ABSTRACT
Human respiratory syncytial virus (RSV) is associated with severe childhood respiratory infections. A clear description of local RSV molecular epidemiology, evolution, and transmission requires detailed sequence data and can inform new strategies for virus control and vaccine development. We have generated 27 complete or nearly complete genomes of RSV from hospitalized children attending a rural coastal district hospital in Kilifi, Kenya, over a 10-year period using a novel full-genome deep-sequencing process. Phylogenetic analysis of the new genomes demonstrated the existence and cocirculation of multiple genotypes in both RSV A and B groups in Kilifi. Comparison of local versus global strains demonstrated that most RSV A variants observed locally in Kilifi were also seen in other parts of the world, while the Kilifi RSV B genomes encoded a high degree of variation that was not observed in other parts of the world. The nucleotide substitution rates for the individual open reading frames (ORFs) were highest in the regions encoding the attachment (G) glycoprotein and the NS2 protein. The analysis of RSV full genomes, compared to subgenomic regions, provided more precise estimates of the RSV sequence changes and revealed important patterns of RSV genomic variation and global movement. The novel sequencing method and the new RSV genomic sequences reported here expand our knowledge base for large-scale RSV epidemiological and transmission studies.
IMPORTANCE The new RSV genomic sequences and the novel sequencing method reported here provide important data for understanding RSV transmission and vaccine development. Given the complex interplay between RSV A and RSV B infections, the existence of local RSV B evolution is an important factor in vaccine deployment.
INTRODUCTION
Human respiratory syncytial virus (RSV) is a leading viral cause of severe respiratory infection during infancy and early childhood and among immunocompromised populations (1, 2). Globally, the virus is estimated to be responsible for 30 million episodes of acute lower respiratory tract infections (RTIs) and more than 50,000 deaths annually in children under 5 years of age (3). RSV infections throughout the world consistently occur as annual or biennial epidemics, and persons of all ages can be infected with diverse clinical outcomes ranging from mild upper RTIs to severe pneumonia or bronchiolitis (2, 4). A vaccine against RSV is not yet available (5). Careful analyses of RSV molecular epidemiology, evolution, and transmission are essential for defining the circulating viruses, for characterizing antigenic variation, and for tracking transmission patterns. The outcome of these studies can support new strategies for RSV control and vaccine use and development.
It has long been known that children suffer repeated RSV infections throughout life (6, 7). The ability of the virus to continue to infect previously exposed individuals is thought to be linked to an ability to bypass preexisting immune responses (8). Sequence variation in attachment (G) protein in consecutive years (9) is thought to be part of this mechanism; also, the global existence of two groups, A and B, and their alternating infection incidences may play a role (10, 11). The transmissibility of RSV group A (RSVA) is estimated to be slightly higher than that of RSV group B (RSVB) (12), and RSVA infections are more frequent than RSVB infections (12). An additional important feature of RSV infection is the apparently rapid global dispersion of new RSV variants (13). Indeed, genetically similar viruses cluster more by time than by location, suggesting rapid global movement of new variants (14). RSV molecular pathology and epidemiology have been reviewed in detail elsewhere (2, 4, 15, 16).
Historically, RSV molecular epidemiology has focused on the 900-bp region encoding the G protein (15, 17). The G protein together with the fusion (F) protein are important targets of human protective antibody responses (8, 15), with changes in this region thought to be driven by pressure to avoid host immune responses. Although studies of the sequence variability of RSV have concentrated on G gene variability, given the rapid infection pace but relatively low evolutionary rate of RSV, transmission studies over short periods require the stronger evolutionary signal provided by the full virus genome sequence (15,200 nucleotides [nt]: 11 open reading frames [ORFs] and noncoding regions). There is also a need to understand the nature of variation of immune targets other than the G protein. Advances in primer design, sequencing technology, and sequence assembly algorithms now allow full-genome sequencing for a number of viruses, including RSV (18–23), norovirus (24), and Middle East respiratory syndrome (MERS) coronavirus (25, 26).
The current work describes RSV genome evolution across a set of clinical samples collected from children who presented with severe RSV disease in a rural coastal Kenyan hospital using a novel RSV whole-genome sequencing (WGS) approach optimized for small amounts of clinical diagnostic samples. The sequence data provide an update of the genome-wide diversity of circulating RSV strains in this part of Kenya, including both RSVA and RSVB and the recently reemerged group B genotype GB3 (27). The novel genomes support previous conclusions on patterns of local RSV variation relative to global RSV diversity and reveal a significant difference in local evolution of RSVA versus RSVB.
MATERIALS AND METHODS
Primer design.
All RSV sequences available (August 2012) with lengths of >14,000 nt were collected and sorted by group, yielding 138 RSVA and 38 RSVB genomes. The sequences for each group were pooled and sliced into 33-nt strings with a 1-nt step size. The 33-mers were filtered to remove sequences with ambiguous nucleotides, and the frequency of each sequence within the set was determined. The 33-nt sequences were then trimmed to a calculated melting temperature (Tm) of 58°C, discarding sequences mapping to human rRNA, with GC contents of <30% or >65%, or with a single nucleotide frequency of >60%. The RSV genome was divided into six 3-kb segments overlapping by 300 nt. All sequences were mapped to an RSVA or RSVB reference strain, and the two most frequent primers mapping within 300 nt of the end of each amplicon were selected. The reverse complement of the downstream sequences was prepared. To ensure amplification of the far ends of the genomes, two additional primers were included from the 5′- and 3′-terminal genomic regions. A summary of the primer sequences and their predicted target sequences across all known RSV genomes is presented in Table 1.
TABLE 1.
Summary of RSV primers used in this study
| Target | Primer | Sequence (5′ to 3′) | Strand | Positiona | Tm (°C)b | % with 0 MMc | % with 0–3 MMd |
|---|---|---|---|---|---|---|---|
| RSVA | rsvas | ACGCGAAAAAATGCGTACAAC | Plus | 1 | 57.13 | 18.28 | 18.97 |
| RSVA | rsva52 | TGTGCATGTTATTACAAGTAGTGATATTTG | Plus | 266 | 56.96 | 95.52 | 98.97 |
| RSVA | rsva50 | GCATGTTATTACAAGTAGTGATATTTGCC | Plus | 269 | 57.51 | 95.17 | 98.97 |
| RSVA | rsva117 | ATAAGAGATGCCATGGTTGGTTTAAGA | Plus | 2849 | 58.44 | 95.86 | 100.00 |
| RSVA | rsva86 | AAGAGATGCCATGGTTGGTTTAAGA | Plus | 2851 | 58.43 | 95.86 | 100.00 |
| RSVA | rsva175 | TTCTCTTAAACCAACCATGGCATCT | Minus | 2878 | 58.43 | 95.86 | 100.00 |
| RSVA | rsva39 | CTTCTCTTAAACCAACCATGGCATC | Minus | 2879 | 58.22 | 95.86 | 100.00 |
| RSVA | rsva1820 | GCAGCATATGCAGCAACAATC | Plus | 5207 | 56.95 | 93.79 | 98.97 |
| RSVA | rsva1914 | CAGCATATGCAGCAACAATCCAA | Plus | 5208 | 58.32 | 93.10 | 98.62 |
| RSVA | rsva1644 | CAACTCCATTGTTATTTGCCCC | Minus | 5674 | 56.05 | 89.66 | 100.00 |
| RSVA | rsva1688 | CAACTCCATTGTTATTTGCCCCA | Minus | 5674 | 57.54 | 89.66 | 100.00 |
| RSVA | rsva704 | ATGTGTTGCCATGAGCAAACTC | Plus | 7893 | 57.95 | 91.03 | 100.00 |
| RSVA | rsva731 | GCCATGAGCAAACTCCTCACT | Plus | 7900 | 58.49 | 71.38 | 99.31 |
| RSVA | rsva341 | TTGTCAGGTAGTATCATTATTTTTGGCATG | Minus | 8196 | 58.53 | 98.97 | 99.31 |
| RSVA | rsva312 | AGGATATTTGTCAGGTAGTATCATTATTTTTGG | Minus | 8203 | 58.08 | 98.97 | 100.00 |
| RSVA | rsva374 | AAGAGAACTCAGTGTAGGTAGAATGTTT | Plus | 10360 | 57.89 | 96.55 | 100.00 |
| RSVA | rsva350 | AGAACTCAGTGTAGGTAGAATGTTTG | Plus | 10363 | 56.64 | 96.55 | 100.00 |
| RSVA | rsva497 | GCTTGATTGAATTTGCTGAGATCTGT | Minus | 10620 | 58.44 | 95.52 | 100.00 |
| RSVA | rsva539 | ATGCTTGATTGAATTTGCTGAGATCTG | Minus | 10622 | 58.68 | 95.52 | 100.00 |
| RSVA | rsva1220 | GATTGGGTGTATGCATCTATAGATAACAAG | Plus | 12386 | 57.94 | 95.86 | 99.31 |
| RSVA | rsva1232 | ATTGGGTGTATGCATCTATAGATAACAAG | Plus | 12387 | 57.17 | 95.86 | 99.31 |
| RSVA | rsva364 | TTATATATCCCTCTCCCCAATCTTTTTCAAA | Minus | 13070 | 58.32 | 96.21 | 100.00 |
| RSVA | rsva385 | ATCAGTTATATATCCCTCTCCCCAATCTT | Minus | 13075 | 58.46 | 96.21 | 100.00 |
| RSVA | rsva4066 | GTTGTATAACAAACTACCTGTGATTTTAATCAG | Minus | 14983 | 57.95 | 88.97 | 99.31 |
| RSVA | rsva5632 | TAACTATAATTGAATACAGTGTTAGTGTGTAGC | Minus | 15063 | 57.95 | 29.31 | 95.17 |
| RSVA | rsvae | ACGAGAAAAAAAGTGTCAAAAACTAATA | Minus | 15223 | 55.09 | 17.59 | 18.28 |
| RSVB | rsvbs | ACGCGAAAAAATGCGTACTACA | Plus | 1 | 57.56 | 43.14 | 43.14 |
| RSVB | rsvb3 | TGGGGCAAATAAGAATTTGATAAGTGC | Plus | 44 | 58.58 | 48.04 | 54.90 |
| RSVB | rsvb1021 | GGGGCAAATAAGAATTTGATAAGTGCTATT | Plus | 45 | 58.75 | 47.06 | 54.90 |
| RSVB | rsvb33 | ATATTAGGAATGCTCCATACATTAGTAGTTG | Plus | 2777 | 57.21 | 88.24 | 100.00 |
| RSVB | rsvb71 | TAAGAGATGCTATGGTTGGTCTAAGAGA | Plus | 2841 | 58.69 | 90.20 | 100.00 |
| RSVB | rsvb50 | AGTCTTGCCATAGCCTCTAACCT | Minus | 2937 | 58.57 | 93.14 | 100.00 |
| RSVB | rsvb95 | CCATTTTTTCGCTTTCCTCATTCCTA | Minus | 2963 | 58.14 | 95.10 | 100.00 |
| RSVB | rsvb7884 | AGTATATGTGGCAACAATCAACTCTG | Plus | 5202 | 57.48 | 81.37 | 100.00 |
| RSVB | rsvb7996 | TATGTGGCAACAATCAACTCTGC | Plus | 5206 | 57.70 | 81.37 | 100.00 |
| RSVB | rsvb7442 | GATGTGGAGGGCTCGGATG | Minus | 5548 | 57.92 | 75.49 | 100.00 |
| RSVB | rsvb7423 | CCATGGTTATTTGCCCCAGATTTAAT | Minus | 5662 | 57.87 | 77.45 | 99.02 |
| RSVB | rsvb3762 | AGAGGTCATTGCTTGAATGGTAGAA | Plus | 7642 | 57.98 | 93.14 | 100.00 |
| RSVB | rsvb3712 | AAGAGCATAGACACTTTGTCTGAAATAAG | Plus | 7762 | 57.89 | 77.45 | 100.00 |
| RSVB | rsvb3652 | GCTTATGGTTATGCTTTTGTGGATATCTAAT | Minus | 8130 | 58.41 | 89.22 | 98.04 |
| RSVB | rsvb3660 | GCAATCATGCTTTCACTTGAGATCAA | Minus | 8247 | 58.67 | 64.71 | 98.04 |
| RSVB | rsvb32 | AAGAAGAGTACTAGAGTATTACTTGAGAGATAA | Plus | 10236 | 57.04 | 90.20 | 100.00 |
| RSVB | rsvb52 | AAATCCAAATCTTAGCAGAGAAAATGATAG | Plus | 10412 | 56.70 | 96.08 | 100.00 |
| RSVB | rsvb47 | CCATGCAGTTCATCTAATACATCACTG | Minus | 10673 | 58.13 | 90.20 | 99.02 |
| RSVB | rsvb168 | TGCATGTCTATATGTACATATTATTGTGACAAG | Minus | 10746 | 58.25 | 91.18 | 99.02 |
| RSVB | rsvb651 | ATCGACATTGTGTTTCAAAATTGCATAAG | Plus | 12640 | 58.40 | 81.37 | 100.00 |
| RSVB | rsvb165 | TTCAAAATTGCATAAGTTTTGGTCTTAGC | Plus | 12653 | 58.06 | 88.24 | 100.00 |
| RSVB | rsvb27 | TTAATGAACATATGATCAGTTATATACCCCTCT | Minus | 13088 | 57.88 | 79.41 | 100.00 |
| RSVB | rsvb60 | AACTTAAAACTGTGACAGCCTTTTATTCT | Minus | 13325 | 58.08 | 89.22 | 100.00 |
| RSVB | rsvb1199 | ATAGTACACTACCTGTTATTTTAATCAGCTTCT | Minus | 14977 | 58.56 | 88.24 | 100.00 |
| RSVB | rsvb989 | TATAGTACACTACCTGTTATTTTAATCAGCTTC | Minus | 14978 | 57.57 | 88.24 | 100.00 |
| RSVB | rsvbe | ACGAGAAAAAAAGTGTCAAAAACTAATGT | Minus | 15216 | 57.47 | 5.88 | 6.86 |
Primer mapping position in RSVA (GenBank accession number FJ948820) or RSVB (GenBank accession number JQ582843).
Tm (melting temperature) calculated using a Python script that approximates the method of Breslauer et al. (51).
Percentage of full-length RSVA genomes (n = 290) or full-length RSVB genomes (n = 102) showing perfect homology to primer, i.e., 0 mismatches (MM).
Percentage of full-length RSVA genomes (n = 290) or full-length RSVB genomes (n = 102) showing the target sequence for the primer with up to 3 mismatches.
Clinical samples.
Viral nucleic acid for sequencing was extracted from RSV-positive clinical specimens (nasopharyngeal swabs [NPS] or washes) collected from children under 5 years old admitted to the Kilifi District Hospital (KDH) with severe or very severe pneumonia between 2002 and 2012. RSV infection was diagnosed with an indirect immunofluorescence antibody technique (IFAT; Light Diagnostics). Details of the study that provided the samples sequenced in this study have been previously provided (28). Informed consent was obtained from a parent or guardian on behalf of each child before specimen collection, and the KEMRI Ethics Review Committee approved all protocols. Additional details on the samples are provided in Table 2.
TABLE 2.
Details for samples used in this study
| MiSeq | Age (mo) | Sample date (day-mo-yr) | Group | Length (nt)a | Coverageb | Present in G setc | Present in F setd | GenBank no.e | ENA no.f |
|---|---|---|---|---|---|---|---|---|---|
| 10028_10 | 0 | 07-Jan-02 | A | 9,346 | 6,401 | Yes | KP317918 | ERR323212 | |
| 10028_11 | 6 | 27-Apr-02 | A | 7,091 | 10,370 | KP317940 | ERR323213 | ||
| 10028_12 | 6 | 28-Jan-03 | A | 9,776 | 5,692 | Yes | KP317955 | ERR323214 | |
| 11866_65 | 5 | 13-Feb-03 | A | 12,151 | 7,347 | Yes | Yes | KP317949 | ERR438932 |
| 11865_75 | 8 | 24-Mar-04 | A | 14,985 | 12,283 | Yes | Yes | KP317956 | ERR438910 |
| 10891_50 | 6 | 21-Jan-05 | A | 5,396 | 3,554 | Yes | Yes | KP317948 | ERR376407 |
| 10891_56 | 0 | 02-Feb-05 | A | 5,396 | 2,369 | Yes | Yes | KP317924 | ERR376413 |
| 9696_45 | 14 | 20-Feb-06 | A | 14,778 | 3,830 | Yes | Yes | KP317944 | ERR303303 |
| 10891_57 | 1 | 23-Feb-06 | A | 14,841 | 4,640 | Yes | Yes | KP317942 | ERR376414 |
| 10891_58 | 0 | 29-Mar-06 | A | 8,864 | 6,016 | KP317943 | ERR376415 | ||
| 10891_59 | 3 | 04-Jan-07 | A | 11,496 | 5,316 | Yes | KP317937 | ERR376416 | |
| 10891_60 | 1 | 05-Jan-07 | A | 14,791 | 4,454 | Yes | Yes | KP317926 | ERR376417 |
| 10891_51 | 0 | 07-Mar-08 | A | 14,967 | 4,882 | Yes | Yes | KP317933 | ERR376408 |
| 10891_52 | 11 | 17-Mar-08 | A | 5,636 | 1,201 | Yes | KP317931 | ERR376409 | |
| 10899_38 | 1 | 22-Feb-09 | A | 14,854 | 8,478 | Yes | Yes | KP317950 | ERR381723 |
| 10899_40 | 4 | 26-Jan-10 | A | 10,113 | 13,351 | Yes | KP317916 | ERR381725 | |
| 10899_41 | 18 | 10-Feb-10 | A | 14,713 | 12,405 | Yes | Yes | KP317935 | ERR381726 |
| 11864_54 | 3 | 29-Apr-10 | A | 14,716 | 7,071 | Yes | Yes | KP317936 | ERR438905 |
| 11862_33 | 3 | 26-Aug-10 | A | 14,719 | 8,961 | Yes | Yes | KP317921 | ERR438868 |
| 11864_53 | 1 | 25-Mar-11 | A | 14,735 | 6,891 | Yes | Yes | KP317951 | ERR438904 |
| 11862_28 | 28 | 13-Apr-11 | A | 15,214 | 10,434 | Yes | Yes | KP317920 | ERR438864 |
| 11862_29 | 4 | 23-Mar-12 | A | 14,950 | 12,922 | Yes | Yes | KP317953 | ERR438865 |
| 11862_32 | 14 | 30-Apr-12 | A | 7,197 | 6,180 | KP317947 | ERR438867 | ||
| 9697_16 | 10 | 06-Jul-02 | B | 15,040 | 5,419 | Yes | Yes | KP317939 | ERR303322 |
| 9697_10 | 8 | 13-Jan-03 | B | 9,790 | 6,853 | Yes | KP317930 | ERR303316 | |
| 10140_1 | 46 | 02-Apr-04 | B | 12,034 | 12,174 | Yes | Yes | KP317919 | ERR331021 |
| 9697_7 | 10 | 22-Dec-04 | B | 15,080 | 4,480 | Yes | Yes | KP317925 | ERR303313 |
| 9697_6 | 2 | 25-Dec-04 | B | 14,998 | 6,523 | Yes | Yes | KP317954 | ERR303312 |
| 9697_5 | 1 | 27-Jan-06 | B | 15,234 | 3,682 | Yes | Yes | KP317917 | ERR303311 |
| 9465_10 | 23 | 27-Feb-09 | B | 14,995 | 16,190 | Yes | Yes | KP317938 | ERR303268 |
| 9465_11 | 31 | 13-Feb-10 | B | 15,004 | 11,722 | Yes | Yes | KP317941 | ERR303269 |
| 9465_12 | 22 | 06-Apr-10 | B | 15,260 | 14,855 | Yes | Yes | KP317932 | ERR303270 |
| 9465_6 | 17 | 09-May-10 | B | 15,333 | 13,719 | Yes | Yes | KP317952 | ERR303264 |
| 9465_7 | 3 | 01-Feb-11 | B | 15,233 | 14,182 | Yes | Yes | KP317927 | ERR303265 |
| 9465_8 | 2 | 14-Apr-11 | B | 15,323 | 15,367 | Yes | Yes | KP317945 | ERR303266 |
| 9465_9 | 1 | 08-Jul-11 | B | 15,237 | 14,709 | Yes | Yes | KP317928 | ERR303267 |
| 9465_3 | 8 | 14-Jan-12 | B | 14,995 | 12,378 | Yes | Yes | KP317946 | ERR303261 |
| 9465_1 | 19 | 13-Feb-12 | B | 15,233 | 12,994 | Yes | Yes | KP317934 | ERR303259 |
| 9465_4 | 14 | 01-Mar-12 | B | 15,179 | 14,802 | Yes | Yes | KP317923 | ERR303262 |
| 10911_9 | 1 | 23-Mar-12 | B | 14,977 | 12,504 | Yes | Yes | KP317929 | ERR376442 |
| 9465_2 | 5 | 16-May-12 | B | 14,941 | 12,906 | Yes | Yes | KP317922 | ERR303260 |
Final sequence length obtained from de novo assembly of short read data (see Materials and Methods).
Coverage calculated by mapping all reads to final assembled contig. Coverage was calculated as the number of mapped reads/(length of the genome fragment/129).
Samples yielding sufficient sequence for G region analysis (Fig. 5).
Samples yielding sufficient sequence for F region analysis (Fig. 5).
The final genome data were deposited in GenBank with the indicated accession numbers.
Short-read data available at European Nucleotide Archive (http://www.ebi.ac.uk/ena).
RNA extraction, RT, and PCR.
Viral RNA was extracted with the QIAmp extraction kit (Qiagen, United Kingdom) from a starting NPS specimen volume of 140 μl and final elution volume of 60 μl. Reverse transcription (RT) of RNA molecules was performed with the forward primers for each of the six amplicons. A separate RT reaction was performed for each amplicon. Typically, the 20-μl reaction mixture contained 2 μl of sample RNA. A 5-μl aliquot of the resulting cDNA was used in each 25-μl PCR mixture. The PCR mixture was incubated at 98°C for 30 s, followed by 40 cycles of 98°C for 10 s, 53°C for 30 s, and 72°C for 3.0 min and a final extension of 72°C for 10.0 min. Following PCR, aliquots of the products were run on a 0.6% agarose gel to monitor amplification success, and the products from the 6 reactions for each sample were pooled for Illumina library preparation.
Deep sequencing.
Sequencing of the pooled amplicons was performed with Illumina MiSeq. Samples were multiplexed at 15 to 20 per MiSeq run and processed as paired-end reads (2 × 149 nt), generating approximately 1.5 million reads per sample. Raw sequence data were processed with QUASR (29) to remove low-quality (< median Phred 35) and adapter-containing reads, and de novo assembly with SPAdes (30) was performed. RSV contigs were identified by BLASTN analysis, and low-coverage contigs were excluded. Where necessary, partial but overlapping genome contigs were combined using Sequencher (v5.2.4). All final viral genomes were examined for appropriate assembly based on length and the presence of the expected intact RSV open reading frames.
Protein changes.
After sorting by virus group (RSV group A or B), the genomic region under investigation was translated, the protein sequence was aligned using MAFFT (31), and protein differences from the consensus sequence of the group were visualized and quantitated using Python scripts.
Reference data set.
A comprehensive RSV genome data set was generated from the GenBank database using as a starting set all reported RSV genomes. The search was conducted on 28 September 2014 using the search term “txid11250 [Organism]) AND 13500[SLEN]: 17000[SLEN].” Genomes with multiple ambiguous bases, lacking country of detection or date of collection (year), or from patent depositions were excluded. The newly sequenced Kilifi RSV genomes for each group were combined with those from GenBank in the subsequent analysis. Thinned representative reference sets were prepared by using the usearch algorithm (32).
Phylogenetic analyses.
Phylogenetic trees of the genome sequences and selected genomic regions were constructed using the Bayesian methods in MrBayes program v3.2.1 (http://mrbayes.sourceforge.net/index.php) under the general time reversible model of evolution. RSVA and RSVB were analyzed separately using both the total data set and the thinned data sets. The viruses within the groups were assigned to genotypes based on the clustering pattern of the G ORF portion sequences with reference sequences representative of the previously described RSV genotypes: for RSVA, strains representing GA1-7, SAA1, and ON1, and for RSVB, strains representing GA1-4, SAB1-SAB4, and BA (11, 33–35). Phylogenetic trees were visualized in FigTree v1.4.2.
Evolutionary analyses.
Nucleotide substitution rates and estimates for time to most recent common ancestor (tMRCA) were obtained from the usearch-thinned data sets, using uclust to remove genomes closer than ID 0.99 (32). The rates and tMRCA estimates were calculated in BEAST v1.7.5 (36) both for full genomes and for the individual ORFs.
Nucleotide sequence accession numbers.
The final set of RSV sequences was deposited in GenBank with the following accession numbers: KP317916 to KP317956.
RESULTS
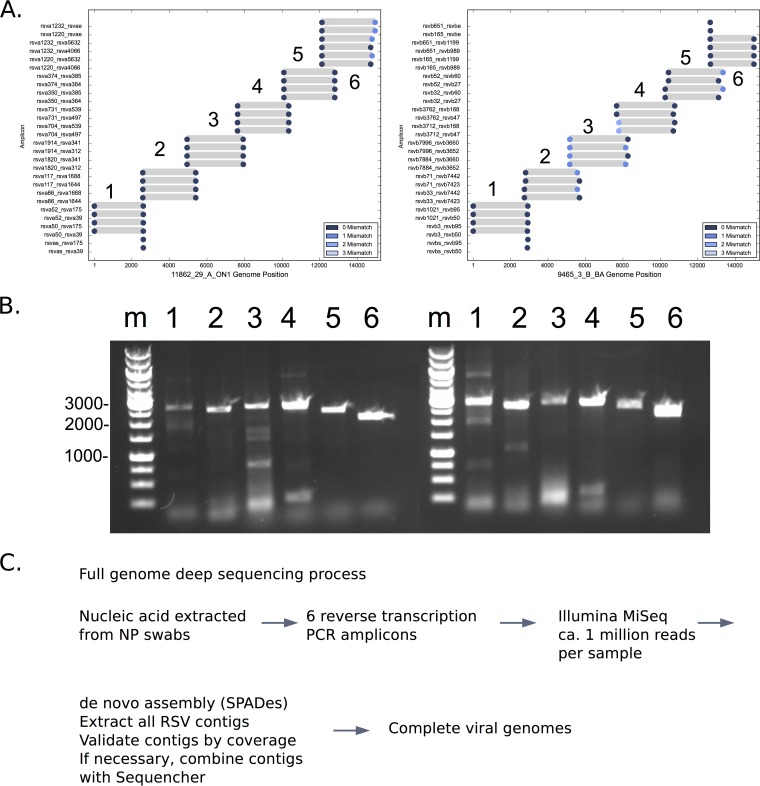
Two sets of reverse transcription and PCR primers were selected from all available RSVA and RSVB genomic sequence data based on frequency, location, and predicted PCR function (see Table 1 for further details). The general pattern of primer sites and the locations of primer targets in RSVA and RSVB genomes are shown in Fig. 1A. Actual PCR results are shown in Fig. 1B for RSVA and RSVB samples, with PCR products of the expected size obtained for all 6 amplicons. These primers were used as part of a deep-sequencing process for RSV combining the full cDNA preparation and genome amplification, deep sequencing with Illumina MiSeq, and de novo assembly (Fig. 1C) to generate 27 complete or nearly complete genomes (11 group A and 16 group B; median length, 14,990 nt; range, 14,666 to 15,232 nt). An additional number of samples yielded RSV contigs of >5,000 nt in length, and these were also retained for further analysis. A summary of the genomic sequences in this study is provided in Table 2.
FIG 1.
(A) PCR primer target sites in RSVA and RSVB. The primer target sequences in representative RSVA (left) and RSVB (right) viruses were determined. Circular markers indicate positions of primer target sites in the test genome color-coded by number of mismatches with the primer; gray bars indicate lengths and positions of the predicted products. (B) Two examples of reverse transcription-PCR function. The DNA products of reverse transcription and PCR amplification of two samples were resolved by agarose gel electrophoresis and visualized by ethidium bromide staining. Sizes of some of the molecular size markers (in base pairs) are indicated to left of the gel. Lane m, molecular size markers; lanes 1 to 6, individual 2- to 3-kb RSV amplicons 1 to 6, respectively. (C) Flowchart of the RSV sequencing process.
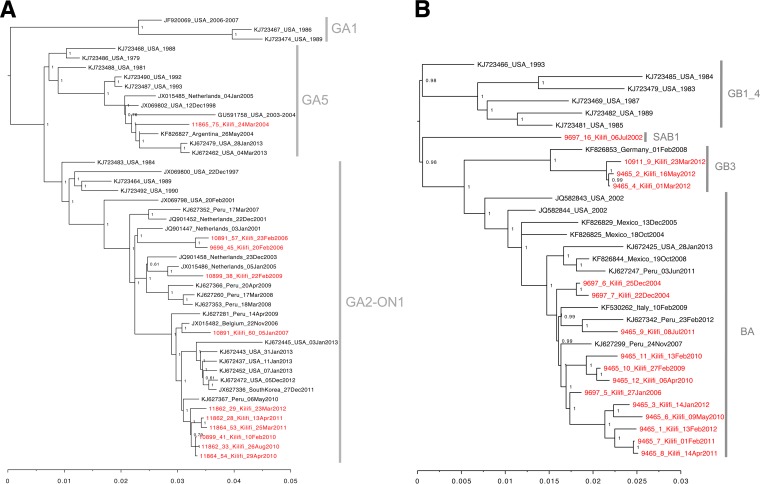
RSV global phylogenetic clustering and placement of Kilifi genomes.
The 27 Kilifi genomes were combined with RSVA and RSVB genomes from 16 countries from specimens collected between the years 1981 and 2013 (see Materials and Methods). The phylogenetic clustering is shown in Fig. 2A (RSVA) and B (RSVB). RSVA forms 3 major clades: GA1 (including strains only from the United States), GA5 (with U.S. and global strains), and a clade with both GA2 and the ON1 viruses with a 72-nucleotide duplication in the G ORF (33), which included nearly all of the new Kilifi RSVA genomes (GA2_ON1). Multiple subclusters showing temporal clustering were detected within each of these clades.
FIG 2.
Phylogenetic analysis of the Kilifi RSVA and RSVB genomes. (A) MrBayes tree of representative global RSVA genome sequences together and the 11 novel Kilifi RSVA genome sequences. (B) MrBayes tree of representative global RSVB genome sequences and the 16 novel Kilifi RSVA genome sequences. Trees were inferred using the Bayesian methods in MrBayes (http://mrbayes.sourceforge.net/index.php) under the GTR model of evolution. The numbers next to the branches indicate the posterior probabilities. The Kilifi taxa are indicated in red font. Thinned global reference sets for RSVA and RSVB were prepared from all available RSV genomes clustering at 0.99% identity using uclust (32). See Materials and Methods for additional details.
Four clades were designated for the RSVB genomes, with BA containing the majority of the Kilifi sequences (Fig. 2B). Clade GB1_GB4 included viruses detected in the United States between 1983 and 1991. Clades SAB1, GB3, and BA included viruses from multiple countries, including the Kilifi RSVB genomes. Similar to that for RSVA, the clustering was more temporal than geographical. Notably, the BA (Buenos Aires) clade viruses are characterized by the presence of a 60-nucleotide duplication within the G ORF. The 4 viruses within clade GB3 (3 from Kilifi and 1 from Germany) lacked the 60-nucleotide duplication. Neither RSVA nor RSVB genomes from Kilifi showed a monophyletic grouping. Instead, the Kilifi genomes were dispersed throughout the observed RSV evolution, clustering with contemporaneous genomes from the other countries. The phylogenetic tree topologies arising from whole-genome and G protein ORF sequences were highly similar (data not shown).
Comparison of genomes of viruses with identical G protein ORFs.
One motivation for developing full-genome methods was to increase the sensitivity for tracking RSV across short-term transmission chains. We asked if viruses identical in their G gene regions had differences elsewhere in their genomes. All RSV genomes (both GenBank or in the new data presented here) with identical G regions were identified, and the number of changes outside the G region were determined. Of 7 sets of viruses with identical G regions, all showed at least 1 but up to 9 nucleotide differences across the full genome (Fig. 3). This increased resolution will be important in future studies examining RSV household transmission patterns to identify who acquires infection from whom.
FIG 3.
Comparison of RSVB genomes with identical G regions. Each panel represents a genome nucleotide alignment of RSVs that had identical G gene sequences. The G protein ORF portions of the genomes are highlighted gray across the panels and were identical. The vertical lines indicate where there are nucleotide substitutions occurring outside the G gene region between the genomes. The blue blocks indicate a gap in the sequence.
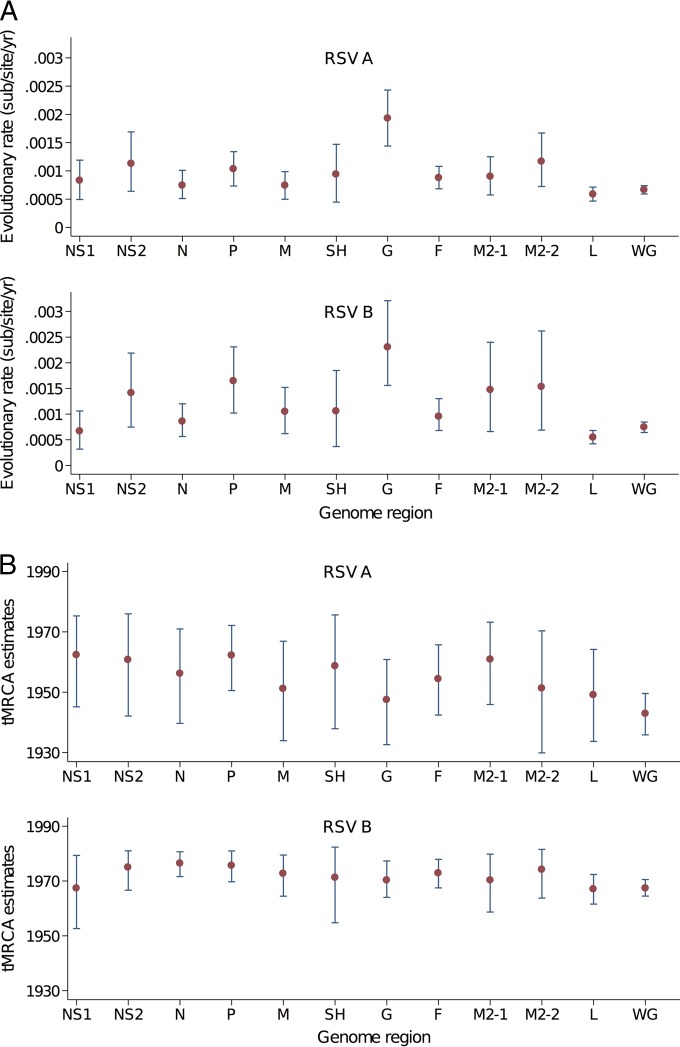
Estimation of RSV tMRCA and evolutionary rates.
Previous data on RSV evolution are largely derived from the G protein coding region. The full genomes generated in this study were combined with the GenBank reference data set, and these allowed an estimation of the global nucleotide substitution rates and the time to most recent common ancestor (tMRCA) for all the recently sequenced RSVA and RSVB viruses. These estimates were calculated for the different ORFs and the whole-genome sequences (Fig. 4). The whole genomes provided more precise estimates of the MRCA, as observed from the interval of lower and upper 95% highest posterior density (HPD) compared to individual ORF data for the same set of viruses (Fig. 4B).
FIG 4.
(A) Estimates of the nucleotide substitution rates for RSVA and RSVB in the individual ORFs and for the whole-genome sequence. (B) Estimates of tMRCA for RSVA and RSVB for the individual ORFs and for the whole-genome sequence. The analysis was undertaken using the usearch-thinned data sets (37 genome sequences for RSVA and 23 sequences for RSVB). The analysis was performed with BEAST (36).
The G protein ORF showed the highest nucleotide substitution rates for both RSVA and RSVB (Fig. 4A). Elevated changes in G and M2-2 were observed previously using RSV full genomes from U.S. and European cohort data (21). Similar to the MRCA estimates, the whole-genome estimates for the evolutionary rates showed narrower confidence intervals than those from the individual ORFs. The two regions considered for vaccine targets, G and F, show a strikingly wide difference in rate, and this may be important for selecting conserved vaccine targets.
Changes in G and F coding regions, comparing local and global viruses.
An important consideration for vaccine development is how representative a vaccine strain is for locally circulating viruses. The transmission patterns of a virus, the evolutionary rate of the virus, and patterns of human movement can strongly influence how quickly global strains reach a rural location. To address this important issue, the amino acid changes encoded by the RSV coding sequences observed in Kilifi were compared to the amino acid changes observed for all known RSV genomes from other parts of the world (Fig. 5; Table 3).
FIG 5.
Kilifi versus global changes in the G, F, and NS2 proteins. (A) Kilifi compared to global G protein changes. For each group, the G protein sequences were identified as Kilifi or non-Kilifi (global) and aligned, and a consensus amino acid sequence was generated (at 60% level). The first portion shows the positions of O-linked (red) and N-linked (blue) glycosylation sites, the second portion shows general features of the G protein, and the third portion shows total changes (Kilifi plus global) at each position. The fourth portion shows amino acid differences in each G sequence from the consensus. Amino acid changes observed only in Kilifi are marked in red, and changes observed either globally or in the Kilifi are marked in gray. Gaps are not indicated. N-linked and O-linked glycosylation sites were determined using NetNGlyc 1.0 and NetOGlyc 3.1 (46–48). (B) Kilifi versus global F protein changes. Changes in F protein were determined and are depicted as in panel A. Known motifs of the F protein (49) include signal peptide (SP), heptad repeat C (HRC), 27-mer fragment (p27), putative fusion peptide (FP), heptad repeat A (HRA), domains 1 and 2 (Dom1&2), heptad repeat B (HRB), transmembrane domain (TM), and cytoplasm domain (CP). Antigenic sites I, II, and IV (ASI, ASII, and ASIV) are sites of neutralizing antibody binding (40, 50). (C) Kilifi versus global NS2 protein changes. Changes in NS2 protein were determined and are depicted as in panel A. Known motifs of the NS2 protein include the TRAF3-interacting domain (TRAF3-ID) and C-terminal tetrapeptide sequence (DLNP) (43).
TABLE 3.
Kilifi versus global evolution
| Protein | No. of distinct changes for all Kilifi and global viruses | No. of distinct changes in Kilifi viruses | No. (%) of distinct changes unique to Kilifi virusesa |
|---|---|---|---|
| RSVA G | 409 | 68 | 7 (11.8)b |
| RSVB G | 299 | 70 | 30 (42.9) |
| RSVA F | 200 | 45 | 9 (22.2)c |
| RSVB F | 81 | 19 | 13 (79.3) |
| RSVA NS2 | 73 | 18 | 3 (16.7)d |
| RSVB NS2 | 38 | 16 | 13 (81.3) |
Number of distinct amino acid changes observed in Kilifi and not in other parts of the world. “Distinct changes” means that the set of changes is reduced to a unique set with multiple occurrences of a change counted only once.
The P value for Fisher's exact test was <0.01 for the number of location-specific distinct changes compared to total distinct changes for RSVA versus RSVB.
The P value for Fisher's exact test was <0.05 for the number of location-specific distinct changes compared to total distinct changes for RSVA versus RSVB.
The P value for Fisher's exact test was <0.05 for the number of location-specific distinct changes compared to total distinct changes for RSVA versus RSVB.
A large percentage of the changes observed in the RSVA G protein were also observed globally, with 88% of the changes seen in Kilifi RSVA G also observed in other parts of the world (Table 3). The Kilifi RSVB viruses appeared to have more local evolution, with only 60% of the observed changes in G shared with global viruses. With reference to the F protein, for Kilifi RSVA viruses, 80% of the observed changes were also found globally, while the Kilifi RSVB viruses showed a higher degree of local evolution, with only 20% of the observed changes specific to Kilifi viruses seen in other locations. To determine if this local evolution of RSVB was observed at other sites, the sequence data were stratified to other locations (the United States, Argentina, and Peru), but no significant local patterns were observed. This suggests that the isolation of the Kilifi site was more pronounced than for other sites. Alternatively, this may reflect more intense sampling of RSVB within a limited area.
The RSV envelope proteins are heavily glycosylated. More than 50% of the G protein mass can be carbohydrate (37), and the potential O-linked glycosylation sites (serine or threonine) comprise up to 30% of the G protein amino acid sequence (38). Changes toward or away from asparagine can be associated with a change in the overall glycosylation of the protein and could be associated with adaptive change to local immune responses. The G protein is subject to heavy O-linked glycosylation in the variable regions, with modification frequently on serine or threonine residues in the vicinity of a proline residue (Fig. 5A). Nearly half of the observed changes in the in G protein affect S, T, or P residues (RSVA GA2 37/81 and RSVB 100/241). This is apparent when potential N- and O-linked glycosylation sites are marked on the G protein region (Fig. 5) and is also facilitated by the single nucleotide changes that distinguish codons for these three amino acids.
In the RSVA G proteins, an N237D polymorphism observed in many of the viruses is within the site NTT and would remove a predicted N-linked glycosylation site. Tan et al. (22) also noted that the RSVA-GA2 group showed a frequent change in two predicted glycosylation sites (N237D and S242N). Within the RSVB viruses, 3 of 15 amino acid changes involve asparagine, but none of these changes are predicted N-linked glycosylation sites (Asn-Xaa-Ser/Thr). Changes in N-linked glycosylation areas are known to effect binding of human convalescent-phase sera to peptides (39).
The RSV F protein contains only 10 to 20% of its mass as carbohydrate, and this is attached exclusively via N-glycosidic bonds (37). For the RSVA viruses, 5 of the protein changes are to or away from asparagine. In RSVB, 3 changes involve asparagine; however, none of these are within predicted glycosylation sites. Many polymorphisms were observed in the F protein p27 domain (Fig. 5B). This peptide is likely to serve as a spacer that is freed by cleavage during F maturation and is not found in the mature protein. The large number of changes may simply reflect the disposable nature of this sequence (40).
The NS2 protein may be important in modulating host innate immune responses (41–43) and may influence movement of infected cells (44). The NS2 showed an elevated level of evolutionary rate (Fig. 4), consistent with a protein interacting with polymorphic host target proteins. Monitoring the local versus global protein changes in NS2 revealed multiple changes occurring in the amino-terminal domain and a portion of the domain important for TRAF3 interactions (43). The majority of changes in the Kilifi RSVA NS2 proteins were also observed in other parts of the world; however, the RSVB NS2 protein showed a significantly high degree of variation only observed in the Kilifi viruses (Fig. 5C).
DISCUSSION
The current work presents a functional approach for community-wide monitoring of RSV whole-genome genetic diversity suitable for detailed transmission studies. A challenge with deep sequencing of large sample sets of RNA viruses is the design of amplification primers. Traditionally, PCR primers were designed using alignments of sequences from the target virus; previous RSV studies with dideoxy sequencing used a greater number of tiled amplicons (2, 24, 25, 38) to cover the whole genome. With larger and more diverse sets, the alignment step becomes problematic. The approach described here bypasses the alignment step and was tailored for deep-sequencing methods. The RSV method uses only 6 amplicons to reduce the amplification costs and the required amount of input RNA. Although two primer sets were designed for RSVA and RSVB, the two sets can be pooled to simplify processing of samples of unknown RSV subtype. The computational methods used for primer selection facilitates updating of the primer sets as additional RSV genome sequence data become available. Frequent updating of these primer sets will help avoid sequence bias that could occur using antiquated primer sets. It is also important that the new full genomes reported here were assembled using de novo assembly methods. Although reference-based methods for assembling genomes from short-read data are rapid and less memory intensive, reference-based methods fail if a close reference genome is not available. The method presented here determines virus genomic sequences directly from patient material and shows sensitivity similar to that of traditional sequencing methods, but it avoids the potential virus selection that may occur if samples are first passaged through cell culture.
The 27 novel Kilifi RSV genomes (11 RSVA and 16 RSVB) generated in this study were used to assess local versus global RSV variety. Similar to the patterns previously observed with G ORF, the full genomic phylogenetic analysis confirmed that Kilifi genomes were interspersed with genomes from other countries, with rapid appearance of variants in Kilifi soon after they are first observed in other parts of the world (45). Kilifi RSV strains are similar to strains that circulate in other regions of the world and reveal only limited local evolution. Phylogenetic clustering appeared to be more influenced by time of virus sample collection than by geographical location, suggesting a fairly rapid global spread of novel RSV variants. It should also be noted that the similarity of the overall topology of phylogenetic trees from whole genomes and G sequences is encouraging and indicates that although full-genome sequences are most useful for detailed transmission studies, the relationships determined with the G region is similar to the patterns observed with the full-genome sequences.
The availability of full genomes allowed a comparison of estimates of the tMRCA of the Kilifi RSV strains. The obtained tMRCAs were broadly similar, although the higher evolutionary rates of the G region lead, as expected, to slightly later tMRCAs. The estimates based on the entire genomes lead to earlier dates and more discrete confidence intervals than estimates from specific genomic regions. Similar observations were made by Tan et al. (22).
Our comparison of genomes determined to be identical in the G region found nucleotide substitutions elsewhere in their genomes. The genomes with identical G regions invariably were from the same geographical region and over the same epidemic, the sample collection date interval ranging from a few days to months. This observation suggests that nucleotide substitution in the RSV genome in the short term is random, i.e., not concentrated in the regions that appear the most variable in the long term, and supports the use of whole-genome sequencing for monitoring viral transmission chains.
The observed sites of change in the G and F proteins were frequently in exposed regions of the proteins; several involved glycosylation site changes suggestive of immune evasion. In addition, similar to previous reports, the NS2 (Fig. 5B) and M2-2 protein coding regions (not shown) were observed to change at rates higher than that for the full genome. Although these changes could be simply the allowed changes of unconstrained proteins, it is also possible that these sites are important for interacting with the host and may be under some pressure to change. Unfortunately, the sequence data set generated in this study was too small to provide statistically supported evidence of positive selection, but future studies with larger data sets will be facilitated by these methods.
The availability of a collection of RSV genome sequences from a single African location allowed a comparison of local versus global RSV evolution patterns. Important for vaccine design, the RSVA variants observed in a small region of Kenya appear to be in equilibrium with global variants. The same was not observed for RSVB. Possibly, RSVB variants may spread less efficiently, with a higher fraction of variants observed to be specific for Kilifi and not detected in other parts of the world. This pattern is consistent with RSVB as a less transmissible infection than RSVA (4, 12). However, there are fewer global sequences available for RSVB, so while the Kilifi RSVB variants appear to be unique, this could be a consequence of less surveillance and documentation of RSVB variation globally. Future work will help clarify this phenomenon, as it may have strong consequences on the efficacy of any RSV vaccine used locally.
ACKNOWLEDGMENTS
The work was supported by the Wellcome Trust Sanger Institute, the Wellcome Trust (084633, 100542, and 102975), and the European Community's Seventh Framework Programme (FP7/2007–2013) under the project EMPERIE, European Community grant agreement 223498. The KEMRI-Wellcome Trust Research Programme is supported by core funding from the Wellcome Trust (077092).
REFERENCES
- 1.Berkley JA, Munywoki P, Ngama M, Kazungu S, Abwao J, Bett A, Lassauniere R, Kresfelder T, Cane PA, Venter M, Scott JA, Nokes DJ. 2010. Viral etiology of severe pneumonia among Kenyan infants and children. JAMA 303:2051–2057. doi: 10.1001/jama.2010.675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Collins PL, Melero JA. 2011. Progress in understanding and controlling respiratory syncytial virus: still crazy after all these years. Virus Res 162:80–99. doi: 10.1016/j.virusres.2011.09.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Nair H, Nokes DJ, Gessner BD, Dherani M, Madhi SA, Singleton RJ, O'Brien KL, Roca A, Wright PF, Bruce N, Chandran A, Theodoratou E, Sutanto A, Sedyaningsih ER, Ngama M, Munywoki PK, Kartasasmita C, Simoes EA, Rudan I, Weber MW, Campbell H. 2010. Global burden of acute lower respiratory infections due to respiratory syncytial virus in young children: a systematic review and meta-analysis. Lancet 375:1545–1555. doi: 10.1016/S0140-6736(10)60206-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cane PA. 2001. Molecular epidemiology of respiratory syncytial virus. Rev Med Virol 11:103–116. doi: 10.1002/rmv.305. [DOI] [PubMed] [Google Scholar]
- 5.Anderson LJ, Dormitzer PR, Nokes DJ, Rappuoli R, Roca A, Graham BS. 2013. Strategic priorities for respiratory syncytial virus (RSV) vaccine development. Vaccine 31(Suppl 2):B209–B215. doi: 10.1016/j.vaccine.2012.11.106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Glezen WP, Taber LH, Frank AL, Kasel JA. 1986. Risk of primary infection and reinfection with respiratory syncytial virus. Am J Dis Child 140:543–546. [DOI] [PubMed] [Google Scholar]
- 7.Henderson FW, Collier AM, Clyde WA Jr, Denny FW. 1979. Respiratory-syncytial-virus infections, reinfections and immunity. A prospective, longitudinal study in young children. N Engl J Med 300:530–534. [DOI] [PubMed] [Google Scholar]
- 8.Sullender WM. 2000. Respiratory syncytial virus genetic and antigenic diversity. Clin Microbiol Rev 13:1–15. doi: 10.1128/CMR.13.1.1-15.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cane PA, Matthews DA, Pringle CR. 1994. Analysis of respiratory syncytial virus strain variation in successive epidemics in one city. J Clin Microbiol 32:1–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Agoti CN, Mwihuri AG, Sande CJ, Onyango CO, Medley GF, Cane PA, Nokes DJ. 2012. Genetic relatedness of infecting and reinfecting respiratory syncytial virus strains identified in a birth cohort from rural Kenya. J Infect Dis 206:1532–1541. doi: 10.1093/infdis/jis570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Peret TC, Hall CB, Schnabel KC, Golub JA, Anderson LJ. 1998. Circulation patterns of genetically distinct group A and B strains of human respiratory syncytial virus in a community. J Gen Virol 79(Part 9):2221–2229. [DOI] [PubMed] [Google Scholar]
- 12.White LJ, Waris M, Cane PA, Nokes DJ, Medley GF. 2005. The transmission dynamics of groups A and B human respiratory syncytial virus (hRSV) in England & Wales and Finland: seasonality and cross-protection. Epidemiol Infect 133:279–289. doi: 10.1017/S0950268804003450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Trento A, Viegas M, Galiano M, Videla C, Carballal G, Mistchenko AS, Melero JA. 2006. Natural history of human respiratory syncytial virus inferred from phylogenetic analysis of the attachment (G) glycoprotein with a 60-nucleotide duplication. J Virol 80:975–984. doi: 10.1128/JVI.80.2.975-984.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.García O, Martin M, Dopazo J, Arbiza J, Frabasile S, Russi J, Hortal M, Perez-Brena P, Martínez I, García-Barreno B,Melero JA. 1994. Evolutionary pattern of human respiratory syncytial virus (subgroup A): cocirculating lineages and correlation of genetic and antigenic changes in the G glycoprotein. J Virol 68:5448–5459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cane P. 2007. Molecular epidemiology and evolution of RSV, p 89–113 InCane P. (ed), Respiratory syncytial virus, vol 1 Elsevier, Amsterdam, The Netherlands. [Google Scholar]
- 16.Collins PL, Graham BS. 2008. Viral and host factors in human respiratory syncytial virus pathogenesis. J Virol 82:2040–2055. doi: 10.1128/JVI.01625-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Katzov-Eckert H, Botosso VF, Neto EA, Zanotto PM. 2012. Phylodynamics and dispersal of HRSV entails its permanence in the general population in between yearly outbreaks in children. PLoS One 7:e41953. doi: 10.1371/journal.pone.0041953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kumaria R, Iyer LR, Hibberd ML, Simoes EA, Sugrue RJ. 2011. Whole genome characterization of non-tissue culture adapted HRSV strains in severely infected children. Virol J 8:372. doi: 10.1186/1743-422X-8-372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lee WJ, Kim YJ, Kim DW, Lee HS, Lee HY, Kim K. 2012. Complete genome sequence of human respiratory syncytial virus genotype A with a 72-nucleotide duplication in the attachment protein G gene. J Virol 86:13810–13811. doi: 10.1128/JVI.02571-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Malboeuf CM, Yang X, Charlebois P, Qu J, Berlin AM, Casali M, Pesko KN, Boutwell CL, DeVincenzo JP, Ebel GD, Allen TM, Zody MC, Henn MR, Levin JZ. 2013. Complete viral RNA genome sequencing of ultra-low copy samples by sequence-independent amplification. Nucleic Acids Res 41:e13. doi: 10.1093/nar/gks794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rebuffo-Scheer C, Bose M, He J, Khaja S, Ulatowski M, Beck ET, Fan J, Kumar S, Nelson MI, Henrickson KJ. 2011. Whole genome sequencing and evolutionary analysis of human respiratory syncytial virus A and B from Milwaukee, WI 1998–2010. PLoS One 6:e25468. doi: 10.1371/journal.pone.0025468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tan L, Coenjaerts FE, Houspie L, Viveen MC, van Bleek GM, Wiertz EJ, Martin DP, Lemey P. 2013. The comparative genomics of human respiratory syncytial virus subgroups A and B: genetic variability and molecular evolutionary dynamics. J Virol 87:8213–8226. doi: 10.1128/JVI.03278-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tan L, Lemey P, Houspie L, Viveen MC, Jansen NJ, van Loon AM, Wiertz E, van Bleek GM, Martin DP, Coenjaerts FE. 2012. Genetic variability among complete human respiratory syncytial virus subgroup A genomes: bridging molecular evolutionary dynamics and epidemiology. PLoS One 7:e51439. doi: 10.1371/journal.pone.0051439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cotten M, Petrova V, Phan MV, Rabaa MA, Watson SJ, Ong SH, Kellam P, Baker S. 2014. Deep sequencing of norovirus genomes defines evolutionary patterns in an urban tropical setting. J Virol 88:11056–11069. doi: 10.1128/JVI.01333-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cotten M, Lam TT, Watson SJ, Palser AL, Petrova V, Grant P, Pybus OG, Rambaut A, Guan Y, Pillay D, Kellam P, Nastouli E. 2013. Full-genome deep sequencing and phylogenetic analysis of novel human betacoronavirus. Emerg Infect Dis 19:736–742. doi: 10.3201/eid1905.130057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cotten M, Watson SJ, Kellam P, Al-Rabeeah AA, Makhdoom HQ, Assiri A, Al-Tawfiq JA, Alhakeem RF, Madani H, AlRabiah FA, Al Hajjar S, Al-nassir WN, Albarrak A, Flemban H, Balkhy HH, Alsubaie S, Palser AL, Gall A, Bashford-Rogers R, Rambaut A, Zumla AI, Memish ZA. 2013. Transmission and evolution of the Middle East respiratory syndrome coronavirus in Saudi Arabia: a descriptive genomic study. Lancet 382:1993–2002. doi: 10.1016/S0140-6736(13)61887-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Agoti CN, Gitahi CW, Medley GF, Cane PA, Nokes DJ. 2013. Identification of group B respiratory syncytial viruses that lack the 60-nucleotide duplication after six consecutive epidemics of total BA dominance at coastal Kenya. Influenza Other Respir Viruses 7:1008–1012. doi: 10.1111/irv.12131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nokes DJ, Ngama M, Bett A, Abwao J, Munywoki P, English M, Scott JA, Cane PA, Medley GF. 2009. Incidence and severity of respiratory syncytial virus pneumonia in rural Kenyan children identified through hospital surveillance. Clin Infect Dis 49:1341–1349. doi: 10.1086/606055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Watson SJ, Welkers MR, Depledge DP, Coulter E, Breuer JM, de Jong MD, Kellam P. 2013. Viral population analysis and minority-variant detection using short read next-generation sequencing. Philos Trans R Soc Lond B Biol Sci 368:20120205. doi: 10.1098/rstb.2012.0205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS, Lesin VM, Nikolenko SI, Pham S, Prjibelski AD, Pyshkin AV, Sirotkin AV, Vyahhi N, Tesler G, Alekseyev MA, Pevzner PA. 2012. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol 19:455–477. doi: 10.1089/cmb.2012.0021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Katoh K, Toh H. 2008. Recent developments in the MAFFT multiple sequence alignment program. Brief Bioinform 9:286–298. doi: 10.1093/bib/bbn013. [DOI] [PubMed] [Google Scholar]
- 32.Edgar RC. 2010. Search and clustering orders of magnitude faster than BLAST. Bioinformatics 26:2460–2461. doi: 10.1093/bioinformatics/btq461. [DOI] [PubMed] [Google Scholar]
- 33.Eshaghi A, Duvvuri VR, Lai R, Nadarajah JT, Li A, Patel SN, Low DE, Gubbay JB. 2012. Genetic variability of human respiratory syncytial virus A strains circulating in Ontario: a novel genotype with a 72 nucleotide G gene duplication. PLoS One 7:e32807. doi: 10.1371/journal.pone.0032807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Peret TC, Hall CB, Hammond GW, Piedra PA, Storch GA, Sullender WM, Tsou C, Anderson LJ. 2000. Circulation patterns of group A and B human respiratory syncytial virus genotypes in 5 communities in North America. J Infect Dis 181:1891–1896. doi: 10.1086/315508. [DOI] [PubMed] [Google Scholar]
- 35.Venter M, Madhi SA, Tiemessen CT, Schoub BD. 2001. Genetic diversity and molecular epidemiology of respiratory syncytial virus over four consecutive seasons in South Africa: identification of new subgroup A and B genotypes. J Gen Virol 82:2117–2124. [DOI] [PubMed] [Google Scholar]
- 36.Drummond AJ, Suchard MA, Xie D, Rambaut A. 2012. Bayesian phylogenetics with BEAUti and the BEAST 1.7. Mol Biol Evol 29:1969–1973. doi: 10.1093/molbev/mss075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wertz GW, Collins PL, Huang Y, Gruber C, Levine S, Ball LA. 1985. Nucleotide sequence of the G protein gene of human respiratory syncytial virus reveals an unusual type of viral membrane protein. Proc Natl Acad Sci U S A 82:4075–4079. doi: 10.1073/pnas.82.12.4075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Collins PL, Mottet G. 1992. Oligomerization and post-translational processing of glycoprotein G of human respiratory syncytial virus: altered O-glycosylation in the presence of brefeldin A. J Gen Virol 73(Part 4):849–863. [DOI] [PubMed] [Google Scholar]
- 39.Melero JA, Garcia-Barreno B, Martinez I, Pringle CR, Cane PA. 1997. Antigenic structure, evolution and immunobiology of human respiratory syncytial virus attachment (G) protein. J Gen Virol 78(Part 10):2411–2418. [DOI] [PubMed] [Google Scholar]
- 40.McLellan JS, Yang Y, Graham BS, Kwong PD. 2011. Structure of respiratory syncytial virus fusion glycoprotein in the postfusion conformation reveals preservation of neutralizing epitopes. J Virol 85:7788–7796. doi: 10.1128/JVI.00555-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Spann KM, Tran KC, Chi B, Rabin RL, Collins PL. 2004. Suppression of the induction of alpha, beta, and lambda interferons by the NS1 and NS2 proteins of human respiratory syncytial virus in human epithelial cells and macrophages [corrected]. J Virol 78:4363–4369. doi: 10.1128/JVI.78.8.4363-4369.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Spann KM, Tran KC, Collins PL. 2005. Effects of nonstructural proteins NS1 and NS2 of human respiratory syncytial virus on interferon regulatory factor 3, NF-kappaB, and proinflammatory cytokines. J Virol 79:5353–5362. doi: 10.1128/JVI.79.9.5353-5362.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Swedan S, Andrews J, Majumdar T, Musiyenko A, Barik S. 2011. Multiple functional domains and complexes of the two nonstructural proteins of human respiratory syncytial virus contribute to interferon suppression and cellular location. J Virol 85:10090–10100. doi: 10.1128/JVI.00413-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Liesman RM, Buchholz UJ, Luongo CL, Yang L, Proia AD, DeVincenzo JP, Collins PL, Pickles RJ. 2014. RSV-encoded NS2 promotes epithelial cell shedding and distal airway obstruction. J Clin Invest 124:2219–2233. doi: 10.1172/JCI72948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Agoti CN, Otieno JR, Gitahi CW, Cane PA, Nokes DJ. 2014. Rapid spread and diversification of respiratory syncytial virus genotype ON1, Kenya. Emerg Infect Dis 20:950–959. doi: 10.3201/eid2006.131438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Gupta R, Brunak S. 2002. Prediction of glycosylation across the human proteome and the correlation to protein function. Pac Symp Biocomput 7:310–322. [PubMed] [Google Scholar]
- 47.Julenius K, Molgaard A, Gupta R, Brunak S. 2005. Prediction, conservation analysis, and structural characterization of mammalian mucin-type O-glycosylation sites. Glycobiology 15:153–164. [DOI] [PubMed] [Google Scholar]
- 48.Steentoft C, Vakhrushev SY, Joshi HJ, Kong Y, Vester-Christensen MB, Schjoldager KT, Lavrsen K, Dabelsteen S, Pedersen NB, Marcos-Silva L, Gupta R, Bennett EP, Mandel U, Brunak S, Wandall HH, Levery SB, Clausen H. 2013. Precision mapping of the human O-GalNAc glycoproteome through SimpleCell technology. EMBO J 32:1478–1488. doi: 10.1038/emboj.2013.79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sun Z, Pan Y, Jiang S, Lu L. 2013. Respiratory syncytial virus entry inhibitors targeting the F protein. Viruses 5:211–225. doi: 10.3390/v5010211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Swanson KA, Settembre EC, Shaw CA, Dey AK, Rappuoli R, Mandl CW, Dormitzer PR, Carfi A. 2011. Structural basis for immunization with postfusion respiratory syncytial virus fusion F glycoprotein (RSV F) to elicit high neutralizing antibody titers. Proc Natl Acad Sci U S A 108:9619–9624. doi: 10.1073/pnas.1106536108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Breslauer KJ, Frank R, Blocker H, Marky LA. 1986. Predicting DNA duplex stability from the base sequence. Proc Natl Acad Sci U S A 83:3746–3750. doi: 10.1073/pnas.83.11.3746. [DOI] [PMC free article] [PubMed] [Google Scholar]