ABSTRACT
Vaccines are used in integrated control strategies to protect poultry against H5N1 high-pathogenicity avian influenza (HPAI). H5N1 HPAI was first reported in Indonesia in 2003, and vaccination was initiated in 2004, but reports of vaccine failures began to emerge in mid-2005. This study investigated the role of Indonesian licensed vaccines, specific vaccine seed strains, and emerging variant field viruses as causes of vaccine failures. Eleven of 14 licensed vaccines contained the manufacturer's listed vaccine seed strains, but 3 vaccines contained a seed strain different from that listed on the label. Vaccines containing A/turkey/Wisconsin/1968 (WI/68), A/chicken/Mexico/28159-232/1994 (Mex/94), and A/turkey/England/N28/1973 seed strains had high serological potency in chickens (geometric mean hemagglutination inhibition [HI] titers, ≥1:169), but vaccines containing strain A/chicken/Guangdong/1/1996 generated by reverse genetics (rg; rgGD/96), A/chicken/Legok/2003 (Legok/03), A/chicken/Vietnam/C57/2004 generated by rg (rgVN/04), or A/chicken/Legok/2003 generated by rg (rgLegok/03) had lower serological potency (geometric mean HI titers, ≤1:95). In challenge studies, chickens immunized with any of the H5 avian influenza vaccines were protected against A/chicken/West Java/SMI-HAMD/2006 (SMI-HAMD/06) and were partially protected against A/chicken/Papua/TA5/2006 (Papua/06) but were not protected against A/chicken/West Java/PWT-WIJ/2006 (PWT/06). Experimental inactivated vaccines made with PWT/06 HPAI virus or rg-generated PWT/06 low-pathogenicity avian influenza (LPAI) virus seed strains protected chickens from lethal challenge, as did a combination of a commercially available live fowl poxvirus vaccine expressing the H5 influenza virus gene and inactivated Legok/03 vaccine. These studies indicate that antigenic variants did emerge in Indonesia following widespread H5 avian influenza vaccine usage, and efficacious inactivated vaccines can be developed using antigenic variant wild-type viruses or rg-generated LPAI virus seed strains containing the hemagglutinin and neuraminidase genes of wild-type viruses.
IMPORTANCE H5N1 high-pathogenicity avian influenza (HPAI) virus has become endemic in Indonesian poultry, and such poultry are the source of virus for birds and mammals, including humans. Vaccination has become a part of the poultry control strategy, but vaccine failures have occurred in the field. This study identified possible causes of vaccine failure, which included the use of an unlicensed virus seed strain and induction of low levels of protective antibody because of an insufficient quantity of vaccine antigen. However, the most important cause of vaccine failure was the appearance of drift variant field viruses that partially or completely overcame commercial vaccine-induced immunity. Furthermore, experimental vaccines using inactivated wild-type virus or reverse genetics-generated vaccines containing the hemagglutinin and neuraminidase genes of wild-type drift variant field viruses were protective. These studies indicate the need for surveillance to identify drift variant viruses in the field and update licensed vaccines when such variants appear.
INTRODUCTION
Since 1959, there have been 35 reported epizootics of high-pathogenicity avian influenza (HPAI) in poultry, of which the majority have been handled using stamping-out (culling) strategies for control, which have mostly led to eradication in less than a year (1, 2). However, vaccines were added as a control tool to augment stamping out in five epizootics: (i) the H5N2 HPAI virus epizootic in Mexico (1995), (ii) the H7N3 HPAI virus epizootic in Pakistan (1995 to present), (iii) the H5N1 HPAI virus epizootic in multiple countries of Asia, Africa, and Europe (2002 to present), (iv) the H7N7 HPAI virus epizootic in North Korea (2005), and (v) the H7N3 HPAI virus epizootic in Mexico (2012 to present). In total, 15 countries have publically utilized poultry vaccination in HPAI control programs either as a preventative measure before HPAI affected poultry in the country, as an emergency measure to limit spread among poultry farms in the face of an acute outbreak, or as a routine nationwide measure when the HPAI virus became endemic (1). Over 113 billion doses of vaccine were used in poultry between 2002 and 2010, with 99% being used in the routine national vaccination programs of China, Vietnam, Indonesia, and Egypt against H5N1 HPAI virus (1, 3). Furthermore, the first outbreaks of H5N1 HPAI virus in China, Indonesia, Vietnam, and Egypt were identified in mid-1996 (4), mid-2003 (5), December 2003 (6), and February 2006 (7), respectively, and the virus became enzootic in national poultry populations before national vaccination programs were implemented in mid-2004, June 2004 (5), October 2005 (8), and March 2006 (7), respectively (1, 5).
Proper application of high-potency vaccines reduces the number of HPAI virus-susceptible poultry; increases their resistance to HPAI virus infection, disease, and death; and reduces the amount of virus that immune but infected poultry excrete (9). In the field, this translates into reduced environmental contamination and reduced farm-to-farm spread (1, 10–13). Furthermore, the integration of H5N1 vaccine usage with other control components in poultry has been associated with a reduction in human cases in Vietnam and Hong Kong and a lack of H5N1 HPAI outbreaks on farms on which poultry were fully vaccinated (10). When initially assessed in the 1990s, diverse H5 and H7 vaccines provided broad protection in poultry against challenge by diverse H5 and H7 HPAI viruses, respectively (14–20). For example, chickens vaccinated with inactivated vaccines using the 1968 H5N9, 1981 H5N2, and 1994 H5N2 low-pathogenicity avian influenza (LPAI) viruses from North America and the 1997 H5N3 virus from Asia as vaccine seed strains were protected from death and had reduced virus replication and shedding from their respiratory and gastrointestinal tracts after challenge by several different H5N1 HPAI viruses (14, 18).
However, by mid-2005, reports of vaccine failures emerged from the field in Indonesia (11). The cause of such failures was unknown, but vaccine quality, seed strain selection, and antigen content, as well as field application issues, could result in vaccine failures. The purpose of this study was to investigate the potential contribution that the 15 different H5 avian influenza vaccines registered in Indonesia during 2007 had in such vaccine failures, specifically, the influence of potency, vaccine seed strain selection, and emergence of variant field viruses, and to determine if serological tests can be used to predict protection in the field.
MATERIALS AND METHODS
Vaccines.
Fifteen commercially available inactivated H5 whole-virus vaccines adjuvanted in proprietary oil emulsions (Table 1), a live recombinant fowl poxvirus (rFPV) vaccine genetically engineered to contain the hemagglutinin (HA) gene insert from A/turkey/Ireland/1378/1983 (H5H9) (rFPV-AI-H5; Trovac; Merial Inc., Gainesville, GA, USA), and two experimental inactivated vaccines were examined. Each different commercial vaccine is represented by a code letter (A to O) in Table 1. The experimental vaccines used the wild-type A/chicken/West Java/PWT-WIJ/2006 (PWT/06) H5N1 HPAI virus field isolate and a strain generated by reverse genetics (rg) containing the HA and neuraminidase (NA) gene segments from PWT/06 and six internal gene segments from the A/Puerto Rico/8/1934(H1N1) (PR8) vaccine strain. The HA gene segment was altered to have a low-pathogenicity (LP) proteolytic cleavage site, as previously described (21). Both of the seed strains were grown in 9- to 11-day-old embryonating chicken eggs. The low-pathogenicity phenotype of the PWT/06 strain generated by rg (the rgPWT/06 strain) was assessed for in vitro plaque-forming ability in cells of the MDCK cell line in the absence and presence of exogenous l-1-tosylamide-2-phenylmethyl chloromethyl ketone (TPCK)-treated trypsin. The pathogenicity potential for specific-pathogen-free (SPF) White Leghorn (WL) chickens was assessed by use of the intravenous pathogenicity index according to World Organization for Animal Health (OIE) guidelines (22–24). Experimental vaccines were prepared by inactivation of seed viruses with 0.1% beta-propiolactone and made into an oil-in-water emulsion (OE) vaccine, as previously described (25). The oil phase consisted of 36 ml mineral oil, 3 ml Span 80 (Sigma-Aldrich, St. Louis, MO), and 1 ml Tween 80 (Sigma-Aldrich, St. Louis, MO).
TABLE 1.
Fifteen commercial inactivated oil-emulsified H5 avian influenza vaccines for poultry
| Vaccine code | Manufacturer's purported seed strain | Most closely related isolatea | % amino acid similarityb |
|---|---|---|---|
| A | A/turkey/England/N28/1973 (H5N2) | A/turkey/England/N28/1973 (H5N2) | 100 |
| B | A/turkey/England/N28/1973 (H5N2) | A/turkey/England/N28/1973 (H5N2) | 99.1 |
| C | rg-generated Indonesian virus | A/chicken/Legok/2003 (H5N1) | 98.2 |
| D | A/turkey/England/N28/1973 (H5N2) | A/goose/Guangdong/1/96 (H5N1) | 99.3 |
| E | A/chicken/Mexico/232/1994 (H5N2) | A/chicken/Hidalgo/28159-232/1994 (H5N2) | 100 |
| F | A/turkey/Wisconsin/1968 (H5N9) (WI/68) | A/turkey/Wisconsin/1968 (H5N9) | 100 |
| G | A/turkey/England/N28/1973 (H5N2) | A/goose/Guangdong/1/1996 (H5N1) | 100 |
| H | A/turkey/England/N28/1973 (H5N2) | A/goose/Guangdong/1/1996 (H5N1) | 99 |
| I | A/chicken/Mexico/232/1994 (H5N2) | Not determined due to the poor quality of the RNA extracted | |
| J | A/chicken/Mexico/232/1994 (H5N2) | A/chicken/Hidalgo/28159-232/1994 (H5N2) | 100 |
| K | A/chicken/Legok/2003 (H5N1) | A/chicken/Legok/2003 (H5N1) | 98.9 |
| L | A/chicken/Legok/2003 (H5N1) | A/chicken/Legok/2003 (H5N1) | 99.4 |
| M | A/chicken/Mexico/232/1994 (H5N2) | A/chicken/Hidalgo/28159-232/1994 (H5N2) | 100 |
| N | A/goose/Guangdong/1/1996 (H5N1) | A/goose/Guangdong/1/1996 (H5N1) | 99.6 |
| O | rg-generated A/duck/Vietnam/2004 (H5N3) | A/chicken/Vietnam/C57/2004 (H5) | 99.3 |
The most closely related isolate was determined on the basis of extraction of RNA from the vaccine, sequencing, and determination of the lineage of HA by BLAST analysis. The indicated strain represents the virus or closely related viruses within the same lineage. Seed strains were (i) the H5N9 USA LPAI virus seed strain A/turkey/Wisconsin/1968 (WI/68), (ii) the H5N2 Mexican LPAI virus seed strain A/chicken/Hidalgo (Mexico)/28159-232/1994 (Mex/94), (iii) the 1973 United Kingdom LPAI virus seed strain A/turkey/England/N28/1973 (Eng/73), (iv) the local Indonesian H5N1 HPAI seed strain A/chicken/Legok/2003 (Legok/03), (v) the Chinese reverse genetics-generated H5N1 LPAI virus seed strain A/goose/Guangdong/1/96 (rgGD/96), (vi) the Indonesian reverse genetics-generated H5N1 LPAI virus strain A/chicken/Legok/2003 (rgLegok/03), and (vii) the Vietnamese reverse genetics-generated H5N3 LPAI strain A/chicken/Vietnam/C57/2004 (rgVN/04).
Percent amino acid similarity between the deduced protein sequence of the vaccine seed strain and the closest relative.
Identification of seed strains in commercial inactivated vaccines.
RNA was extracted from the commercial inactivated vaccines, and then a region of the HA from nucleotides 645 to 1082 was amplified by reverse transcriptase PCR (RT-PCR), gel extracted, and sequenced as previously described (26). The most closely related isolate was identified by a nucleotide BLAST search.
Vaccine potency.
Each vaccine was injected subcutaneously into the nape of the neck between the two scapulae of each of 10 3-week-old SPF WL chickens, using the manufacturer's recommended dose. At 3 weeks postvaccination, blood was collected from the ulnar vein and processed to serum. The serological response was determined by hemagglutination inhibition (HI) assays using individual vaccine viruses as the antigen on the basis of most closely related isolate (Table 1), as previously described (27).
Challenge virus selection: genetic and antigenic methods.
The sequences of available H5N1 HPAI viruses from Indonesia were initially characterized by use of a BLAST search to find the most similar sequences in GenBank. Phylogenetic analysis was performed by aligning the deduced amino acid sequences by use of the Clustal V program (Lasergene, v.7.1; DNAStar, Madison, WI). Bayesian Evolutionary Analysis Sampling trees were constructed with merged duplicate runs with the BEAST (v.1.4.8) program [14] using the Blosum62 substitution, the Gamma + invariant site heterogeneity model, a relaxed lognormal clock, a Yule Process tree prior with default operators with the unweighted-pair group method using average linkages starting tree, and a Markov chain Monte Carlo length of 106. Three H5N1 HPAI viruses were selected for additional antigenic and challenge studies on the basis of sequence diversity and geographic separation: A/chicken/West Java/SMI-HAMD/2006 (H5N1) (SMI-HAMD/06), A/chicken/Papua/TA5/2006 (H5N1) (Papua/06), and PWT/06.
Initial antigenic analysis was performed utilizing sera collected 2 weeks after the last vaccination from groups of three adult SPF WL chickens that had been hyperimmunized with 1 ml of vaccine given subcutaneously in the nape of the neck three times at 3-week intervals. Five vaccines containing one of the following seed strains were used: A/chicken/Legok/2003 (H5N1) (Legok/03), A/chicken/Guangdong/1/1996 (H5N1) generated by rg (rgGD/96), A/turkey/England/N28/1973 (H5N2) (Eng/73), A/chicken/Mexico/23294/1994 (H5N2) (Mex/94), and A/turkey/Wisconsin/1968 (H5N9) (WI/68). Subtype-specific antibodies were assessed in HI tests using the homologous vaccine antigen. One chicken from each group which had an initial HI titer of 1:512 or 1:1,024 was selected for further testing using the HI antigen from the three Indonesian strains selected from the genetic analysis, as described above.
For more detailed examination of the antigenic relationships among the viruses, 3-week-old SPF chickens each received a single immunization with 1 of 14 genetically and antigenically diverse H5 avian influenza viruses, and antisera were collected at 3 weeks postvaccination. Cartographic analysis was conducted as previously described (28–30) using the standard HI assay (31) with all the immunizing antigens and sera. The immunizing viruses included WI/68, Eng/73, A/turkey/1378/Ireland/1983 (H5N8) (Ireland/83), Mex/94, A/mallard/Netherlands/3/1999 (H5N2), A/mallard/Sweden/49/2002 (H5N9), A/mallard/Sweden/21/2002 (H5N2), A/mallard/Sweden/7/2002 (H5N2), and H5N1 HPAI virus strains A/chicken/Hong Kong/220/1997, Legok/03, A/Vietnam/1203/2004, SMI-HAMD/06, Papua/06, and PWT/06.
All H5N1 HPAI viruses were handled in a biosafety level 3 enhanced (BSL-3E) laboratory, and animal studies were conducted under BSL-3E containment in accordance with the guidelines of the U.S. Department of Agriculture (USDA) (available at http://www.afm.ars.usda.gov/ppweb/PDF/242-01M.pdf) and in accordance with the requirements of the USDA-Centers for Disease Control and Prevention select agent programs.
Vaccine efficacy.
To determine the efficacies of the vaccines, three challenge studies were conducted.
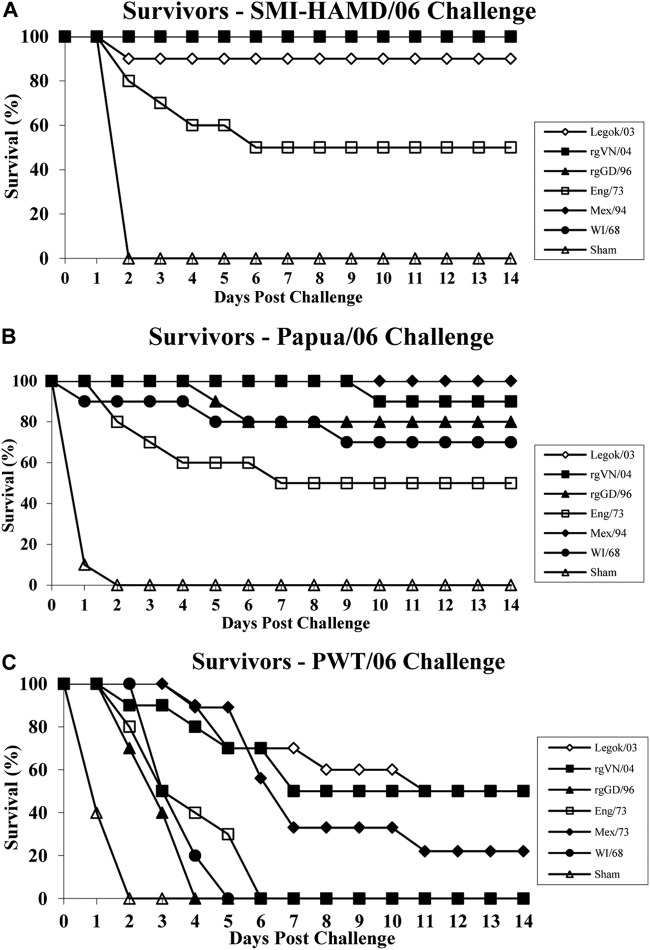
The first challenge study, to determine the protection provided by six vaccine seed strains when chickens were challenged with the three genetically and antigenically divergent field viruses (PWT/06, Papua/06, and SMI-HAMD/06), was performed using 30 3-week-old SPF WL chickens for each seed strain (Legok/03 [vaccine K], A/chicken/Vietnam/C57/2004 [H5N3] generated by rg [rgVN/04; vaccine O], rgGD/96 [vaccine N], Eng/73 [vaccine B], Mex/94 [vaccine E], and WI/68 [vaccine F]). The chickens were divided into groups of 10 birds each and intranasally challenged with 106 mean (50%) chicken embryo infectious doses (EID50s) of SMI-HAMD/06, Papua/06, or PWT/06 virus at 3 weeks postvaccination. Ten sham-vaccinated chickens (which were injected with noninfectious allantoic fluid) were also challenged with each of the three H5N1 HPAI viruses.
A second challenge study was performed to determine if commercial vaccines with the same seed strain but different proprietary emulsion formulations protected the chickens against an antigenic variant H5N1 HPAI virus, PWT/06, differently. Groups of 10 3-week-old SPF WL chickens were immunized with the vaccines containing seed strains Eng/73 (vaccines A and B), rg-generated Legok/03 (rgLegok/03) or Legok/03 (vaccines C, K, and L), rgGD/96 (vaccines D, G, H, and N), and Mex/94 (vaccines E, I, J, and M), and a group that was sham vaccinated (with vaccine R) was included. At 3 weeks postvaccination, the chickens were intranasally challenged with 106 EID50s of PWT/06 virus.
A third challenge study was performed to determine if the PWT/06 homologous vaccine or a combination of existing licensed vaccines would protect against the three Indonesian H5N1 HPAI viruses, PWT/06, Papua/06, and SMI-HAMD/06. Groups of 10 3-week-old SPF WL chickens that were immunized with in-house experimental vaccines containing inactivated wild-type PWT/06 (vaccine P) or rgPWT/06 (vaccine Q) or a combination of commercial live rFPV-AI-H5 (given at 1 day of age by subcutaneous route) and inactivated Legok/03 (vaccine V) (3 weeks of age) vaccines were used. A group that was sham vaccinated (vaccine U) was included. At 3 weeks postvaccination, the wild-type PWT/06 (vaccine P) and rFPV-AI-H5–Legok/03 groups were intranasally challenged with 106 EID50s of SMI-HAMD/06, Papua/06, or PWT/06 HPAI virus, and the rgPWT/06 group was challenged with PWT/06 only.
In all three studies, the chickens were observed daily for 14 days for any clinical signs or deaths. At 2 days postchallenge (DPC), oropharyngeal swabs were collected and processed for virus presence and virus was quantified using quantitative real-time reverse transcriptase PCR as previously described (32). Survivors were terminated by intravenous administration of pentobarbital (5 mg/kg of body weight).
Statistical analysis.
All statistical functions were carried out using the QI Macros for Excel program (KnowWare International, Inc., Denver, CO) and the Statistics Calculator program (StatPac Inc., Bloomington, MN).
RESULTS
Case report of high-pathogenicity avian influenza outbreaks in vaccinated chicken flocks.
The first reports of H5N1 HPAI virus in Indonesia emerged from the field in mid-2003 (5, 33). As the virus spread in commercial operations, vaccination using locally produced vaccines based on Legok/03 or imported available commercial vaccines from Mexico was implemented (34); use of the latter vaccine was based on the broad protection provided by H5 North American vaccine seed strains against Hong Kong H5N1 challenge viruses from 1997 (14). In December 2005, a broiler breeder flock in one production house in West Java, Indonesia, that had been vaccinated four times with inactivated Mex/94 vaccine began to experience increased mortality, and total mortality peaked at over 14%. The remaining chickens were culled. The breeders had been individually handled and vaccinated with inactivated vaccine and at the time of the outbreak had geometric mean titers (GMTs) averaging above 128, signifying that vaccine had been administered to the flock and had produced reasonable titers that should have been protective against H5N1 HPAI virus challenge, suggesting a different cause for the lack of protection.
Vaccine seed strain identification.
Fourteen of 15 vaccines contained a sufficient quantity and good quality of RNA upon extraction for RT-PCR and sequencing to identify the vaccine seed strain. Eleven of these 14 vaccines were formulated with one of seven Indonesian government-licensed seed strain viruses that were consistent with the manufacturers' claims listed on the label or in the company literature (Table 1). However, the remaining three vaccines were purported to contain the Eng/73 H5N2 seed strain, as listed on the label and licensed, but they actually contained the rgGD/96 H5N1 seed strain. Chinese manufacturers produced vaccines with the Eng/73 H5N2 seed strain from 2004 to 2006, while vaccines with rgGD/96 were manufactured from 2006 to 2008 (35), suggesting that the three rgGD/96 vaccines used in this study and manufactured in 2007 were mislabeled with the H5N2 label to meet the licensing and importing requirements of Indonesia.
Potency of commercial vaccines.
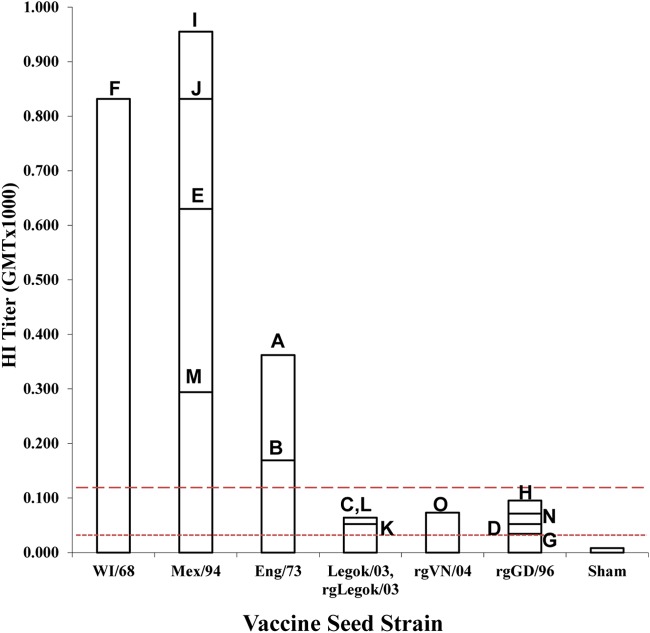
To determine if the 15 inactivated commercial vaccines had the potency or antigenic mass sufficient to be protective, a functional assay of the immune response in SPF WL hens receiving a single vaccination was completed (36). Three weeks after a single immunization, the mean HI serological titers obtained using homologous vaccine seed strain antigen for each vaccine group ranged from 34 to 955 (Fig. 1). The WI/68, Eng/73, and Mex/94 vaccines induced the highest mean titers (GMTs, 169 to 955), while Legok/03, rgLegok/03, rgVN/04, and rgGD/96 induced the lowest titers (GMTs, 34 to 97). Using the minimum titer for protection from homologous lethal challenge (24, 37), all the vaccines met the minimum potency of a GMT of ≥32, which has previously been associated with the prevention of mortality (24). However, a higher minimum potency of a GMT of ≥128 was associated with a reduction in challenge virus replication and shedding, which are necessary to reduce environmental contamination and spread of the virus (24). All vaccines using the WI/68, Mex/94, and Eng/73 seed strains met this higher serological standard, while vaccines using the Legok/03, rgLegok/03, rgVN/04, and rgGD/96 seed strains did not.
FIG 1.
Indirect potency test based on the mean serological titers obtained following a single immunization of 3-week-old chickens with 15 different vaccines and by use of the homologous vaccine antigen in the HI test. Threshold lines for the minimum titers that protect from mortality (dotted line; titer, 1:32) and challenge virus shedding from the oropharynx (dashed line; titer, 1:128) are included.
Challenge virus selection.
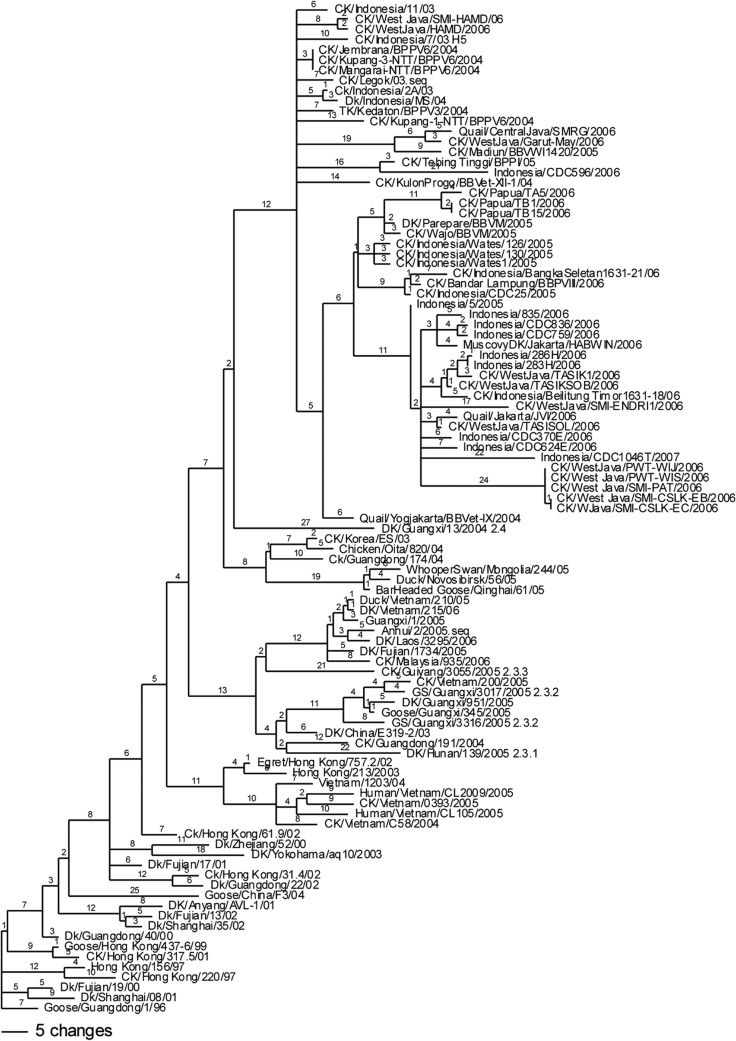
Challenge viruses were selected from available field viruses on the basis of a comparison of the divergence of HA1 sequences and geographic separation (Fig. 2). A phylogenetic tree was constructed with representative H5N1 isolates from Indonesia and other countries (Fig. 2). The earliest reported isolates from Indonesia were recovered in 2003, when there was evidence of only a single introduction of virus that was genetically unique from other outbreak viruses at the time and that was designated a unique genotype, clade 2.1 (33). Because Indonesia is composed of many islands that facilitate geographic isolation of virus populations, the H5N1 virus had further evolved into multiple third-order clades (clades 2.1.1, 2.1.2, and 2.1.3) by 2007 and fourth-order clades (clades 2.1.3.1, 2.1.3.2, 2.1.3.2a, and 2.1.3.3) by 2011, on the basis of pairwise comparisons of nucleotide and amino acid sequences and further support from HI antigenic data (38).
FIG 2.
Phylogenetic tree of representative H5N1 isolates from Indonesia and other countries. DK and Dk, duck; TK, turkey; CK and Ck, chicken; GS, goose. The numbers on nodes are bootstrap values.
Challenge viruses were selected from three different subclades. A/chicken/West Java/SMI-HAMD/06 (SMI-HAMD/06) was selected from the cluster of earlier viruses thought to be closely related to the progenitor of Indonesian H5N1 viruses and that fell within clade 2.1.1 (39, 40). The amino acid sequence of the SMI-HAMD/06 virus had 99.8% identity to that of the local Indonesian H5N1 HPAI vaccine seed strain, Legok/03 (Table 2). An additional two strains were selected from clade 2.1.3, which has emerged to become the predominant clade since 2005 (39). A/chicken/Papua/TA5/2006 (Papua/06) was representative of subclade 2.1.3.1, and A/chicken/West Java/PWT-WIJ/2006 (PWT/06) was representative of subclade 2.1.3.2 (39, 40). Papua/06 and PWT/06 were divergent viruses, having a 3.3% difference in the HA nucleotide sequence. PWT/06 was isolated from vaccinated poultry involved in the H5N1 HPAI outbreak on the broiler breeder farm described in the case report presented above.
TABLE 2.
Pairwise amino acid comparison of full-length and HA1 proteins of the H5 proteins from challenge vaccine seed strains
| Vaccine seed strain | % amino acid sequence identity of full-length protein/HA1 protein for the following challenge viruses: |
||
|---|---|---|---|
| SMI-HAMD/06 | Papua/06 | PWT/06 | |
| Legok/03 | 99.8/100 | 97.5/96.6 | 94.9/92.3 |
| VN/04 | 97.4/97.2 | 94.9/94.1 | 93.5/91.0 |
| GD/96 | 96.0/95.7 | 93.8/93.2 | 92.1/89.5 |
| Eng/73 | 90.1/90.4 | 89.4/89.2 | 86.484.2 |
| Ireland/83 | 89.9/88.2 | 89.1/87.0 | 87.7/84.5 |
| WI/68 | 88.3/86.1 | 88.3/86.4 | 84.8/80.5 |
| Mex/94 | 86.7/85.4 | 86.2/84.8 | 82.8/79.3 |
Pairwise comparison of the three challenge viruses and seven H5 vaccine strains showed amino acid sequence variability of from 0 to 17.2% within the full-length hemagglutinin and 0 to 20.7% within the HA1 segment (Table 2).
Antigenic characterization.
For initial antigenic characterization of vaccine strains compared to Indonesian field viruses, chickens were hyperimmunized with five commercial vaccine seed strains (Legok/03, rgGD/96, Mex/94, Eng/73, and WI/68) and their sera were tested in HI assays with homologous antigens. Individual chicken serum samples with HI titers of 512 or 1,028 were compared for antigenic relatedness in HI tests using the three Indonesia challenge viruses (SMI-HAMD/06, Papua/06, and PWT/06) as the antigen. Only the Legok/03 antisera tested using SMI-HAMD/06 (clade 2.1.1) and Papua/06 (clade 2.1.3.1) as the antigens had titers similar to the titer of the homologous antigen. All other antisera had >3 log2 reductions in titers when the challenge virus antigens rather than homologous antigens were used. The lower that the HA1 relatedness (Table 2) between the viruses used to produce the sera and the test antigen was, the greater that the reduction in HI titers was (Tables 2 and 3). No HI titers were detected for rgGD/96, Mex/94, Eng/73, and WI/68 antisera when tested against the PWT/06 antigen, but low titers (titers, 8 to 32) were seen with Papua/06 antigens and intermediate titers (titers, 32 to 128) were seen with SMI-HAMD/06 antigens.
TABLE 3.
HI GMTs for adult hensa
| Serum | HI GMT for the following antigen (clade): |
|||
|---|---|---|---|---|
| Homologous | SMI-HAMD/06 (2.1.1) | Papua/06 (2.1.3.1) | PWT/06 (2.1.3.2) | |
| Legok/03 | 512 | 2,056 | 512 | 32 |
| rgGD/96 | 512 | 128 | 16 | 0 |
| Eng/73 | 512 | 64 | 32 | 0 |
| Mex/94 | 1,028 | 32 | 16 | 0 |
| WI/68 | 1,028 | 32 | 8 | 0 |
The hens were hyperimmunized with inactivated virus 3 times at 3-week intervals; the resulting sera were tested against the homologous vaccine seed strain antigen and three different challenge viruses.
To provide a more detailed antigenic analysis, sera with antibodies against 14 genetically and antigenically diverse H5 viruses were produced in chickens, the sera were tested in HI assays for activity against all 14 viruses, and a cartographic analysis was conducted (Fig. 3). The antigens from the classic H5 vaccine strains (WI/68, Eng/73, Ireland/83, and Mex/94) antigenically clustered together with the four wild bird H5 viruses. The antigenic relatedness decreased slightly from the eight classical H5 viruses at the root to the A/chicken/Hong Kong/220/1997 virus, and the antigenic relatedness with Legok/03, VN/04, SMI-HAMD/06, and Papua/06 progressively decreased. The virus that was the least antigenically related to the eight root viruses was PWT/06.
FIG 3.
Cartographic map of H5 avian influenza viruses produced with chicken antisera. The colored shapes (viruses) and open shapes (antisera) are relative positions adjusted such that the distances between viruses and antisera in the map represent the corresponding HI measurements with the least error. The blue fill represents the antigenic root from classic H5 influenza viruses, including four viruses of wild bird origin (A/mallard/Netherlands/3/1999 [MA/NETHERLANDS/3/1999], A/mallard/Sweden/49/2002 [MA/SWEDEN/49/2002], A/mallard/Sweden/7/2002 [MA/SWEDEN/7/2002], and A/mallard/Sweden/21/2002 [MA/SWEDEN/21/2002]) and four viruses of poultry origin (A/turkey/Wisconsin/1968 [TU/WISCONSIN/1968], A/turkey/England/N28/1973 (TU/ENGLAND/N28/1973], A/turkey/Ireland/1983 [TU/IRELAND/1983], and A/chicken/Mexico/23294/1994 [CH/Mexico/23294/1994]). The other solid colors represent six H5N1 Guangdong lineage viruses (A/chicken/Hong Kong/220/1997 [CH/HK/220/1997], A/chicken/Legok/2003 [CH/LEGOK/2003], A/Vietnam/1203/2004 [VN/1203/04], A/chicken/West Java/SMI-HAMD/06 [SMI-HAMD/06], A/chicken/Papua/TA5/06 [Papua/06], and A/chicken/West Java/PWT-WIJ/06 [PWT/06]). Since the relative positions of antigens and antisera are determined and both the vertical and horizontal axes represent antigenic distance, the orientation of the map within these axes is free. The spacing between grid lines is 1 unit of antigenic distance, corresponding to a 2-fold dilution of antiserum in the HI assay.
Numerous amino acid differences in the sequences of the HA1 protein were observed among the vaccine and challenge viruses (Fig. 4). Eight amino acid differences were observed at sites known to be important for antigenic recognition (41–48), including sites near the receptor binding site, and as previously reported (48), one potential glycosylation site whose sequence differed among the vaccine and challenge viruses was identified (Table 4). Three of the amino acids were specific to the Indonesian isolates (amino acid residues 124, 189, and 212). Four residues (amino acid residues 94, 140, 141, and 162) were specific to PWT/06 and Papua/06, which were antigenically the most distant from the classical H5 group, and vaccines prepared with classical H5 group strain GD/96 or VN/04 most poorly protected against infection with PWT/06 and Papua/06. The most antigenically distant isolate, PWT/06, had a unique amino acid signature (P74Q). The sequence of PWT/06 was different from that of all the other isolates at a pair of sites, residues 183 and 189, identified to be collectively important for antigenic changes in clade 2.1 (48). The sequence of this isolate was also unique at residue 185, one site of another important pair of residues (residues 133 and 185) previously identified by Koel et al. (48).
FIG 4.
Alignment of the HA1 amino acid sequences of the vaccine and challenge viruses used in these studies. Orange shading, amino acids identified to be important for antigenic recognition by previous studies (41, 42, 44, 45, 75); blue shading, amino acids identified to be part of the receptor binding site; asterisks, potential N-linked glycosylation sites; red arrow, the proteolytic cleavage site; boxes, the loop at position 130, the helix at position 190, and the loop at position 220. Note that amino acids at positions higher than position 138 appear to have numbering 1 greater than their typical numbering due to an insertion in the A/turkey/England/N28/1973 isolate.
TABLE 4.
Amino acid changes between vaccine and challenge viruses at sites critical for H5 antigenic structure
Positions refer to H5 numbering. The results for the initial vaccine strain used in Indonesia, Legok/03, are highlighted.
Efficacy of commercial vaccine seed strains.
Survival is the most common indicator of protection measured for determination of HPAI vaccine efficacy in studies in poultry, but other metrics of protection, such as prevention of challenge virus replication and shedding from respiratory and gastrointestinal tracts, are noteworthy since they are associated with reduced environmental contamination, reductions in contact transmission, and reductions in bird-to-bird and farm-to-farm transmission (1, 13, 49, 50). For the first efficacy study, the vaccine groups with the highest HI titer for each seed strain, Eng/73 (vaccine B), Mex/94 (vaccine E), WI/68 (vaccine F), Legok/03 (vaccine K), rgGD/96 (vaccine N), and rgVN/04 (vaccine O), were intranasally challenged with SMI-HAMD/06, PWT/06, and Papua/06. Using 80% survival as the minimum level of acceptable protection, none of the seed strains provided acceptable protection against PWT/06 challenge, but Legok/03, rgVN/04, rgGD/96, and Mex/94 provided protection resulting in ≥80% survival following virulent challenge with SMI-HAMD/06 and Papua/06, and WI/68 provided protection resulting in ≥80% survival following virulent challenge with SMI-HAMD/06 (Fig. 5). All six vaccine seed strains significantly reduced the number of chickens shedding SMI-HAMD/06 and Papua/06 challenge virus from the respiratory tract (oropharyngeal swab) at 2 DPC and significantly reduced the titers of challenge virus shed (Table 5). However, the reductions in challenge virus replication and shedding were not consistent across the different vaccine seed strains when the PWT/06 challenge virus was used; i.e., Eng/73 provided no measure of protection from shedding, and the other five seed strains provided partial to complete protection from shedding at 2 DPC.
FIG 5.
Survival of 3-week-old chickens vaccinated with each vaccine seed strain and challenged 3 weeks later with H5N1 HPAI viruses SMI-HAMD/06 (A), Papua/06 (B), and PWT/06 (C).
TABLE 5.
Detection of H5N1 HPAI virus shedding from oropharyngeal cavity at 2 DPC in chickens vaccinated with six different vaccine seed strains and challenged with one of three H5N1 HPAI viruses from Indonesia
| Vaccine seed strain | Vaccine code | SMI-HAMD/06 |
Papua/06 |
PWT/06 |
|||
|---|---|---|---|---|---|---|---|
| MDTa | Oral shedding at 2 DPCb | MDT | Oral Shedding at 2 DPC | MDT | Oral shedding at 2 DPC | ||
| Legok/03 | K | 2c | 0/10A (≤3.9A) | 0/10A (≤4.7A) | 6.6AB | 1/10A (5.68A) | |
| VN/04 | O | 1/10A (4.0A) | 6.0d | 0/10A (≤4.7A) | 5ABC | 2/10A (4.5B) | |
| rgGD/96 | N | 0/10A (≤3.9A) | 4.0d | 0/10A (≤4.7A) | 3.1C | NDe | |
| Eng/73 | B | 3.4A | 1/10A (4.0A) | 5.8A | 3/10A (5.1A) | 4BC | 8/10BC (6.13AC) |
| Mex/94 | E | 1/10A (4.0A) | 2/10A (4.8A) | 6.7AC | 0/10A (<4.50B) | ||
| WI/68 | F | 0/10A (≤3.9A) | 5.0AB | 1/10A (4.9A) | 3.7C | 4/10AC (5.40B) | |
| Sham | U | 2.0cA | 10/10B (7.1B) | 1.6cB | 10/10B (7.1B) | 1.7cD | 10/10B (6.3C) |
MDT, mean time to death (in days). Within each column there were statistically significantly differences (analysis of variance, P < 0.05), and different uppercase letters indicate significant differences between individual seeds for each challenge virus (Student's t test, P < 0.05).
Data represent the number of virus-positive chickens/total number of chickens (log10 mean virus titer). For number of virus-positive chickens/total number of chickens, different uppercase letters indicate significant differences (Fisher's exact test, P < 0.05). For oral shedding, there were statistically significant differences in virus titers within each column (analysis of variance, P < 0.05), and for log10 mean virus titers, different uppercase letters indicate statistically significant differences between each vaccine seed strain (Student's t test, P < 0.05). For statistical testing, samples without virus detection were given a titer of the lowest detection limit minus 0.1 log10.
The chickens lacked HI antibody.
Less than 3 chickens died; thus, the numbers were inadequate for statistical analysis.
ND, not done.
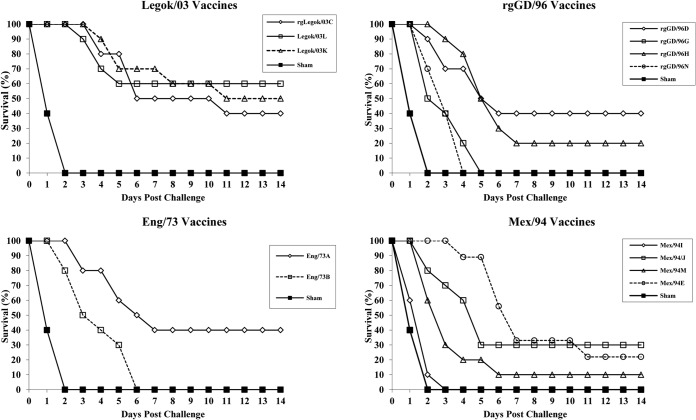
Since individual veterinary vaccine manufacturers use various proprietary oil emulsions and different quantities of hemagglutinin antigen, four vaccines containing seed strains rgLegok/03, Legok/03, Eng/73, and Mex/94 that were made by multiple manufacturers were tested for their ability to provide protection against PWT/06 challenge to determine if such variables would affect protection. The use of none of these additional vaccines resulted in ≥80% survival in vaccinated chickens (Fig. 6). The Legok/03 vaccines provided the best survival (40 to 60%), while all vaccines with rgGD/96, Eng/73, or Mex/94 seed strains provided ≤40% survival. Overall, there was no correlation between survival and HA antigen content, as measured by assay of indirect potency determined from the HI titers obtained using vaccine antigen (P = 0.87) (Table 6). However, a longer mean time to death (MDT) was associated with the presence of H5 HI antibodies, as determined by an HI test against the homologous vaccine seed strain antigen. Specifically, chickens that died and lacked HI antibodies (all sham-vaccinated chickens and one chicken vaccinated with Legok/03) had MDTs of <2 days, while chickens that died but that had measureable HI antibodies (four vaccine groups) exhibited prolonged MDTs (3.1 to 6.7 days), indicating incomplete protection (Table 6). Prolonged MDTs were also noted in vaccinated groups challenged with SMI-HAMD/06 and Papua/06 viruses (Table 5).
FIG 6.
Survival of 3-week-old chickens immunized with vaccines containing seed strains Eng/73, rgLegok/03 or Legok/03, rgGD/96, and Mex/94 and challenged 3 weeks later with antigenic variant H5N1 HPAI virus PWT/06.
TABLE 6.
Detection of H5N1 HPAI virus shedding from oropharyngeal cavity at 2 DPC by chickens vaccinated with four different vaccine seed strains in 2 to 4 different vaccine formulations and challenged with antigenic variant PWT/06 H5N1 HPAI virus from Indonesia
| Vaccine seed strain | Vaccine code | MDTa | Oral shedding at 2 DPCb | HI serology (GMT) with the following antigenc: |
|
|---|---|---|---|---|---|
| Vaccine seed straind | PWT/06 | ||||
| Eng/73 | A | 4.8A | 3/10A (4.83A) | 362A | 0 |
| Eng/73 | B | 4A | 8/10B (6.13B) | 169B | 0 |
| Sham | R | 1.4B | 10/10B (7.6C) | 0C | 0 |
| rgLegok/03 | C | 6.2A | 3/10A (4.80A) | 64A | 0A |
| Legok/03 | K | 6.6A | 2/10A (5.7B) | 52A | 0.3AB |
| Legok/03 | L | 4.0A | NDe | 64A | 1B |
| Sham | R | 1.4B | 10/10B (7.6C) | 0B | 0A |
| rgGD/96 | D | 4.0AB | 5/10A (5.36A) | 52AB | 0 |
| rgGD/96 | G | 3.1B | 5/10A (6.32B) | 34B | 0 |
| rgGD/96 | H | 5.1A | 2/10A (5.63A) | 97A | 0 |
| rgGD/96 | N | 3.1B | ND | 73A | 0 |
| Sham | R | 1.4C | 10/10B (7.6C) | 0C | 0 |
| Mex/94 | E | 6.7A | 0/10A (≤4.5A) | 630A | 0 |
| Mex/94 | I | 1.7B | 10/10B (7.3B) | 955A | 0 |
| Mex/94 | J | 3.7C | 2/10A (5.9C) | 832A | 0 |
| Mex/94 | M | 3.0C | ND | 294B | 0.1 |
| Sham | R | 1.4B | 10/10B (7.5B) | 0C | 0 |
MDT, mean time to death (in days). Within each vaccine seed strain group, MDTs were different (analysis of variance, P < 0.05), and different uppercase letters indicate statistically significant differences between individual vaccines within each vaccine seed strain group (Student's t test, P < 0.05).
Data represent the number of virus-positive chickens/total number of chickens (log10 mean virus titer). Oral shedding data were statistically significantly different within each column (analysis of variance, P < 0.05); different uppercase letters indicate statistically significant differences within the data for each virus strain for number of virus-positive chickens/total number of chickens (Fisher's exact test) and virus titer (Student's t test, P < 0.05). For testing of statistical significance, samples in which virus was not detected were given a titer of the lowest detection limit minus 0.1 log10.
Data were obtained at 3 weeks postvaccination. HI titers were statistically significantly different within each column (analysis of variance, P < 0.05); different uppercase letters indicate statistically significant differences in each vaccine seed strain group for HI titer (Student's t test, P < 0.05).
Correlation of hemagglutinin antigen content of vaccine assayed by an indirect potency test (i.e., HI serology using vaccine seed strain antigen) against survivability (R2 = 0.042, P = 0.87), percent shedding from the oropharynx (R2 = −0.646, P = 0.013), and the titer of challenge virus shed (R2 = −0.296, P < 0.001).
ND, not done.
In the Eng/73-, Legok/03-, and Mex/94-vaccinated groups challenged with PWT/06, there were significant differences in the number of chickens shedding challenge virus and/or the titer of virus shed within each vaccine group. Overall, there was an inverse correlation between the percentage of chickens shedding challenge virus and the antigen content of the vaccine (P = 0.013) and between the titer of the challenge virus being shed and the antigen content (P < 0.001) at 2 DPC (Table 6). These data suggest that any of the vaccine seed strains or vaccines would prolong the time to death, but unexpectedly, neither the different proprietary oil emulsions nor the higher hemagglutinin antigen content significantly improved survival. However, a higher HA antigen content did reduce virus replication and shedding from the oropharynx when measured at 2 DPC.
Experimental vaccines and vaccination protocols.
Because licensed vaccine seed strains provided inconsistent protection against the antigenic variant PWT/06 challenge virus, two experimental inactivated vaccines were made from this virus: the parent HPAI virus strain and a strain generated by rg and developed using the H5 HA gene (altered to contain an LP proteolytic cleavage site), the N1 NA gene from the parent PWT/06 virus, and six internal gene segments of the PR8 influenza A virus strain that allow the rescued virus to grow to high levels when grown in embryonating chicken eggs. The rgPWT/06 vaccine strain did not produce plaques in MDCK cells in the absence of exogenous TPCK-treated trypsin, which is consistent with a low-pathogenic virus. However, supplementation of agar medium with TPCK-treated trypsin resulted in the formation of distinct plaques in MDCK cells. In pathotyping experiments, the chickens inoculated with the rgPWT/06 strain did not show morbidity or mortality, resulting in an intravenous pathogenicity index of 0. In addition, a third group was added. The vaccination scheme used for that group has been implemented in the field and consists of a priming vaccination with a live commercial recombinant fowl poxvirus vaccine containing an insert of the H5 avian influenza virus (A/turkey/Ireland/1378/1983 [H5N9]) HA gene, followed by a booster vaccination with the available commercial inactivated Legok/03 vaccine. In this study, the Legok/03 vaccine provided the best protection among the licensed H5 poultry vaccines. Previously, the synergistic effect of a prime-boost vaccination on protection in poultry was demonstrated for H5N1 Eurasian HPAI viruses in ducks (51). The PWT/06, rgPWT/06, and rFPV-AI-H5–Legok/06 vaccines provided ≥80% survival against lethal challenge by the SMI-HAMD/06, Papua/06, and PWT/06 HPAI viruses (Fig. 7). In addition, all three vaccination groups had significant reductions in the number of chickens shedding challenge virus from the oropharynx, and the titers of virus shed were significantly reduced (Table 7).
FIG 7.
Survival of 3-week-old chickens immunized with experimental vaccines containing inactivated wild-type PWT/06 or rgPWT/06 or a combination of rFPV-AI-H5 (1 day of age) and Legok/03 at 3 weeks of age and challenged 3 weeks later with antigenic variant H5N1 HPAI virus PWT/06.
TABLE 7.
Detection of H5N1 HPAI virus shedding from oropharyngeal cavity at 2 DPC in chickens vaccinated with four different vaccine seed strains and challenged with one of three H5N1 HPAI viruses from Indonesia
| Vaccine seed strain | Vaccine code | SMI-HAMD/06 |
Papua/06 |
PWT/06 |
|||
|---|---|---|---|---|---|---|---|
| MDTa | Oral shedding at 2 DPCb | MDT | Oral Shedding at 2 DPC | MDT | Oral shedding at 2 DPC | ||
| PWT/06 | P | 1/10A (4.0A) | 5.0c | 2/10 (4.7) | NDd | ND | |
| rgPWT/06 | Q | ND | ND | ND | ND | 2.0e | 3/10A (3.5)A |
| rFPV-AI-H5–Legok/03 | V | 0/10A (≤3.9A) | 0/10 (≤4.7) | 9.5c | 1/10A (4.2)B | ||
| Sham | U | 2.0 | 10/10B (7.1B) | 1.6 | 10/10 (7.1) | 1.7 | 10/10B (6.3)C |
MDT, mean time to death (in days).
Data represent the number of virus-positive chickens/total number of chickens (log10 mean virus titer). For number of virus-positive chickens/total number of chickens, different uppercase letters indicate statistically significant differences (Fisher's exact test, P < 0.05). For oral shedding, there were statistically significant differences in virus titers within each column (analysis of variance, P < 0.05), and different uppercase letters indicate statistically significant differences for log10 mean virus titers between each vaccine seed strain (Student's t test, P < 0.05). For testing of statistical significance, samples in which virus was not detected were given a titer of the lowest detection limit minus 0.1 log10.
Two birds died, so the numbers were inadequate for statistical analysis.
ND, not done.
One bird died, so the numbers were inadequate for statistical analysis.
PPV of HI titers.
In predicting vaccine protection in poultry flocks, a quick assay is needed because challenge studies are expensive, cumbersome, and not time responsive. Comparison of the HI titers obtained using the vaccine or challenge virus antigen and protection parameters, such as survival or prevention of oropharyngeal shedding of challenge virus, could be a feasible approach for assessing protection in the field. The ideal serological predictor of protection would have a high sensitivity and a high specificity and would have a high positive predictive value (PPV) and a high negative predictive value (NPV); i.e., the values of all four parameters would be ≥95%. Overall in our study, the presence of HI antibodies using homologous vaccine virus as the HI antigen (GMT, ≥8) was a poor predictor of survival after a virulent challenge with the three antigenically diverse challenge viruses (a low PPV of 43%) (Fig. 3), but a lack of HI antibodies was good at predicting death (NPV = 96%) (Table 8). In contrast, when using the challenge virus as the HI antigen, HI-positive serology was good for predicting survival from a virulent challenge (PPV = 98%) and for preventing oropharyngeal virus shedding (PPV = 94%), but the lack of antibodies did not predict death or the shedding of high titers of virus (NPV = 66% and 42%, respectively). Furthermore, all chickens with HI titers of ≥32 (challenge virus antigen) survived, and those with titers of ≥64 did not shed the challenge virus on day 2 postinfection.
TABLE 8.
HI serological results compared to survival outcomes with sensitivities, specificities, PPVs, and NPVsa
| Test and HI antigen | No. of chickens with the following resultb: |
Sensitivity (%) | Specificity (%) | PPV (%) | NPV (%) | |||
|---|---|---|---|---|---|---|---|---|
| True positive | False negative | False positive | True negative | |||||
| Survival | ||||||||
| Challenge virus | ||||||||
| PWT/06 | 36 | 27 | 1 | 155 | 0.57 | 0.99 | 0.97 | 0.85 |
| SMI-HAMD/06 | 45 | 29 | 0 | 16 | 0.61 | 1.00 | 1.00 | 0.36 |
| Papua/06 | 28 | 43 | 1 | 18 | 0.39 | 0.95 | 0.97 | 0.30 |
| All viruses | 109 | 99 | 2 | 189 | 0.52 | 0.99 | 0.98 | 0.66 |
| Vaccine virus | ||||||||
| PWT/06 challenge | 62 | 1 | 113 | 42 | 0.98 | 0.27 | 0.35 | 0.98 |
| SMI-HAMD/06 challenge | 73 | 1 | 3 | 13 | 0.99 | 0.81 | 0.96 | 0.93 |
| Papua/06 challenge | 8 | 1 | 70 | 11 | 0.89 | 0.14 | 0.10 | 0.92 |
| All viruses | 143 | 3 | 186 | 66 | 0.98 | 0.26 | 0.43 | 0.96 |
| Oropharyngeal shedding of challenge virus at 2 DPC | ||||||||
| Challenge virus | ||||||||
| PWT/06 | 16 | 74 | 4 | 75 | 0.18 | 0.95 | 0.80 | 0.50 |
| SMI-HAMD/06 | 42 | 32 | 2 | 13 | 0.57 | 0.87 | 0.95 | 0.29 |
| Papua/06 | 29 | 43 | 0 | 18 | 0.40 | 1.00 | 1.00 | 0.30 |
| All viruses | 87 | 149 | 6 | 106 | 0.37 | 0.95 | 0.94 | 0.42 |
| Vaccine virus | ||||||||
| PWT/06 challenge | 88 | 1 | 47 | 32 | 0.99 | 0.41 | 0.65 | 0.97 |
| SMI-HAMD/06 challenge | 71 | 4 | 5 | 10 | 0.95 | 0.67 | 0.93 | 0.71 |
| Papua/06 challenge | 67 | 1 | 7 | 11 | 0.99 | 0.61 | 0.91 | 0.92 |
| All viruses | 226 | 6 | 59 | 53 | 0.97 | 0.47 | 0.79 | 0.90 |
The results were obtained at 3 weeks postvaccination. Sensitivity = TP/(TP + FN), specificity = TN/(FP + TN), PPV = TP/(TP + FP), and NPV = TN/(TN + FN), where TP is the number of chickens with a true-positive result, FN is the number of chickens with a false-negative result, TN is the number of chickens with a true-negative result, and FN the number of chickens with a false-negative result.
For survival, a true-positive result was seropositivity and survival, a false-negative result was seronegativity and survival, a false-positive result was seropositivity and death, and a true-negative result was seronegativity and death. For oropharyngeal shedding of challenge virus at 2 DPC, a true-positive result was seropositivity and no virus shedding, a false-negative result was seronegativity and no virus shedding, a false-positive result was seropositivity and virus shedding, and a true-negative result was seronegativity and virus shedding.
More specifically, there was extensive variation in the sensitivity, specificity, PPV, and NPV between the three challenge viruses (Table 8). The data that were closest to optimal for the prediction of survival following SMI-HAMD/06 challenge were obtained from the HI test using vaccine antigen; i.e., of the three challenge viruses, SMI-HAMD/06 was genetically and antigenically the most closely related to all vaccine seed strains (Table 2; Fig. 2 and 3). The presence of an HI titer had a high PPV for survival and reduced virus shedding after challenge with SMI-HAMD/06 irrespective of whether the HI antigen was the vaccine strain or the challenge virus. In comparison, the presence of HI titers had low PPVs for survival after challenge with the genetically and antigenically more distant Papua/06 and PWT/06 challenge viruses when the HI titers obtained using vaccine antigen were evaluated but high PPVs when the HI titers obtained using the respective challenge virus as the HI test antigen were examined.
DISCUSSION
Most H5 and H7 HPAI viruses emerge from H5 and H7 LPAI viruses transmitted from the wild aquatic bird reservoir to terrestrial gallinaceous poultry which are allowed to circulate unimpeded (52). The progenitor LPAI viruses are antigenically similar to the emergent HPAI virus, making them ideal candidates for vaccine seed strains (Fig. 3). Because of prior efforts to vaccinate against low-pathogenic avian influenza virus, several H5 LPAI viruses were previously commercially produced and licensed and were available for use in the initial vaccine campaigns. At first, these LPAI vaccine seed strains were antigenically matched closely enough with field HPAI viruses that experimental studies demonstrated broad protection against homologous subtypes, especially protection from mortality (14, 15, 53–56). However, even with protection from mortality, the reduction of challenge virus replication and shedding from the oropharynx was more difficult to achieve, requiring a close genetic relationship between vaccine and challenge viruses for the antigenic match to be adequate for protection (15). As the H5N1 HPAI virus became endemic in the poultry of some countries, genetic and antigenic drift increased, and with the long-term usage of H5 avian influenza vaccines in poultry and, especially, with the improper usage of vaccines (45), H5 field viruses that were resistant to licensed vaccines appeared in Egypt (45, 57), Hong Kong (58), Mexico (42), and Vietnam (59). In contrast, long-term usage of H7 vaccines against H7N3 HPAI virus in Pakistan did not result in the identification of resistant field viruses (28). In China, the appearance of resistant field viruses has been met with changes in the national vaccine seed strains used in inactivated vaccines, which began with the LPAI virus field strain Eng/73 (H5N2) (2004 to 2006) and progressive replacements with strains generated by rg on the basis of the hemagglutinin and neuraminidase genes of Chinese H5N1 HPAI viruses, i.e., rgGD/96 (clade 0) (2004 to 2008), rg-generated A/duck/Anhui/1/2006 (clade 2.3.4) (2008 to 2012), and rg-generated A/duck/Guangdong/S1322/2010 (clade 2.3.2) (2012 to present) (35, 60). Furthermore, a vaccine seed strain generated by rg, A/chicken/Shanxi/2/2006 (clade 7), began to be used regionally during 2006 in layer chickens, and its limited use continues today. In the current study, we identified two Indonesian H5N1 HPAI viruses that were partially to completely resistant to classic H5 poultry vaccines made from naturally occurring H5 LPAI virus, an early H5N1 LPAI virus generated by rg, or early H5N1 HPAI virus seed strains.
Multiple issues can contribute to vaccine failures, and reports of failures are not proof that all aspects of vaccines and vaccination are a failure. However, when such failures take place, the challenge is to identify the specific factor or factors and make appropriate corrections. In the current study, we conducted a detailed examination into the Indonesian H5 vaccines, including the seed strains used, their serological potency, and the level of protection that they provided to vaccinated chickens following virulent challenge with genetically and antigenically diverse Indonesian field viruses. Of the 15 commercial H5 vaccines tested, RNA extraction and sequencing determined the vaccine seed strains for 14 vaccines. However, the RNA extracted from one purported Mex/94 vaccine was low quality, which prohibited determination of the vaccine seed strain by sequencing (Table 1). When this vaccine was used to immunize chickens, it produced high mean titers of HI antibody against Mex/94 (Table 6) and protected chickens from SMI-HAMD/06 challenge, indicating that H5 antigen was present. The inability to extract usable RNA could be related to the methods used to break the oil emulsion to extract the RNA, or the RNA could have been degraded through the viral inactivation process because chemical inactivating agents like formalin are known to degrade nucleic acid. The 14 vaccines for which the seed strain was determined (Table 1) contained seven licensed seed strains (WI/68 [H5N9], Mex/94 [H5N2], Eng/73 [H5N2], Legok/03 [H5N1], rgGD/96 [H5N1], rgLegok/03 [H5N1], and rgVN/04 [H5N3]). Eleven of the vaccines contained the seed strain listed by the manufacturer. However, three vaccines were labeled to contain Eng/73, which was a seed strain licensed for use in Indonesia beginning in 2006, but on the basis of sequence data, it contained rgGD/96. At the time of the purchase of these three vaccines, rgGD/96 was not licensed in Indonesia, but it did receive licensure for import and use by the end of 2007. A similar issue was reported in Central America, where 7 of 10 H5 poultry vaccines contained the purported and licensed seed strain, but the other 3 vaccines contained H5N2 LPAI virus seed strains other than the licensed Mex/94 seed strain that was reported on the label (26). Interestingly, these three vaccines provided the best protection in chicken studies using a 2003 Guatemalan H5N2 LPAI challenge virus. Such a disparity in licensed versus manufactured seed strains has several positive and negative ramifications. On the negative side, the manufacturer's decision of what strains to include in a vaccine without the authorization of the national veterinary authority could be dangerous because replacing a seed strain without regulatory oversight and scientific support could result in the inclusion of a less efficacious strain, thus reducing protection, and such substitutions are illegal under the laws of national veterinary vaccine authorities. On the positive side, the early replacement of seed strains could provide a more efficacious vaccine, but the regulatory process may be slow, resulting in unreasonable delays in the licensing of more efficacious vaccines that are needed in the field. In the current study, the three vaccines that were labeled to contain Eng/73 but that contained the rgGD/96 seed strain were superior to the licensed Eng/73 vaccines according to protection metrics. In both the current Indonesian study and previous Central American studies, the substituted seed strains improved the efficacy of the vaccine product used in the field, thus emphasizing the need for regulatory reform to achieve a more rapid updating of seed strains using a cassette concept within a streamlined or fast-track regulatory process as field viruses undergo antigenic drift, as is usually the case with human seasonal influenza virus vaccines.
In previous and current studies, data have shown a progressive genetic drift of the clade 2.1 viruses away from the clade 0 progenitor, A/goose/Guangdong/1/1996 (H5N1), and the root of the H5 clade 2.1 virus (Fig. 2 and Table 2) (38, 39, 61, 62). Similarly, the current study demonstrated antigenic drift among the clade 2.1 H5N1 HPAI viruses in Indonesia, with HI test results indicating that strain SMI-HAMD/06 was closely related to the 2003 Indonesian vaccine strain Legok/2003 but progressively less antigenically related to the other vaccine seed strains (Table 4 and Fig. 3). This decline in antigenic relatedness to vaccine seed strains was more evident with Papua/06 and was extreme with PWT/06. The latter had no measurable HI response against the non-Indonesian vaccine seed strains and produced minimal titers against Indonesian Legok/03 (Table 4). Furthermore, the results of the challenge protection study corroborate the moderate and extreme antigenic drift evident in the antigenic map, with partial and poor protection being provided against Papua/06 and PWT/06 challenge, respectively (Fig. 3).
Consistent with the antigenic drift, numerous amino acids that were previously reported to be important for antigenic structure (amino acid residues 41 to 46) and that were distinctive for the Indonesian challenge viruses or commercial vaccine viruses were identified (amino acid residues 124, 189, and 212). Additional amino acids (amino acid residues 94, 140, 141, and 162) were nonsynonymous between PWT/06, Papua/06, and the vaccine viruses, and one amino acid (amino acid residue 74) was unique to PWT/06 compared to the sequences of commercial vaccine viruses Legok/03 and SMI-HAMD/06. PWT/06 and Papua/06 were the most distant antigenically from the classical H5 group, and the vaccines prepared with classical group H5 antigens, rgGD/96 or VN/04, were the most poorly protective. PWT/06 also had some unique amino acid changes near the receptor binding site which have been identified to be antigenically definitive sites in H5 clade 2.1: 183 and 189. Although amino acid 185 was also a unique amino acid change, the change at amino acid 133, which was previously shown to be synergistic in producing antigenic change, was not present; however, the original study did not evaluate amino acid residues 183, 185, and 189 together (48).
Although further investigation would be needed to confirm the role of each of these amino acid changes in the antigenic structure of the virus and the protection that the vaccine provides chickens, it is reasonable to assume that these changes would contribute to the antigenic distance of PWT/06 and Papua/06 as well as the diminished effectiveness of the commercial vaccines prepared with classical H5 group strains and HA strains from older H5N1 clades.
Previous studies have shown that minimum specific HI serological titers were associated with protection in challenge studies when the vaccine and field viruses were genetically and antigenically similar, such that HI serology GMTs of ≥8 (25) or ≥10 (63) were associated with survival, GMTs of ≥40 (63) were associated with the prevention of challenge virus shedding from the oropharynx in the majority of vaccinated chickens, and GMTs of ≥128 (55) were associated with the prevention of challenge virus shedding from the oropharynx in all vaccinated chickens. The OIE terrestrial manual suggests that a serological potency consisting of a GMT of ≥32 offers protection from mortality and that a serological potency consisting of a GMT of ≥128 offers protection from virus shedding (24). Such international serological potency standards provide assurances that commercial vaccines will contain sufficient antigen to elicit a protective immune response in the field when there is a close antigenic match of the vaccine to the challenge strain. The 15 inactivated Indonesian vaccines were examined in the context of serological potency, and all met the international standard of a minimum potency with a GMT of ≥32, while only vaccines using WI/68, Mex/94, and Eng/73 exceeded the international standard consisting of a GMT of ≥128 for reducing challenge virus replication, as has been demonstrated in previous experimental studies that use WL chickens (55). In the current study, all the vaccine strains evaluated were genetically and antigenically the closest to the SMI-HAMD/06 challenge virus, and all the vaccines provided 100% protection from mortality in groups with a GMT of ≥60, while a GMT of between 20 and 30 was associated with 50 to 90% protection when using the vaccine seed strain as the antigen in the HI test (data not shown). In contrast, when the more distantly related challenge viruses Papua/06 and PWT/06 were evaluated, a serological response against the vaccine seed strain did not allow consistent prediction of protection from challenge. In view of these data and previously published data, a minimum protective GMT of ≥1:128 was associated with protection in WL chickens if the field viruses were antigenically closely related to the vaccines, but data on the minimum HI titers associated with protection in other poultry species or breeds within a single poultry species are not available. Generally, WL chickens generate the strongest immune responses among all poultry species following a single immunization with a licensed vaccine, including higher titers than meat-type chickens, turkeys, and domestic ducks, but there is also significant variation in antibody responses by breed (64–67). Some poultry species may require additional booster vaccinations to achieve sufficient levels of protection for their specific type of production system and the length of their production lives. High HI serological titers are associated with protection when the challenge and vaccine viruses are genetically and antigenically closely related. However, in the current study, the high serological titers induced by Mex/94, Eng/73, and WI/68 did not produce protection from the PWT/06 virus, indicating that high HI titers alone do not predict protection against antigenic variant field viruses. However, when using the antigenic variant challenge viruses as the HI test antigen, the presence of HI titers had a high PPV for protection, suggesting that HI antibodies have a strong role in immune protection. However, the presence of survivors among vaccinated chickens which lacked HI antibodies, measured as a low NPV, suggests that non-HI antibodies or nonantibody immunity can provide some heterologous protection. Such protection against influenza A virus which resulted from cell-mediated immunity or IgA mucosal immunity to uncharacterized influenza viral proteins (68–70) or humoral immunity from non-HI antibodies, such as antibodies to conserved regions in the hemagglutinin stalk (71), has been demonstrated in laboratory studies in poultry or mammals.
Unlike human influenza vaccines, where seed strains are changed periodically as field viruses drift (72), frequent changing of seed strains for avian vaccines was historically seen to be unnecessary due to the shorter life span of poultry, less vaccination selection pressure, and strong adjuvant usage (15, 25, 37, 55, 73). However, the recent development of the long-term utilization of vaccines for the control of H5 avian influenza has been associated with the emergence of vaccine-resistant field viruses in Egypt (45, 57), Hong Kong (58), Mexico (42), Vietnam (59), China (35, 60), and, in the current study, Indonesia. This situation indicates the need for ongoing virological surveillance to evaluate field viruses for antigenic drift and update vaccine seed strains to maintain protection in the field. Although naturally occurring LPAI virus seed strains have been efficacious in vaccines used during the initial HPAI outbreaks, as field viruses drift antigenically, new vaccine seed strains will need to be developed by utilizing either HPAI viruses from the field or reverse genetics technologies to produce LPAI viruses as replacement seed strains (35, 53, 74). The rgPWT/06 vaccine strain developed in this study showed exogenous trypsin dependence for the formation of plaques in MDCK cells and was proven safe for SPF chickens. It thus passed the OIE criteria for low-pathogenicity avian influenza viruses (22, 24). In addition, because of manufacturing safety concerns, it is recommended that reverse genetics technologies be used to produce LPAI virus seed strains because of the safety concerns over the use of HPAI viruses because of their zoonotic risk potential (24). Assessment of the protection provided by vaccines can be accomplished by using predominant circulating field viruses as antigens in HI tests and the presence of HI antibodies as a positive predictive value for protection in the field.
ACKNOWLEDGMENTS
We thank J. Beck, K. Moresco, and J. Doster for technical assistance. Cheryl I. Hall and Max Coates of International Services, U.S. Department of Agriculture, are thanked for the assistance provided in Indonesia.
The project was conducted from 2007 to 2014 under the mission of OFFLU, the World Organization for Animal Health (OIE), and the Food and Agriculture Organization (FAO) of the United Nations joint Animal Influenza Network (http://www.offlu.net/).
This work was partially supported by Agricultural Research Service, USDA, Current Research Information Service, projects 6612-32000-048-00D and 6612-32000-064-00D, the U.S. Agency for International Development, FAO project OSRO/INS/703/USA (PR no. 44133), and National Institute of Allergy and Infectious Diseases, National Institutes of Health, contracts HHSN266200700010C and HHSN272201400008C.
The views expressed in this information product are those of the author(s) and do not necessarily reflect the views or policies of FAO.
REFERENCES
- 1.Swayne DE, Pavade G, Hamilton K, Vallat B, Miyagishima K. 2011. Assessment of national strategies for control of high pathogenicity avian influenza and low pathogenicity notifiable avian influenza in poultry, with emphasis on vaccines and vaccination. Rev Sci Tech 30:839–870 http://web.oie.int/boutique/extrait/18swayne839870.pdf. [DOI] [PubMed] [Google Scholar]
- 2.Swayne DE, Spackman E. 2013. Current status and future needs in diagnostics and vaccines for high pathogenicity avian influenza. Dev Biol (Basel) 135:79–94. doi: 10.1159/000325276. [DOI] [PubMed] [Google Scholar]
- 3.FAO. 2013. Highly pathogenic avian influenza H5 events, p 1–7 EMPRES Animal Influenza Update 583. FAO, Rome, Italy. [Google Scholar]
- 4.Xu X, Subbarao K, Cox NJ, Guo Y. 1999. Genetic characterization of the pathogenic influenza A/Goose/Guangdong/1/96 (H5N1) virus: similarity of its hemagglutinin gene to those of H5N1 viruses from the 1997 outbreaks in Hong Kong. Virology 261:15–19. doi: 10.1006/viro.1999.9820. [DOI] [PubMed] [Google Scholar]
- 5.Sawitri Siregar E, Darminto Weaver J, Bouma A. 2007. The vaccination programme in Indonesia. Dev Biol 130:151–158. [PubMed] [Google Scholar]
- 6.OIE. 2011. Animal health data (prior to 2005): handistats II. OIE, Paris, France: http://web.oie.int/hs2/report.asp?lang=en. [Google Scholar]
- 7.Dawson J. 2000. Neuraminidase inhibitor and amantadine. Lancet 355:2254. doi: 10.1016/S0140-6736(05)72755-0. [DOI] [PubMed] [Google Scholar]
- 8.Sims LD, Brown IH. 2008. Multi-continental epidemic of H5N1 high pathogenicity avian influenza (1996-2007), p 251–286 InSwayne DE. (ed), Avian influenza. Blackwell Publishing, Ames, IA. [Google Scholar]
- 9.Swayne DE. 2003. Vaccines for list A poultry diseases: emphasis on avian influenza. Dev Biol (Basel) 114:201–212. [PubMed] [Google Scholar]
- 10.Sims LD. 2007. Lessons learned from Asian H5N1 outbreak control. Avian Dis 51:174–181. doi: 10.1637/7637-042806R.1. [DOI] [PubMed] [Google Scholar]
- 11.Bouma A, Muljono AT, Jatikusumah A, Nell A, Mudjiartiningsih S, Dharmayanti I, Siregar Sawitri E, Claassen I, Koch G, Stegeman JA. 2008. Field trial for assessment of avian influenza vaccination effectiveness in Indonesia. Rev Sci Tech 27:633–642 http://web.oie.int/boutique/extrait/bouma.pdf. [DOI] [PubMed] [Google Scholar]
- 12.van der Goot JA, van Boven M, de Jong MC, Koch G. 2007. Effect of vaccination on transmission of HPAI H5N1: the effect of a single vaccination dose on transmission of highly pathogenic avian influenza H5N1 in Peking ducks. Avian Dis 51(Suppl 1):S323–S324. doi: 10.1637/1933-5334(2007)2[e29:EOVOTO]2.0.CO;2. [DOI] [PubMed] [Google Scholar]
- 13.van der Goot JA, Koch G, de Jong MCM, van Boven M. 2003. Transmission dynamics of low- and high-pathogenicity A/chicken/Pennsylvania/83 avian influenza viruses. Avian Dis 47:939–941. doi: 10.1637/0005-2086-47.s3.939. [DOI] [PubMed] [Google Scholar]
- 14.Swayne DE, Beck JR, Perdue ML, Beard CW. 2001. Efficacy of vaccines in chickens against highly pathogenic Hong Kong H5N1 avian influenza. Avian Dis 45:355–365. doi: 10.2307/1592975. [DOI] [PubMed] [Google Scholar]
- 15.Swayne DE, Garcia M, Beck JR, Kinney N, Suarez DL. 2000. Protection against diverse highly pathogenic avian influenza viruses in chickens immunized with a recombinant fowl pox vaccine containing an H5 avian influenza hemagglutinin gene insert. Vaccine 18:1088–1095. doi: 10.1016/S0264-410X(99)00369-2. [DOI] [PubMed] [Google Scholar]
- 16.Ellis TM, Leung CYHC, Chow MKW, Bissett LA, Wong W, Guan Y, Malik Peiris JS. 2004. Vaccination of chickens against H5N1 avian influenza in the face of an outbreak interrupts virus transmission. Avian Pathol 33:405–412. doi: 10.1080/03079450410001724012. [DOI] [PubMed] [Google Scholar]
- 17.Kodihalli S, Haynes JR, Robinson HL, Webster RG. 1997. Cross-protection among lethal H5N2 influenza viruses induced by DNA vaccine to the hemagglutinin. J Virol 71:3391–3396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Terregino C, Toffan A, Cilloni F, Monne I, Bertoli E, Castellanos L, Amarin N, Mancin M, Capua I. 2010. Evaluation of the protection induced by avian influenza vaccines containing a 1994 Mexican H5N2 LPAI seed strain against a 2008 Egyptian H5N1 HPAI virus belonging to clade 2.2.1 by means of serological and in vivo tests. Avian Pathol 39:215–222. doi: 10.1080/03079451003781858. [DOI] [PubMed] [Google Scholar]
- 19.Robinson HL, Hunt LA, Webster RG. 1993. Protection against a lethal influenza virus challenge by immunization with a haemagglutinin-expressing plasmid DNA. Vaccine 11:957–960. doi: 10.1016/0264-410X(93)90385-B. [DOI] [PubMed] [Google Scholar]
- 20.Crawford J, Wilkinson B, Vosnesensky A, Smith G, Garcia M, Stone H, Perdue ML. 1999. Baculovirus-derived hemagglutinin vaccines protect against lethal influenza infections by avian H5 and H7 subtypes. Vaccine 17:2265–2274. doi: 10.1016/S0264-410X(98)00494-0. [DOI] [PubMed] [Google Scholar]
- 21.Jadhao SJ, Suarez DL. 2010. New approach to delist highly pathogenic avian influenza viruses from BSL3+ select agents to BSL2 non-select status for diagnostics and vaccines. Avian Dis 54:302–306. doi: 10.1637/8926-051509-ResNote.1. [DOI] [PubMed] [Google Scholar]
- 22.Subbarao K, Chen HL, Swayne D, Mingay L, Fodor E, Brownlee G, Xu XY, Lu XH, Katz J, Cox N, Matsuoka Y. 2003. Evaluation of a genetically modified reassortant H5N1 influenza A virus vaccine candidate generated by plasmid-based reverse genetics. Virology 305:192–200. doi: 10.1006/viro.2002.1742. [DOI] [PubMed] [Google Scholar]
- 23.Jadhao SJ, Lee CW, Sylte M, Suarez DL. 2009. Comparative efficacy of North American and antigenically matched reverse genetics derived H5N9 DIVA marker vaccines against highly pathogenic Asian H5N1 avian influenza viruses in chickens. Vaccine 27:6247–6260. doi: 10.1016/j.vaccine.2009.07.110. [DOI] [PubMed] [Google Scholar]
- 24.OIE. 2012. Avian influenza. Manual of diagnostic tests and vaccines for terrestrial animals. OIE, Paris, France: http://www.oie.int/fileadmin/Home/eng/Health_standards/tahm/2.03.04_AI.pdf. [Google Scholar]
- 25.Swayne DE, Beck JR, Garcia M, Stone HD. 1999. Influence of virus strain and antigen mass on efficacy of H5 avian influenza inactivated vaccines. Avian Pathol 28:245–255. doi: 10.1080/03079459994731. [DOI] [PubMed] [Google Scholar]
- 26.Eggert D, Thomas C, Spackman E, Pritchard N, Rojo F, Bublot M, Swayne DE. 2010. Characterization and efficacy determination of commercially available Central American H5N2 avian influenza vaccines for poultry. Vaccine 28:4609–4615. doi: 10.1016/j.vaccine.2010.04.081. [DOI] [PubMed] [Google Scholar]
- 27.Swayne DE, Senne DA, Suarez DL. 2008. Avian influenza, p 128–134 InDufour-Zavala L, Swayne DE, Glisson JR, Pearson JE, Reed WM, Jackwood MW, Woolcock PR (ed), Isolation and identification of avian pathogens, 5th ed American Association of Avian Pathologists, Jacksonville, FL. [Google Scholar]
- 28.Abbas MA, Spackman E, Fouchier R, Smith D, Ahmed Z, Siddique N, Sarmento L, Naeem K, McKinley ET, Hameed A, Rehmani S, Swayne DE. 2011. H7 avian influenza virus vaccines protect chickens against challenge with antigenically diverse isolates. Vaccine 29:7424–7429. doi: 10.1016/j.vaccine.2011.07.064. [DOI] [PubMed] [Google Scholar]
- 29.Fouchier AM, Smith DJ. 2010. Use of antigenic cartography in vaccine seed strain selection. Avian Dis 54:220–223. doi: 10.1637/8740-032509-ResNote.1. [DOI] [PubMed] [Google Scholar]
- 30.Smith DJ, Lapedes AS, de Jong JC, Bestebroer TM, Rimmelzwaan GF, Osterhaus AD, Fouchier RA. 2004. Mapping the antigenic and genetic evolution of influenza virus. Science 305:371–376. doi: 10.1126/science.1097211. [DOI] [PubMed] [Google Scholar]
- 31.Pedersen JC. 2008. Hemagglutination-inhibition test for avian influenza virus subtype identification and the detection and quantitation of serum antibodies to the avian influenza virus. Methods Mol Biol 436:53–66. doi: 10.1007/978-1-59745-279-3_8. [DOI] [PubMed] [Google Scholar]
- 32.Das A, Spackman E, Pantin-Jackwood MJ, Suarez DL. 2009. Removal of real-time reverse transcription polymerase chain reaction (RT-PCR) inhibitors associated with cloacal swab samples and tissues for improved diagnosis of avian influenza virus by RT-PCR. J Vet Diagn Invest 21:771–778. doi: 10.1177/104063870902100603. [DOI] [PubMed] [Google Scholar]
- 33.Wiyono A, Indriani R, Dharmayanti NLPI, Damayanti R, Parede L, Syafriati dan Darminto T. 2004. Isolation and characterization of virus of highly pathogenic avian influenza H5 subtype of chicken from outbreaks in Indonesia. J Ilmu Ternak Vet 9:61–71. [Google Scholar]
- 34.Domenech J, Dauphin G, Rushton J, McGrane J, Lubroth J, Tripodi A, Gilbert J, Sims LD. 2009. Experiences with vaccination in countries endemically infected with highly pathogenic avian influenza: the Food and Agriculture Organization perspective. Rev Sci Tech 28:293–305 http://web.oie.int/boutique/extrait/domenech293306.pdf. [DOI] [PubMed] [Google Scholar]
- 35.Chen H. 2009. Avian influenza vaccination: the experience in China. Rev Sci Tech 28:267–274 http://web.oie.int/boutique/extrait/chen267274.pdf. [DOI] [PubMed] [Google Scholar]
- 36.Swayne DE, Kapczynski DR. 2008. Vaccines, vaccination and immunology for avian influenza viruses in poultry, p 407–451 InSwayne DE. (ed), Avian influenza. Blackwell Publishing, Ames, IA. [Google Scholar]
- 37.Swayne DE. 2006. Principles for vaccine protection in chickens and domestic waterfowl against avian influenza: emphasis on Asian H5N1 high pathogenicity avian influenza. Ann N Y Acad Sci 1081:174–181. doi: 10.1196/annals.1373.021. [DOI] [PubMed] [Google Scholar]
- 38.WHO/OIE/FAO H5N1 Evolution Working Group. 2014. Revised and updated nomenclature for highly pathogenic avian influenza A (H5N1) viruses. Influenza Other Respir Viruses 8:384–388. doi: 10.1111/irv.12230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Takano R, Nidom CA, Kiso M, Muramoto Y, Yamada S, Sakai-Tagawa Y, Macken C, Kawaoka Y. 2009. Phylogenetic characterization of H5N1 avian influenza viruses isolated in Indonesia from 2003-2007. Virology 390:13–21. doi: 10.1016/j.virol.2009.04.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.WHO/OIE/FAO H5N1 Evolution Working Group. 2011. Updated unified nomenclature system for the highly pathogenic H5N1 avian influenza viruses. World Health Organization, Geneva, Switzerland: http://www.who.int/influenza/gisrs_laboratory/h5n1_nomenclature/en/. [Google Scholar]
- 41.Kaverin NV, Rudneva IA, Ilyushina NA, Varich NL, Lipatov AS, Smirnov YA, Govorkova EA, Gitelman AK, Lvov DK, Webster RG. 2002. Structure of antigenic sites on the haemagglutinin molecule of H5 avian influenza virus and phenotypic variation of escape mutants. J Gen Virol 83:2497–2505. [DOI] [PubMed] [Google Scholar]
- 42.Lee CW, Senne DA, Suarez DL. 2004. Effect of vaccine use in the evolution of Mexican-lineage H5N2 avian influenza virus. J Virol 78:8372–8381. doi: 10.1128/JVI.78.15.8372-8381.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hoffmann E, Lipatov AS, Webby RJ, Govorkova EA, Webster RG. 2005. Role of specific hemagglutinin amino acids in the immunogenicity and protection of H5N1 influenza virus vaccines. Proc Natl Acad Sci U S A 102:12915–12920. doi: 10.1073/pnas.0506416102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kaverin N, Rudneva VIA, Govorkova EA, Timofeeva TA, Shilov AA, Kochergin-Nikitsky KS, Krylov PS, Webster RG. 2007. Epitope mapping of the hemagglutinin molecule of a highly pathogenic H5N1 influenza virus by using monoclonal antibodies. J Virol 81:12911–12917. doi: 10.1128/JVI.01522-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Cattoli G, Milani A, Temperton N, Zecchin B, Buratin A, Molesti E, Aly MM, Arafa A, Capua I. 2011. Antigenic drift in H5N1 avian influenza virus in poultry is driven by mutations in major antigenic sites of the hemagglutinin molecule analogous to those for human influenza virus. J Virol 85:8718–8724. doi: 10.1128/JVI.02403-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Li Y, Zhang X, Xu Q, Fu Q, Zhu Y, Chen S, Peng D, Liu X. 2013. Characterisation and haemagglutinin gene epitope mapping of a variant strain of H5N1 subtype avian influenza virus. Vet Microbiol 162:614–622. doi: 10.1016/j.vetmic.2012.11.033. [DOI] [PubMed] [Google Scholar]
- 47.Koel BF, Burke DF, Bestebroer TM, van der Vliet S, Zondag GC, Vervaet G, Skepner E, Lewis NS, Spronken MI, Russell CA, Eropkin MY, Hurt AC, Barr IG, de Jong JC, Rimmelzwaan GF, Osterhaus AD, Fouchier RA, Smith DJ. 2013. Substitutions near the receptor binding site determine major antigenic change during influenza virus evolution. Science 342:976–979. doi: 10.1126/science.1244730. [DOI] [PubMed] [Google Scholar]
- 48.Koel BF, van der Vliet S, Burke DF, Bestebroer TM, Bharoto EE, Yasa IW, Herliana I, Laksono BM, Xu K, Skepner E, Russell CA, Rimmelzwaan GF, Perez DR, Osterhaus AD, Smith DJ, Prajitno TY, Fouchier RA. 2014. Antigenic variation of clade 2.1 H5N1 virus is determined by a few amino acid substitutions immediately adjacent to the receptor binding site. mBio 5(3):e01070-14. doi: 10.1128/mBio.01070-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Capua I, Schmitz A, Jestin V, Koch G, Marangon S. 2009. Vaccination as a tool to combat introductions of notifiable avian influenza viruses in Europe, 2000 to 2006. Rev Sci Tech 28:245–259 http://web.oie.int/boutique/extrait/capua245260.pdf. [DOI] [PubMed] [Google Scholar]
- 50.OIE. 2007. Avian influenza vaccination: OIE information document and Verona recommendations. OIE, Paris, France: http://www.oie.int/eng/info_ev/Other%20Files/A_Guidelines%20on%20AI%20vaccination.pdf. [Google Scholar]
- 51.Steensels M, van Borm S, Lambrecht B, de Vriese J, Le Gros FX, Bublot M, van den Berg T. 2007. Efficacy of an inactivated and a fowlpox-vectored vaccine in Muscovy ducks against an Asian H5N1 highly pathogenic avian influenza viral challenge. Avian Dis 51(Suppl 1):S325–S331 http://www.jstor.org/discover/10.2307/4493219?sid=21105221372471&uid=2&uid=3739704&uid=4&uid=2129&uid=70&uid=3739256. [DOI] [PubMed] [Google Scholar]
- 52.Rohm C, Horimoto T, Kawaoka Y, Suss J, Webster RG. 1995. Do hemagglutinin genes of highly pathogenic avian influenza viruses constitute unique phylogenetic lineages? Virology 209:664–670. doi: 10.1006/viro.1995.1301. [DOI] [PubMed] [Google Scholar]
- 53.Liu M, Wood JM, Ellis T, Krauss S, Seiler P, Johnson C, Hoffmann E, Humberd J, Hulse D, Zhang Y, Webster RG, Perez DR. 2003. Preparation of a standardized, efficacious agricultural H5N3 vaccine by reverse genetics. Virology 314:580–590. doi: 10.1016/S0042-6822(03)00458-6. [DOI] [PubMed] [Google Scholar]
- 54.Rudolf M, Poppel M, Frohlich A, Mettenleiter T, Beer M, Harder T. 2009. Efficacy of a commercial inactivated H5 influenza vaccine against highly pathogenic avian influenza H5N1 in waterfowl evaluated under field conditions. Rev Sci Tech 28:275–291 http://web.oie.int/boutique/extrait/rudolf275292.pdf. [DOI] [PubMed] [Google Scholar]
- 55.Swayne DE, Lee CW, Spackman E. 2006. Inactivated North American and European H5N2 avian influenza virus vaccines protect chickens from Asian H5N1 high pathogenicity avian influenza virus. Avian Pathol 35:141–146. doi: 10.1080/03079450600597956. [DOI] [PubMed] [Google Scholar]
- 56.Bublot M, Manvell RJ, Shell W, Brown IH. 2010. High level of protection induced by two fowlpox vector vaccines against a highly pathogenic avian influenza H5N1 challenge in specific-pathogen-free chickens. Avian Dis 54:257–261. doi: 10.1637/8774-033109-ResNote.1. [DOI] [PubMed] [Google Scholar]
- 57.Grund C, el Abdelwhab SM, Arafa AS, Ziller M, Hassan MK, Aly MM, Hafez HM, Harder TC, Beer M. 2011. Highly pathogenic avian influenza virus H5N1 from Egypt escapes vaccine-induced immunity but confers clinical protection against a heterologous clade 2.2.1 Egyptian isolate. Vaccine 29:5567–5573. doi: 10.1016/j.vaccine.2011.01.006. [DOI] [PubMed] [Google Scholar]
- 58.Leung YHC, Luk G, Sia SF, Wu YO, Ho CK, Chow KC, Tang SC, Guan Y, Malik Peiris JS. 2013. Experimental challenge of chicken vaccinated with commercially available H5 vaccines reveals loss of protection to some highly pathogenic avian influenza H5N1 strains circulating in Hong Kong/China. Vaccine 31:3536–3542. doi: 10.1016/j.vaccine.2013.05.076. [DOI] [PubMed] [Google Scholar]
- 59.Cha RM, Smith D, Shepherd E, Davis CT, Donis R, Nguyen T, Nguyen HD, Do HT, Inui K, Suarez DL, Swayne DE, Pantin-Jackwood M. 2013. Suboptimal protection against H5N1 highly pathogenic avian influenza viruses from Vietnam in ducks vaccinated with commercial poultry vaccines. Vaccine 31:4953–4960. doi: 10.1016/j.vaccine.2013.08.046. [DOI] [PubMed] [Google Scholar]
- 60.Swayne DE. 2012. Impact of vaccines and vaccination on global control of avian influenza. Avian Dis 56:818–828. doi: 10.1637/10183-041012-Review.1. [DOI] [PubMed] [Google Scholar]
- 61.Smith GJ, Naipospos TS, Nguyen TD, de Jong MD, Vijaykrishna D, Usman TB, Hassan SS, Nguyen TV, Dao TV, Bui NA, Leung YH, Cheung CL, Rayner JM, Zhang JX, Zhang LJ, Poon LL, Li KS, Nguyen VC, Hien TT, Farrar J, Webster RG, Chen H, Peiris JS, Guan Y. 2006. Evolution and adaptation of H5N1 influenza virus in avian and human hosts in Indonesia and Vietnam. Virology 350:258–268. doi: 10.1016/j.virol.2006.03.048. [DOI] [PubMed] [Google Scholar]
- 62.Wu WL, Chen Y, Wang P, Song W, Lau SY, Rayner JM, Smith GJ, Webster RG, Peiris JS, Lin T, Xia N, Guan Y, Chen H. 2008. Antigenic profile of avian H5N1 viruses in Asia from 2002 to 2007. J Virol 82:1798–1807. doi: 10.1128/JVI.02256-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Kumar M, Chu HJ, Rodenberg J, Krauss S, Webster RG. 2007. Association of serologic and protective responses of avian influenza vaccines in chickens. Avian Dis 51:481–483. doi: 10.1637/7605-041706R1.1. [DOI] [PubMed] [Google Scholar]
- 64.Westbury HA. 1981. Serological response of chickens, turkeys and ducks to strain V4 of Newcastle disease virus. Aust Vet J 57:328–332. doi: 10.1111/j.1751-0813.1981.tb05838.x. [DOI] [PubMed] [Google Scholar]
- 65.Pfeiffer J, Suarez DL, Sarmento L, To TL, Nguyen T, Pantin-Jackwood MJ. 2010. Efficacy of commercial vaccines in protecting chickens and ducks against H5N1 highly pathogenic avian influenza viruses from Vietnam. Avian Dis 54:262–271. doi: 10.1637/8715-031909-Reg.1. [DOI] [PubMed] [Google Scholar]
- 66.Shany SA, el-Kady MF, Eid BT, Hassan ER, Abdel-Moneim AS. 2011. Humoral antibody responses to different H5N1 and H5N2 vaccination regimes: implications for the development of autogenously based vaccines. Vet Microbiol 153:398–402. doi: 10.1016/j.vetmic.2011.06.006. [DOI] [PubMed] [Google Scholar]
- 67.Chang CS, Tixier-Boichard M, Chazara O, Lee YP, Chen CF, Chang PC, Chen JW, Bed'hom B. 2011. Different immune responses to three different vaccines following H6N1 low pathogenic avian influenza virus challenge in Taiwanese local chicken breeds. BMC Proc 5(Suppl 4):S33. doi: 10.1186/1753-6561-5-S4-S33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Seo SH, Webster RG. 2001. Cross-reactive, cell-mediated immunity and protection of chickens from lethal H5N1 influenza virus infection in Hong Kong poultry markets. J Virol 75:2516–2525. doi: 10.1128/JVI.75.6.2516-2525.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Kapczynski DR, Liljebjelke K, Kulkarni G, Hunt H, Jiang HJ, Petkov D. 2011. Cross reactive cellular immune responses in chickens previously exposed to low pathogenic avian influenza. BMC Proc 5(Suppl 4):S13. doi: 10.1186/1753-6561-5-S4-S13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Price GE, Soboleski MR, Lo CY, Misplon JA, Pappas C, Houser KV, Tumpey TM, Epstein SL. 2009. Vaccination focusing immunity on conserved antigens protects mice and ferrets against virulent H1N1 and H5N1 influenza A viruses. Vaccine 27:6512–6521. doi: 10.1016/j.vaccine.2009.08.053. [DOI] [PubMed] [Google Scholar]
- 71.Krammer F, Palese P. 2013. Influenza virus hemagglutinin stalk-based antibodies and vaccines. Curr Opin Virol 3:521–530. doi: 10.1016/j.coviro.2013.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Smith DJ. 2003. Applications of bioinformatics and computational biology to influenza surveillance and vaccine strain selection. Vaccine 21:1758–1761. doi: 10.1016/S0264-410X(03)00068-9. [DOI] [PubMed] [Google Scholar]
- 73.van den Berg T, Lambrecht B, Marche S, Steensels M, Van Borm S, Bublot M. 2008. Influenza vaccines and vaccination strategies in birds. Comp Immunol Microbiol Infect Dis 31:121–165. doi: 10.1016/j.cimid.2007.07.004. [DOI] [PubMed] [Google Scholar]
- 74.Lee CW, Senne DA, Suarez DL. 2004. Generation of reassortant influenza vaccines by reverse genetics that allows utilization of a DIVA (differentiating infected from vaccinated animals) strategy for the control of avian influenza. Vaccine 22:3175–3181. doi: 10.1016/j.vaccine.2004.01.055. [DOI] [PubMed] [Google Scholar]
- 75.Duvvuri VR, Duvvuri B, Cuff WR, Wu GE, Wu J. 2009. Role of positive selection pressure on the evolution of H5N1 hemagglutinin. Genomics Proteomics Bioinformatics 7:47–56. doi: 10.1016/S1672-0229(08)60032-7. [DOI] [PMC free article] [PubMed] [Google Scholar]