. 1E, left). ACH-2 is an HIV-1 latent T cell line with one copy of integrated proviral DNA per cell, and HIV-1 production can be induced in ACH-2 cells by PMA treatment. Control or DYNLL1-KD ACH-2 cells were incubated with PMA (2 μg/ml). On day 2 or 3 of PMA treatment, the HIV-1 production by ACH-2 cells was determined by measuring the p24Gag concentration from supernatants using HIV-1 p24Gag ELISA. The results revealed no differences in HIV-1 production between control and DYNLL1-KD ACH-2 cells, potentially ruling out a role for DYNLL1 in the late stage of HIV-1 replication (Fig. 1E). In subsequent investigations, we analyzed the total HIV-1 DNA and 2-LTR circle DNA synthesis in DYNLL1-KD C8166T cells at 12 or 24 h p.i. by qPCR, as described in Materials and Methods. Total HIV-1 DNA and 2-LTR circle DNA are the standard markers for HIV-1 reverse transcription and nuclear import, respectively (48). Interestingly, total HIV-1 DNA synthesis was reduced by 3- to 4-fold in DYNLL1-KD cells compared to control cells (Fig. 1F), and a similar loss of 2-LTR circle DNA synthesis was also evident in DYNLL1-KD cells (Fig. 1G). Conversely, the CD4 receptor expression (Fig. 1H) and level of HIV-1 entry (Fig. 1I) were not altered between control and DYNLL1-KD cells, ruling out defective HIV-1 entry as a possible cause for low levels of reverse transcription. Together, the above results suggest that DYNLL1 is specifically involved in efficient HIV-1 reverse transcription at early times postinfection. In order to determine whether the requirement for DYNLL1 is specific to HIV-1 replication, we carried out an MMLV-Luc replication analysis in DYNLL1-KD cells. The replication of MMLV was also reduced by approximately 4-fold in DYNLL1-KD cells (Fig. 1J). Thus, the above data suggest that DYNLL1 may also be essential for the replication of MMLV. To test the effect of p150Glued and DYNLT1-KD on HIV-1 reverse transcription, we performed KD of p150Glued or DYNLT1 in C8166T cells (Fig. 1K), as described in Materials and Methods. However, unlike DYNLL1, DYNLT1- or p150Glued-KD failed to affect total HIV-1 DNA synthesis (Fig. 1L), ruling out their role in HIV-1 reverse transcription. Therefore, we excluded DYNLT1 or p150Glued from further functional analysis in the present study.
FIG 1.
DYNLL1 is required for HIV-1 reverse transcription. (A) DYNLL1 knockdown. DYNLL1 expression in LVP-transduced cells was examined by WB (top panel) using an anti-DYNLL1 antibody. The expression of β-tubulin was included as loading control (bottom panel). (B) A WST-1 cell proliferation assay was performed at the indicated time points following the LVP transduction to determine the proliferation of control and DYNLL1 LVP-transduced cells. (C) Diagram showing LVP transduction and different time intervals for HIV-1 infection and DNA transfection experiments following LVPs transduction. (D) Control or DYNLL1-KD C8166T cells (0.5 × 106) were infected with different doses of HIV-Luc. At 24 h p.i., viral replication was examined by measuring Luc activity. The statistical significance between the control and DYNLL1-KD cells infections was determined by Student's t test. **, P < 0.01 (n = 3). (E) Left, control or DYNLL1-KD ACH-2 cells were incubated with PMA (2 μg/ml). Right, on day 2 or 3 of PMA treatment, the p24Gag concentration in supernatants was determined using HIV-1 p24Gag ELISA (shown on right side). The data were confirmed in two independent experiments. (F and G) Control or DYNLL1-KD C8166T cells (1.5 × 106) were infected with HIV-Luc. At 12 and 24 h p.i., total HIV-1 DNA (F) and 2-LTR circle DNA (G) were quantified by qPCR. The values shown are the averages of triplicates with the indicated standard deviations. A two-way analysis of variance (ANOVA) was performed to determine the statistical significance between the samples. ***, P < 0.001 (n = 3). The data were confirmed in three independent experiments. (H) Control or DYNLL1-KD C8166T cells were stained with anti-CD4-APC or isotype control antibodies and analyzed by flow cytometry. Representative flow cytometric images show the expression of CD4 receptor on the gated live cells. Student's t test was employed to determine the statistical significance. (I) HIV-1 entry into control or DYNLL1-KD cells, as determined by quantifying HIV-1 genomic RNA. (J) Control or DYNLL1-KD cells (0.5 × 106) were infected with equal amounts of MMLV-Luc. At 24 h p.i., the MMLV replication was examined by measuring Luc activity. (K) p150Glued and DYNLT1 knockdown. p150Glued or DYNLT1 expression in the LVP-transduced cells was examined by WB using corresponding antibodies. The expression of β-tubulin was included as loading control. (L) Control, p150Glued-KD, or DYNLT1-KD C8166T cells (1.5 × 106) were infected with HIV-Luc (at 10 ng of HIV-1 p24Gag). At 12 h p.i., total HIV-1 DNA was quantified by qPCR. RLU, relative light units. INA 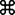 , heat-inactivated virus control.
, heat-inactivated virus control.
HIV-1 integrase, but not capsid, matrix, or reverse transcriptase, interacts with DYNLL1.
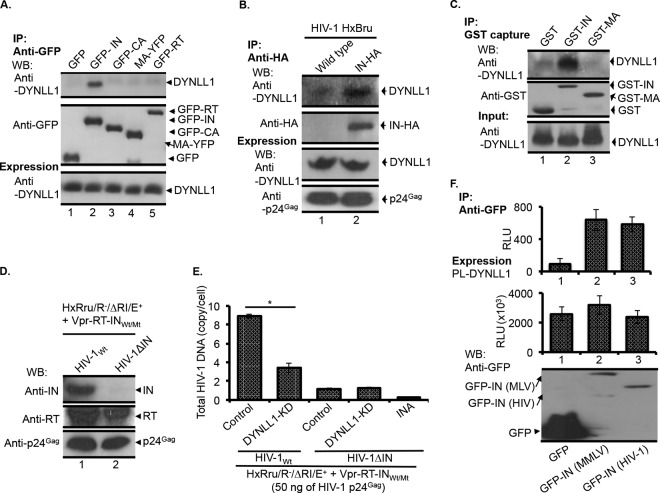
The HIV-1 RTC/PIC contains IN, CA, MA, and RT proteins, which are implicated in reverse transcription (2, 22, 33, 49–54). Thus, we wanted to determine whether DYNLL1 interacts with any or all of these viral proteins. First, we examined the interaction of IN, MA, CA, or RT protein with endogenous DYNLL1 in 293T cells by using a Co-IP assay. In our analysis, the DYNLL1 was specifically coprecipitated with GFP-IN but not with MA-YFP, GFP-CA, or GFP-RT (Fig. 2A, top panel). Importantly, IN was also successfully coprecipitated with endogenous DYNLL1 in a Co-IP performed using HIV-1-infected C8166 CD4+ T cells (Fig. 2B, top panel). To determine if there is a direct interaction between IN and DYNLL1, we performed an in vitro interaction analysis using recombinant GST-IN and DYNLL1 proteins. The results revealed a specific coprecipitation of DYNLL1 with GST-IN but not with GST-MA or GST alone (Fig. 2C, top panel). Together, the above results demonstrate a direct interaction between IN and DYNLL1. Since DYNLL1 interacted specifically with IN, we then wanted to determine whether the requirement for DYNLL1 in HIV-1 reverse transcription is IN dependent. To address this question, we performed an HIV-1ΔIN infection and total HIV-1 DNA analysis in DYNLL1-KD cells. Briefly, equal amounts of HIV-1Wt and HIV-1ΔIN viruses, having equal amounts of viral incorporated RT and p24Gag proteins (Fig. 2D), were used to infect control or DYNLL1-KD C8166T cells. At 12 h p.i., total HIV-1 DNA was quantified by qPCR. Although the total HIV-1 DNA synthesis was significantly reduced in HIV-1Wt-infected DYNLL1-KD cells, the total HIV-1 DNA synthesis in HIV-1ΔIN virus-infected control and DYNLL1-KD cells demonstrated no significant difference (Fig. 2E). Taken together, the above data suggest that the DYNLL1 requirement for HIV-1 reverse transcription is IN dependent. Since DYNLL1 was also required for MMLV replication (Fig. 1J), we examined the MMLV IN interaction with DYNLL1, as described in Materials and Methods. Interestingly, we also observed a positive interaction between GFP-IN (MMLV) and PL-DYNLL1 (Fig. 2F, upper panel), supporting the involvement of DYNLL1 in MMLV replication.
FIG 2.
HIV-1 IN, but not CA, MA, or RT, interacts with DYNLL1. (A) The GFP, GFP-IN, GFP-CA, MA-YFP, or GFP-RT protein was expressed in 293T cells, and coprecipitation of endogenous DYNLL1 (top panel) was detected by Co-IP using an anti-GFP antibody, as described in Materials and Methods. The immunoprecipitation of GFP, GFP-IN, MA-YFP, or GFP-CA (middle panel) was detected by WB using an anti-GFP antibody. DYNLL1 expression in the total cell lysate was detected by WB using an anti-DYNLL1 antibody (bottom panel). The data were confirmed in three independent experiments. (B) IN-DYNLL1 interaction during HIV-1 infection. C8166T cells (15 × 106) were infected with the HxBru or HxBru-IN-HA virus (at 50 ng of virus-associated p24Gag). After 72 h p.i., the cells were subjected to Co-IP using an anti-HA antibody, and coprecipitation of DYNLL1 (top panel) was detected by WB using an anti-DYNLL1 antibody, as described in Materials and Methods. IN-HA immunoprecipitation was determined by probing the WB with an anti-HA antibody (second panel from the top). The expression of DYNLL1 and similar level of HIV-1 infection were determined by probing the total cell lysates for the DYNLL1 and HIV-1 p24Gag proteins by WB (third and fourth panels from the top, respectively) using the corresponding antibodies. The data were confirmed in two independent experiments. (C) IN-DYNLL1 interaction in vitro. GST, GST-IN, or GST-MA protein was incubated with recombinant DYNLL1 protein. The coprecipitation of DYNLL1 (top panel) was detected as described in Materials and Methods. The immunoprecipitation of GST, GST-IN, or GST-MA in elutes was detected by WB using an anti-GST antibody (middle panel). DYNLL1 protein in the supernatants was detected by WB using an anti-DYNLL1 antibody (bottom panel). The data were confirmed in two independent experiments. (D) HIV-1Wt or HIV-1ΔIN virus-incorporated RT (top panel), IN (middle panel), and p24Gag (bottom panel) was detected by WB using the corresponding antibodies, as described in Materials and Methods. (E) Control or DYNLL1-KD C8166T cells (1.5 × 106) were infected with the HIV-1Wt or HIV-1ΔIN virus (at 50 ng of virus-associated p24Gag). At 12 h p.i., total HIV-1 DNA was quantified by qPCR. The values shown are the averages of triplicates with the indicated standard deviations. The data were confirmed in two independent experiments. Statistical significance was determined using Student's t test. *, P < 0.05 (n = 3). (F) DYNLL1 interaction with MMLV IN. GFP, GFP-IN (MMLV), or GFP-IN (HIV-1) was cotransfected with PL-DYNLL1 in 293T cells. After 48 h of transfection, 8/10 of the cells were subjected to Co-IP with the anti-GFP antibody. The coprecipitation of PL-DYNLL1 was detected by measuring the PL activity from the immunoprecipitates (top panel). One-tenth of the cells were subjected to PL-DYNLL1 expression analysis by measuring the PL activity (middle panel). One-tenth of the cells were subjected to GFP, GFP-IN (MMLV), or GFP-IN (HIV-1) expression analysis by WB using the anti-GFP antibody (bottom panel). RLU, relative light units. INA, heat-inactivated virus control.
The conserved motifs in the N- and C-terminal domains of IN are required for DYNLL1 interaction.
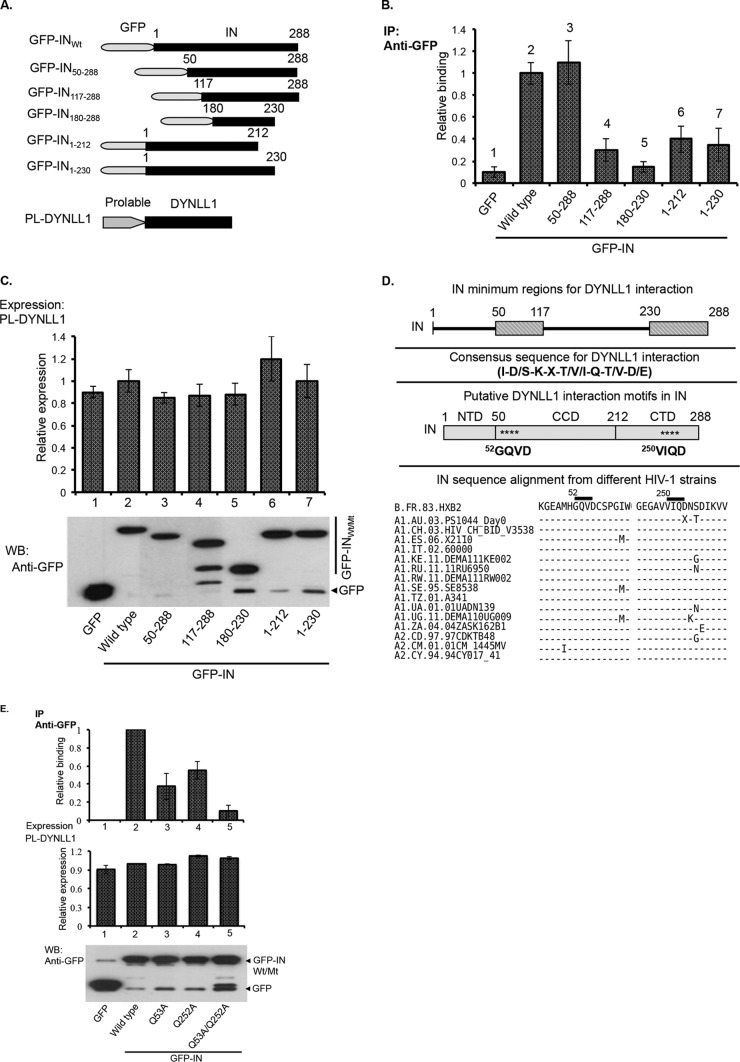
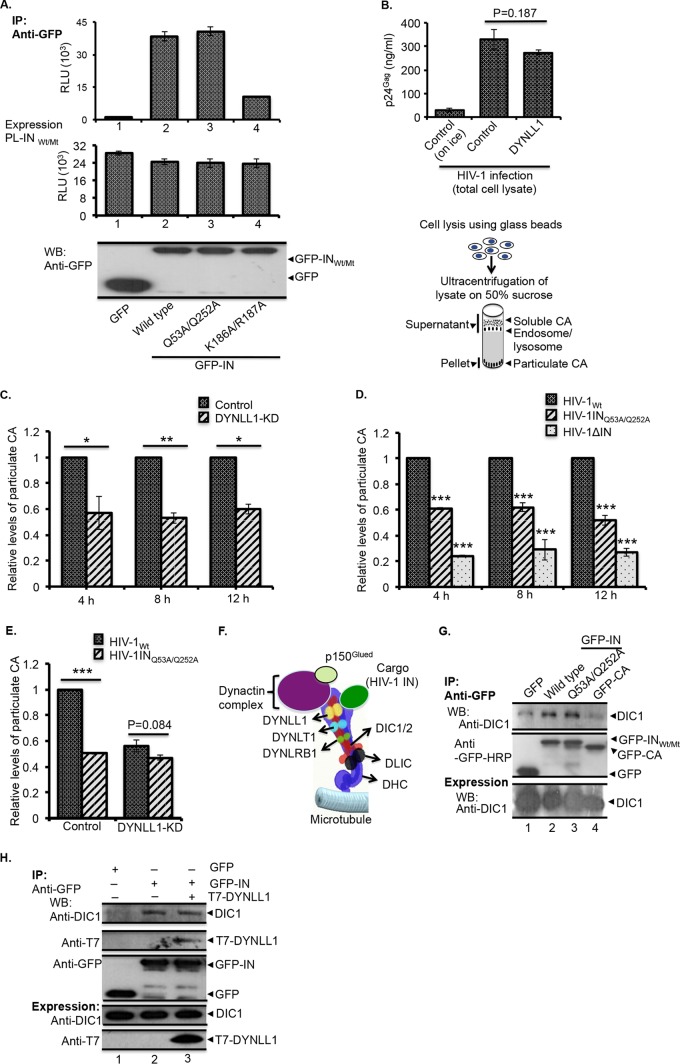
The consensus motifs for DYNLL1 interaction are broadly grouped into the following three classes based on sequence similarity: KXTQTX, XG(I/V)QVD, and noncanonical (55–57) (“X” is any amino acid). In order to elucidate the DYNLL1 interaction motif(s) in IN, we first identified IN minimum regions for DYNLL1 interaction by analyzing DYNLL1 interaction with various IN deletion mutants. Briefly, GFP, GFP-INWt, or different GFP-IN deletion mutant (i.e., amino acids [aa] 50 to 288, 117 to 288, 180 to 230, 1 to 212, and 1 to 230) expression vectors (Fig. 3A) were cotransfected with PL-DYNLL1 expression vector in 293T cells, and the interaction of PL-DYNLL1 with GFP-INWt/Mt was examined by chemiluminescent Co-IP using an anti-GFP antibody, as described in Materials and Methods. IN deletion mutants that lack aa 1 to 117 (i.e., the IN117–288 mutant) or aa 230 to 288 (i.e., the IN1–230 mutant) exhibited attenuated interactions with DYNLL1 (Fig. 3B, bars 4 and 7). Additionally, the IN180–230 deletion mutant, which lacks both aa 1 to 117 and 250 to 288, was severely impaired in its ability to interact with DYNLL1 (Fig. 3B, bar 5). Figure 3C shows the expression levels for PL-DYNLL1 (upper panel) and GFP-INwt or deletion mutants (lower panel). These results led to the conclusion that amino acid residues 50 to 117 and 230 to 288 in IN are essential for interaction with DYNLL1. Interestingly, IN amino acid sequence analysis revealed two motifs (i.e., 52GQVD and 250VIQD) in IN that closely resemble the consensus sequence for DYNLL1 interaction (Fig. 3D) and are also conserved across different HIV-1 strains (representative sequences are shown in Fig. 3D). Because the glutamine (Gln) amino acid in the consensus sequence plays a crucial role in DYNLL1 interactions (56), we introduced Gln (Q)-to-Ala (A) mutations into each of the putative DYNLL1 interaction motifs of IN and examined these INMt protein interactions with PL-DYNLL1 by chemiluminescent Co-IP. Our initial analyses showed attenuated interaction of GFP-INQ53A and GFP-INQ252A with PL-DYNLL1 (data not shown). Subsequently, we introduced a Q53A/Q252A double mutation into GFP-IN (GFP-INQ53A/Q252A) and tested the level of interaction with PL-DYNLL1 by chemiluminescent Co-IP. This GFP-INQ53A/Q252A double mutant was greatly impaired in its ability to interact with PL-DYNLL1 compared to the GFP-INQ53A or GFP-INQ252A single mutant (Fig. 3E, compare bars 3 and 4 with bar 5). Based on the above data, we suggest that the 52GQVD and 250VIQD motifs in IN are involved in the interaction with DYNLL1.
FIG 3.
The consensus motifs in the N- and C-terminal domains of IN are required for DYNLL1 interaction. (A) Schematic diagram of the GFP-IN deletion mutant and PL-DYNLL1 fusion proteins that were used in this study. (B and C) The GFP and GFP-INWt/deletion mutant proteins were coexpressed with PL-DYNLL1 in 293T cells. (B) Coprecipitation of PL-DYNLL1 was detected by Co-IP using an anti-GFP antibody, followed by analysis of PL activity from the immunoprecipitates, as described in Materials and Methods. (C) Upper panel, PL-DYNLL1 expression was detected by measuring PL activity from total cell lysate. The data are presented as fold change in the PL activity with respect to the wild-type control. Lower panel, the expression of GFP-INWt/Mt proteins in the total cell lysate was detected by WB using an anti-GFP antibody. The data shown are the average values from three independent experiments with the indicated standard deviation. (D) Diagram showing the minimum regions in HIV-1IN (i.e., aa 50 to 117 and 230 to 288) for DYNLL1 interaction, the general consensus sequence for DYNLL1 interaction, the putative DYNLL1 interaction motifs in HIV-1IN, and the IN sequence alignment for few representative HIV-1 strains. The full list of HIV-1 sequences, including the accession numbers, can be found in the HIV sequence compendium, 2014. (The IN sequence alignment is adapted from the HIV sequence compendium, 2014.) (E) The GFP-INWt, GFP-INQ53A, GFP-INQ252A, or GFP-INQ53A/Q252A protein was coexpressed with PL-DYNLL1 in 293T cells, and coprecipitation of PL-DYNLL1 (top panels) was detected by Co-IP using an anti-GFP antibody, followed by analysis of PL activity from the immunoprecipitates. PL-DYNLL1 expression was detected by measuring PL activity from total cell lysate (middle panel). The data are presented as fold change in the PL activity with respect to wild-type control. The expression of GFP-INWt/Mt (bottom panel) was detected by WB using an anti-GFP antibody. The data shown are the average values from three independent experiments with the indicated standard deviations.
HIV-1 INQ53A/Q252A mutant virus is defective for reverse transcription.
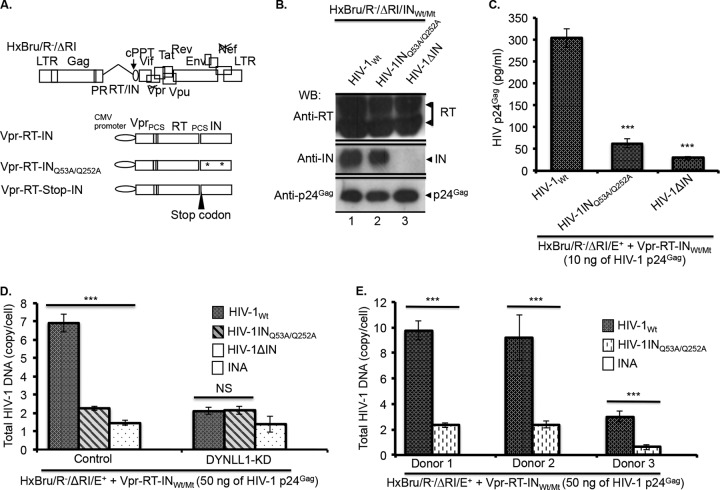
In order to gain more insight into the role of IN-DYNLL1 interaction in HIV-1 reverse transcription, we performed replication or total HIV-1 DNA analysis for DYNLL1 interaction-defective IN mutant HIV-1 (HIV-1INQ53A/Q252A) in the C8166T cell line and primary human CD4+ T lymphocytes. Briefly, HIV-1Wt, HIV-1INQ53A/Q252A, and HIV-1ΔIN viruses were produced by transcomplementation method as described in Materials and Methods (Fig. 4A). Subsequently, similar levels of viral incorporated RT and IN proteins were detected for HIV-1Wt and HIV-1INMt viruses by WB analysis as described in Materials and Methods (Fig. 4B). In order to assess replicative capabilities, 0.5 × 106 C8166T cells were infected with equal amount of the HIV-1Wt, HIV-1INQ53A/Q252A, or HIV-1ΔIN virus. At 72 h p.i., virus replication was detected by examining p24Gag protein production in the supernatants by using an anti-HIV-1p24Gag ELISA. The results demonstrated significantly low levels of virus replication in infections with HIV-1INQ53A/Q252A double mutant or HIV-1ΔIN virus (i.e., approximately 5- to 7-fold reduced) (Fig. 4C). For HIV-1 reverse transcription analysis, we quantified the total HIV-1 DNA levels from HIV-1Wt-, HIV-1INQ53A/Q252A-, or HIV-1ΔIN-infected control and DYNLL1-KD C8166T cells by qPCR. The results illustrated significantly low levels of total HIV-1 DNA in HIV-1INQ53A/Q252A-infected control cells (Fig. 4D). In contrast, there was no significant difference in total HIV-1 DNA levels between HIV-1Wt- and HIV-1INQ53A/Q252A-infected DYNLL1-KD cells (Fig. 4D). Furthermore, we analyzed the total HIV-1 DNA synthesis in activated primary human CD4+ T lymphocytes derived from three independent donors. In all the donors, we detected an approximately 4- to 5-fold-lower total HIV-1 DNA synthesis in HIV-1INQ53A/Q252A mutant virus infection than in HIV-1Wt infection (Fig. 4E). Since IN-RT interaction has been implicated in HIV-1 reverse transcription (50, 54, 58, 59), we analyzed INQ53A/Q252A mutant protein interaction with RT by cell-based Co-IP. However, we found no change in RT interaction with the INWt or the INQ53A/Q252A mutant protein (data not shown). All of these data taken together suggest that DYNLL1 interaction-defective INQ53A/Q252A mutant HIV-1 (HIV-1INQ53A/Q252A) is impaired for reverse transcription.
FIG 4.
HIV-1 INQ53A/Q252A mutant virus is defective for reverse transcription. (A) Diagrammatic representation of the HxBru/R−/ΔRI/E+ provirus and the Vpr-RT-INWt,/Mt fusion protein expression vectors. (Adapted from reference 42.) (B) HIV-1Wt, HIV-1INQ53A/Q252A, or HIV-1ΔIN virus-incorporated RT (top panel), IN (middle panel), and p24Gag (bottom panel), detected as described in Materials and Methods. (C) C8166T cells (0.5 × 106) were infected with the HIV-1Wt, HIV-1INQ53A/Q252A, or HIV-1ΔIN virus (at 10 ng of virus-associated p24Gag), and HIV-1 replication was analyzed by HIV-1 p24Gag ELISA, as described in Materials and Methods. The values shown are the averages of triplicates with the indicated standard deviations. The statistical significance between the infections was determined by using a one-way ANOVA. ***, P < 0.001 (n = 3). The data were confirmed in two independent experiments. (D) Control or DYNLL1-KD C8166T cells (1.5 × 106) were infected with the HIV-1Wt, HIV-1INQ53A/Q252A, and HIV-1ΔIN viruses (at 50 ng of virus-associated p24Gag). At 12 h p.i., the total HIV-1 DNA was quantified by qPCR. The values shown are the averages of triplicates with the indicated standard deviations. One-way ANOVA was performed to determine the statistical significance. The data were confirmed in two independent experiments. ***, P < 0.001 (n = 3); #, nonsignificant (NS). (E) Primary human CD4+ T cells were purified from PBMCs of three healthy adult human volunteers and stimulated with PHA (3 μg/ml) as described in Materials and Methods. CD4+ T cells (1.5 × 106) following stimulation were infected with HIV-1Wt and HIV-1INQ53A/Q252A (at 50 ng of virus-associated p24Gag), and total HIV-1 DNA was quantified at 24 h p.i. as described in Materials and Methods. The Student t test was used to determine the statistical significance. ***, P < 0.001 (n = 3). INA, heat-inactivated virus control.
HIV-1 exhibits premature uncoating in DYNLL1-KD cells or cells infected with HIV-1INQ53A/Q252A mutant virus.
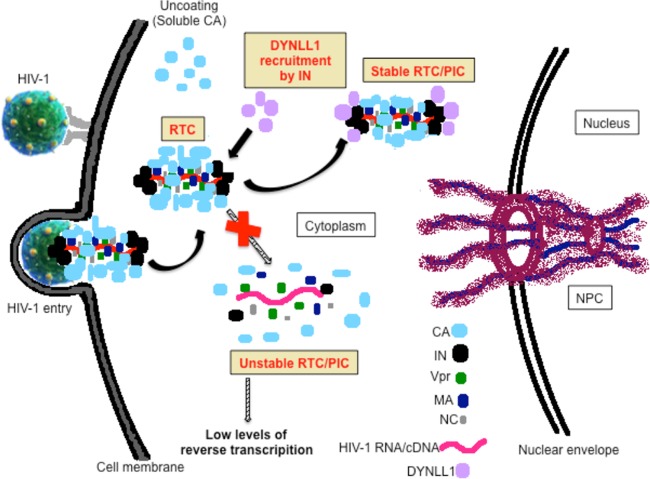
The mechanism by which the interaction between IN and DYNLL1 contributes to HIV-1 reverse transcription remains unknown. Accumulated evidence suggests that proteins undergo conformational change and exhibit a higher α-helical content after binding to DYNLL1 (60–62). Therefore, it is possible that DYNLL1 binding may contribute to a stable IN multimer formation and promote the IN interaction with other cellular and/or viral proteins during infection. Indeed, accumulated evidence suggests that IN functions as a multimer (63–66). However, our data failed to suggest a role for DYNLL1 in IN multimerization (Fig. 5A). Moreover, the coexpression of IN and RT with and without overexpression of DYNLL1 in 293T cells did not alter the interaction between IN and RT (data not shown), suggesting no potential role for DYNLL1 in IN-RT interaction. After ruling out a role for DYNLL1 in IN multimerization and stable IN-RT assembly, we explored the role of DYNLL1 in HIV-1 uncoating. It is now quite evident that both accelerated and delayed uncoating can lead to lower levels of HIV-1 reverse transcription (reviewed in reference 22). We employed the “fate-of-capsid assay” to study HIV-1 uncoating. First, the feasibility of the fate-of-capsid assay for HIV-1 uncoating analysis under our own laboratory conditions was confirmed. In our analysis, the transiently expressed GFP-CA protein, which represents the soluble CA (i.e., CA released from the RTC/PIC), was found exclusively in the supernatant fraction, whereas HIV-1 RTC/PIC-associated IN or CA (i.e., particulate CA) protein was segregated in the pellet fraction (data not shown). Subsequently, 3 × 107 control or DYNLL1-KD C8166T cells were infected with HIV-1 as described in Materials and Methods. Prior to the fate-of-capsid assay, similar levels of HIV-1 infection were detected in control and DYNLL1-KD cells by analyzing p24Gag contents in total cell lysates (Fig. 5B, top). At 4, 8, and 12 h p.i., approximately 107 cells were harvested and subjected to the fate-of-capsid assay as described in Materials and Methods (Fig. 5B, bottom). Interestingly, for all time points of harvest, we detected lower levels of particulate CA in DYNLL1-KD cell infections (i.e., approximately 0.55- ± 0.02-fold) than in empty vector control infections (Fig. 5C). These data for the first time suggested that the lack of DYNLL1 expression would lead to the accelerated (i.e., premature) uncoating of HIV-1. Next, we examined the particulate CA levels in infections with HIV-1Wt or HIV-1INQ53A/Q252A mutant virus. HIV-1ΔIN virus was included as a control. For all time points of harvest, we detected lower levels of particulate CA in infections with HIV-1INQ53A/Q252A (i.e., approximately 0.58- ± 0.05-fold) or HIV-1ΔIN than in HIV-1Wt infections (Fig. 5D). Also, we examined the particulate CA levels following the infection of DYNLL1-KD or control cells with HIV-1Wt and HIV-1INQ53A/Q252A mutant viruses. Even though HIV-1INQ53A/Q252A exhibited lower levels of particulate CA in control cell infections, no significant differences in particulate CA was detected between HIV-1Wt and HIV-1INQ53A/Q252A in DYNLL1-KD cells (P = 0.08, n = 3) (Fig. 5E). A previous study linked the accelerated uncoating of HIV-1ΔIN to a lack of CypA incorporation into the virus (49). However, we did not observe any differences in CypA incorporation between the HIV-1Wt and HIV-1INQ53A/Q252A viruses (data not shown). Together, the above findings support the conclusion that the DYNLL1-IN interaction is required for the proper uncoating of HIV-1. Since some earlier studies implicated DYNLL1 in cargo recruitment to the dynein complex during retrograde transport (17, 18, 67), we were interested in examining the potential role of interaction between IN and DYNLL1 in HIV-1 association with the dynein complex. Since dynein adapter proteins recruit cargo to the dynein complex through the interaction with DIC1/2 (Fig. 5F) (reviewed in reference 68), we examined the interaction of the GFP-INQ53A/Q252A mutant and GFP-INWt proteins with endogenous DIC1 in 293T cells by Co-IP. Interestingly, both GFP-INWt and GFP-INQ53A/Q252A exhibited similar levels of interaction with DIC1 (Fig. 5G, top panel). Moreover, we detected no change in interaction between GFP-IN and DIC1 with or without T7-DYNLL1 overexpression (Fig. 5H, top panel). The above data led us to believe that IN-DYNLL1 interaction is not likely to have a role in HIV-1 association with the dynein complex. Together, these results suggest that IN-DYNLL1 interaction is involved in the proper uncoating of HIV-1. Here, we speculate that the accelerated or premature HIV-1 uncoating in the absence of interaction between IN and DYNLL1 would result in the assembly of unstable RTCs, which would at least partially contribute to the low levels of HIV-1 reverse transcription (Fig. 6). Further investigations are required to precisely understand the role of IN-DYNLL1 interaction in HIV-1 reverse transcription and uncoating.
FIG 5.
HIV-1 exhibits premature uncoating in DYNLL1-KD cells or after infection with HIV-1INQ53A/Q252A mutant virus. (A) 293T cells were cotransfected with various GFP-INWt/Mt or PL-INWt/Mt fusion protein expression vectors as follows: GFP with PL-INWt (bar 1), GFP-INWt with PL-INWt (bar 2), GFP-INQ53A/Q252A with PL-INQ53A/Q252A (bar 3), or GFP-INK186A/R187A with PL-INK186A/R187A (bar 4). After 48 h of transfection, the cells were lysed and the lysates were immunoprecipitated using an anti-GFP antibody. The coprecipitation of PL-INWt/Mt in the immunoprecipitates was examined by measuring the PL activity (top panel). The expression of PL-INWt/Mt was detected by measuring the PL activity (middle panel), and the expression of the GFP-INWt/Mt protein was detected by probing the WB with the anti-GFP antibody (bottom panel). RLU, relative light units. (B) Top, representative data showing the similar levels of HIV-1 infection in control and DYNLL1-KD cells during the fate-of-capsid assay as determined by HIV-1Gag levels in total cell lysates. The statistical significance was analyzed using a Student t test. *, P = 0.187 (n = 3). Bottom, diagram showing the different steps of the fate-of-capsid assay, as described in Materials and Methods. (C) Control or DYNLL1-KD C8166T cells were infected with the HIV-1Wt virus (at 25 ng of virus-associated p24Gag/106 cells). At 4, 8, and 12 h postinfection, the HIV-1 uncoating was analyzed by the fate-of-capsid assay, as described in Materials and Methods. The data were interpreted as the fold difference in particulate CA concentrations between the control and DYNLL1-KD cell infections. The statistical significance of the differences observed between the control and DYNLL1-KD cell infections was determined using a Student t test, *, P < 0.05; **, P < 0.01 (n = 3). (D) C8166T cells were infected with equal amounts of the HIV-1Wt, HIV-1INQ53A/Q252A, or HIV-1ΔIN virus (at 25 ng of virus associated p24Gag/1 × 106 cells). At 4, 8, and 12 h p.i., the HIV-1 uncoating was analyzed by the fate-of-capsid assay. The data were interpreted as the fold differences in particulate CA concentrations between wild-type and mutant virus infections. A two-way ANOVA analysis was performed to determine the statistical significance between the samples. ***, P < 0.001 (n = 3). (E) Control and DYNLL1-KD C8166T cells were infected with the HIV-1Wt or HIV-1INQ53A/Q252A virus (at 25 ng of virus associated p24Gag/1 × 106 cells). At 4 h p.i., HIV-1 uncoating was analyzed by fate-of-capsid assay, and the data were interpreted as fold differences in particulate CA concentrations between wild-type and mutant virus-infected DYNLL1-KD or control cells. The Student t test was used to determine the statistical significance. ***, P < 0.001 (n = 3). (F) Different components of the dynein complex and the recruitment of cargo to the dynein complex through simultaneous interactions of DYNLL1 with DIC1/2 and cargo (HIV-1 IN). DYNLRB1, dynein light chain roadblock-type 1; DLIC, dynein light intermediate chain; DHC, dynein heavy chain. (G) GFP, GFP-INWt, GFP-INQ53A/Q252A, and GFP-CA fusion proteins were expressed in 293T cells, and the coprecipitation of endogenous DIC1 (top panel) was detected by Co-IP using an anti-GFP antibody. The immunoprecipitation of GFP, GFP-INWt, GFP-INQ53A/Q252A, or GFP-CA was detected by WB using an anti-GFP antibody (middle panel). DIC1 expression in the total cell lysate was detected by WB using an anti-DIC1 antibody (bottom panel). (H) GFP or GFP-IN was coexpressed with or without T7-DYNLL1 in 293T cells. The coprecipitation of DIC1 (top panel) or T7-DYNLL1 (second panel from the top) was detected by Co-IP using an anti-GFP antibody. The immunoprecipitation of GFP or GFP-IN was detected by WB (third panel from the top) using an anti-GFP antibody. The expression of T7-DYNLL1 and DIC1 in total cell lysate was detected by WB using the corresponding antibodies (bottom two panels).
FIG 6.
Model depicting the potential mechanism by which the IN-DYNLL1 interaction contributes to efficient reverse transcription and proper uncoating of HIV-1. HIV-1 entry is soon followed by the reorganization of the RTC, uncoating, and reverse transcription. DYNLL1 is recruited to RTC at early times p.i. by interacting with IN and contributes to the RTC reorganization and/or formation of a stable RTC. The process contributes to the efficient reverse transcription and proper uncoating of HIV-1. In the absence of IN-DYNLL1 interaction, HIV-1 exhibits faster (premature) uncoating, which would make the RTC become unstable and contribute to the low levels of HIV-1 reverse transcription. NC, nucleocapsid; NPC, nuclear pore complex.
DISCUSSION
In the present study, we explored the requirement for DYNLL1, DYNLT1, and p150Glued in early steps of HIV-1 replication by a gene KD approach. Our analysis demonstrated the requirement for DYNLL1 in HIV-1 reverse transcription. Subsequently, we elucidated the DYNLL1 interaction with HIV-1 IN and determined that the 52GQVD and 250VIQD motifs in IN were essential for DYNLL1 interaction. Through continued analyses, we have shown the requirement for IN and DYNLL1 in efficient reverse transcription and proper uncoating of HIV-1.
DYNLL1-KD resulted in low levels of luciferase reporter HIV-1 replication (Fig. 1D). Further, we have also demonstrated that the postintegration steps of HIV-1 replication were not affected in DYNLL1-KD cells (Fig. 1E). These data illustrated that DYNLL1 is essential for steps in early-stage HIV-1 replication. In continued investigations, we detected low levels of total HIV-1 DNA synthesis in DYNLL1-KD cells (Fig. 1F). Meanwhile, DYNLL1-KD neither caused loss of CD4 receptor expression (Fig. 1H) nor affected HIV-1 entry into the cell (Fig. 1I). Together, the above data helped us to suggest that DYNLL1 is involved in the reverse transcription step of HIV-1 replication. DYNLL1 was also found to be essential for MMLV replication (Fig. 1J), and MMLV IN was successfully coprecipitated with DYNLL1 in our Co-IP assay (Fig. 2F). These data suggest that DYNLL1 is also involved in MMLV replication and is possibly also engaged in the replication of retroviruses other than HIV-1 or MMLV. Therefore, future investigations exploring the role of DYNLL1 in the replication of other retroviruses are strongly encouraged.
The HIV-1 RTC contains viral proteins such as IN, MA, CA, and RT (69, 70), which are implicated in reverse transcription (reviewed in reference 2). In our Co-IP analyses (Fig. 2A, B, and C), HIV-1 IN but not CA, MA, or RT interacted with DYNLL1, indicating a possible role for IN-DYNLL1 interaction in HIV-1 reverse transcription. In conjunction with our interaction data, a recent study elucidated the HIV-1 IN interaction with yeast dynein light chain (Dyn2p) in Saccharomyces cerevisiae by yeast two-hybrid analysis (71). As Dyn2p is an ortholog of mammalian DYNLL1, that report supports our data on IN interaction with DYNLL1. It has been shown that proteins bind DYNLL1 with the help of a consensus interaction motif(s) [i.e., KXTQTX, XG(I/V)QVD, or noncanonical] (25, 55–57). During our amino acid sequence analysis, we identified two putative DYNLL1 interaction motifs in IN (i.e., 52GQVD and 250VIQD). Interestingly, these motifs were located within the IN minimum regions (aa 50 to 117 and 230 to 288 in IN) for DYNLL1 interaction. It is worth noting that an earlier study, through analysis of IN crystal structure, indicated that the regions of IN that comprise the 52GQVD or 250VIQD motif are solvent exposed (72), indicating that these motifs are able to mediate interaction with partnering proteins. In support of our assumption, Q-to-A mutations in the 52GQVD and 250VIQD motifs of IN (i.e., INQ53A/Q252A) resulted in severely attenuated interaction with DYNLL1 (Fig. 3E). Therefore, we strongly suggest that the 52GQVD and 250VIQD motifs in IN mediate DYNLL1 interaction. This is a first report demonstrating the consensus DYNLL1 interaction motifs in IN and IN interaction with DYNLL.
The DYNLL1 interaction-defective IN mutant HIV-1 (HIV-1INQ53A/Q252A) was impaired in its ability for efficient reverse transcription (Fig. 4D and E). It is known that mutations in IN can cause pleiotropic effects on HIV-1 replication (73). Our data in Fig. 4D, which show no reverse transcription differences between HIV-1Wt and the HIV-1INQ53A/Q252A mutant in DYNLL1-KD cells, would rule out potential pleotropic effects of INQ53A/Q252A mutations on HIV-1 reverse transcription. Meanwhile, these data also highlight the specificity of IN-DYNLL1 interaction for HIV-1 reverse transcription. In addition, HIV-1INQ53A/Q252A was also defective for reverse transcription in primary human CD4+ T cells (Fig. 4E). Even though we noticed donor-dependent variation in the level of HIV-1 infection in donor 3, the reverse transcription differences between different donors were consistent. These data underscore the clinical relevance of IN-DYNLL1 interaction for efficient HIV-1 reverse transcription.
In earlier studies, IN-RT interaction has been implicated in HIV-1 reverse transcription (50, 59). Therefore, we suspected that DYNLL1 binding of IN would favor IN multimerization, which in turn would facilitate IN/RT complex formation. However, in our analysis, DYNLL1 interaction was not essential for IN dimerization (Fig. 5A). Since DYNLL1 failed to interact with RT (Fig. 2A), it is also unlikely that DYNLL1 has a role in de novo HIV-1 reverse transcription. Interestingly, our data suggested that the lack of IN-DYNLL1 interaction leads to a moderate increase in the rate of HIV-1 uncoating (Fig. 5C, D, and E). It is of interest to note that a recent study implicated IN in proper uncoating of HIV-1 based on the finding that HIV-1ΔIN shows accelerated uncoating and lower levels of reverse transcription (49). Although the authors of that paper suggested the lack of CypA incorporation into HIV-1ΔIN as a cause for accelerated uncoating (49), this does not rule out the possibility of alternative mechanisms. Based on our uncoating data, we suggest that DYNLL1 would be recruited to the HIV-1 RTC by interaction with IN and regulate the timely uncoating. We also suggest that this mechanism may at least partially contribute to the lower levels of reverse transcription seen in the absence of IN-DYNLL1 interaction, as HIV-1 reverse transcription has been suggested to be functionally linked with the proper uncoating of the virus (reviewed in reference 22).
In recent studies, the inhibition of dynein function by KD of dynein heavy chain (DYNC1H1) or the disruption of the intact MT filaments in cells by nocodazole treatment delayed the uncoating process during HIV-1 infection (20, 21). Thus, the authors of these papers suggested that the dynein complex and intact MT filament network facilitate HIV-1 uncoating (20, 21). In contrast, neither the inhibition of dynein function nor the disruption of the MT network affected HIV-1 reverse transcription in cells (20). In contrast with these observations, our study showed that the DYNLL1-KD or lack of IN-DYNLL1 interaction resulted in a significant loss of reverse transcription and an increase in rate of HIV-1 uncoating. In addition, our data in Fig. 5G and H also indicated that IN-DYNLL1 interaction probably does not have a role in HIV-1 association with the dynein complex. In support of our observation, a previous study, based on the structural and thermodynamic analysis of DYNLL1, also suggested that DYNLL1 is not able mediate cargo recruitment to the dynein complex (74). Thus, it is possible that DYNLL1 would be contributing to HIV-1 reverse transcription and uncoating from outside the dynein complex. Therefore, we strongly believe that the role of DYNLL1 in HIV-1 reverse transcription or uncoating is distinct from functional dynein complex or intact MT filaments. However, further investigations are certainly needed to precisely define the role of DYNLL1 and the dynein complex or MT network in HIV-1 uncoating.
The findings of this study lead to one important question regarding how IN-DYNLL1 interaction contributes to the proper uncoating of HIV-1 and reverse transcription. DYNLL1 has been shown to interact with a number of different cellular proteins outside the dynein complex and to act as a facilitator for protein complex formation (25). Therefore, cellular proteins may be recruited to the RTC via DYNLL1 interaction, leading to HIV-1 RTC reorganization and/or stabilization, which would in turn contribute to proper uncoating and/or efficient reverse transcription of HIV-1. Conversely, lack of IN-DYNLL1 interaction leads to faster uncoating and formation of unstable RTC, resulting in low levels of HIV-1 reverse transcription (Fig. 6). Alternatively, based on the recent suggestions that CypA-CA interaction in target human cells possibly protects HIV-1 against the rapid uncoating induced by the humanTRIM5α (75, 76), DYNLL1 would contribute to the proper uncoating and reverse transcription of HIV-1 by promoting CypA association with CA or stabilizing CypA-CA interaction in RTCs. Future investigation into both of the above hypotheses would enable us to understand the precise role of IN-DYNLL1 interaction in HIV-1 reverse transcription and uncoating. In conclusion, our study provides novel evidence for the utilization of DYNLL1 by HIV-1 via interaction with its IN protein to promote efficient reverse transcription and proper uncoating. This work, in addition to evaluating the involvement of HIV-1 IN in proper uncoating, provides evidence for the existence of a possible alternative mechanism by which IN facilitates the proper uncoating and efficient reverse transcription of HIV-1. The findings of this study may have implications for development of therapeutic strategies against HIV-1 infection.
ACKNOWLEDGMENTS
We thank Gary Kobinger for valuable discussions and Alexander Bello for technical assistance. We also greatly appreciate the editorial review of Carmen Lopez and Lindsay Aboud. We thank Vicente Planelles for providing the pLET-Lai vector.
Kallesh Danappa Jayappa is a recipient of Manitoba Health Research Council/Manitoba Institute of Child Health (MHRC/MICH) and CIHR International Infectious Disease & Global Health Training Program scholarships. Xiaojian Yao holds a Manitoba Research Chair Award from the Manitoba Health Research Council. This work was supported by the Canadian Foundation for AIDS Research (CANFAR grant 023-013), the National Sciences and Engineering Research Council of (NSERC)Canada (grant 418543-2013), the Canadian Institute of Health Research (CIHR) HIV/AIDS Bridge Funding Biomed/Clini stream (HBF-131553), and a CIHR/MHRC RPP grant (RPA-132176) to Xiaojian Yao.
REFERENCES
- 1.Nisole S, Saib A. 2004. Early steps of retrovirus replicative cycle. Retrovirology 1:9. doi: 10.1186/1742-4690-1-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Jayappa KD, Ao Z, Yao X. 2012. The HIV-1 passage from cytoplasm to nucleus: the process involving a complex exchange between the components of HIV-1 and cellular machinery to access nucleus and successful integration. Int J Biochem Mol Biol 3:70–85. [PMC free article] [PubMed] [Google Scholar]
- 3.Bieniasz PD. 2012. An overview of intracellular interactions between immunodeficiency viruses and their hosts. AIDS 26:1243–1254. doi: 10.1097/QAD.0b013e328353bd04. [DOI] [PubMed] [Google Scholar]
- 4.Friedrich BM, Dziuba N, Li G, Endsley MA, Murray JL, Ferguson MR. 2011. Host factors mediating HIV-1 replication. Virus Res 161:101–114. doi: 10.1016/j.virusres.2011.08.001. [DOI] [PubMed] [Google Scholar]
- 5.Kok KH, Lei T, Jin DY. 2009. siRNA and shRNA screens advance key understanding of host factors required for HIV-1 replication. Retrovirology 6:78. doi: 10.1186/1742-4690-6-78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Van Maele B, Busschots K, Vandekerckhove L, Christ F, Debyser Z. 2006. Cellular co-factors of HIV-1 integration. Trends Biochemi Sci 31:98–105. doi: 10.1016/j.tibs.2005.12.002. [DOI] [PubMed] [Google Scholar]
- 7.Hamamoto S, Nishitsuji H, Amagasa T, Kannagi M, Masuda T. 2006. Identification of a novel human immunodeficiency virus type 1 integrase interactor, Gemin2, that facilitates efficient viral cDNA synthesis in vivo. J Virol 80:5670–5677. doi: 10.1128/JVI.02471-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nishitsuji H, Hayashi T, Takahashi T, Miyano M, Kannagi M, Masuda T. 2009. Augmentation of reverse transcription by integrase through an interaction with host factor, SIP1/Gemin2 Is critical for HIV-1 infection. PLoS One 4:e7825. doi: 10.1371/journal.pone.0007825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mascarenhas AP, Musier-Forsyth K. 2009. The capsid protein of human immunodeficiency virus: interactions of HIV-1 capsid with host protein factors. FEBS J 276:6118–6127. doi: 10.1111/j.1742-4658.2009.07315.x. [DOI] [PubMed] [Google Scholar]
- 10.Misumi S, Inoue M, Dochi T, Kishimoto N, Hasegawa N, Takamune N, Shoji S. 2010. Uncoating of human immunodeficiency virus type 1 requires prolyl isomerase Pin1. J Biol Chem 285:25185–25195. doi: 10.1074/jbc.M110.114256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Aiken C. 2006. Viral and cellular factors that regulate HIV-1 uncoating. Curr Opin HIV AIDS 1:194–199. doi: 10.1097/01.COH.0000221591.11294.c1. [DOI] [PubMed] [Google Scholar]
- 12.Fricke T, White TE, Schulte B, de Souza Aranha Vieira DA, Dharan A, Campbell EM, Brandariz-Nunez A, Diaz-Griffero F. 2014. MxB binds to the HIV-1 core and prevents the uncoating process of HIV-1. Retrovirology 11:68. doi: 10.1186/PREACCEPT-6453674081373986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Guth CA, Sodroski J. 2014. Contribution of PDZD8 to stabilization of the human immunodeficiency virus type 1 capsid. J Virol 88:4612–4623. doi: 10.1128/JVI.02945-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Warren K, Warrilow D, Meredith L, Harrich D. 2009. Reverse transcriptase and cellular factors: regulators of HIV-1 reverse transcription. Viruses 1:873–894. doi: 10.3390/v1030873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dohner K, Wolfstein A, Prank U, Echeverri C, Dujardin D, Vallee R, Sodeik B. 2002. Function of dynein and dynactin in herpes simplex virus capsid transport. Mol Biol Cell 13:2795–2809. doi: 10.1091/mbc.01-07-0348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lo KW, Kogoy JM, Pfister KK. 2007. The DYNLT3 light chain directly links cytoplasmic dynein to a spindle checkpoint protein, Bub3. J Biol Chem 282:11205–11212. doi: 10.1074/jbc.M611279200. [DOI] [PubMed] [Google Scholar]
- 17.Navarro C, Puthalakath H, Adams JM, Strasser A, Lehmann R. 2004. Egalitarian binds dynein light chain to establish oocyte polarity and maintain oocyte fate. Nat Cell Biol 6:427–435. doi: 10.1038/ncb1122. [DOI] [PubMed] [Google Scholar]
- 18.Su Y, Qiao W, Guo T, Tan J, Li Z, Chen Y, Li X, Li Y, Zhou J, Chen Q. 2010. Microtubule-dependent retrograde transport of bovine immunodeficiency virus. Cell Microbiol 12:1098–1107. doi: 10.1111/j.1462-5822.2010.01453.x. [DOI] [PubMed] [Google Scholar]
- 19.Mallik R, Gross SP. 2004. Molecular motors: strategies to get along. Curr Biol 14:R971–R982. doi: 10.1016/j.cub.2004.10.046. [DOI] [PubMed] [Google Scholar]
- 20.Lukic Z, Dharan A, Fricke T, Diaz-Griffero F, Campbell EM. 2014. HIV-1 uncoating is facilitated by dynein and kinesin 1. J Virol 88:13613–13625. doi: 10.1128/JVI.02219-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pawlica P, Berthoux L. 2014. Cytoplasmic dynein promotes HIV-1 uncoating. Viruses 6:4195–4211. doi: 10.3390/v6114195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Arhel N. 2010. Revisiting HIV-1 uncoating. Retrovirology 7:96. doi: 10.1186/1742-4690-7-96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pawlica P, Le Sage V, Poccardi N, Tremblay MJ, Mouland AJ, Berthoux L. 2014. Functional evidence for the involvement of microtubules and dynein motor complexes in TRIM5alpha-mediated restriction of retroviruses. J Virol 88:5661–5676. doi: 10.1128/JVI.03717-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Merino-Gracia J, Garcia-Mayoral MF, Rodriguez-Crespo I. 2011. The association of viral proteins with host cell dynein components during virus infection. FEBS J 278:2997–3011. doi: 10.1111/j.1742-4658.2011.08252.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rapali P, Szenes A, Radnai L, Bakos A, Pal G, Nyitray L. 2011. DYNLL/LC8: a light chain subunit of the dynein motor complex and beyond. FEBS J 278:2980–2996. doi: 10.1111/j.1742-4658.2011.08254.x. [DOI] [PubMed] [Google Scholar]
- 26.Poisson N, Real E, Gaudin Y, Vaney MC, King S, Jacob Y, Tordo N, Blondel D. 2001. Molecular basis for the interaction between rabies virus phosphoprotein P and the dynein light chain LC8: dissociation of dynein-binding properties and transcriptional functionality of P. J Gen Virol 82:2691–2696. [DOI] [PubMed] [Google Scholar]
- 27.Raux H, Flamand A, Blondel D. 2000. Interaction of the rabies virus P protein with the LC8 dynein light chain. J Virol 74:10212–10216. doi: 10.1128/JVI.74.21.10212-10216.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tan GS, Preuss MA, Williams JC, Schnell MJ. 2007. The dynein light chain 8 binding motif of rabies virus phosphoprotein promotes efficient viral transcription. Proc Natl Acad Sci U S A 104:7229–7234. doi: 10.1073/pnas.0701397104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Schneider MA, Spoden GA, Florin L, Lambert C. 2011. Identification of the dynein light chains required for human papillomavirus infection. Cell Microbiol 13:32–46. doi: 10.1111/j.1462-5822.2010.01515.x. [DOI] [PubMed] [Google Scholar]
- 30.Schroer TA. 2004. Dynactin. Annu Rev Cell Dev Biol 20:759–779. doi: 10.1146/annurev.cellbio.20.012103.094623. [DOI] [PubMed] [Google Scholar]
- 31.Alonso C, Miskin J, Hernaez B, Fernandez-Zapatero P, Soto L, Canto C, Rodriguez-Crespo I, Dixon L, Escribano JM. 2001. African swine fever virus protein p54 interacts with the microtubular motor complex through direct binding to light-chain dynein. J Virol 75:9819–9827. doi: 10.1128/JVI.75.20.9819-9827.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Engelke MF, Burckhardt CJ, Morf MK, Greber UF. 2011. The dynactin complex enhances the speed of microtubule-dependent motions of adenovirus both towards and away from the nucleus. Viruses 3:233–253. doi: 10.3390/v3030233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ao Z, Fowke KR, Cohen EA, Yao X. 2005. Contribution of the C-terminal tri-lysine regions of human immunodeficiency virus type 1 integrase for efficient reverse transcription and viral DNA nuclear import. Retrovirology 2:62. doi: 10.1186/1742-4690-2-62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ao Z, Huang G, Yao H, Xu Z, Labine M, Cochrane AW, Yao X. 2007. Interaction of human immunodeficiency virus type 1 integrase with cellular nuclear import receptor importin 7 and its impact on viral replication. J Biol Chem 282:13456–13467. doi: 10.1074/jbc.M610546200. [DOI] [PubMed] [Google Scholar]
- 35.Wang X, Ao Z, Chen L, Kobinger G, Peng J, Yao X. 2012. The cellular antiviral protein APOBEC3G interacts with HIV-1 reverse transcriptase and inhibits its function during viral replication. J Virol 86:3777–3786. doi: 10.1128/JVI.06594-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ao Z, Danappa Jayappa K, Wang B, Zheng Y, Kung S, Rassart E, Depping R, Kohler M, Cohen EA, Yao X. 2010. Importin alpha3 interacts with HIV-1 integrase and contributes to HIV-1 nuclear import and replication. J Virol 84:8650–8663. doi: 10.1128/JVI.00508-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ao Z, Jayappa KD, Wang B, Zheng Y, Wang X, Peng J, Yao X. 2012. Contribution of host nucleoporin 62 in HIV-1 integrase chromatin association and viral DNA integration. J Biol Chem 287:10544–10555. doi: 10.1074/jbc.M111.317057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ho SN, Hunt HD, Horton RM, Pullen JK, Pease LR. 1989. Site-directed mutagenesis by overlap extension using the polymerase chain reaction. Gene 77:51–59. doi: 10.1016/0378-1119(89)90358-2. [DOI] [PubMed] [Google Scholar]
- 39.Berthoux L, Sebastian S, Muesing MA, Luban J. 2007. The role of lysine 186 in HIV-1 integrase multimerization. Virology 364:227–236. doi: 10.1016/j.virol.2007.02.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Jayappa KD, Ao Z, Yang M, Wang J, Yao X. 2011. Identification of critical motifs within HIV-1 integrase required for importin alpha3 interaction and viral cDNA nuclear import. J Mol Biol 410:847–862. doi: 10.1016/j.jmb.2011.04.011. [DOI] [PubMed] [Google Scholar]
- 41.Zheng Y, Ao Z, Wang B, Jayappa KD, Yao X. 2011. Host protein Ku70 binds and protects HIV-1 integrase from proteasomal degradation and is required for HIV replication. J Biol Chem 286:17722–17735. doi: 10.1074/jbc.M110.184739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ao Z, Yao X, Cohen EA. 2004. Assessment of the role of the central DNA flap in human immunodeficiency virus type 1 replication by using a single-cycle replication system. J Virol 78:3170–3177. doi: 10.1128/JVI.78.6.3170-3177.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mouland AJ, Mercier J, Luo M, Bernier L, DesGroseillers L, Cohen EA. 2000. The double-stranded RNA-binding protein Staufen is incorporated in human immunodeficiency virus type 1: evidence for a role in genomic RNA encapsidation. J Virol 74:5441–5451. doi: 10.1128/JVI.74.12.5441-5451.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Janas AM, Dong C, Wang JH, Wu L. 2008. Productive infection of human immunodeficiency virus type 1 in dendritic cells requires fusion-mediated viral entry. Virology 375:442–451. doi: 10.1016/j.virol.2008.01.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Marechal V, Prevost MC, Petit C, Perret E, Heard JM, Schwartz O. 2001. Human immunodeficiency virus type 1 entry into macrophages mediated by macropinocytosis. J Virol 75:11166–11177. doi: 10.1128/JVI.75.22.11166-11177.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Nobile C, Moris A, Porrot F, Sol-Foulon N, Schwartz O. 2003. Inhibition of human immunodeficiency virus type 1 Env-mediated fusion by DC-SIGN. J Virol 77:5313–5323. doi: 10.1128/JVI.77.9.5313-5323.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Stremlau M, Perron M, Lee M, Li Y, Song B, Javanbakht H, Diaz-Griffero F, Anderson DJ, Sundquist WI, Sodroski J. 2006. Specific recognition and accelerated uncoating of retroviral capsids by the TRIM5alpha restriction factor. Proc Natl Acad Sci U S A 103:5514–5519. doi: 10.1073/pnas.0509996103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Butler SL, Hansen MS, Bushman FD. 2001. A quantitative assay for HIV DNA integration in vivo. Nat Med 7:631–634. doi: 10.1038/87979. [DOI] [PubMed] [Google Scholar]
- 49.Briones MS, Dobard CW, Chow SA. 2010. Role of human immunodeficiency virus type 1 integrase in uncoating of the viral core. J Virol 84:5181–5190. doi: 10.1128/JVI.02382-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Dobard CW, Briones MS, Chow SA. 2007. Molecular mechanisms by which human immunodeficiency virus type 1 integrase stimulates the early steps of reverse transcription. J Virol 81:10037–10046. doi: 10.1128/JVI.00519-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Frankel AD, Young JA. 1998. HIV-1: fifteen proteins and an RNA. Annu Rev Biochem 67:1–25. doi: 10.1146/annurev.biochem.67.1.1. [DOI] [PubMed] [Google Scholar]
- 52.Koh K, Miyaura M, Yoshida A, Sakurai A, Fujita M, Adachi A. 2000. Cell-dependent gag mutants of HIV-1 are crucially defective at the stage of uncoating/reverse transcription in non-permissive cells. Microbes Infect 2:1419–1423. doi: 10.1016/S1286-4579(00)01295-8. [DOI] [PubMed] [Google Scholar]
- 53.Van Maele B, Debyser Z. 2005. HIV-1 integration: an interplay between HIV-1 integrase, cellular and viral proteins. AIDS Rev 7:26–43. [PubMed] [Google Scholar]
- 54.Zhu K, Dobard C, Chow SA. 2004. Requirement for integrase during reverse transcription of human immunodeficiency virus type 1 and the effect of cysteine mutations of integrase on its interactions with reverse transcriptase. J Virol 78:5045–5055. doi: 10.1128/JVI.78.10.5045-5055.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Fan J, Zhang Q, Tochio H, Li M, Zhang M. 2001. Structural basis of diverse sequence-dependent target recognition by the 8 kDa dynein light chain. J Mol Biol 306:97–108. doi: 10.1006/jmbi.2000.4374. [DOI] [PubMed] [Google Scholar]
- 56.Lo KW, Naisbitt S, Fan J S, Sheng M, Zhang M. 2001. The 8-kDa dynein light chain binds to its targets via a conserved (K/R)XTQT motif. J Biol Chem 276:14059–14066. doi: 10.1074/jbc.M010320200. [DOI] [PubMed] [Google Scholar]
- 57.Rodriguez-Crespo I, Yelamos B, Roncal F, Albar JP, Ortiz de Montellano PR, Gavilanes F. 2001. Identification of novel cellular proteins that bind to the LC8 dynein light chain using a pepscan technique. FEBS Lett 503:135–141. doi: 10.1016/S0014-5793(01)02718-1. [DOI] [PubMed] [Google Scholar]
- 58.Hehl EA, Joshi P, Kalpana GV, Prasad VR. 2004. Interaction between human immunodeficiency virus type 1 reverse transcriptase and integrase proteins. J Virol 78:5056–5067. doi: 10.1128/JVI.78.10.5056-5067.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wilkinson TA, Januszyk K, Phillips ML, Tekeste SS, Zhang M, Miller JT, Le Grice SF, Clubb RT, Chow SA. 2009. Identifying and characterizing a functional HIV-1 reverse transcriptase-binding site on integrase. J Biol Chem 284:7931–7939. doi: 10.1074/jbc.M806241200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Benison G, Nyarko A, Barbar E. 2006. Heteronuclear NMR identifies a nascent helix in intrinsically disordered dynein intermediate chain: implications for folding and dimerization. J Mol Biol 362:1082–1093. doi: 10.1016/j.jmb.2006.08.006. [DOI] [PubMed] [Google Scholar]
- 61.Makokha M, Hare M, Li M, Hays T, Barbar E. 2002. Interactions of cytoplasmic dynein light chains Tctex-1 and LC8 with the intermediate chain IC74. Biochemistry 41:4302–4311. doi: 10.1021/bi011970h. [DOI] [PubMed] [Google Scholar]
- 62.Nyarko A, Hare M, Hays TS, Barbar E. 2004. The intermediate chain of cytoplasmic dynein is partially disordered and gains structure upon binding to light-chain LC8. Biochemistry 43:15595–15603. doi: 10.1021/bi048451+. [DOI] [PubMed] [Google Scholar]
- 63.Bao KK, Wang H, Miller JK, Erie DA, Skalka AM, Wong I. 2003. Functional oligomeric state of avian sarcoma virus integrase. J Biol Chem 278:1323–1327. doi: 10.1074/jbc.C200550200. [DOI] [PubMed] [Google Scholar]
- 64.Faure A, Calmels C, Desjobert C, Castroviejo M, Caumont-Sarcos A, Tarrago-Litvak L, Litvak S, Parissi V. 2005. HIV-1 integrase crosslinked oligomers are active in vitro. Nucleic Acids Res 33:977–986. doi: 10.1093/nar/gki241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Li M, Mizuuchi M, Burke TR Jr., Craigie R. 2006. Retroviral DNA integration: reaction pathway and critical intermediates. EMBO J 25:1295–1304. doi: 10.1038/sj.emboj.7601005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Ren G, Gao K, Bushman FD, Yeager M. 2007. Single-particle image reconstruction of a tetramer of HIV integrase bound to DNA. J Mol Biol 366:286–294. doi: 10.1016/j.jmb.2006.11.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Lo KW, Kan HM, Chan LN, Xu WG, Wang KP, Wu Z, Sheng M, Zhang M. 2005. The 8-kDa dynein light chain binds to p53-binding protein 1 and mediates DNA damage-induced p53 nuclear accumulation. J Biol Chem 280:8172–8179. doi: 10.1074/jbc.M411408200. [DOI] [PubMed] [Google Scholar]
- 68.Kardon JR, Vale RD. 2009. Regulators of the cytoplasmic dynein motor. Nat Rev Mol Cell Biol 10:854–865. doi: 10.1038/nrm2804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Fassati A, Goff SP. 2001. Characterization of intracellular reverse transcription complexes of human immunodeficiency virus type 1. J Virol 75:3626–3635. doi: 10.1128/JVI.75.8.3626-3635.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Miller MD, Farnet CM, Bushman FD. 1997. Human immunodeficiency virus type 1 preintegration complexes: studies of organization and composition. J Virol 71:5382–5390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.de Soultrait VR, Caumont A, Durrens P, Calmels C, Parissi V, Recordon P, Bon E, Desjobert C, Tarrago-Litvak L, Fournier M. 2002. HIV-1 integrase interacts with yeast microtubule-associated proteins. Biochim Biophys Acta 1575:40–48. doi: 10.1016/S0167-4781(02)00241-5. [DOI] [PubMed] [Google Scholar]
- 72.Wang JY, Ling H, Yang W, Craigie R. 2001. Structure of a two-domain fragment of HIV-1 integrase: implications for domain organization in the intact protein. EMBO J 20:7333–7343. doi: 10.1093/emboj/20.24.7333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Lu R, Ghory HZ, Engelman A. 2005. Genetic analyses of conserved residues in the carboxyl-terminal domain of human immunodeficiency virus type 1 integrase. J Virol 79:10356–10368. doi: 10.1128/JVI.79.16.10356-10368.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Williams JC, Roulhac PL, Roy AG, Vallee RB, Fitzgerald MC, Hendrickson WA. 2007. Structural and thermodynamic characterization of a cytoplasmic dynein light chain-intermediate chain complex. Proc Natl Acad Sci U S A 104:10028–10033. doi: 10.1073/pnas.0703614104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Sokolskaja E, Sayah DM, Luban J. 2004. Target cell cyclophilin A modulates human immunodeficiency virus type 1 infectivity. J Virol 78:12800–12808. doi: 10.1128/JVI.78.23.12800-12808.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Towers GJ, Hatziioannou T, Cowan S, Goff SP, Luban J, Bieniasz PD. 2003. Cyclophilin A modulates the sensitivity of HIV-1 to host restriction factors. Nat Med 9:1138–1143. doi: 10.1038/nm910. [DOI] [PubMed] [Google Scholar]