ABSTRACT
Flavivirus RNA synthesis is mediated by a multiprotein complex associated with the endoplasmic reticulum membrane, named the replication complex (RC). Within the flavivirus RC, NS4B, an integral membrane protein with a role in virulence and regulation of the innate immune response, binds to the NS3 protease-helicase. NS4B modulates the RNA helicase activity of NS3, but the molecular details of their interaction remain elusive. Here, we used dengue virus (DENV) to map the determinants for the NS3-NS4B interaction. Coimmunoprecipitation and an in situ proximity ligation assay confirmed that NS3 colocalizes with NS4B in both DENV-infected cells and cells coexpressing both proteins. Surface plasmon resonance demonstrated that subdomains 2 and 3 of the NS3 helicase region and the cytoplasmic loop of NS4B are required for binding. Using nuclear magnetic resonance (NMR), we found that the isolated cytoplasmic loop of NS4B is flexible, with a tendency to form a three-turn α-helix and two short β-strands. Upon binding to the NS3 helicase, 12 amino acids within the cytoplasmic loop of NS4B exhibited line broadening, suggesting a participation in the interaction. Sequence alignment showed that 4 of these 12 residues are strictly conserved across different flaviviruses. Mutagenesis analysis showed that three (Q134, G140, and N144) of the four evolutionarily conserved NS4B residues are essential for DENV replication. The mapping of the NS3/NS4B-interacting regions described here can assist the design of inhibitors that disrupt their interface for antiviral therapy.
IMPORTANCE NS3 and NS4B are essential components of the flavivirus RC. Using DENV as a model, we mapped the interaction between the viral NS3 and NS4B proteins. The subdomains 2 and 3 of NS3 helicase as well as the cytoplasmic loop of NS4B are critical for the interaction. Functional analysis delineated residues within the NS4B cytoplasmic loop that are crucial for DENV replication. Our findings reveal molecular details of how flavivirus NS3 protein cooperates with NS4B within the RC. In addition, this study has established the rationale and assays to search for inhibitors disrupting the NS3-NS4B interaction for antiviral drug discovery.
INTRODUCTION
Dengue is a major and emerging public health problem: 50 to 100 million dengue cases occur every year, and over 40% of the world population is at risk (1). According to a 2014 update by WHO, “an estimated 500,000 people with severe dengue are hospitalized each year including a large proportion of children,” and of those, about 2.5% die (2). The disease is caused by infection with one of the four serotypes of dengue virus (DENV), transmitted by Aedes mosquitoes. There is currently no clinically approved antiviral therapeutics. The recent phase IIb clinical trials in Thailand revealed that the CYD tetravalent vaccine failed to protect against DENV serotype 2 (DENV2) although the vaccine showed efficacy against the other three serotypes of DENV (3). Before effective vaccine and antiviral treatments become available, dengue patient care has to rely on supportive treatment, and vector control still remains an important means of reducing outbreaks.
DENV belongs to the Flavivirus genus within the family Flaviviridae. In addition to DENV, several other flaviviruses are important human pathogens, such as yellow fever virus (YFV), West Nile virus (WNV), Japanese encephalitis virus (JEV), and tick-borne encephalitis virus (TBEV). DENV is a small enveloped virus (4) containing a single plus-strand RNA genome that comprises approximately 10,700 nucleotides. The DENV genome encodes a total of 10 proteins, consisting of three structural proteins (capsid [C], premembrane [prM], and envelope [E]) and seven nonstructural proteins (NS1, NS2A, NS2B, NS3, NS4A, NS4B, and NS5). The structural proteins form the viral particle, while the nonstructural proteins are responsible for RNA replication in the host cell and evasion from the innate immune responses (5). In particular, glycoprotein NS1 plays a role in viral RNA synthesis at an early step of the viral infection cycle (6); in conjunction with the cofactor NS2B, NS3 acts as a viral serine protease (7) and also as an RNA triphosphatase (8) and RNA helicase (9). NS5 functions as a methyltransferase (10–12) and also as an RNA-dependent RNA polymerase (13).
Flavivirus RNA replication occurs in close association with host cytoplasmic membranes, where both viral and cellular factors interact and cooperate within an organelle-like replication factory called the replication complex (RC) (for reviews, see references 14 and 15). Several nonstructural proteins of the RC constitute validated drug targets because of their essential functions during viral replication (for a recent review, see reference 16). However, a major impediment in developing drugs targeting the DENV RC is that its detailed morphology and composition, the interplay between its molecular constituents, and the precise molecular mechanisms for viral RNA replication are still elusive (15). Over the last decade, individual protein components of the DENV RC, such as NS1, NS2B-NS3, and NS5, were characterized at both the structural and functional levels (16); moreover, topology models for integral membrane proteins NS2A, NS4A, and NS4B were proposed (17–19). However, because the integral membrane proteins NS2A, NS4A, and NS4B lack enzymatic activities, it has been challenging to understand their precise roles and also to develop in vitro functional assays to screen inhibitors directed against these potentially attractive viral targets (16). Thus, defining the molecular interactions between the RC components has both fundamental interest and potential applicability for drug discovery.
Flavivirus NS4B (28 kDa) is a valid target for antiviral development. Small-molecule inhibitors targeting YFV and DENV NS4B have been identified by both academic and industry groups (20–22). A better understanding of the structure, function, and interactions of NS4B is required to support further antiviral development. A model for the DENV2 NS4B structure was proposed with two N-terminal amphipathic helices, three transmembrane helices, and a cytoplasmic region spanning residues 129 to 165 of the polypeptide chain (18). The flavivirus NS4B protein oligomerizes when expressed in vitro and in vivo (23). In infected cells, NS4B colocalizes with NS3 and double-stranded RNA (dsRNA) in endoplasmic reticulum (ER) vesicles predicted to constitute sites of RNA replication (15). Moreover, NS4B from DENV specifically binds to NS3, enhancing its helicase activity by reducing the affinity of NS3 for single-stranded RNA (24). Crystal structures of the ATPase/helicase region of NS3 from DENV4 have been reported bound to ATP and to single-stranded RNA (25, 26). Here, using biochemical, biophysical, and genetic approaches, we characterized at the molecular level the NS3-NS4B interaction in DENV. We have located the major determinants for the interaction within NS3 helicase subdomains 2 and 3 and the cytoplasmic loop of NS4B. Nuclear magnetic resonance (NMR) studies revealed 12 amino acids within the NS4B cytoplasmic loop that may contribute to the NS3-NS4B interaction. Mutagenesis analysis identified three evolutionarily conserved NS4B amino acids within the cytoplasmic loop that are important determinants for DENV replication. Our results provide a potential approach to design antiviral compounds by targeting the flavivirus NS3/NS4B interface.
MATERIALS AND METHODS
Cloning of the NS4B protein.
The cDNA encoding the NS4B protein from DENV2 strain TSV01 (GenBank accession number AY037116) with an N-terminal decahistidine tag was amplified by fusion PCR, digested with NcoI and NotI, and inserted into the pET28a vector to generate a pET28a-NS4B plasmid. The construct was confirmed by cDNA sequencing and transformed into BL21(DE3) Escherichia coli cells (Stratagene).
Expression and purification of NS4B.
Cells were grown in 2× yeast extract-tryptone (YT; BD Difco) medium supplemented with 50 μg ml−1 kanamycin (Sigma) to an optical density at 600 nm (OD600) of 0.8 to 1.0. Expression of the recombinant proteins was induced by the addition of 0.2 mM isopropyl β-d-1-thiogalactopyranoside (IPTG) and incubation at 16°C for 16 h. Cells were pelleted by centrifugation at 7,000 × g for 7 min at 15°C, resuspended in lysis buffer (20 mM Tris-HCl, 300 mM NaCl, 10% glycerol, pH 8.0), and disrupted by sonication using a Digital Sonifier 450 (Branson) at 40% amplitude for 10 min. The cell lysate was clarified by centrifugation at 10,000 × g for 10 min at 4°C to remove cell debris. The supernatants were centrifuged at 35,000 rpm for 1 h at 4°C using a Beckman type 70 Ti rotor to pellet membranes and their associated proteins. The membrane pellets were then resuspended thoroughly in lysis buffer supplemented with 1% n-dodecyl-β-d-maltopyranoside (DDM; Anatrace) by stirring at 4°C for 2 h. The solubilized lysates were clarified by ultracentrifugation at 35,000 rpm for 1 h at 4°C. Supernatants were purified by immobilized metal affinity chromatography (IMAC) using a HisTrap Fast Flow column (GE Healthcare) equilibrated with buffer A [20 mM Tris-HCl, 300 mM NaCl, 20 mM imidazole, 2 mM tris(2-carboxyethyl)phosphine (TCEP), 0.05% DDM, pH 8.0]. After the sample was loaded, the column was washed with 10 column volumes of buffer A. The bound protein was eluted using a linear gradient of 5 mM to 300 mM imidazole. The fractions containing NS4B proteins were pooled, concentrated, and injected onto a HiLoad Superdex 200 16/60 column (GE Healthcare) preequilibrated in 20 mM Tris-HCl, 300 mM NaCl, 10% glycerol, 2 mM TCEP, and 0.05% DDM, pH 8.0. Fractions containing NS4B proteins were pooled and concentrated to approximately 5 to 10 mg ml−1 for storage at −80°C. The histidine tag could not be cleaved even following extensive incubation with tobacco etch virus (TEV) protease. The circular dichroism (CD) spectrum for NS4B was collected on a Jasco J810 spectro-polarimeter and was smoothed using the routines provided with the instrument.
SPR.
Surface plasmon resonance (SPR) measurements were performed using a Biacore 3000 instrument (GE Healthcare) in 20 mM Tris-HCl, pH 8.0, 150 mM NaCl, 10% glycerol, and 5 mM dithiothreitol (DTT) at 25°C. The purified recombinant NS4B protein was directly immobilized onto CM5 sensor chips (GE Healthcare) using standard amine-coupling chemistry at a flow rate of 10 ml/min. Carboxyl derivatives on a CM5 dextran surface were activated with a 10-min injection of a 1:1 mixture of 0.2 M N-ethyl-N′-(3-(diethylamino)propyl)-carbodiimide (EDC) and 50 mM N-hydroxysuccinimide (NHS). Recombinant NS4B was diluted into 10 mM sodium acetate, pH 4.5, and injected across the activated dextran surface until an immobilization level of about 11,000 resonance units (RU) was reached. Unreacted activated carboxyl derivatives were blocked with 0.5 M ethanolamine-HCl at pH 8.5 for 10 min. Binding partners consisting of either the full-length NS3 protein (NS3FL), its protease domain, its helicase domain, or helicase subdomains 1 or 2 or 2 plus 3 (see Fig. 2 and 3), were diluted into running buffer at concentrations ranging from 3.125 nM to 1 μM as 2-fold serial dilutions and injected across the immobilized surface in replicates. Raw sensorgrams were double referenced, and responses were corrected using a reference flow cell and a blank buffer injection. For each corrected sensorgram, the zero for the x and y axes was taken at the point of sample injection. Responses for the varied concentrations were overlaid and fit to a 1:1 simple binding model to obtain binding affinities.
FIG 2.
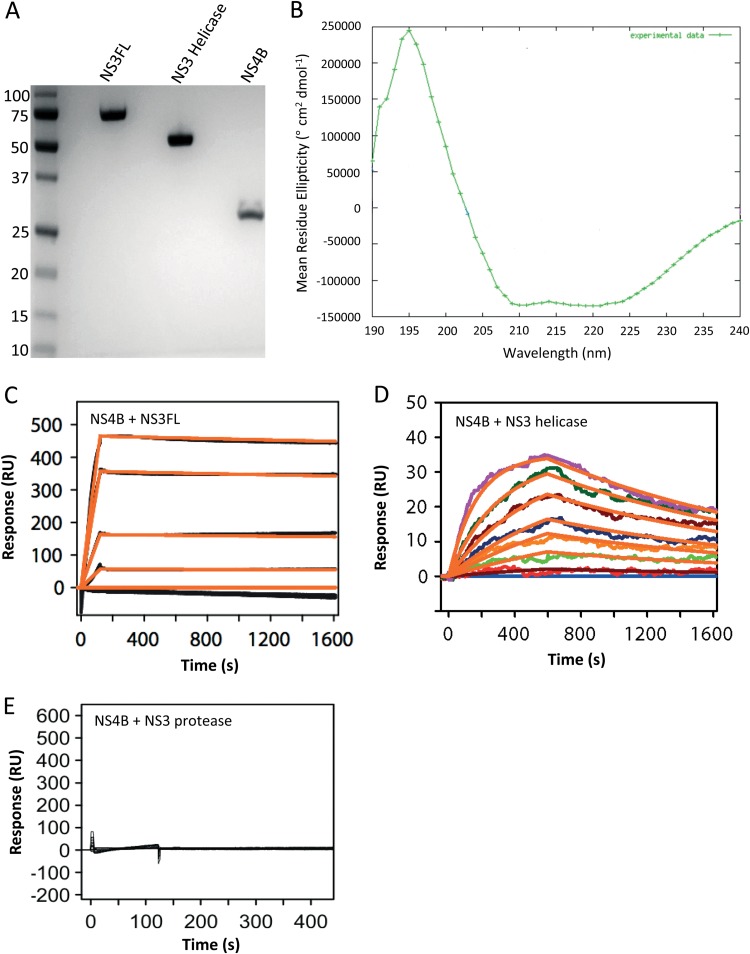
Kinetic analysis of the interaction between NS4B and NS3 from DENV2. (A) SDS-PAGE of the purified NS3 full-length protein (NS3 FL), NS3 helicase domain, and NS4B from DENV2 used for the SPR experiment. (B) The CD spectrum of the recombinant NS4B protein from DENV2 displays two minima at 210 nm and 220 nm, indicative of the presence of predominant helical structures (80% α-helices, 4% β-strands, 6% turns, and 10% disordered). (C) Sensorgrams showing the interaction between 2-fold serially diluted NS3 protease-helicase (NS3FL) ranging from 53 nM to 3.4 μM (black curves) and immobilized NS4B. (D) Sensorgrams showing the interaction between the 2-fold serially diluted NS3 helicase domain ranging from 53 nM to 3.4 μM (colored curves) and immobilized NS4B. Only one replicate is shown for clarity. A simple 1:1 binding model fit to all replicates is shown (orange curve). The NS4B-NS3 helicase interaction is estimated to have the following kinetic constants: dissociation rate (ka) of (1.58 ± 0.03) × 103 M−1s−1 and kd of (3.51 ± 0.02) × 10−4 s−1, giving a KD of 222 ± 4 nM. (E) Sensorgram demonstrating the absence of interaction between NS3 protease domain (the construct includes 18 residues of the NS2B cofactor) (8, 28) at 3.4 μM (black curve) and immobilized NS4B. Only one replicate is shown for clarity.
FIG 3.
Mapping of the NS3 binding domain using an SPR biosensor. (A) Schematic representation of the 3D structure of the NS3 helicase domain; its three subdomains are shown in red (subdomain 1), green (subdomain 2), and 3 (yellow). The single-stranded RNA binding tunnel and ATP binding sites are indicated. (B) Schematic representation of the helicase protein fragments used for the SPR experiment. The color code is the same as described in panel A. Subdomain boundaries were defined according to Xu et al. (25). (C) SDS-PAGE showing the purified helicase domain and the three helicase fragments from DENV4 used for the SPR experiment. (D) The CD spectra of the recombinant NS3 helicase protein subdomains 1, 2, and 2 plus 3 from DENV4 are compared with those of the DENV4 NS3 helicase and are indicative of folded proteins. (E) Fragments of the DENV4 NS3 helicase (7.8 nM to 1 μM) were injected across the immobilized NS4B from DENV2 in replicate; however, only one set of replicates is shown for figure clarity. Overlaid is a simple 1:1 binding model shown in orange. The affinity constants obtained are as follows: for subdomains 1 plus 2 plus 3 (entire helicase domain of NS3 from DENV4), KD of ∼ 530 nM; subdomain 1, negligible response; subdomain 2, KD of ∼4.8 μM; subdomains 2 plus 3, KD of ∼340 nM.
For the NS4B cytoplasmic loop peptide-NS4B interactions (see Fig. 4), the responses were fit using the steady-state affinity model in BIAevaluation, version 4.1, software. Experimental values of the response at equilibrium (Req) for the corrected sensorgrams were compared to the simulated values for Kd (dissociation constant) of 1 μM, 2 μM, and 3 μM for this interaction.
FIG 4.
Mapping of the NS4B binding motif using an SPR biosensor. (A) Sequence alignment of the cytoplasmic loop region of NS4B for various flaviviruses. Strictly conserved residues are starred, while conservative amino acid substitutions are indicated by colons. (B) Three biotinylated peptides with amino acid sequences encompassing the NS4B cytoplasmic loop from DENV2 (residues 130 to 167, 130 to 148, and 149 to 167, respectively) were captured on preimmobilized NeutrAvidin on CM5 sensor chips. The DENV2 NS3 helicase domain (1.6 to 200 μM; 2-fold serially diluted) was injected across all peptide surfaces in duplicate except for the highest concentration. Binding responses for NS4B peptides consisting of residues 130 to 167 and 149 to 167 were at least twice the responses for the peptide consisting of residues 130 to 148. (C) Percentages of NS3 helicase binding for the three peptides from the NS4B cytoplasmic loop. (D) Experimental values of Req for the corrected sensorgrams were compared to the simulated Kd values of 1 μM, 2 μM, and 3 μM for this interaction. The values for the dissociation constants are between 1 μM and 3 μM.
Cells, viruses, and antibodies.
BHK-21 cells were purchased from the American Type Culture Collection (ATCC) and maintained in high-glucose Dulbecco's modified Eagle medium (DMEM) (Invitrogen, Carlsbad, CA) supplemented with 10% fetal bovine serum (FBS) (HyClone Laboratories, Logan, UT) and 1% penicillin-streptomycin (Invitrogen). 293T cells were grown in low-glucose DMEM (Invitrogen) containing 10% FBS and 1% penicillin-streptomycin. DENV2 (strain New Guinea C [NGC]; GenBank accession number AF038403) was generated from its corresponding infectious cDNA clone (pACYC-NGC FL). The following antibodies were used in this study: mouse monoclonal antibody (MAb) 44-4-7 against DENV2 NS4B (27), rabbit polyclonal antibody (pAb) against DENV2 NS3 (Genetex), goat anti-rabbit antibody conjugated to horseradish peroxidase (HRP) (Sigma), mouse IgG2b negative-control antibody and protein A conjugated to HRP (Sigma), mouse MAb 4G2 against DENV envelope protein (ATCC), and Alexa Fluor 488 goat anti-mouse IgG (Life Technologies).
Plasmid construction.
Standard molecular biology procedures were performed for all plasmid constructions. The fragments encoding the NS2B-NS3 construct and a construct consisting of a peptide with a molecular weight of 2,000 (2K peptide) and NS4B (2K-NS4B) were amplified from pACYC-DENV2 TSVO1 FL by PCR using corresponding primer pairs. The PCR products were digested by restriction enzymes and cloned into the pXJ expression vector. All constructs were validated by DNA sequencing. Primer sequences are available upon request.
Coimmunoprecipitation.
293T cells in a 10-cm dish were transfected with various constructs using X-tremeGENE 9 DNA transfection reagent (Roche). At 48 h posttransfection (p.t.), cells were lysed in 1 ml of immunoprecipitation (IP) buffer (20 mM Tris HCl, pH 7.5, 100 mM NaCl, 0.5% DDM, and EDTA-free protease inhibitor cocktail [Roche]) by rotation at 4°C for 1 h. Lysates were clarified by centrifugation at 20,000 × g at 4°C for 30 min and subjected to coimmunoprecipitation (co-IP) using protein A-conjugated magnetic beads according to the manufacturer's instructions (Millipore). Briefly, immune complexes were formed at 4°C overnight by mixing 200 μl of cell lysates with mouse anti-NS4B MAb (4 μg) in a 500-μl reaction volume containing 250 mM sodium chloride. Subsequently, the complexes were precipitated with protein A-conjugated magnetic beads at 4°C for 1 h with rotation, followed by extensive washing with IP buffer (250 mM sodium chloride). Finally, proteins (NS3, NS4B, and the antibody against NS4B) were eluted with 4× lithium dodecyl sulfate (LDS) sample buffer (Life Technologies) supplemented with 100 mM DTT, heated at 70°C for 10 min, and analyzed by Western blotting. The 10-min 70°C treatment was used to dissociate NS3, NS4B, and NS4B antibody from the protein A-conjugated magnetic beads. NS4B was detected by the primary MAb 44-4-7 against NS4B (27) and protein A conjugated to HRP.
BHK-21 cells in a T-175 flask were infected with DENV2 NGC (multiplicity of infection [MOI] of 1). At 48 h postinfection (p.i.), cells were lysed in 1 ml of IP buffer. Clarified supernatant was subjected to co-IP using protein A-Sepharose (GE). Immune complexes were formed at 4°C overnight by mixing 100 μl of cell lysates with mouse anti-NS4B MAb (4 μg) or mouse IgG2b negative-control antibody (4 μg) in a 500-μl reaction volume containing 250 mM sodium chloride. Subsequently, complexes were precipitated with protein A-Sepharose at 4°C for 1 h with rotation.
In situ PLA.
A proximity of ligation assay (PLA) was performed according to the manufacturer's instructions (Olink Bioscience). Briefly, BHK-21 cells were seeded in an eight-well chamber (Nunc). The next day cells were infected with DENV2 NGC (MOI of 1) or transiently transfected with pXJ-NS2B-NS3 DNA and pXJ-2K-NS4B-HA DNA. At 48 h p.i. or p.t., cells were fixed by 4% paraformaldehyde at room temperature for 30 min and then permeabilized by phosphate-buffered saline (PBS) supplemented with 0.1% Triton X-100 at room temperature for 10 min. Subsequently, cells were incubated with PBS containing 1% FBS and 0.05% Tween 20 (PBST) at room temperature for 1 h and then incubated with the primary antibodies anti-NS4B mouse MAb 44-4-7 (27) and anti-NS3 rabbit pAb. The secondary antibodies supplied by the kit (PLA probe anti-mouse Plus and anti-rabbit Minus) were used at a 1:5 dilution and incubated at 37°C for 1 h. Afterwards, ligation-ligase solution was added and incubated at 37°C for 30 min. Signal amplification was performed by the addition of amplification-polymerase solution and incubation at 37°C for 100 min. Finally, chamber slides were mounted by Duolink in situ mounting medium containing 4′,6-diamidino-2-phenylindole (DAPI). Signals were observed on a Leica DM4000 B system at 598 nm. Images were merged using the Adobe Photoshop CS3 software.
RNA transcription, electroporation, immunofluorescence assay (IFA), and virus production.
DENV2 NS4B mutations were engineered using a QuikChange II XL site-directed mutagenesis kit (Stratagene). Wild-type (WT) and mutant DENV2 genome-length RNAs were transcribed in vitro using a T7 mMessage mMachine kit (Ambion) from cDNA plasmids prelinearized by XbaI. For RNA electroporation, 8 × 106 BHK-21 cells were washed once before being resuspended in 0.8 ml of cold PBS containing 10 μg of RNA. The cells were pulsed three times at 0.85 kV/25 μF in 0.4-cm cuvettes with a 3-s interval using a GenePulser apparatus (Bio-Rad). BHK-21 cells electroporated with viral RNA were seeded in an eight-well Lab-Tek chamber slide (Thermo Fisher Scientific). At time points indicated in Fig. 7, the cells were fixed in 100% ethanol at −20°C for 30 min. After 1 h of blocking in PBS supplemented with 1% FBS at room temperature, the cells were treated with mouse MAb 4G2 for 1 h, followed by three washes with PBS. Cells were then incubated with Alexa Fluor 488 goat anti-mouse IgG for 1 h. After three washes, the cells were mounted in a mounting medium with DAPI (Vector Laboratories, Inc.). Fluorescence images were taken and processed as described above.
FIG 7.
Analysis of NS4B mutations in DENV2 genome-length RNA. (A) IFA. BHK-21 cells were electroporated with equal amounts (10 μg) of WT or mutant genome-length RNAs (containing the indicated amino acid substitutions in NS4B). The expression of viral E protein (indicated in green) was monitored on day 1 and day 4 p.t. The nuclei were stained with DAPI. (B) Plaque morphologies of mutant and WT viruses. Culture fluids derived from the transfected cells in the experiment shown in panel A were analyzed for plaque morphology and virus yields on day 5 p.t. Virus titers were quantified by plaque assay. (C) Sequencing analysis of the recovered viruses. The NS4B region of the viruses recovered on day 5 p.t. from the experiment shown in panel A was sequenced. Mutant G140A virus reverted to the wild-type sequence, whereas mutant P124A, N144A, and D154A viruses retained the engineered substitutions.
BHK-21 cells electroporated with genome-length RNA were seeded in one T-75 flask. After incubation at 37°C for 1 day, the medium was replaced with fresh DMEM supplemented with 2% FBS, and cells were cultured at 30°C for another 4 days. On day 5 p.t., culture fluids were collected, and the virus titer was quantified by plaque assay as described previously (23).
Protein expression and purification.
To express and purify a polypeptide chain that includes the entire NS4B cytoplasmic loop region, the cDNA encoding residues I121 to V171 of NS4B from DENV2 (note that the cytoplasmic loop defined by Miller et al. [18] comprises only residues 129 to 165) was cloned into the PNIC28-Bsa4 vector. The resulting plasmid was transformed into E. coli BL21(DE3) competent cells. The recombinant protein was soluble and was first purified using metal affinity chromatography. The N-terminal hexahistidine tag was then removed using TEV protease. The protein was further purified using gel filtration chromatography using a Superdex-200 column (GE Healthcare). The expression and purification of the protease domain from DENV2 and the complete helicase region of NS3 from the DENV2 and DENV4 serotypes (residues 168 to 618) were conducted as previously reported (25, 26). Expression and purification of the full-length NS3 protease-helicase from DENV4 (fused to 40 hydrophilic residues from the NS2B cofactor) were done as described previously (28). Expression of the individual subdomains of the NS3 helicase gave poorly soluble proteins using the DENV2 serotype but soluble proteins for the DENV4 serotype. Protein fragments encompassing subdomain 1 (residues 168 to 322), subdomain 2 (residues 323 to 484), and subdomains 2 plus 3 (residues 323 to 618) from DENV4 were expressed and purified using metal affinity chromatography followed by size exclusion chromatography. The N-terminal hexahistidine tags of all NS3 protein constructs were removed using TEV protease.
NMR measurements.
Prior to the titration experiments, uniformly 15N- and 13C-labeled NS4B residues I121 to V171 as expressed above were purified in an NMR buffer containing 20 mM sodium phosphate, pH 6.5, 150 mM NaCl, and 1 mM DTT. All NMR spectra were acquired at 298 K on a Bruker Avance II 600 MHz or 700 MHz spectrometer equipped with a triple-resonance cryo-probe. Spectra were processed with Topspin, version 2.1 (Bruker), and NMRPipe (29) and visualized with NMRView (30) or Sparky (http://www.cgl.ucsf.edu/home/sparky/). Uniformly 13C/15N-labeled protein (0.5 mM) in the NMR buffer was used for data collection. The backbone 1HN, 15N, 13Cα, and 13C′ resonances were assigned using two-dimensional (2D) and three-dimensional (3D) experiments that included 1H-15N-heteronuclear single-quantum coherence (HSQC), 3D-HNCO, HNCACB, CBCA(CO)NH, and HNCA (31). All pulse programs were from the Bruker standard library (Topspin, version 2.1). The protein secondary structure was analyzed using the Cα chemical shift (32). Steady-state {1H}-15N nuclear Overhauser effects (NOEs) were obtained using two data sets that were collected with and without initial proton saturation for a period of 3 s (33). For the titration experiment, 2D-1H-15N-HSQC spectra were collected for the 15N- and 13C-labeled protein (I121 to V17) in the absence and presence of different amounts of unlabeled helicase.
RESULTS
NS3-NS4B interaction in virus-infected cells.
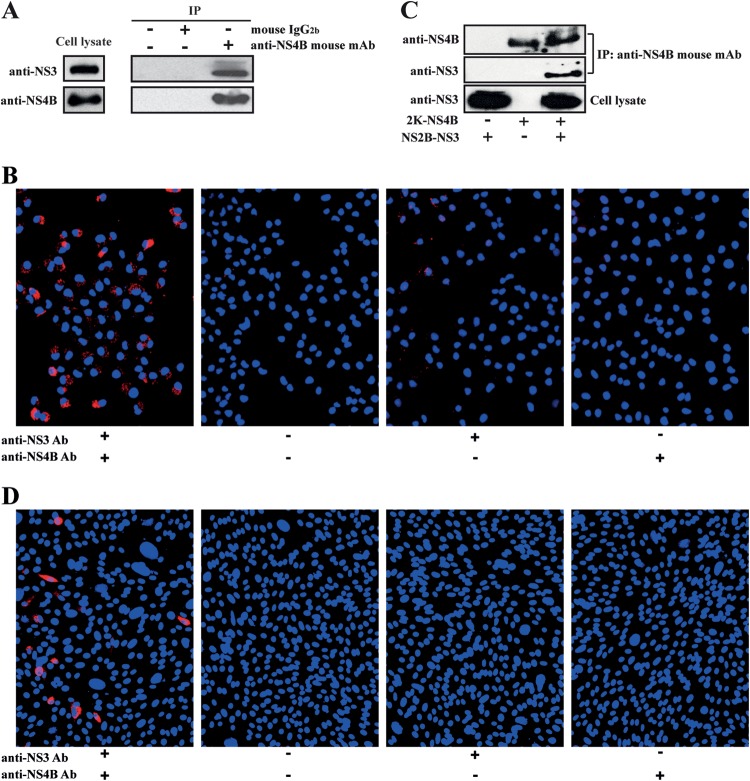
Using DENV2, we performed two sets of experiments to demonstrate that NS3 interacts with NS4B when they are expressed inside cells. The first set of experiments aimed to probe the NS3/NS4B association in the context of viral infection. BHK-21 cells were infected with DENV2 virus (MOI of 1). The cells were harvested at 48 h p.i., and cellular lysates were subjected to immunoprecipitation using anti-NS4B monoclonal antibody 44-4-7 (27). Viral NS3 was coimmunoprecipitated with NS4B from the infected cellular lysates (Fig. 1A). Approximately 9% of the NS3 protein detected in cell lysates was pulled down by the NS4B protein. As negative controls, no NS3 was precipitated with Sepharose-protein A beads alone or with the beads supplemented with mouse IgG2b (Fig. 1A).
FIG 1.
Analysis of the interaction between NS4B and NS3 by coimmunoprecipitation (co-IP) and proximity ligation assay (PLA). (A and B) BHK-21 cells were infected with DENV-2 at an MOI of 1. At 48 h p.i., the infected cells were subjected to co-IP (A) and PLA (B) as described in Materials and Methods. (C) 293T cells were cotransfected with expression vectors pXJ-NS2B-NS3 and/or pXJ-2K-NS4B. At 48 h p.t., the transfected cells were subjected to co-IP. (D) BHK-21 cells were cotransfected with expression vectors pXJ-NS2B-NS3 and/or pXJ-2K-NS4B. At 48 h p.t., the transfected cells were analyzed by PLA. BHK-21 cells, rather than 293T cells, were chosen for the PLA because PLA requires many washing steps, and 293T cells are easily washed off during these steps. In the PLA, red fluorescent signals indicate NS3-NS4B proximity. Nuclei were stained in blue by 4′,6-diamidino-2-phenylindole (DAPI).
Next, we applied an in situ proximity ligation assay (PLA) to confirm the NS3-NS4B proximity in DENV2-infected BHK-21 cells. PLA is a well-proven method to indicate close proximity of two molecules inside cells (34). As shown in Fig. 1B, red PLA fluorescent signals were detected in DENV2-infected cells (far left panel), suggesting that NS3 colocalizes with NS4B. In contrast, no red fluorescent signal was detected in various negative-control experiments, including PLA without one or both NS3 and NS4B antibodies in the infected cells (Fig. 1B, right three panels). These results indicate that NS3 specifically colocalizes with NS4B during DENV2 replication.
NS3-NS4B interaction using transient protein expression.
The second experiment aimed to demonstrate that DENV2 NS3 interacts with NS4B in the absence of viral infection. 293T cells were cotransfected with two plasmids: one expressing NS2B-NS3 and another expressing 2K-NS4B. The NS2B and 2K segments were included in the plasmids to ensure membrane association of NS3 and the correct membrane topology of NS4B, respectively. Cells were harvested at 48 h p.t., and cellular lysates were used for coimmunoprecipitation. As shown in Fig. 1C, NS2B-NS3 was coimmunoprecipitated with NS4B using anti-NS4B monoclonal antibody 44-4-7 (27); no NS2B-NS3 was pulled down in the absence of 2K-NS4B expression. We also performed PLA of cells cotransfected with the NS2B-NS3 and 2K-NS4B expression plasmids. In the PLA experiment, we cotransfected BHK-21 cells, rather than 293T cells, with the NS2B-NS3 plasmid and the 2K-NS4B plasmid (described above). BHK-21 cells were chosen for the PLA experiment because PLA requires many washing steps, and 293T cells easily detach from the culture surface during these washing steps. As shown in Fig. 1D, red fluorescent signals were detected in cotransfected cells (far left panel), suggesting that NS2B-NS3 colocalizes with 2K-NS4B inside cells. No red fluorescent signals were detected in negative-control experiments, which excluded the use of one or both NS3 and NS4B antibodies (Fig. 1D, right three panels). Taken together, these data indicate that NS3 is colocalized with NS4B in the absence of other viral proteins. However, these data do not indicate whether the association is direct or indirect.
Subdomains 2 and 3 of the NS3 helicase are determinants for NS4B interaction.
To demonstrate a direct NS3-NS4B interaction inside cells, as opposed to an interaction mediated by a host or viral protein, we expressed and purified the full-length NS3 protein, its helicase domain, and NS4B from DENV2 (Fig. 2A) and examined their interaction using SPR. The structural integrity of NS4B was assessed by CD spectroscopy (Fig. 2B), which indicated a predominantly α-helical structure consistent with the published topology model (18). The SPR sensorgram showed that the NS3 protease-helicase interacts with NS4B at a stoichiometry of 1:1 with an equilibrium dissociation constant Kd of 163 nM (Fig. 2C). To determine whether the protease or helicase domain of NS3 is responsible for the interaction with NS4B, we prepared a recombinant helicase domain of DENV2 (Fig. 2A). SPR analysis showed that the DENV2 helicase domain could bind NS4B with a KD of 222 ± 4 nM (Fig. 2D). A recombinant fragment of NS3 containing the protease domain alone (and a 40-residue segment of the NS2B cofactor) did not bind NS4B in the SPR assay (Fig. 2E). These results indicate that NS4B binds the helicase domain of NS3.
The flavivirus NS3 helicase region comprises 450 residues and is composed of three subdomains (Fig. 3A). Both subdomains 1 (residues 168 to 322) and 2 (residues 323 to 484) adopt a RecA-like fold between which ATP is bound (25, 26). Single-stranded RNA binds within a tunnel formed at the interface between subdomains 1 and 2 on one side and subdomain 3 (residues 485 to 618) on the other (Fig. 3A). To map the helicase subdomains interacting with NS4B, we expressed and purified individual subdomains 1 and 2 from DENV4 (Fig. 3B). Subdomain 3 could not be prepared due to its poor solubility in both DENV2 and DENV4; however, we successfully expressed and purified a recombinant protein containing both subdomains 2 and 3 (Fig. 3C). Proper folding for subdomains 1, 2, and 2 plus 3 was checked using CD (Fig. 3D). We used the DENV4 serotype to prepare the helicase fragments listed above because the corresponding subdomains of DENV2 helicase were poorly soluble. The ability of each helicase subdomain to bind the NS4B protein was tested by SPR by passing them through a CM5 chip to which NS4B (from DENV2) was covalently attached. As shown in Fig. 3E, the helicase fragments spanning its subdomains 2 and 3 bind to NS4B with a KD of 340 nM, which is comparable to the KD derived from the complete DENV4 helicase-NS4B interaction (530 nM). In contrast, negligible binding was observed for subdomain 1, whereas subdomain 2 alone has a lower binding affinity, with a KD of 4.84 μM. These results suggest that NS4B binds to subdomains 2 and 3 of the NS3 helicase.
The cytoplasmic loop of NS4B binds to the NS3 helicase region.
Based on the membrane topology model of DENV2 NS4B, only amino acids 129 to 165 and the C-terminal four amino acids (245 to 248) of NS4B are oriented toward the cytoplasmic side of the ER membrane (18). Since NS3 is located at the cytoplasmic side during flaviviral replication, we hypothesized that the cytoplasmic loop of NS4B (we used residues 130 to 167 because residues 166 and 167 are hydrophilic and 129 is hydrophobic) could form a direct interaction with NS3. Three peptides were synthesized to test this hypothesis (Fig. 4A): one spanning the complete cytoplasmic loop of NS4B (residues 130 to 167), one representing its N-terminal half (residues 130 to 148), and one consisting of the C-terminal half (residues 149 to 167) (Fig. 4A). SPR analysis (Fig. 4B, C, and D) revealed that the C-terminal part of the cytoplasmic loop has an affinity of 1.6 μM for the NS3 helicase, slightly lower than that of the entire cytoplasmic loop (Kd of 1.3 μM) and higher than the affinity displayed by the N-terminal segment (Kd of 2.8 μM).
Structural analysis of the NS4B cytoplasmic loop.
We used solution NMR spectroscopy to further explore the NS4B-NS3 interaction. The solution structure of the cytoplasmic loop of NS4B from DENV2 was probed first. A fragment of NS4B comprising its cytoplasmic loop (residues 121 to 171 of NS4B) was expressed in E. coli and purified (Fig. 5A). The identity of the purified NS4B fragment was verified by mass spectroscopy (Fig. 5B). A 2D 1H-15N-HSQC spectrum of the NS4B cytoplasmic loop was obtained (Fig. 5C). The narrow dispersion of the cross-peaks suggests that the purified fragment is not highly structured. We also conducted backbone resonance assignments for the purified NS4B fragment using multidimensional heteronuclear NMR spectroscopy. Secondary structure analysis based on the Cα chemical shift indicates the presence of several secondary elements under the experimental conditions used: residues 127 to 135 tend to fold into a three-turn α-helix, while residues 141 to 144 and 150 to 153 have a tendency to form two short β-strands (Fig. 5D). To further explore the dynamic nature of the cytoplasmic NS4B loop, we conducted backbone 1H-15N heteronuclear NOE (HNOE) experiments. The N- and C-terminal ends of the NS4B fragment as well as most of the residues located at positions 135 to 152 displayed negative NOEs. Conversely, residues 127 to 134 and 153 to 164 (excluding Pro159 and Pro162) showed positive NOE values (less than 0.4) with no cross-peaks and overlapped cross-peaks in the 1H-15N-HSQC spectrum (Fig. 5E). These results suggest that the overall structure adopted by the cytoplasmic loop of NS4B is flexible in solution and that only small segments of the loop have a tendency to form regular secondary structures.
FIG 5.
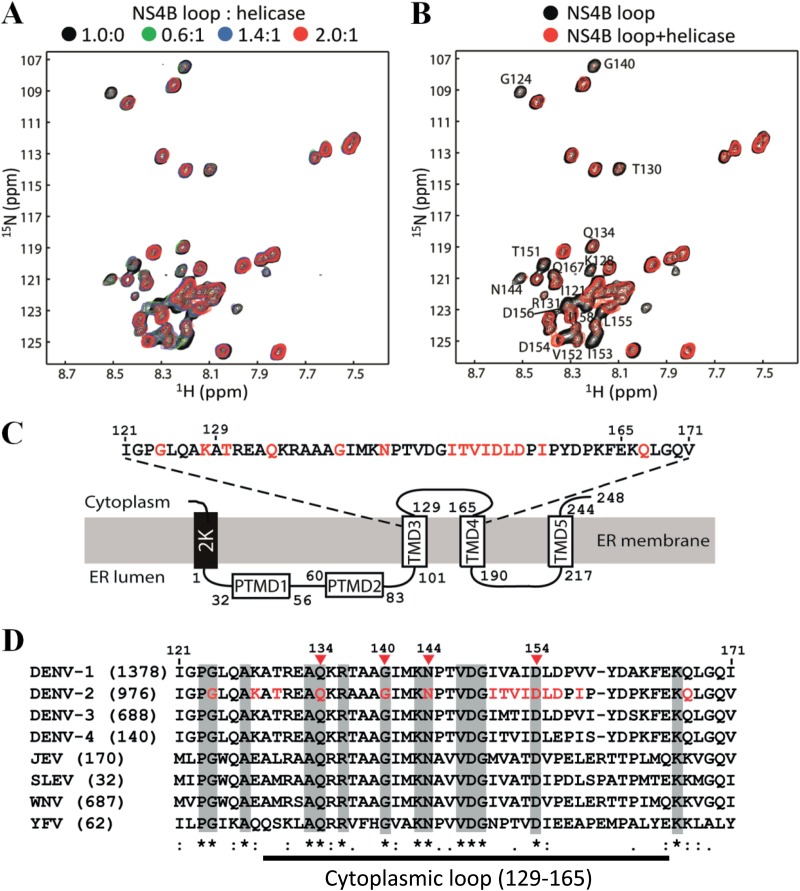
NMR analysis of the cytoplasmic loop of NS4B. (A) Purification of a recombinant protein that includes the NS4B cytoplasmic loop. A fragment of NS4B (residues 121 to 171) spanning the entire cytoplasmic loop of NS4B (residues 129 to 165 in DENV2 [18]) was expressed, purified, and analyzed by SDS-PAGE. The protein was purified through a nickel column followed by gel filtration. Lane 1, molecular mass markers; lane 2, NS4B loop after nickel-NTA column purification and cleavage of the N-terminal His tag with TEV protease; lane 3, purified NS4B loop after gel filtration. (B) Mass spectrometry analysis of the purified protein. The molecular weight of the purified protein agrees well with its theoretical mass. (C) Assignment of the 1H-15N-HSQC spectrum. The assignments of the cross-peaks in the spectrum are shown with residue names and sequence numbers. (D) Secondary structure analysis of NS4B (residues 121 to 171). The Cα chemical shift difference (ΔCα) was obtained by comparing the Cα chemical shift values with those of a random coil. Positive values suggest that the residue may form a helix, and negative values suggest that the residue may form a β-strand. (E) {1H}-15N HNOE of NS4B (residues 121 to 171). For clarity, the HNOE values of proline residues having no cross-peaks and overlapped residues in the HSQC spectrum are shown as 0.
Interaction between the NS4B cytoplasmic loop and the NS3 helicase.
To identify residues within the NS4B cytoplasmic loop that directly interact with the NS3 helicase, we measured chemical shift perturbations of a 2D 1H-15N-HSQC spectrum of the NS4B cytoplasmic loop induced by the addition of the NS3 helicase from DENV2. In the presence of the NS3 helicase, some resonances of the NS4B cytoplasmic loop are broadened (Fig. 6A and B), indicating binding to the NS3 helicase. The perturbed resonances were assigned to the 15 following residues of NS4B from DENV2: G124, K128, T130, Q134, G140, N144, I150 to D156, I158, and Q167 (Fig. 6C). Note that some of these residues may not be directly involved in helicase binding, and the chemical shift perturbations observed could be caused by an indirect conformational change upon helicase binding. Nonetheless, the results are consistent with the SPR data showing that both the N-terminal and C-terminal halves of the NS4B cytoplasmic loop participate in the interaction with the NS3 helicase (Fig. 4).
FIG 6.
Chemical shift perturbation analysis of the NS3-NS3B interaction. The recombinant NS3 helicase domain (Fig. 2A) and NS4B (residues 121 to 171) (Fig. 5A) were used to perform chemical shift perturbation. (A) Overlay of the 1H-15N-HSQC spectra of the NS4B cytoplasmic loop in the absence and presence of different amounts of helicase. A uniformly 13C/15N-labeled NS4B loop at a 0.2 mM concentration was used in the study. All the experiments were conducted at 298 K. (B) Residues affected by helicase binding. Overlay of 2D 1H-15N-HSQC of labeled NS4B (residues 121 to 171) in the absence (black) and presence (red) of helicase/NS4B loop at a molar ratio of 1:2 is shown. The affected residues are shown with residue name and number. (C) Schematic diagram of the topology of NS4B from DENV2. Residues identified from the chemical shift perturbation experiment are indicated in red. (D) Sequence alignment of the NS4B fragment (residues 121 to 171) among different flaviviruses. The numbers in parentheses are the numbers of NS4B sequences that were used for the alignment. Sequences were downloaded from the National Center for Biotechnology Information (NCBI) protein database. The alignment was performed using CLC Main Workbench software (CLC bio). The amino acid positions of NS4B are numbered according to the DENV2 NGC strain (GenBank number AF038403). The flavivirus-conserved residues are shaded in gray. The amino acids identified from the chemical shift perturbation analysis are shown in red in the DENV2 sequence, among which four residues (indicated by arrowheads), belonging to the cytoplasmic loop (defined as residues 129 to 165, according to Miller et al. [18]), are strictly conserved across flaviviruses.
Genetic analysis.
To examine the functional relevance of the residues identified from the chemical shift perturbation experiment, we performed a mutagenesis analysis using a full-length DENV2 infectious clone. We selected residues G124, Q134, G140, N144, and D154 for Ala substitution because these amino acids are strictly conserved across various flavivirus NS4B proteins (Fig. 6D). Equal amounts of wild-type and mutant genome-length RNAs were electroporated into BHK-21 cells. The expression levels of viral E protein in the transfected cells were monitored by IFA at time points indicated on Fig. 7A. No E-positive cells were detected for the Q134A or G140A mutant on day 1 and day 4 p.t.; fewer E-positive cells were observed for the N144A mutant; similar numbers of E-positive cells were observed for the mutants G124 and D154A and the wild-type viruses. In agreement with the IFA results, the plaque assay indicates that no infectious virus was produced for the mutant Q134A (Fig. 7B). Mutant G140A produced a small amount of virus at 10 PFU/ml on day 5 p.t.; sequencing of the recovered G140A virus revealed a reversion of the engineered Ala (GCA) to the wild-type Gly (GGA) (Fig. 7C). Mutant G124A, N144A, and D154A viruses exhibited plaques slightly smaller than those of the wild-type virus; sequencing analysis showed that the recovered viruses retained the engineered mutations (Fig. 7C). Continuous passaging of mutant viruses G124A, N144A, and D154A for 10 rounds (3 to 4 days per round) did not reveal any adaptive mutations; similar passaging of the culture fluid from the Q134A RNA-transfected cells did not yield any viable virus (data not shown), suggesting that no revertant could be selected. Collectively, these results indicate that, within the evolutionarily conserved residues of the NS4B cytoplasmic loop, Q134 and G140 are essential for DENV2 replication, while mutant N144A produced a 1,000-fold reduction in virus yield.
DISCUSSION
Temporal and spatial regulations of molecular interactions among viral proteins and RNA as well as between viral and host cell proteins are required to orchestrate the distinct steps of viral replication. In principle, compounds disrupting such protein/protein or protein/RNA interfaces could be developed as probes of the flavivirus RC and also for antiviral therapy. The present study aimed to characterize the interaction between the flavivirus NS3 and NS4B proteins. Previously, a yeast two-hybrid study reported that the helicase domain of NS3 from DENV2 interacts with NS4B (24). The interaction was further confirmed by biochemical pulldown and immunoprecipitation, both with partially purified proteins and with DENV-infected cellular lysates (24). The interaction enhances the helicase activity of NS3 by stimulating the dissociation of single-stranded RNA (24). Here, we confirmed that DENV NS3 binds to NS4B regardless of the presence of other viral or cellular proteins. Importantly, we found that subdomains 2 and 3 of the helicase domain of NS3 are required for binding to the cytoplasmic loop of NS4B. Line-broadening NMR analysis identified 12 amino acids within the cytoplasmic loop of NS4B (spanning residues 129 to 165 as defined by Miller et al. [18]) that may interact with the NS3 helicase. Four of the 12 identified residues in the cytoplasmic loop (Q134, G140, N144, and D154) are strictly conserved among various flavivirus NS4B proteins. Mutagenesis studies in a DENV infectious clone showed that substitution Q134A was lethal; mutation G140A greatly reduced viral replication but rapidly reverted to WT, while mutation N144A produced a 1,000-fold reduction in virus titer. However, the replication defects observed in the mutant NS4B viruses could result from malfunctions of NS4B other than through a direct interaction with NS3. Whether Q134, G140, and N144 play a role in the NS3-NS4B interaction was therefore tested using peptides bearing the corresponding mutations in an SPR binding assay with NS3 helicase. The NS4B peptides with the corresponding mutations retain the ability to bind to NS3 (data not shown). This result might simply be due to a nonoptimal mimicry of the NS4B loop structure when it is presented as shorter peptides lacking the transmembrane anchoring points of the native protein. It also has to be reconciled with the observation that NS4B dimerizes (23). Previous studies showed that NS4B dimerization is mediated by the cytosolic loop as well as by the C-terminal region (amino acids 166 to 248) of NS4B and that the C-terminal region alone is sufficient to induce NS4B dimer formation (23). Thus, the replication defects observed in these three viruses mutated in the cytoplasmic loop region could result from impairing the dimerization of NS4B or other interactions within the flavivirus RC rather than from the NS4B-NS3 interaction.
NMR analysis of the cytoplasmic loop of NS4B showed that the peptide is not highly structured in solution, with a tendency to form only short secondary structures. The flexibility of the NS4B cytoplasmic loop is consistent with its multifunctional roles: it contributes to NS4B dimerization (23), and it binds NS3 (28). This flexibility could allow NS4B to interact with different protein partners at various stages of viral replication.
Our results provide a potential antiviral approach through the disruption of the flavivirus NS3/NS4B interface. The SPR-based assay reported in this study could be optimized for this purpose. The biophysical SPR assay screens for compounds that directly interfere with the NS3-NS4B interaction. This assay, together with the possibility to solve the cocrystal structure of helicase (25, 26, 28) in complex with the NS4B cytoplasmic loop, will inform future screening for inhibitors of the NS3-NS4B interaction.
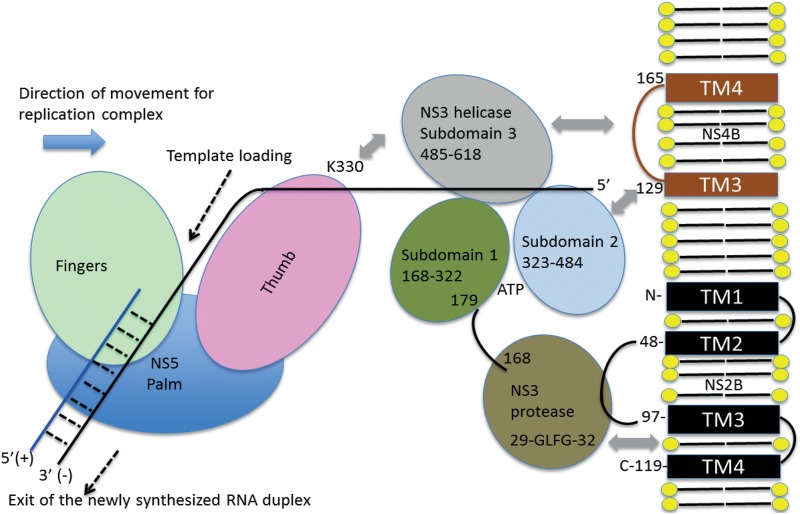
A model for the interaction between NS4B (via its cytoplasmic loop), NS3 (through its helicase subdomains 2 and 3), and NS5 is shown in Fig. 8. This working model is consistent with the available mapping data (35) and also the elongated molecular shape observed in the crystal structure of the NS2B-NS3 protease-helicase from DENV (28). In this structure, the protease region sits beneath subdomains 1 and 2 of the helicase and is tethered to the membrane by both the essential cofactor residues 49 to 96 of NS2B and an exposed hydrophobic loop of NS3 (G29-L30-F31-G32) (36) (Fig. 8). These contacts would leave the concave surface between subdomains 2 and 3 of the helicase region (where the 5′ end of the RNA is emerging from the RNA binding tunnel) available for interaction with both the cytoplasmic loop of NS4B and the NS5 methyltransferase-polymerase. Residue K330 from the thumb region of the NS5 polymerase was shown to interact with the NS3 helicase (35). The accompanying paper by Zou et al. (37) characterizes the NS4A and NS4B protein interaction, and further studies in the presence of RNA are now needed to clarify the detailed network of intermolecular contacts established between NS2B, NS4A, NS4B, and NS5 at various stages of RNA replication.
FIG 8.
A model for the NS4B-NS2B-NS3-NS5 interactome during positive-strand RNA synthesis. This schematic representation summarizes intermolecular interactions (gray arrows) between NS4B and NS3 that are mediated by the cytoplasmic loop (residues 129 to 165) of NS4B (in blue) and both subdomains 2 and 3 of the NS3 helicase domain (this work). The back of the thumb subdomain of NS5 interacts with subdomain 3 of the NS3 helicase region, and K330 plays a crucial role in this interaction (35). The orientation of the protease domain of NS3 (that is connected to the helicase via a flexible linker region encompassing residues 168 to 179) with respect to the membrane is depicted according to Luo et al. (36). An exposed hydrophobic loop of NS3 protease (Gly29-Leu30-Phe31-Gly32), labeled as GLFG, interacts with the membrane (36). The replicative enzymes NS3 and NS5 progress along the negative RNA template strand from the 3′ end toward the 5′ end with a polarity of the single strand in the helicase tunnel compatible with the crystallographic results (26).
ACKNOWLEDGMENTS
We thank Lijian Yap, Hongping Dong, Ying Lei Wong, Qiwei Huang, Young Mee Kim, Michelle Yueqi Lee, Rong Li, and other colleagues for technical support and helpful discussions during the course of the project.
C.K.'s team is supported by the A*STAR BMRC IAF grant (111105). J.L.'s group was supported by grant CRP2008 from the National Research Foundation and a Collaborative Basic Research grant from the Singapore National Medical Research Council (May14). This project was supported by a National Medical Research Council Translational Clinical Research STOP dengue grant (NMRC/TCR/005/2008; to P.-Y.S.).
REFERENCES
- 1.Gubler DJ. 2011. Dengue, urbanization and globalization: the unholy trinity of the 21st century. Trop Med Health 39(4 Suppl):3–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.World Health Organization. 2014. Dengue fact sheet. World Health Organization, Manila, the Philippines: http://www.wpro.who.int/mediacentre/factsheets/fs_09032012_Dengue/en/. [Google Scholar]
- 3.Sabchareon A, Wallace D, Sirivichayakul C, Limkittikul K, Chanthavanich P, Suvannadabba S, Jiwariyavej V, Dulyachai W, Pengsaa K, Wartel TA, Moureau A, Saville M, Bouckenooghe A, Viviani S, Tornieporth NG, Lang J. 2012. Protective efficacy of the recombinant, live-attenuated, CYD tetravalent dengue vaccine in Thai schoolchildren: a randomized, controlled phase 2b trial. Lancet 380:1559–1567. doi: 10.1016/S0140-6736(12)61428-7. [DOI] [PubMed] [Google Scholar]
- 4.Kuhn RJ, Zhang W, Rossmann MG, Pletnev SV, Corver J, Lenches E, Jones CT, Mukhopadhyay S, Chipman PR, Strauss EG, Baker TS, Strauss JH. 2002. Structure of dengue virus: implications for flavivirus organization, maturation, and fusion. Cell 108:717–725. doi: 10.1016/S0092-8674(02)00660-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lindenbach BD, Thiel HJ and Rice CM. 2007. Flaviviridae: the viruses and their replication, p 1101–1152 InKnipe DM, Howley PM, Griffin DE, Lamb RA, Martin MA, Roizman B, Straus SE (ed), Fields virology, 5th ed Lippincott Williams & Wilkins, Philadelphia, PA. [Google Scholar]
- 6.Lindenbach B, Rice C. 1997. trans-Complementation of yellow fever virus NS1 reveals a role in early RNA replication. J Virol 71:9608–9617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chambers TJ, Grakoui A, Rice CM. 1991. Processing of the yellow fever virus nonstructural polyprotein: a catalytically active NS3 proteinase domain and NS2B are required for cleavages at dibasic sites. J Virol 65:6042–6050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Falgout B, Miller RH, Lai CJ. 1993. Deletion analysis of dengue virus type 4 nonstructural protein NS2B: identification of a domain required for NS2B-NS3 protease activity. J Virol 67:2034–2042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Li H, Clum S, You S, Ebner KE, Padmanabhan R. 1999. The serine protease and RNA-stimulated nucleoside triphosphatase and RNA helicase functional domains of dengue virus type 2 NS3 converge within a region of 20 amino acids. J Virol 73:3108–3116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Egloff MP, Benarroch D, Selisko B, Romette J L, Canard B. 2002. An RNA cap (nucleoside-2′-O-)-methyltransferase in the flavivirus RNA polymerase NS5: crystal structure and functional characterization. EMBO J 21:2757–2768. doi: 10.1093/emboj/21.11.2757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ray D, Shah A, Tilgner M, Guo Y, Zhao Y, Dong H, Deas T, Zhou Y, Li H, Shi P-Y. 2006. West Nile virus 5′-cap structure is formed by sequential guanine N-7 and ribose 2′-O methylations by nonstructural protein 5. J Virol 80:8362–8370. doi: 10.1128/JVI.00814-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dong H, Chang DC, Hua MH, Lim SP, Chionh YH, Hia F, Lee YH, Kukkaro P, Lok SM, Dedon PC, Shi PY. 2012. 2′-O methylation of internal adenosine by flavivirus NS5 methyltransferase. PLoS Pathog 8:e1002642. doi: 10.1371/journal.ppat.1002642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yap TL, Xu T, Chen YL, Malet H, Egloff M-P, Canard B, Vasudevan SG, Lescar J. 2007. The crystal structure of the dengue virus RNA-dependent RNA polymerase at 1.85-angstrom resolution. J Virol 81:4753–4765. doi: 10.1128/JVI.02283-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.den Boon JA, Ahlquist P. 2010. Organelle-like membrane compartmentalization of positive-strand RNA virus replication factories. Annu Rev Microbiol 64:241–256. doi: 10.1146/annurev.micro.112408.134012. [DOI] [PubMed] [Google Scholar]
- 15.Paul D, Bartenschlager R. 2013. Architecture and biogenesis of plus-strand RNA virus replication factories. World J Virol 2:32–48. doi: 10.5501/wjv.v2.i2.32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lim SP, Wang Q-Y, Noble CG, Chen Y-L, Dong H, Zou B, Yokokawa F, Nilar S, Smith P, Beer D, Lescar J, Shi P-Y. 2013. Ten years of dengue drug discovery: progress and prospects. Antiviral Res 100:500–519. doi: 10.1016/j.antiviral.2013.09.013. [DOI] [PubMed] [Google Scholar]
- 17.Miller S, Kastner S, Krijnse-Locker J, Buhler S, Bartenschlager R. 2007. The non-structural protein 4A of dengue virus is an integral membrane protein inducing membrane alterations in a 2K-regulated manner. J Biol Chem 282:8873–8882. doi: 10.1074/jbc.M609919200. [DOI] [PubMed] [Google Scholar]
- 18.Miller S, Sparacio S, Bartenschlager R. 2006. Subcellular localization and membrane topology of the dengue virus type 2 non-structural protein 4B. J Biol Chem 281:8854–8863. doi: 10.1074/jbc.M512697200. [DOI] [PubMed] [Google Scholar]
- 19.Xie X, Gayen S, Kang C, Yuan Z, Shi PY. 2013. Membrane topology and function of dengue virus NS2A protein. J Virol 87:4609–4622. doi: 10.1128/JVI.02424-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Xie X, Wang QY, Xu HY, Qing M, Kramer L, Yuan Z, Shi PY. 2011. Inhibition of dengue virus by targeting viral NS4B protein. J Virol 85:11183–11195. doi: 10.1128/JVI.05468-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.van Cleef KW, Overheul GJ, Thomassen MC, Kaptein SJ, Davidson AD, Jacobs M, Neyts J, van Kuppeveld FJ, van Rij RP. 2013. Identification of a new dengue virus inhibitor that targets the viral NS4B protein and restricts genomic RNA replication. Antiviral Res 99:165–171. doi: 10.1016/j.antiviral.2013.05.011. [DOI] [PubMed] [Google Scholar]
- 22.Patkar CG, Larsen M, Owston M, Smith JL, Kuhn RJ. 2009. Identification of inhibitors of yellow fever virus replication using a replicon-based high-throughput assay. Antimicrob Agents Chemother 53:4103–4114. doi: 10.1128/AAC.00074-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zou J, Xie X, Lee LT, Chandrasekaran R, Reynaud A, Yap L, Wang Q-Y, Dong HP, Kang C, Yuan Z, Lescar J, Shi P-Y. 2014. Dimerization of flavivirus NS4B protein. J Virol 88:3379–3391. doi: 10.1128/JVI.02782-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Umareddy I, Chao A, Sampath A, Gu F, Vasudevan SG. 2006. Dengue virus NS4B interacts with NS3 and dissociates it from single-stranded RNA. J Gen Virol 87:2605–2614. doi: 10.1099/vir.0.81844-0. [DOI] [PubMed] [Google Scholar]
- 25.Xu T, Sampath A, Chao A, Wen D, Nanao M, Chene P, Vasudevan SG, Lescar J. 2005. Structure of the dengue virus helicase/nucleoside triphosphatase catalytic domain at a resolution of 2.4 Å. J Virol 79:10278–10288. doi: 10.1128/JVI.79.16.10278-10288.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Luo D, Xu T, Watson RP, Scherer-Becker D, Sampath A, Jahnke W, Yeong SS, Wang CH, Lim SP, Strongin A, Vasudevan SG, Lescar J. 2008. Insights into RNA unwinding and ATP hydrolysis by the flavivirus NS3 protein. EMBO J 27:3209–3219. doi: 10.1038/emboj.2008.232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Xie X, Zou J, Wang Q-Y, Noble CG, Lescar J, Shi P-Y. 2014. Generation and characterization of two mouse monoclonal antibodies against NS4B protein of dengue virus. Virology 450–451:250–257. doi: 10.1016/j.virol.2013.12.025. [DOI] [PubMed] [Google Scholar]
- 28.Luo D, Xu T, Hunke C, Gruber G, Vasudevan SG, Lescar J. 2008. Crystal structure of the NS3 protease-helicase from dengue virus. J Virol 82:173–183. doi: 10.1128/JVI.01788-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Delaglio F, Grzesiek S, Vuister GW, Zhu G, Pfeifer J, Bax A. 1995. NMRPipe: a multidimensional spectral processing system based on UNIX pipes. J Biomol NMR 6:277–293. [DOI] [PubMed] [Google Scholar]
- 30.Johnson BA.2004. Using NMRView to visualize and analyze the NMR spectra of macromolecules. Methods Mol Biol 278:313–352. doi: 10.1385/1-59259-809-9:313. [DOI] [PubMed] [Google Scholar]
- 31.Li Q, Raida M, Kang C. 2010. 1H, 13C and 15N chemical shift assignments for the N-terminal domain of the voltage-gated potassium channel-hERG. Biomol NMR Assign 4:211–213. doi: 10.1007/s12104-010-9248-3. [DOI] [PubMed] [Google Scholar]
- 32.Andersen NH, Cao B, Chen C. 1992. Peptide/protein structure analysis using the chemical shift index method: upfield α-CH values reveal dynamic helices and αL sites. Biochem Biophys Res Commun 184:1008–1014. doi: 10.1016/0006-291X(92)90691-D. [DOI] [PubMed] [Google Scholar]
- 33.Kay LE, Torchia DA, Bax A. 1989. Backbone dynamics of proteins as studied by 15N inverse detected heteronuclear NMR spectroscopy: application to staphylococcal nuclease. Biochemistry 28:8972–8979. doi: 10.1021/bi00449a003. [DOI] [PubMed] [Google Scholar]
- 34.Söderberg O, Gullberg M, Jarvius M, Ridderstrale K, Leuchowius KJ, Jarvius J, Wester K, Hydbrid P, Bahram F, Larsson LG, Landegren U. 2006. Direct observation of individual endogenous protein complexes in situ by proximity ligation. Nat Methods 3:995–1000. doi: 10.1038/nmeth947. [DOI] [PubMed] [Google Scholar]
- 35.Zou G, Chen Y-L, Dong H, Lim CC, Yap LJ, Yau YH, Geifman Shochat S, Lescar J, Shi P-Y. 2011. Functional analysis of two cavities in flavivirus NS5 polymerase. J Biol Chem 286:14362–14372. doi: 10.1074/jbc.M110.214189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Luo D, Wei N, Doan D, Paradkar P, Chong Y, Davidson A, Kotaka M, Lescar J, Vasudevan SG. 2010. Flexibility between the protease and helicase domains of the dengue virus NS3 protein conferred by the linker region and its functional implications. J Biol Chem 285:18817–18827. doi: 10.1074/jbc.M109.090936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zou J, Xie X, Wang Q-Y, Dong H, Lee MY, Kang C, Yuan Z, Shi P-Y. 2015. Characterization of dengue virus NS4A and NS4B protein interaction. J Virol 89:3455–3470. doi: 10.1128/JVI.03453-14. [DOI] [PMC free article] [PubMed] [Google Scholar]