ABSTRACT
Two rhadinovirus lineages have been identified in Old World primates. The rhadinovirus 1 (RV1) lineage consists of human herpesvirus 8, Kaposi's sarcoma-associated herpesvirus (KSHV), and closely related rhadinoviruses of chimpanzees, gorillas, macaques and other Old World primates. The RV2 rhadinovirus lineage is distinct and consists of closely related viruses from the same Old World primate species. Rhesus macaque rhadinovirus (RRV) is the RV2 prototype, and two RRV isolates, 26-95 and 17577, were sequenced. We determined that the pig-tailed macaque RV2 rhadinovirus, MneRV2, is highly associated with lymphomas in macaques with simian AIDS. To further study the role of rhadinoviruses in the development of lymphoma, we sequenced the complete genome of MneRV2 and identified 87 protein coding genes and 17 candidate microRNAs (miRNAs). A strong genome colinearity and sequence homology were observed between MneRV2 and RRV26-95, although the open reading frame (ORF) encoding the KSHV ORFK15 homolog was disrupted in RRV26-95. Comparison with MneRV2 revealed several genomic anomalies in RRV17577 that were not present in other rhadinovirus genomes, including an N-terminal duplication in ORF4 and a recombinative exchange of more distantly related homologs of the ORF22/ORF47 interacting glycoprotein genes. The comparison with MneRV2 has revealed novel genes and important conservation of protein coding domains and transcription initiation, termination, and splicing signals, which have added to our knowledge of RV2 rhadinovirus genetics. Further comparisons with KSHV and other RV1 rhadinoviruses will provide important avenues for dissecting the biology, evolution, and pathology of these closely related tumor-inducing viruses in humans and other Old World primates.
IMPORTANCE This work provides the sequence characterization of MneRV2, the pig-tailed macaque homolog of rhesus rhadinovirus (RRV). MneRV2 and RRV belong to the rhadinovirus 2 (RV2) rhadinovirus lineage of Old World primates and are distinct but related to Kaposi's sarcoma-associated herpesvirus (KSHV), the etiologic agent of Kaposi's sarcoma. Pig-tailed macaques provide important models of human disease, and our previous studies have indicated that MneRV2 plays a causal role in AIDS-related lymphomas in macaques. Delineation of the MneRV2 sequence has allowed a detailed characterization of the genome structure, and evolutionary comparisons with RRV and KSHV have identified conserved promoters, splice junctions, and novel genes. This comparison provides insight into RV2 rhadinovirus biology and sets the groundwork for more intensive next-generation (Next-Gen) transcript and genetic analysis of this class of tumor-inducing herpesvirus. This study supports the use of MneRV2 in pig-tailed macaques as an important model for studying rhadinovirus biology, transmission and pathology.
INTRODUCTION
Kaposi's sarcoma-associated herpesvirus/human herpesvirus 8 (KSHV) was first detected in 1994 in Kaposi's sarcoma (KS) lesions of individuals with human immunodeficiency virus (HIV)-AIDS (1). Subsequent studies have shown that KSHV is etiologically associated with all forms of KS, including classical (HIV-negative), AIDS-related, endogenous, and iatrogenic KS (2). In addition, KSHV plays a role in the pathogenesis of two B-cell lymphoproliferative disorders, primary effusion lymphoma (PEL) and multicentric Castleman's disease (MCD/MCD-associated plasmablastic lymphoma) and is associated with HIV-related diffuse large B-cell lymphoma (3). Sequence analysis of the KSHV genome revealed a close similarity in sequence and gene organization with herpesvirus saimiri (HVS), the prototype of the Rhadinovirus genus of gammaherpesviruses found in the New World squirrel monkey (4). The KSHV genes were annotated using the HVS nomenclature for conserved genes (open reading frames [ORFs] 2 to 75), and novel genes present in KSHV were given a K designation (K1 to -15). A more distant evolutionary relationship was noted to Epstein-Barr virus (EBV), the prototype of the Lymphocryptovirus genus of gammaherpesviruses. While the KSHV genome contained a large set of core herpesvirus genes conserved between the alpha-, beta-, and gammaherpesviruses, the KSHV-specific genes showed homology to a variety of cellular host genes that have been captured during virus evolution (5). These viral homologs of cellular genes are believed to contribute unique biological properties to KSHV functioning to disrupt antiviral responses, cytokine-regulated cell growth, apoptosis, and cell cycle control (6).
Two distinct lineages of rhadinoviruses related to KSHV have been identified in Old World primates (7, 8). The rhadinovirus 1 (RV1) rhadinovirus lineage consists of KSHV and its homologs in different nonhuman Old World primate species, including macaques, monkeys, drills, gorillas, and chimpanzees (7, 9–11). The prototype macaque RV1 rhadinovirus is RFHVMn, the retroperitoneal fibromatosis herpesvirus (RFHV) from the pig-tailed macaque (Macaca nemestrina). Closely related RFHV variants have been detected in other macaque species, including RFHVMm (rhesus macaque; Macaca mulatta) and RFHVMf (long-tailed macaque; Macaca fascicularis). The RV2 rhadinovirus lineage consists of a group of more distantly related viruses, which coinfect the same Old World primate species (7, 8, 10, 12–14). Rhesus rhadinovirus (RRV) is the prototype macaque RV2 rhadinovirus, although closely related RV2 rhadinoviruses have also been detected in other macaque species, including MneRV2/PRV (pig-tailed macaque), MfaRV2 (long-tailed macaque), and MfuRV2/JMRV (Japanese macaque; Macaca fuscata) (8, 15–17). The macaque RV1 and RV2 rhadinoviruses are highly prevalent and persistent in captive macaque populations with infection rates exceeding 90% (16, 18, 19). A high proportion of macaques show evidence of coinfection with both RV1 and RV2 rhadinoviruses (16, 19).
RFHVMn and other RFHV variants were initially detected in retroperitoneal fibromatosis (RF) lesions, KS-like tumors in macaques with simian AIDS (9). Subsequent studies showed that the RF spindloid tumor cells were infected with RFHV as antibodies to the RFHV ORF73 latency-associated nuclear antigen (LANA) strongly reacted with the tumor cell nuclei. High levels of RFHV DNA were detected by quantitative PCR (qPCR) in the tumor lesions, consistent with a widespread latent infection with approximately 3 to 4 RFHV genomes/cell (20–22). RV2 rhadinoviruses, including RRV, MneRV2, and MfaRV2, have been detected in T- and B-cell lymphomas in different macaque species with simian immunodeficiency virus (SIV)-associated simian AIDS. Early and late cycle gene products were detected by immunohistochemistry (IHC) in the lymphoma tumor cells, and up to 800 RV2 genomes/cell were detected by qPCR, consistent with a widespread RV2 lytic infection in the lymphoma (23). The combination of quantitative PCR and IHC detection of RV1 and RV2 rhadinovirus infection in the macaque RF and lymphoma tumor cells provides strong evidence for a causative association (21, 23). Other studies have detected RRV in SIV-associated experimentally induced B-cell hyperplasia and lymphoproliferative lesions (24, 25), although the extent of the infection was not quantitated. Furthermore, MfuRV2, the Japanese macaque RV2 rhadinovirus, was identified in animals with a spontaneous multiple sclerosis-like disease (17), but no viral quantitation or IHC analysis was performed to provide evidence for a biological role in the disease.
The complete genome sequences have been determined for the pig-tailed macaque RV1 rhadinovirus RFHVMn (26), multiple variants of the rhesus macaque RV2 rhadinoviruses RRV17577 (27) and RRV26-95 (28), and the Japanese macaque RV2 rhadinovirus MfuRV2 (29). Comparison of genome sequences demonstrated that the RFHVMn and KSHV genomes were highly conserved. RFHVMn contained homologs of all of the KSHV-specific genes, except ORFs K5 and K6 (26), but lacked a homolog of ORF11, which is distantly related to ORF10. The RRV and MfuRV2 genomes showed a high conservation of herpesvirus core gene sequences with the KSHV and RFHVMn genomes; however, homologs of most of the KSHV-specific ORFs, including K3, K4.1, K4.2, K5, K6, K7, K10, K10.5, K11, and K12, were not detected. The RRV and MfuRV2 genomes were colinear with KSHV and RFHVMn, except that the ORF2 viral dihydrofolate reductase (vDHFR) homolog was positioned near the far left end of the RV2 genomes, and the genomes contained an 8-fold duplication of the KSHV ORFK9 viral interferon. Evolutionary analysis suggests that the position of the RV2 vDHFR homolog represents the original capture event of a cellular DHFR gene and that the RV1 vDHFR homologs subsequently relocated (28, 30). The two sequenced RRV variants, RRV17577 and RRV26-95, showed very strong sequence similarity. Only four of the identified ORFs, including glycoproteins H (ORF22) and L (ORF47), uracil DNA glucosidase (ORF46), and the tegument protein (ORF67), showed less than 95% identity (28). It was also noted that the ORF4 homolog in RRV17577 was considerably longer than the ORF4 homolog in RRV26-95. Unique ORFs were identified in the repetitive regions of both RRV strains which were not detected in the KSHV genome.
Phylogenetic analysis of a set of conserved core herpesvirus genes, including ORF8 (glycoprotein B; UL27 family), ORF9 (DNA polymerase; UL30 family), ORF37 (SOX shutoff exonuclease; UL12 family), and ORF64 (large tegument protein; UL36 family), has confirmed the separation of the Old World primate rhadinoviruses into two distinct lineages, RV1 and RV2 (26). These studies suggest that a nonspeciative divergence after divergence of the Old and New World primates resulted in the RV1 and RV2 lineages, each populated by viruses from a range of Old World primate species. The macaque RFHVMn and the human KSHV are examples of the RV1 rhadinovirus lineage, while MneRV2, RRV, MfaRV2, and MfuRV2 from different macaque species are examples of the RV2 rhadinovirus lineage. Although the existence of a human RV2 rhadinovirus is suspected, there is no evidence, as yet, for its presence in the current human population. A tentative evolutionary time scale suggests that the RV1/RV2 divergence occurred about 10 million years before divergence of the human and macaque homologs within each lineage (26). Although the biological basis for the divergence is not known, novel gene capture events and mutational changes that affect tropism, host immune evasion, and/or biological processes are possible driving forces. Phylogenetic analysis showed longer branch lengths within RV1 rhadinoviruses than within RV2 rhadinoviruses, indicating an enhanced rate of evolution within the RV1 lineage (26). The characterization and comparison of members of these two rhadinovirus lineages will provide insight into the nature of important evolutionary changes in these tumor-associated herpesviruses.
The pig-tailed macaque, Macaca nemestrina, is the only Old World primate that can be infected with HIV-1, and numerous pig-tailed macaque models have been developed for AIDS-related research. The pig-tailed macaque RV1 (RFHVMn) and RV2 (MneRV2) rhadinoviruses have been detected in AIDS-related malignancies, with strong evidence of an etiological association (21, 23). We have recently determined the complete genome sequence of RFHVMn to study its role in KS-like malignancies in the pig-tailed macaque and its evolutionary relationship with KSHV and other RV1 rhadinoviruses (26). To compare the RV1 and RV2 rhadinoviruses coinfecting the same primate species and to provide further information on the evolution of the RV2 rhadinovirus lineage, we have isolated an MneRV2 variant from a pig-tailed macaque (J97167) at the Washington National Primate Research Center (WaNPRC) and have determined the complete sequence of the MneRV2 genome. We report here the analysis of the MneRV2 genome with evolutionary comparisons to known RV1 and RV2 rhadinoviruses.
MATERIALS AND METHODS
Purification of MneRV2 DNA.
An MneRV2 clone was originally isolated from peripheral blood leukocytes of pig-tailed macaque J97167 from the WaNPRC by coculture on rhesus primary fetal fibroblasts (RPFF) (31). To produce viral DNA for sequencing, confluent cultures of RPFF were infected with MneRV2 and cultured until marked cytopathic effects were observed. Culture supernatant was centrifuged at low speed to remove cell debris and filtered through a 0.45-μm filter. Virus particles were pelleted by high-speed centrifugation, and contaminating cellular DNA was digested with DNase I. Purified viral DNA was isolated by phenol-chloroform extraction of the treated particle pellet. The ratio of MneRV2 genomic and contaminating cellular DNA was determined to be 26,000 to 1, using qPCR assays specific for macaque RV2 rhadinovirus and oncostatin M, a single-copy cellular gene, as described previously (16).
Next-generation (Next-Gen) sequence analysis and de novo assembly.
A DNA library was prepared from 5 μg of purified MneRV2 viral DNA using the Roche kit as described previously (32). The sequencing was carried out on a Genome Sequencer 20 (GS20) system (454 Life Sciences Corporation). De novo assembly was performed using Newbler (32). Contig gaps were closed by PCR with Invitrogen Platinum Taq with 2.5× enhancer using PCR primers designed from the contig ends. The PCR products were blunt cloned into pJET2.1, and the resulting plasmids were sequenced with pJET primers. These sequences were used to assemble a complete genomic DNA contig.
Gene annotations.
The annotation of the MneRV2 genome was initially performed using the Gene Annotation Transfer Utility (GATU) provided at the Viral Bioinformatics Resource Center (33) based on the reference sequence for RRV26-95 (AF210726). Additional open reading frames were identified using BLASTX (NCBI) and manual analysis.
Dot plot alignments.
Alignments of the complete genome sequences of MneRV2, RRV26-95, and RRV17577 were performed using the Java Dot Plot Alignment program (JDotter) provided at the Viral Bioinformatics Resource Center (34).
Comparative and phylogenetic analyses.
Sequence alignments were performed using GenePro software (Riverside Scientific) or MUSCLE (35). Phylogenetic analyses were performed using protein maximum likelihood PhyML 3.0, implemented at LIRMM (36).
miRNA prediction.
Ab initio prediction of pre-microRNA (miRNA) hairpin structures was carried out as described previously (37, 38) using the VMir software package with default settings.
Nucleotide sequence accession numbers.
The MneRV2 genome was submitted to GenBank under accession number KP265674, and nucleotide and protein sequences analyzed in this report were derived from this accession record. Sequences used for comparison were derived from the GenBank accession records for KSHV (NC_009333), RFHVMn (KF703446), RRV17577 (NC_003401), RRV26-95 (AF201726), MfuRV2/JMRV (AY528864), and EBV (NC_007605). Additional specific protein accession records are indicated in the text.
RESULTS AND DISCUSSION
454-based Next-Gen sequence analysis of the genome of MneRV2, the pig-tailed macaque RV2 rhadinovirus.
We have previously isolated MneRV2 from a pig-tailed macaque (J97167) from the WaNPRC (16). Using PCR-based approaches, we and others have obtained sequences of several MneRV2 genes, including ORF8 glycoprotein B, ORF9 DNA polymerase, ORF59 DNA polymerase processivity factor, and ORF73 latency-associated nuclear antigen (LANA) (8, 22, 31, 39). To further characterize MneRV2, we purified viral DNA from culture supernatants of rhesus primary fetal fibroblasts lytically infected with MneRV2 (J97167). A DNA library was prepared and sequenced using 454 technology. This resulted in 17,510 reads comprising 8,706,350 bases. After removing sequences shorter than 486 bp, a de novo assembly was performed using Newbler (32). The assembly resulted in three large contigs with an average read depth of 200 separated by two gaps. By comparing the contigs to the published sequence of RRV, the gaps were determined to be ∼800 and ∼1,000 bp. Primers were designed from the ends of the contigs and used in PCRs to obtain a complete sequence of the MneRV2 (J97167) genome.
Genome organization of MneRV2.
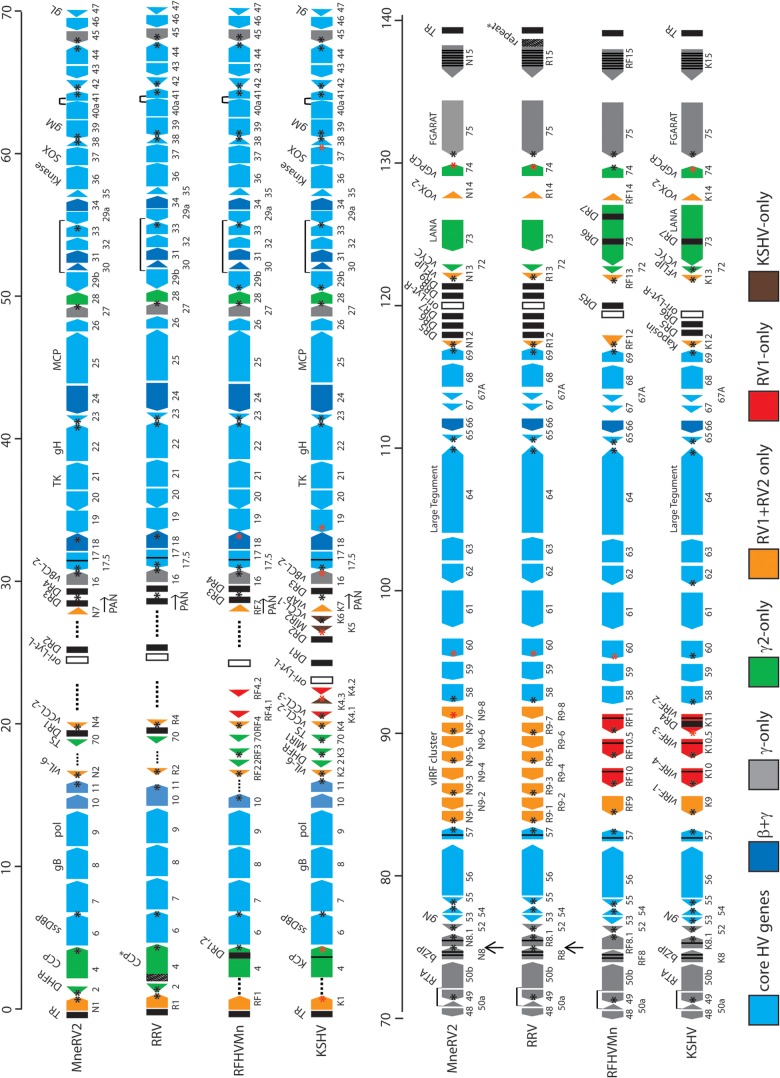
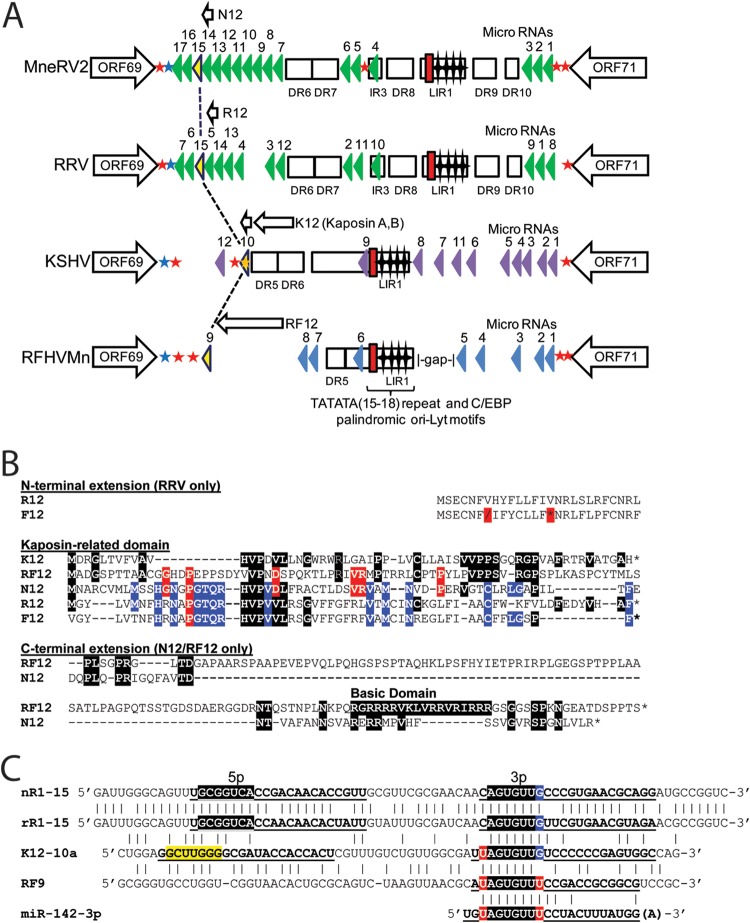
The final sequence of the pig-tailed macaque MneRV2 genome contained 129,494 bp with an overall G+C content of 53.8%, which is similar to that of the KSHV genome at 53.5%. Sequence analysis identified 87 ORFs with similarity to other herpesvirus genes (Fig. 1). Previous analysis of individual MneRV2 genes or gene fragments revealed a close similarity with corresponding gene homologs of the rhesus macaque RV2 rhadinovirus, RRV (8, 22, 31, 39). The genomes of two closely related variants of rhesus macaque RV2 rhadinovirus, RRV17577 (27) and RRV26-95 (28), have been completely sequenced. The genome organizations of the two RRV variants were essentially identical and were closely but not entirely colinear with that of the human RV1 rhadinovirus, KSHV. Eighty-four open reading frames (ORFs) were identified in the original report of RRV26-95, while only 80 ORFs were identified in the original RRV17577 report. Three of the RRV ORFs that were not reported in the original RRV17577 manuscript have been subsequently identified in the NCBI accession record (NC_003401) and include R8, a homolog of KSHV K8/bZIP (NP_598360); R8.1, a homolog of KSHV envelope glycoprotein K8.1 (NP_598361); and ORF67.5, a homolog of KSHV ORF67.5 (NP_598363). The remaining unreported gene in RRV17577, RK15, is a homolog of the terminal KSHV spliced K15 gene that has been identified (40) but is not present in the current NCBI accession record (NC_003401). The nomenclature used to identify the RRV ORFs was not consistent between the two publications. The RRV26-95 manuscript patterned the gene names after the KSHV nomenclature, whereas the RRV17577 manuscript devised a new nomenclature, especially for the RRV-specific ORFs. We have used the RRV26-95 nomenclature to annotate the MneRV2 genome, since this corresponds to that used previously for KSHV (4) and its pig-tailed macaque homolog RFHVMn (26).
FIG 1.
Comparative map of the MneRV2, RRV, RFHVMn, and KSHV genomes. The positions and transcription directions of the ORFs identified in the MneRV2 genome are compared to the corresponding ORFs in RRV (strain 26-95 and strain 17577), RFHVMn, and KSHV (see Materials and Methods for accession records). A composite map of the two RRV strains is shown, as RRV17577 has a duplicated N-terminal domain in CCP/ORF4 (indicated as CCP*) while RRV26-95 has a large repeated domain disrupting the R15 homolog (indicated as R15-repeat*), as discussed in the text. The numbering of the MneRV2, RRV, RFHVMn, and KSHV ORFs is patterned after the genome structure of the prototype rhadinovirus, herpesvirus saimiri (HVS), with KSHV-specific ORFs designated K1 to K15 and their homologs in MneRV2, RRV, and RFHVMn designated with an N, R, or RF prefix, respectively. This follows the nomenclature convention established for the RRV26-95 and RFHVMn genome sequences. The 8-fold duplications of the K9/RF9 vIRF-1 gene found in the vIRF cluster in MneRV2 and both RRV strains are labeled as N9-1 to -8 and R9-1 to -8, respectively. The approximate positions of the ORFs are shown with regard to their position within the KSHV genome (i.e., 1 to 140 kb). Note the conserved location of the ORF2/DHFR gene upstream of ORF4 in MneRV2 and both RRV strains. The sizes of the ORF markers are approximately consistent with the sizes of the encoded proteins. Vertical black lines within an ORF indicate splicing events or internal initiations, while longer-range splices of protein coding exons are indicated with a bar. Transcription-terminating poly(A) signals identified in KSHV (42) and corresponding conserved signals in the other genomes are indicated as a black (AAUAAA) or red (AUUAAA) asterisk. The poly(A) signals downstream of MneRV2 and RRV ORFs N1/R1, N8/R8, 48, 60, and N15/R15, which are not present in KSHV or RFHVMn, as discussed in the text, are indicated with arrows. The ORFs are color coded with regard to their conservation in other herpesvirus genomes, showing core HV genes conserved in most herpesvirus families, both beta- and gammaherpesviruses, gammaherpesviruses only, gamma-2-herpesviruses only, RV1 and RV2 rhadinoviruses only, RV1 rhadinoviruses only, and KSHV only, as indicated. The basic design of this map was patterned after Fig. 1 in reference 28 and is updated from Fig. 1 in reference 26.
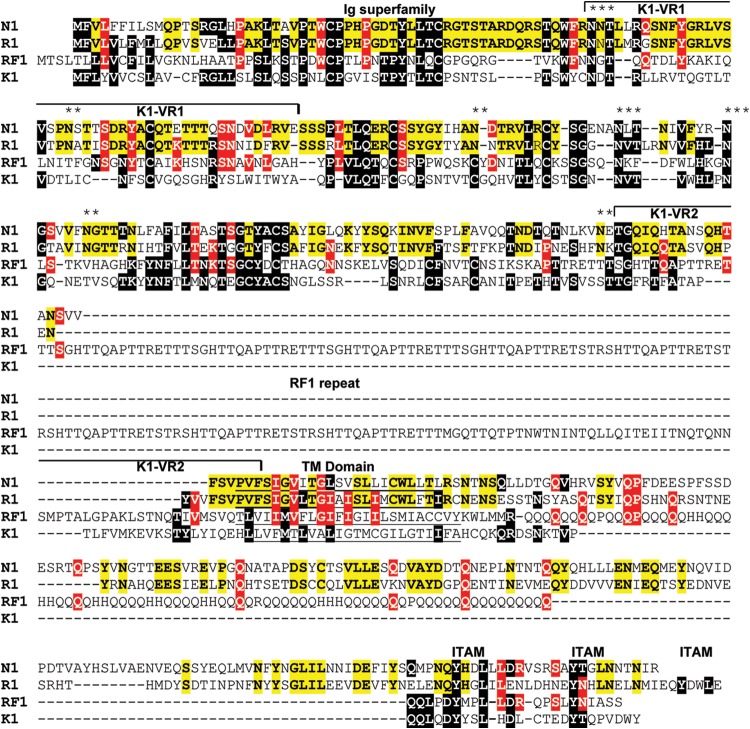
The MneRV2 sequence initiated 451 bp upstream of the leftmost identified ORF, annotated as N1 due to the sequence similarity with the leftmost genes K1 in the KSHV genome, RF1 in the RFHVMn genome, and R1 in the RRV genomes. The 451-bp upstream sequence was 60.5% AT with 37.7% T residues and was homologous (>70% identical) to the corresponding sequences at the left ends of both RRV variants. Although the original report of the RRV17577 genome found no sequence homology between the leftmost RRV R1 and KSHV K1 genes, the R1 sequences from the two RRV variants were 98% identical, aligned closely with the MneRV2 N1 (53% identity; Table 1), and showed homology to the KSHV K1 and RFHVMn RF1 sequences (Fig. 2). All four proteins contained two immunoglobulin-like (Ig-like) domains of Fc gamma receptor-like proteins with a putative transmembrane-spanning domain and a C-terminal domain containing multiple immunoglobulin receptor tyrosine-based activation motif (ITAM)-like motifs.
TABLE 1.
Characteristics of ORFs
| ORF | Function | MneRV2,a size (aa) | RRV,b size (aa) | % identity | KSHV,c size (aa) | % identity |
|---|---|---|---|---|---|---|
| N1 | ITAM-containing signal-transducing membrane protein; KSHV K1 | 432 | 423 | 53.0 | 289 | 16.6 |
| 2 | Dihydrofolate reductase (vDHFR) | 185 | 188 | 66.5 | 210d | 47.0 |
| 4 | Complement control protein; membrane protein; KSHV KCP | 381 | 395 | 76.6 | 550 | 67.7 |
| 6 | Single-strand DNA binding protein (ssDBP); UL29 | 1,132 | 1,132 | 94.1 | 1,133 | 37.6 |
| 7 | Subunit of terminase; UL28 | 684 | 686 | 86.4 | 695 | 51.6 |
| 8 | Glycoprotein B; UL27 | 826 | 829 | 91.0 | 845 | 65.7 |
| 9 | DNA polymerase; UL30 | 1,013 | 1,014 | 93.7 | 1,012 | 66.8 |
| 10 | Derived from dUTPase; CMVk UL82/83/84 | 416 | 416 | 81.0 | 418 | 32.9 |
| 11 | Derived from dUTPase; CMV UL82/83/84 | 410 | 409 | 86.6 | 407 | 31.0 |
| N2 | vIL-6; KSHV K2 | 207 | 207 | 82.1 | 204 | 14.5 |
| -e | (Position of vDHFR in KSHV) | - | - | - | 210 | |
| - | (E3 ubiquitin ligase membrane protein; KSHV K3; MIR1; ORF12) | - | - | - | 333 | |
| 70 | Thymidylate synthase (vTS) | 333 | 333 | 93.1 | 337 | 65.8 |
| N4 | Interleukin 8-like CC chemokine; KSHV K4; vMIP-II; vCCL-2 | 120 | 118 | 75.0 | 94 | 23.3 |
| - | (Interleukin 8-like CC chemokine; KSHV K4.1; vMIP-Iβ; vCCL-3) | - | - | - | 114 | |
| - | (Contains hydrophobic domains; KSHV K4.2) | - | - | - | 182 | |
| - | (E3 ubiquitin ligase membrane protein; KSHV K5; MIR2) | - | - | - | 256 | |
| - | (Interleukin 8-like CC chemokine; KSHV K6; vMIP-I; vCCL-1) | - | - | - | 95 | |
| N7 | Related to KSHV K7; RFHVMn RF7 | 116 | ***f | ***f | 126 | 7.8 |
| 16 | BCL-2-like inhibitor of apoptosis; vBCL-2 | 184 | 187 | 90.2 | 175 | 44.6 |
| 17 | Minor capsid scaffold protein; UL26 | 537 | 536 | 82.7 | 553 | 58.3 |
| 17.5 | Major capsid scaffold protein; UL26.5 | 292 | 291 | 77.7 | 288 | 30.0 |
| 18 | Related to CMV UL79 | 299 | 257 | 73.6 | 257 | 56.0 |
| 19 | Putative portal-capping protein; UL25 | 547 | 547 | 84.3 | 549 | 51.7 |
| 20 | Nuclear protein; UL24 | 350 | 350 | 83.1 | 320 | 33.4 |
| 21 | Thymidine kinase; UL23 | 556 | 557 | 87.0 | 580 | 41.4 |
| 22 | Glycoprotein H; UL22 | 726 | 726 | 81.8 | 730 | 38.0 |
| 23 | Tegument protein; UL21 | 419 | 402 | 80.2 | 404 | 45.6 |
| 24 | Related to CMV UL87 | 733 | 732 | 85.9 | 752 | 58.5 |
| 25 | Major capsid protein; UL19 | 1,378 | 1,378 | 95.6 | 1,376 | 72.3 |
| 26 | Capsid triplex protein; UL18 | 305 | 305 | 93.8 | 305 | 64.3 |
| 27 | Related to EBV BDLF2 | 273 | 269 | 76.9 | 290 | 27.1 |
| 28 | Membrane protein; related to KSHV ORF28 | 91 | 91 | 59.3 | 102 | 26.4 |
| 29B | Putative ATPase subunit of terminase; UL15; unspliced | 348 | 348 | 94.3 | 351 | 65.5 |
| 29 | Putative ATPase subunit of terminase; UL15; 29A, 29B spliced | 681 | 681 | 91.9 | 687 | 64.4 |
| 29A | Putative ATPase subunit of terminase; UL15; unspliced | 327 | 327 | 86.9 | 312 | 60.3 |
| 30 | Related to CMV UL91 | 76 | 76 | 72.4 | 77 | 44.7 |
| 31 | Related to CMV UL92 | 217 | 217 | 86.2 | 224 | 41.9 |
| 32 | Tegument protein; DNA packaging; UL17 | 464 | 464 | 80.8 | 454 | 40.1 |
| 33 | Tegument protein; UL16 | 336 | 336 | 85.1 | 312 | 39.3 |
| 34 | Related to CMV UL95 | 328 | 327 | 82.3 | 327 | 47.9 |
| 35 | Tegument protein; UL14 | 149 | 149 | 90.6 | 151 | 35.6 |
| 36 | Serine-threonine protein kinase; UL13 | 435 | 435 | 90.3 | 444 | 45.1 |
| 37 | SOX; host shutoff; DNase; UL12 | 480 | 472 | 92.7 | 486 | 63.8 |
| 38 | Alkaline exonuclease; UL11 | 70 | 69 | 75.7 | 61 | 44.3 |
| 39 | Glycoprotein M; UL10 | 378 | 378 | 95.0 | 399 | 60.1 |
| 40A | Subunit of helicase-primase complex; UL8; unspliced | 460 | 468 | 80.7 | 457 | 32.0 |
| 40 | Subunit of helicase-primase complex; UL8; 40A, 41 spliced | 638 | 638 | 78.8 | 669 | 27.1 |
| 41 | Subunit of helicase-primase complex; UL8; unspliced | 203 | 203 | 75.4 | 205 | 24.1 |
| 42 | Tegument protein; UL7 | 272 | 272 | 90.8 | 278 | 44.1 |
| 43 | Portal protein; UL6 | 581 | 576 | 93.3 | 605 | 61.8 |
| 44 | Helicase subunit of helicase-primase complex; UL5 | 790 | 790 | 93.5 | 788 | 66.7 |
| 45 | Inhibition of IRF-7; virion phosphoprotein; | 304 | 353 | 67.4 | 407 | 22.4 |
| 46 | Uracil-DNA glycosylase; UL2 | 255 | 255g | 82.0 | 255 | 61.2 |
| 47 | Glycoprotein gL; UL1 | 163 | 163 | 81.0 | 167 | 28.2 |
| 48 | Unknown function; related to BRRF2 EBV | 389 | 389 | 82.5 | 402 | 26.0 |
| 49 | Unknown function; related to BRRF1 EBV | 301 | 301 | 94.0 | 302 | 53.2 |
| 50 | RTA; transactivator; related to BRLF1 EBV; exons 1 and 2 spliced | 575 | 577 | 83.9 | 691 | 37.7 |
| N8γ | bZIP transcription factor; KSHV K8γ; exon 1 unspliced | 162 | 177 | 50.9 | 239 | 13.0 |
| N8α | bZIP transcription factor; KSHV K8α; exons 1, 2, and 3 spliced | 222 | 234 | 62.2 | 237 | 19.6 |
| N8.1γ | Glycoprotein; ORF51; KSHV K8.1γ; exon 1 unspliced | 195 | 230 | 56.8 | 197 | 12.6 |
| N8.1α | Glycoprotein; ORF51; KSHV K8.1α; exons 1 and 2 spliced | 274 | 275 | 61.8 | 228 | 11.8 |
| 52 | Related to BLRF2 EBV | 138 | 139 | 87.0 | 131 | 43.5 |
| 53 | Glycoprotein N; UL49A | 96 | 104 | 87.5 | 110 | 47.9 |
| 54 | Deoxyuridine triphosphatase; UL50 | 290 | 290 | 90.3 | 318 | 40.7 |
| 55 | Tegument protein; UL51 | 210 | 210 | 95.7 | 227 | 56.2 |
| 56 | Primase subunit of helicase-primase complex; UL52 | 839 | 828 | 88.4 | 843 | 51.6 |
| 57 | Posttranscriptional regulator; UL54; exons 1 and 2 spliced | 447 | 446 | 87.9 | 455 | 38.2 |
| 57A | Posttranscriptional regulator; UL54; unspliced | 443 | 442 | 87.1 | - | - |
| N9-1 | Related to KSHV vIRF-1/K9; unspliced | 414 | 415 | 69.8 | 449h | 19.3 |
| N9-2 | Related to KSHV vIRF-1/K9; unspliced | 413 | 415 | 70.7 | 449h | 17.2 |
| N9-3 | Related to KSHV vIRF-1/K9; unspliced | 351 | 351 | 82.6 | 449h | 20.5 |
| N9-4 | Related to KSHV vIRF-1/K9; unspliced | 322 | 361 | 74.8 | 449h | 19.6 |
| N9-5 | Homolog of N9-1; related to KSHV vIRF-1/K9; unspliced | 386 | 385 | 81.3 | 449h | 19.9 |
| N9-6 | Homolog of N9-2; related to KSHV vIRF-1/K9; unspliced | 394 | 390 | 77.4 | 449h | 20.6 |
| N9-7 | Homolog of N9-3; related to KSHV vIRF-1/K9; unspliced | 355 | 355 | 81.7 | 449h | 19.2 |
| N9-8 | Homolog of N9-4; related to KSHV vIRF-1/K9; unspliced | 364 | 364 | 80.5 | 449h | 14.6 |
| - | (KSHV vIRF-4/K10; spliced) | - | - | - | 911 | |
| - | (KSHV vIRF-3/K10.5; spliced) | - | - | - | 566 | |
| - | (KSHV vIRF-2/K11; spliced) | - | - | - | 680 | |
| 58 | Multiple transmembrane protein; UL43 | 360 | 360 | 88.9 | 357 | 36.1 |
| 59 | DNA polymerase processivity factor; UL42 | 394 | 394 | 88.3 | 396 | 51.8 |
| 60 | Ribonucleotide reductase small subunit; UL40 | 314 | 304 | 91.4 | 305 | 67.8 |
| 61 | Ribonucleotide reductase large subunit; UL39 | 790 | 788 | 91.0 | 792 | 61.5 |
| 62 | Capsid triplex protein; UL38 | 331 | 331 | 92.7 | 331 | 57.1 |
| 63 | Tegument protein; UL37 | 938 | 939 | 83.5 | 927 | 41.3 |
| 64 | Large tegument protein; UL36 | 2,554 | 2,548 | 74.5 | 2,635 | 17.3 |
| 65 | Small capsid protein on hexon tips; UL35 | 169 | 169 | 82.8 | 170 | 36.1 |
| 66 | BFRF2 EBV homolog; related to CMV UL49 | 443 | 448 | 77.4 | 429 | 43.3 |
| 67 | Capsid docking protein on nuclear lamina; UL34 | 268 | 268i | 79.9 | 271 | 58.2 |
| 67A | DNA packaging; UL33 | 89 | 86 | 82.0 | 80 | 57.3 |
| 68 | Nuclear localization of capsids and DNA packaging; UL32 | 457 | 457 | 82.1 | 545 | 50.3 |
| 69 | Egress of capsids from nucleus; UL31 | 297 | 297 | 89.9 | 225 | 48.8 |
| N12 | Homolog of KSHV K12; Kaposin A | 109 | 89 | 32.2 | 60 | 11.9 |
| N13 | FLIP-like inhibitor of apoptosis; vFLIP; KSHV K13; ORF71 | 174 | 174 | 81.0 | 188 | 33.9 |
| 72 | Cyclin D-like protein; KSHV vCyc | 250 | 254 | 90.4 | 257 | 35.2 |
| 73 | Latency-associated nuclear antigen; KSHV LANA | 438 | 448 | 59.0 | 1,162 | 16.2 |
| N14 | vOX2; contains two Ig domains; KSHV K14 | 254 | 253 | 83.9 | 348 | 31.5 |
| 74 | vGPCR; G-protein-coupled receptor; similar to IL-8 receptor | 437 | 342 | 66.6 | 342 | 32.5 |
| 75 | Tegument protein; related to EBV FGARAT | 1,296 | 1,298 | 84.7 | 1,296 | 43.5 |
| N15 | Signal-transducing membrane protein; K15; EBV LAMP; spliced | 529 | ***j | ***j | 490 | 13.0 |
MneRV2 genome (KP265674).
RRV26-95 (AF10726).
KSHV genome (NC_009333).
The KSHV vDHFR is not located at this position within the KSHV genome.
KSHV-specific ORFs missing in RFHVMn and/or RRV are indicated in parentheses, and their relative positions within the macaque viral genomes are indicated by a hyphen.
The homologous ORF in RRV17577 and RRV26-95 is disrupted.
The 255-aa RRV26-95 ORF46 sequence is corrected from the published genomic sequence, which encoded an erroneous 230-aa ORF due to two single-base-pair insertion sequencing errors at bp 62577 (extra T) and bp 62668 (extra A), as described in the text.
KSHV K9.
The 268-aa RRV26-95 ORF67 sequence is corrected from the published genomic sequence, which encoded an erroneous 222-aa ORF due to a base pair insertion sequencing error at bp 107619 (extra C), as described in the text. An additional sequencing error was identified in the RRV17577 ORF67 at bp 109544 (missing C), by sequence comparison. The corrected RRV17577 ORF67 sequence was 268 aa and 100% identical to the corrected RRV26-95 sequence.
R15 gene in RRV26-95 is disrupted as indicated in the text. The homologous gene in RRV 17577 (532 aa) has 61.1% identity with N15.
CMV, cytomegalovirus.
FIG 2.
Alignment of the left-end K1 homologs. The leftmost ORFs in the genomes of MneRV2, RRV26-95, RFHVMn, and KSHV, corresponding to ORFs N1, R1, RF1, and K1, respectively, were aligned. The RRV17577 R1 sequence varies from the RRV26-95 sequence by only 2 aa. K1 residues conserved in other sequences are highlighted in black. Residues conserved between RV1 and RV2 rhadinovirus homologs are highlighted in red, and residues conserved between the macaque RV2 viruses MneRV2 and RRV26-95 are highlighted in yellow. The N-terminal domain related to the immunoglobulin receptor and the immunoglobulin receptor tyrosine-based activation motifs (ITAM) (61) are indicated, and the variable regions (VR1 and VR2) identified in KSHV subtypes (62) are shown. Residues predicted to form a hydrophobic transmembrane (TM) domain and the position of the repeat domain within the RF1 sequence are indicated. Conserved potential N-linked glycosylation sites are shown with asterisks (**, conserved between MneRV2 and RRV; ***, conserved between RV1 and RV2 rhadinoviruses).
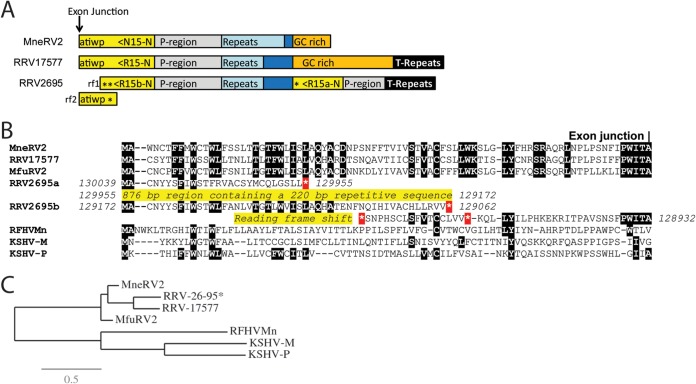
The MneRV2 genome sequence ended 1,143 bp downstream of the rightmost identified ORF, annotated as N15 due to the sequence similarity with the rightmost genes K15 in the KSHV genome and RF15 in the RFHVMn genome (Fig. 3A). N15 was encoded on the DNA strand opposite that encoding N1, translated leftward in the MneRV2 genome (Fig. 1). Although sequences homologous to the spliced KSHV K15 gene were observed in the RRV26-95 genome (annotated R15) (28), no analysis of the potential spliced R15 ORF was provided in the original manuscript, nor was a K15 homolog identified in the original manuscript describing the RRV17577 genome (28). A K15 homolog was subsequently identified in RRV17577, and the sequence of the spliced gene product, termed RK15, was determined (40). The putative MneRV2 N15 homolog of KSHV K15, RFHVMn RF15, and RRV17577 RK15 contained an analogous spliced gene structure with conserved splice donor and acceptor sites (not shown), with sequence identities of 13%, 17%, and 61%, respectively. Analysis of the homologous R15 sequences present in the RRV26-95 genome revealed a number of unexpected stop codons and reading frameshifts, as well as a duplication of the N-terminal domain upstream, indicating that the RRV26-95 R15 gene was disrupted compared to the other K15 homologs (Fig. 3B). Phylogenetic analysis revealed that the MneRV2 N15 sequence clustered with the RRV26-95 R15 (partial) and RRV17577 RK15 sequences in a branch separate from the two K15 isoforms, K15M and K15P, and the RFHVMn RF15 (Fig. 3C). A comparison of the nucleotide sequences of MneRV2, RRV26-95, and RRV17577 revealed a common N15/R15 proximal promoter region (Fig. 3A; P-region, gray), a common repeat region containing variable numbers of repeats (MneRV2 DR10; repeat size 10, count 30, light blue), and an additional common region (dark blue). The common region was flanked by a GC-rich region (∼90% GC, orange) 83% conserved between MneRV2 and RRV17577, which flanked the terminal repeat region at the right end of the genomes (Fig. 3A).
FIG 3.
Comparison of the right-end K15 homologs. (A) Graphical representation of the structure at the right end of the MneRV2, RRV26-95, and RRV17577 genomes with the N-terminal exon of the K15 homolog shown (yellow), with the direction of translation and the amino acid sequence immediately upstream of the exon junction. The presumed promoter region (P-region; gray), repeat region (light blue), GC-rich region (orange), and terminal repeat (T-repeat) region (black) are indicated. The duplicated N-terminal domains of RRV26-95 R15 are shown (asterisk indicates a stop codon). Reading frames 1 and 2 (rf1 and rf2) of the N-terminal domain of RRV26-95 R15b are shown, indicating the reading frameshift disrupting the R15b sequence. (B) Alignment of the N-terminal sequences of the K15 homologs of KSHV (M and P variants), RFHVMn, RRV17577, RRV26-95, and MfuRV2. The conserved exon junction is indicated. The duplicated N-terminal domains of RRV26-95 R15 are shown with the disrupted reading frames and stop codons (*). (C) Maximum likelihood analysis of the complete amino acid sequences of the fully spliced K15 homologs: KSHV-M (AAD45296), KSHV-P (AAK72632), RFHVMn (AGY30764), RRV17577 (40), MfuRV2 (AY528864), RRV26-95 (AF210726; lacks the disrupted N-terminal exon), and MneRV2. The numbers of substitutions per site are indicated.
The MneRV2 genome was colinear with the genomes of RRV26-95 and RRV17577 and contained 46 herpesvirus core genes (Fig. 1; light blue), 8 genes conserved in beta- and gammaherpesviruses (Fig. 1; dark blue), and 11 genes conserved only in gammaherpesviruses, including ORFs 16 (vBCL-2), 27, 45, 48, 49, and 50 (RTA); K8 (bZIP); K8.1, 52, and 75 (FGARAT); and K15 (LMP2A) (Fig. 1; gray) (Table 1). In addition, MneRV2 had homologs of genes conserved within the rhadinovirus gamma-2-herpesvirus lineage, including ORF2 (vDHFR), ORF4 (complement control protein [CCP]), ORF70 (vTS), ORF28 (membrane protein), ORF72 (vCyclin), ORF73 (LANA), and ORF74 (viral G-protein-coupled receptor [vGPCR]) (Fig. 1; green). Finally, MneRV2 genes conserved with KSHV and other Old World primate rhadinoviruses included homologs of the ORFs K1, K2 (viral interleukin-6 [vIL-6]), K4 (vCCL-2), K7 (vIAP), K9 (vIRF-1), K12 (Kaposin), K13 (vFLIP), and K14 (vOX-2) (Fig. 1; orange). Like both RRV variants, MneRV2 lacked genes conserved within the RV1 rhadinoviruses KSHV and RFHVMn, including ORFs K3 (MIR1), K4.1 (vCCL-3), K4.2, K10, K10.5, and K11 and genes found uniquely in KSHV, including ORFs K5 (MIR2) and K6 (vCCL-1). Although an RRV homolog of KSHV ORFK12 had not been detected previously, our analysis revealed positional homologs of ORFK12 in the MneRV2 and RRV genomes, as described below. We have also identified an MneRV2 homolog of KSHV ORFK7 and RFHVMn ORFRF7, as described below, although no intact homologous ORFs were detected in either RRV strain or in MfuRV2. Like both RRV strains, MneRV2 contained an 8-fold duplication of homologs of the KSHV ORFK9 and RFHVMn ORFRF9 vIRF-1 genes (Fig. 1). Amino acid identities between the proteins encoded by MneRV2 and RRV26-95 ranged from 32% (N12 and R12 homologs of the K12 Kaposin) to 95.7% (ORF55 tegument protein homologs), with a median of 82.8%. In contrast, a comparison of the MneRV2 and KSHV ORFs showed amino acid identities ranging from 11.0% (K8.1 virion envelope protein) to 72.3% (ORF25 major capsid protein) with a median of 41% (Table 1).
Nine regions with tandem direct repeats were identified in the MneRV2 genome, clustering between ORF70 and ORF16 at the left end of the genome and between ORF69 and ORFN13 at the right end of the genome, which ranged in size from 65 bp (DR2; 89% AT) to 579 bp (DR6; 79% GC) (Fig. 1). These correspond to similar repeat regions within the RRV genomes with nucleotide identities ranging from 71% (DR3; 50% GC) to 97% (DR7; 48% GC) between the corresponding MneRV2 and RRV26-95 direct repeats (Fig. 1). Fourteen small unique ORFs with no homology to existing cellular or viral sequences have been identified previously in the RRV26-95 genome, which span or are adjacent to these direct repeat regions (28). The RRV26-95 RU-1 ORF (102 amino acids [aa]), which is immediately upstream of the DR2 region, overlaps the predicted Ori-Lyt-L regulatory domain that is conserved in MneRV2 (see below). No positional or sequence homolog of RU-1 was detected in the MneRV2 sequence. A number of unique ORFs were identified at the right end of the RRV26-95 genome within the DR5-7 and DR8-9 tandem repeat regions. While the nucleotide sequence similarity with the MneRV2 sequence was very high in these regions, the potential ORFs corresponding to the RRV unique ORFs (leftward, RU-8L, -10L, -12L, -13L, and -14L; rightward, RU-1R, -2R, -3R, -4R, -5R, and -7R) either had no initiating ATG in the corresponding MneRV2 sequence or contained internal stops or frameshifts disrupting potential conserved coding sequences. RU-11L and RU-6R ORFs, which are derived from opposite strands of the same genomic sequence, and RU-9L were conserved in the MneRV2 genome. As indicated below, RU-9L is a distantly related positional homolog of the MneRV2 N12 homolog of KSHV ORFK12 Kaposin. None of the putative RRV unique ORFs were conserved intact in the closely related MfuRV2 genome sequence, which also lacked ATG initiators or contained internal stops or multiple reading frameshifts. The lack of sequence homology of the RRV unique ORFs to corresponding repetitive regions of MneRV2 and MfuRV2 genomes, coupled with the fact that the RRV unique ORFs were detected in regions of the RRV, MneRV2, and MfuRV2 genomes that encode the set of latent microRNAs or contain the potential Ori-Lyt regulatory sequences, suggests that the putative RRV unique ORFs, except RU-9L, may not be protein coding.
Alternate initiation sites.
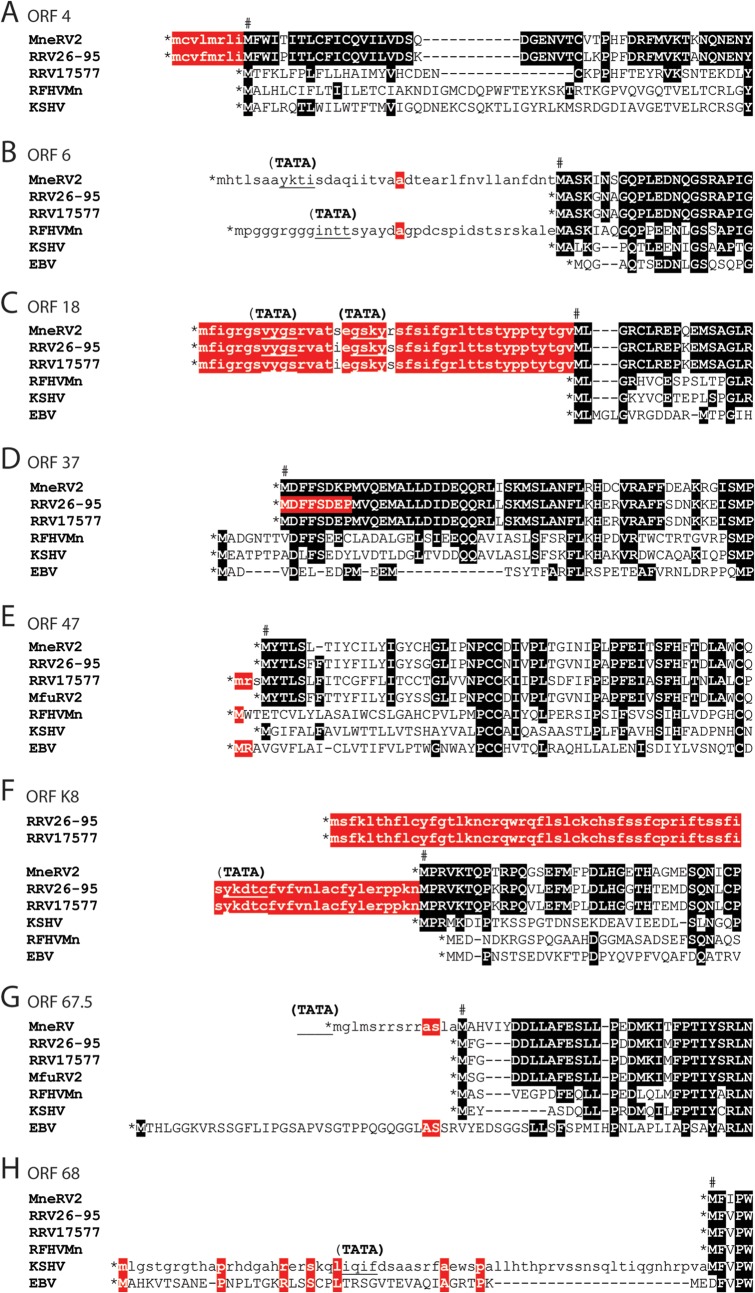
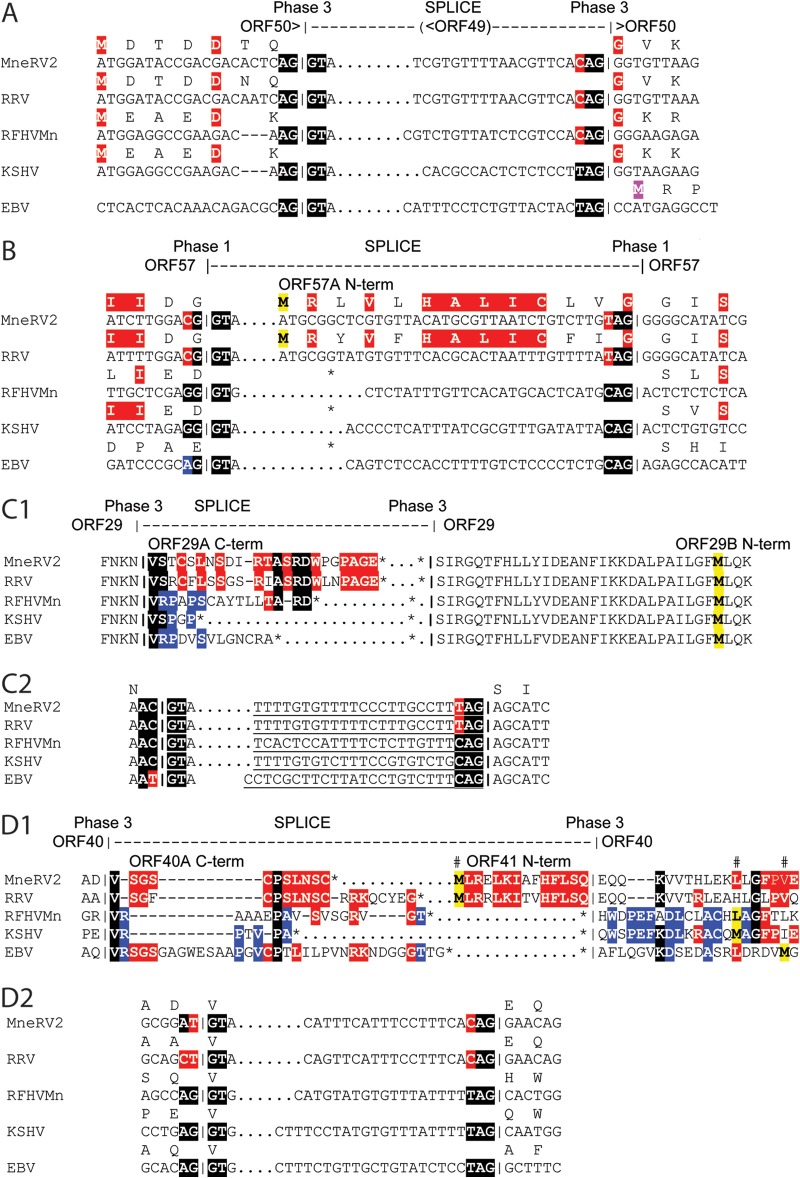
Analysis of the MneRV2 genome revealed a number of ORFs with methionine codons in frame and upstream of the protein initiation site predicted by sequence conservation, including ORFs 4, 6, 18, and 67.5, suggesting possible alternate sites of translation initiation. For the MneRV2 and RRV26-95 ORF4 and ORF18, highly conserved sequences extended downstream from the first ATG codon in the open reading frame (Fig. 4A and C, lowercase red highlight) to a second ATG codon, which was conserved with the translation initiation sites in the ORF4 and ORF18 homologs of RFHVMn and KSHV (Fig. 4A and C, indicated by #). The ORF4 and ORF18 homologs in RFHVMn, KSHV, and EBV initiated at the site of the second ATG codon in the MneRV2 and RRV26-95 sequences, as a translation stop codon occurred immediately upstream (Fig. 4A and C, indicated by *). While the RRV17577 ORF4 sequence initiated at the same position as RFHVMn and KSHV ORF4 homologs (Fig. 4A), the RRV17577 ORF18 sequence initiated at the same upstream ATG codon as the MneRV2 and RRV26-95 ORF18 sequences, and the encoded alternate N-terminal sequence was identical to that seen in RRV26-95 (Fig. 4C). An upstream ATG codon was also present within the ORF6 homologs of MneRV2 and RFHVMn; however, little sequence conservation was apparent in the upstream coding sequences (Fig. 4B, lowercase). In ORF67.5, the N-terminal extension of the coding sequence in MneRV2 corresponded to an N-terminal extension of the EBV homolog BFRF1A (Fig. 4G). Alternate N-terminal initiations of ORF37 in RRV26-95 and RRV17577 were reported previously (AAF60016 and NP_570778, respectively). Alignment of the RRV, MneRV2, RFHVMn, KSHV, and EBV ORF37 homologs indicated that the ORF37 sequence predicted for RRV26-95 (AAF60016) was only a partial sequence, as the open reading frame extended 8 additional amino acids upstream to the first ATG codon in the open reading frame, which were identical to the predicted N-terminal sequences of the ORF37 homologs of MneRV2 and RRV17577 (Fig. 4D). While the ORFK8 homologs of MneRV2 and KSHV showed a single conserved ATG initiation site in the open reading frame (Fig. 4F), the open reading frames of the two RRV homologs extended much further upstream to a conserved alternate ATG initiation site and encoded 69 identical amino acids (Fig. 4F). The open reading frames encoding ORF47 were different between the two RRV homologs. The initiation of the MneRV2 ORF47 homolog was identical to the RRV26-95 and MfuRV2 ORF47 homologs and corresponded to the same position in the KSHV ORF47. In contrast, the RRV17577 ORF47 open reading frame initiated three codons further upstream at a position analogous to the initiation of the ORF47 homologs of RFHVMn and EBV (BKRF2) (Fig. 4E). Note that the RRV17577 ORF47 appears to be evolutionarily distinct from the ORF47 homologs of MneRV2 and RRV26-95 (see below). Analysis of the ORF68 homologs revealed a conserved Met initiator in MneRV2, RRV, and RFHVMn downstream of a stop codon. While KSHV ORF68 encoded a conserved Met initiator at this position, a long upstream ORF extension contained an additional possible Met initiator that was homologous to the initiator in the EBV BFLF1 homolog (Fig. 4H). The KSHV ORF17 reading frame contained an additional Met initiator and 18 amino acids encoded upstream of the proposed Met initiators conserved in ORF17 homologs of RFHVMn, MneRV2, RRV, and EBV (data not shown).
FIG 4.
Comparison of ORF initiation. An alignment of the predicted MneRV2 ORFs and gammaherpesvirus homologs showing alternate initiation sites: ORF4 (A), ORF6/BALF2 (B), ORF18/BVLF1 (C), ORF37/BGLF5 (D), ORF47/BKRF2 (E), ORFK8/bZIP (F), ORF67.5/BFRF1A (G), and ORF68/BFLF1 (H). Predicted initiating methionines in the MneRV2 ORFs, based on sequence homology, are indicated (#), and encoded amino acids sequences conserved with other ORFs are highlighted black; alternate coding sequences initiating upstream are in lowercase and highlighted red when conserved. Upstream translational stop codons limiting the N-terminal coding sequences are indicated (*), the locations of potential TATA promoter motifs within or adjacent to the upstream ORF that could influence the mRNA start site are shown (TATA), and the position is indicated by underlining.
In many of the cases of N-terminal extension of open reading frames upstream of a conserved initiation site, analysis of the underlying DNA sequence revealed the presence of a putative TATA transcriptional promoter element within the alternate upstream coding sequence (Fig. 4B, C, F, G, and H; indicated as TATA). If transcription initiated downstream of these promoter elements, then translation of the transcribed mRNA would initiate at the conserved ATG initiation codon (labeled # in Fig. 4). If, however, transcription initiated further upstream, then the encoded proteins could contain the additional N-terminal amino acid sequences indicated in Fig. 4. An example of this is evident for the RRV26-95 R8 homolog of K8, which is transcribed in two mRNA transcripts (41). One large bicistronic mRNA encodes the complete R8 ORF downstream of the ORF50 coding sequences, including the alternate N-terminal 69-amino-acid extension shown in Fig. 4F, suggesting that ribosomal initiation within the bicistronic mRNA could produce an R8 variant containing the N-terminal extension. A second monocistronic mRNA initiating downstream within the DNA corresponding to the R8 N-terminal extension (as indicated in Fig. 4F) (TATA) would encode only the R8 ORF from the downstream conserved ATG initiation site. Since the K8 homologs of MneRV2, KSHV, RFHVMn, and EBV have a stop translation codon immediately upstream of the conserved ATG initiation site (Fig. 4F; *), bicistronic mRNAs would not encode a longer K8 variant in these viruses.
Conservation of transcription termination sites.
Similar to mammalian transcripts, herpesvirus RNA polyadenylation employs two major polyadenylation [poly(A)] signals, “AAUAAA” and “AUUAAA,” and flanking sequences to determine the 3′ end of the mRNA transcript. The presence of putative poly(A) signals defining transcript termination in KSHV has been determined both bioinformatically and experimentally (42). Examination of the MneRV2 and RRV genome sequences revealed a high level of conservation of poly(A) signals at the termini of the same open reading frames that have validated poly(A) signals in KSHV, thus defining the 3′ end of similar mRNA transcripts. In the majority of cases, the same poly(A) signal, either AAUAAA or AUUAAA, was detected in MneRV2, RRV, and KSHV, including the homologs of KSHV ORFs N1, 6, 11, K2, K4, PAN, 16, 17, 22, 23, 27, 33, 38, 39, 41, 42, 44, 45, 49, K8.1, 52, 54, 55, 57, 58, 64, 65, 69, K12, K13, 74, and 75 (Fig. 1, black and red asterisks, respectively). In only a few instances, including the homologs of KSHV ORFs 4, 16, and 18, did the MneRV2 or RRV ORF terminate with AAUAAA while the KSHV ORF terminated with AUUAAA. Additional poly(A) signals were identified in the KSHV ORFs 70, 19, 29B, 37, 53, 62, and 72 which were not found in the MneRV2 or RRV homologs, suggesting a difference in gene regulation between KSHV and the macaque RV2 rhadinoviruses. In most cases, the macaque RV1 rhadinovirus, RFHVMn, showed poly(A) signals similar to those of KSHV, although RFHVMn lacked signals present in the KSHV homologs of ORFs K1, 70, 19, 53, and 58 or had different poly(A) signals in ORFs 4, K11, and 74 (Fig. 1). The RFHVMn genome lacks a homolog of ORF11 and instead has a poly(A) signal in ORF10. A putative poly(A) signal unique to MneRV2 was identified for ORF67A. Notably, both MneRV2 and RRV had unique poly(A) signals after the ORFs 2, 18, 48, K8, 60, and N15/R15 homologs (Fig. 1, arrows). The poly(A) signal in ORF N8/R8 was shown to terminate RRV R8 transcription (41, 43), yielding bicistronic ORF50/R8 or monocistronic R8 transcripts in RRV rather than the tricistronic ORF50/K8/K8.1, bicistronic K8/K8.1 transcripts identified in KSHV. A tricistronic RRV transcript depicted in the study by DeWire et al. (41) is not supported by published data.
A major difference between the RV2 rhadinoviruses (MneRV2 and RRV) and the RV1 rhadinoviruses (KSHV and RFHVMn) is the presence of an amplified set of 8 homologs of the unspliced K9 vIRF gene in MneRV2 and RRV (N/R9-1, N/R9-2, N/R9-3, N/R9-4, N/R9-5, N/R9-6, N/R9-7, and N/R9-8), while KSHV and RFHVMn have 3 additional distinct vIRF homologs (K/RF10, K/RF10.5, and K/RF11) which are spliced (26). All of the RV1 vIRF homologs have conserved AAUAAA poly(A) signals, while conserved AAUAAA poly(A) signals are detected only in N/R9-1, N/R9-3, N/R9-5, and N/R9-7 (Fig. 1), suggesting that these vIRF genes are expressed either as monocistronic or as bicistronic transcripts with the next adjacent vIRF gene. MneRV2 ORFN9-8 contains additional AAUAAA and AUUAAA poly(A) signals that are not conserved in RRV ORFR9-8. While additional variant poly(A) signals may exist, the overall high conservation of AAUAAA and AUUAAA poly(A) signals indicates strong similarities in mRNA transcript production and regulation between the RV1 and RV2 rhadinoviruses.
Conservation of spliced transcripts.
Comparative analysis of the MneRV2 genome identified a strong conservation of donor and acceptor sites within the coding regions for the characteristic spliced gene transcripts identified in KSHV and related rhadinoviruses, which included ORF29, ORF40, ORF50, K8, K8.1, ORF57, and K15 (Fig. 1; spliced sites indicated with a thin black bar within colored ORFs or as connections between colored ORFs). While alternate splicing has been detected in the KSHV ORF4 KCP (44) and RRV17577 ORF4 CCP (45), no conserved splice donor or acceptor sites were unambiguously identified by sequence analysis within the ORF4 homologs of the MneRV2 or RRV26-95 genomes. Additional spliced transcripts have been detected in noncoding regions of the KSHV K13, ORF72, and ORF73, but lack of sequence similarity within the MneRV2 homologs of these genes precluded the identification of conserved splice sites.
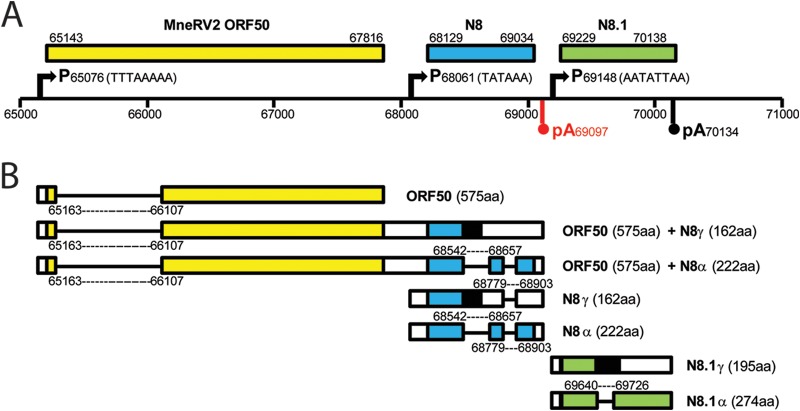
ORF50/K8/K8.1 homologs.
Major immediate early transcripts of KSHV initiating from a “TTTAAAAA” TATA-like element upstream of ORF50 are tricistronic with alternate splicing across the ORF50/K8/K8.1 region of the genome, encoding the replication transactivator ORF50 and multiple variants of the ORFK8 homolog of bZIP (46). Alternative promoter elements, “TATAA” upstream of ORFK8 and “AATATTAA” upstream of ORFK8.1, specify bicistronic ORFK8/K8.1 and monocistronic ORFK8.1 transcripts, respectively (47–49). All transcripts terminate at an “AAUAAA” poly(A) signal immediately downstream of the K8.1 region. A complete conservation of the ORF50, K8, and K8.1 promoter elements was observed in the MneRV2 and RRV genomes (Fig. 5A). However, an additional “AAUAAA” poly(A) site was detected downstream of the N8 and R8 genes that was not conserved in the KSHV K8 homolog (Fig. 1, vertical arrows, and Fig. 5A, highlighted in red). Only bicistronic ORF50/R8 and monocistronic transcripts of ORF50, R8, or R8.1 have been identified in RRV (41, 43) (Fig. 5B). For KSHV ORF50, an initial exon located leftward of ORF49, which encodes the initiating methionine and 6 other amino acids, is spliced to an exon rightward of ORF49 that encodes the remainder of the ORF50 protein (Fig. 1 and 5). A similar splice event has been detected in RRV (41, 43) (Fig. 5B). Analysis of the MneRV2 sequence revealed splice donor (AGGT) and acceptor (pyrimidine stretch-T/CAG) sites in the MneRV2 ORF50 gene that were highly conserved in the ORF50 homologs of RRV, RFHVMn, KSHV, and EBV (BRLF1) (Fig. 6A), which would yield a 575-aa spliced MneRV2 ORF50 gene product (Fig. 5). Alternately spliced transcripts of the RRV R8 gene have been detected (41, 43), and the splice donor and acceptor sites were completely conserved in the MneRV2 N8 gene (Fig. 5B). Bicistronic and monocistronic transcripts from the N8 gene would yield ORFN8γ (162 aa) from an unspliced transcript and ORFN8α (222 aa) from a doubly spliced transcript (Fig. 5B), showing strong homology to similar ORFs R8γ (177 aa) and R8α (234 aa) derived from corresponding RRV transcripts (Table 1). Alternatively spliced monocistronic transcripts have been detected for KSHV K8.1 (47) and RRV R8.1 (49) genes from the immediate upstream promoter “AATATTAA.” This promoter and the splice donor and acceptor sites were highly conserved in the MneRV2 N8.1 gene, specifying transcripts encoding N8.1γ (195 aa) from an unspliced transcript and N8.1α (274 aa) from a singly spliced transcript (Fig. 5B) showing strong homology to similar ORFs R8.1γ (230 aa) and R8.1α (275 aa) derived from corresponding transcripts of RRV (Table 1). Both N8.1 variants would contain a putative N-terminal hydrophobic signal sequence with predicted cleavage between S26 and V27 and a highly glycosylated N-terminal domain. N8.1α would also contain a C-terminal hydrophobic transmembrane-associated domain from the downstream spliced exon, similar in structure to the R8.1α and the K8.1α virion glycoprotein (50, 51).
FIG 5.
Structure of the MneRV2 genomic region homologous to KSHV ORF50/K8/K8.1. (A) Graphical representation of the genomic structure of MneRV2 from bp 65000 to 71000 containing the MneRV2 homologs of the KSHV ORFs 50, K8, and K8.1. The positions of the TATA promoter (P) motifs of KSHV ORF50 (TTTAAAAA), K8 (TATAAA), and K8.1 (AATATTAA) conserved in MneRV2 and RRV are indicated. The polyadenylation (pA) site downstream of the K8.1 homologs, which is conserved between KSHV, MneRV2, and RRV, is shown in black, while the polyadenylation site downstream of the MneRV2 and RRV homologs of K8, which is not conserved in KSHV, is shown in red. (B) Graphical representations of the potential spliced transcripts from this region in MneRV2 are shown based on homology to KSHV and RRV (see the text) and the presence of conserved splice donor and acceptor sites. The putative encoded ORFs and their sizes are shown.
FIG 6.
Conservation of translation initiation in the MneRV2 spliced genes. Comparison of the nucleotide and encoded amino acid sequences surrounding the splice donor and acceptor sites for the spliced gene homologs of ORF50/BRLF1 (A), ORF57/(BSLF2/BMLF1) (B), ORF29/(BGRF1/BDRF1) (C), and ORF40/(BBLF2/BBLF3) (D) from the rhadinoviruses MneRV2, RRV, RFHVMn, and KSHV and the lymphocryptovirus EBV. The position and phasing of the conserved splice sites are indicated with highlighting of residues flanking the splice sites to show conservation. Putative initiating methionines are highlighted in purple. (A) The initiating methionine in ORF50 of the rhadinoviruses is present in a short exon upstream of ORF49/BRRF1, while the initiating methionine in the lymphocryptovirus BRLF1 homolog is present in the downstream spliced exon. (B) While an initiating methionine is present in the upstream exon of spliced transcripts of the rhadinovirus ORF57 and lymphocryptovirus BSLF2 homologs, both MneRV2 and RRV encode an alternate highly conserved N-terminal sequence upstream of the ORF57 coding sequence from a potential nonspliced transcript. The other genomes lack a potential initiating codon in the downstream exon (*, stop codon within the ORF). (C1) The rhadinovirus ORF29A and ORF29B, like the homologous lymphocryptovirus BGRF1 and BDRF1, can be spliced, producing a longer transcript encoding a fusion protein, ORF29 or BGRF1-BDRF1, as indicated. However, strong sequence conservation is seen within the C-terminal domain of ORF29A of MneRV2, RRV, and RFHVMn that would be encoded by an unspliced ORF29A transcript. Furthermore, a highly conserved initiating methionine is present in the ORF29B homologs of MneRV2, RRV, RFHVMn, KSHV, and EBV (red highlight). This methionine initiates translation of KSHV ORF29B from a bicistronic spliced transcript containing ORF48 and ORF29B (48), suggesting that the same could occur in the other viruses. (C2) Analysis of the ORF29A donor site in the five viruses reveals a highly conserved atypical donor site, AYGT, suggesting common regulation of splicing. (D1) The rhadinovirus ORF40A and ORF41, like the homologous lymphocryptovirus BBLF2 and BBLF3, can be spliced, producing a longer transcript encoding a fusion protein, ORF40, as indicated. However, strong sequence conservation is seen within the C-terminal domain of ORF40A of MneRV2 and RRV that would be encoded by an unspliced ORF40A transcript (red highlight). Whereas possible translation initiators were identified in the ORF41/BBLF3 exons of RFHVMn, KSHV, and EBV (purple highlight) allowing for the separate expression of distinct ORF41/BBLF3 proteins, both MneRV2 and RRV encode an alternate highly conserved N-terminal sequence upstream of ORF41 from a potential nonspliced transcript (red highlight). The other genomes contain a stop codon (*) in the sequence upstream flanking the acceptor site. (D2) Analysis of the ORF40A donor site in the five viruses reveals a conserved atypical donor site, DYGT, in the MneRV2 and RRV sequences that is not found in the other viral genomes, suggesting differential regulation of splicing.
ORF57.
In KSHV, ORF57 is produced from a spliced transcript containing a small upstream exon encoding a methionine-rich 15-aa N-terminal sequence and a large downstream exon encoding the remainder of the 455-aa protein. Conserved splice donor and acceptor sites were identified in the ORF57 genes of MneRV2 and RRV predicting spliced transcripts encoding 447-aa and 446-aa ORF57 homologs, respectively, that were 87.9% identical (Fig. 6B; Table 1). Unlike the ORF57 homologs of KSHV and RFHVMn, the open reading frame in the large downstream exons of MneRV2 and RRV initiated with a methionine codon within the spliced intron region such that an unspliced transcript would encode 443-aa and 442-aa MneRV2 and RRV ORF57A homologs, respectively, containing 13 highly conserved N-terminal amino acids encoded within the intron region (Fig. 6B). Thus, MneRV2 and RRV could encode ORF57 homologs with alternate N-terminal sequences derived from spliced or unspliced transcripts.
ORF29.
In KSHV, ORF29 is split into two distinct coding sequences, ORF29A (312 aa) and ORF29B (351 aa), separated by ORFs 30 to 33, which can be spliced into a single transcript in which the coding sequence in ORF29A continues in ORF29B (Fig. 6C1), creating a 687-aa spliced ORF29 immediate early gene product (YP_001129382). MneRV2 encodes homologs of ORF29A (327 aa) and ORF29B (348 aa), which are homologous to similar ORFs in KSHV and RRV (Table 1). The splice donor and acceptor sites for the ORF29 spliced gene product are highly conserved in MneRV2, RRV, RFHVMn, KSHV, and EBV (Fig. 6C2) and would yield a fused ORF29 gene product in MneRV2 (681 aa) (Table 1). An additional KSHV transcript has been identified in which an exon encoding ORF48 is spliced into the ORF29B exon after the ORF48 stop codon, creating a bicistronic mRNA encoding ORF48 and ORF29B as single protein products (48). The ORF29B homologs of KSHV, MneRV2, RRV, RFHVMn, and EBV all contain a conserved Met initiator (Fig. 6C1, highlighted in yellow), suggesting that the ORF29B exon can also be expressed as a single gene product in these related viruses. Examination of the downstream C-terminal extension of the MneRV2, RRV, and RFHVMn ORF29A homologs within the intron region revealed substantial sequence similarity (Fig. 6C1, highlighted), suggesting that the ORF29A homologs could also be expressed as single gene products from an unspliced transcript with a conserved C-terminal domain encoded within the intron, which is lacking from the spliced ORF29 sequence.
ORF40/41.
In KSHV, ORF40A (457 aa) and ORF41 (205 aa) can be spliced into a single transcript producing ORF40 (669 aa), a fusion of the coding sequences of ORFs 40A and 41 (52). MneRV2 encodes homologs of ORF40A (460 aa) and ORF41 (203 aa), similar to RRV (Table 1). A conserved splice donor site at the end of ORF40A and acceptor site at the beginning of ORF41 are also present in the ORF40/41 homologs of MneRV2, RRV, RFHVMn, and EBV (BBLF2/BBLF3) (Fig. 6D1 and D2). This suggests that MneRV2, like the other viruses, could produce a single spliced transcript encoding an ORF40 homolog (638-aa) fusion of ORF40A and ORF41 (Table 1). It is interesting that the donor sites of MneRV2 and RRV utilize the more variable XTGT sequence rather than the more common XGGT sequence found in the other viral sequences (Fig. 6D2, highlighted in red). A smaller monocistronic transcript has been identified in KSHV encoding only ORF41 (52), which initiates at an ATG codon that is not conserved in MneRV2, RRV, RFHVMn, or EBV (Fig. 6D1; see Met highlighted yellow). Analysis of the ORF41 reading frames in MneRV2 and RRV revealed N-terminal amino acid sequences upstream of the splice acceptor site that encode 14 additional highly conserved amino acids (Fig. 6D1; ORF41 N-term), which were not detected in ORF41 homologs of KSHV, RFHVMn, or EBV, as a translational stop codon in the reading frame was immediately upstream of the splice acceptor site (Fig. 6D1; stop codon shown by *). The coupling of the variable XTGT donor site with the presence of alternate N-terminal ATG initiation sites in MneRV2 and RRV ORF41 homologs suggests a difference in the regulation of the spliced and nonspliced variants of ORF41 between the RV2 rhadinoviruses, MneRV2 and RRV, and the other gammaherpesviruses.
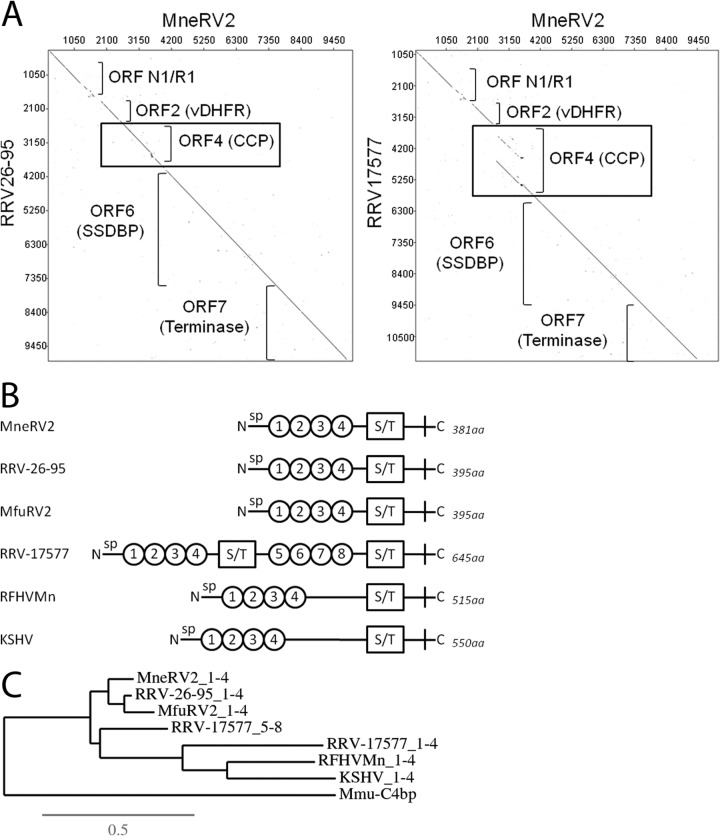
The MneRV2 ORF4 CCP homolog is structurally similar to ORF4 of RRV26-95 and not RRV17577.
A DNA dot plot analysis of the first 10,000 bp of the MneRV2 genome revealed a close sequence similarity and colinearity with the RRV26-95 genome encoding ORFN1/R1, ORF2 (DHFR), ORF4 (complement control protein [CCP] homolog), ORF6 (SSDB), and ORF7 (terminase) (Fig. 7A). The same dot plot analysis between MneRV2 and the RRV17577 genome sequence revealed a marked discontinuity within the ORF4 region. Previous studies have shown that the ORF4 homolog of RRV is different in the two RRV strains. RRV26-95 encodes a protein (AAF59982) with four CCP N-terminal domains, while RRV17577 encodes a larger protein (NP_570746) with eight CCP domains (45). Analysis of the encoded amino acid sequences revealed that the MneRV2 ORF4 was closely related to the RRV26-95 and MfuRV2 ORF4 sequences with an N-terminal signal peptide, four CCP domains, and a conserved serine/threonine (S/T) domain prior to the first cysteine residue (Fig. 7B). This gene structure was similar to that seen in the ORF4 KCP homologs of KSHV and RFHVMn, except that these latter proteins had larger regions between the CCP and S/T domains (Fig. 7B). Phylogenetic analysis of the CCP domains revealed a close clustering of the CCP domains 1 to 4 of RRV26-95 and MfuRV2, consistent with the close evolutionary relationship between the rhesus and fuscata macaque hosts (Fig. 7C). The phylogenetic analysis revealed that the MneRV2 CCP domains 1 to 4 were more distantly related but clustered together with the other RV2 rhadinovirus RRV26-95 and MfuRV2 homologs. The CCP domains of the rhesus macaque complement 4 binding protein (Mmu-C4BP; XP_001082210) were used as an outgroup. The CCP domains 1 to 4 of the ORF4 homologs of KSHV and RFHVMn clustered together, separately from the RV2 rhadinovirus proteins, confirming the close evolutionary relationship between these RV1 rhadinoviruses. Interestingly, the RRV17577 CCP domains 1 to 4 and 5 to 8 branched separately from the RV1 and RV2 rhadinovirus CCP domains. The RRV17577 CCP domains 5 to 8 were more closely related to the RV2 rhadinovirus CCP domains 1 to 4, while the RRV17577 CCP domains 1 to 4 were more closely related to the RV1 rhadinovirus CCP sequences. This suggests that the additional N-terminal CCP domains 1 to 4 and adjacent S/T domain of RRV17577 could have derived through recombination with an ancestral RV1 rhadinovirus at some point in evolution, resulting in a larger protein with a duplication of the four CCP and one S/T domain. MneRV2, like RRV26-95, shows no evidence for this potential recombination event.
FIG 7.
The ORF4 homolog of MneRV2, like RRV26-95, lacks the gene duplication present in RRV17577. (A) Dot plot of the left end of the MneRV2 genome sequence versus the RRV26-95 and RRV17577 sequences. The approximate positions of the different genes in this region are shown. The analogous position of the gene duplication identified in the ORF4 CCP homolog of RRV17577 is boxed. (B) A graphical representation of the N-terminal domains of the ORF4 homologs of MneRV2, RRV26-95, MfuRV2, RRV17577, RFHVMn, and KSHV, showing the presence of the N-terminal signal peptide (sp), the conserved complement control protein (CCP) domains (numbered circles), the serine/threonine-rich domains (S/T), and the sizes of the complete encoded proteins. (C) Phylogenetic relationship between the ORF4 CCP domain of MneRV2 (aa 27 to 271) and analogous domains in other ORF4 homologs (see panel B), determined by maximum likelihood analysis. The macaque C4BP alpha chain isoform 2 CCP domain (XP_001082210) was included as an outgroup. The numbers of substitutions per site are indicated.
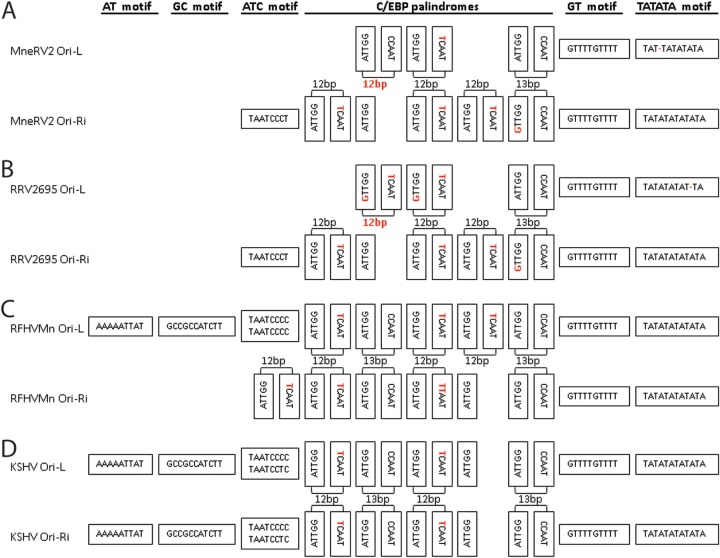
Conservation of nucleotide motifs in the Ori-Lyt regions of MneRV2.
KSHV contains two duplicated origins of lytic replication (Ori-Lyt) within an ∼1-kb inverted repeat. Ori-Lyt-L is located between ORFs 11 and 16, while Ori-Lyt-R is located between ORFs 69 and 71 (Fig. 1). A sequence alignment of the KSHV Ori-Lyt-L and Ori-Ly-R (inverted) revealed ∼90% average nucleotide identity across the 1-kb region interrupted only by the kshv-miR-K12-9 gene, with 500 bp of identical sequence (26). Previous analysis of the homologous region in RFHVMn revealed little nucleotide similarity between the left and right Ori-Lyt regions except a remarkable conservation of a number of sequence motifs, which were also conserved in both KSHV Ori-Lyt-L and Ori-Lyt-R (26). A number of these sequence motifs have been identified as critical for KSHV lytic DNA replication, including eight CCAAT/enhancer binding protein (C/EBP) binding motifs required for binding of the ORFK8/bZIP homolog and an 18-bp AT palindrome essential for Ori-Lyt function (53, 54). The palindromes in RFHVMn and KSHV consist of conserved “CCAAT/TCAAT” and “ATTGG” motifs separated by a 12- or 13-bp loop region (Fig. 8C and D, respectively). Analysis of the homologous regions in the MneRV2 genome revealed a 15-bp TATATA motif homologous to the 18-bp AT palindrome in KSHV and RFHVMn and a set of conserved C/EBP palindromic motifs, whose structure and sequence varied slightly from that seen in KSHV (Fig. 8A). A functional lytic origin has been identified between ORFs 69 and 71 in RRV (55), and analysis of the RRV sequence in this region revealed a 15-bp TATATA motif and a set of conserved C/EBP motifs with sequences nearly identical to the MneRV2 motifs (Fig. 8B). Like RFHVMn, the MneRV2 and RRV Ori-L and Ori-R regions are part of an inverted repeat structure that shows little similarity between the two regions other than the conserved motifs, in contrast to KSHV. While the KSHV and RFHVMn Ori regions contain additional motifs not conserved in MneRV2 and RRV (Fig. 8C and D; AT and GC motifs), the MneRV2 and RRV Ori regions contain two other motifs flanking the C/EBP motifs that are conserved in RFHVMn and KSHV (Fig. 8A to D; ATC and GT motifs), which are predicted to play critical roles in replication due to their evolutionary conservation.
FIG 8.
The left and right Ori-Lyt regions of MneRV2 are divergent but contain motifs conserved in RRV, RFHVMn, and KSHV. Schematic representation of the nucleotide sequence motifs conserved between the Ori-Lyt-Left and Ori-Lyt-Right lytic origins of DNA replication of MneRV2 (A), RRV26-95 (B), RFHVMn (C), and KSHV (D), including the TATATA, GT, and C/EBP palindrome motifs conserved in both left and right origins of all four viruses and the AT, GC, and ATC motifs, which are differentially conserved, as described in the text. Nucleotide changes from the common ATTGG-CCATT palindrome sequences involved in binding C/EBP during virus replication (see the text) and changes in the length of the palindromic sequence loops in MneRV2 and RRV are colored red. The Ori-Lyt-Right sequences are inverted.
MneRV2 PAN locus.
The PAN locus in KSHV, located between ORFs K6 and 16, encodes PAN, the major lytic polyadenylated nuclear RNA (T1.1), and ORFK7, an inhibitor of apoptosis with sequence similarity to a splice variant of human survivin (56). Using transcriptome sequencing (RNA-seq) analysis of RNA transcripts from spleen tissue from an MneRV2-infected macaque, we identified a homolog of KSHV PAN in the MneRV2 genome downstream of ORF16 (Fig. 1), which is similarly positioned, with close sequence homology to the PAN RNA identified in RRV (data not shown). Analysis of the sequence immediately preceding the 5′ end of the MneRV2 PAN RNA revealed a previously unidentified ORF encoding a 116-aa protein with sequence similarity to KSHV ORFK7 (7.8% identity) and RFHVMn ORFRF7 (29.3% identity) (Table 1). Like ORFK7, MneRV2 ORFN7 initiates upstream of the transcription start site of the PAN RNA, suggesting that ORFN7 and PAN are expressed as separate transcripts terminating at the same poly(A) site (Fig. 1). ORFs K7 and RF7 encode N-terminal hydrophobic signal sequences and show sequence homology to human survivin (26). Despite obvious sequence homology with ORFs K7 and RF7, ORFN7 lacks an obvious signal sequence and has only minimal sequence similarity with survivin (Fig. 9). While the RRV and MfuRV2 PAN regions encode homologous sequences, there is no obvious translation initiator, and the RRV PAN ORF is disrupted with a stop codon (Fig. 9). Both MneRV2 and RRV PAN RNAs contain a region of repetitive DNA upstream of the polyadenylation site (Fig. 1, DR3), with close sequence similarity between the repeats.
FIG 9.
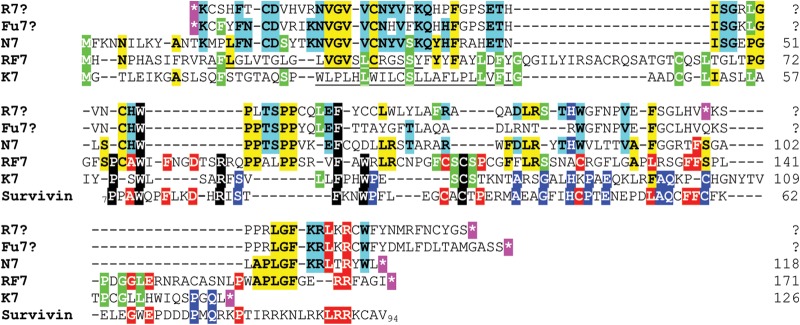
The MneRV2 PAN locus encodes a homolog of KSHV ORFK7. The sequences of MneRV2 ORFN7, KSHV ORFK7 (YP_001129367), and RFHVMn ORFRF7 (AGY30697) positional homologs were aligned. Since the K7 sequence was previously shown to have sequence similarity to human survivin, the sequence of survivin IAP repeat-containing protein 5 isoform 2 (NP_001012270) from aa 7 to aa 94 was also aligned. The amino acid sequences of the homologous regions within RRV (R7?) and MfuRV2 (Fu7?), which lack obvious ATG or CTG initiation codons and are disrupted by stop codons (*, highlighted in purple), were also aligned. Amino acid residues conserved among K7, RF7, and survivin are highlighted in black; those conserved between K7, RF7, N7, R7?, and/or Fu7? are highlighted in green; those conserved between RF7 and survivin are highlighted in red; those conserved between RF7 and N7 are highlighted in yellow; those conserved between K7 and survivin are highlighted in blue with white text; and those conserved between N7, R7?, and Fu7? are highlighted in blue with black text. Putative N-terminal hydrophobic transmembrane domains missing in N7, R7?, and Fu7? are underlined.
MneRV2 latency locus.
Like KSHV and RRV, MneRV2 contains a genomic region homologous to the KSHV latency locus at the right end of the genome and encodes homologs of vFLIP-K13 (ORF71), vCyc (ORF72), and LANA (ORF73) (Fig. 1). We have previously published the sequence of MneRV2 LANA and showed that, like RRV LANA, it lacks the characteristic large acidic repeat region that is found in KSHV and its RV1 macaque homolog, RFHVMn (22). MneRV2 and RRV LANA displayed ∼60% amino acid identity and contained a similar serine-proline-rich N-terminal region and a C-terminal domain that was homologous to KSHV LANA. However, there was only 16.2% amino acid identity with KSHV LANA (Table 1). The MneRV2 and RRV vFLIP-K13 and vCyc homologs were highly conserved, with 90.4 and 83.9% amino acid identity, respectively, and showed strong similarity to the related KSHV sequences (Table 1).
The 60-aa KSHV ORFK12 Kaposin A is encoded downstream of the KSHV Ori-R, while the larger Kaposin B is encoded within the adjacent DR5/6 direct repeats (Fig. 10A). The RFHVMn genome lacks direct repeats in this region but encodes RF12 (212 aa), a larger homolog of Kaposin A with a C-terminal extension containing a highly basic domain (26) (Fig. 10B). Analysis of the MneRV2 genome sequence identified an open reading frame encoding a distantly related 109-aa Kaposin homolog that we have designated N12, which contains an “HVPV-L” motif conserved in Kaposin A (Fig. 10B). Analysis of the two RRV genomes revealed an 89-aa ORF, designated R12, which is homologous to N12 containing the “HVPV-L” motif and an adjacent “PGTQR” motif conserved in N12. The R12 ORF could initiate either at a Met codon conserved with the Kaposin initiator, encoding 63 aa, or at an upstream Met initiating a longer ORF encoding an additional N-terminal hydrophobic domain reminiscent of a signal peptide (AF210726, 89 aa) (Fig. 10B). While a homologous sequence, here designated F12, was detected in the Japanese macaque rhadinovirus (MfuRV2) genome, the N-terminal sequence contained a stop codon and a reading frameshift, with no obvious Met initiator (Fig. 10B). Unlike K12 and R12, the N12 ORF contained a C-terminal extension, similar to that seen in RF12, which also contained an arginine-rich C-terminal basic domain (Fig. 10B).
FIG 10.
The latency regions of MneRV2 and RRV are highly homologous and encode protein and miRNAs conserved with KSHV and RFHVMn. (A) Schematic representation of the genomic regions of MneRV2, RRV, RFHVMn, and KSHV between ORFs 69 and 71; the presence of encoded proteins and miRNAs is shown. The positions of the TATATA motif (red box) and C/EBP palindromic motifs (stars) within the long inverted repeat (LIR) regions are indicated (Fig. 8). The known KSHV (purple) and RRV (green) miRNA genes are indicated, as are the predicted MneRV2 (green) and RFHVMn (blue) candidate miRNAs. Homologous miRNAs with identical seed sequences conserved across the four viruses are colored yellow (see panel C). Direct repeats (DR) and putative protein coding ORFs are shown. Poly(A) signals “AAUAAA” (blue star, left to right transcription; red star, right to left transcription) and “AAAUCA” (orange star, right to left transcription) identified in KSHV (42) are shown with homologous signals in the other viruses. (B) Alignment of KSHV ORFK12 Kaposin A (YP_001129428) and positional homologs MneRV2 ORFN12, RRV ORFR12 (AF210726 or, without N-terminal extension, NP_598364), and RFHVMn ORFRF12 (AGY30757). The translated sequence of Fu12 from the MfuRV2 genome sequence is shown but lacks a methionine initiator and contains a shift (\) and stop codon (*) in the reading frame. Residues conserved with K12 are highlighted black; residues conserved with RF12 are highlighted red; residues conserved with N12 are highlighted blue. (C) Alignment of the kshv-miR-K12-10a, rfhvmn-miRc-RF9, mnerv2-miRc-N12-15, and rrv-miR-rR1-15 viral pre-miRNAs with the human cellular miR-142-3p. The 5p and 3p mature miRNAs are indicated (bold, underlined). The conserved seed sequences are highlighted in black with 5′ and 3′ residues highlighted in red or blue that would constitute part of the seed sequence depending on processing. The nonconserved K12-10-5p seed is highlighted in yellow.
The genomic region of KSHV flanking Ori-R between ORFs 69 and 71 encodes a set of 12 microRNA (miRNA) sequences (Fig. 10) adjacent to ORFK12. Fifteen miRNAs have been identified in RRV flanking the homologous Ori-R, with the majority (12/15) positioned downstream between ORF69 and Ori-R. Analysis of the MneRV2 genome revealed 17 putative miRNA sequences adjacent to ORFN12 with 15 positional homologs of the RRV miRNAs (Fig. 10A). High levels of sequence similarity were detected between the MneRV2 and RRV positional homologs, with more than half having >90% sequence conservation (Table 2). Of the 15 positional homologs, 13 had at least one mature miRNA with a conserved 7-bp seed, and one had a conserved 6-bp seed (mnerv2-miRc-N12-12). Only one MneRV2 candidate, mnerv2-miRc-N12-2, had no conserved seed sequence with its positional homolog. Of the 27 known RRV mature miRNAs, 20 had a conserved 7-bp seed, 4 had a conserved 6-bp seed, and only three were not conserved in the positional homolog in MneRV2. Independent studies previously identified a seed sequence conservation between KSHV miR-K12-10a-3p and either RRV miR-rR1-15-3p (57) or RFHVMn miRc-RF9-3p (26). The MneRV2 miR-N12-15 sequence was closely related to RRV rR1-15 with identical 5p and 3p seed sequences (Fig. 10C). The 3p seed sequences of both MneRV2 miRc-N12-15 and RRV rR1-15 aligned with the 3p seed sequences of KSHV K12-10a and RFHV RF9, indicating an important conservation during the miRNA evolution of these primate rhadinoviruses. The viral miRNAs maintained the same seed sequence as the human cellular miRNA 142-3p (Fig. 10C), which is known to antagonize growth factor signaling influencing cellular proliferation (58).
TABLE 2.
Nucleotide similarities of candidate MneRV2 and RRV miRNA species
| MneRV2 pre-miRNA candidate | RRV positional homolog | % overall similarity (length)a |
Seed conservationb |
||
|---|---|---|---|---|---|
| 5p miRNA | 3p miRNA | 5p miRNA | 3p miRNA | ||
| N12-1 | R1-8 | 100 (20) | 100 (19) | 7mer | 7mer |
| N12-2 | R1-1 | 55 (20) | 71 (21) | NCc | NC |
| N12-3 | R1-9 | 84 (19) | 91 (22) | 6mer | 7mer |
| N12-4 | R1-10 | NAd | 100 (18) | NA | 7mer |
| N13-5 | R1-11 | 100 (23) | 81 (21) | 7mer | 7mer |
| N12-6 | R1-2 | NA | 100 (22) | NA | 7mer |
| N12-7 | R1-12 | 95 (20) | 91 (22) | 7mer | 7mer |
| N12-8 | R1-3 | 84 (22) | 79 (19) | 7mer | NC |
| N12-9 | None | NA | NA | NA | NA |
| N12-10 | None | NA | NA | NA | NA |
| N12-11 | R1-4 | 84 (19) | 77 (22) | 7mer | 7mer |
| N12-12 | R1-13 | 86 (22) | 91 (22) | NC | 6mer |
| N12-13 | R1-5 | 100 (20) | NA | 7mer | NA |
| N12-14 | R1-14 | 100 (22) | 94 (18) | 7mer | 6mer |
| N12-15 | R1-15 | 86 (22) | 82 (22) | 7mer | 7mer |
| N12-16 | R1-6 | 67 (18) | 100 (13) | 7mer | 7mer |
| N12-17 | R1-7 | 91 (22) | 95 (21) | 7mer | 7mer |
Percent nucleotide identity between the published mature RRV miRNA species and the homologous region of the MneRV2 pre-miRNA candidate. The length of the mature RRV miRNA region (in base pairs) is shown in parentheses.
Conservation between seed sequences of known mature RRV miRNAs and the homologous region of MneRV2 miRNA candidates. 6mer or 7mer matches indicate perfect conservation of nucleotides 2 to 7 or 2 to 8, respectively, of the mature RRV miRNA.
NC, not conserved.
NA, not applicable (RRV does not encode a pre-miRNA at or produce mature miRNAs from the homologous locus).
Phylogenetic relationship of MneRV2 to other Old World primate rhadinoviruses.
Previous studies analyzing the genes of the MneRV2 ORF9 DNA polymerase and ORF59 DNA polymerase processivity factor, identified by the consensus-degenerate hybrid oligonucleotide primer (CODEHOP) PCR technique, have shown that MneRV2 (host = pig-tailed macaque) clusters with RRV (host = rhesus macaque) within the RV2 lineage of Old World primate rhadinoviruses (8, 16, 31). ORF59 from MfaRV2, the RV2 rhadinovirus of the long-tailed macaque, clustered closely with the RRV ORF59, while the MneRV2 ORF59 branched separately. As the rhesus and long-tailed macaques are more closely related to each other than to the pig-tailed macaque due to geographical separation (59), the evolutionary relationship of the viral ORF59 homologs correlates with the evolutionary relationship of their macaque hosts (31). To examine in more detail the phylogenetic relationship of the MneRV2-encoded proteins to the other Old World primate rhadinoviruses, we aligned and performed maximum likelihood analysis examining several conserved homologs of the KSHV glycoproteins ORFK1, ORFK8.1, and ORF8 (gB) encoded within the genomes of MneRV2, RFHVMn (RV1 rhadinovirus from the pig-tailed macaque), and the two RRV variants, 26-95 and 17577, isolated from rhesus macaques at the New England and Oregon National Primate Research Centers, respectively. As the genome of MfaRV2 has not been completely characterized, we also examined the phylogenetic relationship of the glycoprotein homologs of MfuRV2 (host = Japanese macaque), which has been completely sequenced. Like the long-tailed macaque, the Japanese macaque is more closely related to the rhesus macaque than to the pig-tailed macaque. As seen in Fig. 11A, the ORFK1, K8.1, and ORF8 gB homologs of MneRV2, RRV, and MfuRV2 phylogenetically clustered in a pattern consistent with the evolutionary relationship of their respective hosts. The homologs from the two RRV strains clustered together. While the MfuRV2 homologs clustered with those of the two RRV strains, the MneRV2 homologs branched separately, showing a more distant relationship. In all cases, the homologs of the RV1 rhadinoviruses KSHV and RFHVMn clustered separately from the macaque RV2 rhadinovirus homologs. For ORF8 gB, the homologs of pig-tailed macaque rhadinovirus (PRV), an additional strain of MneRV2 identified in the New England primate center, and RFHVMm from a rhesus macaque were included in the analysis. These sequences clustered in a manner consistent with the evolutionary relationship of their hosts.
FIG 11.
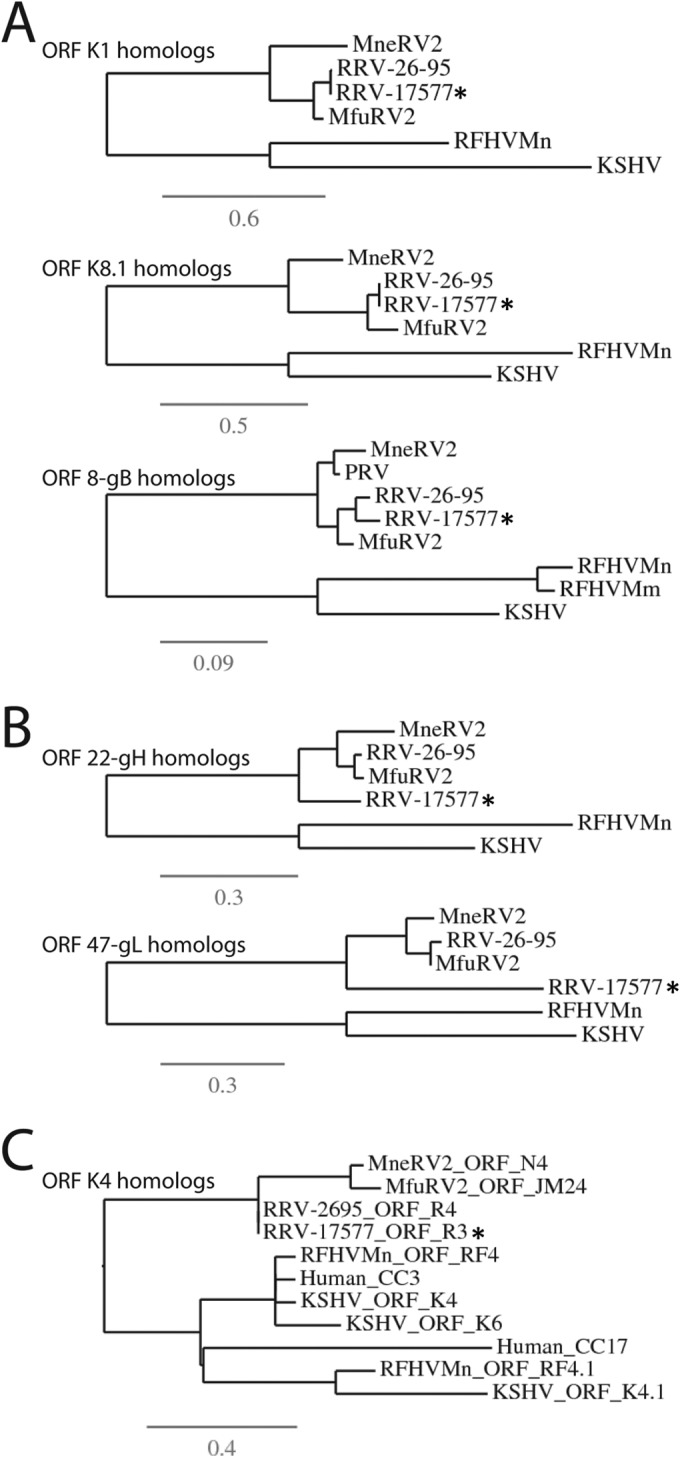
Phylogenetic analysis of select MneRV2 proteins. The encoded amino acids of selected MneRV2 ORFs were compared to the corresponding homologs in Old World primate rhadinoviruses of the RV1 lineage, including KSHV (human) and RFHVMn (pig-tailed macaque), and the RV2 lineage, including two RRV strains, 26-95 and 17577, isolated at the New England and Oregon National Primate Research Centers, respectively (rhesus macaque), and MfuRV2 (Japanese macaque). The protein sequences were aligned using MUSCLE, and phylogeny was determined by protein maximum likelihood. The variant phylogenetic clustering of RRV17577 proteins is indicated (*). (A) Homologs of glycoproteins ORFK1, ORFK8.1, and ORF8 (gB), including the gB homolog of RFHVMm (rhesus macaque; AAF78826) and PRV, an additional MneRV2 strain from a pig-tailed macaque at the New England National Primate Research Center (63). (B) Homologs of glycoproteins ORF22 (gH) and ORF47 (gL). (C) Homologs of ORFK4 and ORFK4.1 interleukin 8-like CC chemokines. The RRV26-95 ORFR4 and the RRV17577 ORFR3 are homologs of the same gene but were numbered differently, in their respective initial publications. The related human CC3 (NP_002974) and CC17 (NP_002978) chemokines were included for comparison purposes. The numbers of substitutions per site are indicated.
It was previously noted that phylogenetic clustering of the interacting glycoprotein pairs ORF22 (gH) and ORF47 (gL) was distinct between RRV isolates similar to RRV26-95 and isolates similar to RRV17577. Furthermore, the phylogenetic clustering of the independent glycoprotein ORF8 gB sequences was not associated with the gH/gL paired clustering (60). An alignment and protein maximum likelihood analysis of the ORF22 gH and ORF47 gL homologs of MneRV2 showed a similar phylogenetic relationship to the homologs of RRV26-95 and MfuRV2, as seen with ORFK1, ORFK8.1, and ORF8 gB above (Fig. 11B), in which the RRV26-95 and MfuRV2 sequences clustered together, separate from the MneRV2 sequences. In contrast, the ORF22 gH and ORF47 gL of RRV17577 both branched separately from the MneRV2/RRV26-95/MfuRV2 cluster, indicating an atypical evolutionary relationship compared to the other RRV17577 glycoprotein and core genes (Fig. 11B). The gH and gL genes from the RRV isolates clustering with RRV17577 lacked sequence variation, showing no evidence of unusual evolutionary pressure (60). This suggests that unlike MneRV2 and RRV26-95, the RRV17577 gH and gL variants were derived from a gene capture event of one or both of the interacting genes from a more distantly related herpesvirus. Since the ORF22 gH and ORF47 gL genes are separated by 30,000 bp of the genome, we examined the evolutionary relationship of the immediate flanking genes to determine the extent of a potential capture anomaly. The sequences of ORFs 21 and 23 flanking ORF22 and ORF48 flanking the right side of ORF47 were 99 to 100% identical between RRV26-95 and RRV17577 and showed the expected evolutionary relationship with MneRV2 typified by the genes in Fig. 11A. While the C-terminal 230 aa of the 255-aa ORF46 were 99% identical between RRV26-95 and RRV17577, the N-terminal 25 aa adjacent to the C terminus of ORF47 on the left side were only 50% identical, indicating that the genomic anomaly present in RRV17577 ORF47 gL continued upstream into the N terminus of ORF46. In this 25-aa region, the ORF46 homologs of MfuRV2 and RRV26-95 were 92% identical. Thus, the MneRV2 gene structure in these regions resembled RRV26-95 and MfuRV2 and not RRV17577. Note that a 255-aa corrected version of RRV26-95 ORF46 was used in this analysis (Table 1), as obvious sequence insertion errors were present in the RRV26-95 genomic sequence (AF201726) at positions 62577 and 62668, encoding an erroneous 230-aa ORF46 sequence indicated in the original manuscript (28).
Summary.
We have sequenced the complete genome of the pig-tailed macaque RV2 rhadinovirus, MneRV2, and have identified 87 protein coding genes and 17 candidate miRNAs. Phylogenetic analysis of common herpesvirus core genes demonstrated that MneRV2 was closely related to but distinct from the other known macaque RV2 rhadinoviruses, including the two RRV strains, RRV26-95 and RRV17577, from rhesus macaques; MfuRV2/JMRV from the Japanese macaque; and MfaRV2 from the long-tailed macaque, which all clustered in a separate phylogenetic grouping. This relationship mirrored the evolutionary relationship between the macaque host species. MneRV2 appears to have evolved in long-term synchrony with its pig-tailed macaque host. However, obvious genomic changes have occurred recently in the two RRV strains since the separation of pig-tailed and rhesus macaques and the divergence of the two RRV strains (Fig. 12). While the structure of the MneRV2 genome closely matched the RRV26-95 genome with a high degree of protein and RNA sequence conservation, R15, the rightmost gene of RRV26-95, was disrupted compared to MneRV2, with an insertion of a repetitive sequence and a duplication of the N terminus. The RRV17577 genome contained several variations compared to MneRV2 and RRV26-95, including a duplication within ORF4 and a potential recombination event from a divergent rhadinovirus involving the interacting glycoprotein partners ORF22 (gH) and ORF47 (gL). Like RRV26-95, RRV17577 lacked the intact homolog of ORFK7 that was identified in MneRV2 (N7). Thus, MneRV2 exhibits a more typical rhadinovirus genomic structure than either of the two known RRV strains, RRV26-95 and RRV17577. Although different subspecies of rhesus macaques are known, it is not clear whether the two RRV strains derive from different host subspecies. The comparison of MneRV2 with other rhadinoviruses has revealed important conservation of protein coding domains and transcription initiation, termination, and splicing signals, which have added to our knowledge of the genetics of the RV2 rhadinoviruses. Our analysis of the MneRV2 genome has detected previously unknown rhadinovirus gene conservation. The comparison of the MneRV2 genome sequence with the published sequences of RRV17577 and RRV26-95 revealed probable sequencing errors in RRV26-95 ORF46 (2 single-base-pair insertions) and ORF67 (1-bp insertion) and RRV17577 ORF67 (1-bp deletion) (Table 1), which changed the C-terminal ends of these ORFs from highly conserved to nonsense unconserved. While these ORFs were identified as the least conserved between RRV26-95 and RRV17577 in the original publication (28), the revised C-terminal sequences after elimination of the sequencing errors were 100% identical between the two viruses. Further comparisons with KSHV and other RV1 rhadinoviruses will provide important avenues for dissecting the biology and pathology of these closely related viruses in humans and other Old World primates. It is interesting that the evolutionary studies suggest that the human lineage was also host to an RV2 rhadinovirus related to MneRV2 and RRV, but the search for such a virus has been unsuccessful to this point.
FIG 12.
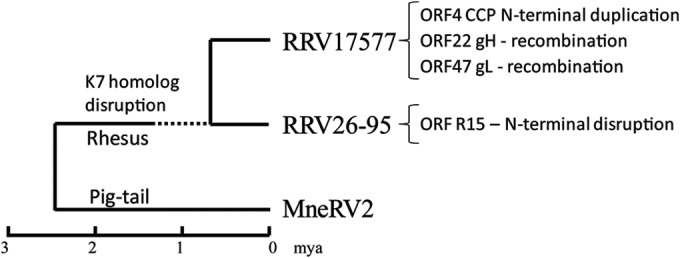
Evolution of macaque RV2 rhadinoviruses. The proposed evolutionary relationship of the pig-tailed macaque MneRV2 and rhesus macaque RRV17577 and RRV26-95 is shown in relationship to a tentative evolutionary time scale (mya, millions of years ago) based on the proposed divergence of rhesus and pig-tailed macaques and the divergence of different rhesus macaque subspecies (64). The genomic changes identified in the two RRV strains are indicated (see the text).
ACKNOWLEDGMENTS
This work was supported by the National Center for Research Resources (NCRR) and the Office of Research Infrastructure Programs (ORIP) of the National Institutes of Health (NIH) through grant RR023343 (T. M. Rose).
We acknowledge the help of David Blackbourne in establishing the collaboration with the Birmingham Genomics Group and Chris Upton for help with viral bioinformatics.
REFERENCES
- 1.Chang Y, Cesarman E, Pessin MS, Lee F, Culpepper J, Knowles DM, Moore PS. 1994. Identification of herpesvirus-like DNA sequences in AIDS-associated Kaposi's sarcoma. Science 266:1865–1869. doi: 10.1126/science.7997879. [DOI] [PubMed] [Google Scholar]
- 2.Ganem D. 2010. KSHV and the pathogenesis of Kaposi sarcoma: listening to human biology and medicine. J Clin Invest 120:939–949. doi: 10.1172/JCI40567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Du MQ, Bacon CM, Isaacson PG. 2007. Kaposi sarcoma-associated herpesvirus/human herpesvirus 8 and lymphoproliferative disorders. J Clin Pathol 60:1350–1357. doi: 10.1136/jcp.2007.047969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Russo JJ, Bohenzky RA, Chien MC, Chen J, Yan M, Maddalena D, Parry JP, Peruzzi D, Edelman IS, Chang Y, Moore PS. 1996. Nucleotide sequence of the Kaposi sarcoma-associated herpesvirus (HHV8). Proc Natl Acad Sci U S A 93:14862–14867. doi: 10.1073/pnas.93.25.14862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nicholas J, Zong JC, Alcendor DJ, Ciufo DM, Poole LJ, Sarisky RT, Chiou CJ, Zhang X, Wan X, Guo HG, Reitz MS, Hayward GS. 1998. Novel organizational features, captured cellular genes, and strain variability within the genome of KSHV/HHV8. J Natl Cancer Inst Monogr 1998(23):79–88. [DOI] [PubMed] [Google Scholar]
- 6.Moore PS, Chang Y. 2001. Molecular virology of Kaposi's sarcoma-associated herpesvirus. Philos Trans R Soc Lond B Biol Sci 356:499–516. doi: 10.1098/rstb.2000.0777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Greensill J, Sheldon JA, Renwick NM, Beer BE, Norley S, Goudsmit J, Schulz TF. 2000. Two distinct gamma-2 herpesviruses in African green monkeys: a second gamma-2 herpesvirus lineage among old world primates? J Virol 74:1572–1577. doi: 10.1128/JVI.74.3.1572-1577.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Schultz ER, Rankin GW Jr, Blanc MP, Raden BW, Tsai CC, Rose TM. 2000. Characterization of two divergent lineages of macaque rhadinoviruses related to Kaposi's sarcoma-associated herpesvirus. J Virol 74:4919–4928. doi: 10.1128/JVI.74.10.4919-4928.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rose TM, Strand KB, Schultz ER, Schaefer G, Rankin GW Jr, Thouless ME, Tsai CC, Bosch ML. 1997. Identification of two homologs of the Kaposi's sarcoma-associated herpesvirus (human herpesvirus 8) in retroperitoneal fibromatosis of different macaque species. J Virol 71:4138–4144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lacoste V, Mauclere P, Dubreuil G, Lewis J, Georges-Courbot MC, Gessain A. 2000. KSHV-like herpesviruses in chimps and gorillas. Nature 407:151–152. doi: 10.1038/35025145. [DOI] [PubMed] [Google Scholar]
- 11.Lacoste V, Mauclere P, Dubreuil G, Lewis J, Georges-Courbot MC, Rigoulet J, Petit T, Gessain A. 2000. Simian homologues of human gamma-2 and betaherpesviruses in mandrill and drill monkeys. J Virol 74:11993–11999. doi: 10.1128/JVI.74.24.11993-11999.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lacoste V, Kadyrova E, Chistiakova I, Gurtsevitch V, Judde JG, Gessain A. 2000. Molecular characterization of Kaposi's sarcoma-associated herpesvirus/human herpesvirus-8 strains from Russia. J Gen Virol 81:1217–1222 http://vir.sgmjournals.org/content/81/5/1217.long. [DOI] [PubMed] [Google Scholar]
- 13.Lacoste V, Mauclere P, Dubreuil G, Lewis J, Georges-Courbot MC, Gessain A. 2001. A novel gamma 2-herpesvirus of the Rhadinovirus 2 lineage in chimpanzees. Genome Res 11:1511–1519. doi: 10.1101/gr.158601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Whitby D, Stossel A, Gamache C, Papin J, Bosch M, Smith A, Kedes DH, White G, Kennedy R, Dittmer DP. 2003. Novel Kaposi's sarcoma-associated herpesvirus homolog in baboons. J Virol 77:8159–8165. doi: 10.1128/JVI.77.14.8159-8165.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mansfield KG, Westmoreland SV, DeBakker CD, Czajak S, Lackner AA, Desrosiers RC. 1999. Experimental infection of rhesus and pig-tailed macaques with macaque rhadinoviruses. J Virol 73:10320–10328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bruce AG, Bakke AM, Thouless ME, Rose TM. 2005. Development of a real-time QPCR assay for the detection of RV2 lineage-specific rhadinoviruses in macaques and baboons. Virol J 2:2. doi: 10.1186/1743-422X-2-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Axthelm MK, Bourdette DN, Marracci GH, Su W, Mullaney ET, Manoharan M, Kohama SG, Pollaro J, Witkowski E, Wang P, Rooney WD, Sherman LS, Wong SW. 2011. Japanese macaque encephalomyelitis: a spontaneous multiple sclerosis-like disease in a nonhuman primate. Ann Neurol 70:362–373. doi: 10.1002/ana.22449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Desrosiers RC, Sasseville VG, Czajak SC, Zhang X, Mansfield KG, Kaur A, Johnson RP, Lackner AA, Jung JU. 1997. A herpesvirus of rhesus monkeys related to the human Kaposi's sarcoma-associated herpesvirus. J Virol 71:9764–9769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.White JA, Todd PA, Yee JL, Kalman-Bowlus A, Rodgers KS, Yang X, Wong SW, Barry P, Lerche NW. 2009. Prevalence of viremia and oral shedding of rhesus rhadinovirus and retroperitoneal fibromatosis herpesvirus in large age-structured breeding groups of rhesus macaques (Macaca mulatta). Comp Med 59:383–390 http://www.ncbi.nlm.nih.gov/pmc/articles/PMC2779215/. [PMC free article] [PubMed] [Google Scholar]
- 20.Bielefeldt-Ohmann H, Barouch DH, Bakke AM, Bruce AG, Durning M, Grant R, Letvin NL, Ryan JT, Schmidt A, Thouless ME, Rose TM. 2005. Intestinal stromal tumors in a simian immunodeficiency virus-infected, simian retrovirus-2 negative rhesus macaque (Macaca mulatta). Vet Pathol 42:391–396. doi: 10.1354/vp.42-3-391. [DOI] [PubMed] [Google Scholar]
- 21.Bruce AG, Bakke AM, Bielefeldt-Ohmann H, Ryan JT, Thouless ME, Tsai CC, Rose TM. 2006. High levels of retroperitoneal fibromatosis (RF)-associated herpesvirus in RF lesions in macaques are associated with ORF73 LANA expression in spindleoid tumour cells. J Gen Virol 87:3529–3538. doi: 10.1099/vir.0.82339-0. [DOI] [PubMed] [Google Scholar]
- 22.Burnside KL, Ryan JT, Bielefeldt-Ohmann H, Gregory Bruce A, Thouless ME, Tsai CC, Rose TM. 2006. RFHVMn ORF73 is structurally related to the KSHV ORF73 latency-associated nuclear antigen (LANA) and is expressed in retroperitoneal fibromatosis (RF) tumor cells. Virology 354:103–115. doi: 10.1016/j.virol.2006.06.022. [DOI] [PubMed] [Google Scholar]
- 23.Bruce AG, Bielefeldt-Ohmann H, Barcy S, Bakke AM, Lewis P, Tsai CC, Murnane RD, Rose TM. 2012. Macaque homologs of EBV and KSHV show uniquely different associations with simian AIDS-related lymphomas. PLoS Pathog 8:e1002962. doi: 10.1371/journal.ppat.1002962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wong SW, Bergquam EP, Swanson RM, Lee FW, Shiigi SM, Avery NA, Fanton JW, Axthelm MK. 1999. Induction of B cell hyperplasia in simian immunodeficiency virus-infected rhesus macaques with the simian homologue of Kaposi's sarcoma-associated herpesvirus. J Exp Med 190:827–840. doi: 10.1084/jem.190.6.827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Orzechowska BU, Powers MF, Sprague J, Li H, Yen B, Searles RP, Axthelm MK, Wong SW. 2008. Rhesus macaque rhadinovirus-associated non-Hodgkin lymphoma: animal model for KSHV-associated malignancies. Blood 112:4227–4234. doi: 10.1182/blood-2008-04-151498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bruce AG, Ryan JT, Thomas MJ, Peng X, Grundhoff A, Tsai CC, Rose TM. 2013. Next-generation sequence analysis of the genome of RFHVMn, the macaque homolog of Kaposi's sarcoma (KS)-associated herpesvirus, from a KS-like tumor of a pig-tailed macaque. J Virol 87:13676–13693. doi: 10.1128/JVI.02331-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Searles RP, Bergquam EP, Axthelm MK, Wong SW. 1999. Sequence and genomic analysis of a rhesus macaque rhadinovirus with similarity to Kaposi's sarcoma-associated herpesvirus/human herpesvirus 8. J Virol 73:3040–3053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Alexander L, Denekamp L, Knapp A, Auerbach MR, Damania B, Desrosiers RC. 2000. The primary sequence of rhesus monkey rhadinovirus isolate 26-95: sequence similarities to Kaposi's sarcoma-associated herpesvirus and rhesus monkey rhadinovirus isolate 17577. J Virol 74:3388–3398. doi: 10.1128/JVI.74.7.3388-3398.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Estep RD, Hansen SG, Rogers KS, Axthelm MK, Wong SW. 2013. Genomic characterization of Japanese macaque rhadinovirus, a novel herpesvirus isolated from a nonhuman primate with a spontaneous inflammatory demyelinating disease. J Virol 87:512–523. doi: 10.1128/JVI.02194-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.McGeoch DJ. 2001. Molecular evolution of the gamma-Herpesvirinae. Philos Trans R Soc Lond B Biol Sci 356:421–435. doi: 10.1098/rstb.2000.0775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bruce AG, Bakke AM, Gravett CA, DeMaster LK, Bielefeldt-Ohmann H, Burnside KL, Rose TM. 2009. The ORF59 DNA polymerase processivity factor homologs of Old World primate RV2 rhadinoviruses are highly conserved nuclear antigens expressed in differentiated epithelium in infected macaques. Virol J 6:205. doi: 10.1186/1743-422X-6-205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Margulies M, Egholm M, Altman WE, Attiya S, Bader JS, Bemben LA, Berka J, Braverman MS, Chen YJ, Chen Z, Dewell SB, Du L, Fierro JM, Gomes XV, Godwin BC, He W, Helgesen S, Ho CH, Irzyk GP, Jando SC, Alenquer ML, Jarvie TP, Jirage KB, Kim JB, Knight JR, Lanza JR, Leamon JH, Lefkowitz SM, Lei M, Li J, Lohman KL, Lu H, Makhijani VB, McDade KE, McKenna MP, Myers EW, Nickerson E, Nobile JR, Plant R, Puc BP, Ronan MT, Roth GT, Sarkis GJ, Simons JF, Simpson JW, Srinivasan M, Tartaro KR, Tomasz A, Vogt KA, Volkmer GA, Wang SH, Wang Y, Weiner MP, Yu P, Begley RF, Rothberg JM. 2005. Genome sequencing in microfabricated high-density picolitre reactors. Nature 437:376–380. doi: 10.1038/nature03959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tcherepanov V, Ehlers A, Upton C. 2006. Genome Annotation Transfer Utility (GATU): rapid annotation of viral genomes using a closely related reference genome. BMC Genomics 7:150. doi: 10.1186/1471-2164-7-150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Brodie R, Roper RL, Upton C. 2004. JDotter: a Java interface to multiple dotplots generated by dotter. Bioinformatics 20:279–281. doi: 10.1093/bioinformatics/btg406. [DOI] [PubMed] [Google Scholar]
- 35.Edgar RC. 2004. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res 32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Dereeper A, Guignon V, Blanc G, Audic S, Buffet S, Chevenet F, Dufayard JF, Guindon S, Lefort V, Lescot M, Claverie JM, Gascuel O. 2008. Phylogeny.fr: robust phylogenetic analysis for the non-specialist. Nucleic Acids Res 36:W465–W469. doi: 10.1093/nar/gkn180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Grundhoff A, Sullivan CS, Ganem D. 2006. A combined computational and microarray-based approach identifies novel microRNAs encoded by human gamma-herpesviruses. RNA 12:733–750. doi: 10.1261/rna.2326106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sullivan CS, Grundhoff AT, Tevethia S, Pipas JM, Ganem D. 2005. SV40-encoded microRNAs regulate viral gene expression and reduce susceptibility to cytotoxic T cells. Nature 435:682–686. doi: 10.1038/nature03576. [DOI] [PubMed] [Google Scholar]
- 39.Strand K, Harper E, Thormahlen S, Thouless ME, Tsai C, Rose T, Bosch ML. 2000. Two distinct lineages of macaque gamma herpesviruses related to the Kaposi's sarcoma associated herpesvirus. J Clin Virol 16:253–269. doi: 10.1016/S1386-6532(99)00080-3. [DOI] [PubMed] [Google Scholar]
- 40.Wang L, Pietrek M, Brinkmann MM, Havemeier A, Fischer I, Hillenbrand B, Dittrich-Breiholz O, Kracht M, Chanas S, Blackbourn DJ, Schulz TF. 2009. Identification and functional characterization of a spliced rhesus rhadinovirus gene with homology to the K15 gene of Kaposi's sarcoma-associated herpesvirus. J Gen Virol 90:1190–1201. doi: 10.1099/vir.0.007971-0. [DOI] [PubMed] [Google Scholar]
- 41.DeWire SM, McVoy MA, Damania B. 2002. Kinetics of expression of rhesus monkey rhadinovirus (RRV) and identification and characterization of a polycistronic transcript encoding the RRV Orf50/Rta, RRV R8, and R81 genes. J Virol 76:9819–9831. doi: 10.1128/JVI.76.19.9819-9831.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Majerciak V, Ni T, Yang W, Meng B, Zhu J, Zheng ZM. 2013. A viral genome landscape of RNA polyadenylation from KSHV latent to lytic infection. PLoS Pathog 9:e1003749. doi: 10.1371/journal.ppat.1003749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lin SF, Robinson DR, Oh J, Jung JU, Luciw PA, Kung HJ. 2002. Identification of the bZIP and Rta homologues in the genome of rhesus monkey rhadinovirus. Virology 298:181–188. doi: 10.1006/viro.2002.1490. [DOI] [PubMed] [Google Scholar]
- 44.Spiller OB, Robinson M, O'Donnell E, Milligan S, Morgan BP, Davison AJ, Blackbourn DJ. 2003. Complement regulation by Kaposi's sarcoma-associated herpesvirus ORF4 protein. J Virol 77:592–599. doi: 10.1128/JVI.77.1.592-599.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mark L, Spiller OB, Okroj M, Chanas S, Aitken JA, Wong SW, Damania B, Blom AM, Blackbourn DJ. 2007. Molecular characterization of the rhesus rhadinovirus (RRV) ORF4 gene and the RRV complement control protein it encodes. J Virol 81:4166–4176. doi: 10.1128/JVI.02069-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zhu FX, Cusano T, Yuan Y. 1999. Identification of the immediate-early transcripts of Kaposi's sarcoma-associated herpesvirus. J Virol 73:5556–5567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tang S, Zheng ZM. 2002. Kaposi's sarcoma-associated herpesvirus K8 exon 3 contains three 5′-splice sites and harbors a K8.1 transcription start site. J Biol Chem 277:14547–14556. doi: 10.1074/jbc.M111308200. [DOI] [PubMed] [Google Scholar]
- 48.Saveliev A, Zhu F, Yuan Y. 2002. Transcription mapping and expression patterns of genes in the major immediate-early region of Kaposi's sarcoma-associated herpesvirus. Virology 299:301–314. doi: 10.1006/viro.2002.1561. [DOI] [PubMed] [Google Scholar]
- 49.Tang S, Yamanegi K, Zheng ZM. 2004. Requirement of a 12-base-pair TATT-containing sequence and viral lytic DNA replication in activation of the Kaposi's sarcoma-associated herpesvirus K8.1 late promoter. J Virol 78:2609–2614. doi: 10.1128/JVI.78.5.2609-2614.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Chandran B, Bloomer C, Chan SR, Zhu L, Goldstein E, Horvat R. 1998. Human herpesvirus-8 ORF K8.1 gene encodes immunogenic glycoproteins generated by spliced transcripts. Virology 249:140–149. doi: 10.1006/viro.1998.9316. [DOI] [PubMed] [Google Scholar]
- 51.Raab MS, Albrecht JC, Birkmann A, Yaguboglu S, Lang D, Fleckenstein B, Neipel F. 1998. The immunogenic glycoprotein gp35-37 of human herpesvirus 8 is encoded by open reading frame K8.1. J Virol 72:6725–6731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.AuCoin DP, Pari GS. 2002. The human herpesvirus-8 (Kaposi's sarcoma-associated herpesvirus) ORF 40/41 region encodes two distinct transcripts. J Gen Virol 83:189–193 http://vir.sgmjournals.org/content/83/1/189.long. [DOI] [PubMed] [Google Scholar]
- 53.Wu FY, Wang SE, Tang QQ, Fujimuro M, Chiou CJ, Zheng Q, Chen H, Hayward SD, Lane MD, Hayward GS. 2003. Cell cycle arrest by Kaposi's sarcoma-associated herpesvirus replication-associated protein is mediated at both the transcriptional and posttranslational levels by binding to CCAAT/enhancer-binding protein alpha and p21(CIP-1). J Virol 77:8893–8914. doi: 10.1128/JVI.77.16.8893-8914.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wang Y, Li H, Chan MY, Zhu FX, Lukac DM, Yuan Y. 2004. Kaposi's sarcoma-associated herpesvirus ori-Lyt-dependent DNA replication: cis-acting requirements for replication and ori-Lyt-associated RNA transcription. J Virol 78:8615–8629. doi: 10.1128/JVI.78.16.8615-8629.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Pari GS, AuCoin D, Colletti K, Cei SA, Kirchoff V, Wong SW. 2001. Identification of the rhesus macaque rhadinovirus lytic origin of DNA replication. J Virol 75:11401–11407. doi: 10.1128/JVI.75.23.11401-11407.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wang HW, Sharp TV, Koumi A, Koentges G, Boshoff C. 2002. Characterization of an anti-apoptotic glycoprotein encoded by Kaposi's sarcoma-associated herpesvirus which resembles a spliced variant of human survivin. EMBO J 21:2602–2615. doi: 10.1093/emboj/21.11.2602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Umbach JL, Strelow LI, Wong SW, Cullen BR. 2010. Analysis of rhesus rhadinovirus microRNAs expressed in virus-induced tumors from infected rhesus macaques. Virology 405:592–599. doi: 10.1016/j.virol.2010.06.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Lei Z, Xu G, Wang L, Yang H, Liu X, Zhao J, Zhang HT. 2014. MiR-142-3p represses TGF-beta-induced growth inhibition through repression of TGFbetaR1 in non-small cell lung cancer. FASEB J 28:2696–2704. doi: 10.1096/fj.13-247288. [DOI] [PubMed] [Google Scholar]
- 59.Morales JC, Melnick DJ. 1998. Phylogenetic relationships of the macaques (Cercopithecidae: Macaca), as revealed by high resolution restriction site mapping of mitochondrial ribosomal genes. J Hum Evol 34:1–23. doi: 10.1006/jhev.1997.0171. [DOI] [PubMed] [Google Scholar]
- 60.Shin YC, Jones LR, Manrique J, Lauer W, Carville A, Mansfield KG, Desrosiers RC. 2010. Glycoprotein gene sequence variation in rhesus monkey rhadinovirus. Virology 400:175–186. doi: 10.1016/j.virol.2010.01.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Lee H, Guo J, Li M, Choi JK, DeMaria M, Rosenzweig M, Jung JU. 1998. Identification of an immunoreceptor tyrosine-based activation motif of K1 transforming protein of Kaposi's sarcoma-associated herpesvirus. Mol Cell Biol 18:5219–5228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Zong JC, Ciufo DM, Alcendor DJ, Wan X, Nicholas J, Browning PJ, Rady PL, Tyring SK, Orenstein JM, Rabkin CS, Su IJ, Powell KF, Croxson M, Foreman KE, Nickoloff BJ, Alkan S, Hayward GS. 1999. High-level variability in the ORF-K1 membrane protein gene at the left end of the Kaposi's sarcoma-associated herpesvirus genome defines four major virus subtypes and multiple variants or clades in different human populations. J Virol 73:4156–4170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Auerbach MR, Czajak SC, Johnson WE, Desrosiers RC, Alexander L. 2000. Species specificity of macaque rhadinovirus glycoprotein B sequences. J Virol 74:584–590. doi: 10.1128/JVI.74.1.584-590.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Hayasaka K, Fujii K, Horai S. 1996. Molecular phylogeny of macaques: implications of nucleotide sequences from an 896-base pair region of mitochondrial DNA. Mol Biol Evol 13:1044–1053. doi: 10.1093/oxfordjournals.molbev.a025655. [DOI] [PubMed] [Google Scholar]