ABSTRACT
JC polyomavirus (JCPyV) infection of immunocompromised individuals results in the fatal demyelinating disease progressive multifocal leukoencephalopathy (PML). The viral capsid of JCPyV is composed primarily of the major capsid protein virus protein 1 (VP1), and pentameric arrangement of VP1 monomers results in the formation of a pore at the 5-fold axis of symmetry. While the presence of this pore is conserved among polyomaviruses, its functional role in infection or assembly is unknown. Here, we investigate the role of the 5-fold pore in assembly and infection of JCPyV by generating a panel of mutant viruses containing amino acid substitutions of the residues lining this pore. Multicycle growth assays demonstrated that the fitness of all mutants was reduced compared to that of the wild-type virus. Bacterial expression of VP1 pentamers containing substitutions to residues lining the 5-fold pore did not affect pentamer assembly or prevent association with the VP2 minor capsid protein. The X-ray crystal structures of selected pore mutants contained subtle changes to the 5-fold pore, and no other changes to VP1 were observed. Pore mutant pseudoviruses were not deficient in assembly, packaging of the minor capsid proteins, or binding to cells or in transport to the host cell endoplasmic reticulum. Instead, these mutant viruses were unable to expose VP2 upon arrival to the endoplasmic reticulum, a step that is critical for infection. This study demonstrated that the 5-fold pore is an important structural feature of JCPyV and that minor modifications to this structure have significant impacts on infectious entry.
IMPORTANCE JCPyV is an important human pathogen that causes a severe neurological disease in immunocompromised individuals. While the high-resolution X-ray structure of the major capsid protein of JCPyV has been solved, the importance of a major structural feature of the capsid, the 5-fold pore, remains poorly understood. This pore is conserved across polyomaviruses and suggests either that these viruses have limited structural plasticity in this region or that this pore is important in infection or assembly. Using a structure-guided mutational approach, we showed that modulation of this pore severely inhibits JCPyV infection. These mutants do not appear deficient in assembly or early steps in infectious entry and are instead reduced in their ability to expose a minor capsid protein in the host cell endoplasmic reticulum. Our work demonstrates that the 5-fold pore is an important structural feature for JCPyV.
INTRODUCTION
The JC polyomavirus (JCPyV) is a common human pathogen that causes severe disease in immunocompromised individuals. Serological studies have demonstrated that primary infection occurs early in childhood, and it is estimated that over 50% of the adult population is seropositive for JCPyV (1). In healthy individuals, JCPyV has been reported to establish a persistent infection in tissues of the kidney, bone marrow, and B cells. However, conditions of immunosuppression result in dissemination of JCPyV to the central nervous system, leading to a lytic infection of astrocytes and oligodendrocytes (reviewed in reference 2). Destruction of oligodendrocytes results in demyelination of neurons and causes the fatal demyelinating disease progressive multifocal leukoencephalopathy (PML) (3). There are currently no effective treatments for PML.
JCPyV is a small nonenveloped double-stranded DNA virus with a capsid diameter of approximately 50 nm. The viral capsid is composed of the major capsid protein viral protein 1 (VP1) and minor capsid proteins VP2 and VP3 (4). The exterior surface of the virus is formed by 360 copies of VP1 that are arrayed in T=7d pentameric symmetry to form 72 pentamers (5). VP1 also contains residues that are necessary for binding to the sialic acid-containing motif lactoseries tetrasaccharide c (LSTc) (6). A prominent feature of the JCPyV pentamer is a cavity located at the 5-fold axis of symmetry (6). One copy of either VP2 or VP3 is incorporated into each VP1 pentamer and is sequestered within the viral capsid. In the X-ray costructure of mouse polyomavirus (MPyV) VP1 and a VP2 fragment, electron density corresponding to the C terminus of VP2 was observed bound to the wall of the interior axial cavity of VP1 (7). Thus, it is likely that VP2 occupies much of the space of the interior axial cavity of the VP1 pentamer. The axial cavity connects to the exterior of the viral capsid through a 12.5-Å opening, referred to in this paper as the 5-fold pore. The importance of this pore in entry or assembly of JCPyV is unknown.
JCPyV utilizes complex intracellular transport pathways to gain access to the host cell nucleus for viral replication. JCPyV initiates infection by binding to LSTc on the surface of cells. This virus-receptor interaction is highly specific, as closely related molecules such as LSTb do not bind to JCPyV (6). JCPyV then enters cells by clathrin-mediated endocytosis, and the human serotonin receptor 2A family has been reported to enhance uptake of virus (8–11). After endocytosis, JCPyV enters the endocytic system and associates with Rab5-positive endosomes prior to retrograde transport to the endoplasmic reticulum (ER) (12, 13). Upon delivery to the ER, it is likely that JCPyV depends on isomerization of intrapentameric disulfide bonds to allow partial uncoating of the virus, as pharmacological treatments that reduce ER function or knockdown of the ER chaperones PDI and ERp57 decrease infectivity (13). The VP2 minor capsid protein of JCPyV is also exposed upon delivery to the ER (14). From there, JCPyV is likely retrotranslocated into the cytoplasm prior to nuclear transport and the initiation of viral replication.
Exposure of VP2 is a critical aspect of viral entry, and JCPyV mutants that lack VP2 or VP3 are noninfectious (15). Studies in MPyV or simian virus 40 (SV40) have demonstrated that VP2 and VP3 bind to cellular membranes or liposomes following exposure (16, 17). Additionally, VP2 is able to perforate membranes, likely by the insertion of additional predicted transmembrane regions or the myristolyated N terminus of VP2 (18). Antibodies raised to the N terminus of VP2 were able to prevent association of virions with liposomes, suggesting that the N terminus of VP2 mediates association of SV40 with membranes (16). Despite this importance, the mechanism by which VP2 is exposed from within the viral capsid remains unclear.
In this study, we sought to define the role of the 5-fold pore in JCPyV infection. JCPyV mutants were generated that contained amino acid substitutions to residues lining this pore and were expected to reduce the diameter of this pore. We demonstrated that alteration of this pore significantly reduces viral infectivity but does not interfere with viral assembly or packaging of VP2 and VP3. Using JCPyV pseudoviruses (PSVs) that contain these mutations, we show that these mutant PSVs are defective in transducing cells, despite showing no morphological changes compared to wild-type PSVs. Finally, these mutant PSVs are defective in exposing VP2 in the ER of cells, despite being transported to these compartments. These data suggest that amino acid substitutions to residues lining the 5-fold pore prevent exposure of the minor capsid proteins.
MATERIALS AND METHODS
Viruses, cells, and plasmids.
The Mad-1 strain of JCPyV and the human fetal glial (SVGA) cells used in these experiments have been previously described (19, 20). SVGA cells were maintained in complete media (minimum essential media [MEM] supplemented with 10% fetal bovine serum, 0.5% penicillin, and 0.5% streptomycin). 293TT cells were maintained in Dulbecco's modified Eagle medium that was supplemented with 10% fetal bovine serum and MEM nonessential amino acids (Cellgro).
Western blotting.
Protein samples were loaded onto 12% SDS-PAGE gels (Mini-Protean TGX; Bio-Rad) and transferred to nitrocellulose membranes (Trans-Blot SD semidry transfer cell; Bio-Rad). Membranes were blocked with 1% (wt/vol) bovine serum albumin–phosphate-buffered saline (PBS). VP1 was then detected using a mouse monoclonal antibody to VP1 (PAB597; a kind gift from Ed Harlow), and VP2 and VP3 were detected using a rabbit polyclonal antibody to SV40 VP2 that is cross-reactive with VP3 (ab53983; Abcam). Both antibodies were diluted in PBS containing 0.05% Tween 20 (PBS-T) and incubated overnight at 4°C. Membranes were washed in PBS-T and then incubated with secondary antibodies to mouse (goat anti-mouse Alexa Fluor 800; Life Technologies) or rabbit (goat anti-rabbit Alexa Fluor 680; Life Technologies). Antibodies were diluted 1:5,000 in PBS-T and incubated for 1 h at 21°C. Membranes were washed three times in PBS-T and once in PBS, and fluorescence was visualized and analyzed using an Odyssey infrared imaging system (Li-COR).
PSV production.
Viral genes were codon optimized for optimal expression in the 293TT human-derived cell line using the National Cancer Institute Center for Cancer Research Laboratory of Cellular Oncology Technical Files (http://home.ccr.cancer.gov/LCO/production.asp). Codon-optimized sequences were based on the Mad-1 Polyomaviridae orthopolyomavirus strain (NC_001699.1) and commercially synthesized (Blue Heron Biotech). These genes were then subcloned into pwP expression vector in place of the MPyV VP1 gene (22519; Addgene). PSVs were produced by transfection of the codon-optimized VP1, VP2, VP3, and phGluc plasmids into 293TT cells using FuGENE 6 transfection reagent (Promega) in a 3:1:1:2 ratio. Cells were split 1:3 at 48 h posttransfection (hpi) and harvested at 7 d posttransfection by scraping and then pelleted and resuspended in buffer A with EDTA-free protease inhibitors (Roche Applied Science). Cells were lysed by three rounds of freezing and thawing, sonicated and treated with 0.25% deoxycholic acid at 37°C for 30 min, and then sonicated three times on ice (Branson) (50% amplitude, 50% duty cycle, power 4, 1 min). The pH was lowered to 6.0, and the lysates were treated with type V neuraminidase (Sigma) at 37°C for 1 h to release JCPyV still bound to cells. The pH was then raised to 7.5, cellular debris was pelleted by centrifugation, and the viral supernatant was pelleted through a 20% sucrose cushion in a SW40ti rotor (Beckman Coulter) at 150,000 × g at 4°C for 3 h. The viral pellet was resuspended into buffer A (10 mM Tris-HCl [pH 7.4], 50 mM NaCl, 0.1 mM CaCl2) and sonicated 3 times (30% amplitude, 50% duty cycle, power 3, 1 min). The resuspended pellet was loaded onto a CsCl step gradient (1.29 to 1.35 g/ml) and spun at 115,000 × g at 4°C for 18 h in a SW55ti rotor (Beckman Coulter). The band corresponding to DNA-containing virions was isolated and dialyzed against buffer A.
PSVs were quantified by absolute genome copy numbers. Purified virions were treated with DNase I, and encapsidated phGluc plasmid was extracted using a DNeasy blood and tissue kit (Qiagen). Absolute quantification was performed using TaqMan quantitative PCR (Applied Biosystems), based on a standard curve of phGluc plasmid. The number of copies for the known plasmid was plotted in a scatter plot against the threshold cycle (CT) value determined for each dilution. A best-fit line was generated, and the trend line equation from regression analysis was used to calculate the absolute number of genomes in each virus sample.
PSV luciferase infectivity assay.
SVGA cells were seeded in 96-well plates at 70% confluence and infected with 1 × 107 genomes/well of wild-type and mutant PSV in 35 μl of serum-free medium. Cells were incubated at 37°C for 1 h and washed with PBS, complete medium without phenol red was added, and cells were incubated at 37°C for 72 h.
Secreted luciferase was quantitated in 50 μl of cellular supernatants using the BioLux Gaussia luciferase assay (New England BioLabs) according to the manufacturer's instructions using an opaque 96-well microplate in a GloMax multidetection system luminometer (Promega) equipped with an autoinjector.
Mutagenesis.
An infectious clone of JCPyV (Mad-1 strain, pUC19 backbone) was mutagenized by Phusion site-directed mutagenesis according to the protocol of the manufacturer (New England BioLabs). The mutagenic primers used in this study are listed in Table 1. The entire genome of each mutant was sequenced to ensure that no erroneous point mutations were generated during the PCR amplification. Mutagenesis of VP1 pentamers was performed using a pET15b plasmid encoding JCPyV VP1 (residues 22 to 289) from the Mad-1 strain and an N-terminal hexahistidine tag (6). The same primers were used to mutagenize this pentamer expression plasmid as were used for the infectious clone. Mutants were maintained in the JM109 cells, and Escherichia coli BL21(DE3) cells were used for pentamer expression (Invitrogen).
TABLE 1.
Primers used to generate pore mutations
| Mutation | Orientation | Primer sequence (5′–3′) |
|---|---|---|
| P223V | F | GGAGAAAATGTTGUACCAGTTCTTC |
| R | TCCTGTTAGTGTCCCAAAATATCTTG | |
| P223I | F | GGAGAAAATGTTATCCCAGTTCTTC |
| R | TCCTGTTAGTGTCCCAAAATATCTTG | |
| P223L | F | GGAGAAAATGTTCTACCAGTTCTTC |
| R | TCCTGTTAGTGTCCCAAAATATCTTG | |
| P223M | F | GGAGAAAATGTTATGCCAGTTCTTC |
| R | TCCTGTTAGTGTCCCAAAATATCTTG | |
| N221Q | F | GGAGGAGAACAAGTTCCTCCAG |
| R | TGTTAGTGTCCCAAAATATCTTGTG | |
| N221Y | F | GGAGGAGAATACGTTCCTCCAG |
| R | TGTTAGTGTCCCAAAATATCTTGTG | |
| N221W | F | GGAGGAGAATGGGTTCCTCCAG |
| R | TGTTAGTGTCCCAAAATATCTTGTG | |
| Q137Y | F | GCCAGTGTACGGCACCAG |
| R | TTCCCTGCACCATTGTCATGAG | |
| 137W | F | GCCAGTGTGGGGCACCAG |
| R | TTCCCTGCACCATTGTCATGAG |
Pentamer expression and purification.
Purification of JCPyV pentamers has previously been described (6). Briefly, 2 liters of Luria broth was inoculated with E. coli BL21(DE3) cells containing a pET15b-derived plasmid expressing wild-type or mutant VP1. These cultures were grown at 37°C to an optical density of 0.8. These cultures were then induced with 0.2 mM isopropyl β-d-1-thiogalactopyranoside and grown at 21°C for 18 h. Cultures were pelleted by centrifugation, resuspended in 20 ml of loading buffer (50 mM Tris-HCl [pH 7.5], 250 mM NaCl, 10 mM imidazole, 5% [vol/vol] glycerol), and frozen in liquid nitrogen. After thawing, the pellet was sonicated for 1 min on ice using a 50% duty cycle and 50% amplitude followed by a 5-min cooldown. After five rounds of sonication, bacterial debris was separated from the supernatant centrifugation at 17,640 × g for 20 min at 4°C. The supernatant was filtered through a 0.22-μm-pore-size filter and purified by immobilized metal ion affinity chromatography (IMAC) using a HisTrap 5-ml column and an Akta Purifier system (GE Healthcare). Pentamers were eluted from the column with a linear gradient of 10 to 500 mM imidazole, and fractions corresponding to pentamers were collected and dialyzed against a reaction mixture consisting of 20 mM Tris base (pH 7.5), 5% (vol/vol) glycerol, 250 mM NaCl, and 10 mM dithiothreitol (DTT). The sample was then concentrated, and nonaggregated VP1 pentamers were purified by size exclusion chromatography (SEC) using a Superdex S200 column and eluted in 20 mM HEPES (pH 7.5)–150 mM NaCl–50 mM DTT (GE Healthcare). All samples were concentrated to 1 mg/ml using a molar extinction coefficient of 28,030 M−1 cm−1 at 280 nm. For crystallization, the N-terminal hexahistidine tag was cleaved off in solution by incubation with 10 U thrombin (GE Healthcare) per mg VP1 for 20 h at 20°C prior to size exclusion chromatography (Superdex S200 column).
Coexpression and purification of VP1 and VP2 pentamers.
Full-length JCPyV VP2 lacking affinity tags was cloned into the pACYC-duet plasmid. E. coli BL21(DE3) cells were then cotransformed with full-length VP2 and wild-type or mutant VP1. VP1 and VP2 pentamers were then purified as described for the VP1-only pentamers.
Reconstitution of mutants and multicycle growth assay.
The infectious clone of each mutant was digested with EcoRI to liberate the viral genome from the plasmid backbone. SVGA cells were seeded onto 18-mm-diameter coverslips, and 1 μg of this digestion product was transfected using Fugene 6, according to the instructions of the manufacturer (Roche). Wild-type JCPyV was included as a positive control, and pUC19 alone was included as a negative control. Replicate samples were then assayed every 3 days, and the number of infected cells was scored to determine viral spread. At the time points specified, replicates for each mutant were fixed with methanol and stained with a monoclonal antibody against JCPyV VP1 (PAb 597). A goat anti-mouse Alexa 488 antibody was used as a secondary antibody, and nuclei were counterstained with DAPI (4′,6-diamidino-2-phenylindole; Life Technologies). Positive cells were counted on a T2000E inverted fluorescence microscope (Nikon), using the DAPI filter block to find a field of representative cells. Five representative fields for each replicate were counted, and samples were processed in a blind manner to minimize bias.
Crystallization.
JCPyV VP1 pentamers with an N221W, N221Q, or P223M mutation were concentrated to 4.5 mg/ml in 20 mM HEPES (pH 7.5)–150 mM NaCl. Crystals were set up at 20°C using the sitting-drop vapor diffusion technique and a reservoir solution containing 100 mM HEPES (pH 7.5), 200 mM KSCN, and 12% (wt/vol) polyethylene glycol (PEG) 3350. Drops (1 μl protein solution and 1 μl reservoir solution) were cross-seeded by adding microseeds obtained from previously grown JCPyV Mad-1 VP1 crystals. Crystals were flash-cooled in liquid nitrogen using a harvesting solution supplemented with 30% (vol/vol) glycerol.
Data collection and structure determination.
Diffraction data were collected at beamline X06DA at the Swiss Light Source (Villigen, Switzerland) and processed with XDS (21). Structures were solved by molecular replacement with Phaser (22) in CCP4 (23) using the JCPyV Mad-1 VP1 pentamer structure (PDB: 3NXG) lacking a solvent molecule(s) as a search model. In order to remove model bias, rigid-body and simulated annealing refinement was carried out with Phenix (24). Crystals of mutant VP1 pentamers have the same space group and very similar unit cell parameters. Mutations were introduced into the respective structural models, water molecules were added, and alternating rounds of model building in Coot (25) were performed. Refmac5 (26) was used for restrained refinement, including 5-fold NCS restraints and the translation-libration-screw (TLS) method (27). Simulated annealing Fobs-Fcalc omit electron density maps were calculated after refinement with Phenix by omitting the respective mutated amino acid residue for the validation of the final structural models. Coordinates and structure factor amplitudes have been deposited with the RCSB Protein Data Bank (www.pdb.org). Structural characteristics and radii of the 5-fold VP1 pore tunnels were visualized and analyzed with CAVER 3.0 (28). Structure figures were prepared with PyMOL (The PyMOL Molecular Graphics System, version 1.3; Schrödinger LLC). All amino acids are numbered without the inclusion of the N-terminal methionine, consistent with the previously described JCPyV VP1 structures (6, 29).
For the P223L mutant, a homology model was manually prepared in the Coot program by selecting possible rotamer conformations of P223L. Additionally, homology modeling was carried out with the program MODELLER (30). MODELLER implements comparative protein structure modeling via energy minimization and spatial restraints. The two models are highly similar and feature a diameter more similar to those seen with the wild type, N221W, and N221Q. However, due to the surrounding of the P223L, only a more uncommon side chain rotamer of L223 is allowed in the context of VP1 without steric hindrance.
Differential scanning fluorometry.
In order to assess the stability of wild-type and mutant pentamers, differential scanning fluorometry was performed using Sypro Orange, as previously described (31). Briefly, 4 μM VP1 monomers (0.8 μM VP1 pentamers) and 20 mM HEPES (pH 7.5)–150 mM NaCl–50 mM dithiothreitol (DTT) were mixed with a 10× solution of Sypro Orange in a final volume of 50 μl (Sypro Orange is provided as a 5,000× solution). Fluorescence was then monitored at the indicated temperatures in a Bio-Rad IQ5 real-time PCR machine, using the excitation filter for 6-carboxyfluorescein (FAM) and emission filter for ROX (Bio-Rad). A blank sample containing buffer but not pentamers was used as a negative control. The fluorescence from the negative control was subtracted from that measured for each sample. Six replicates were performed for each sample.
Negative-staining electron microscopy.
PSV virion formation was determined using negative-staining transmission electron microscopy (TEM). A 5-μl sample of each purified PSV was adsorbed onto carbon-coated Formvar copper grids for 2 min. Grids were washed with water, and 5 μl of nanoW, a tungsten-based pH neutral stain, was added to each grid for 1 min. Excess stain was blotted, and samples were visualized on a Phillips 410 transmission electron microscope. Micrographs were acquired at 80 kV and ×112,000 magnification.
Flow cytometry.
SVGA cells were seeded into 6-well plates at a density of 5 × 105 cells per well and incubated overnight. The following day, cells were detached using a nonenzymatic dissociation agent (Cellstripper; Cellgro) and incubated on ice for 30 min. For each PSV, an equivalent number of genomes was then added to each sample and allowed to incubate on ice for 2 h. Samples were then washed in cold PBS and immunostained with a fluorescence-conjugated monoclonal antibody to VP1 (PAB597-488). After incubation for 1 h on ice, excess antibody was washed away with PBS and samples were read on a BD FACSCalibur II flow cytometer (Becton Dickinson).
PLA.
Colocalization between JCPyV and the ER was performed as previously described (14). The primary antibodies used for the proximity ligation assay (PLA) were rabbit anti-VP1 and (in the case of JCPyV) mouse anti-PDI. Cells were either left untreated or pretreated for 30 min with 0.1 mM Retro-2cycl (Chembridge) and inoculated with 2 × 105 genome equivalents of JCPyV per cell. Virus was removed by aspiration, and any unbound virus was removed by washing with media. Fresh media containing 0.1 mM Retro-2cycl or media alone were added. Cells were then incubated at 37°C for 8 h prior to fixation with 4% paraformaldehyde (PFA). Cells were permeabilized with PBS containing 0.5% Triton X-100 for 0.5 h, washed three times in PBS, and then blocked in 5% donkey serum for 1 h at 37°C and stained for VP1 (ab53977; Abcam) (1:1,000 dilution) and PDI (ab2792; Abcam) (1:100 dilution) by overnight incubation at 4°C. Colocalization between VP1 and PDI was then assessed using the proximity ligation assay (PLA), following the manufacturer's instructions (Bethyl Labs). Cells were washed, and the cell nuclei were counterstained using DAPI. Fluorescence micrographs were collected by confocal microscopy, and maximal z-projections were displayed. Fluorescent foci were quantified using the Blobfinder program (http://www.cb.uu.se/∼amin/BlobFinder/). Threshold levels were set using a PLA-processed uninfected sample, and the same threshold levels were used across all samples. Blobfinder analysis was performed using a 7-by-7-pixel blob size, and at least 50 cells per sample were analyzed. The average number of blobs per cell was normalized to the wild-type PSV sample. Experiments were performed in triplicate, and error bars denote standard deviations.
VP2 exposure assays.
SVGA cells were seeded in 12-well plates at a concentration of 5 × 104 cells per well and incubated overnight. Cells were either left untreated or pretreated with 0.1 mM Retro-2cycl for 30 min. Samples were then inoculated with equivalent numbers of genome copies of JCPyV PSVs for 1 h at 37°C. Excess virus was washed off, and samples were incubated in either complete media or complete media with 0.1 mM Retro-2cycl. The cells were then stained with a polyclonal antibody to VP2 (ab53983; Abcam) and counterstained with DAPI. Randomized fields of view were imaged, and the number of cells with exposed VP2 was determined. Results were normalized to the wild-type PSV sample. Experiments were performed in triplicate, and error bars denote standard deviations.
Protein structure accession numbers.
Coordinates and structure factor amplitudes have been deposited with the RCSB Protein Data Bank (www.pdb.org) under accession codes 4WDY (N221Q), 4WDZ (N221W), and 4WE0 (P223M).
RESULTS
The 5-fold pore is conserved among all polyomaviruses.
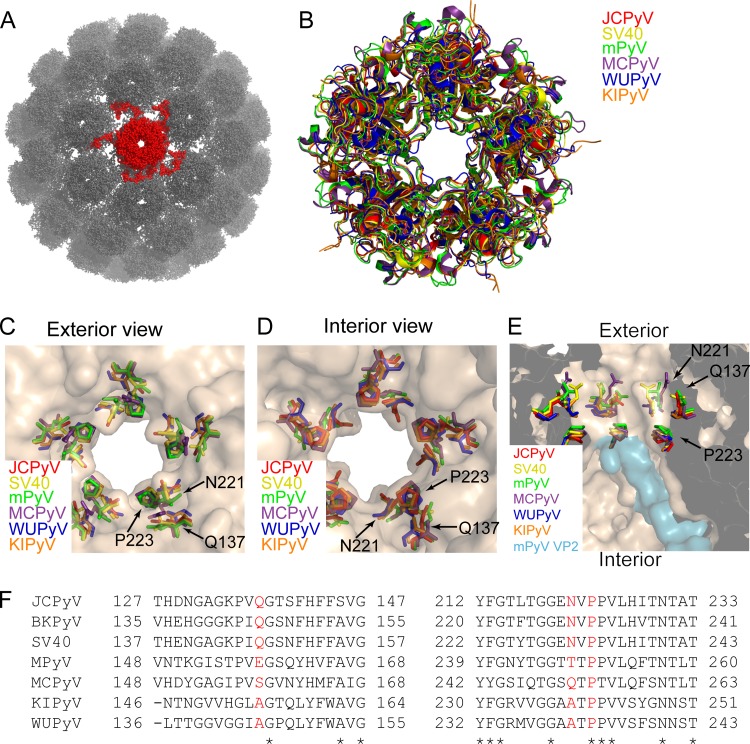
The polyomavirus 5-fold pore is formed by pentameric arrangement of VP1 monomers around the 5-fold axis of symmetry, as shown in the crystal structure of SV40 (Fig. 1A). Alignment of six divergent polyomavirus VP1 crystal structures demonstrates that the architecture of the 5-fold pore is conserved across polyomaviruses (Fig. 1B). In JCPyV, the N221 and P223 residues line the surface of the pore at its narrowest point, and Q137 is in close proximity to the 5-fold pore (Fig. 1C and D). This narrow constriction is in close proximity to VP2, as seen in the crystal structure of the VP1-VP2 complex in MPyV (Fig. 1E). The diameter of this pore is well conserved across structurally characterized polyomaviruses, with diameters ranging from 7.6 Å (WUPyV) to 8.6 Å (MPyV) at the respective bottlenecks describing the accessibility of the pore. The diameter of the pore was calculated by employing the Caver 3.0 algorithm (28). The sequence identity of amino acids lining the VP1 pore, including the highly conserved proline at position 223 in JCPyV VP1, which is present at corresponding positions in all analyzed polyomaviruses (Fig. 1F), is rather high among polyomaviruses.
FIG 1.
The VP1 5-fold pore is conserved across polyomaviruses. (A) The X-ray crystal structure of SV40 (PDB: 1SVA), represented by a ball and stick rendering. A single VP1 pentamer, containing the 5-fold pore, is colored red. (B) Alignment of the VP1 structures from six divergent polyomaviruses: JCPyV (PDB: 3NXG), SV40 (PDB: 3BWQ), MPyV (PDB: 1VPN), Merkel cell polyomavirus (MCPyV) (PDB: 4FMG), unassembled KI polyomavirus (KIPyV) (PDB: 3S7V), and unassembled Washington University polyomavirus (WUPyV) (PDB: 3S7X). (C) Exterior view of the 5-fold pore, illustrating that residues Q137, N221, and P223 line the pore at its narrowest point. (D) Interior view of the 5-fold pore. (E) Cross-sectional view of the interior axial cavity, showing the relationship of the indicated residues to VP2 in the MPyV VP1-VP2 complex structure (PDB: 1CN3). (F) Alignment of VP1 amino acid residues from seven different polyomaviruses. Red letters indicate residues that were substituted in this study; asterisks indicate residues that are conserved in all seven polyomaviruses.
Amino acid substitutions to residues lining the pore restrict growth.
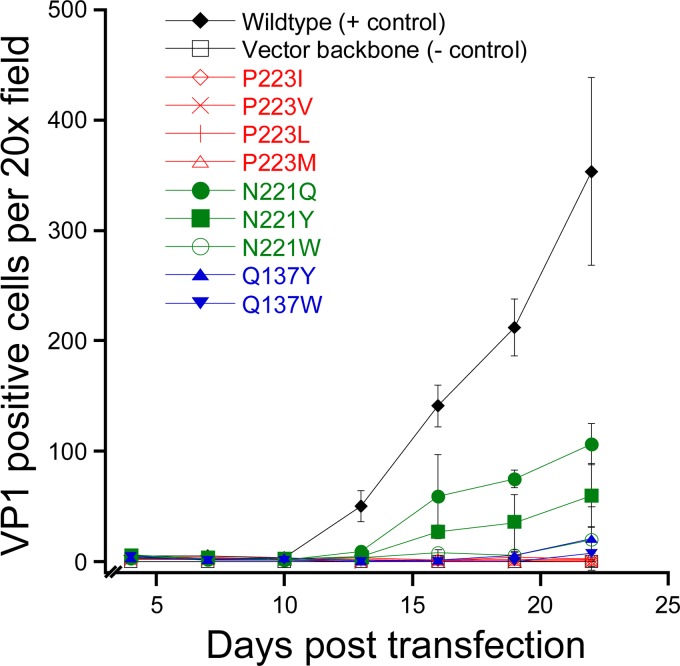
Using mutational analysis, we first sought to determine whether the 5-fold pore played a role in viral assembly or infectious cellular entry. A panel of mutant viruses were generated that contained amino acid substitutions to residues lining or near the 5-fold pore at its narrowest point. These residues, P223, N221, and Q137, were replaced with bulky amino acids that were predicted to reduce the diameter of the 5-fold pore. In addition, one mutation, N221W, was predicted to increase the hydrophobicity of this pore. Infectious clones of wild-type or mutant JCPyV were transfected into SVGA cells, and viral replication was assessed by multicycle growth assays. Transfection efficiencies were equivalent, and all samples expressed VP1, with an average of 4 VP1-positive cells per ×20 field (data not shown). This result indicated that these mutants were not defective in expressing the viral capsid protein. Whereas wild-type JCPyV spread throughout the tissue culture monolayer over a time period of 22 days, the 5-fold pore mutants were severely restricted in viral spread (Fig. 2). Only N221Q and N221Y, with 30% and 17% of wild-type levels, respectively, demonstrated limited viral spread. In order to verify that reversion mutations had not occurred in N221Q and N221Y, PCR and sequencing were performed on samples from the day 22 time point. VP1 sequencing demonstrated that these viruses had not reverted to a wild-type sequence. Thus, substitution of residues lining the 5-fold pore significantly reduced viral spread.
FIG 2.
Amino acid substitutions to residues lining the 5-fold pore reduce infectivity. Infectious clones of wild-type JCPyV or pore mutants were transfected into SVGA cells, and viral spread was monitored over 22 days. Data represent the means of the results of three independent experiments, and the error bars indicate standard deviations.
Substitutions to the 5-fold pore do not inhibit assembly of the VP1 pentamer.
Since Q137, N221, and P223 are spatially close to the 5-fold axis, it is likely that any steric clashes resulting from amino acid substitutions would be amplified due to the close proximity of symmetry-related VP1 monomers also containing these substitutions. We therefore sought to determine whether amino acid substitutions to residues lining the 5-fold pore would interfere with assembly of the VP1 pentamer. Five-fold pore mutations were introduced into a bacterial expression plasmid encoding a truncated VP1 pentamer that has been used extensively to characterize the structures of polyomaviruses (6). Recombinant VP1 pentamers containing these 5-fold pore mutations were expressed in bacteria and purified by affinity and size exclusion chromatography to determine if these substitutions either inhibited protein expression or resulted in pentamers of altered size. Size exclusion chromatography demonstrated that the elution times of all pentamers were similar, indicating that substitution of the 5-fold pore does not result in larger structural defects in the VP1 pentamer (Fig. 3A). A secondary peak is seen in some chromatographs and is due to the formation of interpentameric disulfide bonds.
FIG 3.
VP1 pentamers with altered 5-fold pores are not deficient in assembly. (A) A truncated VP1 containing a hexahistidine tag was expressed in bacteria and purified in order to assess the ability of pore mutants to form pentamers. (B) Truncated VP1 containing a hexahistidine tag and full-length VP2 lacking affinity tags were expressed in bacteria and purified by IMAC and SEC. Western blotting was used to determine whether the indicated pore mutants were able to copurify VP2. (C) Differential scanning fluorometry of VP1 pore mutants. The indicated VP1 was mixed with Sypro Orange, and fluorescence was monitored over the indicated range of temperatures. Max, maximum.
Pentamers containing pore mutations package the minor capsid proteins.
Previous studies have shown that a segment of VP2 is close to the 5-fold pore in MPyV (7). To address the possibility that substitutions to residues lining the 5-fold pore inhibit association with VP2, wild-type or mutant VP1 pentamers were coexpressed with full-length VP2 lacking any affinity tags. Therefore, VP2 should copurify with VP1 only when these proteins can still interact. After coexpression and purification, immunoblotting was used to assay for the presence of VP2 (Fig. 3B). The results demonstrate that wild-type VP1 and each mutant VP1 are able to bind to VP2 (Fig. 3B).
Pore mutant pentamers are not reduced in thermal stability.
We next sought to determine whether substitutions to residues lining the 5-fold pore reduced the stability of recombinant pentamers. To test this, differential scanning fluorometry was employed. This method involves the use of Sypro Orange, a fluorophore that fluorescences brightly when bound to nonpolar and, usually, buried residues. Assaying Sypro Orange fluorescence at various temperatures should therefore give an indication of the relative stability of each mutant pentamer. The results show an increase in Sypro Orange binding at between 40 and 50°C and no subsequent fluorescence at higher temperatures (Fig. 3C). Increased fluorescence was also seen for some mutants below 35°C. The cause of this increased fluorescence at low temperatures is not readily apparent but may be due to an increase in the levels of surface-accessible nonpolar residues in some mutants. The results were similar when VP1 and VP2 pentamers were used (data not shown). These results show that the wild-type VP1 pentamer and most 5-fold pore mutants are very similar with respect to thermal stability, with the exception of the Q137W mutant, which exhibits increased resistance to thermal denaturation.
VP1 mutations alter the pore morphology.
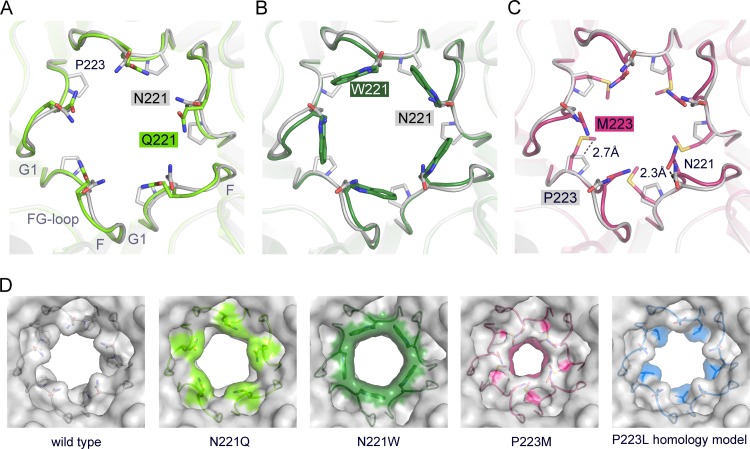
Next, in order to assess structural effects of the pore mutations on the atomic level, we solved X-ray structures of VP1 pentamers containing N221Q, N221W, and P223M substitutions at resolutions of 1.9, 1.8, and 2.1 Å, respectively (Table 2). Well-defined electron density was observed in all cases in the simulated annealing Fobs-Fcalc omit maps for the respective mutations. VP1 pentamer structures superpose with equivalent Cα atoms with root mean square deviation (RMSD) values of about 0.2 Å and are, as expected, very similar. A detailed analysis of the 5-fold pore shows very minor local structural changes due to the introduced mutations (Fig. 4). The diameter of the accessible pore of wild-type JCPyV VP1 was calculated to be about 8.2 Å at the narrowest point formed by residue N221 (28). Very minor local changes of the VP1 backbone were seen for N221Q and N221W (Fig. 4A and B). The glutamine alters the pore morphology and slightly narrows the accessible bottleneck diameter to about 7.1 Å, which is the tightest constriction of the pore, whereas the rather drastic N221W mutation does not reduce the pore diameter (the bottleneck diameter at W221 was calculated to be 8.6 Å) (Fig. 4D). Additional bulky electron density was observed within the pore of the N221W VP1 pentamer around the 5-fold axis. This density is unique for the N221W VP1 structure. It source could not be conclusively identified, but it is likely a hydrophobic solvent molecule that has no physiological relevance. The 5-fold cluster of hydrophobic tryptophans within the narrow pore favors hydrophobic interactions, including small hydrophobic solvent molecules also. The P223M substitution constrains the pore to a reduced diameter of about 5.2 Å at the bottleneck, where methionines face toward the 5-fold axis (Fig. 4C and D). Furthermore, additional minor structural rearrangements of the Cα backbone within the FG1-loop, such that N221 moves by about 2.3 Å toward the center of the pore (Fig. 4C), are seen. Because the P223L mutation was to be used in later experiments, a homology model of this mutant was generated. The diameter of the pore in the P223L homology model is much less restricted than in the P223M model and contains an accessible diameter of about 8.3 Å that is more similar to those seen with the wild type and the N221W and N221Q mutants (Fig. 4D).
TABLE 2.
Data collection and structural refinement statisticsa
| Statistic | Result |
||
|---|---|---|---|
| VP1 N221Q | VP1 N221W | VP1 P223M | |
| Space group | C2 | C2 | C2 |
| Unit cell dimensions | |||
| a, b, c (Å) | 149.71, 95.79, 128.51 | 149.34, 95.98, 128.5 | 149.72, 95.63, 128.51 |
| α, β, γ (°) | 90.0, 110.4, 90.0 | 90.0, 110.5, 90.0 | 90.0, 110.40, 90.0 |
| Resolution (Å) | 50–1.90 (1.95–1.90) | 50–1.8 (1.85–1.80) | 50–2.10 (2.15–2.10) |
| No. of unique reflections | 131,987 (8,272) | 155,280 (10,329) | 98,969 (7,258) |
| No. of observed reflections | 538,018 (24,462) | 633,087 (29,258) | 378,369 (27,027) |
| Rmeas (%) | 11.9 (91.0) | 9.2 (84.9) | 17.3 (96.6) |
| I/σI | 9.43 (1.7) | 11.7 (1.5) | 9.2 (1.7) |
| CC1/2 (%) | 99.7 (82.1) | 99.8 (59.1) | 99.2 (65.2) |
| Completeness (%) | 98.1 (83.5) | 98.8 (88.5) | 99.8 (99.3) |
| Wilson B-factor (Å2) | 31.9 | 30.7 | 29.8 |
| Refinement | |||
| Rwork, Rfree (%) | 19.1, 22.9 | 16.0, 18.6 | 18.0, 21.8 |
| No. of atoms | |||
| Protein | 10,061 | 10,096 | 10,058 |
| Water | 724 | 873 | 530 |
| Avg B-factor (Å2) | |||
| Protein | 31.2 | 26.7 | 29.1 |
| Water | 35.8 | 33.7 | 30.8 |
| RMS deviations | |||
| Bond length (Å) | 0.008 | 0.007 | 0.008 |
| Bond angle (°) | 1.315 | 1.269 | 1.308 |
Values for the highest resolution bins are given in parentheses. meas, measured; RMS, root mean square.
FIG 4.
Crystal structures of VP1 pentamers with N221Q, N221W, and P223M pore mutations. (A to C) Structural superposition of pore mutants N221Q (A), N221W (B), and P221M (C) with wild-type Mad-1 VP1. VP1 pentamers are shown in cartoon representations with side chains of key amino acid residues highlighted in stick representations and colored according to atom type (oxygens are shown in red, nitrogens in blue, and carbons in the colors assigned for the respective mutants). Wild-type VP1 is shown in gray. The accessible diameter at the narrowest constriction of the 5-fold pore was determined (28). The diameter was calculated to be 8.2 Å for wild-type VP1 and to be 7.1 Å and 8.6 Å for N221Q and N222W, respectively. The P223M mutation results in a constrained accessible pore diameter of 5.2 Å. (D) Surface representations of the 5-fold pore. Views are equivalent in all cases, and mutated amino acids are colored on the surface. A VP1 homology model for P223L was generated, and the most favorable rotamer conformation of Leu is shown in a surface representation using sticks.
JCPyV PSVs containing 5-fold pore substitutions are not morphologically altered or deficient in packaging VP2 or VP3.
In order to study the effect of these pore mutations on infectious entry of JCPyV, we next generated wild-type or mutant JCPyV PSVs. PSVs have been extensively used to characterize events in infectious entry of polyomaviruses and additionally allow the study of mutants that could not otherwise be propagated. We selected two pore mutants, N221W and P223L, for further study in order to directly assess the effect of amino acid substitutions of residues lining the surface of the 5-fold pore. PSVs were generated for Q137W, but large variations in the number of infectious particles recovered between preparations were observed, suggesting that amino acid substitutions at this residue may alter virion stability. Additionally, since the Q138W pentamer appeared to have altered stability in our thermal denaturation experiments compared to wild-type or other mutant pentamers, Q137W was excluded from further analysis.
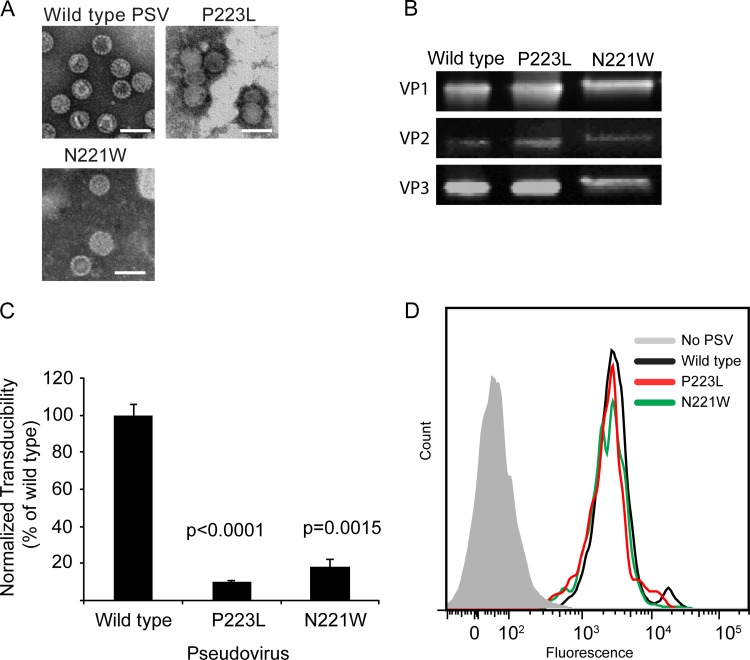
After purification, these PSVs were examined by negative-staining transmission electron microscopy to determine the effect of these substitutions on virion assembly. Wild-type and mutant PSVs appeared to be similar in morphology, demonstrating that PSVs containing these substitutions to the 5-fold pore are not grossly distorted or deficient in assembly (Fig. 5A). Immunoblotting also demonstrated that N221W and P223L PSVs packaged the minor capsid protein, further suggesting that substitution of residues lining the 5-fold pore does not interfere with minor capsid protein packaging (Fig. 5B).
FIG 5.
PSVs containing pore mutations are not deficient in assembly or binding to cells. (A) Wild-type or mutant PSVs were produced, and assembly was assessed by TEM. Scale bars denote 50 nm. (B) Western blotting was performed to demonstrate that all of the mutants were able to package VP2 and VP3. (C) Pore mutant PSVs are deficient in viral entry. SVGA cells were inoculated with equivalent numbers of genome copies of the indicated PSV, and luciferase expression was determined 72 hpi. (D) Pore mutant PSVs are not deficient in binding to cells. SVGA cells were inoculated with equivalent numbers of genome copies of the indicated PSV, and binding was determined by flow cytometry.
Pore mutant PSVs are deficient in transduction.
To determine whether modulation of residues surrounding the 5-fold pore affected viral entry, we performed single-cycle growth assays using wild-type or mutant PSVs that package a reporter plasmid expressing luciferase. Previous studies have demonstrated that expression of this reporter is directly proportional to the number of cells infected (32). Equivalent numbers of genome copies of each PSV were added to SVGA cells, and transduction efficiency was determined by determinations of relative luciferase expression levels. The results of these studies demonstrated that both N221W and P223L PSVs were deficient in transducing cells, suggesting a defect in viral entry (Fig. 5C).
Mutants are not defective in binding to host cells.
We next sought to determine what step in the infectious life cycle is inhibited as a result of amino acid substitutions in residues lining the 5-fold pore. Binding assays were used to determine whether mutant PSVs were deficient in binding to cells. Equivalent numbers of genome copies of each PSV were added to SVGA cells, and binding was determined using a fluorescently labeled monoclonal antibody to VP1. This analysis demonstrated that wild-type, N22Q, and P223L PSVs all bound to cells equivalently and suggests that these amino acid substitutions do not reduce the ability of these mutant virions to bind to cells (Fig. 5D).
Pore mutant PSVs are transported to the ER but are deficient in exposing VP2.
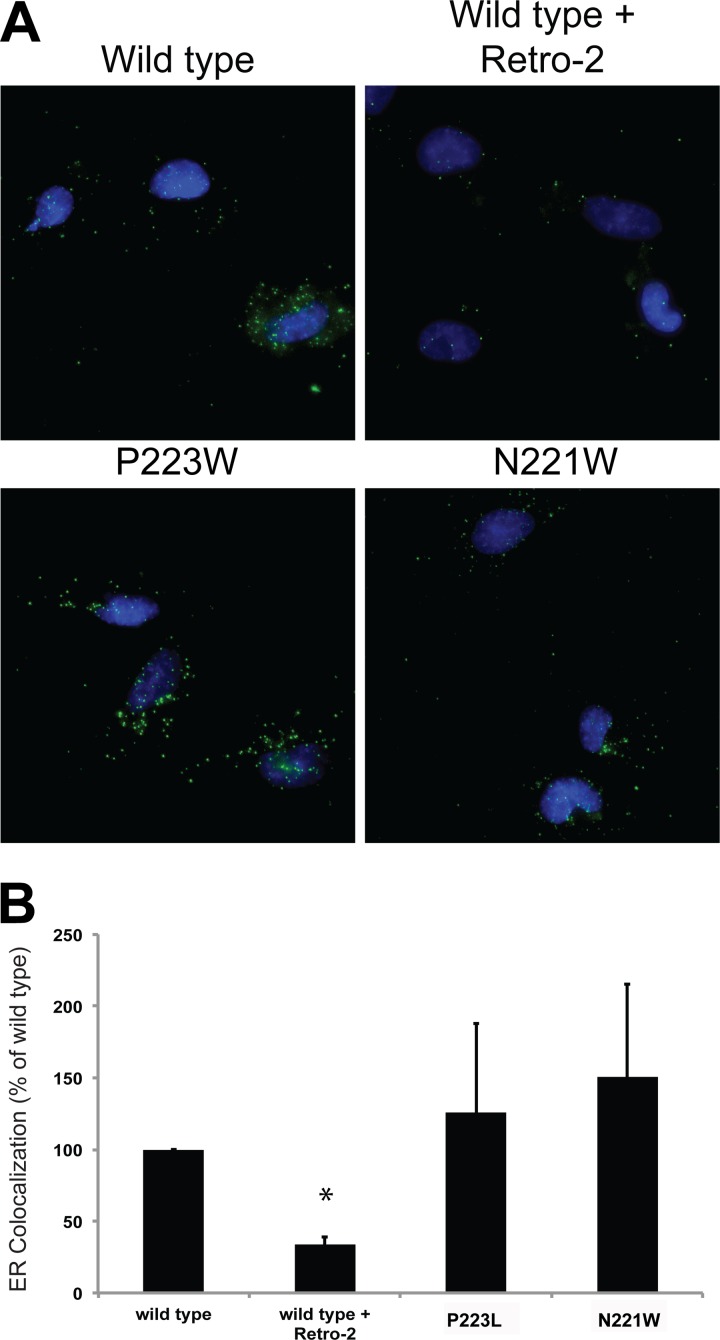
Transport of JCPyV to the ER is a critical step in infectious entry of JCPyV. Using proximity ligation assays, a method for reporting specific colocalization, we sought to determine whether these pore mutants were defective in ER transport. Cells were inoculated with equivalent numbers of genome copies of each virus, and samples were fixed and stained at 8 h postinfection, a time point that has been shown to correspond to the arrival of JCPyV at the ER (14). PLA using antibodies to VP1 and PDI demonstrated that the pore mutants were transported to the ER as efficiently as wild-type viruses. Conversely, treatment of cells with Retro-2, a recently described inhibitor of JCPyV retrograde transport, prevented colocalization of JCPyV and PDI (Fig. 6).
FIG 6.
Pore mutant PSVs are transported to the ER as efficiently as wild-type PSVs. (A) SVGA cells were inoculated with equivalent numbers of genome copies of each PSV, and ER colocalization was assessed at 8 hpi by proximity ligation assays. (B) Quantification of the data presented in panel A. Results represent the means of the results of three independent experiments, and error bars denote standard deviations. *, P < 0.05.
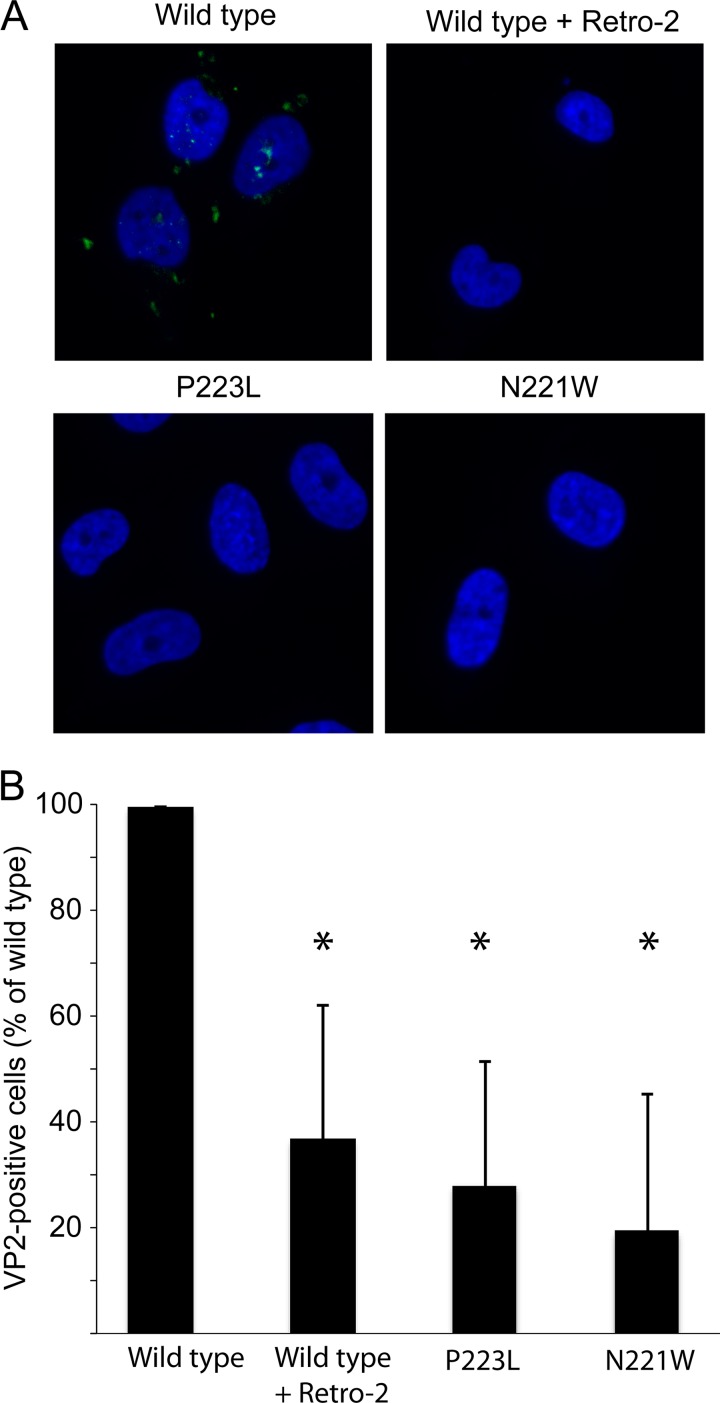
Upon delivery to the ER, JCPyV interacts with ER resident chaperones and exposes the VP2 minor capsid protein. Since these pore mutations are in close proximity to the packaging site of VP2, we next determined whether these mutants are able to expose VP2 in the ER. Cells were inoculated with equivalent numbers of genome copies of each virus, and samples were fixed and immunostained for VP2 at 8 h hpi. This analysis revealed that VP2 foci could be observed for wild-type PSVs. In contrast, Retro-2 treatment of cells reduced the level of VP2 exposure. Surprisingly, both of the pore mutants failed to expose VP2, suggesting that these mutants were deficient in undergoing productive conformational changes in the ER needed to expose VP2 (Fig. 7).
FIG 7.
Pore mutants are deficient in exposing VP2 in the ER. (A) SVGA cells were inoculated with equivalent numbers of genome copies of each PSV, and VP2 exposure was assessed at 10 hpi by immunostaining for VP2. (B) Quantification of the data presented in panel A. Results represent the means of the results of three independent experiments, and error bars denote standard deviations. *, P < 0.05.
DISCUSSION
Our report demonstrates that relatively subtle changes in residues lining the 5-fold pore of JCPyV result in dramatic decreases in infectivity. We show that these mutants are not deficient in assembly, binding to cells, or intercellular transport. Instead, these pore mutants are unable to expose the VP2 minor capsid protein in the ER during viral entry. This report suggests that contacts between the 5-fold pore and VP2 residues are a critical feature in the JCPyV capsid.
We initially suspected that this 5-fold pore was merely generated by the capsid symmetry and that residues occupying this space would result in significant structural clashes due to the close proximity of symmetry-related monomers. However, our results demonstrate that none of these substitutions results in assembly defects. In the case of N221Q, N221W, and P223M, only subtle changes were seen in the X-ray structure of VP1 pentamers, and similar minor changes were seen in the homology model of P223L. N221Q and N221W alter the pore diameter to only a very minor extent, but (in particular) the bulky N221W substitution mutation and, likely, also N221Y drastically change the molecular surface within the narrow pore and, likely, the interaction between VP1 and VP2. The structural impact of the P223M mutation on the FG1-loop conformation might explain why the prolines at equivalent VP1 positions are highly conserved. Although P223I, P223V, and P223L were not analyzed by X-ray crystallography, one could speculate about similar minor changes within the FG-loop. Leucine, valine, and isoleucine are smaller than methionine, so that the bottleneck radius might be less reduced; however, the nonpolar character at position 223 is preserved in all cases. Substitutions at amino acid position 137 (Q137Y and Q137W) were not reviewed by structural analysis, so the possibility cannot be ruled out that local structural rearrangement may occur. Indeed, differential scanning fluorometry analysis suggested that VP1 pentamers containing the Q137W substitution were altered in their thermal stability. The amino acid at position 137 does not directly line the 5-fold pore.
Our structural studies, demonstrating no significant structural differences between wild-type and mutant pentamers, coupled with the ability of both pentamers and PSVs to incorporate the minor capsid proteins, suggest that the mutant viruses do not display any drastic overall structural defects. This interpretation is additionally supported by the lack of differences in pentamer stability, as measured by differential scanning fluorometry. However, we cannot rule out the possibility that subtle structural changes in the capsid of these mutant viruses are responsible for the decrease seen in our multicycle growth assays.
PSVs containing these pore mutants were severely restricted in their ability to transduce susceptible cells, and the results suggested that these mutants were deficient at some step in viral entry. P223L and N221W were as efficient as wild-type PSVs in binding to cells and in transport to the ER, suggesting that that these pore mutants are still able to productively interact with cellular receptors and host transport factors. Instead, our VP2 exposure experiments demonstrate that these pore mutants do not expose VP2 in the ER. At present, the exact mechanism which causes these mutants to be deficient in exposure of VP2 is unclear.
The viral capsid of polyomaviruses is stabilized by interpentameric disulfide bonds, coordination of calcium ions, and hydrophobic interactions between the C terminus of VP1 and residues on the neighboring VP1 pentamer. It is likely that complete disassembly of the viral capsid requires disruption of the majority of these stabilizing factors. In SV40, virions remain largely intact following disulfide bond isomerization in the ER and retrotranslocation across the ER membrane occurs with virions in the form of large and intact particles (33). In vitro studies support this notion and demonstrate that treatment with the disulfide bond-reducing agent DTT is not sufficient for shedding of viral pentamers. Virions are seen to shed VP1 pentamers only after treatment with DTT and the calcium-sequestering molecule EGTA (34). Thin-section transmission electron microscopy studies have demonstrated that SV40 virions in the ER appear smaller in diameter and show visible VP2 in the form of extrusions (16). These studies suggested that VP2 is exposed in the absence of capsid disassembly. One possibility would be that the isomerization of intrapentameric disulfide bonds loosens the contacts between neighboring pentamers enough to allow exposure of the N terminus of VP2 though this opening. Alternatively, the 5-fold pore is situated near VP2, and previous studies have postulated that the 5-fold pore may provide a site of egress for the N terminus of VP2 (7). However, it should be noted that in vitro treatment of purified SV40 with ER luminal extracts results in distorted particles from which viral genomic DNA can be extracted, which suggests that capsid disassembly has occurred (16). It is unclear whether an analogous situation occurs during normal infectious entry of polyomaviruses.
At least four distinct possibilities exist for this observed defect in VP2 exposure. First, it is possible that these amino acids alter the interaction of VP1 with VP2, thereby interfering with the ability of the virion to expose VP2 during infectious cellular entry. Second, it is possible that these substitutions alter a recognition site for an as-yet-unidentified ER-resident chaperone or inhibit productive association with ER-residing chaperones, thereby inhibiting the conformational changes needed to expose the minor capsid proteins. Third, while these mutant viruses do not appear to be defective in ER transport, recent studies have shown that only a small percentage of virions that are transported to the ER actually expose VP2 or VP3. The identity of these parts of the ER is not well understood, but they appear to recruit chaperones such as BIP and DNAJ proteins to the foci where VP2 is exposed (35). It is therefore possible that these pore mutants are successfully transported to the ER but are deficient in transport to critical regions of the ER necessary for VP2 exposure and retrotranslocation. Finally, it is possible that the 5-fold pore of VP1 serves as a conduit for egress of the N terminus of VP2.
Definitively determining the site of VP2 egress from JCPyV is challenging. VP2 must be exposed for productive infection, and yet this exposure does not appear to be accompanied by complete disassembly of the viral capsid (16, 33). If disassembly of the viral capsid prior to VP2 exposure does not occur, then it is possible that VP2 is externalized either through the interpentameric gaps between monomers or through the pore present at the 5-fold axis of symmetry. While our results do not definitively determine the site of VP2 egress, they clearly demonstrate that modulation of this pore hampers exposure of VP2. Increased hydrophobicity within the pore and minor alterations of the pore diameter might alter, and perhaps strengthen, the contacts with VP2. Most of VP2 and VP3 is not resolved in the atomic resolution structures of MPyV, and it is therefore difficult to determine the extent to which the minor capsid proteins interacted with either the wild-type or the pore mutant in these studies. Further studies will be needed to definitively determine the mechanism by which these minor capsid proteins are exposed.
In conclusion, we demonstrate that the 5-fold pore of JCPyV is an important structural feature and that modification of this pore results in a severe reduction in infectivity. These mutations do not hamper assembly of the virion but instead prevent the exposure of the minor capsid proteins. This work suggests that the 5-fold pore is important for the function of infectious cellular entry and that implementation of small molecules targeting this pore may prove to be an effective antiviral therapy.
ACKNOWLEDGMENTS
We thank members of the Atwood laboratory for critical discussions and reviews of the manuscript.
This work was supported by R01NS043097 (W.J.A.), P01NS065719 (W.J.A. and T.S.), and a Ruth L. Kirschstein National Research Service award (F32NS070687) (C.D.S.N.).
We thank the members of the DiMaio laboratory at Yale University for helpful discussions and for their assistance in performing proximity ligation assays. Quantitative PCR and imaging were performed in the Genomics Core at Brown, which is supported by P30GM103410 (W.J.A.).
REFERENCES
- 1.Kean JM, Rao S, Wang M, Garcea RL. 2009. Seroepidemiology of human polyomaviruses. PLoS Pathog 5:e1000363. doi: 10.1371/journal.ppat.1000363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ferenczy MW, Marshall LJ, Nelson CD, Atwood WJ, Nath A, Khalili K, Major EO. 2012. Molecular biology, epidemiology, and pathogenesis of progressive multifocal leukoencephalopathy, the JC virus-induced demyelinating disease of the human brain. Clin Microbiol Rev 25:471–506. doi: 10.1128/CMR.05031-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zurhein G, Chou SM. 1965. Particles resembling papova viruses in human cerebral demyelinating disease. Science 148:1477–1479. [DOI] [PubMed] [Google Scholar]
- 4.Knipe DM, Howley PM, Griffin DE, Lamb RA, Martin MA, Roizman B, Straus SE (ed). 2007. Fields virology, 5th ed Lippincott Williams & Wilkins, Philadelphia, PA. [Google Scholar]
- 5.Liddington RC, Yan Y, Moulai J, Sahli R, Benjamin TL, Harrison SC. 1991. Structure of simian virus 40 at 3.8-A resolution. Nature 354:278–284. doi: 10.1038/354278a0. [DOI] [PubMed] [Google Scholar]
- 6.Neu U, Maginnis MS, Palma AS, Stroh LJ, Nelson CD, Feizi T, Atwood WJ, Stehle T. 2010. Structure-function analysis of the human JC polyomavirus establishes the LSTc pentasaccharide as a functional receptor motif. Cell Host Microbe 8:309–319. doi: 10.1016/j.chom.2010.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chen XS, Stehle T, Harrison SC. 1998. Interaction of polyomavirus internal protein VP2 with the major capsid protein VP1 and implications for participation of VP2 in viral entry. EMBO J 17:3233–3240. doi: 10.1093/emboj/17.12.3233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Elphick GF, Querbes W, Jordan JA, Gee GV, Eash S, Manley K, Dugan A, Stanifer M, Bhatnagar A, Kroeze WK, Roth BL, Atwood WJ. 2004. The human polyomavirus, JCV, uses serotonin receptors to infect cells. Science 306:1380–1383. doi: 10.1126/science.1103492. [DOI] [PubMed] [Google Scholar]
- 9.Assetta B, Maginnis MS, Gracia Ahufinger I, Haley SA, Gee GV, Nelson CD, O'Hara BA, Allen Ramdial SA, Atwood WJ. 2013. 5-HT2 receptors facilitate JC polyomavirus entry. J Virol 87:13490–13498. doi: 10.1128/JVI.02252-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pho MT, Ashok A, Atwood WJ. 2000. JC virus enters human glial cells by clathrin-dependent receptor-mediated endocytosis. J Virol 74:2288–2292. doi: 10.1128/JVI.74.5.2288-2292.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Querbes W, Benmerah A, Tosoni D, Di Fiore PP, Atwood WJ. 2004. A JC virus-induced signal is required for infection of glial cells by a clathrin- and eps15-dependent pathway. J Virol 78:250–256. doi: 10.1128/JVI.78.1.250-256.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Querbes W, O'Hara BA, Williams G, Atwood WJ. 2006. Invasion of host cells by JC virus identifies a novel role for caveolae in endosomal sorting of noncaveolar ligands. J Virol 80:9402–9413. doi: 10.1128/JVI.01086-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nelson CD, Derdowski A, Maginnis MS, O'Hara BA, Atwood WJ. 2012. The VP1 subunit of JC polyomavirus recapitulates early events in viral trafficking and is a novel tool to study polyomavirus entry. Virology 428:30–40. doi: 10.1016/j.virol.2012.03.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nelson C, Carney D, Derdowski A, Lipovsky A, Gee G, O'Hara B, Williard P, DiMaio D, Sello J, Atwood W. 2013. A retrograde trafficking inhibitor of ricin and Shiga-like toxins inhibits infection of cells by human and monkey polyomaviruses. mBio 4:e00729–13. doi: 10.1128/mBio.00729-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gasparovic ML, Gee GV, Atwood WJ. 2006. JC virus minor capsid proteins Vp2 and Vp3 are essential for virus propagation. J Virol 80:10858–10861. doi: 10.1128/JVI.01298-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Geiger R, Andritschke D, Friebe S, Herzog F, Luisoni S, Heger T, Helenius A. 2011. BAP31 and BiP are essential for dislocation of SV40 from the endoplasmic reticulum to the cytosol. Nat Cell Biol 13:1305–1314. doi: 10.1038/ncb2339. [DOI] [PubMed] [Google Scholar]
- 17.Magnuson B, Rainey EK, Benjamin T, Baryshev M, Mkrtchian S, Tsai B. 2005. ERp29 triggers a conformational change in polyomavirus to stimulate membrane binding. Mol Cell 20:289–300. doi: 10.1016/j.molcel.2005.08.034. [DOI] [PubMed] [Google Scholar]
- 18.Rainey-Barger EK, Magnuson B, Tsai B. 2007. A chaperone-activated nonenveloped virus perforates the physiologically relevant endoplasmic reticulum membrane. J Virol 81:12996–13004. doi: 10.1128/JVI.01037-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Vacante DA, Traub R, Major EO. 1989. Extension of JC virus host range to monkey cells by insertion of a simian virus 40 enhancer into the JC virus regulatory region. Virology 170:353–361. doi: 10.1016/0042-6822(89)90425-X. [DOI] [PubMed] [Google Scholar]
- 20.Aksamit AJ, Mourrain P, Sever JL, Major EO. 1985. Progressive multifocal leukoencephalopathy: investigation of three cases using in situ hybridization with JC virus biotinylated DNA probe. Ann Neurol 18:490–496. doi: 10.1002/ana.410180412. [DOI] [PubMed] [Google Scholar]
- 21.Kabsch W. 2010. XDS. Acta Crystallogr D Biol Crystallogr 66(Pt 2):125–132. doi: 10.1107/S0907444909047337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.McCoy AJ, Grosse-Kunstleve RW, Adams PD, Winn MD, Storoni LC, Read RJ. 2007. Phaser crystallographic software. J Appl Crystallogr 40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Winn MD, Ballard CC, Cowtan KD, Dodson EJ, Emsley P, Evans PR, Keegan RM, Krissinel EB, Leslie AG, McCoy A, McNicholas SJ, Murshudov GN, Pannu NS, Potterton EA, Powell HR, Read RJ, Vagin A, Wilson KS. 2011. Overview of the CCP4 suite and current developments. Acta Crystallogr D Biol Crystallogr 67:235–242. doi: 10.1107/S0907444910045749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Adams PD, Afonine PV, Bunkoczi G, Chen VB, Davis IW, Echols N, Headd JJ, Hung LW, Kapral GJ, Grosse-Kunstleve RW, McCoy AJ, Moriarty NW, Oeffner R, Read RJ, Richardson DC, Richardson JS, Terwilliger TC, Zwart PH. 2010. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr 66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Emsley P, Lohkamp B, Scott WG, Cowtan K. 2010. Features and development of Coot. Acta Crystallogr D Biol Crystallogr 66:486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Murshudov GN, Vagin AA, Dodson EJ. 1997. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr D Biol Crystallogr 53:240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- 27.Painter J, Merritt EA. 2006. Optimal description of a protein structure in terms of multiple groups undergoing TLS motion. Acta Crystallogr D Biol Crystallogr 62(Pt 4):439–450. doi: 10.1107/S0907444906005270. [DOI] [PubMed] [Google Scholar]
- 28.Chovancova E, Pavelka A, Benes P, Strnad O, Brezovsky J, Kozlikova B, Gora A, Sustr V, Klvana M, Medek P, Biedermannova L, Sochor J, Damborsky J. 2012. CAVER 3.0: a tool for the analysis of transport pathways in dynamic protein structures. PLoS Comput Biol 8:e1002708. doi: 10.1371/journal.pcbi.1002708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Maginnis MS, Stroh LJ, Gee GV, O'Hara BA, Derdowski A, Stehle T, Atwood WJ. 2013. Progressive multifocal leukoencephalopathy-associated mutations in the JC polyomavirus capsid disrupt lactoseries tetrasaccharide c binding. mBio 4:e00247–13. doi: 10.1128/mBio.00247-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Eswar N, Webb B, Marti-Renom MA, Madhusudhan MS, Eramian D, Shen MY, Pieper U, Sali A. 2006. Curr Protoc Bioinformatics 15:5.6:5.6.1–5.6.30. doi: 10.1002/0471250953.bi0506s15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Niesen FH, Berglund H, Vedadi M. 2007. The use of differential scanning fluorimetry to detect ligand interactions that promote protein stability. Nat Protoc 2:2212–2221. doi: 10.1038/nprot.2007.321. [DOI] [PubMed] [Google Scholar]
- 32.Schowalter R, Reinhold W, Buck C. 2012. Entry tropism of BK and Merkel cell polyomaviruses in cell culture. PLoS One 7:e42181. doi: 10.1371/journal.pone.0042181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Inoue T, Tsai B. 2011. A large and intact viral particle penetrates the endoplasmic reticulum membrane to reach the cytosol. PLoS Pathog 7:e1002037. doi: 10.1371/journal.ppat.1002037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Schelhaas M, Malmstrom J, Pelkmans L, Haugstetter J, Ellgaard L, Grunewald K, Helenius A. 2007. Simian virus 40 depends on ER protein folding and quality control factors for entry into host cells. Cell 131:516–529. doi: 10.1016/j.cell.2007.09.038. [DOI] [PubMed] [Google Scholar]
- 35.Goodwin EC, Lipovsky A, Inoue T, Magaldi TG, Edwards AP, Van Goor KE, Paton AW, Paton JC, Atwood WJ, Tsai B, Dimaio D. 2011. BiP and multiple DNAJ molecular chaperones in the endoplasmic reticulum are required for efficient simian virus 40 infection. mBio 2:e00101–11. doi: 10.1128/mBio.00101-11. [DOI] [PMC free article] [PubMed] [Google Scholar]