ABSTRACT
Ranaviruses (Iridoviridae) are posing an increasing threat to amphibian populations, with anuran tadpoles being particularly susceptible to these viral infections. Moreover, amphibians are the most basal phylogenetic class of vertebrates known to possess both type I and type III interferon (IFN)-mediated immunity. Moreover, little is known regarding the respective roles of the IFN mediators in amphibian antiviral defenses. Accordingly, we transcriptionally and functionally compared the amphibian Xenopus laevis type I (IFN) and III (IFN-λ) IFNs in the context of infections by the ranavirus frog virus 3 (FV3). X. laevis IFN and IFN-λ displayed distinct tissue expression profiles. In contrast to our previous findings that X. laevis tadpoles exhibit delayed and modest type I IFN responses to FV3 infections compared to the responses of adults, here we report that tadpoles mount timely and robust type III IFN gene responses. Recombinant forms of these cytokines (recombinant X. laevis IFN [rXlIFN] and rXlIFN-λ) elicited antiviral gene expression in the kidney-derived A6 cell line as well as in tadpole leukocytes and tissues. However, rXlIFN-λ was less effective than rXlIFN in preventing FV3 replication in A6 cells and tadpoles and inferior at promoting tadpole survival. Intriguingly, FV3 impaired A6 cell and tadpole kidney type III IFN receptor gene expression. Furthermore, in A6 cultures rXlIFN-λ conferred equal or greater protection than rXlIFN against recombinant viruses deficient for the putative immune evasion genes, the viral caspase activation and recruitment domain (vCARD) or a truncated vIF-2α gene. Thus, in contrast to previous assumptions, tadpoles possess intact antiviral defenses reliant on type III IFNs, which are overcome by FV3 pathogens.
IMPORTANCE Anuran tadpoles, including those of Xenopus laevis, are particularly susceptible to infection by ranavirus such as FV3. We investigated the respective roles of X. laevis type I and type III interferons (IFN and IFN-λ, respectively) during FV3 infections. Notably, tadpoles mounted timely and more robust IFN-λ gene expression responses to FV3 than adults, contrasting with the poorer tadpole type I IFN responses. However, a recombinant X. laevis IFN-λ (rXlIFN-λ) conferred less protection to tadpoles and the A6 cell line than rXlIFN, which may be explained by the FV3 impairment of IFN-λ receptor gene expression. The importance of IFN-λ in tadpole anti-FV3 defenses is underlined by the critical involvement of two putative immune evasion genes in FV3 resistance to IFN- and IFN-λ-mediated responses. These findings challenge the view that tadpoles have defective antiviral immunity and suggest, rather, that their antiviral responses are predominated by IFN-λ responses, which are overcome by FV3.
INTRODUCTION
Vertebrate antiviral immunity relies heavily on the interferon (IFN) response, which in mammals is comprised of three classes of cytokines, type I, II, and III IFNs (1). IFN-γ, the only mammalian type II IFN (bony fish possess multiple type II IFNs [2]) has a plethora of immune and antiviral roles, whereas type I and III IFNs function predominantly as antiviral molecules. While type I IFNs affect a broad range of cell types, the type III IFNs (also known as IFN-λ or interleukin-28 [IL-28] and IL-29) act on a limited range of cell subsets (3, 4). These differences are dictated at the receptor level, where the type I IFN receptors, IFNAR1 and IFNAR2, are ubiquitously expressed (5). In contrast, the type III receptor complex consists of the ligand-binding and IFN-λ-specific IFNLR1 chain (interferon lambda receptor 1), which is expressed on a select subset of cells (chiefly among these are epithelial cells [6]), and the cell-signal propagating IL-10R2 chain (shared with IL-10, IL-22, and IL-26) (7, 8). Despite these differences, type I and type III IFN cytokines utilize the same downstream signaling pathways, culminating in comparable antiviral outcomes, including increased gene expression of antiviral cellular mediators such as protein kinase R (PKR) and myxovirus resistance (Mx) proteins (1).
While the mammalian IFN responses have been relatively well characterized, the IFN immunity of phylogenetically more ancestral ectothermic vertebrate species appears to be distinct. At present, only the type I IFN systems of bony fish have been explored in detail, and it is thought that teleosts do not possess type III IFNs. The fish type I IFNs are subdivided into four groups (IFNa to IFNd) according to phylogeny (9, 10), and unlike the single cognate type I IFN receptor complex of mammals (11, 12), fish group I and II IFNs signal through distinct receptor complexes (13). We have recently demonstrated that the amphibian Xenopus laevis type I IFN is a potent antiviral mediator, conferring considerable protection against the emerging ranaviral pathogen frog virus 3 (FV3) (14).
The mammalian type III IFNs (including interferon lambda 1[IFN-λ1], IFN-λ2, and IFN-λ3; also designated IL-28A, IL-28B, and IL-29, respectively) are encoded by five exon/four intron gene transcripts reminiscent of the fish type I IFNs. Intriguingly, although bona fide type III IFNs either do not exist or have not yet been identified in bony fish, amphibians possess both type I IFNs with the five-exon/four-intron gene organization of their fish counterparts, as well as true type III IFNs (15). There has been considerable debate regarding the precise phylogenetic relationships of the teleost type I IFNs to the higher vertebrate type I and III cytokines. In this context and given the key phylogenetic position as intermediate between fish and mammals, together with the possession of fish-like type I and mammalian-like type III IFN genes (15), amphibians are particularly interesting for studying the evolution of antiviral immunity (10, 16, 17).
Aside from the inherent fundamental value, a greater understanding of amphibian antiviral IFN defenses is important in the context of emerging infectious diseases caused by ranavirus pathogens (family Iridoviridae), which are decimating amphibian populations worldwide. Indeed, the worldwide decline in nearly one-third (32%) of all amphibian species represents an imminent threat to the extinction of these organisms (18). Moreover, while these die-offs may be attributed to a range of underlying causes (19, 20), the dramatic increase in ranavirus infections and the resulting mortalities suggest that these pathogens are a significant contributing force behind amphibian declines (18–20). Ranaviruses are large, icosahedral, double-stranded DNA (dsDNA) viruses that manifest in systemic diseases, hemorrhaging, and necrotic cell death within multiple afflicted organs (18). Typically, amphibian tadpoles are more susceptible to, and succumb from, these infections, whereas mature adults are usually more resistant to these pathogens (14, 21–24). Frog virus 3 (FV3) is the type species of the ranavirus genus, and thus FV3 infection of the amphibian Xenopus laevis presents a pertinent research platform for studying the interface between the ranavirus and the amphibian host immune response.
Most notably, considering that the frog kidney epithelium is believed to be a primary site of ranaviral replication (25) and that the mammalian type III IFNs specifically target epithelial cells (6) raises the question of the roles of the functionally uncharacterized amphibian type III IFNs in the context of anti-ranaviral immunity. Accordingly, we utilized the X. laevis FV3 infection model to address the roles of frog type III IFNs in antiviral immunity.
MATERIALS AND METHODS
Animals.
Outbred premetamorphic (developmental stage 54, according to Nieuwkoop and Faber [39]) tadpoles and metamorphic (stage 64) and adult (2 years old) frogs were obtained from our X. laevis research resource for immunology at the University of Rochester (http://www.urmc.rochester.edu/mbi/resources/xenopus-laevis/). All animals were handled under strict laboratory and University Committee on Animal Resources (UCAR) regulations (approval number 100577/2003-151).
Identification of X. laevis type III IFN.
The X. laevis IFN-λ cDNA corresponding to the open reading frame (ORF) was cloned using primers (Table 1) against the Xenopus tropicalis IFN-λ. Briefly, the full-length X. laevis IFN-λ was amplified by reverse transcription-PCR (RT-PCR) using cDNA derived from FV3-infected X. laevis adult spleen as the template. The resulting amplicon was cloned into the pGEM-T sequencing vector (Promega), and five individual clones were sequenced.
TABLE 1.
List of primer sequences
| Primer target (function) | Sequence (5′–3′)a |
|---|---|
| IFN-λ (cloning) | F, ATGGAAATTCCTATCAGACTGGCCGCCATG |
| R, TTCATTATTAGCCCAACACATTACATC | |
| IFN-λ (insect expression) | F, GCTAAGCTTTCCACACAGAAGGCACTGCCACAT |
| R, AGACTCGAGTTCATTATTAGCCCAACACATTAC | |
| DNA Pol II | F, ACGAGCCCGACGAAGACTACA |
| R, TGGTGGTCCTCAGCATCCT | |
| GAPDH | F, GACATCAAGGCCGCCATTAAGACT |
| R, AGATGGAGGAGTGAGTGTCACCAT | |
| IFN | F, GCTGCTCCTGCTCAGTCTCA |
| R, GAAAGCCTTCAGGATCTGTGTGT | |
| IFN-λ | F, TCCCTCCCAACAGCTCATG |
| R, CCGACACACTGAGCGGAAA | |
| IFNLR1 | F, GGAGCCTGATCCCAATGAATTA |
| R, TCTCAAAGCGCACACTAAGG | |
| IL-10R2 | F, TCACCAGCATGGACTCTTTAC |
| R, CTCACAAATGGCTTGGCTTAAT | |
| Mx1 | F, AGCAGTGGTCAACAGGAGCC |
| R, TGTTCCGCCGCTGTTCCTCT | |
| Mx2 | F, GGAACGCCGCACTTGCAGAA |
| R, CGATTAATCCTGGCACCTCC | |
| PKR | F, GCTCACCGGCGGGATTA |
| R, TTCAACTTTATTCATGCGTGCTATC |
F, forward; R, reverse.
Frog virus 3 stocks and animal infections.
Fathead minnow (FHM) cells (American Type Culture Collection; ATCC CCL-42) were maintained in Dulbecco's modified Eagle's medium (DMEM; Invitrogen) supplemented with 10% fetal bovine serum (Invitrogen), penicillin (100 U/ml), and streptomycin (100 μg/ml) at 30°C with 5% CO2. FV3 was grown by a single passage in FMH cells and purified via ultracentrifugation on a 30% sucrose cushion. Tadpole kidneys and A6 cells to be assessed for FV3 loads by plaque assays were subjected to three rounds of sequential freeze-thaw lysis and repeated passages through a 24-gauge needle. All plaque assays were performed on BHK monolayers under an overlay of 1% methylcellulose, as previously described (26).
The production and characterization of recombinant FV3 bearing site-specific deletions of the 18K (ORF 82R) and vIF-2 genes has been previously described (27), while the characterization of ΔvCARD FV3 (where vCARD is viral caspase activation and recruitment domain; open reading frame 64R, nucleotides 75529 to 75816) is presently in review as a separate manuscript. The two recombinant FV3s were generated by homologous recombination; target genes (FV3 genomic location for ORF 52L, nucleotides 57481 to 58548; ORF 64R, 75529 to 75816) were PCR amplified from the FV3 genome and cloned into right (restriction sites XhoI and ClaI) and left (restriction sites SacI and SpeI) sides of cassettes bearing a puromycin (Puro) resistance gene fused with the coding sequence of enhanced green fluorescent protein (EGFP) under the control of FV3 immediate early (IE) 18K gene promoter (18Kprom-Puro-EGFP cassette). Both recombinants were shown to have growth kinetics similar to those of the wild-type (WT) virus when cultured in BHK cells, and both have been confirmed to be of high purity by monitoring fluorescence signal in plaque assays and by diagnostic PCR.
All tadpole infections were achieved by intraperitoneal (i.p.) injection of 1 × 104 FV3 PFU in 10-μl volumes. All adult frog infections were performed i.p. with 5 × 106 FV3 PFU in 100-μl volumes. At 0, 1, 3, and 6 days postinfection, animals were euthanized by immersion in 0.5% tricaine methane sulfonate (MS-222), and tissues and cells were removed and processed for RNA and DNA isolation and PFU analysis to determine respective FV3 loads.
Quantitative-PCR gene expression analysis.
Total RNA and DNA were extracted from frog tissues and cells using TRIzol reagent according to the manufacturer's directions (Invitrogen). All cDNA synthesis was performed using an iScript cDNA synthesis kit according to manufacturer's directions (Bio-Rad, Hercules, CA) using 500 ng of total DNase-treated (Ambion) RNA. Quantitative PCR (qPCR) analysis was performed using 2.5 μl of cDNA templates and 50 ng of DNA templates.
Relative qPCR gene expression analyses of IFN, IFN-λ, Mx1, Mx2, PKR, IFNLR1, and IL-10R2 were performed via the ΔΔCT method (where CT is threshold cycle), with expression examined relative to the glyceraldehyde-3-phosphate dehydrogenase (GAPDH) endogenous control and normalized against the lowest observed expression. To measure FV3 viral loads, absolute quantitative RT-PCR (qRT-PCR) was performed on DNA using a serially diluted standard curve. Briefly, an FV3 viral DNA (vDNA) polymerase II (Pol II) PCR fragment was cloned into the pGEM-T vector (Promega), amplified in bacteria, quantified, and serially diluted to yield 1010 to 101 vDNA Pol II fragment-containing plasmid copies. These dilutions were employed as the standard curve in subsequent absolute qPCR assays of FV3 DNA quantities. All experiments were performed using an ABI 7300 real-time PCR system and PerfeCta SYBR green FastMix, ROX (Quanta). ABI sequence detection system software (SDS) was employed for all expression analysis. All primers were validated prior to use (Table 1).
Generation of rXlIFN and rXlIFN-λ insect expression constructs.
The production of the X. laevis recombinant IFN (rXlIFN) has been previously described (14), and the rXlIFN-λ was generated in the same manner. Briefly, full-length X. laevis IFN and IFN-λ sequences without the signal peptide were PCR amplified from FV3-infected adult X. laevis spleen cDNA using iProof high-fidelity DNA polymerase (Bio-Rad) and primers containing HindIII and XhoI restriction sites, designed to meet the requirements of the pMIB/V5-His A insect expression vector (Invitrogen). PCR products were double digested with HindIII and XhoI and ligated into the pMIB/V5-His A. In-frame insertions of X. laevis IFN and IFN-λ were confirmed by sequencing from both directions.
Production of rXlIFN and rXlIFN-λ.
The expression plasmids were transfected into Sf9 insect cells using Lipofectamine (Invitrogen), and their expression was confirmed by RT-PCR and Western blotting using the V5 epitopes. Sf9 insect cells transfected with rXlIFN- and rXlIFN-λ-pMIB/V5-His A were selected using 10 μg/ml blasticidin, scaled up into 500-ml liquid cultures, and grown for 5 days under blasticidin selection. Culture supernatants were dialyzed overnight at 4°C (150 mM sodium phosphate), concentrated against polyethylene glycol flakes (8 kDa), and dialyzed again. Recombinant proteins were purified by Ni-nitrilotriacetic acid (NTA) agarose chromatography (Qiagen). Bound proteins were washed at high stringency (20 volumes of 0.5% Tween 20, 50 mM sodium phosphate, 500 mM sodium chloride, 100 mM imidazole), followed by washing at low stringency (5 volumes of 0.5% Tween 20, 50 mM sodium phosphate, 500 mM sodium chloride, 100 mM imidazole), and then eluted with 250 mM imidazole. Purity was determined by SDS-PAGE and Western blotting using the V5 epitope. Protein concentration was determined by a Bradford protein assay (Bio-Rad). Protein preparations were aliquoted and stored at 4°C in the presence of a protease inhibitor cocktail (Roche).
The vector control samples were obtained by transfecting Sf9 cells with an empty expression vector and following the same cell culture and protein purification steps as described above.
Cell culture medium.
The ASF culture medium used in these studies has been previously described (28). All cell cultures were established using ASF medium supplemented with 10% fetal bovine serum, 20 μg/ml kanamycin, and 100 U/ml penicillin–100 μg/ml streptomycin (Gibco). Amphibian phosphate-buffered saline (APBS) has been previously described (28).
A6 cell stimulation and infection.
A6 cells (5 × 105 per well of 48-well plates), incubated for 6 h with 100 ng/ml of either rXlIFN, rXlIFN-λ, or an equal volume of vector control, were infected at a multiplicity of infection (MOI) of 0.5 with FV3 for an additional 16 h. Then RNA and DNA were isolated, and cDNA was synthesized. To assess dose-dependent effects of rXlIFN and rXlIFN-λ, 5 × 105 A6 cells were treated with 0.5, 5, 50, 500, or 5,000 ng/ml of either recombinant cytokine for 6 h, infected at an MOI of 0.5 with FV3 for 16 h, and harvested for plaque assays.
Tadpole cytokine stimulation and FV3 infections.
For tadpole gene expression analysis, tadpoles were injected i.p. with 1 μg of rXlIFN, 1 μg of rXlIFN-λ, or an equal volume of a vector control. The following day, tadpoles were euthanized in 0.5% tricaine methane sulfonate (MS-222), and cells and tissues were isolated and processed for RNA.
For short-term protection assays, stage 54 tadpoles (4/treatment group; n = 4 groups) were injected i.p. with 1 μg of rXlIFN, 1 μg of rXlIFN-λ, or an equal volume of the vector control and 6 h later infected with 104 PFU of FV3 in APBS. Plaque assays were performed for peritoneal leukocytes (PLs), kidney, spleen, and liver at 3 and 9 days post-FV3 infection.
For tadpole survival studies, stage 50 tadpoles (12/treatment group; n = 12) were infected as described above and monitored over the course of 60 days. Stage 50 tadpoles were used to ensure that animals did not reach metamorphosis during the experimental period. Tadpoles were checked twice daily, and dead animals were immediately frozen and stored at −20°C for DNA isolation.
Statistical analysis.
Statistical analysis was performed using one-way analysis of variance (ANOVA) and Tukey's post hoc test. A two-sample F test was performed on the A6 cell Mx1 gene expression data. A probability (P) level of <0.05 was considered significant. Vassar Stat was used for statistical computation (http://faculty.vassar.edu/lowry//anova1u.html).
Nucleotide sequence accession number.
The Xenopus tropicalis IFN-λ sequence was submitted to GenBank under accession number KP325221.
RESULTS
Gene expression analysis of X. laevis type I and type III IFNs.
To investigate the biological roles of type III interferons in ectothermic vertebrates, we identified an X. laevis IFN-λ gene homolog and compared its expression by qPCR with the previously identified X. laevis type I IFN, here referred to as IFN (Fig. 1). X. laevis tadpoles (developmental stage 54) exhibited significantly greater IFN-λ gene expression than that of IFN in all examined tissues, with the exception of kidney and intestine (Fig. 1A). IFN-λ transcript levels were highest in the spleen, liver, thymus, and lungs; it was more modest in the kidney and gills and lowest in the intestine. Similar expression patterns were observed in metamorphs (stage 64), with the exception of significantly elevated kidney and decreased thymic IFN-λ gene expression (Fig. 1B and D). The intestinal gene expression levels of the metamorphic type I and type III IFNs were comparable (Fig. 1B). The adult frog type III IFN gene expression was also significantly higher than that of the type I IFN for all tissues examined, excluding intestine (Fig. 1C).
FIG 1.
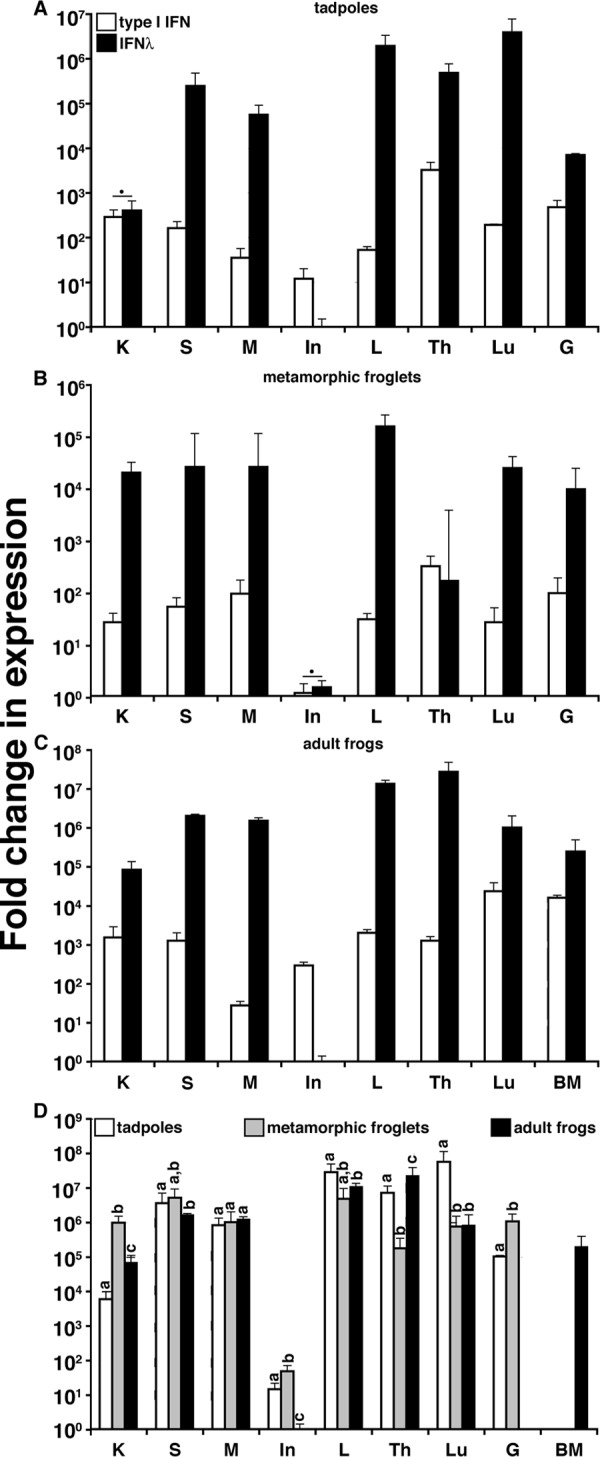
Analysis of X. laevis type I and type III IFN gene expression in tissue. Fold change in IFN expression in tadpoles (stage 54) (A), metamorphic froglets (stage 64) (B), and adult frogs (2 years old) (C) was determined. (D) Comparison of IFN-λ gene expression in tissue of premetamorphic, metamorphic, and postmetamorphic X. laevis. Tissues from three individuals of each stage were examined (n = 3). Letters at the top of the bars indicate tissues exhibiting significantly different (P < 0.05) gene expression levels. IFN-λ gene expression was significantly greater for all tissues with the exception of those marked with a filled circle (P < 0.05). Gene expression was examined relative to the level of the GAPDH endogenous control, and all results are depicted as means ± standard errors of the means. K, kidney; S, spleen; M, muscle; In, intestine; L, liver; Th, thymus; Lu, lung; G, gill; BM, bone marrow.
A comparison of type III IFN gene expression during X. laevis development revealed marked increases of IFN-λ kidney and gill expression of this gene during metamorphosis relative to levels of the larval and adult stages (Fig. 1D). The considerable decrease in thymic IFN-λ gene expression during metamorphosis, followed by its restoration in adult frogs, is consistent with the death of most larval thymocytes during metamorphosis and the differentiation of adult thymocytes after the metamorphic completion (29). In contrast, the decreased metamorphic lung IFN-λ transcript levels persisted into frog adulthood (Fig. 1D).
Kidney IFN-λ gene expression analysis in FV3-infected X. laevis tadpoles and adults.
In our previous efforts to investigate the inefficiency in X. laevis tadpole antiviral immunity during FV3 infections, we were perplexed to find that, despite a meager and delayed type I IFN gene expression response (compared to adult frogs), tadpoles concomitantly exhibit significantly lower FV3 loads than X. laevis adults (14). Given the overall greater expression of the IFN-λ gene than the type I IFN gene in tadpole tissues, we hypothesized that IFN-λ may play a more prominent role in tadpole antiviral immune responses. Accordingly, we examined IFN-λ transcript levels during FV3 infection in tadpole and adult frog kidneys (primary site of FV3 replication). Notably, although adult frogs displayed greater basal kidney IFN-λ transcript levels than tadpoles, IFN-λ gene expression markedly increased (2 logs) as early as 24 h post-FV3 infection, whereas no significant expression increase was detected in infected adult kidneys (Fig. 2A). IFN-λ gene expression in tadpole kidneys remained elevated at 3 days postinfection (p.i.) and returned close to basal levels at 6 days p.i. (Fig. 2A). As previously observed, the FV3 genomic DNA copy number (as assessed by absolute qPCR) substantially increased in virally infected adult kidneys from 1 to 6 days p.i., whereas the tadpole kidney FV3 loads were significantly more modest and did not increase from day 1 to 6 p.i. (Fig. 2B).
FIG 2.
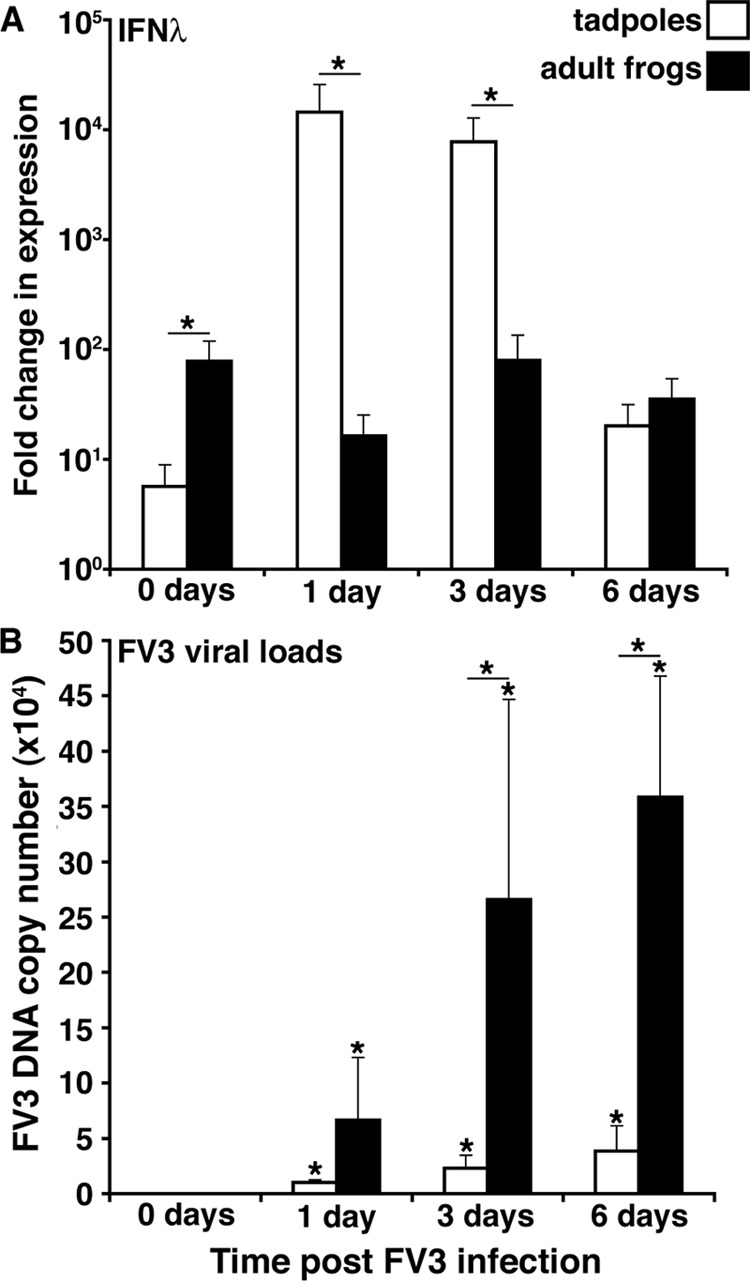
Quantitative analysis of IFN-λ gene expression in tadpole and adult X. laevis (A) and of kidney FV3 DNA loads at 0, 1, 3 and 6 days postinfection (B). X. laevis tadpoles and adults were infected with 1 × 104 and 5 × 106 PFU of FV3, respectively. Tissues were isolated at the indicated times, and qPCR analysis was performed to determine IFN-λ gene expression relative to the level of the GAPDH endogenous control and to determine the FV3 loads in relation to an FV3 vDNA Pol II standard curve. Tissues from five individual animals (n = 5) were assessed for each time point. Results are means ± standard errors of the means. Significant differences in results relative to the control level and between treatment groups (as denoted with a horizontal bar) are indicated (*, P < 0.05).
Analysis of antiviral gene expression and anti-FV3 protection of A6 cultures stimulated with rXlIFN or rXlIFN-λ.
To determine whether the tadpole induction of IFN-λ gene expression during FV3 infections could account for the relatively low FV3 loads, we generated a recombinant form of this cytokine (rXlIFN-λ) and compared its antiviral activity in vitro to that of the previously characterized recombinant X. laevis type I IFN (rXlIFN) (14). To assess the relative antiviral efficacies of rXlIFN-λ and rXlIFN across a range of concentrations, we pretreated the kidney-derived A6 cell line cultures for 6 h with 0.5, 5, 50, 500, and 5,000 ng/ml of either cytokine, infected the cells with FV3, and assessed the viral loads within these cultures by plaque assays (Fig. 3A). With the exception of the lowest tested doses, rXlIFN proved to be more effective than rXlIFN-λ at preventing viral replication across all other tested concentrations (Fig. 3A). Notably, the trend line for the dose-dependent antiviral effects of rXlIFN is substantially steeper (R2 = 0.9692) than that for rXlIFN-λ (R2 = 9457) (Fig. 3A). Based on our previous rXlIFN studies (14) and in accordance with the dose-dependent antiviral effects of rXlIFN and rXlIFN-λ presented here (Fig. 3A), we employed the intermediate 100 ng/ml dose of either cytokine for all subsequent in vitro studies. At this dose, qPCR analysis of FV3 DNA viral loads confirmed that although both recombinant cytokines markedly decreased viral loads in A6 cells, rXlIFN was significantly more protective than rXlIFN-λ (Fig. 3B).
FIG 3.
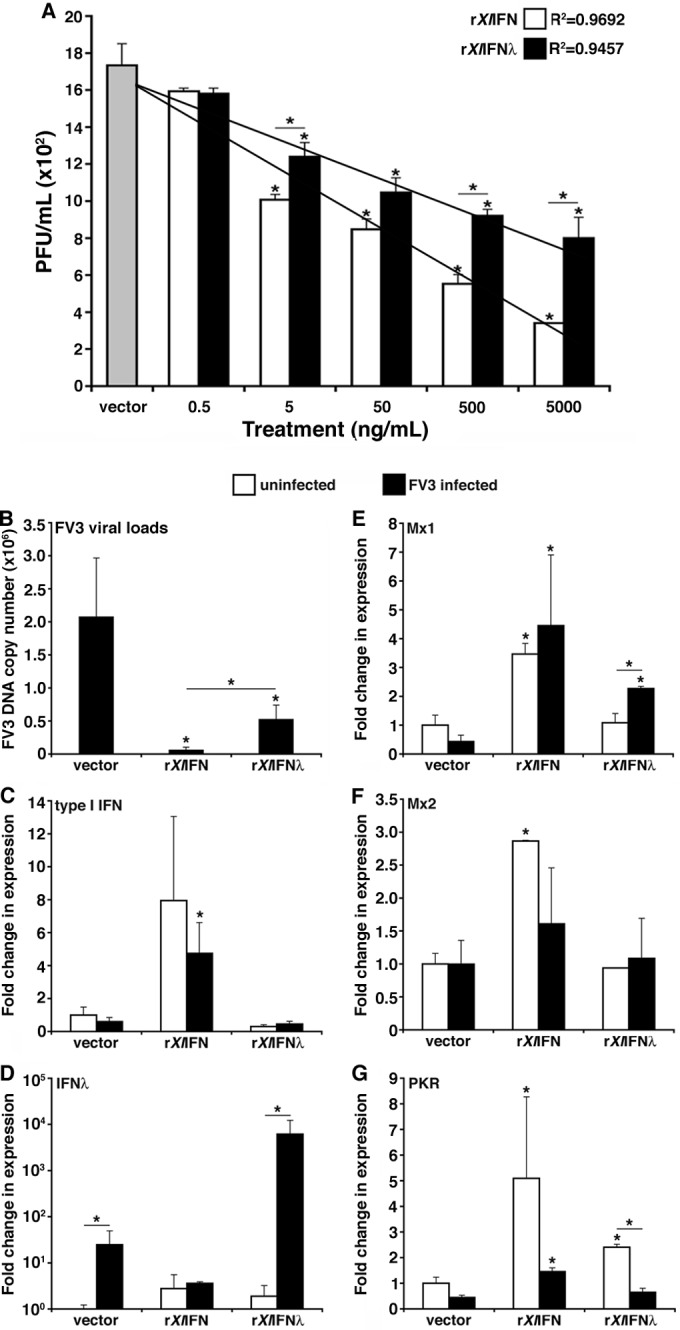
Assessment of the antiviral effects of rXlIFN and rXlIFN-λ on the kidney-derived A6 cell line. (A) A6 cells were pretreated for 6 h with 0.5, 5, 50, 500, or 5,000 ng/ml of either rXlIFN or rXlIFN-λ, infected with FV3 at an MOI of 0.5 for 16 h, and assessed for viral loads by plaque assays. (B to G) A6 cultures were treated with the vector control or 100 ng/ml of either rXlIFN or rXlIFN-λ for 6 h and infected with FV3 at an MOI of 0.5 for an additional 16 h. The FV3 DNA copy number was assessed by absolute qPCR against the FV3 vDNA Pol II (using a vDNA Pol II standard curve) (B). Antiviral qPCR gene expression analysis was performed for type I IFN (C), type III IFN (IFN-λ) (D), Mx1 (E), Mx2 (F), and PKR (G). Gene expression was analyzed relative to the level of the GAPDH endogenous control. Three A6 cell cultures were subjected to each of the experimental conditions (n = 3). Results are means ± standard errors of the means. Significant differences in the results relative to those with the vector control and between treatment groups (as denoted with a horizontal bar) are indicated (*, P < 0.05).
To account for the differences in anti-FV3 protection, we assessed antiviral gene expression in A6 cultures stimulated by either cytokine during steady state and following FV3 infection (Fig. 3C to G). A6 cells treated with rXlIFN but not with rXlIFN-λ exhibited increased type I IFN gene expression, and this was not significantly altered by FV3 infections (Fig. 3C). Remarkably, IFN-λ but not type I IFN gene expression was induced by FV3 infection of A6 cells (Fig. 3C and D). Moreover, pretreatment of A6 cells with rXlIFN-λ resulted in further increases in IFN-λ gene expression following FV3 infection (Fig. 3D). Conversely, although rXlIFN pretreatment induced rXlIFN gene expression, FV3 infection did not significantly increase this rXlIFN-mediated expression (Fig. 3C). It is of note that rXlIFN-λ pretreatment did not induce type I IFN gene expression and vice versa (Fig. 3C and D).
The functional differences between the two IFNs were further evidenced by the distinct IFN-induced changes in Mx1, Mx2, and PKR gene expression responses (Fig. 3E to G). Pretreatment of A6 cultures with rXlIFN considerably increased the expression of the antiviral Mx1 and Mx2 genes without further significant expression changes observed following FV3 infection (Fig. 3E and F). In contrast, rXlIFN-λ pretreatment resulted in significantly increased Mx1 but not Mx2 gene expression upon FV3 infection (Fig. 3E and F). Interestingly, FV3 infections dramatically ablated the gene expression of protein kinase R (PKR) induced by both rXlIFN-λ and rXlIFN pretreatments (Fig. 3G).
Assessment of short-term rXlIFN-λ anti-FV3 protection in X. laevis tadpoles.
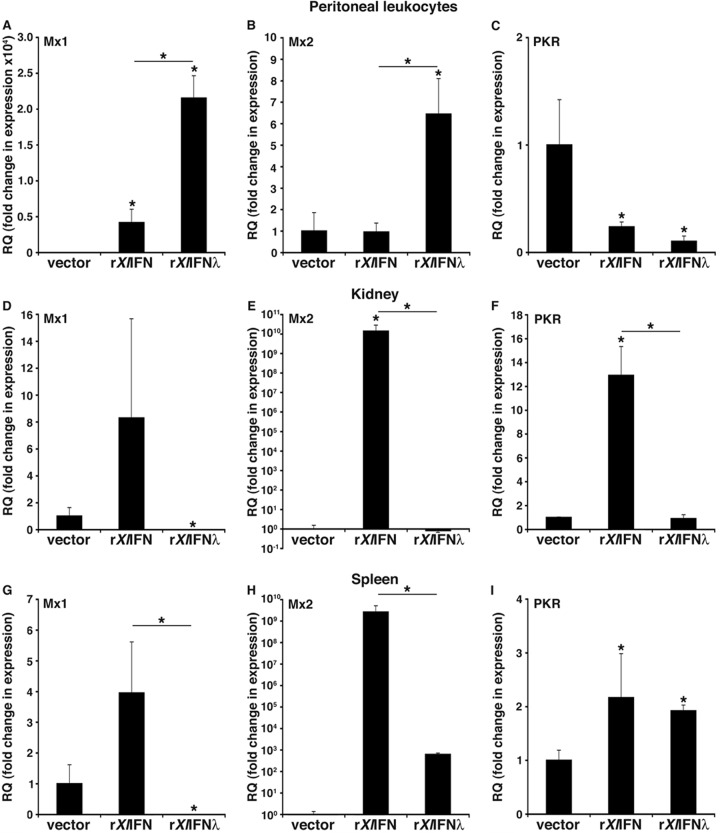
To extend our in vitro findings, we administered rXlIFN, rXlIFN-λ, or the vector control intraperitoneally to X. laevis tadpoles and examined antiviral gene expression in peritoneal leukocytes (PLs), kidney (primary FV3 target), and spleen (central immune organ) 24 h later (Fig. 4). Interestingly, rXlIFN-λ elicited robust Mx1 and Mx2 gene expression responses in PLs, whereas rXlIFN induced only a modest increase of Mx1 and no change in Mx2 mRNA levels (Fig. 4A and B, respectively). Surprisingly, PKR gene expression was decreased in PLs from both rXlIFN- and rXlIFN-λ-treated tadpoles (Fig. 4C).
FIG 4.
Assessment of the antiviral effects of rXlIFN and rXlIFN-λ on X. laevis tadpole peritoneal leukocytes (A to C), kidneys (D to F), and spleens (G to I). Stage 54 tadpoles were i.p. injected with 1 μg of rXlIFN, 1 μg of rXlIFN-λ, or an equal volume of the vector control, and antiviral gene expression was assessed 24 h later in peritoneal leukocytes (PLs), kidneys, and spleens, as indicated at the top of each panel. Gene expression was examined relative to the level of the GAPDH endogenous control, and all results are means ± standard errors of the means. Significant differences in the results relative to those with the vector control and between treatment groups (as denoted with a horizontal bar) are indicated (*, P < 0.05). RQ, relative quantification.
In kidneys, rXlIFN treatments induced marked increases in Mx1, Mx2, and PKR gene expression, whereas rXlIFN-λ administration decreased Mx1 transcript levels and had no significant effect on Mx2 and PKR expression (Fig. 4D to F). Finally, rXlIFN treatment significantly increased the splenic expression of Mx1, Mx2, and PKR, whereas rXlIFN-λ decreased Mx1 but induced Mx2 (albeit significantly less so than rXlIFN) and PKR expression (Fig. 4G to I, respectively). These results further substantiate the functional differences between X. laevis IFN-λ and IFN in antiviral immune responses.
To further compare the antiviral effects of rXlIFN-λ and rXlIFN, we next pretreated tadpoles as described above, infected them with FV3, and assessed FV3 viral loads in kidneys, PLs, spleens, and livers at 3 and 9 days p.i. by plaque assays (Fig. 5). As expected, FV3 replication was markedly higher in kidneys (over 1 log) than in PLs, spleen, or liver (Fig. 5), underlining the importance of this organ for FV3 infections and thus tadpole anti-FV3 protection. Although pretreatment with either recombinant cytokine resulted in similar protective effects in kidneys at 3 days p.i. (2-fold decrease in virus load), prevention of viral replication by rXlIFN was significantly more effective than that with rXlIFN-λ at 9 days p.i. (Fig. 5A). For PLs, the protective effect of pretreatment was detected only at 9 days p.i., and the effects were not significantly different between pretreatment with the two recombinant cytokines (Fig. 5B). In the liver, FV3 loads were significantly diminished by rXlIFN but not rXlIFN-λ pretreatment although viral load also decreased in vector-treated control animals at 9 days p.i. compared to the level at 3 days p.i., suggesting the development of a tadpole immune response more potent at limiting viral dissemination (Fig. 5C). Finally, in the spleen, only rXlIFN-pretreated animals showed significantly decreased FV3 loads at 9 days p.i., whereas animals stimulated with rXlIFN-λ possessed significantly lower spleen viral loads at 3 and 9 days p.i. than the levels detected in the rXlIFN-treated cohorts (Fig. 5D).
FIG 5.
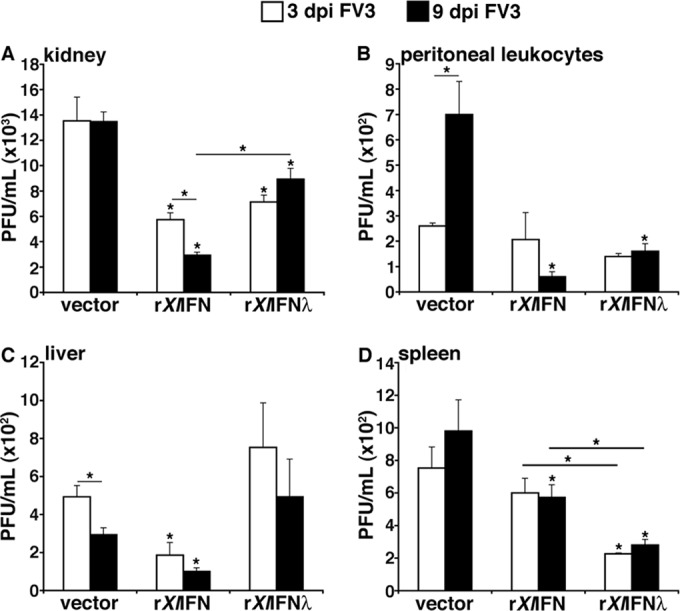
Comparison of rXlIFN and rXlIFN-λ anti-FV3 protection in tadpoles. Stage 54 tadpoles were i.p. injected with 1 μg of rXlIFN, 1 μg of rXlIFN-λ, or an equal volume of the vector control and infected 6 h later with 104 PFU of FV3. Viral loads were determined by plaque assays at 3 and 9 days p.i. (dpi) for kidneys (A), peritoneal leukocytes (B), livers (C), and spleens (D). Four tadpoles (n = 4) were employed for each treatment group. All viral loads are depicted as means ± standard errors of the means. Significant differences in the results relative to those with the vector control and between treatment groups (as denoted with a horizontal bar) are indicated (*, P < 0.05).
It is noteworthy that viral loads in kidney, liver, spleen, and peritoneal leukocytes of FV3-infected tadpoles pretreated with equal doses of the two recombinant cytokines were comparable to those following rXlIFN treatments alone (data not shown), suggesting the absence of additive antiviral effects.
Assessment of long-term rXlIFN-λ anti-FV3 protection of X. laevis tadpoles.
To further compare the antiviral effects of rXlIFN-λ and rXlIFN, we next monitored tadpole survival following FV3 infection of control-, rXlIFN-, and rXlIFN-λ-stimulated animals (Fig. 6). Notably, and consistent with the observed reduction of viral loads, both rXlIFN-λ and rXlIFN treatments resulted in significant increases in tadpole survival, especially during the initial 25 days post-FV3 challenge. However, whereas the survival of rXlIFN-stimulated tadpoles remained greater than that of control animals for the remainder of the 60-day study, after 25 days p.i. the survival of rXlIFN-λ-treated tadpoles drastically decreased to levels comparable to those of vector control-treated animals (Fig. 6A). Furthermore, while the rXlIFN-treated animals had significantly decreased postmortem FV3 DNA loads, rXlIFN-λ-treated tadpoles possessed modestly, but not significantly, diminished FV3 loads compared to those of the vector control animals (Fig. 6B). These results suggest that the anti-FV3 protection conferred by rXlIFN-λ is both less effective and shorter lasting than that of rXlIFN.
FIG 6.

Survival of FV3-infected tadpoles pretreated with either rXlIFN-λ or rXlIFN. Stage 50 tadpoles (12/treatment group; n = 12) were preinjected with 1 μg of rXlIFN, 1 μg of rXlIFN-λ, or an equal volume of the vector control and 6 h later infected with FV3 (104 PFU) or mock infected by APBS injection. Animal survival was monitored over the course of 60 days post-FV3 infection (A), and postmortem viral loads were determined by absolute qPCR against FV3 vDNA Pol II (using a vDNA Pol II standard curve) (B). Results in panel B are means ± standard errors of the means. Results that are significantly different from those of the vector control are indicated (*, P < 0.05).
Analysis of IFN-λ receptor gene expression in healthy and FV3-infected animals.
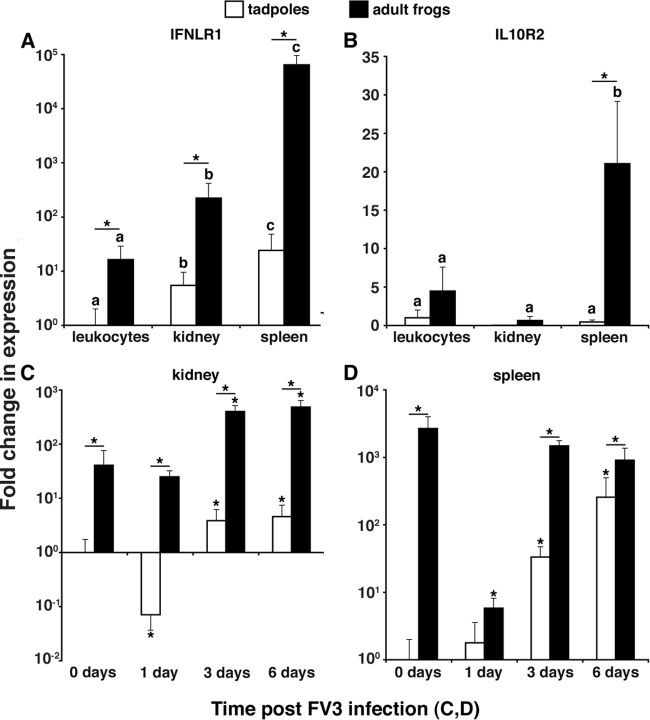
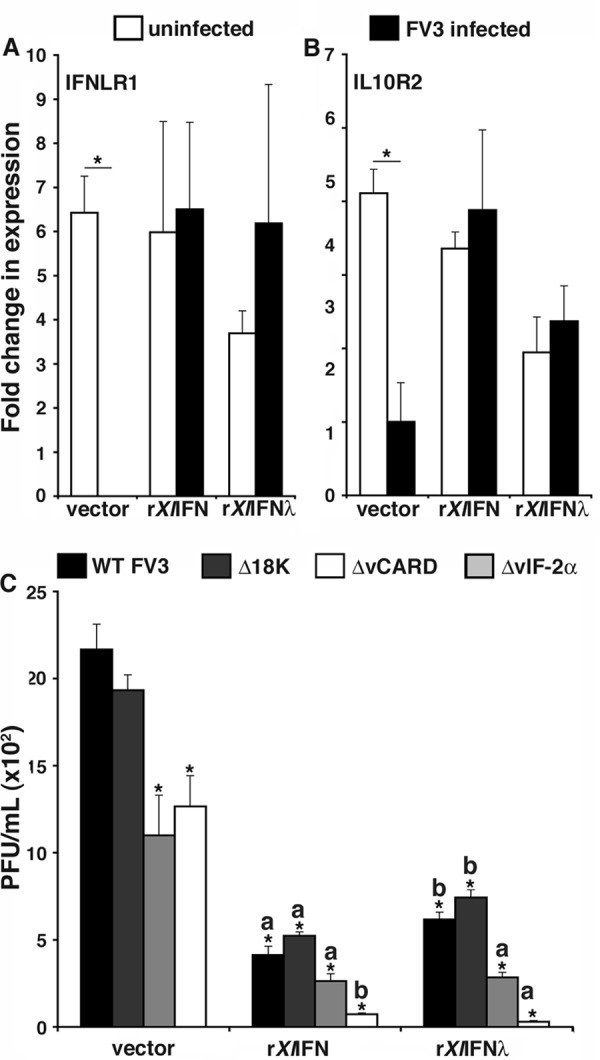
It is well established that mammalian type III IFNs signal by ligating the interferon lambda receptor 1 (IFNLR1), subsequently complexed by the interleukin-10 receptor 2 (IL-10R2), which propagates the cellular signaling (6). To more comprehensively define amphibian type III IFN antiviral immunity, we examined the gene expression of the X. laevis IFN-λ receptors in healthy and FV3-infected X. laevis tadpoles and adults (Fig. 7). The expression levels of both the IFN-λ ligand-binding and signal-propagating chains (IFNLR1 and IL-10R, respectively) were significantly greater in adult PLs, kidneys, and especially spleens than in the respective tadpole tissues (Fig. 7A and B).
FIG 7.
Gene expression analysis of the X. laevis IFN-λ receptors, IFNLR1 and IL-10R2. (A and B) Analysis of IFNLR1 and IL-10R2 gene expression was performed in healthy (stage 54) tadpoles and adults (2 years old). (C and D) Analysis of IFNLR1 expression in tadpole and adult kidney and spleen was performed at 0, 1, 3 and 6 days post-FV3 challenge. Five animals (n = 5) were used for each experimental group. Expression levels were determined relative to the level of the GAPDH endogenous control, and all results are presented as means ± standard errors of the means. Significant differences in the results relative to those with the vector control and between treatment groups (as denoted with a horizontal bar) are indicated (*, P < 0.05).
Intriguingly, IFNLR1 gene expression was significantly decreased in tadpole but not adult frog kidneys at 1 day p.i., whereas at 3 and 6 days p.i., both tadpoles and adults exhibited increased IFNLR1 expression (Fig. 7C). This presumably reflects the previously observed timely leukocyte infiltration of infected kidneys (25). It is noteworthy that the increased kidney IFNLR1 gene expression at 3 and 6 days p.i. was markedly lower in tadpoles than that in adult frogs (1 to 2 logs) (Fig. 7C). Interestingly, while tadpole spleen IFNLR1 gene expression significantly increased with infection progression, the relatively robust adult splenic IFNLR1 levels significantly declined at 1 day p.i. and were restored by 3 days p.i. (Fig. 7D). Whether these splenic gene expression changes are due to gene regulation and/or cell migration is currently unknown.
IL-10R2 gene expression levels in kidneys and spleens of tadpoles and adults were not significantly altered under these experimental conditions and at the times examined (data not shown).
Since FV3 infections resulted in decreased tadpole kidney IFNLR1 gene expression (Fig. 7C), we also examined the IFN-λ receptor gene expression in recombinant cytokine-stimulated, FV3-infected A6 cultures (Fig. 8A). Notably, while FV3 infection significantly decreased A6 cell expression of IFNLR1 and IL-10R2, pretreatment of parallel cultures with either rXlIFN or rXlIFN-λ restored the expression levels of these two receptors in the face of FV3 challenge (Fig. 8A).
FIG 8.
Assessment of A6 cell IFN-λ receptor gene expression and rXlIFN-/rXlIFN-λ antiviral protection against recombinant FV3. (A and B) A6 cells were pretreated with 100 ng/ml of rXlIFN, 100 ng/ml of rXlIFN-λ, or an equal volume of the vector control for 6 h and infected at an MOI of 0.5 with WT FV3 for 16 h before IFNLR1 and IL-10R2 gene expression was assessed by qPCR, using GAPDH as an endogenous control. (C) A6 cells were pretreated with 100 ng/ml of rXlIFN, 100 ng/ml of rXlIFN-λ, or an equal volume of the vector control for 6 h and infected for 16 h at an MOI of 0.5 with either WT FV3, FV3-Δ18K, FV3-ΔvCARD, or FV3-ΔvIF-2α. Cells were subsequently harvested, processed, and assessed for respective viral burdens by plaque assays. All experiments described above employed three A6 cultures per treatment group (n = 3), and all of the results are presented as means ± standard errors of the means. Significant differences in the results relative to those with the vector control and between treatment groups (as denoted with a horizontal bar) are indicated (*, P < 0.05). The statistically different protective effects conferred by rXlIFN and rXlIFN-λ against distinct recombinant FV3 are designated by the letters a and b above the bars, representing relatively more and less significant protection, respectively (P < 0.05).
Susceptibility of recombinant FV3 mutants deficient for putative virulence genes to type I and III IFNs.
It stands to reason that the less effective antiviral capacity of rXlIFN-λ, as observed in our studies, may be specific to FV3, a virus that has coevolved with the amphibian immune system. This notion is supported by our findings that FV3 infections decreased IFNLR1 gene expression (Fig. 7C and 8A). To begin to address this issue, we took advantage of several FV3 recombinants bearing site-specific deletions of putative virulence and/or immune evasion genes. These knockout mutant viruses included deletions of the conserved ranavirus immediate early 18K gene (ORF 82R), a truncated viral homolog of the alpha subunit of eukaryotic initiation factor 2 (eIF-2), vIF-2α (ORF 26R), and a viral protein with a caspase activation and recruitment domain, vCARD (ORF 64R). Both FV3-Δ18K and FV3-ΔvIF-2α recombinants were previously described and shown to contribute to FV3 virulence in vivo in tadpoles (27). We have recently generated an FV3-ΔvCARD recombinant that shows unaffected growth kinetics in vitro in BHK cells (F. De Jesús Andino, L. Grayfer, G. Chen, V. Chinchar, and J. Robert, unpublished data). We hypothesized that one or several of these deleted FV3 genes may target the antiviral effects elicited by IFN-λ. Accordingly, A6 cultures were pretreated with rXlIFN-λ, rXlIFN, or a vector control and then infected with the WT or one of the recombinant viruses (Fig. 8B). Notably, FV3-ΔvIF-2α and FV3-ΔvCARD but not FV3-Δ18K showed a partial replication defect in A6 cells, and this defect was more pronounced after pretreatment with either rXlIFN-λ or rXlIFN (Fig. 8B). Interestingly, rXlIFN-λ was significantly more effective (P = 0.008) against FV3-ΔvIF-2α and was as potent as rXlIFN at inhibiting FV3-ΔvCARD replication (Fig. 8B). These results strongly suggest that the vIF-2α and vCARD FV3 genes are critically involved in resistance to IFN-λ- and IFN-mediated antiviral responses, whereas 18K-mediated virulence is IFN independent and here serves as an additional control.
DISCUSSION
This report marks the first functional characterization of a type III IFN in an ectothermic vertebrate, the amphibian Xenopus laevis. Our findings are particularly relevant, considering the key position of amphibians in vertebrate phylogeny and evolution of antiviral interferon immunity. In this regard, a hallmark characteristic of fish and amphibian type I IFNs is the five-exon/four-intron genomic organization, not shared by the distinct intronless avian, mammalian, and reptilian type I IFNs (10, 16, 17). Moreover, in light of the complex evolutionary relationships of the teleost type I IFNs to higher vertebrate type I and/or type III IFNs (4, 15, 16, 30), the fact that amphibians possess both fish-like type I IFNs and bona fide type III IFNs (15) is particularly compelling. Provided that teleosts indeed do not possess type III IFNs, this implies that the divergence of type I and III IFNs took place prior to or during the emergence of tetrapods (15) and brings into question the relative biological roles of the amphibian type I IFNs compared to those of fish. Here, we report that while an amphibian type III IFN appears to be less effective than a type I IFN in antiviral defense, this inefficiency may stem from an immune evasion strategy specific to FV3. Since rapid and robust IFN-λ gene expression is induced in X. laevis tadpoles in response to FV3, this cytokine may predominate antiviral defenses during early amphibian life. Moreover, our findings indicate that FV3 not only decreases kidney IFNLR1 gene expression early on during infection but also counteracts the downstream antiviral cascades initiated by IFN-λ. Thus, it is possible that, in comparison to the delayed and modest FV3-induced tadpole type I IFN expression (14), the prompt and robust IFN-λ response in tadpoles but not adults may reduce the initial FV3 expansion prior to FV3 host evasion, explaining the relatively modest tadpole FV3 loads. The current absence of X. laevis-specific anti-IFN and anti-IFN-λ antibodies has prevented us from addressing whether the differences in gene expression levels correspond to differences in the respective IFN cytokine protein levels. It will be interesting to revisit this notion upon reagent availability.
It is interesting that that while both rXlIFN and rXlIFN-λ elicited changes in antiviral gene expression in the kidney-derived A6 cell line, the magnitudes of these expression changes were more prominent following rXlIFN stimulation. Similarly, tadpole kidney and spleen expression of antiviral genes was more robust following rXlIFN than that following rXlIFN-λ stimulation. In contrast, peritoneal leukocytes from rXlIFN-λ-administered animals exhibited substantially greater expression of Mx1 and Mx2. This is a bit paradoxical, considering that our expression studies indicate that tadpole kidney and spleen tissues possessed greater IFNLR1 expression levels. Possibly, the kinetics of rXlIFN- and rXlIFN-λ-elicited antiviral gene expression are distinct, whereby rXlIFN-λ may actually induce greater antiviral gene expression at distinct times. In support of this notion and in corroboration of the high splenic IFNLR1 expression, it is noteworthy that rXlIFN-λ-treated tadpoles actually exhibited significantly lower FV3 loads than the rXlIFN-administered animals. Again, this brings into question the absolute efficacies of the X. laevis type I and type III IFNs since we observed FV3-induced downregulation of IFNLR1 expression in the tadpole kidney but not spleen, which correlates with the relatively less effective rXlIFN-λ protection of tadpole kidneys and more effective splenic protection.
Our previous investigations suggested that susceptibility of X. laevis tadpoles to FV3 was marked by delayed and meager antiviral (14) and inflammatory (31) responses compared to those of adults. The present evidence of rapid and greater IFN-λ gene expression in response to FV3 infection warrants a reevaluation of this hypothesis. It stands to reason that tadpoles have an intact and timely antiviral response in the form of IFN-λ, which may be effective against less proficient pathogens than ranaviruses. Indeed, rXlIFN-λ was as potent as rXlIFN in inhibiting FV3-ΔvCARD and even more potent at inhibiting the FV3-ΔvIF-2α recombinants. The sensitivity of these two FV3 mutants to the IFN response is also supported by their partially defective replication in vector control-treated A6 cells compared to that in wild-type or 18K knockout FV3. In this regard, it is interesting that FV3 infection of A6 cells results is greater gene expression of IFN-λ than of IFN.
These results are also interesting since the FV3 vIF-2α gene is truncated and lacks the protein kinase R N-terminal binding and central helicase domains (27). Nonetheless, FV3-ΔvIF-2α exhibits reduced replication and lower mortality in infected X. laevis tadpoles (27) and here is severely impaired in overcoming the antiviral effects of IFN and especially IFN-λ. Notably, several other ranaviruses including the epizootic haematopoietic necrosis virus (EHNV) (32), the Ambystoma tigrinum virus (ATV) (33), and the Rana catesbeiana virus Z (RCV-Z) (34), encode full-length vIF-2α genes. Moreover, both the ATV and the RCV-Z vIF-2α gene products are thought to function as pseudosubstrates for the cellular protein kinase R by inhibiting its phosphorylation of the cellular eIF-2α translation factor. While it remains unclear whether the truncated vIF-2α may be expressed as a chimeric product with an adjacent ORF or whether it is capable of blocking PKR phosphorylation as a truncated protein, it is clear that this truncated FV3 vIF-2α gene is critical for overcoming the IFN-induced antiviral state.
Substantially less is known regarding the ranavirus vCARD genes. The 10-kDa vCARD gene product contains a caspase activation and recruitment domain (CARD) motif that impairs interactions between other CARD-containing cellular proteins (35, 36). Known cellular signaling moieties possessing such domains include proapoptotic proteins, proinflammatory molecules, and, most notably, proteins participating in cellular interferon responses (37, 38). It has been postulated that the ranavirus vCARD interacts with one or more of these signaling molecules to abrogate cellular antiviral responses, and indeed our results indicate that the FV3 vCARD is crucial to overcoming cellular antiviral states induced by type I and type III IFNs.
It is interesting to consider the possibility that, since tadpoles do not readily upregulate type I IFN expression but undergo such drastic type III IFN gene responses to a viral infection, ranaviruses coevolved to dampen the tadpole type III responses and the adult frog type I IFN immunity through virulence determinants such as vIF-2α and vCARD. Both the relative antiviral efficacy of rXlIFN and the inefficiency of rXlIFN-λ against tadpole FV3 infections may reflect this. Indeed, our observations that both cytokines are nearly equally effective at inhibiting FV3 kidney replication and tadpole survival early in infection support this notion. Gaining further insights into the amphibian type I and type III IFN responses is imperative not only to defining the limitations within these immune mechanisms during ranaviral infections but also to gaining a greater appreciation for the evolutionary origins of our own antiviral defenses.
ACKNOWLEDGMENTS
This work was supported by R24-AI-059830. L.G. was supported by an LSRF PDF from the Howard Hughes Medical Institute.
We thank Tina Martin for animal husbandry.
REFERENCES
- 1.Sadler AJ, Williams BR. 2008. Interferon-inducible antiviral effectors. Nat Rev Immunol 8:559–568. doi: 10.1038/nri2314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Grayfer L, Garcia EG, Belosevic M. 2010. Comparison of macrophage antimicrobial responses induced by type II interferons of the goldfish (Carassius auratus L.). J Biol Chem 285:23537–23547. doi: 10.1074/jbc.M109.096925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Levraud JP, Boudinot P, Colin I, Benmansour A, Peyrieras N, Herbomel P, Lutfalla G. 2007. Identification of the zebrafish IFN receptor: implications for the origin of the vertebrate IFN system. J Immunol 178:4385–4394. doi: 10.4049/jimmunol.178.7.4385. [DOI] [PubMed] [Google Scholar]
- 4.Zou J, Tafalla C, Truckle J, Secombes CJ. 2007. Identification of a second group of type I IFNs in fish sheds light on IFN evolution in vertebrates. J Immunol 179:3859–3871. doi: 10.4049/jimmunol.179.6.3859. [DOI] [PubMed] [Google Scholar]
- 5.Donnelly RP, Kotenko SV. 2010. Interferon-lambda: a new addition to an old family. J Interferon Cytokine Res 30:555–564. doi: 10.1089/jir.2010.0078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hamming OJ, Gad HH, Paldan S, Hartmann R. 2010. Lambda interferons: new cytokines with old functions. Pharmaceuticals 3:795–809. doi: 10.3390/ph3040795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jaks E, Gavutis M, Uze G, Martal J, Piehler J. 2007. Differential receptor subunit affinities of type I interferons govern differential signal activation. J Mol Biol 366:525–539. doi: 10.1016/j.jmb.2006.11.053. [DOI] [PubMed] [Google Scholar]
- 8.Uze G, Schreiber G, Piehler J, Pellegrini S. 2007. The receptor of the type I interferon family. Curr Top Microbiol Immunol 316:71–95. doi: 10.1007/978-3-540-71329-6_5. [DOI] [PubMed] [Google Scholar]
- 9.Chang M, Nie P, Collet B, Secombes CJ, Zou J. 2009. Identification of an additional two-cysteine containing type I interferon in rainbow trout Oncorhynchus mykiss provides evidence of a major gene duplication event within this gene family in teleosts. Immunogenetics 61:315–325. doi: 10.1007/s00251-009-0366-y. [DOI] [PubMed] [Google Scholar]
- 10.Sun B, Robertsen B, Wang Z, Liu B. 2009. Identification of an Atlantic salmon IFN multigene cluster encoding three IFN subtypes with very different expression properties. Dev Comp Immunol 33:547–558. doi: 10.1016/j.dci.2008.10.001. [DOI] [PubMed] [Google Scholar]
- 11.Li Z, Strunk JJ, Lamken P, Piehler J, Walz T. 2008. The EM structure of a type I interferon-receptor complex reveals a novel mechanism for cytokine signaling. J Mol Biol 377:715–724. doi: 10.1016/j.jmb.2007.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Samuel CE. 2001. Antiviral actions of interferons. Clin Microbiol Rev 14:778–809. doi: 10.1128/CMR.14.4.778-809.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Aggad D, Mazel M, Boudinot P, Mogensen KE, Hamming OJ, Hartmann R, Kotenko S, Herbomel P, Lutfalla G, Levraud JP. 2009. The two groups of zebrafish virus-induced interferons signal via distinct receptors with specific and shared chains. J Immunol 183:3924–3931. doi: 10.4049/jimmunol.0901495. [DOI] [PubMed] [Google Scholar]
- 14.Grayfer L, De Jesus Andino F, Robert J. 2014. The amphibian (Xenopus laevis) type I interferon response to frog virus 3: new insight into ranavirus pathogenicity. J Virol 88:5766–5777. doi: 10.1128/JVI.00223-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Qi Z, Nie P, Secombes CJ, Zou J. 2010. Intron-containing type I and type III IFN coexist in amphibians: refuting the concept that a retroposition event gave rise to type I IFNs. J Immunol 184:5038–5046. doi: 10.4049/jimmunol.0903374. [DOI] [PubMed] [Google Scholar]
- 16.Robertsen B. 2006. The interferon system of teleost fish. Fish Shellfish Immunol 20:172–191. doi: 10.1016/j.fsi.2005.01.010. [DOI] [PubMed] [Google Scholar]
- 17.Robertsen B, Bergan V, Rokenes T, Larsen R, Albuquerque A. 2003. Atlantic salmon interferon genes: cloning, sequence analysis, expression, and biological activity. J Interferon Cytokine Res 23:601–612. doi: 10.1089/107999003322485107. [DOI] [PubMed] [Google Scholar]
- 18.Chinchar VG. 2002. Ranaviruses (family Iridoviridae): emerging cold-blooded killers. Arch Virol 147:447–470. doi: 10.1007/s007050200000. [DOI] [PubMed] [Google Scholar]
- 19.Williams T, Barbosa-Solomieu V, Chinchar VG. 2005. A decade of advances in iridovirus research. Adv Virus Res 65:173–248. doi: 10.1016/S0065-3527(05)65006-3. [DOI] [PubMed] [Google Scholar]
- 20.Chinchar VG, Hyatt A, Miyazaki T, Williams T. 2009. Family Iridoviridae: poor viral relations no longer. Curr Top Microbiol Immunol 328:123–170. doi: 10.1007/978-3-540-68618-7_4. [DOI] [PubMed] [Google Scholar]
- 21.Bayley AE, Hill BJ, Feist SW. 2013. Susceptibility of the European common frog Rana temporaria to a panel of ranavirus isolates from fish and amphibian hosts. Dis Aquat Organ 103:171–183. doi: 10.3354/dao02574. [DOI] [PubMed] [Google Scholar]
- 22.Hoverman JT, Gray MJ, Miller DL. 2010. Anuran susceptibilities to ranaviruses: role of species identity, exposure route, and a novel virus isolate. Dis Aquat Organ 89:97–107. doi: 10.3354/dao02200. [DOI] [PubMed] [Google Scholar]
- 23.Landsberg JH, Kiryu Y, Tabuchi M, Waltzek TB, Enge KM, Reintjes-Tolen S, Preston A, Pessier AP. 2013. Co-infection by alveolate parasites and frog virus 3-like ranavirus during an amphibian larval mortality event in Florida, USA. Dis Aquat Organ 105:89–99. doi: 10.3354/dao02625. [DOI] [PubMed] [Google Scholar]
- 24.Reeve BC, Crespi EJ, Whipps CM, Brunner JL. 2013. Natural stressors and ranavirus susceptibility in larval wood frogs (Rana sylvatica). Ecohealth 10:190–200. doi: 10.1007/s10393-013-0834-6. [DOI] [PubMed] [Google Scholar]
- 25.Morales HD, Robert J. 2007. Characterization of primary and memory CD8 T-cell responses against ranavirus (FV3) in Xenopus laevis. J Virol 81:2240–2248. doi: 10.1128/JVI.01104-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Morales HD, Abramowitz L, Gertz J, Sowa J, Vogel A, Robert J. 2010. Innate immune responses and permissiveness to ranavirus infection of peritoneal leukocytes in the frog Xenopus laevis. J Virol 84:4912–4922. doi: 10.1128/JVI.02486-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chen G, Ward BM, Yu KH, Chinchar VG, Robert J. 2011. Improved knockout methodology reveals that frog virus 3 mutants lacking either the 18K immediate-early gene or the truncated vIF-2α gene are defective for replication and growth in vivo. J Virol 85:11131–11138. doi: 10.1128/JVI.05589-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Robert J, Abramowitz L, Gantress J, Morales HD. 2007. Xenopus laevis: a possible vector of ranavirus infection? J Wildl Dis 43:645–652. doi: 10.7589/0090-3558-43.4.645. [DOI] [PubMed] [Google Scholar]
- 29.Robert J, Ohta Y. 2009. Comparative and developmental study of the immune system in Xenopus. Dev Dyn 238:1249–1270. doi: 10.1002/dvdy.21891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lutfalla G, Roest Crollius H, Stange-Thomann N, Jaillon O, Mogensen K, Monneron D. 2003. Comparative genomic analysis reveals independent expansion of a lineage-specific gene family in vertebrates: the class II cytokine receptors and their ligands in mammals and fish. BMC Genomics 4:29. doi: 10.1186/1471-2164-4-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.De Jesus Andino F, Chen G, Li Z, Grayfer L, Robert J. 2012. Susceptibility of Xenopus laevis tadpoles to infection by the ranavirus frog-virus 3 correlates with a reduced and delayed innate immune response in comparison with adult frogs. Virology 432:435–443. doi: 10.1016/j.virol.2012.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Essbauer S, Bremont M, Ahne W. 2001. Comparison of the eIF-2α homologous proteins of seven ranaviruses (Iridoviridae). Virus Genes 23:347–359. doi: 10.1023/A:1012533625571. [DOI] [PubMed] [Google Scholar]
- 33.Jancovich JK, Jacobs BL. 2011. Innate immune evasion mediated by the Ambystoma tigrinum virus eukaryotic translation initiation factor 2α homologue. J Virol 85:5061–5069. doi: 10.1128/JVI.01488-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rothenburg S, Chinchar VG, Dever TE. 2011. Characterization of a ranavirus inhibitor of the antiviral protein kinase PKR. BMC Microbiol 11:56. doi: 10.1186/1471-2180-11-56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kawai T, Akira S. 2009. The roles of TLRs, RLRs and NLRs in pathogen recognition. Int Immunol 21:317–337. doi: 10.1093/intimm/dxp017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kawai T, Akira S. 2010. The role of pattern-recognition receptors in innate immunity: update on Toll-like receptors. Nat Immunol 11:373–384. doi: 10.1038/ni.1863. [DOI] [PubMed] [Google Scholar]
- 37.Besch R, Poeck H, Hohenauer T, Senft D, Hacker G, Berking C, Hornung V, Endres S, Ruzicka T, Rothenfusser S, Hartmann G. 2009. Proapoptotic signaling induced by RIG-I and MDA-5 results in type I interferon-independent apoptosis in human melanoma cells. J Clin Invest 119:2399–2411. doi: 10.1172/JCI37155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Meylan E, Curran J, Hofmann K, Moradpour D, Binder M, Bartenschlager R, Tschopp J. 2005. Cardif is an adaptor protein in the RIG-I antiviral pathway and is targeted by hepatitis C virus. Nature 437:1167–1172. doi: 10.1038/nature04193. [DOI] [PubMed] [Google Scholar]
- 39.Nieuwkoop PD, Faber J. 1967. Normal tables of Xenopus laevis (Daudin). North-Holland, Amsterdam, The Netherlands. [Google Scholar]