ABSTRACT
Saccharomyces cerevisiae and Saccharomyces paradoxus lack the conserved RNA interference pathway and utilize a novel form of copy number control (CNC) to inhibit Ty1 retrotransposition. Although noncoding transcripts have been implicated in CNC, here we present evidence that a truncated form of the Gag capsid protein (p22) or its processed form (p18) is necessary and sufficient for CNC and likely encoded by Ty1 internal transcripts. Coexpression of p22/p18 and Ty1 decreases mobility more than 30,000-fold. p22/p18 cofractionates with Ty1 virus-like particles (VLPs) and affects VLP yield, protein composition, and morphology. Although p22/p18 and Gag colocalize in the cytoplasm, p22/p18 disrupts sites used for VLP assembly. Glutathione S-transferase (GST) affinity pulldowns also suggest that p18 and Gag interact. Therefore, this intrinsic Gag-like restriction factor confers CNC by interfering with VLP assembly and function and expands the strategies used to limit retroelement propagation.
IMPORTANCE Retrotransposons dominate the chromosomal landscape in many eukaryotes, can cause mutations by insertion or genome rearrangement, and are evolutionarily related to retroviruses such as HIV. Thus, understanding factors that limit transposition and retroviral replication is fundamentally important. The present work describes a retrotransposon-encoded restriction protein derived from the capsid gene of the yeast Ty1 element that disrupts virus-like particle assembly in a dose-dependent manner. This form of copy number control acts as a molecular rheostat, allowing high levels of retrotransposition when few Ty1 elements are present and inhibiting transposition as copy number increases. Thus, yeast and Ty1 have coevolved a form of copy number control that is beneficial to both “host and parasite.” To our knowledge, this is the first Gag-like retrotransposon restriction factor described in the literature and expands the ways in which restriction proteins modulate retroelement replication.
INTRODUCTION
Retrovirus-like retrotransposons and their long terminal repeat (LTR) derivatives inhabit the genomes of many organisms, including the budding yeast Saccharomyces cerevisiae and its closest relative, Saccharomyces paradoxus. The Ty1 family is active and related to the LTR retrotransposons Ty2 to Ty5 in budding yeast (1). Variations in Ty1 copy number can be attributed to the relative rates of transposition, loss by LTR-LTR recombination, or additional types of genome rearrangements, all of which can impact fitness (2–5). Ty1 resembles retroviruses in genome organization and replication (1). These elements consist of two overlapping open reading frames (ORFs), GAG and POL, which are flanked by LTRs. Ty1 genomic RNA is translated or packaged as a dimer into virus-like particles (VLPs). The primary translation products are Gag (p49) and Gag-Pol (p199) precursors, the latter resulting from a +1 ribosomal frameshift during translation. Mature Gag (p45) is the major structural component of VLPs. POL encodes the enzymes required for proteolytic processing of Gag and Gag-Pol (protease [PR]), cDNA integration (integrase [IN]), and reverse transcription (reverse transcriptase [RT]). Ty1 and Ty3 VLPs assemble within cytoplasmic foci, termed retrosomes or T bodies, which contain Ty proteins and RNA (6–9). Once VLPs undergo maturation via the action of PR, Ty1 genomic RNA is reverse transcribed to form a linear cDNA. A protein/DNA complex minimally containing Ty1 cDNA and IN is imported into the nucleus, where integration usually occurs near genes transcribed by RNA polymerase III.
S. cerevisiae and S. paradoxus laboratory strains and natural isolates contain fewer than 40 copies of Ty1 per haploid genome, and several strains contain few if any elements (5, 10–14). Although budding yeast genomes characterized to date tend to have low Ty1 copy numbers, fertile S. cerevisiae strains containing more than 100 Ty1 insertions have been created artificially by numerous rounds of induction of a multicopy plasmid containing an active Ty1 element (Ty1H3) fused to the GAL1 promoter (pGTy1) (15, 16). Host cofactor and restriction genes involved in modulating Ty1 retrotransposition are diverse and encompass different steps in the replication cycle, ranging from transcription to integration site preference (17–21). For example, SPT3 is required for transcription of full-length Ty1 mRNA (22) and encodes a component of the SAGA chromatin-remodeling complex (23), and XRN1 is an important Ty1 cofactor implicated in transcription (24, 25) and assembly of functional VLPs (7, 8) and encodes a 5′-3′ exonuclease required for mRNA turnover (26).
Transposon-derived regulatory factors are critically important for keeping transposition at a low level. Forms of RNA interference (RNAi) affect the level or utilization of transposon mRNA, and the source of the interfering RNAs can be the transposons themselves (27). A unique form of copy number control (CNC) minimizes Ty1 transposition in S. cerevisiae and S. paradoxus (28) in the absence of dicer and argonaute genes that comprise a functional RNAi pathway in a distantly related species, Saccharomyces castellii (29, 30). Ty1 CNC is defined by a copy number-dependent decrease in Ty1 retrotransposition and is especially robust in a “Ty1-less” strain of S. paradoxus (28) that may have lost Ty1 elements by LTR-LTR recombination (13). Ty1 CNC acts posttranslationally and in trans, can be overcome by pGTy1 expression, and is characterized by lower levels of mature IN and reverse transcripts (28, 31, 32). Reduced levels of endogenous Ty1 IN, PR, cDNA, and VLPs are also present in S. cerevisiae (33–35), which displays CNC (28). These results suggest that Ty1 produces a titratable factor that inhibits transposition in a copy number-dependent manner.
Ty1 antisense (Ty1AS) RNAs have been implicated in silencing Ty1 expression by alterations in chromatin function (24) or when RNAi is reconstituted in S. cerevisiae (30). We reported evidence suggesting that Ty1AS RNAs interfere with Ty1 transposition posttranslationally (31). Inhibition occurs in a copy number-dependent manner, and the antisense transcripts map to a region within GAG that confers CNC (28). Deleting the common 3′ end of the antisense transcripts abolishes CNC and decreases the level of Ty1AS RNAs. However, ectopic expression of individual antisense transcripts does not restore CNC, suggesting that either multiple antisense transcripts or additional factors are required (24, 31). Also, nuclease protection and structural probing analyses suggest that although Ty1AS RNAs specifically associate with VLPs from CNC+ strains, these transcripts are not packaged into VLPs and do not interact with Ty1 mRNA (32).
Here, further characterization of the minimal Ty1 sequence that confers CNC has led to the discovery of p22, an N-terminally truncated form of Gag that is likely encoded by an internally initiated Ty1 mRNA. Importantly, p22 is both necessary and sufficient for CNC. Coexpression of p22 and Ty1 interferes with assembly of functional VLPs, which is conceptually similar to the inhibition displayed by Gag-like restriction factors derived from endogenous retroviruses in mammals (36, 37).
MATERIALS AND METHODS
Genetic techniques, media, and strain construction.
Strains are listed in Table 1. Strains repopulated with Ty1 elements were obtained following pGTy1 induction as described previously (28). Standard yeast genetic and microbiological procedures were used in this work (38).
TABLE 1.
Yeast strains
| Strain | Genotype | Plasmid(s) | Reference or source |
|---|---|---|---|
| DG2196 | MATa his3-Δ200hisG ura3 trp1 Ty1-less Ty1his3-AI(96) | 28 | |
| DG2254 | DG2196 | pGAL/2μ-URA3 | 28 |
| DG2255 | DG2196 | pGTy1/2μ-URA3 | 28 |
| DG2411 | DG2196 | Empty/2μ-URA3 | 28 |
| DG2374 | DG2196 | pBDG1130 (pGPOLΔ/2μ-URA3) | This study |
| YAS73 | DG2196 | pBAS39 (pGPOLΔ-T399C) | This study |
| YAS69 | DG2196 | pBAS38 (pGPOLΔ-T1108C) | This study |
| YAS71 | DG2196 | pBAS35 (pGPOLΔ-A1123G) | This study |
| YAS72 | DG2196 | pBAS36 (pGPOLΔ-A1296G) | This study |
| YAS70 | DG2196 | pBAS34 (pGPOLΔ-ΔA1456) | This study |
| YAS74 | DG2196 | pBAS43 (pGPOLΔ-Δ238-281) | This study |
| YAS75 | DG2196 | pBAS44 (pGPOLΔ-Δ238-353) | This study |
| JM321 | DG2196 | pBJM79 (pGPOLΔ-ΔC1071) | This study |
| JM320 | DG2196 | pBJM78 (pGPOLΔ-+A1303) | This study |
| DG2511 | DG2196 plus 12 Ty1 | This study | |
| DG3856 | DG2196 | pBDG1595 (pGPOLΔ-GCG1GCG2) | This study |
| DG2512 | DG2196 plus 9 Ty1 | This study | |
| DG3798 | DG2196 plus 7 Ty1-A1123G | This study | |
| DG1768 | MATα his3-Δ200hisG ura3 Ty1-less | 28 | |
| DG2533 | DG1768, Ty1–4253his3-AI | 3 | |
| DG2634 | DG1768, Ty1–4253his3-AI, plus 37 Ty1 | 31 | |
| YEM515 | DG2634, spt3-ΔkanMX4 | E. Matsuda | |
| GRF167 | MATα ura3-167 his3-Δ200 | 15 | |
| DG789 | GRF167, spt3-101 | 50 | |
| BY4742 | MATα his3-Δ1 leu2-Δ0 lys2-Δ0 ura3-Δ0 | 96 | |
| DG2247 | BY4742, spt3-ΔkanMX4 | 31 | |
| MAC103 | BY4742, xrn1/kem1-ΔkanMX4 | 7 | |
| DG3582 | DG1768, trp1 | This study | |
| DG3753 | DG3582 | pGAL/Cen-URA3, pBDG1534 (pGTy1his3-AI/Cen-TRP1) | This study |
| DG3751 | DG3582 | pBDG1354 (pGAL:1042-5889/Cen-URA3), pBDG1534 | This study |
| DG3739 | DG3582 | pGAL-Yes2/2μ-URA3, pBDG1534 | This study |
| DG3774 | DG3582 | pBDG1565 (pGAL-Yes2:1038-1616), pBDG1534 | This study |
| DG3784 | DG3582 | pBDG1565, empty/Cen-TRP1 | This study |
| JM367 | DG3582 | pBDG1568 (pGAL-Yes2:1038-V5-1616), pBDG1534 | This study |
| DG3791 | DG3582 | pBDG1571 (pGAL-Yes2:1038-1496), pBDG1534 | This study |
| JM399 | DG3582 | pBJM90 (pGAL-Yes2:1038-1616Gag*PR), pBDG1534 | This study |
| DG3808 | BY4742 | pEG(KT): pGAL:GST/2μ-URA3 | This study |
| DG3809 | BY4742 | pBDG1576 (pGAL:GST-1038-1496) | This study |
| DG3810 | DG3582 | pBDG1576 | This study |
Plasmids.
All nucleotide information used here corresponds to the Ty1H3 sequence (39) (GenBank M18706.1). pGPOLΔ derivatives of pGTy1 were generated by digestion with BglII and ligation. pBJM78, pBJM79, and pBDG1595 were constructed by overlap PCR using flanking primers (Ty335F, 5′-TGGTAGCGCCTGTGCTTCGGTTAC-3′; TyRP1, 5′-CATTGATAGTCAATAGCACTAGACC-3′) and overlapping primers (DELC1071b, 5′-GGTATCAGATTCATTTTTTCAATACTTTTGGAAAGAATTTTC-3′; DELC1071c, 5′-GTATTGAAAAAATGAATCTGATACCCAAGAGGCAAACGAC-3′; ADDA1303b, 5′-GAACAGTTCATGCGACTGTCATATTTAGATGTCGATGACGTG-3′; ADDA1303c, 5′-CTAAATATGACAGTCGCATGAACTGTTCTTAGATATCCATGC-3′; B-AUG1Ala-R, 5′-AAAGAATTTTCGCGATATCCGTATAATCAACG-3′; C-AUG1Ala-F, 5′-GGATATCGCGAAAATTCTTTCCAAAAGTATTG-3′; B-AUG2Ala-R, 5′-TATCAGATTGCGCTTTTTCAATACTTTTGG-3′; C-AUG2Ala-F, 5′-TGAAAAAGCGCAATCTGATACCCAAGAGGC-3′), and Ty1H3 as the template. Final PCR products were subcloned into pGPOLΔ using BstXI and BglII restriction sites. Plasmid pBDG1534 was generated from plasmid pBDG606 (pGTy1his3-AI/Cen-URA3) (18) by replacing the URA3 marker for TRP1. Briefly, TRP1 was amplified from BY4742 with primers containing flanking URA3 sequence (20718uratrpfwd, 5′-ATGTCGAAAGCTACATATAAGGAACGTGCTGCTACTCATCAATTCGGTCGAAAAAAGAAA-3′; 20916uratrprev, 5′-AGCTTTTTCTTTCCAATTTTTTTTTTTTCGTCATTATAATATGCTTGCTTTTCAAAAGGC-3′), and the PCR product was cotransformed into yeast with pBDG606 linearized within URA3 with ApaI. Transformants were selected on synthetic complete medium with Trp (SC-Trp medium), and plasmids were verified phenotypically and by restriction mapping. Plasmid pBDG1565 was created by PCR amplifying the Ty1 GAG coding sequence (nucleotides [nt] 1038 to 1613; EcoRIstartF, 5′-CATGTTTCGAATTCATGAAAATTCTTTCCAAAAGTATTG-3′; Xho1stopR, 5′-CATGTTTCCTCGAGTTAGTAAGTTTCTGGCCTAAGATGAAG-3′) using Ty1H3 as a template and cloning into pYES2 (Life Technologies, Carlsbad, CA) using EcoRI and XhoI. Plasmid pBDG1568 was made in a similar manner as pBDG1565, except that an initial PCR step was performed to insert V5 coding sequence (underlined) in frame between Ty1 GAG nt 1442 and 1443 (V51442b, 5′-CGTAGAATCGAGACCGAGGAGAGGGTTAGGGATAGGCTTACCTATAACTTTGGGTTTGGT-3′; V51442c, 5′-GGTAAGCCTATCCCTAACCCTCTCCTCGGTCTCGATTCTACGGCTCGGAATCCTCAAAAA-3′). For plasmid pBDG1571, GAG coding sequence cloned into pYES2 ended at nt 1496 (1496XhoI, 5′-CATGTTTCCTCGAGTTAGTGAGCCCTGGCTGTTTCG-3′). The GAG*PR mutation was created by mutating the Gag-PR cleavage site (RAHNVS) to AAGSAA (40) using overlapping primers (Gag*PRb, 5′-AGCCGCTGCTGGATCCGCTGCTACATCTAATAACTCTCCCAGC-3′; Gag*PRc, 5′-GATGTAGCAGCGGATCCAGCAGCGGCTGTTTTCGATTTCGAAT-3′). To construct the GAL1-promoted glutathione S-transferase (GST)–p18 protein fusion, the coding region for p18 (1038 to 1496) was amplified with XbaI and HindIII primer sets (1038XbaI, 5′-CTAGTCTAGACATGAAAATTCTTTCCAAAAGTATTG-3′; 1496XbaI, 5′-CCCAAGCTTTTAGTGAGCCCTGGCTGTTTTCG-3′). The PCR fragment was cloned into pEG(KT) (41), yielding pBDG1576. All plasmids generated by PCR cloning were verified by DNA sequencing. Phusion DNA polymerase, T4 DNA ligase, and restriction enzymes were obtained from New England BioLabs (Ipswich, MA).
Random mutagenesis and gap repair.
The Ty1 CNC region was mutagenized by amplification with Taq DNA polymerase (Thermo Fisher Scientific, Waltham, MA) using PCR forward primer FP1 (5′-CTCCGTGCGTCCTCGTCTTCACC-3′) and reverse primer RP1 (5′-CATTGATAGTCAATAGCACTAGACC-3′). Gel-purified PCR product was cotransformed into DG2196 along with a multicopy pGTy1 plasmid gapped with XhoI and BstEII. Gap-repaired transformants were selected on SC-Ura medium. Plasmids recovered from the CNC− strains were introduced into DG2196 to verify loss of CNC and then subjected to DNA sequencing. Aligning mutant sequences with Ty1H3 using ClustalW2 identified point mutations.
Ribo-seq analysis of chromosomal Ty1 elements.
Samples were prepared, and ribosome footprint profiling (Ribo-seq) was performed as previously described (42). Briefly, S. cerevisiae strain Sigma 1278b (YWG025; MATa ura3 leu2 trp1 his3) was grown to an optical density at 600 nm (OD600) of ∼1.0 to 1.1 at 30°C in yeast extract-peptone-dextrose (YEPD) medium, spun down, and resuspended in prewarmed yeast-peptone-adenine hemisulfate (YPA) (no glucose) medium. After 3 h in YPA medium, cycloheximide was added to a final concentration of 0.1 mg/ml and cells were harvested by centrifugation. Cells were lysed in 1× PLB {20 mM HEPES-KOH, pH 7.4, 2 mM magnesium acetate [Mg(OAc)2], 100 mM potassium acetate [KOAc], 1% Triton X-100, 0.1 mg/ml cycloheximide, 3 mM dithiothreitol [DTT]}, and libraries were prepared essentially as described previously (43). Reads were mapped to Ty1H3 using the STAR transcriptome sequencing (RNA-seq) alignment (44), allowing for zero mismatches. The Ty1 reads represent a composite of all Ty1 elements in the genome, including partial elements such as solo LTRs. No attempts were made to sort multiple mapping reads. The abundance of 5′-end reads was displayed over Ty1 using custom scripts available upon request. Libraries used for these analyses include NCBI GEO accession numbers SRX264202 and SRX366898 (Sigma Ribo-seq).
Isolation of cDNA clones.
An S. cerevisiae cDNA expression library fused to the GAL1 promoter on a centromere-based URA3 vector (45) was introduced into DG2196. Approximately 5,000 primary transformants were replica plated to SC-Ura plus 2% galactose and incubated for 3 days at 30°C. Colonies were then replica plated to SC-His-Ura, and Ty1HIS3 papillae were scored after incubation for 3 days at 30°C. Galactose induction was performed at a suboptimal temperature for transposition to sensitize the screen, since induction at 22°C resulted in too many Ty1HIS3 papillae. Most transformants yielded about 5 Ty1HIS3 mobility events/colony. Thirty-three transformants that had a lower level of Ty1 mobility were retested. Plasmids from 7 transformants were recovered in Escherichia coli and sequenced from their 5′ and 3′ ends using GAL1- and vector-specific primers. CCW12 (cell wall mannoprotein), MSS4 (phosphatidylinositol-4-phosphate 5-kinase), MRH1 (membrane protein), RGD1 (GTPase-activating protein), TIR1 (cell wall mannoprotein), and WHI5 (repressor of G1 transcription) were recovered as partial or complete cDNA clones and were not studied further. One clone (pBDG1354) contained Ty1 sequences from nt 1042 to 5889 and conferred a strong trans-dominant negative inhibition of Ty1his3-AI mobility.
RNA isolation.
Cultures were grown at 22°C for 24 h in SC or YEPD medium. Total RNA was extracted using the MasterPure yeast RNA purification kit (Epicentre Biotechnologies, Madison, WI) according to the manufacturer's protocol with minor modifications; 400 μl RNA extraction reagent and 200 μl of protein precipitation reagent were used instead of 300 μl and 160 μl, respectively. Poly(A)+ RNA was isolated from ∼250 μg total RNA using the NucleoTrap mRNA purification kit (Clontech, Mountain View, CA).
Northern blotting.
RNA was resolved on a 1.2% agarose-formaldehyde gel at 120 V for 2 h and blotted onto Hybond-XL N membranes (GE Healthcare, Little Chalfont, United Kingdom). Riboprobes were transcribed in vitro from Ty1 GAG and ACT1 coding sequence using a MAXIscript kit (Life Technologies) and uniformly labeled with [α-32P]UTP (3,000 Ci/mmol; PerkinElmer, Waltham, MA). Hybridization and phosphorimage analysis were carried out as previously described (19, 28).
5′ RACE.
Two hundred nanograms of poly(A)+ RNA was used for synthesis of the cDNA library using the SMARTer PCR cDNA amplification kit (Clontech). This method is 5′ cap independent, and the library included cDNA from all poly(A)+ transcripts. Ty1-specific cDNA was amplified with the gene-specific primer GSP1_3389 (5′-GACATGGGAGCAAGTAAAGGAAC-3′) and the universal primer mix from the supplier. Rapid amplification of cDNA ends (RACE) products were resolved on a 1% agarose gel. Gel-purified DNA fragments were TA cloned into pCR2.1-TOPO vector (Life Technologies). Plasmid DNA was subjected to DNA sequencing using Ty1-specific sequencing primer (Ty1new2rev; 5′-GAGAATCATTCTTCTCATCACTCG-3′).
qPCR.
The number of Ty1A1123G transposition events in strain DG3798 was estimated by quantitative PCR (qPCR). Strains DG2196 (Ty1his3-AI), DG2512 (Ty1his3-AI plus 9 additional Ty1 elements), and DG2511 (Ty1his3-AI plus 12 additional Ty1 elements) were used as standards, based on results from Southern analysis (28; H. W. Ahn and D. J. Garfinkel, unpublished results). Duplicate samples were subjected to qPCR using IQ SYBR green Supermix (Bio-Rad Laboratories, Hercules, CA) and two different primer pairs from Ty1 POL (4681F, 5′-GAAATTCAATATGACATACTTGGC-3′; +4851R, 5′-GTTCATCCTGGTCTATATATAAAGA-3′; 3251F, 5′-GAGAAGTTGACCCCAACATATCTG-3′; +3480R, 5′-TGTATGATTAGTCTCATTTTCAC-3′).
Ty1his3-AI mobility.
The frequency of Ty1his3-AI mobility was determined as described previously (28, 46) with minor modifications. For transposition assays involving strains containing pGPOLΔ, a single colony from an SC-Ura medium plate incubated at 30°C was resuspended in 1 ml of water and 5 μl of cells was added to quadruplicate 1-ml cultures of SC-Ura liquid medium. The cultures were grown for 3 days at 22°C, washed, diluted, and spread onto SC-Ura and SC-His-Ura medium plates to calculate Ty1 mobility. For mobility assays with strains repopulated with Ty1 elements, a single colony from a YEPD plate incubated at 30°C was diluted into 10 ml of water, and 1 μl of cell suspension was added to quadruplicate 1-ml YEPD cultures. The cultures were incubated for 2 to 3 days at 22°C, washed, diluted, and then spread onto YEPD and SC-His plates. Plates were incubated for 4 days at 30°C. For mobility assays involving strains expressing pGTy1his3-AI and GAL1-p22 or related plasmids, a single colony was resuspended in 1 ml SC-Ura-Trp medium plus 2% raffinose, grown for 16 h at 30°C, and then diluted 25-fold into quadruplicate 1-ml cultures of SC-Ura-Trp medium plus 2% galactose. Cultures were grown at 22°C for 2 days, washed, diluted, and spread onto SC-Ura-Trp and SC-Ura-Trp-His medium plates. Qualitative Ty1his3-AI mobility assays were performed as described previously (28, 46). For qualitative mobility assays involving strains containing pGPOLΔ, single colonies patched onto SC-Ura medium plates were incubated at 22°C for 2 days. To detect Ty1HIS3 mobility events, cells were replica plated onto SC-Ura-His medium plates and incubated at 30°C for 3 days. For strains expressing pGTy1his3-AI and pGAL-p22 or related plasmids, single colonies patched onto SC-Ura-Trp medium plates were incubated for 2 days at 30°C. The resulting patches were replica plated to plates of SC-Ura-Trp medium plus 2% galactose followed by incubation at 22°C for 2 to 4 days. To detect Ty1HIS3 mobility events, galactose-induced cells were replica plated to SC-Ura-Trp-His medium plates followed by incubation for 3 days at 30°C.
p18 antiserum.
Ty1 (1068 to 1496) was amplified with primers (1068NdeI, 5′-CATGTTCCATATGCAATCTGATACCCAAGAGGCAA-3′; 1496XhoI, 5′-CATGTTTCCTCGAGTTAGTGAGCCCTGGCTGTTTCG-3′) and cloned into pET15bTEV vector (Novagen EMD, San Diego, CA). An 800-ml culture of E. coli BL21(DE3) cells containing the expression plasmid in LB plus 100 μg/ml ampicillin (Sigma-Aldrich, St. Louis, MO) was induced by 0.15 mM isopropyl-β-d-thiogalactopyranoside (IPTG) (Sigma-Aldrich) at 37°C. When cells reached an OD600 of 0.6 to 0.8, the temperature was reduced to 16°C, and the cells were incubated for an additional 24 h. The cells were resuspended in 50 ml lysis buffer A (50 mM phosphate buffer, pH 7.8, 1 M NaCl) and harvested by sonication. The His-tagged Ty1 product was purified with Talon affinity resin (Clontech) and eluted with 300 mM imidazole (Sigma-Aldrich). The elution product was dialyzed against storage buffer (10% glycerol, 1 M NaCl, and 25 mM Tris-HCl [pH 8.0]) overnight. A rabbit polyclonal antibody was raised against the truncated Ty1 Gag protein by Bio-synthesis Inc. (Lewisville, TX).
Protein isolation and immunoblotting.
To detect protein expression from pGPOLΔ in the absence of galactose induction, 5 ml of SC-Ura medium was inoculated with a single colony and grown at 22°C for 24 h. For coexpression of independent pGAL expression plasmids, 1 ml of SC-Ura-Trp medium plus 2% raffinose was inoculated with a single colony and grown at overnight at 30°C. The overnight culture was diluted 25-fold into SC-Ura-Trp medium plus 2% galactose and grown for 2 days at 22°C. Five milliliters of culture was processed by trichloroacetic acid (TCA) extraction as described previously (47), except that cells were broken by vortexing in the presence of glass beads, and 10 μl of the supernatant was separated by electrophoresis. For sucrose fractions, equal volumes of each fraction were analyzed. For P40 and VLP samples, 5 μg of P40 was used to detect p22/p18 and 10 μg of P40 was used for RT and IN. Samples were separated on 10% (for RT and IN detection) or 15% (Gag p49/p45 and p22/p18 detection) SDS-PAGE gels. For optimal detection of p22/p18, proteins were transferred to a polyvinylidene difluoride (PVDF) membrane at 100 V for 90 min. The membranes were blocked in 5% milk–Tris-buffered saline–Tween (TBST) (500 mM NaCl, 20 mM Tris-HCl, 0.1% Tween 20, pH 7.6) and then incubated with rabbit polyclonal antisera at the following dilutions: anti-p18, 1:5,000 in 2.5% milk-TBST; anti-RT/B8, 1:5,000 in TBST; and anti-IN/B2, 1:2,500 in TBST (48). Immune complexes were detected with enhanced chemiluminescence (ECL) reagent (GE Healthcare, Little Chalfont, United Kingdom).
VLP isolation.
VLP purification from DG3739, DG3774, and DG3784 (Table 1) and reverse transcriptase assays were performed as described previously (49, 50) with the following modifications. Briefly, 40-ml cultures (SC-Ura-Trp medium plus 2% raffinose) of strains used for VLP analysis were grown overnight at 30°C with shaking. Each culture was diluted 25-fold into 1 liter of SC-Ura-Trp medium plus 3% galactose and grown at 21°C to an OD600 of 1 to 1.2. Cells were harvested by centrifugation at 6,000 rpm and homogenized with acid-washed glass beads in buffer B (15 mM KCl, 10 mM HEPES-KOH, pH 7, 5 mM EDTA) containing protease inhibitor cocktail (0.125 mg/ml aprotinin, leupeptin, and pepstatin A and 1.6 mg/ml phenylmethylsulfonyl fluoride [PMSF]). The crude lysate was centrifuged at 10,000 rpm, and the supernatant was loaded onto a step gradient of 20%, 30%, 45%, and 75% sucrose in buffer B. The step gradient was centrifuged at 25,000 rpm in an SW28 rotor for 3 h. Four milliliters of the gradient at the junction of the 30% and the 45% sucrose layers was withdrawn, diluted to 10% sucrose with buffer B, and pelleted by centrifugation at 55,000 rpm in a Ti 70.1 rotor for 45 min. The resulting crude VLP pellet (P40) was suspended in buffer B and centrifuged through a 20 to 60% continuous sucrose gradient in buffer B at 25,000 rpm in an SW41 rotor for 3 h. The entire gradient was dripped into 19 equal fractions using an ISCO Foxy Jr. fraction collector (Lincoln, NE). All steps were carried out 4°C unless specified. Fractions were assayed for Ty1 reverse transcriptase activity as described previously (34, 49), except that 10-μl samples were incubated with exogenous reverse transcriptase mix [50 mM Tris-HCl, pH 8, 10 mM MgCl2, 20 mM DTT, 15 μM dGTP, 10.7 μg poly(rC-dG)] and [α-32P]dGTP (3,000 Ci/mmol; PerkinElmer).
Electron microscopy.
Three sucrose gradient fractions with the highest reverse transcriptase activity from DG3739 (fractions 5 to 7) and DG3774 (fractions 4 to 6) were pooled, diluted with buffer B, and pelleted as described above. The sample was allowed to bind for 15 min to Formvar- and carbon-coated 400-mesh copper grids. Grids were stained with 2% ammonium molybdate, pH 6.5, for 10 s and visualized with a JEM-1210 transmission electron microscope (JEOL USA Inc., Peabody, MA) equipped with an XR41C bottom-mount charge-coupled device (CCD) camera (Advanced Microscopy Techniques, Woburn, MA). Approximately 100 VLPs were analyzed to determine the percentage of closed versus open particles. VLP diameter was measured with closed VLPs only using ImageJ (51), and the two data sets were compared using an unpaired t test.
FISH/IF.
Two-milliliter cultures of SC-Ura-Trp medium plus 3% raffinose were inoculated with a single colony and grown for 16 h at 30°C. The overnight cultures were diluted 10-fold into SC-Ura-Trp medium plus 3% galactose and grown at 22°C for 24 to 30 h until an OD600 of 0.8 to 1.0 was reached. Formaldehyde was added directly to the culture at a final concentration of 4% and allowed to fix for 1.75 h. Processing of the cells for fluorescence in situ hybridization-indirect immunofluorescence (FISH/IF) was performed as described previously (7). For Gag/p22-V5 colocalization experiments, primary antibodies were anti-VLP (rabbit polyclonal, 1:2,000; a kind gift from Alan and Susan Kingsman) and anti-V5 (Life Technologies; 1:4,000) and secondary antibodies used were anti-rabbit-AF488 (Life Technologies; 1:200) and anti-mouse-AF594 (Life Technologies; 1:400). Image acquisition was carried out using a Zeiss Axio Observer microscope equipped with an AxioCam HSm camera, and images were analyzed with AxioVision v4.6 software (Carl Zeiss Microscopy, LLC, North America). Exposure times used to capture fluorescent and 4′,6-diamidino-2-phenylindole (DAPI) images were kept consistent throughout each experiment. Figures were constructed with Adobe Photoshop software (Adobe Systems, San Jose, CA).
GST pulldown.
One milliliter of SC-Ura medium plus 2% raffinose was inoculated with a single colony at 30°C overnight and was then diluted 1:25 into 5 ml SC-Ura medium plus 2% galactose and grown for 2 days at 22°C. A 2.5-ml amount of galactose-induced cells was suspended in 150 μl lysis buffer C (20 mM Tris-HCl, pH 7.5, 100 mM NaCl, 10 mM MgCl2, 1 mM EDTA, 1 mM DTT, 0.1% Triton X-100, 1 mM PMSF, 1 μg/ml aprotinin, 0.5 μg/ml leupeptin, and 1 μg/ml pepstatin A) and homogenized with the same volume of acid-washed glass beads. The crude lysate was centrifuged at 10,000 rpm at 4°C for 10 min, and 500 μl supernatant containing 300 μg of protein was gently mixed with 20 μl glutathione-coated resin (GenScript, Piscataway, NJ) at 4°C for 2 h. The resin was washed three times with 1 ml lysis buffer C and then suspended in 40 μl SDS loading buffer. After boiling for 10 min, 5 to 8 μl per lane was loaded onto a 12% SDS-polyacrylamide gel. Immunoblotting was performed as described above, and membranes were incubated with mouse monoclonal antibody anti-GST/B-14 (Santa Cruz Biotech) at 1:1,000 or anti-TY tag (a kind gift from Stephen Hajduk) at 1:50,000 in TBST.
RESULTS
An internal Ty1 sense-strand RNA is required for CNC.
The CNC region of Ty1 spans the 5′ untranslated region (UTR) and all of GAG, and a multicopy pGTy1 expression plasmid confers CNC in trans even when GAL1-promoted transcription is repressed (28, 31). To identify sequences necessary for CNC (Fig. 1A), a genetic screen for CNC− mutations was performed in a Ty1-less S. paradoxus strain repopulated with a single chromosomal Ty1 insertion containing the selectable indicator gene his3-AI (46) (Table 1). Ty1HIS3 insertions usually occur by retrotransposition following splicing of the artificial intron. Since Ty1HIS3 cDNA can also undergo homologous recombination with genomic Ty1 elements or solo LTRs (52, 53), the term Ty1 “mobility” is used to describe both types of insertion. Ty1 mobility was followed using a qualitative papillation assay for His+ cells in a Ty1-less test strain containing a chromosomal Ty1his3-AI element and an empty vector, wild-type pGTy1 plasmid, or randomly mutagenized pGTy1. Cells were grown under repressive conditions for GAL1 expression. To generate point mutations, the CNC region was amplified using Taq DNA polymerase and PCR products were recombined into pGTy1 in vivo by gap repair. Approximately 3,500 pGTy1 recombinants were screened for loss of Ty1 CNC, and recovered plasmids were reintroduced to confirm the CNC− phenotype. Although pGTy1 plasmids with one to four base changes in the CNC region were identified, only plasmids carrying single mutations (Fig. 1A and Table 2) were analyzed further.
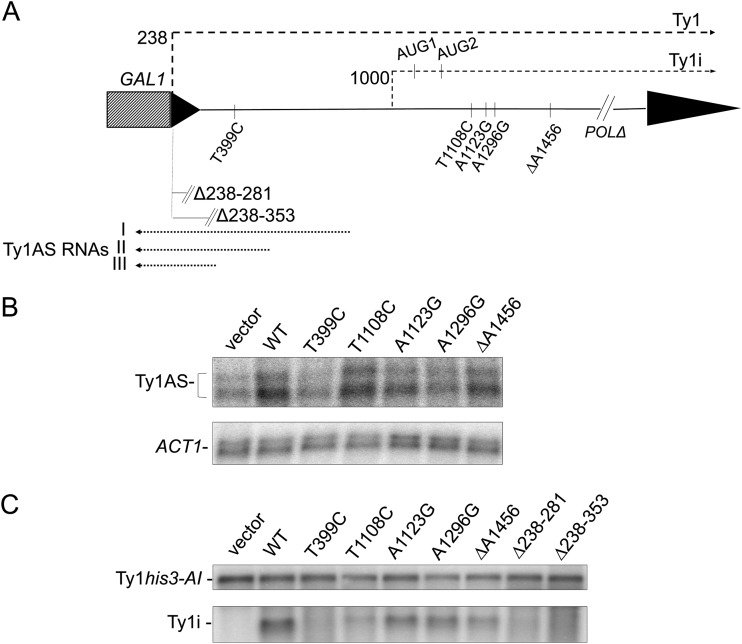
FIG 1.
An internal Ty1i transcript is involved in CNC. (A) Functional organization of the Ty1 CNC region, which covers GAG and the beginning of POL. Locations of the GAL1 promoter (hatched rectangle), LTR (solid triangle), Ty1 transcripts (Fig. 4), candidate initiation codons present on Ty1i RNA, and CNC− defective deletions and point mutations are noted. Ty1AS RNAs I, II, and III are shown with dotted lines. Ty1AS RNAs share a 3′ end at nt 136 but have different 5′ ends, nt 760 for II and 594 for III. The exact 5′ end of Ty1AS RNA I has not been determined (31). (B) Total RNA from a Ty1-less strain with a single chromosomal Ty1his3-AI element containing empty vector, wild-type (WT) pGPOLΔ (DG2374), or mutant plasmids T399C (YAS73), T1108C (YAS69), A1123G (YAS71), A1296G (YAS72), and ΔA1456 (YAS70) was analyzed by Northern blotting to detect Ty1AS RNAs. Cells were grown in glucose, and Ty1 strand-specific (nt 238 to 1702) and ACT1 32P-labeled riboprobes were used. (C) Total RNA from the strains in panel B, plus two additional strains containing mutant plasmids Δ238-281 (YAS74) and Δ238-353 (YAS75), was probed for Ty1i transcripts. Ty1his3-AI served as a loading control.
TABLE 2.
Ty1his3-AI mobility
| Group | Strain | Relevant genotype | Ty1his3-AI mobility, 10−6 (SD) | Fold decreasea |
|---|---|---|---|---|
| A | DG2254 | Ty1-less Ty1his3-AI pGAL/2μ | 220 (69) | 1 |
| DG2374 | pGPOLΔ | 7 (1.8) | 31 | |
| YAS73 | pGPOLΔ-T399C (Gag: Ser36Pro) | 24 (5.7) | 9.2 | |
| YAS69 | pGPOLΔ-T1108C (Gag: Leu272Pro) | 100 (27) | 2.2 | |
| YAS71 | pGPOLΔ-A1123G (Gag: Tyr277Cys) | 82 (11) | 2.7 | |
| YAS72 | pGPOLΔ-A1296G (Gag: Thr335Ala) | 42 (3.4) | 5.2 | |
| YAS70 | pGPOLΔ-ΔA1456 | 31 (9.7) | 7 | |
| B | DG2196 | Ty1-less Ty1his3-AI | 120 (14) | 1 |
| DG2511 | Plus 12 Ty1 | 3.6 (0.63) | 33 | |
| DG3798 | Plus 7 Ty1A1123G | 570 (150) | 4.8↑ | |
| C-1 | DG2411 | Ty1-less Ty1his3-AI empty/2μ | 140 (30) | 1 |
| DG2374 | pGPOLΔ | 2 (0.36) | 74 | |
| JM321 | pGPOLΔ-ΔC1071 | 61 (39) | 2.3 | |
| JM320 | pGPOLΔ-+A1303 | 58 (6.2) | 2.4 | |
| C-2 | JM321 | pGPOLΔ-ΔC1071 | 65 (22) | 2.1 |
| DG3856 | pGPOLΔ-GCG1GCG2 | 137 (23) | 1 | |
| D | DG3753 | Ty1-less pGTy1his3-AI/Cen pGAL/Cen | 21,000 (2,600) | 1 |
| DG3751 | pGAL:1042-5889 | 37 (8.8) | 570 | |
| E | DG3739 | Ty1-less pGTy1his3-AI/Cen pYES2/2μ | 60,000 (4,700) | 1 |
| DG3774 | pYES2: 1038-1616 | 1.7 (0.57) | 35,000 | |
| JM367 | pYES2: 1038-V5-1616 | 2 (0.65) | 32,000 |
Fold changes represent decreases, except where indicated by an upward arrow.
To minimize the possibility that sequence changes outside the gap-repaired region influence CNC and to facilitate molecular analyses, most of POL was deleted from the mutant pGTy1 plasmids to generate plasmid pGPOLΔ. Quantitative Ty1his3-AI mobility assays were performed with five mutants from the screen (Table 2). Mutations T399C, ΔA1456, and A1296G conferred moderate decreases in CNC compared with the CNC+ control, while T1108C and A1123G conferred low levels of CNC. Furthermore, the T1108C mutation affected CNC the most and was obtained from four independent isolates, suggesting that T1108 is part of an important sequence motif involved in CNC. Since Ty1AS RNAs were reported to be necessary for CNC (31), Northern blotting was performed with total RNA from the five single mutants. All of the mutants except T399C contained a level of Ty1AS RNAs similar to that produced from a wild-type pGPOLΔ plasmid, compared with the ACT1 loading control (Fig. 1B). Surprisingly, four of the five CNC− mutations do not map in the Ty1AS RNA transcription units and instead are located in an adjacent segment of the CNC region (Fig. 1A), and all change GAG's coding potential (Table 2).
A 5′-truncated Ty1 sense RNA can be detected in wild-type cells and is enriched in an spt3 mutant (22, 51). A similar observation was reported for an xrn1 mutant, where the RNA was termed Ty1SL (Ty1 short-length RNA) (24). Therefore, the point mutations identified in the screen could map in a shorter Ty1 sense RNA that initiates in GAG independently of normal Ty1 transcription, and this transcript could be involved in CNC. To determine if a shorter Ty1 sense RNA was produced from the pGPOLΔ plasmids, total RNA was subjected to Northern blotting using a strand-specific 32P-labeled riboprobe from GAG. Cells containing pGPOLΔ and mutant derivatives were used in the Northern blotting since deleting POL results in the synthesis of Ty1 transcripts that are clearly distinguishable from Ty1his3-AI RNA. All point mutants except T399C made a shorter sense-strand Ty1 RNA, termed Ty1 internal (Ty1i) RNA, whereas cells containing an empty vector control lacked this transcript (Fig. 1C). Two additional mutants, Δ238-281 and Δ238-353, were derived in the pGPOLΔ context and included in this analysis. Originally described in the work of Matsuda and Garfinkel (31), pGTy1 plasmids with short deletions in the 5′ LTR abolished CNC. The loss of CNC was attributed to a decrease in the level of the Ty1AS RNAs due to deletion of their 3′ ends. However, the lack of detectable Ty1i RNA in the Δ238-281 and Δ238-353 mutants may now explain their CNC− phenotype. These results also suggest that sequences near the 5′ LTR, which contains the enhancer required for Ty1 transcription (5), may also be important for synthesizing Ty1i RNA.
Chromosomal Ty1A1123G insertions fail to confer CNC.
The genetic screen identified several missense mutations in GAG that weakened CNC and were present on both Ty1 mRNA and the Ty1i transcript. To determine if CNC− mutations impacted Gag function, full-length pGTy1 plasmids containing A1123G (Tyr277Cys) or A1296G (Thr335Ala) mutations were compared with wild-type pGTy1 for their ability to stimulate or trans-activate movement of a chromosomal Ty1his3-AI element (33). The A1296G mutation likely affects both CNC and transposition, since pGTy1A1296G expression did not stimulate Ty1 mobility. However, induction of pGTy1A1123G increased Ty1his3-AI mobility in trans to levels similar to those observed with wild-type pGTy1, suggesting that pGTy1A1123G encodes functional Gag and yet is defective for CNC (data not shown). To determine if Ty1A1123G conferred CNC in a natural chromosomal context (Fig. 2), a strain with a single chromosomal Ty1his3-AI (Fig. 2A) was repopulated with wild-type (Fig. 2B) or Ty1A1123G (Fig. 2C) elements. As expected, Ty1 mobility decreased 33-fold in a strain repopulated with 12 wild-type Ty1 elements compared with the starting strain (Fig. 2B; Table 2). However, Ty1 mobility increased almost 5-fold in a strain containing 7 copies of Ty1A1123G, indicating that A1123G abolishes CNC without disrupting the function of Gag (Fig. 2C; Table 2). The separation of function phenotype displayed by Ty1A1123G raised the possibility that an altered form of Gag encoded by Ty1i RNA mediates CNC.
FIG 2.
Chromosomal Ty1A1123G insertions do not confer CNC. Ty1-less S. paradoxus containing a single chromosomal Ty1his3-AI (A) was repopulated with unmarked, wild-type (B), or A1123G (C) Ty1 elements. Genome repopulation with 12 wild-type Ty1 elements resulted in an overall decrease in Ty1his3-AI mobility, while repopulation with 7 CNC− mutant Ty1A1123G elements resulted in an overall increase in Ty1his3-AI mobility. Also refer to Table 2.
Expression of Ty1i RNA.
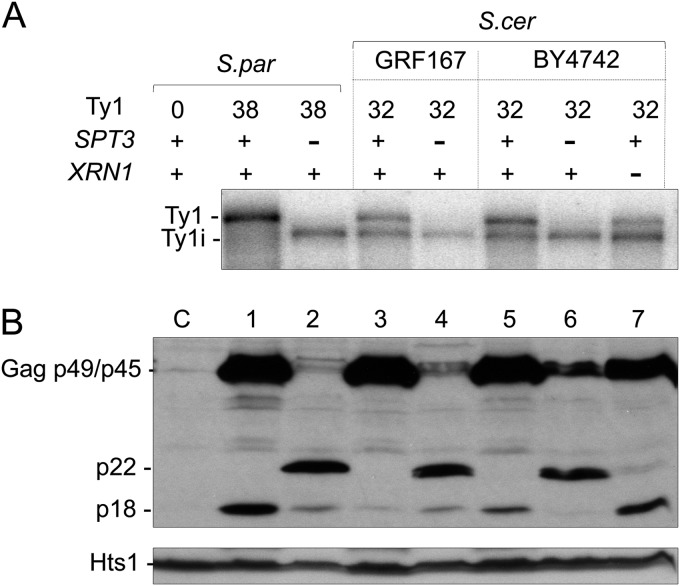
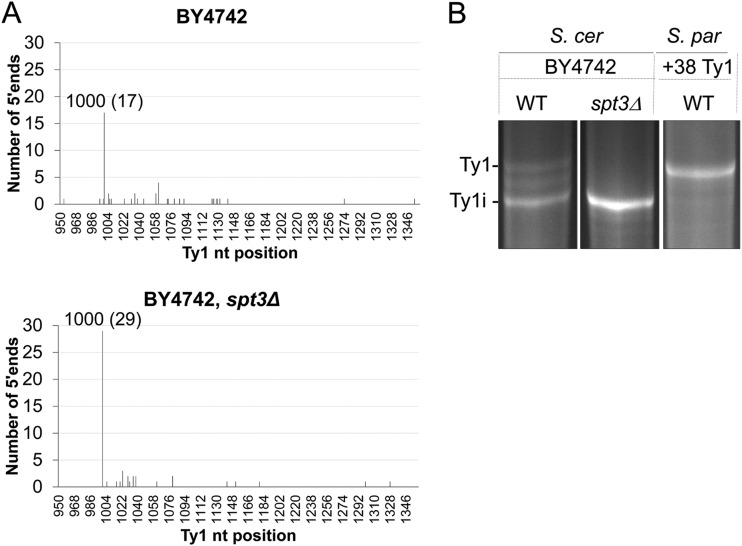
Since multicopy Ty1 plasmids were used as the source of trans-acting factors required for CNC, it was important to determine if chromosomal elements also synthesized Ty1i RNA and truncated forms of Gag (Fig. 3). To detect Ty1i RNA in repopulated S. paradoxus as well as S. cerevisiae strains, poly(A)+ RNA was subjected to Northern blotting using a 32P-labeled riboprobe from GAG-POL (nt 1266 to 1601) (Fig. 3A). Three S. paradoxus strains were analyzed: the Ty1-less strain (Fig. 3A, lane C), a derivative repopulated with 38 Ty1 elements (lane 1), and an isogenic spt3Δ mutant (lane 2). Five S. cerevisiae strains were also analyzed: GRF167 (Fig. 3A, lane 3) and an isogenic spt3Δ mutant (lane 4), BY4742 (lane 5), and isogenic spt3Δ (lane 6) and xrn1Δ (lane 7) mutant derivatives. A discrete subgenomic Ty1 RNA of 4.9 kb was detected below the full-length transcript (5.7 kb) in all strains except repopulated S. paradoxus (Fig. 3A, lane 1). The failure to detect a distinct transcript in this strain was unexpected but may result from 5′ heterogeneity of the 4.9-kb transcript. The 4.9-kb Ty1 RNA comigrated with the truncated transcripts detected in spt3Δ and xrn1Δ mutants. To determine the 5′ end of the 4.9-kb transcript in BY4742 and an isogenic spt3Δ mutant, poly(A)+ RNA was subjected to cap-independent 5′ RACE (Fig. 4). In both strains, the majority of the 5′ ends from the 4.9-kb transcript mapped to nucleotide 1000 of Ty1H3 (Fig. 4A). These results indicate that the 4.9-kb RNA observed in the wild-type and an spt3Δ mutant share the same 5′ ends. However, 5′-RACE analysis of the wild-type repopulated S. paradoxus strain showed heterogeneous amplification products (Fig. 4B) rather than discrete bands, supporting the results from Northern blotting (Fig. 3). Although our results suggest that the 4.9-kb Ty1 RNA contains the Ty1i transcript in S. cerevisiae, other truncated forms of Ty1 RNA may be present (54, 55).
FIG 3.
Detecting Ty1i RNA and p22/p18 from chromosomal Ty1 elements. (A) Northern blotting of poly(A)+ RNA from S. paradoxus and S. cerevisiae (GRF167 and BY4742) wild-type and spt3Δ (DG789 and DG2247) and xrn1Δ (MAC103) mutant strains. Ty1 32P-labeled riboprobe (nt 1266 to 1601) hybridized with full-length Ty1 and Ty1i transcripts. (B) Total protein extracts were immunoblotted with the p18 antiserum to detect full-length Gag p49/p45 and p22/p18. A Ty1-less S. paradoxus strain (DG1768) and cellular histidyl tRNA synthetase (Hts1) served as negative (lane C) and loading controls, respectively.
FIG 4.
The major 5′ end of the 4.9-kb Ty1i RNA maps to nt 1000. (A) Cap-independent 5′ RACE was performed with poly(A)+ RNA from wild-type BY4742 and an isogenic spt3Δ mutant (DG2247). The number of 5′ termini was plotted against the Ty1H3 sequence, and that and the distribution of the termini are on the x and y axes, respectively. The tallest peak represents the total number of 5′ ends captured at nt 1000 and is shown in parentheses. (B) 5′ RACE cDNA libraries from the wild-type and spt3Δ strains mentioned above and a repopulated S. paradoxus strain (DG2634) were amplified using a universal primer mix and a Ty1-specific primer, GSP1_3389. The amplification reaction mixtures were separated by agarose gel electrophoresis to demonstrate the presence of cDNA products corresponding to the 5′ ends of the full-length (5.7-kb) Ty1 and the truncated (4.9-kb) Ty1i RNAs.
Ty1i RNA encodes Gag proteins p22 and p18.
Two closely spaced AUG codons are present 38 (AUG1) and 68 (AUG2) nucleotides downstream of the transcription start site for Ty1i RNA, and one or both may be utilized to initiate synthesis of a truncated form of Gag (Fig. 1A). However, neither the predicted 22-kDa (p22) Gag-like protein nor its processed product (p18), if p22 is cleaved by Ty1 PR, has been reported to date, and a commonly used VLP antiserum (56) failed to detect p22/p18 reproducibly (data not shown). Therefore, we purified recombinant p18 and generated a new antiserum to determine if Ty1i RNA is translated to produce an N-terminally truncated form of Gag (Fig. 3B). Whole-cell extracts from the strains described above (Fig. 3A) were immunoblotted with p18 antiserum to detect endogenous Gag and additional Gag-related proteins. As expected, normal levels of Ty1 Gag p49/p45 were detected in wild-type strains (Fig. 3B, lanes 1, 3, and 5) while reduced levels were observed in spt3Δ (lanes 2, 4, and 6) mutants. Importantly, p22 was detected in the spt3Δ mutants (lanes 2, 4, and 6), whereas p18 was detected only in the wild-type strains (lanes 1, 3, and 5) and the xrn1Δ mutant (lane 7). The increase in p18 observed in the xrn1Δ mutant likely results from an increase in Ty1i RNA level, since Xrn1 is the major 5′-3′ exonuclease involved in RNA decay. Taken together, not only do our results suggest that Ty1i RNA encodes p22, but the striking relationship between expression of full-length Ty1 mRNA, and hence Ty1 PR, and detection of p22 versus p18 suggests that p22 is cleaved by Ty1 PR to form p18. Furthermore, processing of p22 to p18 raises the possibility that p22 associates with VLPs to gain access to PR. As expected, Gag proteins were not detected in the Ty1-less strain (Fig. 3B, lane C).
Ribosome footprint profiling reveals an internal AUG as a potential translation start for p22.
To determine if the candidate AUG1 or AUG2 translation start sites on Ty1i RNA (Fig. 1A) were present in genomic sequencing analyses, we turned to ribosome footprint profiling (Ribo-seq) (Fig. 5). In Ribo-seq, ribosomes in the act of translating an mRNA are treated with RNase I, leaving a ∼28-nt ribosome footprint, which is harvested for high-throughput sequencing to provide a snapshot of the abundance and distribution of ribosomes on mRNAs (43). Yeast starved for glucose for 3 h accumulate as much as 10% of Ribo-seq reads at the start codon of open reading frames (ORFs) (42), providing a sensitive method for detecting initiation codons in vivo. We utilized a published data set to analyze the Ribo-seq read distribution at the 5′ end of Ty1i RNA (57). The most abundant read in this region corresponded to a ribosome footprint located on AUG1, which is the first start codon downstream of the Ty1i transcription start site (Fig. 4A). Also, the density of Ribo-seq reads increased downstream of AUG1, consistent with translation of the downstream ORF under glucose starvation. Additional mutational analysis of AUG1 and AUG2 will be required to verify the translation start of p22.
FIG 5.
Whole-genome analysis of internal translation initiation sites. Ribosome footprint profiling (Ribo-seq) was performed to detect translation initiation at internal AUG codons, two of which (AUG1 and AUG2 [Fig. 1]) are located immediately downstream of the Ty1i RNA transcription start site. Reads per million (rpm) were placed on the Ty1H3 sequence, and the 5′ ends of ribosome footprints aligned downstream of the Ty1i transcription start are shown. Ribo-seq reads with 5′ ends 12 to 13 nt upstream of AUG1 and AUG2 are highlighted in orange and green, respectively. The position ∼12 nt downstream of the 5′ end corresponds to the ribosomal P site. Because these libraries were prepared with poly(A) tailing, the exact 3′ end of the footprint, and thus the footprint size at AUG1, is ambiguous but within the range of 26 to 30 nt, inclusive.
p22/p18 encoded by Ty1i RNA is necessary for CNC.
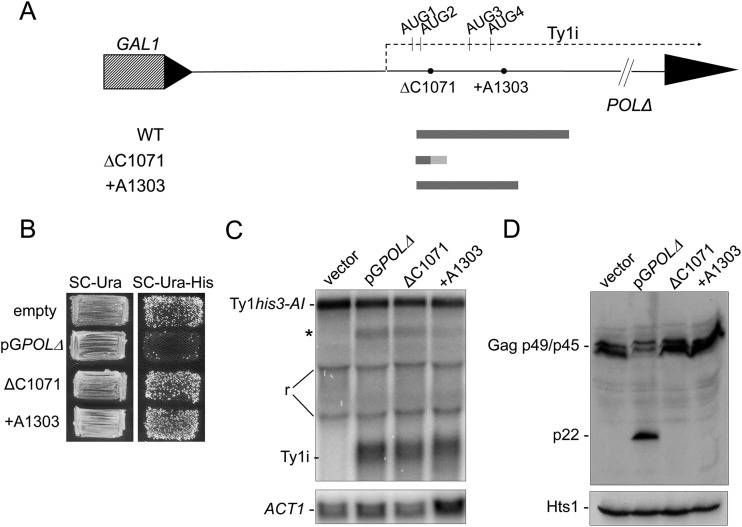
To establish that p22/p18, rather than the Ty1i transcript itself, is responsible for CNC, we analyzed frameshift mutations in the pGPOLΔ construct (Fig. 6A) for alterations in Ty1his3-AI mobility (Fig. 6B; Table 2), Ty1i RNA levels (Fig. 6C), and protein levels (Fig. 6D). Cells containing pGPOLΔ decreased the mobility of a chromosomal Ty1his3-AI element up to 74-fold (Fig. 6B; Table 2) compared to an empty vector control and produced Ty1i RNA (Fig. 6C) and p22 (Fig. 6D). Two frameshift mutations were placed downstream of AUG1 and AUG2 that introduce premature termination codons, ΔC1071 and +A1303. +A1303 was created to eliminate the possibility that downstream in-frame AUGs (AUG3 and AUG4, Fig. 6A) could be utilized to produce a trans-dominant factor. Both frameshift mutations caused an increase in Ty1his3-AI mobility to almost the same level as that obtained in a strain lacking CNC (Fig. 6B; Table 2). Cells containing the mutant plasmids produced Ty1i RNA (Fig. 6C) but not wild-type p22 (Fig. 6D). The residual level of CNC conferred by the plasmids carrying the frameshift mutations may be caused by truncated protein synthesized prior to encountering the mutations; however, immunoblotting using the p18 antiserum did not detect these smaller proteins (Fig. 6A and D). To fully eliminate protein production from AUG1 and AUG2, we replaced both initiation codons with the alanine codon GCG in pGPOLΔ. In cells carrying pGPOLΔ-GCG1GCG2, transposition frequency was fully restored and about 2-fold higher than the ΔC1071 frameshift (Table 2). These results show that AUG1 and/or AUG2 is necessary for CNC and reinforce the observation that ΔC1071 confers a very low level of CNC. Taken together, our results identify p22 as a trans-dominant negative inhibitor of Ty1 retrotransposition and the intrinsic factor responsible for CNC.
FIG 6.
p22 is necessary for CNC. (A) Ty1 sequence present on pGPOLΔ illustrating the Ty1i RNA transcription start site (nt 1000), location of in-frame AUGs, and frameshift mutations (ΔC1071 and +A1303, black circles). Proteins encoded by wild-type (WT) or mutant plasmids are shown (wild-type sequence, solid; nonsense sequence, dashed) based on predicted usage of AUG1 by Ribo-seq (Fig. 5). ΔC1071 and +A1303 are predicted to synthesize truncated p22 peptides of 11 and 89 residues, respectively, before encountering the frameshift mutation. (B) An S. paradoxus strain with a single chromosomal Ty1his3-AI carrying an empty vector (DG2411), pGPOLΔ (DG2374), or the mutant plasmids ΔC1071 (JM321) and +A1303 (JM320) was assessed for Ty1 mobility using a qualitative assay. Cell patches grown on SC-Ura medium at 22°C were replica plated to SC-Ura-His medium to select for cells that contain at least one Ty1HIS3 insertion. The number of His+ papillae that grew on SC-Ura-His medium is a readout for Ty1 mobility. Also refer to Table 2. (C) Total RNA from the strains described above was subjected to Northern blotting to detect Ty1his3-AI and Ty1i transcripts as described for Fig. 1. The band labeled with an asterisk is a pervasive transcript approximately 4.5 kb in length and contains both Ty1 and non-Ty1 sequences from the pGPOLΔ. The “r” represents compression bands formed by two main species of rRNA in yeast, the 26S (3.8-kb) and 18S (2-kb) rRNAs. (D) Total cell extracts were analyzed for the presence of p22/p18 as described in the legend to Fig. 3.
Ectopic expression of p22/p18 is sufficient to inhibit Ty1 movement.
To determine if p22/p18 reduces Ty1 transposition, a cDNA expression library (45) was screened for clones that inhibited Ty1his3-AI mobility, and p22/p18 was ectopically expressed from the GAL1 promoter. One clone was obtained from the GAL1-driven cDNA library that contained Ty1 sequences 1042 to 5889 and inhibited chromosomal Ty1his3-AI mobility. The 5′ end of the cDNA included AUG2 and 26 additional nucleotides upstream but did not contain AUG1. The 3′ end terminated in the R region (3′ LTR) of Ty1 RNA, which is similar to the 3′ ends mapped previously (15, 55). Therefore, an almost-full-length 4.9-kb Ty1i transcript from a chromosomal element was likely captured as this cDNA clone and contains coding sequence for p22, as well as the POL coding sequence for PR, IN, and RT. When the cDNA clone and pGTy1his3-AI were coexpressed, Ty1 mobility decreased 570-fold compared with a control strain expressing only pGTy1his3-AI (Table 2). These results support the idea that a truncated Gag protein likely utilizing AUG2 inhibits Ty1 mobility, although initiation from AUG2 occurs less frequently than that from AUG1 based on ribosome profiling in the Sigma 1278b strain (Fig. 5).
The following segments of GAG sequence starting with AUG1 were fused to the GAL1 promoter on a multicopy expression plasmid and analyzed for trans-dominance (Table 2) or protein expression (Fig. 7 and data not shown) in the Ty1-less S. paradoxus strain: p22, p22 containing an internal V5 epitope, p18, and p22Gag*PR containing a previously characterized mutation that disrupts Gag-PR cleavage by PR (40). Ty1his3-AI mobility decreased more than 32,000-fold in cells coexpressing pGTy1his3-AI and p22 or p22-V5 compared with the control strain expressing pGTy1his3-AI (Table 2). Both p22 and p18 are present, again suggesting that some p22 is incorporated into VLPs and cleaved by Ty1 PR (Fig. 7). Coexpression of p18 and pGTy1his3-AI reduced Ty1 mobility to levels similar to those observed with p22 (Fig. 7). GAL1-promoted expression of p22 or p18 also inhibited pGTy1his3-AI mobility in S. cerevisiae strains BY4742 and GRF167 (data not shown).
FIG 7.
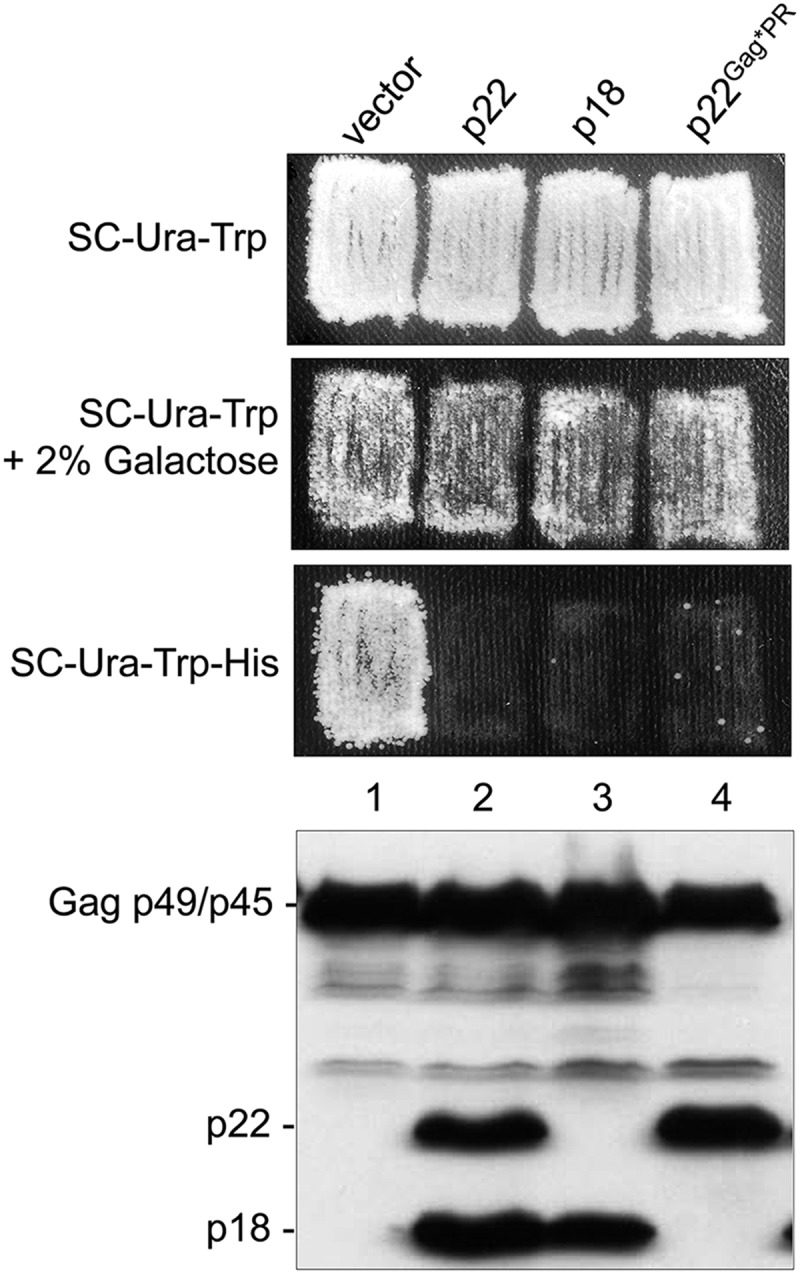
Cleavage of p22 to p18 does not disturb trans-dominant inhibition of Ty1 mobility. A mutant Gag-PR cleavage site, AAGSAA (Gag*PR) (40), was inserted into p22, replacing the normal Gag-PR cleavage site, RAHNVS. A Ty1-less strain containing pGTy1his3-AI and an empty vector (DG3739; lane 1), GAL1-p22 (DG3774; lane 2), GAL1-p18 (DG3791; lane 3), or GAL1-p22Gag*PR (JM399; lane 4) was analyzed for Ty1his3-AI mobility using a qualitative assay. Cell patches from a single colony were induced for pGTy1 expression by replica plating from SC-Ura-Trp medium to SC-Ura-Trp medium plus 2% galactose for 2 days at 22°C. To detect Ty1his3-AI mobility, galactose-induced cells were replica plated to SC-Ura-His medium. Below is an immunoblot assay using total cell extracts from the same strains and the p18 antiserum to detect Gag-p49/p45 and p22/p18.
To determine if p22 alone inhibited Ty1 mobility, we coexpressed p22Gag*PR and pGTy1his3-AI (Fig. 7) in the Ty1-less S. paradoxus strain. Results from qualitative mobility assays indicated that p22Gag*PR retained most if not all of its inhibitory function compared with wild-type p22 or p18 and the empty vector control, even though processing of p22 to p18 was blocked (Fig. 7). The level of p22Gag*PR when expressed from the GAL1 promoter was also comparable to that obtained with p22, p18, or full-length Gag. Together, these results show that p22 and p18 are potent trans-dominant inhibitors of Ty1 transposition.
p22/p18 cofractionates with VLPs and alters Ty1 proteins.
One possibility to account for the dramatic decrease in Ty1 mobility is that p22 associates with assembling VLPs in the cell, leading to abnormal VLP function. Therefore, crude VLP preparations from Ty1-less strains expressing pGTy1his3-AI alone (Fig. 8A), p22 and pGTy1his3-AI (Fig. 8B), or p22 alone (Fig. 8C) were separated by centrifugation through 20 to 60% continuous sucrose gradients. Fractions were assayed for reverse transcriptase activity using an exogenous primer/template and immunoblotted for Gag, IN, RT, and p22/p18 (Fig. 8A and B) or p22/p18 alone (Fig. 8C). As expected, a peak of reverse transcriptase activity coincided with the highest concentrations of mature Gag, RT, and IN proteins in the strain expressing just pGTy1his3-AI (Fig. 8A). When pGTy1his3-AI and p22 were coexpressed, Gag and p22/p18 displayed a similar fractionation pattern across the gradient (Fig. 8B). p18 appeared to be the predominant form present in crude VLP preparations, which is likely due to processing by Ty1 PR in VLPs. To further investigate if the cofractionation of Gag and p22/p18 resulted from an association between VLPs and p22, rather than comigration of a protein complex containing p22 that had a density similar to that of VLPs, an identical fractionation was performed in a strain expressing only GAL1-promoted p22. When expressed alone, p22 was detected near the top of the gradient (Fig. 8C) and, therefore, had a different fractionation profile than that observed when pGTy1his3-AI and p22 were coexpressed (Fig. 8B). Furthermore, we detected p18 in the CNC+ VLPs (data not shown) used for structural probing of packaged Ty1 RNA (32). These results support an interaction between Ty1 VLPs and p22/p18.
FIG 8.
Cofractionation of p22/p18 with Ty1 VLPs. Crude VLP pellets (P40) prepared from galactose-induced Ty1-less strains expressing pGTy1his3-AI alone (A) (DG3739), pGTy1his3-AI and p22 (B) (DG3774), or p22 alone (C) (DG3784) were fractionated through a 20 to 60% continuous sucrose gradient. VLP pellets (P40) and equal volumes from collected fractions were analyzed by immunoblotting with p18 antiserum and IN and RT antisera. Ty1 proteins are labeled, brackets indicate known Ty1 processing intermediates, and the asterisk indicates aberrant Ty1 proteins (estimated sizes, 65 and 90 kDa). Reverse transcriptase activity was detected using an exogenous poly(rC)-oligo(dG) template and [α-32P]dGTP.
Comparing the strain expressing pGTy1his3-AI (Fig. 8A) with one expressing pGTy1his3-AI and p22 (Fig. 8B), several differences in the fractionation patterns, protein composition and distribution, and reverse transcriptase activity were evident. First, cells expressing only pGTy1his3-AI yielded a higher concentration of Gag, IN, RT, and reverse transcriptase-catalyzed incorporation of [α-32P]dGTP in the peak fractions. Second, cells coexpressing pGTy1his3-AI and p22 showed a broader distribution of Ty1 proteins and reverse transcriptase activity. Third, the VLPs formed in the presence of p22 had a lower level of [α-32P]dGTP incorporation throughout the gradient. Fourth, Ty1 protein processing or stability was altered when pGTy1his3-AI and p22 were coexpressed. There was an accumulation of the PR-IN precursor (p91), and there was much less mature IN (p71), which is similar to results obtained previously (31). Ty1 RT (p63) now appeared as a doublet with an additional higher-molecular-weight protein that reacted with the RT antibody (Fig. 8B, denoted by an asterisk). Fifth, Ty1 Gag appeared to undergo more proteolysis overall when p22 was present, as evidenced by multiple lower-molecular-weight Gag-related proteins, which cofractionated with full-length Gag. These unusual Ty1 proteins may result from aberrant processing by Ty1 PR, from cleavage by a cellular protease, or from differences in posttranslational modification of Ty1 proteins brought about by a VLP-p22 interaction. Therefore, the mechanism of CNC involves differences in the physical and biochemical properties of VLPs assembled in the presence of p22.
p22/p18 changes VLP morphology.
Since p22/p18 affected the fractionation of Ty1 VLPs and appearance of Ty1 proteins (Fig. 8), we examined the size and morphology of VLPs assembled in the presence or absence of p22 by electron microscopy (Fig. 9). Equivalent sucrose gradient fractions with the highest level of [α-32P]dGTP incorporation (Fig. 8) were pooled, diluted, and concentrated by ultracentrifugation prior to staining with 2% ammonium molybdate. Ty1 VLPs formed in the absence of p22 (Fig. 9A) were mostly intact with an average diameter of 37.4 ± 2.7 nm, and only 13% of wild-type VLPs appeared malformed. In contrast, almost half of Ty1 VLPs formed in the presence of p22 (Fig. 9B) appeared open or incomplete, suggesting that these VLPs either are not formed properly or are less stable during sample preparation. The diameter of intact VLPs assembled in the presence of p22 was 39.2 ± 3.1 nm. Although the difference in diameters of the two batches of VLPs is statistically significant (P = 0.0005), further analyses will be required to determine if this difference is functionally relevant.
FIG 9.
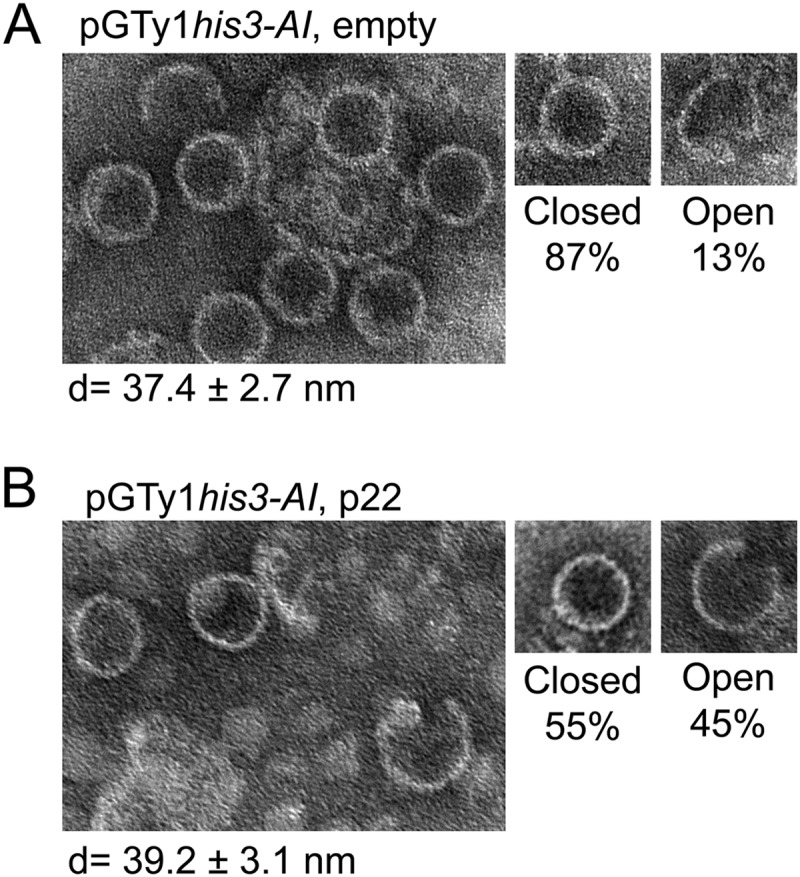
Electron microscopy of Ty1 VLPs assembled in the presence of p22/p18. VLP pellets were collected from sucrose gradient fractions with peak reverse transcriptase activity from experiments similar to those shown in Fig. 8. VLPs from pGTy1his3-AI alone (A) (DG3739) or pGTy1his3-AI and p22 (B) (DG3774) were stained with 2% ammonium molybdate and examined by transmission electron microscopy. Approximately 100 VLPs were analyzed for closed versus open particles, and representative images are shown. The diameter (d) was measured with closed VLPs only.
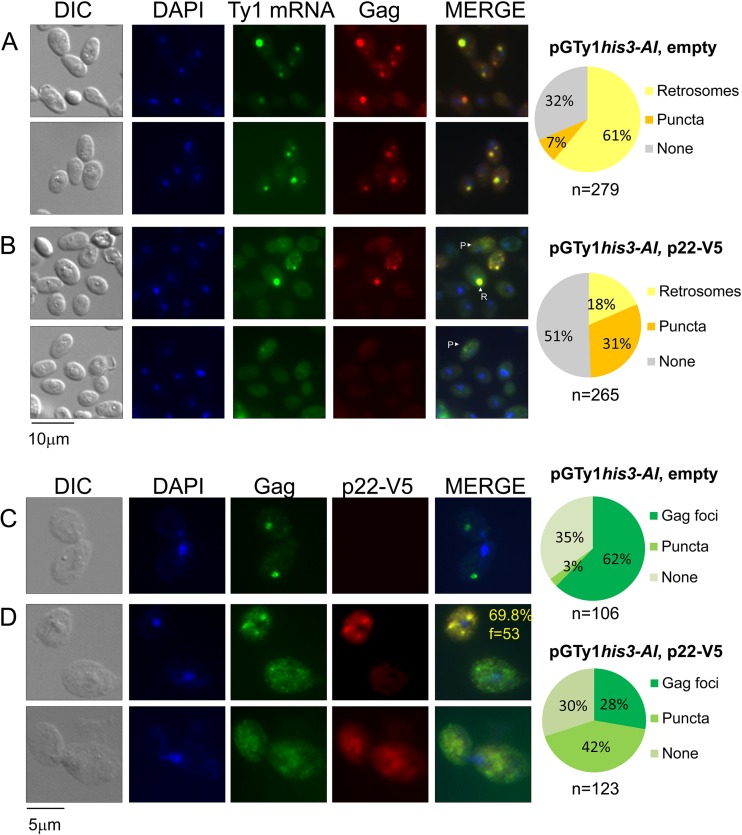
p22-V5 disrupts pGTy1-induced retrosomes and colocalizes with Gag.
Since p22 altered the fractionation pattern and morphology of VLPs and the processing or stability of Ty1 proteins, we examined whether p22 influenced the appearance of retrosomes, which are sites for VLP assembly. Ty1-less strains expressing p22-V5 and pGTy1his3-AI alone or together were subjected to indirect immunofluorescence (IF) and fluorescence in situ hybridization (FISH) to visualize retrosomes (Fig. 10). VLP or V5 antibodies were used to detect Ty1 proteins, and a GAG-digoxigenin (DIG) probe was used to detect full-length Ty1 mRNA. The internal V5 tag in p22-V5 did not disrupt trans-dominance (Table 2), and retrosome analysis of cells expressing untagged p22 was identical to that from strains expressing p22-V5. Three types of staining were observed: (i) large, distinct foci that costained for Ty1 mRNA and Gag were defined as retrosomes (R); (ii) nondistinct, punctate staining for both Ty1 mRNA and Gag was termed “puncta” (P); and (iii) lack of staining for Ty1 mRNA, Gag, or both was designated “none.” In cells containing puncta, colocalization between Ty1 mRNA and Gag could not be confidently determined in the majority of cells. In a control strain expressing pGTy1his3-AI alone, retrosomes were observed in 61% of cells, while only 7% of cells showed a punctate localization of Ty1 mRNA and Gag proteins (Fig. 10A). When p22-V5 and pGTy1his3-AI were coexpressed, the percentage of cells containing normal retrosomes decreased to 18% while Ty1 puncta were observed in 31% of cells. Thus, p22-V5 disrupts Ty1 retrosomes in a large fraction of cells. In addition, cells were analyzed for Ty1 Gag and p22-V5 colocalization using VLP and V5 antibodies, respectively (Fig. 10C and D). As expected, a similar percentage of cells exhibited retrosomes (61%) (Fig. 10A) and Gag foci in the absence of p22-V5 (62%) (Fig. 10C). In the presence of p22-V5, a comparable fraction of cells displaying Gag foci (28%) and puncta (42%) was observed (Fig. 10D) compared to the staining observed using FISH/IF analysis (Fig. 10B). Interestingly, we detected colocalization of p22-V5 and Gag in almost 70% of Gag foci (Fig. 10D, inset). p22-V5 colocalized with endogenous retrosomes in S. cerevisiae, suggesting the possibility that p22 can associate with VLP preassembly intermediates (J. A. Mitchell and D. J. Garfinkel, unpublished results).
FIG 10.
p22-V5 disrupts retrosomes and colocalizes with Gag. Ty1-less strains expressing pGTy1his3-AI alone (A and C) (DG3739) or pGTy1his3-AI and p22-V5 together (B and D) (JM367) were galactose induced and analyzed for Ty1 mRNA and Gag colocalization via FISH/IF (A and B). Pie charts depict cells examined for the appearance of retrosomes (R), puncta (P), or no staining (None). Refer to the text for additional details. In a separate experiment, cells were analyzed for Ty1 Gag and p22-V5 colocalization via IF using VLP and V5 antibodies, respectively (C and D). The experiment in panel D was additionally analyzed for the percentage of Gag foci that colocalize with p22-V5 (yellow; f, total Gag foci analyzed). For both experiments, DNA was stained with DAPI and representative images are shown (n, number of cells analyzed). DIC, differential interference contrast.
GST pulldowns support an interaction between Gag and p18.
To provide additional evidence for an interaction between p22/p18 and Gag, a fusion protein consisting of p18 tagged at its N terminus with glutathione S-transferase (41) was expressed from the GAL1 promoter in BY4742 or a Ty1-less strain (Fig. 11). Free glutathione S-transferase was expressed alone as a negative control. Protein extracts were immunoblotted using antisera specific for GST, Gag p49/p45, or Hts1 prior to mixing with the glutathione-coated resin (input) or released from the GST complexes bound to resin after several washes with lysis buffer (pulldown). Fusions between GST and full-length Gag were insoluble under a variety of conditions and, therefore, could not be analyzed further. The GST-p18 fusion protein was soluble under the conditions used for the pulldown; however, partial degradation of GST-p18 resulted in free GST protein. GST-p18 formed a complex containing Gag p45 and p49 encoded by the genomic Ty1 elements in BY4742, whereas GST expressed alone did not. Ty1 Gag-p18 complexes were also not detected in the Ty1-less strain. Hts1 was used to control for nonspecific trapping of cellular proteins in the Ty1 complexes and, as expected, was detected only in the input samples. Together, these results suggest that p18 and Gag interact.
FIG 11.
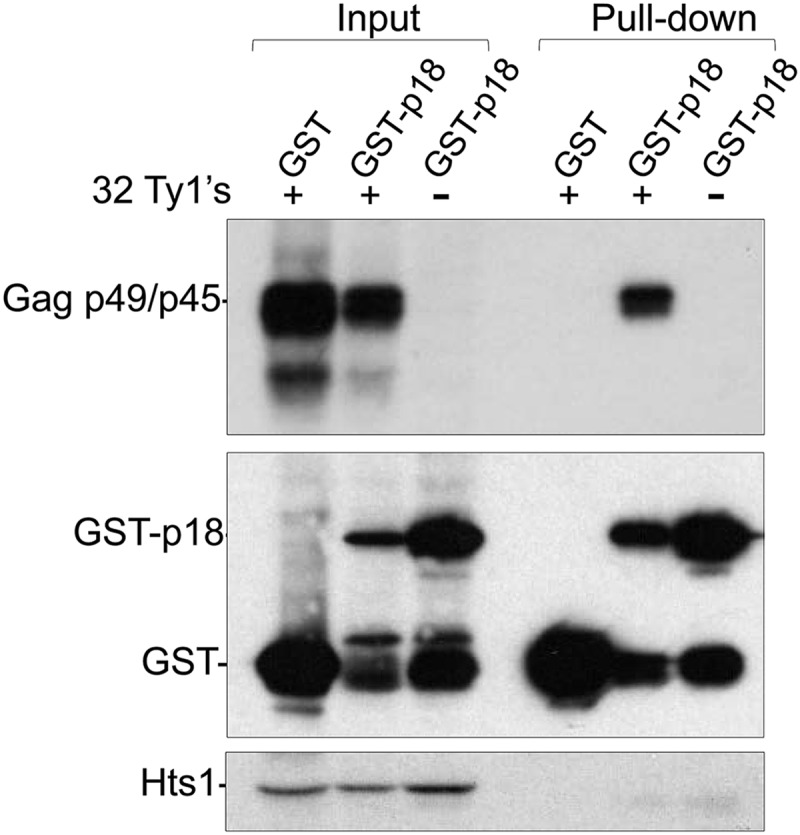
GST-p18 interacts with endogenous Ty1 Gag. Protein extracts (Input) from BY4742 induced for expression of GST (DG3808) or GST-p18 (DG3809) were incubated with glutathione-coated resin. Bound proteins were analyzed by immunoblotting to detect Gag, GST-p18, and p18/Ty1 Gag complexes (Pull-down) after extensive washing with lysis buffer. A Ty1-less strain expressing GST-p18 (DG3810) and the presence of Hts1 served as negative controls. Gag was detected with TY tag monoclonal antibody, which recognizes p49/p45 but not p22/p18 due to the location of the epitope. GST proteins and Hts1 were detected with GST and Hts1 antibodies, respectively.
DISCUSSION
Here, we characterize a restriction factor derived from Ty1 GAG that confers CNC by perturbing VLP assembly and function. This unique form of transposon CNC (28) may have evolved after an ancestral S. cerevisiae/paradoxus lineage lost the evolutionarily conserved RNAi pathway used to silence Ty1 expression (29, 30). Noncoding antisense transcripts from Ty1 have been implicated in repressing transcription (24), RNAi in budding yeast (30), and CNC (31). The identification of mutations that abrogated both CNC and Ty1AS RNA expression implicated Ty1AS RNAs in CNC. Additionally, the association of Ty1AS RNAs with VLPs further supported models of AS RNA-based CNC. Here, we show that additional mutations in the CNC region of Ty1 fail to confer CNC and yet do not perturb Ty1AS RNA expression. One GAG mutation in particular abolished CNC but did not affect transposition or AS RNA production. The behavior of this separation of function mutation suggested that a Ty1 protein might contribute to CNC. Evaluation of these mutants, along with those previously reported, helped reveal p22, a Gag-like restriction factor encoded by a 5′-truncated sense RNA (Ty1i) that likely forms the basis of CNC. The role of Ty1AS RNAs in Ty1 CNC, if any, remains to be determined.
We detect differences in the transcripts encoding the p22 restriction factor and how these transcripts are utilized for protein synthesis. In S. cerevisiae, a 4.9-kb Ty1i RNA is detected in wild-type strains both in our work and in previous studies when poly(A)+ RNA is subjected to Northern blotting (22, 54) but is rarely detected in numerous studies when total RNA is analyzed (7, 8, 18, 58–60). Perhaps, the level of RNA degradation observed with the abundant 5.7-kb Ty1 genomic RNA obscures the 4.9-kb Ty1i transcript when total RNA is analyzed by Northern blotting, because we can detect a shorter Ty1i transcript produced from a pGPOLΔ plasmid with total RNA from S. paradoxus. Alternatively, it has been reported that only 15% of Ty1 mRNA transcripts are polyadenylated (9). Hence, it is possible that Ty1i RNA is readily detected by Northern blotting of poly(A)+ RNA because the majority of Ty1i transcripts are polyadenylated, whereas the majority of Ty1 mRNA is not. However, in an isogenic repopulated S. paradoxus strain, a discrete Ty1i transcript is not detected from chromosomal Ty1 elements even when poly(A)+ RNA is analyzed by Northern blotting or cap-independent 5′ RACE. Ty1i RNA is present in both species when full-length Ty1 transcription is altered by deleting the Spt3 subunit of SAGA and related complexes (22, 23). Spt3 helps modulate the recruitment of the TATA-binding protein to the TATA box of SAGA-dependent promoters (61–63) and, therefore, can specify transcriptional initiation. However, the initiation site for Ty1i RNA within GAG predominates in an spt3Δ mutant, which is similar to the activation of cryptic intragenic promoters observed in a variety of chromatin and transcription-related mutants (64). Although our results are consistent with the idea that transcription of Ty1i RNA responds differently to the complexes containing Spt3, such as SAGA, in S. cerevisiae versus S. paradoxus, detailed functional comparisons between Spt3/SAGA from these species will be required to resolve this question.
Surprisingly, appreciable levels of p22/p18 are present in wild-type S. paradoxus repopulated with Ty1H3 in the absence of detectable 4.9-kb Ty1i RNA. This result raises the possibility that full-length Ty1 and Ty1i transcripts may utilize an internal ribosome entry site (65) upstream of AUG1 or AUG2 to drive synthesis of p22. Other mechanisms by which p22 could be translated from full-length Ty1 mRNA are leaky scanning, where scanning ribosomes sometimes initiate translation from an alternate AUG codon (66–69) or translation reinitiation in which translation starts at a downstream AUG after translation of an ORF situated upstream (70, 71). Although leaky scanning and translation reinitiation remain possible mechanisms, both require closely spaced AUGs. However, seven in-frame and seven out-of-frame AUGs are present in the 745 bases between the Gag initiation codon (nt 293) and p22 AUG1 (nt 1038), making leaky scanning or translation reinitiation unlikely. Alternatively, exceptional forms of translation initiation may not be required to synthesize p22 if heterogeneous Ty1i transcripts that contain AUG1 or AUG2 in the repopulated S. paradoxus strain remain translatable. In support of this view, we show that Ty1i RNA is a functional template for translation of p22 in S. paradoxus and S. cerevisiae spt3Δ mutants in the absence of full-length Ty1 mRNA. Although it is possible that there are two modes of p22 production in yeast (Ty1 mRNA and Ty1i RNA mediated), production of p22 from internal Ty1 RNA products alone is an attractive idea.
Once synthesized, p22 profoundly inhibits retrotransposition by altering VLP assembly and function. Earlier work as well as our mutational analysis of the CNC region demonstrates that Ty1 produces a trans-dominant inhibitor, now identified as p22, that decreases Ty1his3-AI mobility 20- to >340-fold depending on the relative expression of Ty1 and p22 (28, 31, 32, 72). However, when a cDNA derived from Ty1i RNA or p22 and Ty1his3-AI are coexpressed from the GAL1 promoter in a Ty1-less strain, mobility decreases 570- and 32,000-fold, respectively, indicating that p22 is necessary and sufficient for inhibition. The extreme inhibitory effect and broad dynamic range raise the possibility that the process of retrotransposition is very sensitive to the level of p22, with increasingly severe defects appearing as the level of p22 increases. Conversely, the relative amount of Ty1 versus p22 expression can likely saturate the inhibitor, as is evident from previous studies utilizing GAL1-promoted Ty1 induction (15, 16, 33, 34). In fact, Ty1 “transpositional dormancy,” which was described upon the discovery of Ty1 retrotransposition (15, 34), may result from an inhibitor that is saturated or overcome when Ty1 is induced via the GAL1 promoter (73–75). The work presented here supports this hypothesis and identifies p22 as the intrinsic inhibitor at least partly responsible for Ty1 dormancy.
When crude VLPs from the Ty1-less strain expressing Ty1 and p22 are analyzed by sucrose gradient sedimentation, both p22 and its processed product p18 cofractionate with Ty1 VLPs. p22 does not exhibit the same fractionation pattern in the absence of pGTy1 expression. Furthermore, analysis of a p22Gag*PR cleavage site mutant shows that p22 as well as p18 effectively inhibits transposition, and cleavage of p22 does not play a major role in CNC. The sucrose gradient fractions have also been assayed for reverse transcriptase activity and subjected to additional immunoblotting to detect Gag, IN, RT, and p22/p18. Expression of p22 causes a moderate decrease in the level of reverse transcriptase activity when assayed using an exogenous primer/template; prevents the accumulation of mature IN, a finding which reinforces previous work (31, 32); and broadens the peak containing VLP proteins. In addition, an overall degradation of Gag and the presence of aberrant RT proteins are indicative of proteolysis of the Gag-Pol precursor by Ty1 PR, increased proteolysis by cellular enzymes, or possible posttranslational modifications. Furthermore, the excessive proteolysis of IN could explain the appearance of higher-molecular-weight, RT antibody-reactive proteins and the absence of mature IN. Our results suggest that p22 interacts with and inhibits VLP functionality during assembly or in association with fully formed VLPs and also is processed by Ty1 PR to form p18.
Since these results suggest that VLP structure may be altered by p22, peak sucrose gradient fractions have been concentrated and visualized by electron microscopy. Most of the VLPs (87%) isolated from the control strain lacking p22/p18 are completely spherical with similar curvatures; however, almost half (46%) of the VLPs formed in the presence of p22/p18 are aberrant and have an open or incomplete morphology. VLPs analyzed from CNC+ cells containing much less p22/p18 do not appear malformed, but when extracts containing these VLPs are treated with the endonuclease Benzonase, less protection of packaged Ty1 mRNA is observed (32). Our results suggest that VLP integrity is compromised in the presence of higher levels of p22/p18 and that normal assembly of functional VLPs is inhibited by an interaction between Gag and p22.
To further investigate if Gag and p22 interact, cells expressing Ty1 and p22/p18 have been subjected to FISH/IF microscopy and GST pulldown analysis. The number of cells with aberrant retrosomes increases more than 3-fold when Ty1 and p22-V5 are coexpressed, and 70% of Gag foci also stain for p22-V5. In addition, GST pulldown analysis suggests that endogenous Gag can interact with GST-p18. Although p22 engages Gag during active VLP assembly, p22 may also interact with Gag in endogenous retrosomes, which contain few if any VLPs (7). Ty1 GAG is necessary for retrosome formation (7, 76), and certain Ty3 GAG mutations alter retrosome appearance or location (77, 78). Interestingly, cellular mutations that alter retrograde movement of Gag from the endoplasmic reticulum (ER) destabilize Gag and abolish nucleation of retrosomes (79). Whether p22 enters the ER remains to be determined. Ty1 GAG mutations that confer a trans-dominant negative phenotype (80–82) or affect VLP assembly (83) have been isolated, and some of these mutations map in p22. A synthetic peptide containing sequences within p22 also displays RNA chaperone activity (84), which is required for specific RNA transactions during the retroviral life cycle such as virion assembly, RNA packaging, primer annealing, and reverse transcription (85). Thus, p22 may inhibit multiple functions carried out by Gag.
Certain retroelement genes have undergone purifying selection in mammals, suggesting that these elements have been domesticated or exapted by their host (86). To date, domesticated GAG and POL genes either have evolved a new function used in normal cellular processes or have been incorporated into an innate defense pathway used to inhibit retroviral propagation. The prototypic Gag-like restriction factors Fv1 and enJS56A1 block replication of murine leukemia virus (MLV) and Jaagsiekte sheep retrovirus (JSRV), respectively, by interacting with viral proteins during infection (87–89) and share features in common with CNC of Ty1 by p22/p18. Fv1 is derived from the GAG gene of a member of the HERV-L family of human and murine endogenous retroviruses (87, 90, 91). Fv1 inhibits progression of the MLV life cycle following infection and reverse transcription but prior to integration. Although the infecting viral Gag protein as well as Fv1 determines the level of restriction, an ordered assembly of Gag is required for efficient Fv1 binding (88, 92). Our results suggest that Ty1 Gag interacts with p22/p18; however, the polymerization state of Gag and p22 required for maximum restriction of retrotransposition remains an open question. In addition, p22/p18 affects VLP assembly and function, whereas Fv1 inhibits a different step in the replication cycle that occurs postinfection. Conceptually similar to MLV-Fv1 restriction, the sheep genome harbors about 20 copies of endogenous (en) JSRVs and these sequences are homologous with exogenous JSRV that can cause lung cancer. Certain endogenous copies have evolved a trans-dominant Gag protein, enJS56A, which like Ty1-p22 blocks replication at a step soon after protein synthesis. The JSRV-enJS56A interaction prevents Gag from entering into an endosome trafficking pathway and results in aggregation and turnover by the proteasome (89, 93).
The MLV-Fv1 and JSRV-enJS56A restriction systems contain two components, raising the possibility of an arms race between the infecting retrovirus and the domesticated chromosomal GAG gene (94). In contrast, the many retrotransposition-competent Ty1 elements inhabiting Saccharomyces genomes encode their own inhibitor and, therefore, must balance mutations altering p22 potency with those affecting GAG fitness. Since Ty1 GAG or p22 coding regions have not been detected as an exapted gene capable of inhibiting Ty1 movement, the graduated retrotransposition rate provided by CNC may benefit Saccharomyces and Ty1, as suggested by recent work relating increases in Ty1 copy number with longer chronological life span (95). The Ty1-p22 interaction appears to directly block assembly of functional VLPs in a dose-dependent manner and to our knowledge represents a novel and effective way to allow some but not rampant retroelement movement. Further understanding of the molecular events underlying Ty1 Gag-p22 interaction, including the characterization of CNC-resistant mutants and the role that cellular genes have in modulating p22 expression or function, should reveal additional similarities and differences between Ty1 and retroviral restriction factors.
ACKNOWLEDGMENTS
We acknowledge Karen Stefanisko for technical help at the beginning of this project and Mary Ard (University of Georgia, CAUR) for assistance with electron microscopy. We thank Thomas Mason (Hts1), Stephen Hajduk (TY tag), and Alan and Susan Kingsman (Ty1 VLP) for providing antisera and Emiko Matsuda and Mary Ann Checkley for yeast strains. We thank Claiborne Glover, Steven Hajduk, William Lanzilotta, Michael Terns, Zachary Wood, and Shaying Zhang for sharing equipment. We thank Hyo Won Ahn and Katarzyna Purzycka for useful discussions.
This work was supported by NIH grants GM095622 (D.J.G.) and GM081399 (W.V.G.). J.A.M. was supported in part by the National Science Foundation graduate research fellowship 1011RH25213, and J.A.A. was supported by an NRSA postdoctoral fellowship, 1F32GM112474-01, through NIGMS.
REFERENCES
- 1.Voytas DF, Boeke JD. 2002. Ty1 and Ty5 of Saccharomyces cerevisiae, p 614–630 InCraig NL, Craigie R, Gellert M, Lambowitz AM (ed), Mobile DNA II. ASM Press, Washington, DC. [Google Scholar]
- 2.Dunham M, Badrane H, Ferea T, Adams J, Brown P, Rosenzweig F, Botstein D. 2002. Characteristic genome rearrangements in experimental evolution of Saccharomyces cerevisiae. Proc Natl Acad Sci U S A 99:16144–16149. doi: 10.1073/pnas.242624799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Garfinkel DJ. 2005. Genome evolution mediated by Ty elements in Saccharomyces. Cytogenet Genome Res 110:63–69. doi: 10.1159/000084939. [DOI] [PubMed] [Google Scholar]
- 4.Wilke C, Adams J. 1992. Fitness effects of Ty transposition in Saccharomyces cerevisiae. Genetics 131:31–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wilke CM, Maimer E, Adams J. 1992. The population biology and evolutionary significance of Ty elements in Saccharomyces cerevisiae. Genetica 86:155–173. doi: 10.1007/BF00133718. [DOI] [PubMed] [Google Scholar]
- 6.Beliakova-Bethell N, Beckham C, Giddings TH, Winey M, Parker R, Sandmeyer S. 2006. Virus-like particles of the Ty3 retrotransposon assemble in association with P-body components. RNA 12:94–101. doi: 10.1261/rna.2264806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Checkley MA, Nagashima K, Lockett SJ, Nyswaner KM, Garfinkel DJ. 2010. P-body components are required for Ty1 retrotransposition during assembly of retrotransposition-competent virus-like particles. Mol Cell Biol 30:382–398. doi: 10.1128/MCB.00251-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dutko JA, Kenny AE, Gamache ER, Curcio MJ. 2010. 5′ and 3′ mRNA decay factors colocalize with Ty1 Gag and human APOBEC3G and promote Ty1 retrotransposition. J Virol 84:5052–5066. doi: 10.1128/JVI.02477-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Malagon F, Jensen TH. 2008. The T body, a new cytoplasmic RNA granule in Saccharomyces cerevisiae. Mol Cell Biol 28:6022–6032. doi: 10.1128/MCB.00684-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bleykasten-Grosshans C, Friedrich A, Schacherer J. 2013. Genome-wide analysis of intraspecific transposon diversity in yeast. BMC Genomics 14:399. doi: 10.1186/1471-2164-14-399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Carr M, Bensasson D, Bergman CM. 2012. Evolutionary genomics of transposable elements in Saccharomyces cerevisiae. PLoS One 7:e50978. doi: 10.1371/journal.pone.0050978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Liti G, Peruffo A, James SA, Roberts IN, Louis EJ. 2005. Inferences of evolutionary relationships from a population survey of LTR-retrotransposons and telomeric-associated sequences in the Saccharomyces sensu stricto complex. Yeast 22:177–192. doi: 10.1002/yea.1200. [DOI] [PubMed] [Google Scholar]
- 13.Moore SP, Liti G, Stefanisko KM, Nyswaner KM, Chang C, Louis EJ, Garfinkel DJ. 2004. Analysis of a Ty1-less variant of Saccharomyces paradoxus: the gain and loss of Ty1 elements. Yeast 21:649–660. doi: 10.1002/yea.1129. [DOI] [PubMed] [Google Scholar]
- 14.Liti G, Carter DM, Moses AM, Warringer J, Parts L, James SA, Davey RP, Roberts IN, Burt A, Koufopanou V, Tsai IJ, Bergman CM, Bensasson D, O'Kelly MJT, Van Oudenaarden A, Barton DBH, Bailes E, Nguyen AN, Jones M, Quail MA, Goodhead I, Sims S, Smith F, Blomberg A, Durbin R, Louis EJ. 2009. Population genomics of domestic and wild yeasts. Nature 458:337–341. doi: 10.1038/nature07743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Boeke JD, Garfinkel DJ, Styles CA, Fink GR. 1985. Ty elements transpose through an RNA intermediate. Cell 40:491–500. doi: 10.1016/0092-8674(85)90197-7. [DOI] [PubMed] [Google Scholar]
- 16.Scheifele LZ, Cost GJ, Zupancic ML, Caputo EM, Boeke JD. 2009. Retrotransposon overdose and genome integrity. Proc Natl Acad Sci U S A 106:13927–13932. doi: 10.1073/pnas.0906552106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Maxwell PH, Curcio MJ. 2007. Host factors that control long terminal repeat retrotransposons in Saccharomyces cerevisiae: implications for regulation of mammalian retroviruses. Eukaryot Cell 6:1069–1080. doi: 10.1128/EC.00092-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dakshinamurthy A, Nyswaner KM, Farabaugh PJ, Garfinkel DJ. 2010. BUD22 affects Ty1 retrotransposition and ribosome biogenesis in Saccharomyces cerevisiae. Genetics 185:1193–1105. doi: 10.1534/genetics.110.119115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nyswaner KM, Checkley MA, Yi M, Stephens RM, Garfinkel DJ. 2008. Chromatin-associated genes protect the yeast genome from Ty1 insertional mutagenesis. Genetics 178:197–214. doi: 10.1534/genetics.107.082602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Risler JK, Kenny AE, Palumbo RJ, Gamache ER, Curcio MJ. 2012. Host co-factors of the retrovirus-like transposon Ty1. Mobile DNA 3:12. doi: 10.1186/1759-8753-3-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Suzuki K, Morimoto M, Kondo C, Ohsumi Y. 2011. Selective autophagy regulates insertional mutagenesis by the Ty1 retrotransposon in Saccharomyces cerevisiae. Dev Cell 21:358–365. doi: 10.1016/j.devcel.2011.06.023. [DOI] [PubMed] [Google Scholar]
- 22.Winston F, Durbin KJ, Fink GR. 1984. The SPT3 gene is required for normal transcription of Ty elements in S. cerevisiae. Cell 39:675–682. doi: 10.1016/0092-8674(84)90474-4. [DOI] [PubMed] [Google Scholar]
- 23.Grant PA, Duggan L, Côté J, Roberts SM, Brownell JE, Candau R, Ohba R, Owen-Hughes T, Allis CD, Winston F, Berger SL, Workman JL. 1997. Yeast Gcn5 functions in two multisubunit complexes to acetylate nucleosomal histones: characterization of an Ada complex and the SAGA (Spt/Ada) complex. Genes Dev 11:1640–1650. doi: 10.1101/gad.11.13.1640. [DOI] [PubMed] [Google Scholar]
- 24.Berretta J, Pinskaya M, Morillon A. 2008. A cryptic unstable transcript mediates transcriptional trans-silencing of the Ty1 retrotransposon in S. cerevisiae. Genes Dev 22:615–626. doi: 10.1101/gad.458008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Haimovich G, Medina DA, Causse SZ, Garber M, Millán-Zambrano G, Barkai O, Chávez S, Pérez-Ortín JE, Darzacq X, Choder M. 2013. Gene expression is circular: factors for mRNA degradation also foster mRNA synthesis. Cell 153:1000–1011. doi: 10.1016/j.cell.2013.05.012. [DOI] [PubMed] [Google Scholar]
- 26.Parker R. 2012. RNA degradation in Saccharomyces cerevisae. Genetics 191:671–702. doi: 10.1534/genetics.111.137265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dumesic PA, Madhani HD. 2014. Recognizing the enemy within: licensing RNA-guided genome defense. Trends Biochem Sci 39:25–34. doi: 10.1016/j.tibs.2013.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Garfinkel DJ, Nyswaner K, Wang J, Cho J-Y. 2003. Post-transcriptional cosuppression of Ty1 retrotransposition. Genetics 165:83–99. doi: 10.1534/genetics.107.082602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Drinnenberg IA, Fink GR, Bartel DP. 2011. Compatibility with killer explains the rise of RNAi-deficient fungi. Science 333:1592. doi: 10.1126/science.1209575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Drinnenberg IA, Weinberg DE, Xie KT, Mower JP, Wolfe KH, Fink GR, Bartel DP. 2009. RNAi in budding yeast. Science 326:544–550. doi: 10.1126/science.1176945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Matsuda E, Garfinkel DJ. 2009. Posttranslational interference of Ty1 retrotransposition by antisense RNAs. Proc Natl Acad Sci U S A 106:15657–15662. doi: 10.1073/pnas.0908305106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Purzycka KJ, Legiewicz M, Matsuda E, Eizentstat LD, Lusvarghi S, Saha A, Le Grice SFJ, Garfinkel DJ. 2013. Exploring Ty1 retrotransposon RNA structure within virus-like particles. Nucleic Acids Res 41:463–473. doi: 10.1093/nar/gks983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Curcio MJ, Garfinkel DJ. 1992. Posttranslational control of Ty1 retrotransposition occurs at the level of protein processing. Mol Cell Biol 12:2813–2825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Garfinkel DJ, Boeke JD, Fink GR. 1985. Ty element transposition: reverse transcriptase and virus-like particles. Cell 42:502–517. [DOI] [PubMed] [Google Scholar]
- 35.Curcio MJ, Garfinkel DJ. 1999. New lines of host defense: inhibition of Ty1 retrotransposition by Fus3p and NER/TFIIH. Trends Genet 15:43–45. doi: 10.1016/S0168-9525(98)01643-6. [DOI] [PubMed] [Google Scholar]
- 36.Sanz-Ramos M, Stoye JP. 2013. Capsid-binding retrovirus restriction factors: discovery, restriction specificity and implications for the development of novel therapeutics. J Gen Virol 94:2587–2598. doi: 10.1099/vir.0.058180-0. [DOI] [PubMed] [Google Scholar]
- 37.Spencer TE, Palmarini M. 2012. Endogenous retroviruses of sheep: a model system for understanding physiological adaptation to an evolving ruminant genome. J Reprod Dev 58:33–37. doi: 10.1262/jrd.2011-026. [DOI] [PubMed] [Google Scholar]
- 38.Guthrie C, Fink GR (ed). 1991. Methods in enzymology, vol 194 Guide to yeast genetics and molecular biology. Academic Press, Inc., San Diego, CA. [PubMed] [Google Scholar]
- 39.Boeke JD, Eichinger D, Castrillon D, Fink GR. 1988. The Saccharomyces cerevisiae genome contains functional and nonfunctional copies of transposon Ty1. Mol Cell Biol 8:1432–1442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Merkulov GV, Lawler JF, Eby Y, Boeke JD. 2001. Ty1 proteolytic cleavage sites are required for transposition: all sites are not created equal. J Virol 75:638–644. doi: 10.1128/JVI.75.2.638-644.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mitchell DA, Marshall TK, Deschenes RJ. 1993. Vectors for the inducible overexpression of glutathione S-transferase fusion proteins in yeast. Yeast 9:715–722. doi: 10.1002/yea.320090705. [DOI] [PubMed] [Google Scholar]
- 42.Vaidyanathan PP, Zinshteyn B, Thompson MK, Gilbert WV. 2014. Protein kinase A regulates gene-specific translational adaptation in differentiating yeast. RNA 20:912–922. doi: 10.1261/rna.044552.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ingolia NT, Ghaemmaghami S, Newman JR, Weissman JS. 2009. Genome-wide analysis in vivo of translation with nucleotide resolution using ribosome profiling. Science 324:218–223. doi: 10.1126/science.1168978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, Batut P, Chaisson M, Gingeras TR. 2013. STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29:15–21. doi: 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Liu H, Krizek J, Bretscher A. 1992. Construction of a GAL1-regulated yeast cDNA expression library and its application to the identification of genes whose overexpression causes lethality in yeast. Genetics 132:665–673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Curcio MJ, Garfinkel DJ. 1991. Single-step selection for Ty1 element retrotransposition. Proc Natl Acad Sci U S A 88:936–940. doi: 10.1073/pnas.88.3.936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lawler JF, Merkulov GV, Boeke JD. 2002. A nucleocapsid functionality contained within the amino terminus of the Ty1 protease that is distinct and separable from proteolytic activity. J Virol 76:346–354. doi: 10.1128/JVI.76.1.346-354.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Garfinkel DJ, Hedge AM, Youngren SD, Copeland TD. 1991. Proteolytic processing of Pol-TYB proteins from the yeast retrotransposon Ty1. J Virol 65:4573–4581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Eichinger DJ, Boeke JD. 1988. The DNA intermediate in yeast Ty1 element transposition copurifies with virus-like particles: cell-free Ty1 transposition. Cell 54:955–966. doi: 10.1016/0092-8674(88)90110-9. [DOI] [PubMed] [Google Scholar]
- 50.Youngren SD, Boeke JD, Sanders NJ, Garfinkel DJ. 1988. Functional organization of the retrotransposon Ty from Saccharomyces cerevisiae: Ty protease is required for transposition. Mol Cell Biol 8:1421–1431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Schneider CA, Rasband WS, Eliceiri KW. 2012. NIH Image to ImageJ: 25 years of image analysis. Nat Methods 9:671–675. doi: 10.1038/nmeth.2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Melamed C, Nevo Y, Kupiec M. 1992. Involvement of cDNA in homologous recombination between Ty elements in Saccharomyces cerevisiae. Mol Cell Biol 12:1613–1620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sharon G, Burkett TJ, Garfinkel DJ. 1994. Efficient homologous recombination of Ty1 element cDNA when integration is blocked. Mol Cell Biol 14:6540–6551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Winston F, Dollard C, Malone EA, Clare J, Kapakos JG, Farabaugh P, Minehart PL. 1987. Three genes are required for trans-activation of Ty transcription in yeast. Genetics 115:649–656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Elder RT, Loh EY, Davis RW. 1983. RNA from the yeast transposable element Ty1 has both ends in the direct repeats, a structure similar to retrovirus RNA. Proc Natl Acad Sci U S A 80:2432–2436. doi: 10.1073/pnas.80.9.2432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Adams SE, Mellor J, Gull K, Sim RB, Tuite MF, Kingsman SM, Kingsman AJ. 1987. The functions and relationships of Ty-VLP proteins in yeast reflect those of mammalian retroviral proteins. Cell 49:111–119. doi: 10.1016/0092-8674(87)90761-6. [DOI] [PubMed] [Google Scholar]
- 57.Arribere JA, Gilbert WV. 2013. Roles for transcript leaders in translation and mRNA decay revealed by transcript leader sequencing. Genome Res 23:977–987. doi: 10.1101/gr.150342.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Curcio MJ, Hedge AM, Boeke JD, Garfinkel DJ. 1990. Ty RNA levels determine the spectrum of retrotransposition events that activate gene expression in Saccharomyces cerevisiae. Mol Gen Genet 220:213–221. [DOI] [PubMed] [Google Scholar]
- 59.Servant G, Pennetier C, Lesage P. 2008. Remodeling yeast gene transcription by activating the Ty1 long terminal repeat retrotransposon under severe adenine deficiency. Mol Cell Biol 28:5543–5554. doi: 10.1128/MCB.00416-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Servant G, Pinson B, Tchalikian-Cosson A, Coulpier F, Lemoine S, Pennetier C, Bridier-Nahmias A, Todeschini A-L, Fayol H, Daignan-Fornier B, Lesage P. 2012. Tye7 regulates yeast Ty1 retrotransposon sense and antisense transcription in response to adenylic nucleotides stress. Nucleic Acids Res 40:5271–5282. doi: 10.1093/nar/gks166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Belotserkovskaya R, Sterner DE, Deng M, Sayre MH, Lieberman PM, Berger SL. 2000. Inhibition of TATA-binding protein function by SAGA subunits Spt3 and Spt8 at Gcn4-activated promoters. Mol Cell Biol 20:634–647. doi: 10.1128/MCB.20.2.634-647.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Eisenmann DM, Arndt KM, Ricupero SL, Rooney JW, Winston F. 1992. Spt3 interacts with TFIID to allow normal transcription in Saccharomyces cerevisiae. Genes Dev 6:1319–1331. doi: 10.1101/gad.6.7.1319. [DOI] [PubMed] [Google Scholar]
- 63.Mohibullah N, Hahn S. 2008. Site-specific cross-linking of TBP in vivo and in vitro reveals a direct functional interaction with the SAGA subunit Spt3. Genes Dev 22:2994–3006. doi: 10.1101/gad.1724408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Cheung V, Chua G, Batada NN, Landry CR, Michnick SW, Hughes TR, Winston F. 2008. Chromatin and transcription-related factors repress transcription from within coding regions throughout the Saccharomyces cerevisiae genome. PLoS Biol 6:e277. doi: 10.1371/journal.pbio.0060277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Gilbert WV. 2010. Alternative ways to think about cellular internal ribosome entry. J Biol Chem 285:29033–29038. doi: 10.1074/jbc.R110.150532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Harrison PM, Kumar A, Lang N, Snyder M, Gerstein M. 2002. A question of size: the eukaryotic proteome and the problems in defining it. Nucleic Acids Res 30:1083–1090. doi: 10.1093/nar/30.5.1083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Kochetov AV. 2008. Alternative translation start sites and hidden coding potential of eukaryotic mRNAs. Bioessays 30:683–691. doi: 10.1002/bies.20771. [DOI] [PubMed] [Google Scholar]
- 68.Kochetov AV, Sarai A, Rogozin IB, Shumny VK, Kolchanov NA. 2005. The role of alternative translation start sites in the generation of human protein diversity. Mol Genet Genomics 273:491–496. doi: 10.1007/s00438-005-1152-7. [DOI] [PubMed] [Google Scholar]
- 69.Wang XQ, Rothnagel JA. 2004. 5′-untranslated regions with multiple upstream AUG codons can support low-level translation via leaky scanning and reinitiation. Nucleic Acids Res 32:1382–1391. doi: 10.1093/nar/gkh305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Kochetov AV. 2005. AUG codons at the beginning of protein coding sequences are frequent in eukaryotic mRNAs with a suboptimal start codon context. Bioinformatics 21:837–840. doi: 10.1093/bioinformatics/bti136. [DOI] [PubMed] [Google Scholar]
- 71.Porras P, Padilla CA, Krayl M, Voos W, Barcena JA. 2006. One single in-frame AUG codon is responsible for a diversity of subcellular localizations of glutaredoxin 2 in Saccharomyces cerevisiae. J Biol Chem 281:16551–16562. doi: 10.1074/jbc.M600790200. [DOI] [PubMed] [Google Scholar]
- 72.Garfinkel DJ, Nyswaner KM, Stefanisko KM, Chang C, Moore SP. 2005. Ty1 copy number dynamics in Saccharomyces. Genetics 169:1845–1857. doi: 10.1534/genetics.104.037317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Curcio MJ, Garfinkel DJ. 1994. Heterogeneous functional Ty1 elements are abundant in the Saccharomyces cerevisiae genome. Genetics 136:1245–1259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Farabaugh P. 1995. Post-transcriptional regulation of transposition by Ty retrotransposons of Saccharomyces cerevisiae. J Biol Chem 270:10361–10364. doi: 10.1074/jbc.270.18.10361. [DOI] [PubMed] [Google Scholar]
- 75.Fink G, Boeke J, Garfinkel D. 1986. The mechanism and consequences of retrotransposition. Trends Genet 2:118–123. doi: 10.1016/0168-9525(86)90200-3. [DOI] [Google Scholar]
- 76.Malagon F, Jensen TH. 2011. T-body formation precedes virus-like particle maturation in S. cerevisiae. RNA Biol 8:184–189. doi: 10.4161/rna.8.2.14822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Clemens K, Larsen L, Zhang M, Kuznetsov Y, Bilanchone V, Randall A, Harned A, DaSilva R, Nagashima K, McPherson A, Baldi P, Sandmeyer S. 2011. The Ty3 Gag3 spacer controls intracellular condensation and uncoating. J Virol 85:3055–3066. doi: 10.1128/JVI.01055-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Larsen LS, Beliakova-Bethell N, Bilanchone V, Zhang M, Lamsa A, Dasilva R, Hatfield GW, Nagashima K, Sandmeyer S. 2008. Ty3 nucleocapsid controls localization of particle assembly. J Virol 82:2501–2514. doi: 10.1128/JVI.01814-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Doh JH, Lutz S, Curcio MJ. 2014. Co-translational localization of an LTR-retrotransposon RNA to the endoplasmic reticulum nucleates virus-like particle assembly sites. PLoS Genet 10:e1004219. doi: 10.1371/journal.pgen.1004219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Braiterman LT, Monokian GM, Eichinger DJ, Merbs SL, Gabriel A, Boeke JD. 1994. In-frame linker insertion mutagenesis of yeast transposon Ty1: phenotypic analysis. Gene 139:19–26. doi: 10.1016/0378-1119(94)90518-5. [DOI] [PubMed] [Google Scholar]
- 81.Checkley MA, Mitchell JA, Eizenstat LD, Lockett SJ, Garfinkel DJ. 2013. Ty1 Gag enhances the stability and nuclear export of Ty1 mRNA. Traffic 14:57–69. doi: 10.1111/tra.12013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Monokian GM, Braiterman LT, Boeke JD. 1994. In-frame linker insertion mutagenesis of yeast transposon Ty1: mutations, transposition and dominance. Gene 139:9–18. doi: 10.1016/0378-1119(94)90517-7. [DOI] [PubMed] [Google Scholar]
- 83.Martin-Rendon E, Marfany G, Wilson S, Ferguson DJ, Kingsman SM, Kingsman AJ. 1996. Structural determinants within the subunit protein of Ty1 virus-like particles. Mol Microbiol 22:667–679. doi: 10.1046/j.1365-2958.1996.d01-1716.x. [DOI] [PubMed] [Google Scholar]
- 84.Cristofari G, Ficheux D, Darlix J-L. 2000. The Gag-like protein of the yeast Ty1 retrotransposon contains a nucleic acid chaperone domain analogous to retroviral nucleocapsid proteins. J Biol Chem 275:19210–19217. doi: 10.1074/jbc.M001371200. [DOI] [PubMed] [Google Scholar]
- 85.Rein A, Datta SAK, Jones CP, Musier-Forsyth K. 2011. Diverse interactions of retroviral Gag proteins with RNAs. Trends Biochem Sci 36:373–380. doi: 10.1016/j.tibs.2011.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Kaneko-Ishino T, Ishino F. 2012. The role of genes domesticated from LTR retrotransposons and retroviruses in mammals. Front Microbiol 3:262. doi: 10.3389/fmicb.2012.00262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Best S, Le Tissier P, Towers G, Stoye JP. 1996. Positional cloning of the mouse retrovirus restriction gene Fv1. Nature 382:826–829. doi: 10.1038/382826a0. [DOI] [PubMed] [Google Scholar]
- 88.Hilditch L, Matadeen R, Goldstone DC, Rosenthal PB, Taylor IA, Stoye JP. 2011. Ordered assembly of murine leukemia virus capsid protein on lipid nanotubes directs specific binding by the restriction factor, Fv1. Proc Natl Acad Sci U S A 108:5771–5776. doi: 10.1073/pnas.1100118108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Murcia PR, Arnaud F, Palmarini M. 2007. The transdominant endogenous retrovirus enJS56A1 associates with and blocks intracellular trafficking of Jaagsiekte sheep retrovirus Gag. J Virol 81:1762–1772. doi: 10.1128/JVI.01859-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Bénit L, De Parseval N, Casella JF, Callebaut I, Cordonnier A, Heidmann T. 1997. Cloning of a new murine endogenous retrovirus, MuERV-L, with strong similarity to the human HERV-L element and with a gag coding sequence closely related to the Fv1 restriction gene. J Virol 71:5652–5657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Qi CF, Bonhomme F, Buckler-White A, Buckler C, Orth A, Lander MR, Chattopadhyay SK, Morse HC. 1998. Molecular phylogeny of Fv1. Mamm Genome 9:1049–1055. doi: 10.1007/s003359900923. [DOI] [PubMed] [Google Scholar]
- 92.Goldstone DC, Walker PA, Calder LJ, Coombs PJ, Kirkpatrick J, Ball NJ, Hilditch L, Yap MW, Rosenthal PB, Stoye JP, Taylor IA. 2014. Structural studies of postentry restriction factors reveal antiparallel dimers that enable avid binding to the HIV-1 capsid lattice. Proc Natl Acad Sci U S A 111:9609–9614. doi: 10.1073/pnas.1402448111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Arnaud F, Murcia PR, Palmarini M. 2007. Mechanisms of late restriction induced by an endogenous retrovirus. J Virol 81:11441–11451. doi: 10.1128/JVI.01214-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Jern P, Coffin J. 2008. Effects of retroviruses on host genome function. Annu Rev Genet 42:709–732. doi: 10.1146/annurev.genet.42.110807.091501. [DOI] [PubMed] [Google Scholar]
- 95.VanHoute D, Maxwell PH. 2014. Extension of Saccharomyces paradoxus chronological lifespan by retrotransposons in certain media conditions is associated with changes in reactive oxygen species. Genetics 198:531–545. doi: 10.1534/genetics.114.168799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Brachmann C, Davies A, Cost G, Caputo E, Li J, Hieter P, Boeke J. 1998. Designer deletion strains derived from Saccharomyces cerevisiae S288C: a useful set of strains and plasmids for PCR-mediated gene disruption and other applications. Yeast 14:115–132. doi:. [DOI] [PubMed] [Google Scholar]