ABSTRACT
The replication of hepatitis C virus (HCV) is uniquely dependent on a host microRNA, miR-122. Previous studies using genotype 1a H77S.3 virus demonstrated that miR-122 acts in part by protecting the RNA genome from 5′ decay mediated by the cytoplasmic 5′ exoribonuclease, Xrn1. However, this finding has been challenged by a recent report suggesting that a predominantly nuclear exoribonuclease, Xrn2, mediates the degradation of genotype 2a JFH1 RNA. Here, we dissect the roles of these two 5′ exoribonucleases in restricting the replication of different HCV strains and mediating the decay of HCV RNA. Small interfering RNA (siRNA) depletion experiments indicated that Xrn1 restricts replication of all HCV strains tested: JFH1, H77S.3, H77D (a robustly replicating genotype 1a variant), and HJ3-5 (a genotype 1a/2a chimeric virus). In contrast, the antiviral effects of Xrn2 were limited to JFH1 and H77D viruses. Moreover, such effects were not apparent in cells infected with a JFH1 luciferase reporter virus. Whereas Xrn1 depletion significantly slowed decay of JFH1 and HJ3-5 RNAs, Xrn2 depletion marginally enhanced the JFH1 RNA half-life and had no effect on HJ3-5 RNA decay. The positive effects of Xrn1 depletion on JFH1 replication were largely redundant and nonadditive with those of exogenous miR-122 supplementation, whereas Xrn2 depletion acted additively and thus independently of miR-122. We conclude that Xrn1 is the dominant 5′ exoribonuclease mediating decay of HCV RNA and that miR-122 provides protection against it. The restriction of JFH1 and H77D replication by Xrn2 is likely indirect in nature and possibly linked to cytopathic effects of these robustly replicating viruses.
IMPORTANCE HCV is a common cause of liver disease both within and outside the United States. Its replication is dependent upon a small, liver-specific noncoding RNA, miR-122. Although this requirement has been exploited for the development of an anti-miR-122 antagomir as a host-targeting antiviral, the molecular mechanisms underpinning the host factor activity of miR-122 remain incompletely defined. Conflicting reports suggest miR-122 protects the viral RNA against decay mediated by distinct cellular 5′ exoribonucleases, Xrn1 and Xrn2. Here, we compare the roles of these two exoribonucleases in HCV-infected cells and confirm that Xrn1, not Xrn2, is primarily responsible for decay of RNA in cells infected with multiple virus strains. Our results clarify previously published research and add to the current understanding of the host factor requirement for miR-122.
INTRODUCTION
Persistent infections with hepatitis C virus (HCV) are a common cause of chronic and life-threatening liver diseases, including cirrhosis and hepatocellular carcinoma (1). A positive-strand RNA virus classified within the genus Hepacivirus in the family Flaviviridae, HCV typically infects surreptitiously, causing little overt disease until late in the course of the infection, often several decades later. The virus is largely, if not exclusively, hepatotropic, but viral RNA and proteins are present only at low abundance in hepatocytes, making their detection difficult by conventional methods (2). HCV also replicates very inefficiently in cell culture in the absence of adaptive mutations, even in cells lacking strong innate immune responses (3, 4). In part, this reflects an unusual sensitivity to oxidative membrane damage that downregulates replication and maintains HCV gene expression at low levels (4). A notable exception is JFH1 virus, a genotype 2a isolate that is resistant to lipid peroxidation and replicates robustly in Huh-7 human hepatoma cells without cell culture-adaptive mutations (4–6). To a much greater extent than other HCV strains, infection with JFH1 causes significant cell injury, resulting in G1 cell cycle arrest and apoptosis (7–9). Nonetheless, because of its robust replication phenotype, many laboratories have come to rely on JFH1 for cell-based studies of virus-host interactions.
Many host factors are usurped by HCV to support its replication (10–13). Among these is a liver-specific microRNA (miRNA), miR-122 (14, 15). miR-122 binds two sites in the viral genome close to its 5′ terminus, recruiting argonaute 2 protein (AGO2) to the viral RNA (16, 17). In addition to acting directly to stimulate viral RNA synthesis (18), miR-122 forms a ternary complex with AGO2 that protects the viral genome from 5′ exonucleolytic decay in infected cells (17, 19). Thus, supplementing the endogenous abundance of miR-122 in hepatoma cells by transfection of synthetic duplex miR-122 increases the half-life (t1/2) of viral RNA following pharmacologic arrest of new viral RNA synthesis in infected cells, while the opposite effect, a reduction in the HCV RNA t1/2, can be induced by transfection of antisense oligoribonucleotides complementary to the miR-122 guide strand (17).
Previous studies in our laboratory demonstrated that the major cytosolic 5′ exoribonuclease, Xrn1, is responsible for the degradation of HCV RNA in infected cells (19). These studies used a cell culture-adapted genotype 1a virus, H77S.3 (20), that replicates less efficiently than JFH1. H77S.3 virus does not induce G1 arrest and also causes substantially less apoptosis in Huh-7 cells than JFH1 (9). RNA interference (RNAi)-mediated depletion of Xrn1 slowed the decay of H77S.3 RNA in infected cells and also ablated the effect of miR-122 supplementation on the stability of the genome (19). In contrast, depletion of components of the 3′ exosome complex, also involved in host mRNA decay, was without effect. Because Xrn1 depletion and miR-122 supplementation similarly, but nonadditively, enhanced the stability of the HCV genome, we concluded that miR-122 protects the viral RNA from 5′ decay mediated primarily by Xrn1 (19).
This conclusion was recently challenged by Sedano and Sarnow (21), who suggested that Xrn2, a well-studied nuclear 5′ exoribonuclease (22), is responsible for the degradation of HCV RNA. These investigators adopted an experimental approach similar to that we had described previously (17, 19), examining the decay of viral RNA in Xrn2-depleted cells after arresting viral RNA synthesis with a potent NS5B polymerase inhibitor. However, they used the more cytopathic JFH1 strain of HCV for their studies (21). The identification of Xrn2 was intriguing, as it has not previously been shown to have a cytoplasmic function, whereas the HCV life cycle is thought to be exclusively cytoplasmic (22, 23). However, Sedano and Sarnow (21) did not compare the effects of Xrn1 versus Xrn2 depletion on JFH1 replication or the rate of decay of JFH1 RNA, leaving unresolved the relative contributions of the two 5′ exoribonucleases to decay of viral RNA in JFH1-infected cells.
Here, we compare the effects of Xrn1 and Xrn2 depletion on replication of JFH1 and other HCV strains, including HJ3-5, a chimeric virus with the structural proteins of H77S.3 and the nonstructural proteins and noncoding RNA sequences of JFH1 (24, 25). We measure the impact of depleting these host exoribonucleases on the decay of the HCV genome in infected cells following arrest of viral RNA synthesis. While we confirm that Xrn2 restricts JFH1 replication, we show Xrn1 to be the dominant 5′ exoribonuclease responsible for decay of viral RNA in both JFH1- and HJ3-5-infected cells. Our data indicate that the antiviral effects of Xrn2 are limited to more cytopathic HCV strains and, unlike Xrn1, are not countered by miR-122 supplementation.
MATERIALS AND METHODS
Cells, viruses, plasmids, and reagents.
Huh-7.5 and Huh-7 cells were maintained as described previously (17, 19). HJ3-5 and JFH viruses (the latter containing a single, cell culture-adaptive Gln1251-Leu mutation in NS3) have been described previously (5, 25, 26). Plasmids containing infectious molecular clones of H77D virus and reporter viruses expressing Gaussia princeps luciferase (GLuc), pH77S.3/GLuc2A, pHJ3-5/GLuc2A, and pJFH1/GLuc2A, have also been described (4, 20). RNA transcripts were prepared from these plasmid DNAs as reported previously (19). Sofosbuvir was a gift from Ann Sluder, Scynexis, Inc.
Transfections.
Small interfering RNA (siRNA) pools targeting Xrn1, Xrn2, and a control siRNA (siCtrl) pool (Dharmacon) were transfected using siLentfect Lipid Reagent (Bio-Rad). Duplex synthetic miRNAs (50 nM) (4, 15, 27) were also transfected using siLentfect Lipid Reagent where indicated. For reporter virus experiments, in vitro-transcribed HCV RNA (1.25 μg) (15) was transfected into 5 × 105 Huh-7.5 cells using the TransIT mRNA kit (Mirus Bio).
Preparation of cytoplasmic and nuclear lysates.
Huh-7.5 cells were harvested in lysis buffer A (150 mM KCl, 25 mM Tris-HCl [pH 7.5], 5 mM EDTA, 1% Triton X-100, 2 mM dithiothreitol [DTT], Complete Protease Inhibitor Mixture [Roche]). The lysates were centrifuged for 5 min at 1,000 × g at 4°C. The supernatants were collected as cytoplasmic lysates. The nuclear pellet was washed with phosphate-buffered saline (PBS) and lysed in buffer B (500 mM KCl, 25 mM Tris-HCl [pH 7.5], 2 mM EDTA, 1% NP-40, 0.1% SDS, Complete Protease Inhibitor Mixture [Roche]). The cytoplasmic and nuclear lysates were clarified by centrifugation at 17,000 × g for 10 min at 4°C.
RT-qPCR.
To quantify HCV RNA, cDNA was generated by reverse transcription (RT) using oligo(dT) and an HCV-specific primer (5′-GAAGAGATATCGGCCGCAAA-3′) targeting the NS5B region of the genome and SuperScript III Reverse Transcriptase (Invitrogen). Quantitative PCR (qPCR) analysis was carried out using iTaq SYBR green Supermix with the CFX96 system (Bio-Rad). HCV RNA abundance was determined using PCR primers targeting the 5′ untranslated region (UTR) (5′-CATGGCGTTAGTATGAGTGTCGT-3′ and 5′-CCCTATCAGGCAGTACCACAA-3′, and normalized to the abundance of β-actin mRNA (primers: 5′-GTCACCGGAGTCCATCACG-3′ and 5′-GACCCAGATCATGTTTGAGACC-3′).
Gaussia luciferase assay.
Cell culture supernatant fluids were collected at intervals following transfection of reporter virus RNAs or infection with reporter viruses, and the cells were refed with fresh medium. Secreted GLuc activity was measured using the Biolux Gaussia Luciferase assay kit (New England BioLabs) as previously described (19).
Immunoblots.
Immunoblotting was carried out using standard methods with the following antibodies: mouse monoclonal antibody (MAb) to HCV core protein (Pierce), rabbit polyclonal antibody (PAb) to Xrn1 (Bethyl Laboratories), rabbit polyclonal antibody to Xrn2 (ProteinTech), and goat PAb to lamin A (Santa Cruz). Protein bands were visualized with an Odyssey Infrared Imaging System (Li-Cor Biosciences).
Confocal immunofluorescence microscopy.
Cells were fixed with 4% paraformaldehyde for 30 min and then stained with rabbit PAb specific for Xrn1 (Bethyl Laboratories) or Xrn2 (ProteinTech) and a secondary Alexa-488-conjugated goat anti-rabbit antibody (Invitrogen). HCV core protein was labeled with mouse anti-HCV core MAb (Pierce) and Alexa-568-conjugated goat anti-mouse antibody (Invitrogen). Nuclei were counterstained with DAPI (4′,6-diamidino-2-phenylindole). Confocal microscopy was carried out using a Leica SP2 laser scanning confocal microscope.
Cell proliferation assay.
The WST-1 (Roche) tetrazolium salt cleavage assay for cell proliferation was carried out following the manufacturer's suggested procedure. Huh-7.5 cells (1 × 104) were grown in 96-well plates and infected with virus at a multiplicity of infection (MOI) of 0.1. On the day of infection and at 24-h intervals thereafter, 10 μl WST-1 reagent was added to the culture medium, and the cells were incubated for 45 min prior to determining absorbance at 450 nm.
Statistical analysis.
Statistical significance was assessed using Prism 6.0e for Mac OS X software (GraphPad Software, Inc.).
RESULTS
Intracellular localization of Xrn1 and Xrn2.
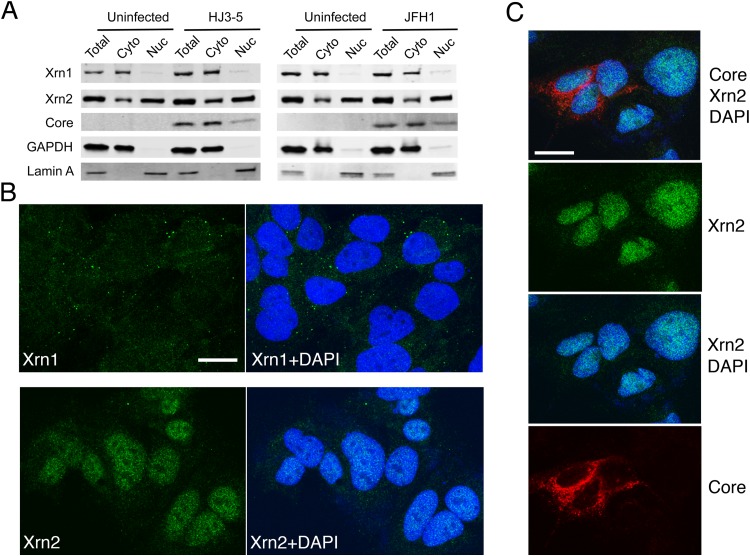
Xrn1 is known to be ubiquitously expressed within the cytoplasm of cells, where it plays a central role in the 5′ exonucleolytic mRNA decay pathway, whereas Xrn2 possesses similar 5′ exoribonuclease activity but is expressed primarily in the nucleus (22, 28). Xrn2 has been implicated in maintaining the quality of cellular transcripts by regulating the termination of RNA polymerase II-mediated transcription. We confirmed the cytoplasmic localization of Xrn1 versus the predominantly nuclear localization of Xrn2 in immunoblots of subcellular fractions prepared from uninfected Huh-7.5 cells (Fig. 1A). However, as described by Sedano and Sarnow (21), a significant amount of Xrn2 was also detected in cytosolic fractions. Since this could reflect contamination due to a small degree of nuclear membrane rupture, largely unavoidable during the preparation of such fractions, we also ascertained the intracellular localization of these proteins using confocal immunofluorescence microscopy (Fig. 1B). These studies confirmed the cytoplasmic localization of Xrn1, which was primarily present in small punctate structures that most likely represent P bodies. In contrast, Xrn2 was predominantly nuclear in its localization, with little definite cytoplasmic signal evident. Sedano and Sarnow suggested that JFH1 virus infection induced a small increase in expression of Xrn2 (21). However, we observed neither a quantitative change in its expression nor a qualitative change in its intracellular localization in cells infected with either JFH1 or HJ3-5 virus, a chimera in which sequence encoding the genotype 1a H77 structural proteins is placed in the genotype 2a JFH1 background (Fig. 1A and C).
FIG 1.
Xrn1 and Xrn2 expression and intracellular localization in uninfected and HCV-infected Huh-7.5 cells. (A) Immunoblots of Xrn1 and Xrn2 in whole-cell lysates and isolated nuclear (Nuc) and cytoplasmic (Cyto) fractions from Huh-7.5 cells with and without infection by the indicated viruses. GAPDH (glyceraldehyde-3-phosphate dehydrogenase) and lamin A were included as controls for cytosolic and nuclear proteins. (B) Immunostaining of Xrn1 (top) and Xrn2 (bottom) in uninfected Huh-7.5 cells. On the right are merged images with nuclei counterstained with DAPI. Xrn1 labeling is cytoplasmic and associated with puncta representing P bodies (19); Xrn2 labeling is predominantly nuclear. (C) Xrn2 localization (green) in JFH1-infected Huh-7.5 cells. Dual labeling with antibody to HCV core protein (red) identifies infected cells. Nuclei are counterstained with DAPI. (B and C) The size markers represent 20 μm.
Xrn1 and Xrn2 restriction of HCV replication.
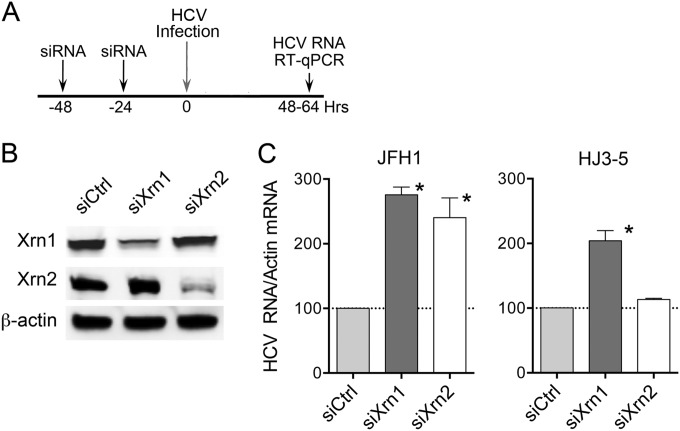
In initial experiments, we individually knocked down the expression of Xrn1 and Xrn2 in Huh-7.5 cells by repetitive transfection of gene-specific siRNAs (Fig. 2A and B). We then infected the cells with HJ3-5 or JFH1 virus at comparable multiplicities and quantified the abundance of intracellular viral RNA 48 to 64 h later by RT-qPCR. Depletion of either Xrn1 or Xrn2 (Fig. 2B) significantly enhanced the abundance of JFH1 RNA compared to that present in cells transfected with a nontargeting control siRNA, siCtrl (Fig. 2C). Xrn1 depletion had a similar effect in HJ3-5-infected cells, but Xrn2 depletion had no discernible effect on the abundance of replicating HJ3-5 RNA. The difference evident between these two viruses was reproduced in multiple independent experiments. Similar results were also obtained in Huh-7 cells, from which Huh-7.5 cells are derived (not shown). Thus, Xrn1 is a host restriction factor for both JFH1 and HJ3-5 viruses, but Xrn2 restricts only JFH1 replication. Since the 5′ UTRs of JFH1 and HJ3-5 RNAs are identical in sequence (as are the 3′ UTRs) (24), the restriction of JFH1 virus by Xrn2 is unlikely to reflect a difference in the capacity of the 5′ exoribonuclease to degrade these genomes. For the same reason, miR-122 should interact with the 5′ binding sites in these genomes in a similar fashion (27) and protect the two RNAs equivalently against 5′ exonuclease-mediated decay. Thus, Xrn2 most likely restricts JFH1 replication by a mechanism that is unrelated to miR-122 or 5′ exonucleolytic decay of the viral genome.
FIG 2.
Impact of Xrn1 or Xrn2 depletion on replication of JFH1 and HJ3-5 viruses. (A) Experimental scheme. Huh-7 or Huh-7.5 cells were transfected with Xrn1- or Xrn2-specific siRNAs (or control siCtrl) twice prior to infection with either JFH1 or HJ3-5 virus at an MOI of 0.1. The cells were lysed 48 to 64 h later, and viral RNA was quantified by RT-qPCR. (B) Immunoblots of Xrn1 and Xrn2 72 h after the second siRNA transfection. Actin was included as a loading control. (C) Relative intracellular HCV RNA abundances 48 to 64 h after infection, normalized to β-actin mRNA, with the mean abundance in siCtrl-transfected cell set at 100. The results shown represent the means and SEM from 4 (JFH1) or 2 (HJ3-5) independent experiments, each including 2 to 4 technical replicates. *, P < 0.05 compared to siCtrl-transfected cells by a Mann-Whitney test.
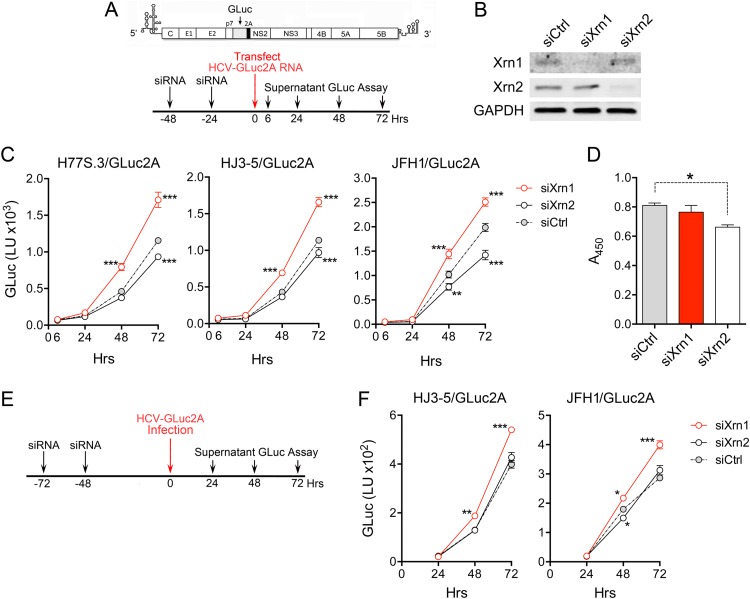
To confirm these results, we assessed the impacts of Xrn1 and Xrn2 depletion on replication of JFH1- and HJ3-5-based reporter virus RNAs that express GLuc from sequence inserted between the p7 and NS2 segments of the genome (JFH/GLuc2A and HJ3-5/GLuc2A, respectively) (4). We also tested in parallel the impact of Xrn1 or Xrn2 depletion on replication of a purely genotype 1a reporter virus RNA (H77S.3/GLuc2A) (20). In these experiments, replication was initiated by transfection of synthetic viral RNA following two sequential transfections of siRNA specific for Xrn1 or Xrn2 (Fig. 3A and B). As with the infection experiments shown in Fig. 2, Xrn1 depletion significantly enhanced the replication of all three RNAs, as determined by increases in GLuc secreted into supernatant culture fluids over a period of 72 h (Fig. 3C). In contrast, Xrn2 depletion resulted in a significant decrease in the replication of all three viral RNAs, especially JFH1/GLuc2A. In part, this may have been due to a small but significant effect of Xrn2 depletion on cell proliferation (Fig. 3D). These data confirm that Xrn1 is a restriction factor for JFH-1 and HJ3-5, as well as H77S.3 virus (19), and that Xrn2 does not restrict replication of either HJ3-5 or H77S.3.
FIG 3.
Replication of GLuc reporter virus RNAs in Xrn1- and Xrn2-depleted cells. (A) Experimental scheme. The reporter virus genome organization is shown at the top, with GLuc sequence inserted between p7 and NS2A. Huh-7.5 cells were transfected with Xrn1- or Xrn2-specific siRNA (or siCtrl) 48 and 24 h prior to transfection with the indicated GLuc reporter virus RNA. Supernatant culture fluids were subsequently sampled at intervals for GLuc activity. (B) Immunoblot of Xrn1 and Xrn2. Cell lysates were prepared 72 h after the final siRNA transfection. GAPDH was assessed as a loading control. (C) GLuc activities in supernatant fluids of cells transfected with H77S.3/GLuc2A, HJ3-5/GLuc2A, or JFH/GLuc2A RNA. The data represent means ± SEM from triplicate cultures and are representative of multiple independent experiments. Secreted GLuc activities diverged from siCtrl-treated cells as shown. The 6-h GLuc value represents translation of input RNA and thus serves as a transfection control. It did not differ significantly between experimental groups (P = 0.95 to 0.99). **, P < 0.01, and ***, P < 0.001 by two-way analysis of variance (ANOVA) with Holm-Sidak correction for multiple comparisons. (D) WST-1 assay showing differences in cell mass 4 days after siRNA transfection. *, P < 0.05 by one-way ANOVA with Dunnett's multiple-comparison test. (E) Design of experiments involving infection of Xrn1- and Xrn2-depleted Huh-7.5 cells with HJ3-5/GLuc2A and JFH1/GLuc2A viruses. (F) GLuc activities in supernatant fluids of cells infected with HJ3-5/GLuc2A or JFH/GLuc2A viruses at an MOI of 0.02. The data shown represent means ± SEM from triplicate cell cultures. *, P < 0.05; **, P < 0.01; ***, P < 0.001 by two-way ANOVA with correction for multiple comparisons.
The substantial reduction we observed in replication of JFH/GLuc2A RNA (Fig. 3C) contrasts sharply with the enhanced replication we observed in JFH1-infected cells after Xrn2 depletion (Fig. 2). To determine whether this reflects the presence of the GLuc2A insertion or the use of RNA transfection rather than virus infection in the experiments shown in Fig. 3C, we infected Xrn1- and Xrn2-depleted cells with cell-free HJ3-5/GLuc2A or JFH1/GLuc2A virus and then followed replication of the viruses by monitoring secreted GLuc activity (Fig. 3E). Similar to the results obtained in the transfection experiments, Xrn1 depletion significantly boosted the replication of both viruses (Fig. 3F). In contrast, the only apparent effect of Xrn2 depletion was a minor but statistically significant reduction in GLuc secretion 48 h after infection with JFH1/GLuc2A virus. Thus, the insertion of the GLuc2A sequence in JFH1 virus eliminates the Xrn2-mediated restriction of JFH1 replication shown in Fig. 2C.
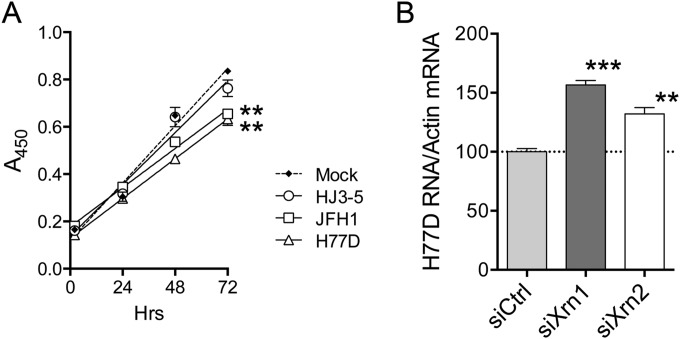
The insertion of the GLuc2A sequence in JFH/GLuc2A reduces the robust replication of JFH1, similar to other foreign-gene insertions in the HCV genome (20, 29). Similarly, although they share identical nonstructural proteins and 5′ and 3′ noncoding RNA sequences, the chimeric construction of HJ3-5 virus renders its replication phenotype less robust than that of JFH1 (4, 24). It also reduces its associated cytopathic effects, as reflected in the absence of G1 arrest and less induction of apoptosis in HJ3-5-infected cells (9). Consistent with these previously reported cytopathic effects, we confirmed that JFH1 infection significantly slowed the proliferation of JFH1-infected Huh-7.5 cells, whereas HJ3-5 infection had little effect on cell proliferation (Fig. 4A). Thus, we considered the possibility that cytopathic effects associated with robust replication of unmodified JFH1 virus account for the differences we observed in the impacts of Xrn2 depletion on JFH1 versus JFH1/GLuc2A and HJ3-5 replication. To test this hypothesis, we assessed the effects of Xrn1 versus Xrn2 depletion on replication of H77D virus, a lipid peroxidation-resistant variant of H77S.3 with a robust replication phenotype similar to that of JFH1 virus (4). Like JFH1 virus, H77D generates visible cytopathic effects and significantly slows the proliferation of Huh-7.5 cells (Fig. 4A). Also similar to JFH1 virus, Xrn2 depletion significantly boosted H77D replication, although once again, this effect was less than that observed with Xrn1 depletion (Fig. 4B). Thus, the ability of Xrn2 to restrict HCV replication appears to be limited to cytopathic virus strains with robust replication phenotypes and is not linked to specific 5′-UTR or replicase protein sequence.
FIG 4.
HCV effects on cell proliferation and Xrn2 restriction of H77D virus. (A) Proliferation of Huh-7.5 cells following infection with various HCV strains (MOI = 0.1). The cells were not transfected with siRNAs. Proliferation was assessed by WST-1 assay (see Materials and Methods). The data were fitted to a linear regression model, with differences in rates of proliferation (slopes) of mock-infected cells assessed by a two-tailed run test. **, P < 0.01. (B) Impact of Xrn1 or Xrn2 depletion on replication of H77D virus. See the legend to Fig. 2C for details. ***, P < 0.001, and **, P < 0.01 by one-way ANOVA with Dunnett's multiple-comparison test.
Impacts of Xrn1 and Xrn2 depletion on HCV RNA decay.
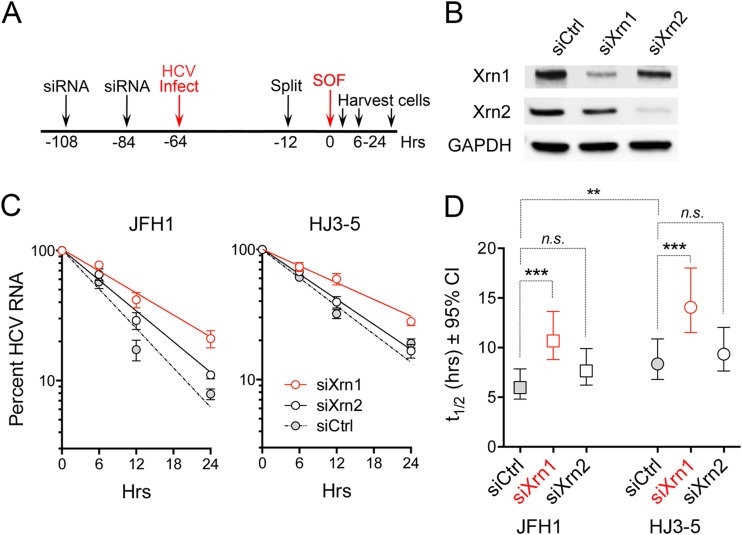
We next compared how the depletion of Xrn1 or Xrn2 influences the rate of decay of viral RNA in infected cells. Previous studies of the effects of Xrn2 depletion did not include such a comparison (21). To assess RNA decay, we adopted the approach taken originally by Shimakami et al. (17) (Fig. 5A). Cells were transfected with specific siRNAs to deplete Xrn1 or Xrn2 (Fig. 5B) and then infected with either JFH1 or HJ3-5 virus. Once infection was well established (64 h), the cells were treated with a high concentration of a nucleotide inhibitor of NS5B (sofosbuvir) to arrest viral RNA synthesis, and the rate of decay of intracellular HCV RNA was subsequently assessed by RT-qPCR. Relative HCV RNA levels were fitted to a one-phase decay model (R2 = 0.90 to 0.97) (Fig. 5C), and differences in the estimated decay constant [k, where k = ln(2)/t1/2] were assessed using the extra sum-of-squares F test (Fig. 5D). Xrn1 depletion increased the t1/2 of HJ3-5 virus RNA from 8.4 to 14.1 h (P < 0.0001), whereas Xrn2 depletion resulted in no change in the t1/2 of the viral RNA (Fig. 5D). Xrn1 depletion similarly slowed the decay of JFH1 RNA (t1/2 increased from 6.0 to 10.7 h; P < 0.0001), whereas Xrn2 depletion resulted in only a marginally significant increase (t1/2 increased from 6.0 to 7.7 h; P = 0.053) (Fig. 5D). Thus, Xrn1 depletion causes a statistically significant lowering of the rates of decay of both HJ3-5 and JFH1 virus RNAs in infected cells. Xrn2 depletion has at most only a modest effect on JFH1 RNA decay and no effect on HJ3-5 virus RNA.
FIG 5.
Impacts of Xrn1 and Xrn2 depletion on decay of HCV RNA following antiviral shutdown of new RNA synthesis. (A) Experimental scheme. Huh-7.5 cells were transfected twice with siRNAs specific for Xrn1 or Xrn2 (or siCtrl) 48 and again 24 h prior to infection with the indicated viruses (MOI = 0.1). The cells were treated with sofosbuvir (SOF) (5.0 μM) 64 h after infection (0 h) to arrest viral RNA synthesis and then harvested at intervals, and RNA was extracted for quantitation of HCV RNA by RT-qPCR. (B) Immunoblots showing abundances of Xrn1 and Xrn2 in siRNA-transfected cells. (C) HCV RNA decay following addition of sofosbuvir to cultures. The data represent means ± SEM from 5 biological replicates studied in 2 independent experiments. The lines were fitted to a one-phase decay model (R2 = 0.90 to 0.97). (D) Half-life of HCV RNA ± 95% confidence intervals (CI) estimated from the one-phase decay model. ***, P < 0.001; **, P < 0.01; n.s., not significant by the extra sum-of-squares F test. Note that JFH1 RNA decays significantly more rapidly than HJ3-5 RNA (t1/2 = 6.4 versus 8.4 h; P < 0.01). The results shown are derived from multiple independent experiments, each with technical replicates. The data from one of these experiments for the HJ3-5 virus appeared previously (18).
Xrn2 depletion promotes JFH1 replication independently of miR-122.
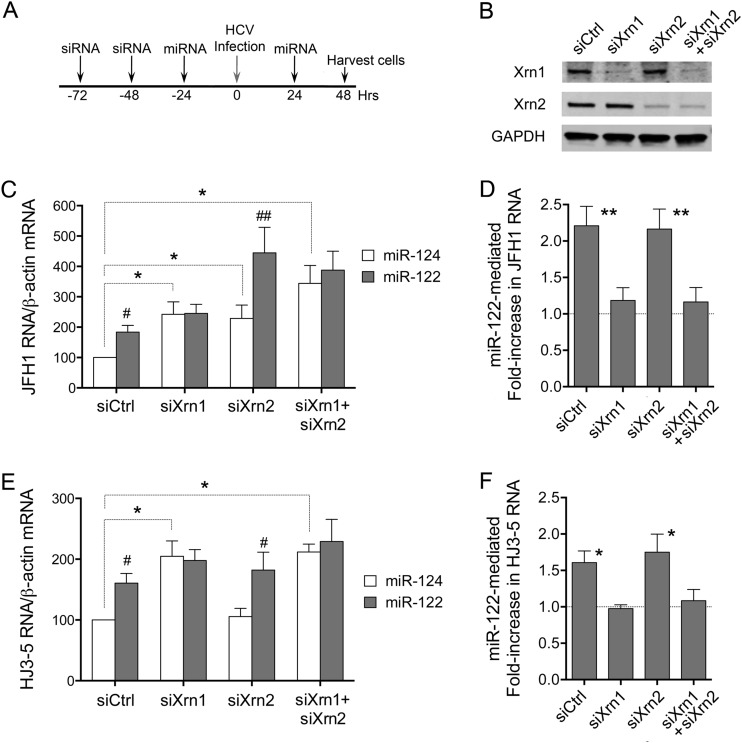
Our previous studies showed that miR-122 supplementation and Xrn1 depletion act similarly and nonadditively to enhance the stability and slow decay of HCV RNA in cells infected with the genotype 1a H77S.3 virus (19). Thus, in Xrn1-depleted cells, miR-122 supplementation has no effect on viral RNA stability, indicating that the stabilizing effect of miR-122 is due to its ability to block Xrn1-mediated decay. We carried out similar studies in JFH1- and HJ3-5-infected cells, asking whether Xrn2 depletion similarly alters the response to supplementation with exogenous miR-122 versus a control, brain-specific miRNA, miR-124 (Fig. 6). Under the control conditions (miR-124 supplementation), JFH1 replication was increased in cells depleted of either Xrn1 or Xrn2 compared with siCtrl-transfected cells (Fig. 6C). Interestingly, the combined depletion of both Xrn1 and Xrn2 had an additive effect, further boosting replication and enhancing JFH1 RNA abundance (P = 0.05 by paired t test). Compared with miR-124, supplementing cells with duplex miR-122 resulted in an increase in JFH1 RNA in control siCtrl- and siXrn2-depleted cells, but not Xrn1-depleted or Xrn1- plus Xrn2-doubly depleted cells (Fig. 6C and D). This difference was highly significant and reproducible in multiple experiments. Thus, Xrn1 depletion and miR-122 supplementation stimulate JFH1 replication in a nonadditive, redundant fashion, consistent with a common underlying mechanism and miR-122 protection against Xrn1-mediated RNA decay, as reported previously (19). In contrast, Xrn2 depletion acts additively, suggesting that Xrn2 restricts JFH1 virus replication by an independent mechanism. Since Xrn2 depletion has only a marginal effect on JFH1 RNA stability (Fig. 5C), it is likely to impact other aspects of the HCV life cycle.
FIG 6.
Impact of miR-122 supplementation on replication of JFH1 and HJ3-5 viruses in Xrn1- and Xrn2-depleted cells. (A) Experimental scheme. Huh-7.5 cells were transfected with siRNAs specific for Xrn1 or Xrn2, both Xrn1 and Xrn2, or siCtrl 72 and 48 h prior to infection with virus (MOI = 0.1). The cells were supplemented with exogenous duplex miR-122 or miR-124 24 h prior to and again 24 h following infection. Cells were harvested, and HCV RNA was quantified 48 h after infection. (B) Immunoblots showing Xrn1 and Xrn2 protein expression. (C) Relative JFH1 RNA abundances in cells transfected with miR-124 versus miR-122, with that in siCtrl/miR-124 transfected cells set arbitrarily to 100. #, P < 0.05, and ##, P < 0.01 by two-tailed paired t test (n = 4). The dashed lines indicate comparisons between miR-124-transfected siCtrl cells and Xrn1-, Xrn-2-, and Xrn1- plus Xrn2-depleted cells; *, P < 0.05. (D) Fold increases in JFH1 RNA mediated by miR-122 versus miR-124 supplementation. Deviations above a hypothetical value of 1.0 (no increase) were assessed by a one-sample t test. **, P < 0.01 (n = 8 for single depletions and 4 for Xrn1 plus Xrn2 depletion). (E) Relative HCV RNA abundances in cells infected with HJ3-5 virus under the conditions shown in panel A. The details are as in the legend to panel C. (F) Fold increase in HJ3-5 RNA mediated by miR-122 versus miR-124 supplementation. Deviations above a hypothetical value of 1.0 (no increase) were assessed by a one-sample t test; *, P < 0.05 (n = 3). All data shown are representative of multiple independent experiments.
Similar experiments in HJ3-5-infected cells confirmed nonadditive, redundant effects of miR-122 supplementation and Xrn1 depletion on replication of the virus (Fig. 6D and E). Again, Xrn2 depletion did not enhance replication of the virus. As with JFH1 virus, miR-122 supplementation significantly stimulated replication in Xrn2-depleted cells, but not in Xrn1-depleted cells or in cells depleted of both Xrn1 and Xrn2 (Fig. 6E).
DISCUSSION
miR-122 plays an essential role in the HCV life cycle, directly stimulating viral RNA synthesis while also stabilizing the positive-strand HCV RNA genome and slowing its decay (17, 18). Xrn1 and Xrn2 share limited sequence homology restricted to their N-terminal exoribonuclease domains but have very distinct subcellular localization and functions (28). We have shown previously that Xrn1 and miR-122 have competing effects on the stability and decay of the HCV genome consistent with a dominant role for this cytoplasmic 5′ exoribonuclease in degradation of HCV RNA (19). RNAi-mediated depletion of Xrn1 increased the t1/2 of genotype 1a H77S.3 RNA following antiviral arrest of new viral RNA synthesis, and by eliminating 5′ exoribonuclease activity ablated the stabilizing effects of miR-122 on the viral genome (19). Sedano and Sarnow (21) carried out similar experiments with the genotype 2a JFH1 virus but concluded that Xrn2 (not Xrn1) is largely responsible for HCV RNA decay. They found that Xrn2 restricts replication of JFH1 virus in cell culture (21), a finding we have confirmed here (Fig. 2). However, while they demonstrated that miR-122 can protect JFH1 RNA from recombinant Xrn2-mediated degradation in a cell-free reaction (21), they did not compare the effects of Xrn1 versus Xrn2 depletion on the stability of JFH1 RNA in infected cells.
In the experiments we describe here, high-grade Xrn2 depletion only marginally enhanced the stability of JFH1 RNA in infected cells, whereas Xrn1 depletion slowed the decay of the RNA to a much greater extent (Fig. 5C). These results indicate that Xrn1, not Xrn2, is the primary exoribonuclease responsible for degradation of JFH1 RNA and are consistent with our earlier studies with H77S.3 virus (19). We also demonstrated that miR-122 supplementation and Xrn1 depletion have largely redundant, nonadditive effects on JFH1 replication, whereas Xrn2 depletion acts in an additive and thus likely miR-122-independent fashion (Fig. 6C and D). Importantly, whereas Xrn2 restricts replication of JFH1, it does not restrict HJ3-5 virus, and its depletion has no measurable impact on the stability of HJ3-5 RNA (Fig. 2C and 5D). In contrast, Xrn1 restricts replication of all 4 viruses we tested: H77S.3, H77D, HJ3-5, and JFH1 (19) (Fig. 2, 3C, and 4B).
Surprisingly, Sedano and Sarnow (21) reported that the t1/2 of JFH1 RNA was only 1.2 h after arrest of RNA synthesis in infected cells. This compares with a t1/2 of 6.0 h in the studies we describe here (Fig. 5D), indicating a much faster turnover of JFH1 RNA in the cells studied by Sedano and Sarnow (21). The very short RNA t1/2 reported by Sedano and Sarnow (21) in the absence of Xrn2 depletion thus contrasts sharply with what we observed. By way of comparison, we found the t1/2 of HJ3-5 virus RNA to be about 8.4 h (Fig. 5D), while our previous studies with the genotype 1a H77S.3 virus suggested a t1/2 of about 10 h (19). Multiscale modeling of the response to antiviral drugs in patients with chronic hepatitis C suggested that the t1/2 of HCV RNA in the liver is approximately 11 h (30).
Although Xrn2 depletion has very different effects on JFH1 and HJ3-5 replication, these viruses are very closely related. HJ3-5 is a chimera in which sequences encoding the structural proteins of H77c virus have been placed in the background of JFH1 (24, 25). HJ3-5 and JFH1 thus share identical nontranslated RNA and replicase sequences, despite the absence of any effect of Xrn2 on HJ3-5 replication. These viruses do differ in their abilities to induce cytopathic effects, however. JFH1 is atypical in its capacity to replicate robustly in cell culture without adaptive mutations (5, 6). Unlike most other HCV strains, it exerts a distinct cytopathic effect in infected cell cultures, inducing apoptosis, as well as G1 cell cycle arrest (7–9). HJ3-5 replicates less robustly than JFH1 because of its chimeric construction and does not induce G1 arrest; it also causes significantly less apoptosis than JFH1 (9). Also, as indicated above and in Fig. 5D, HJ3-5 RNA decays more slowly than JFH1 RNA (P < 0.01). Given these multiple differences, we suspect that the restriction of JFH1 replication by Xrn2 may be related to its robust replication and cytopathic effects. This would also explain why Xrn2 fails to restrict replication of JFH1/GLuc2A virus (Fig. 3), the sequence of which differs from that of JFH1 only in the GLuc2A insertion between the p7 and NS2 regions of the genome. This insertion impairs the efficiency of replication, making JFH1/GLuc2A less cytopathic than its JFH1 parent. A relationship between cytopathic effect and Xrn2 restriction is also consistent with Xrn2 restriction of H77D, a virus with a replication phenotype rivaling that of JFH1 (4) and that, like JFH1, slows proliferation of infected cells (Fig. 4). Thus, the restriction of HCV replication by Xrn2 is not linked to specific virus sequence, but rather, to the robustness of replication and induced cytopathic effects. This suggests that it is likely to be an artifact of the JFH1 or H77D cell culture system.
Could it be argued instead that Xrn2 restriction of the robustly replicating JFH1 and H77D viruses reflects the situation in the liver during natural infection and that this effect is simply missed with the less efficiently replicating H77S.3, HJ3-5, and JFH1/GLuc2A viruses? While we cannot absolutely exclude such a possibility, it seems unlikely for several reasons. First, multiple modalities, including two-photon fluorescence microscopy (2), fluorescent in situ RNA hybridization (31), and single-cell laser capture microdissection coupled with qRT-PCR (32), all indicate that HCV replicates very inefficiently with limited expression of viral proteins and RNA within individual hepatocytes in the infected human liver. Infected hepatocytes are estimated to contain no more than 100 copies of the HCV genome (33), far fewer than JFH1 (or even H77S.3) virus-infected Huh-7.5 cells. Thus, viruses that replicate less robustly in cell culture, such as H77S.3, are likely to more closely mimic virus-host cell interactions in vivo. Second, JFH1 virus represents a clear outlier among HCV strains, demonstrating a strong resistance to lipid peroxidation that is not evident in wild-type genotype 1 HCV or in cell culture-adapted viruses of multiple genotypes (4). H77D virus shares that resistance (4). A third point to consider, in addition to the marked cytopathic effects induced by JFH1 (9), is that JFH1 virus produced in cell culture does not appear capable of establishing persistent infection, a hallmark of hepatitis C virus, in chimpanzees, whereas cell culture-produced H77S.3 virus does (34).
Because Xrn2 depletion and miR-122 supplementation have additive effects on JFH1 replication (Fig. 6), it is likely that Xrn2 acts via an indirect mechanism to restrict JFH1 replication rather than by direct 5′ exonucleolytic decay of the genome. This would explain why Xrn2 depletion marginally affects the stability of JFH1 RNA (Fig. 5C and D), despite promoting JFH1 replication to almost the same extent as Xrn1 depletion (Fig. 2C). Although we did not observe measurable leakage of nuclear Xrn2 into the cytoplasm in JFH1-infected cells (Fig. 1), it is possible that this occurs in association with the cytopathic effects of JFH1. Xrn2 has multiple functions tied to RNA metabolism (22, 28), and aberrant localization of the protein could have negative consequences for the cell, resulting in reduced permissiveness for HCV replication. Importantly, Xrn2 has no influence on the replication of HCV strains that have less robust cell culture phenotypes than JFH1 or H77D and little or no cytopathic effect and that are likely to mirror more closely the biology of HCV in the liver.
ACKNOWLEDGMENTS
We thank Michael Chua for assistance in collection of confocal microscopic images and Ann Sluder, Charles Rice, and Takaji Wakita for reagents and cells.
This work was supported by grants from the National Institutes of Health (R01-AI095690 and R01-CA164029) and the University of North Carolina Cancer Research Fund.
REFERENCES
- 1.Thomas DL. 2013. Global control of hepatitis C: where challenge meets opportunity. Nat Med 19:850–858. doi: 10.1038/nm.3184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Liang Y, Shilagard T, Xiao SY, Snyder N, Lau D, Cicalese L, Weiss H, Vargas G, Lemon SM. 2009. Visualizing hepatitis C virus infections in human liver by two-photon microscopy. Gastroenterology 137:1448–1458. doi: 10.1053/j.gastro.2009.07.050. [DOI] [PubMed] [Google Scholar]
- 3.Ploss A, Khetani SR, Jones CT, Syder AJ, Trehan K, Gaysinskaya VA, Mu K, Ritola K, Rice CM, Bhatia SN. 2010. Persistent hepatitis C virus infection in microscale primary human hepatocyte cultures. Proc Natl Acad Sci U S A 107:3141–3145. doi: 10.1073/pnas.0915130107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yamane D, McGivern DR, Wauthier E, Yi M, Madden VJ, Welsch C, Antes I, Wen Y, Chugh PE, McGee CE, Widman DG, Misumi I, Bandyopadhyay S, Kim S, Shimakami T, Oikawa T, Whitmire JK, Heise MT, Dittmer DP, Kao CC, Pitson SM, Merrill AH Jr, Reid LM, Lemon SM. 2014. Regulation of the hepatitis C virus RNA replicase by endogenous lipid peroxidation. Nat Med 20:927–935. doi: 10.1038/nm.3610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wakita T, Pietschmann T, Kato T, Date T, Miyamoto M, Zhao Z, Murthy K, Habermann A, Krauslich HG, Mizokami M, Bartenschlager R, Liang TJ. 2005. Production of infectious hepatitis C virus in tissue culture from a cloned viral genome. Nat Med 11:791–796. doi: 10.1038/nm1268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lindenbach BD, Evans MJ, Syder AJ, Wolk B, Tellinghuisen TL, Liu CC, Maruyama T, Hynes RO, Burton DR, McKeating JA, Rice CM. 2005. Complete replication of hepatitis C virus in cell culture. Science 309:623–626. doi: 10.1126/science.1114016. [DOI] [PubMed] [Google Scholar]
- 7.Walters KA, Syder AJ, Lederer SL, Diamond DL, Paeper B, Rice CM, Katze MG. 2009. Genomic analysis reveals a potential role for cell cycle perturbation in HCV-mediated apoptosis of cultured hepatocytes. PLoS Pathog 5:e1000269. doi: 10.1371/journal.ppat.1000269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhong J, Gastaminza P, Chung J, Stamataki Z, Isogawa M, Cheng G, McKeating JA, Chisari FV. 2006. Persistent hepatitis C virus infection in vitro: coevolution of virus and host. J Virol 80:11082–11093. doi: 10.1128/JVI.01307-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kannan RP, Hensley LL, Evers L, Lemon SM, McGivern DR. 2011. Hepatitis C virus infection causes cell cycle arrest at the level of entry to mitosis. J Virol 85:7989–8001. doi: 10.1128/JVI.00280-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Randall G, Panis M, Cooper JD, Tellinghuisen TL, Sukhodolets KE, Pfeffer S, Landthaler M, Landgraf P, Kan S, Lindenbach BD, Chien M, Weir DB, Russo JJ, Ju J, Brownstein MJ, Sheridan R, Sander C, Zavolan M, Tuschl T, Rice CM. 2007. Cellular cofactors affecting hepatitis C virus infection and replication. Proc Natl Acad Sci U S A 104:12884–12889. doi: 10.1073/pnas.0704894104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kaul A, Stauffer S, Berger C, Pertel T, Schmitt J, Kallis S, Zayas M, Lohmann V, Luban J, Bartenschlager R. 2009. Essential role of cyclophilin A for hepatitis C virus replication and virus production and possible link to polyprotein cleavage kinetics. PLoS Pathog 5:e1000546. doi: 10.1371/journal.ppat.1000546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gao L, Aizaki H, He JW, Lai MM. 2004. Interactions between viral nonstructural proteins and host protein hVAP-33 mediate the formation of hepatitis C virus RNA replication complex on lipid raft. J Virol 78:3480–3488. doi: 10.1128/JVI.78.7.3480-3488.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ariumi Y, Kuroki M, Abe K, Dansako H, Ikeda M, Wakita T, Kato N. 2007. DDX3 DEAD-box RNA helicase is required for hepatitis C virus RNA replication. J Virol 81:13922–13926. doi: 10.1128/JVI.01517-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jopling CL, Yi M, Lancaster AM, Lemon SM, Sarnow P. 2005. Modulation of hepatitis C virus RNA abundance by a liver-specific microRNA. Science 309:1577–1581. doi: 10.1126/science.1113329. [DOI] [PubMed] [Google Scholar]
- 15.Jangra RK, Yi M, Lemon SM. 2010. Regulation of hepatitis C virus translation and infectious virus production by the microRNA miR-122. J Virol 84:6615–6625. doi: 10.1128/JVI.00417-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jopling CL, Schutz S, Sarnow P. 2008. Position-dependent function for a tandem microRNA miR-122-binding site located in the hepatitis C virus RNA genome. Cell Host Microbe 4:77–85. doi: 10.1016/j.chom.2008.05.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shimakami T, Yamane D, Jangra RK, Kempf BJ, Spaniel C, Barton DJ, Lemon SM. 2012. Stabilization of hepatitis C virus RNA by an Ago2-miR-122 complex. Proc Natl Acad Sci U S A 109:941–946. doi: 10.1073/pnas.1112263109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Masaki T, Arend KC, Li Y, Yamane D, McGivern DR, Kato T, Wakita T, Moorman NJ, Lemon SM. 2015. miR-122 stimulates hepatitis C virus RNA synthesis by altering the balance of viral RNAs engaged in replication versus translation. Cell Host Microbe 17:217–228. doi: 10.1016/j.chom.2014.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Li Y, Masaki T, Yamane D, McGivern DR, Lemon SM. 2013. Competing and noncompeting activities of miR-122 and the 5′ exonuclease Xrn1 in regulation of hepatitis C virus replication. Proc Natl Acad Sci U S A 110:1881–1886. doi: 10.1073/pnas.1213515110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Shimakami T, Welsch C, Yamane D, McGivern D, Yi M, Zeuzem S, Lemon SM. 2011. Protease inhibitor-resistant hepatitis C virus mutants with reduced fitness from impaired production of infectious virus. Gastroenterology 140:667–675. doi: 10.1053/j.gastro.2010.10.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sedano CD, Sarnow P. 2014. Hepatitis C virus subverts liver-specific miR-122 to protect the viral genome from exoribonuclease Xrn2. Cell Host Microbe 16:257–264. doi: 10.1016/j.chom.2014.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Miki TS, Grosshans H. 2013. The multifunctional RNase XRN2. Biochem Soc Trans 41:825–830. doi: 10.1042/BST20130001. [DOI] [PubMed] [Google Scholar]
- 23.Scheel TK, Rice CM. 2013. Understanding the hepatitis C virus life cycle paves the way for highly effective therapies. Nat Med 19:837–849. doi: 10.1038/nm.3248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yi M, Ma Y, Yates J, Lemon SM. 2007. Compensatory mutations in E1, p7, NS2 and NS3 enhance yields of cell culture-infectious inter-genotypic chimeric hepatitis C virus. J Virol 81:629–638. doi: 10.1128/JVI.01890-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.McGivern DR, Villanueva RA, Chinnaswamy S, Kao CC, Lemon SM. 2009. Impaired replication of hepatitis C virus containing mutations in a conserved NS5B retinoblastoma protein-binding motif. J Virol 83:7422–7433. doi: 10.1128/JVI.00262-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ma Y, Yates J, Liang Y, Lemon SM, Yi M. 2008. NS3 helicase domains involved in infectious intracellular hepatitis C virus particle assembly. J Virol 82:7624–7639. doi: 10.1128/JVI.00724-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shimakami T, Yamane D, Welsch C, Hensley L, Jangra RK, Lemon SM. 2012. Base pairing between hepatitis C virus RNA and microRNA 122 3′ of its seed sequence is essential for genome stabilization and production of infectious virus. J Virol 86:7372–7383. doi: 10.1128/JVI.00513-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nagarajan VK, Jones CI, Newbury SF, Green PJ. 2013. XRN 5′→3′ exoribonucleases: structure, mechanisms and functions. Biochim Biophys Acta 1829:590–603. doi: 10.1016/j.bbagrm.2013.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jones CT, Murray CL, Eastman DK, Tassello J, Rice CM. 2007. Hepatitis C virus p7 and NS2 proteins are essential for production of infectious virus. J Virol 81:8374–8383. doi: 10.1128/JVI.00690-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Guedj J, Dahari H, Rong L, Sansone ND, Nettles RE, Cotler SJ, Layden TJ, Uprichard SL, Perelson AS. 2013. Modeling shows that the NS5A inhibitor daclatasvir has two modes of action and yields a shorter estimate of the hepatitis C virus half-life. Proc Natl Acad Sci U S A 110:3991–3996. doi: 10.1073/pnas.1203110110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wieland S, Makowska Z, Campana B, Calabrese D, Dill MT, Chung J, Chisari FV, Heim MH. 2014. Simultaneous detection of hepatitis C virus and interferon stimulated gene expression in infected human liver. Hepatology 59:2121–2130. doi: 10.1002/hep.26770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kandathil AJ, Graw F, Quinn J, Hwang HS, Torbenson M, Perelson AS, Ray SC, Thomas DL, Ribeiro RM, Balagopal A. 2013. Use of laser capture microdissection to map hepatitis C virus-positive hepatocytes in human liver. Gastroenterology 145:1404–1413. doi: 10.1053/j.gastro.2013.08.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Graw F, Balagopal A, Kandathil AJ, Ray SC, Thomas DL, Ribeiro RM, Perelson AS. 2014. Inferring viral dynamics in chronically HCV infected patients from the spatial distribution of infected hepatocytes. PLoS Comput Biol 10:e1003934. doi: 10.1371/journal.pcbi.1003934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yi M, Hu F, Joyce M, Saxena V, Welsch C, Chavez D, Guerra B, Yamane D, Veselenak R, Pyles R, Walker CM, Tyrrell L, Bourne N, Lanford RE, Lemon SM. 2014. Evolution of a cell culture-derived genotype 1a hepatitis C virus (H77S.2) during persistent infection with chronic hepatitis in a chimpanzee. J Virol 88:3678–3694. doi: 10.1128/JVI.03540-13. [DOI] [PMC free article] [PubMed] [Google Scholar]