ABSTRACT
A major hurdle to killing Epstein-Barr virus (EBV)-infected tumor cells using oncolytic therapy is the presence of a substantial fraction of EBV-infected cells that does not support the lytic phase of EBV despite exposure to lytic cycle-promoting agents. To determine the mechanism(s) underlying this refractory state, we developed a strategy to separate lytic from refractory EBV-positive (EBV+) cells. By examining the cellular transcriptome in separated cells, we previously discovered that high levels of host STAT3 (signal transducer and activator of transcription 3) curtail the susceptibility of latently infected cells to lytic cycle activation signals. The goals of the present study were 2-fold: (i) to determine the mechanism of STAT3-mediated resistance to lytic activation and (ii) to exploit our findings to enhance susceptibility to lytic activation. We therefore analyzed our microarray data set, cellular proteomes of separated lytic and refractory cells, and a publically available STAT3 chromatin immunoprecipitation sequencing (ChIP-Seq) data set to identify cellular PCBP2 [poly(C)-binding protein 2], an RNA-binding protein, as a transcriptional target of STAT3 in refractory cells. Using Burkitt lymphoma cells and EBV+ cell lines from patients with hypomorphic STAT3 mutations, we demonstrate that single cells expressing high levels of PCBP2 are refractory to spontaneous and induced EBV lytic activation, STAT3 functions via cellular PCBP2 to regulate lytic susceptibility, and suppression of PCBP2 levels is sufficient to increase the number of EBV lytic cells. We expect that these findings and the genome-wide resources that they provide will accelerate our understanding of a longstanding mystery in EBV biology and guide efforts to improve oncolytic therapy for EBV-associated cancers.
IMPORTANCE Most humans are infected with Epstein-Barr virus (EBV), a cancer-causing virus. While EBV generally persists silently in B lymphocytes, periodic lytic (re)activation of latent virus is central to its life cycle and to most EBV-related diseases. However, a substantial fraction of EBV-infected B cells and tumor cells in a population is refractory to lytic activation. This resistance to lytic activation directly and profoundly impacts viral persistence and the effectiveness of oncolytic therapy for EBV+ cancers. To identify the mechanisms that underlie susceptibility to EBV lytic activation, we used host gene and protein expression profiling of separated lytic and refractory cells. We find that STAT3, a transcription factor overactive in many cancers, regulates PCBP2, a protein important in RNA biogenesis, to regulate susceptibility to lytic cycle activation signals. These findings advance our understanding of EBV persistence and provide important leads on devising methods to improve viral oncolytic therapies.
INTRODUCTION
Oncogenic gammaherpesviruses such as Epstein-Barr virus (EBV) and Kaposi's sarcoma herpesvirus are causally linked to cancers such as Burkitt lymphoma (BL), nasopharyngeal cell carcinoma, posttransplant lymphoproliferative diseases, Kaposi's sarcoma, and primary effusion lymphomas (1–3). EBV genomes have also been identified in other types of cancer such as breast and gastric carcinomas (4, 5). While other herpesviruses, such as cytomegalovirus, are not known to cause cancers, they can nevertheless be present in cancers such as glioblastomas (6). Herpesviruses are therefore recognized as attractive therapeutic targets, potentially for a broad range of cancers.
Herpesvirus-directed oncolytic therapy involves pharmacologically switching the latent virus to its lytic phase in cancer cells, thereby making such cancer cells susceptible to killing by antiviral agents such as ganciclovir. Indeed, a phase 1/2 trial of butyrate, a short-chain fatty acid, to induce the EBV lytic cycle, and ganciclovir, a nucleoside-type antiviral agent, to then kill cells supporting the lytic phase of EBV, showed promise in patients with refractory EBV-positive (EBV+) lymphomas (7). However, with this approach, killing of cancer cells is restricted by the ability of cells to support the lytic phase of the viral life cycle. Studies have shown that only about half the number of latently infected cells in a population responds to lytic cycle-activating agents (8, 9). Consequently, a substantial fraction of cells in a population is refractory to oncolytic killing. We reasoned that to improve cell killing, the susceptibility of latently infected cancer cells to lytic activating signals would need to be increased.
In our efforts to improve the susceptibility of latently infected cells to lytic cycle-inducing stimuli, we developed a strategy to robustly detect and separate cells lytically infected with EBV from refractory (latently infected) cells (10). By probing a cellular microarray using RNA from separated cells, we then identified STAT3 (signal transducer and activator of transcription 3) as a key regulator of the refractory state. Specifically, we found that high levels of cellular STAT3 restrict the susceptibility of latently infected cells to lytic cycle activation signals (8, 9). In this study, we examined the proteome of EBV+ sorted refractory and lytic cells to identify PCBP2 [poly(C)-binding protein 2], an RNA-binding protein, as a transcriptional target of STAT3 in refractory cells. We also show that EBV-positive Burkitt lymphoma cells expressing high levels of PCBP2 are resistant to lytic cycle-inducing stimuli, that the manipulation of PCBP2 levels impacts the number of lytic cells, and that STAT3 exploits cellular PCBP2 to regulate the refractory state.
MATERIALS AND METHODS
Cell lines.
EBV lymphoblastoid cell lines (EBV-LCLs) derived from 3 healthy subjects and 3 patients with autosomal dominant hyper-IgE syndrome (AD-HIES) were described previously (9). Two AD-HIES patients had a mutation in the SH2 domain and the third patient had a mutation in the DNA-binding domain of their STAT3 gene. The study of cell lines derived from human subjects was approved by the Institutional Review Board (IRB) at Stony Brook University. The EBV-infected HH514-16 cell line is a subclone of the P3J-HR1 Burkitt lymphoma cell line (11).
Chemical treatment of cell lines.
The EBV lytic cycle was activated as previously described (10). Briefly, HH514-16 cells were subcultured at 3 × 105 cells/ml; 48 h later, cells were treated with 3 mM sodium butyrate (NaB; Sigma) and harvested at the indicated times.
Transfection of cell lines.
HH514-16 cells were subcultured at 3 × 105 to 5 × 105 cells/ml 24 h prior to transfection, washed twice with phosphate-buffered saline (PBS), and transfected with 100 μM small interfering RNA (siRNA) (targeting STAT3 [catalog number sc-29493] or PCBP2 [catalog number sc-38270], scrambled [sc-37007], or fluorescein isothiocyanate [FITC] scrambled [catalog number sc-36869]; Santa Cruz Biotechnology) or 3 μg of plasmid DNA (pCMV-STAT3, pCMV-Neo [both gifts from Nancy Reich], pPCBP2, or pCDNA3 [the latter 2 plasmids were gifts from Ken Fujimura]) per million cells via nucleofection, as described previously (9). For the flow cytometry experiment depicted in Fig. 5B, combinations of siRNA (targeted or scrambled) and FITC-scrambled siRNA were transfected at a 3:1 ratio to identify transfected cells (12).
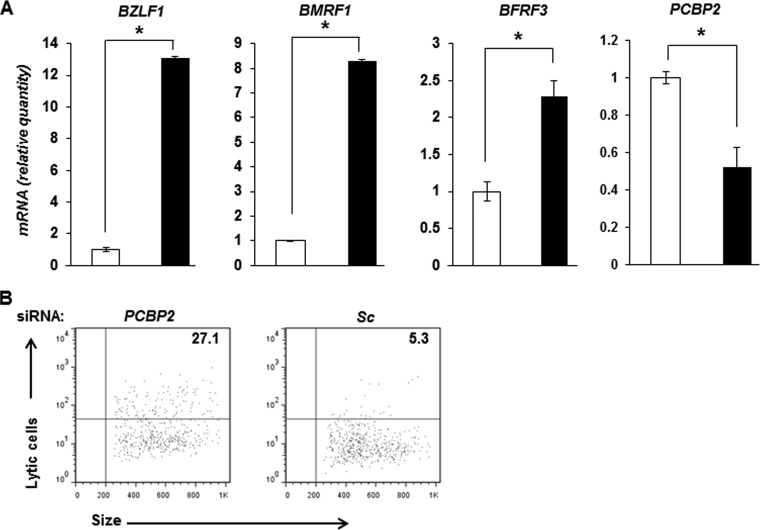
FIG 5.
Suppression of PCBP2 results in EBV lytic activation. (A) HH514-16 cells were transfected with scrambled siRNA (open bars) or siRNA targeting PCBP2 (black bars) and harvested 24 h later for determination of relative amounts of transcripts from 3 EBV lytic genes, BZLF1, BMRF1, and BFRF3, as well as PCBP2 by qRT-PCR after normalization to 18S rRNA levels by using the ΔΔCT method. Error bars indicate standard errors of the means of data from 3 technical replicates from each of 2 transfection experiments (*, P < 0.05). (B) HH514-16 cells were transfected with either PCBP2 siRNA and FITC+ scrambled siRNA (PCBP2) at a 3:1 ratio (the latter to mark transfected cells) or FITC-negative and FITC+ scrambled siRNA at a 3:1 ratio (Sc). After 24 h, the FITC+ (i.e., transfected) population was examined for lytic cells by flow cytometry using reference EBV-seropositive and -seronegative human sera. Numbers indicate the percentages of transfected cells that were spontaneously lytic. Data are representative of two experiments.
Immunoblotting.
Total cell extracts were electrophoresed in 10% SDS-polyacrylamide gels, transferred onto nitrocellulose membranes, and reacted with antibodies (Abs) as described previously (9). Mouse monoclonal Abs were used to detect PCBP2 (1/500 dilution) (catalog number sc-101136; Santa Cruz Biotechnology) and β-actin (1/3,000 dilution) (catalog number AC-15; Sigma). Signals were detected by using extended chemiluminescence.
Flow cytometry.
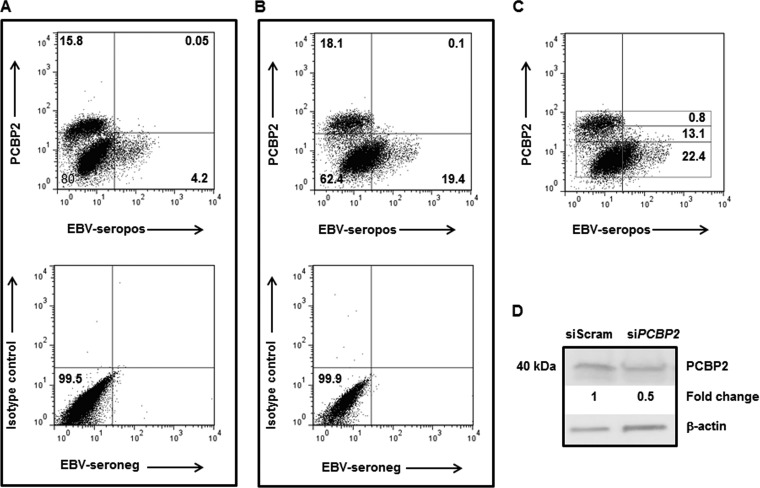
Cells were stained in PBS containing 5% fetal bovine serum (FBS) with reference human sera (EBV positive or negative) followed by FITC-conjugated anti-human IgG (1/200 dilution) (BD), as described previously (8, 10). Cells were fixed and permeabilized by using Cytofix/Cytoperm (BD) and incubated with anti-PCBP2 Ab (1/50 dilution) (catalog number sc-101136; Santa Cruz Biotechnology) or a mouse IgG isotype control followed by allophycocyanin (APC)-conjugated anti-mouse IgG (1/500 dilution) (Life Technologies). Specific detection of lytic antigens by reference EBV-positive serum was determined over background detection of similarly treated cells by reference EBV-negative serum (0.5 to 1% detection as a cutoff) using dot plot analyses. Cells shown in Fig. 3C were divided into PCBP2hi, PCBP2int, and PCBP2lo/− subpopulations after comparison with cells stained with the isotype control antibody. Events (100,000) were acquired by using a FACSCalibur flow cytometer (BD), and data were analyzed by using FlowJo software (Tree Star).
FIG 3.
Cells expressing high levels of PCBP2 protein are refractory to spontaneous and induced EBV lytic activation. (A to C) HH514-16 cells were untreated (A) or treated with NaB (B and C). Cells were harvested 48 h later and evaluated for lytic cells (using reference EBV-seropositive serum [top] or reference EBV-seronegative serum [bottom] in panels A and B) and PCBP2 expression by flow cytometry. Numbers in panels A and B represent the percentages of cells in quadrants; numbers in rectangular gates in panel C represent the percentages of PCBP2hi (top), PCBP2int (middle), and PCBP2lo/− (bottom) cells undergoing lytic activation. Staining with a matched isotype control antibody for PCBP2 is shown at the bottom of panels A and B. (D) HH514-16 cells were transfected with control scrambled siRNA or siRNA targeting PCBP2, harvested 24 h later, and subjected to immunoblotting with anti-PCBP2 and anti-β-actin antibodies. Numbers indicate the relative amounts of PCBP2 protein after normalization to β-actin levels. These experiments were performed at least twice. Data for representative experiments are shown.
Immunofluorescence microscopy.
Staining and microscopy were performed as described previously (9, 12). Briefly, cells were stained as described above for flow cytometry by using rabbit anti-human STAT3 (Santa Cruz Biotechnology) followed by FITC–anti-rabbit IgG (Life Technologies), washed, cytospun onto glass slides, air dried, and mounted with 4′,6-diamidino-2-phenylindole (DAPI) Prolong Gold Antifade (Life Technologies). All images were acquired under identical settings at a ×40 magnification on an AxioScope A1 microscope (Zeiss) with SPOT v4.0 software. Images were deidentified, and nuclei were counted by two individuals.
Quantitative reverse transcription-PCR (qRT-PCR).
Total RNA was isolated from EBV-LCL and HH514-16 cells by using an RNeasy kit (Qiagen), followed by DNase digestion (Promega). RNA was quantitated by using a NanoDrop instrument (Thermo Scientific). RNA (500 ng) was converted to cDNA by using qScript cDNA SuperMix (Quanta BioSciences). Relative transcript levels of selected cellular and viral genes were determined with gene-specific primers by using Fast SYBR green master mix on a StepOne Plus thermocycler (Applied Biosystems). Sequences of primers for 18S rRNA, STAT3, and BMRF1 were described previously (9). Sequences of other primers are as follows: forward primer TTCCACAGCCTGCACCAGTG and reverse primer GGCAGCCACCTCACGGT for BZLF1, forward primer ACTCCCGGCTGTAAATTCCT and reverse primer CCATACAGGACACAACACCTCA for BRLF1, forward primer TTCTGACATCCCAGTTCTGC and reverse primer TGTCTATGGCGCGTTGG for BFRF3, forward primer CGGAAAGGAAGTAGGAAGC and reverse primer GTGGAGTTGGGCAGCATA for PCBP1, forward primer AGGCAGGTTACCATCACTGG and reverse primer CATTGTTCTAGCTGCTCCCC for PCBP2, forward primer CGCTAGGATGAAGCTCGTG and reverse primer CCACACCTGTGATTGTTCCA for SNRPD1, forward primer TAGAGGAATTCGCAGAGGGA and reverse primer TTGTATCCCAGCTCAAGCCT for HNRPDL, and forward primer TGAACATGTGCTCAGTGGCT and reverse primer AAAGGGTCTGTCACTGGCTG for UTP14A.
Relative expression levels were calculated by using the ΔΔCT method, normalized to 18S rRNA levels, and compared to those of untreated but mock-sorted (see Fig. 2), healthy subject-derived LCL (Fig. 4B), or scrambled siRNA-transfected (see Fig. 4 and 5) cells with StepOne software v2.2. Individual samples were assayed in triplicate.
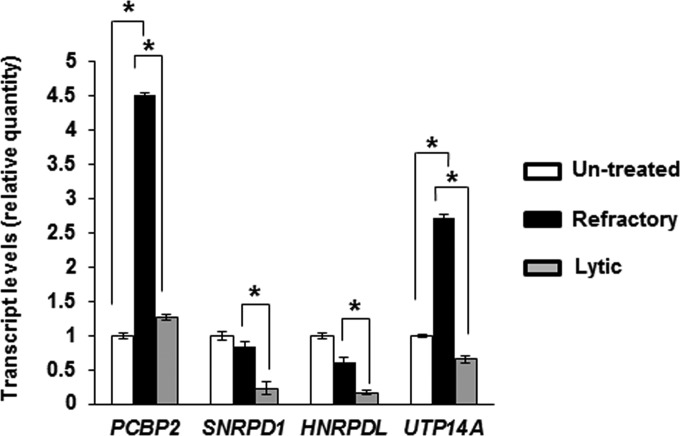
FIG 2.
Candidate genes contributing to the RNA processing GO term show increased transcript levels in refractory cells. Untreated but mock-sorted HH514-16 BL cells, NaB-treated sorted refractory cells, and NaB-treated sorted lytic cells were subjected to qRT-PCR using primers targeting four candidate genes that are transcriptional targets of STAT3 and contribute to the “RNA processing” GO biological process pathway. Results represent means of relative amounts of RNA normalized to the level of 18S rRNA ± standard errors of the means from three technical replicates from each of three independent sorting experiments (*, P < 0.05).
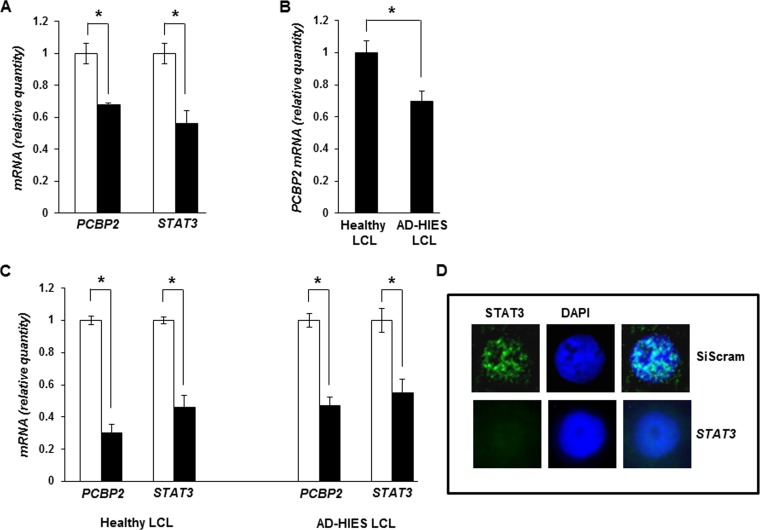
FIG 4.
Impairment of STAT3 results in suppression of PCBP2 transcript levels. (A) HH514-16 cells were transfected with scrambled siRNA (open bars) or siRNA targeting STAT3 (black bars) and harvested 24 h later for determination of relative amounts of PCBP2 and STAT3 mRNAs by qRT-PCR after normalization to 18S rRNA levels using the ΔΔCT method. (B) Seven-week-old LCLs derived from 3 healthy subjects and 3 AD-HIES patients were examined for relative levels of PCBP2 transcripts by qRT-PCR as described above for panel A. (C) Healthy subject-derived and AD-HIES patient-derived LCLs were transfected with scrambled siRNA (open bars) or siRNA targeting STAT3 (black bars) and harvested 24 h later for determination of the relative amounts of PCBP2 and STAT3 mRNAs by qRT-PCR as described above for panel A. Error bars are standard errors of the means of data from 3 technical replicates from each of 2 transfection experiments in panels A and C (*, P < 0.05). (D) Healthy subject-derived LCLs were transfected with scrambled siRNA (SiScram) (top) or STAT3 siRNA (bottom), harvested 24 h later, and examined for STAT3 expression by immunofluorescence. Representative nuclei (∼15% of cells transfected with siRNA targeting STAT3, consistent with our typical transfection efficiency) are shown. Experiments were performed twice.
FACS sorting of lytic and refractory cells.
HH514-16 cells were treated with NaB (or untreated), stained for lytic and refractory cells, and sorted by fluorescence-activated cell sorting (FACS), as described previously (10).
SILAC labeling of cells for proteomic analysis.
HH514-16 cells were metabolically labeled with [13C6]lysine and [13C6]arginine (catalog number 88210; Pierce) by using the SILAC (stable isotope labeling by amino acids in cell culture) method according to the manufacturer's instructions (catalog number 1860971; Pierce). Compared to unlabeled cells, 99% of proteins in labeled cells had incorporated “heavy” amino acids after 48 h of culture. Labeled cells were treated with NaB for 24 h and sorted into lytic and refractory populations by FACS. One million each of lytic and refractory cells were obtained from each sort. Pooling of cells from 2 independent sorting experiments yielded 60 μg of total protein from each population; these proteins were subjected to mass spectrometry (MS).
Multidimensional protein identification technology and tandem mass spectrometry.
After trypsin digestion, peptide mixtures were subjected to multidimensional protein identification technology (MudPIT) by using an LTQ-Orbitrap XL mass spectrometer equipped with a nano-liquid chromatography (LC) electrospray ionization source (ThermoFinnigan, San Jose, CA) (13). Full MS spectra of the peptides were recorded over a range of 400 to 2,000 m/z by using the Orbitrap spectrometer, followed by five tandem mass spectrometry (MS/MS) events sequentially generated by linear trap quadrupole (LTQ) in a data-dependent manner on the first, second, third, and fourth most intense ions selected from the full MS spectrum (at 35% collision energy). Mass spectrometer scan functions and high-performance liquid chromatography (HPLC) solvent gradients were controlled by the Xcalibur data system (ThermoFinnigan).
Interpretation of MS/MS data sets.
Tandem mass spectra were extracted from raw files, and low-quality MS/MS spectra were removed based on an empirical classifier. The remaining spectra were searched against the Uniprot human protein database (released February 2011). To calculate confidence levels and false-positive rates, we used a decoy database containing the reverse sequences of annotated protein sequences and the SEQUEST algorithm (14) to find the best-matching sequences using the Integrated Proteomics Pipeline (IP2; Integrated Proteomics Inc., CA). The peptide mass search tolerance was set to 50 ppm. No differential modifications were considered. At least half-tryptic status was imposed on the database search, so the search space included all candidate peptides whose theoretical mass fell within the 50-ppm mass tolerance window. The validity of peptide/spectrum matches was assessed with DTASelect2 (15) using SEQUEST-defined parameters, the cross-correlation score (XCorr), and the normalized difference in cross-correlation scores (DeltaCN). The search results were grouped by charge state (+1, +2, and +3) and tryptic status (fully tryptic, half-tryptic, and nontryptic), resulting in 9 distinct subgroups. For each subgroup, the distribution of XCorr and DeltaCN values for (i) direct and (ii) decoy database hits was obtained, and the two subsets were separated by quadratic discriminant analysis. Outlier points in the two distributions (for example, matches with very low Xcorr but very high DeltaCN values) were discarded. The discriminant score was set such that a false-positive rate of 1% was determined based on the number of accepted decoy database peptides. In addition, a minimum sequence length of 7 amino acid residues was required, and each protein on the final list was supported by at least two independent peptide identifications unless otherwise specified. After this last filtering step, the false identification rate was reduced to <1%. SILAC ratios were derived by using CenSus (16).
The mass spectrometry proteomics data were deposited in the ProteomeXchange Consortium.
Bioinformatic analysis.
Raw data from microarray experiments (GEO accession number GSE49568) were normalized and analyzed by using dChip software (17 December 2010 build) (17). Comparison of lytic and refractory samples was performed under default dChip settings (fold change of >2 with 90% confidence). For genes with multiple probes selected by dChip, the average fold change was used. In a few cases, different probes from the same gene had conflicting up- or downregulation information; these genes were omitted from the differentially expressed gene lists. The present/absent calls were generated from Affymetrix GCOS software using the MAS5 algorithm. A gene was considered absent if all its probes had an absent flag. A 2-fold cutoff (by total ionic counts or spectral counts) was also used to select the changes for proteins in lytic and refractory comparisons. Potential STAT3 target genes were identified by using the ENCODE GM12878 cell line STAT3 antibody track and PeakSeq software, as described previously (see the supplemental material in reference 9). Common genes/proteins between differentially expressed genes, proteins, and potential STAT3 targets were identified by using gene symbols. Gene Ontology (GO) enrichment analysis was performed by using DAVID Bioinformatics Resources version 6.7 (18). For gene lists generated from microarray and potential STAT3 targets, the whole genome was used as the background; for protein lists, all proteins detected in the mass spectrum were used as the background. A false discovery rate (FDR) of <25% was used to determine significantly enriched GO terms.
Statistical analyses.
P values were calculated by comparing the means for two groups of interest using the unpaired Student t test.
Accession number.
Raw files generated by the mass spectrometer and searched files with protein identifications are available at the ProteomeXchange Consortium (http://proteomecentral.proteomexchange.org/) via the PRIDE partner repository with the data set identifier PXD001838.
RESULTS
A multipronged approach narrows the list of STAT3-responsive genes likely to regulate susceptibility of EBV-infected B cells to lytic activation signals.
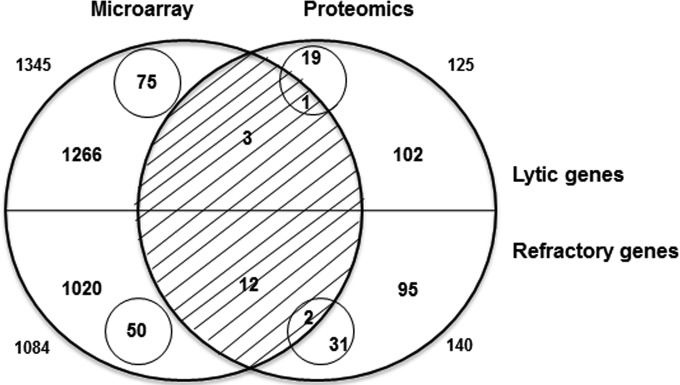
The central function of STAT3 is transcriptional activation. Therefore, to determine the mechanism by which STAT3 contributes to the maintenance of the EBV refractory state, we needed to identify the gene(s) transcriptionally activated by STAT3 in refractory compared to lytic cells. However, because STAT3 can potentially regulate the transcription of several thousand genes (9, 19), this made the identification of the downstream target(s) of STAT3 challenging. We therefore exploited two genome-wide analysis strategies: a transcriptome analysis of human genes in sorted refractory and sorted lytic cell populations that we have previously described (9) and a proteomic analysis of human genes in the same 2 populations, presented here. To identify cellular genes whose mRNA and protein levels were both increased by at least 2-fold, we compared relative expression levels of all cellular gene products in lytic and refractory cells in the microarray and proteomic data sets (Fig. 1). As we have shown previously, lytic cells demonstrated 1,345 transcripts with levels that were higher than those of the corresponding transcripts in refractory cells (9). Of these, 75 transcripts were detected only in lytic cells. In refractory cells, 1,084 transcripts showed higher levels than those in lytic cells (9). Of these, 50 transcripts were detected only in refractory cells. For proteins, 125 were detected at higher levels in lytic than in refractory cells; of these, 20 were detected only in lytic cells. In comparison, 140 proteins were present at higher levels in refractory cells than in lytic cells; of these, 33 were detected only in refractory cells. The overlap of the two analyses revealed that both transcript and protein levels were elevated for 4 genes in lytic cells and 14 genes in refractory cells. The lists of 125 and 140 proteins with increased levels in lytic and refractory cells, respectively, are shown in Table S1 in the supplemental material. Genes whose products were observed only in lytic or refractory cells by microarray and proteomic analyses, respectively, are shown in Table S2 in the supplemental material. As expected, we observed an enrichment of EBV lytic proteins in the lytic fraction (see Table S3 in the supplemental material).
FIG 1.
Comparison of gene expression levels in sorted lytic and refractory cells by using microarray and proteomic approaches. (A) Venn diagram showing the overlap between the numbers of genes with at least 2-fold increased levels of gene products in lytic cells (top half) and those in refractory cells (bottom half). Numbers outside circles indicate the total number of genes with increased levels of mRNA (by microarray analysis [left]) or protein (by proteomic analysis [right]). Numbers inside small circles indicate the number of genes whose products were observed in lytic or refractory cells only.
To determine which of the 4 and 14 genes with increased product levels in lytic and refractory cells, respectively, were transcriptional targets of STAT3, we used another genome-wide analysis measure: a publically available STAT3 chromatin immunoprecipitation sequencing (ChIP-Seq) data set from the EBV-positive lymphoblastoid cell line (LCL) GM12878 (19). As described previously, our analysis of this STAT3 ChIP-Seq peak set produced a list of >8,000 potential STAT3-regulated genes (9). Intersection of the gene lists from Fig. 1 with the list of STAT3-regulated genes revealed that 2 of the 4 genes whose levels were increased in lytic cells were potential transcriptional targets of STAT3. In comparison, 12 of the 14 genes with increased expression levels in refractory cells were potential transcriptional targets of STAT3. A list of these genes is provided in Table S4 in the supplemental material.
The “RNA processing” Gene Ontology term is a biological target of STAT3 in refractory cells.
Our strategy for identifying a mechanism by which STAT3 promotes the refractory state involved identifying genes whose mRNA and protein levels were both increased, whose expression(s) might be regulated by STAT3, and whose functions may be biologically linked. We therefore converted lytic and refractory gene lists with increased levels from microarray (9) and proteomic (see Table S1 in the supplemental material) approaches and the list of genes in the STAT3 ChIP-Seq peak set to GO terms. In the lytic population, only 2 microarray-derived but 25 proteomics-derived terms reached statistical significance (hypergeometric distribution, with an FDR of <25%). In the refractory population, 33 microarray-derived and 6 proteomics-derived terms were statistically significant. In the STAT3 ChIP-Seq peak set, 124 terms were statistically significant. When these terms were overlapped, no common GO terms were identified in the lytic population. In contrast, a single common GO term (GO:0006396; RNA processing) was identified in the refractory population. Of the genes in the RNA processing pathway, 5 (PCBP2, SNRPD1, HNRPDL, UTP14A, and RBM17) demonstrated elevated mRNA and protein levels. Importantly, these genes were also among the 14-member list of refractory genes shown in Table S4 in the supplemental material. Of the 5 genes, 4 (PCBP2, SNRPD1, HNRPDL, and RBM17) were also potential transcriptional targets of STAT3. Thus, 4 genes with increased mRNA and protein levels in refractory cells met the criteria of being STAT3 targets and being biologically linked. In contrast, none of the genes with increased mRNA and protein levels in lytic cells met these criteria.
Components of the “RNA processing” biological process are involved in resistance to lytic activation.
To further narrow the list of candidate genes, we sorted EBV-positive HH514-16 Burkitt lymphoma (BL) cells into refractory and lytic cells 24 h after exposure to the lytic cycle-inducing histone deacetylase (HDAC) inhibitor sodium butyrate (NaB). We were able to amplify transcripts from 4 genes (PCBP2, SNRPD1, HNRPDL, and UTP14A). Levels of mRNA for all 4 genes were significantly higher in refractory cells than in lytic cells (Fig. 2). However, mRNA levels for only 2 genes (PCBP2 and UTP14A) were significantly elevated in refractory cells compared to cells unexposed to lytic activation signals. Thus, 2 candidate genes contributing to the “RNA processing” biological process demonstrated increased transcript levels in refractory cells compared to lytic cells and cells unexposed to lytic signals.
EBV-infected B cells expressing high levels of the PCBP2 protein are refractory to EBV lytic activation.
Of PCBP2 and UTP14A, the 2 genes with increased mRNA levels in refractory cells compared to both lytic cells and cells unexposed to lytic signals, only PCBP2 was a transcriptional target of STAT3 (based on the results of the STAT3 ChIP-Seq peak set). We therefore focused our investigation on PCBP2 [poly(C)-binding protein 2] to determine the relationship between PCBP2 protein levels and the refractory state, whether its expression was regulated by STAT3, and whether manipulation of its levels affected lytic susceptibility. PCBP2 is a member of the PCBP family of proteins that bind to C-rich single-strand RNA motifs (20). PCBP proteins have been found to mediate a multitude of functions ranging from mRNA stabilization, translational enhancement, and translational silencing to transcriptional regulation, antiviral activities, and cell survival (20–24).
Since refractory cells that have high levels of STAT3 (8) also demonstrate high levels of PCBP2 mRNA (Fig. 2) and since high levels of STAT3 restrain EBV lytic activation (8, 9), we examined the relationship between PCBP2 levels and susceptibility to lytic activation signals at the single-cell level. As shown by using cells not treated with NaB (Fig. 3A), spontaneously lytic cells were derived primarily from cells expressing low levels of PCBP2, and cells expressing high levels of PCBP2 were refractory to lytic activation signals in culture. As expected, treatment of cells with NaB resulted in an increase in the percentage of lytic cells, and these lytic cells were again derived from cells expressing low levels of PCBP2, with those expressing high levels of PCBP2 remaining refractory to the lytic activation signal (Fig. 3B). Furthermore, single cells expressing larger amounts of PCBP2 protein demonstrated progressively increasing resistance to lytic activation signals such that 22.4% of PCBP2lo cells, 13.1% of PCBP2int cells, and 0.8% of PCBP2hi cells were lytic (Fig. 3C). As expected, the anti-PCBP2 antibody detected endogenous PCBP2, as demonstrated by a decrease in the amount of the target protein of the expected size following the introduction of siRNA targeting PCBP2 (Fig. 3D). Thus, cells expressing high levels of PCBP2 are refractory to spontaneous and induced EBV lytic activation.
Impairment of STAT3 causes suppression of PCBP2 transcript levels.
Since PCBP2 is predicted to be a STAT3-responsive gene, we next examined the effect of STAT3 on PCBP2 transcript levels. Figure 4A shows a decrease in the PCBP2 transcript level in the presence of siRNA targeting STAT3 compared to BL cells transfected with a scrambled siRNA. In a complementary approach, we examined EBV-positive B cell lines (LCLs) derived from patients with autosomal dominant hyper-IgE syndrome (AD-HIES), each carrying a distinct, naturally occurring hypomorphic mutation in the STAT3 gene. Patients with AD-HIES have a dominant negative mutation in their STAT3 gene that renders the majority of the protein nonfunctional. As shown in Fig. 4B, we found that the baseline levels of the PCBP2 transcript were lower in AD-HIES LCLs than in EBV-LCLs derived from healthy subjects. This result is consistent with our previously reported observation that AD-HIES LCLs are more lytically active at baseline than are healthy subject-derived LCLs (9). Furthermore, transfection of siRNA targeting STAT3 in both AD-HIES LCLs and healthy subject-derived LCLs resulted in a significant reduction in PCBP2 mRNA levels compared to those in scrambled siRNA-transfected LCLs (Fig. 4C). As expected, compared to cells transfected with scrambled siRNA, transfection of siRNA targeting STAT3 resulted in decreases in STAT3 mRNA levels (Fig. 4A and C) and STAT3 protein levels in ∼15% of cells (Fig. 4D [representative nuclei are shown]). Thus, paralleling the STAT3 ChIP-Seq results, our findings indicate that PCBP2 transcript levels are regulated by the transcription factor STAT3.
Inhibition of PCBP2 is sufficient for EBV lytic activation.
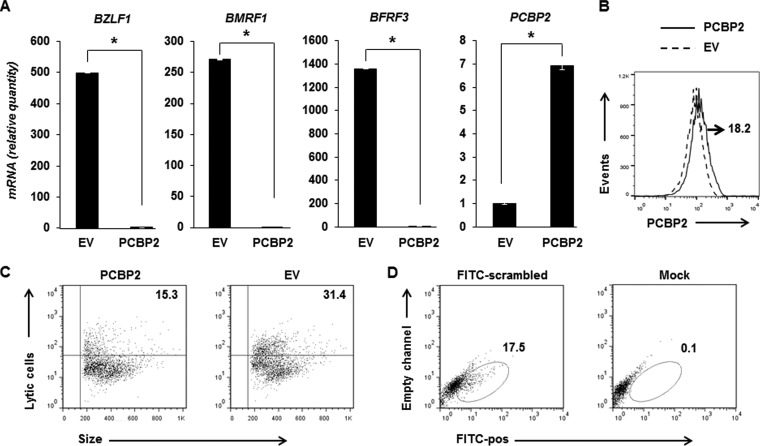
The correlation between high levels of PCBP2 and the refractory state prompted us to examine the effect of PCBP2 suppression on EBV lytic activation. As shown in Fig. 5A, siRNA-mediated suppression of PCBP2 resulted in 13-fold, 8-fold, and 2.3-fold increases in the relative levels of transcripts of the lytic genes BZLF1 (immediate early), BMRF1 (early), and BFRF3 (late), respectively. As expected, the PCBP2 transcript level was suppressed in the presence of siRNA targeting PCBP2 compared to scrambled siRNA. To determine if this increase in lytic gene transcript levels indicated a corresponding increase in the number of cells supporting lytic activation, we evaluated single cells by flow cytometry. Figure 5B shows that there were 5.1-fold more lytic cells when BL cells were transfected with siRNA targeting PCBP2 than when BL cells were transfected with scrambled siRNA. Thus, suppression of PCBP2 is sufficient to increase susceptibility to EBV lytic activation.
Overexpression of PCBP2 restricts EBV lytic cycle activation.
To determine if an increase in the PCBP2 level results in an opposite effect on susceptibility to EBV lytic activation signals, we transiently overexpressed PCBP2 in BL cells (Fig. 6). Following exposure to NaB, cells transfected with a PCBP2 plasmid, compared to those transfected with an empty vector, demonstrated a significant repression of representative lytic genes of all kinetic classes (Fig. 6A). As expected, transfection of the PCBP2 plasmid resulted in increases in the levels of PCBP2 mRNA (Fig. 6A) and PCBP2 protein at the single-cell level in ∼18% of cells (Fig. 6B). Importantly, there were 16.1% fewer lytic cells following the overexpression of PCBP2 than in empty vector-transfected cells (Fig. 6C). Consistent with our typical nucleofection efficiency of 15% (9), 17.4% of cells underwent transfection in this experiment, as demonstrated by the uptake of FITC+ scrambled siRNA (Fig. 6D). It was not possible to fluorochrome mark PCBP2-overexpressing cells while simultaneously evaluating their response to NaB, because cotransfection with FITC+ scrambled siRNA and the PCBP2 plasmid followed by NaB treatment resulted in extensive cell death. These experiments demonstrate that an increase in the PCBP2 level curbs lytic gene expression and limits the number of cells susceptible to a lytic activation signal.
FIG 6.
Overexpression of PCBP2 results in repression of the EBV lytic cycle. (A to C) HH514-16 cells were transfected with an empty vector (EV) or the PCBP2 plasmid, exposed to NaB after 12 h, and harvested after another 24 h for determination of the relative amounts of transcripts from 3 EBV lytic genes, BZLF1, BMRF1, and BFRF3, as well as PCBP2 by qRT-PCR after normalization to 18S rRNA levels by using the ΔΔCT method (A) and examined for PCBP2 expression (after comparison with cells stained with the isotype control antibody) (B) or for lytic cells by using reference EBV-seropositive and -seronegative human sera after gating on “live cells” based on forward- and side-scatter distribution by flow cytometry (C). Error bars in panel A indicate standard errors of the means of data from 3 technical replicates from each of 2 transfection experiments (*, P < 0.05). The number in panel B indicates the percentage of live, PCBP2-transfected cells expressing high levels of PCBP2. Numbers in panel C indicate the percentages of live cells that were lytic. (D) HH514-16 cells were transfected at the same time as in panels A and B with FITC+ scrambled siRNA (or were mock transfected as a control), harvested 36 h later, and examined for the percentage of FITC-positive cells (numbers in dot plots) by flow cytometry. Data are representative of two experiments.
STAT3 functions via PCBP2 to limit EBV lytic cycle activation.
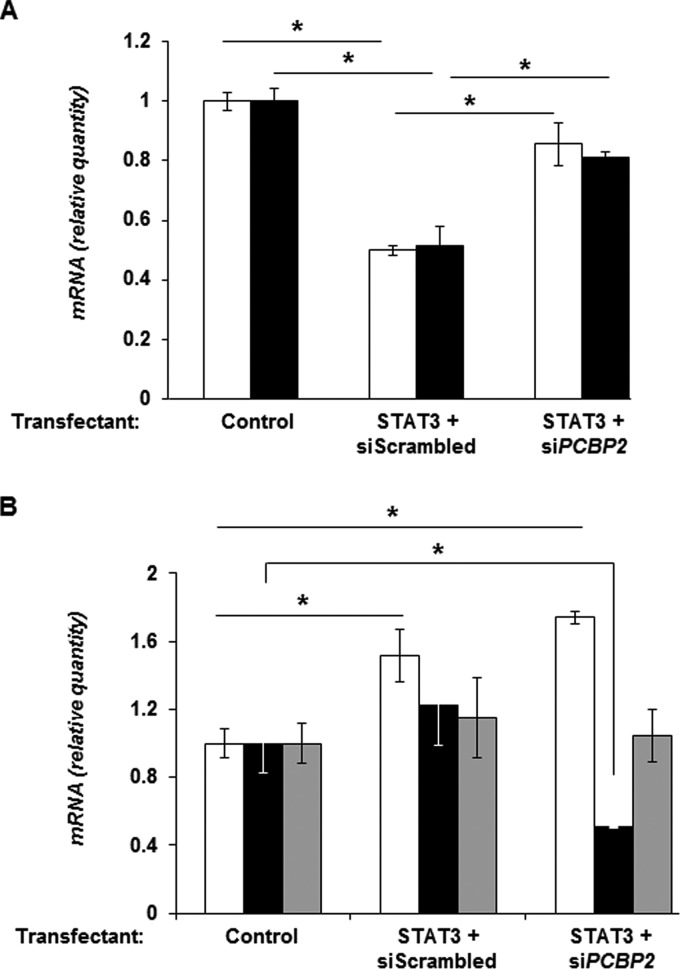
We reasoned that if STAT3 functions via PCBP2 to repress lytic susceptibility, then experimentally depleting PCBP2 while simultaneously overexpressing STAT3 should impair the ability of STAT3 to restrict lytic gene expression in response to NaB. As expected, cotransfection of the STAT3 plasmid and scrambled siRNA significantly reduced the levels of BZLF1 and BRLF1 transcripts in response to NaB exposure; in contrast, but as predicted, cotransfection of the STAT3 plasmid and siRNA targeting PCBP2 did not have a similar repressive effect on the levels of lytic gene transcripts (Fig. 7A). Figure 7B shows that transfection of the STAT3 plasmid resulted in an increase in the level of STAT3 mRNA, while the introduction of siRNA targeting PCBP2 caused a fall in the level of the PCBP2 transcript but not the PCBP1 transcript. In summary, STAT3 regulates the expression of PCBP2 to regulate the susceptibility of EBV-infected cells to lytic activation signals.
FIG 7.
STAT3 functions via PCBP2 to restrict EBV lytic cycle activation. HH514-16 cells were transfected with the empty vector and scrambled siRNA (control), the STAT3 plasmid and scrambled siRNA, or the STAT3 plasmid and siRNA targeting PCBP2; exposed to NaB after 12 h; and harvested after another 24 h for determination of the relative amounts of transcripts from the EBV immediate early lytic genes BZLF1 (open bars) and BRLF1 (black bars) (A) as well as STAT3 (open bars), PCBP2 (black bars), and PCBP1 (gray bars) (B) by qRT-PCR after normalization to 18S rRNA levels by using the ΔΔCT method. Error bars are standard errors of the means of data from 3 technical replicates from each of 2 transfection experiments (*, P < 0.05).
DISCUSSION
Our efforts to delineate the mechanism(s) by which STAT3 regulates EBV lytic susceptibility have identified the cellular protein PCBP2 as a prime candidate. We have confirmed the biological relevance of PCBP2 by investigating its levels in single lytic and refractory cells and determining the functional consequence of impairing (or increasing) PCBP2 expression on lytic activation, also at the single-cell level. Furthermore, our experiments mechanistically link STAT3, a transcription factor overactive in many human cancers (25–27), to PCBP2, a protein important in RNA biogenesis (20). The involvement of these proteins in EBV lytic susceptibility expands our understanding of how EBV manipulates the cellular machinery to regulate its persistence in humans. This study now also provides the EBV scientific community with a proteomic data set from lytic and refractory cells to unravel additional mechanisms underlying susceptibility to EBV lytic activation signals.
Analysis of the cellular transcriptome using messages from sorted refractory and lytic cells revealed differential levels of gene products from several thousand candidate genes (9). A combined informatics analysis of this data set, a proteomic data set from similarly sorted populations, and a publically available STAT3 ChIP-Seq peak set resulted in causally linking STAT3 and PCBP2 to the refractory state. Although it was somewhat surprising that the levels of STAT3 protein in refractory cells did not meet the 2-fold increase cutoff, this result was consistent with our observation of a small increase in the STAT3 mRNA level in the transcriptome analysis (8, 9). These data are furthermore consistent with STAT3 being a transcription factor, a group of proteins that can effect considerable downstream changes without large changes in their own levels. Furthermore, because it potentially regulates the transcription of thousands of genes (9), STAT3 may function to maintain the refractory state not by one but by multiple mechanisms. Indeed, we have discovered that STAT3 may also restrain lytic susceptibility by upregulating members of a transcriptional corepressor family in refractory cells (9).
PCBP2 belongs to the KH (hnRNP K homology) domain superfamily of RNA-binding proteins that bind to C-rich single-strand motifs (20). PCBPs are defined by their triple-KH-domain structure and their poly(C)-binding specificity. A variety of functions, including stabilization of human globin mRNA, translational silencing of the human 15-lipoxygenase mRNA, and enhancement of the antiviral activity of alpha interferon (IFN-α), have been attributed to PCBPs (20, 24). PCBP2 can also stabilize poliovirus RNA, increase/facilitate the translation of poliovirus and hepatitis A virus RNAs, silence human papillomavirus 16 L2 mRNA, and suppress the transcription of vesicular stomatitis virus genes (20, 21). How PCBP2 regulates EBV lytic susceptibility remains to be determined. In this context, we have found that STAT3 also regulates the transcriptional repressors SZF1/ZNF589 and ZNF557 such that the levels of their gene products are increased in refractory cells (9). It is therefore tempting to speculate that STAT3-mediated coregulation of PCBP2 and the repressors SZF1 and ZNF557 may promote the stabilization of SZF1 and ZNF557 gene products by PCBP2 in refractory cells, thereby enhancing the repression of viral lytic genes compared to repression by only SZF1 and ZNF557. This would be consistent with the greater-than-expected change in lytic RNA levels that we observed following the introduction of the PCBP2 plasmid compared to the empty vector (Fig. 6A). Alternatively, PCBP2 may destabilize lytic mRNAs in refractory cells. Whether products of the SNRPD1, HNRBPL, and UTP14A genes, also important in RNA biogenesis (28–31) and with elevated levels in refractory cells, coregulate EBV lytic susceptibility remains to be determined.
Advances in proteomic approaches have prompted several recent studies of different aspects of the EBV life cycle. Some of these studies have evaluated changes in expression levels of cellular proteins as well as interactions of the latency protein EBNA1 with the cellular proteins nucleophosmin and hnRNP in EBV-infected epithelial cell lines (32, 33). Other studies have identified modifications of cellular proteins in response to EBV infection of B cells (34) and identified cellular proteins that are incorporated into herpesvirus virions (35). Yet others have used proteomics to examine the modulation of protein content in exosomes secreted by gammaherpesvirus-infected B cells (36). To our knowledge, the proteomic data set presented here is the first of its kind comparing cellular protein levels in separated latent cells to those in separated lytic cells—lytic cells that were identified based on the simultaneous expression of multiple lytic genes without the introduction of exogenous genetic material. This data set of protein (and cellular mRNA) levels under conditions of EBV latency and lytic cycles provides a subset of genes to be investigated further. Such investigations are likely to identify additional STAT3-dependent and -independent mechanisms of regulation of the latency-to-lytic cycle switch.
While the correlation between transcripts and proteins appeared to be low in our data sets (Fig. 1), it is unclear what the degree of correlation should be in our system. Studies with cancer cells at steady state have shown correlations ranging from 29% to 42% (37–40). Our analysis is further complicated by the facts that our cells were virus infected and exposed to an HDAC inhibitor known to have wide-ranging effects on the genome and that the metabolic state of cells responding (or not) to a lytic inducing agent is not likely to be at steady state. Furthermore, we had prespecified a minimum cutoff of a 2-fold increase to limit the candidates to only those proteins with the greatest magnitudes of change between lytic and refractory populations; this would also contribute to the low correlation. Notwithstanding the complicating factors mentioned above, further analysis of our data sets without a prespecified fold change showed that ∼78% of all proteins whose levels were increased in the refractory population also showed increases in RNA levels; in contrast, only ∼21% of all proteins with increased levels in the lytic population showed increases at the RNA level (data not shown). This difference between refractory and lytic populations is consistent with host cell shutoff resulting in RNA degradation in lytic cells (41).
This study demonstrates that high levels of PCBP2, a molecule which is transcriptionally regulated by STAT3, limit the susceptibility of latently infected cells to spontaneous lytic cycle-inducing signals in culture. Upon exposure to another lytic cycle-inducing signal such as NaB, a fraction of cells with lower levels of PCBP2 responds by supporting EBV lytic replication. However, another fraction of cells responds to NaB by further increasing the levels of intracellular PCBP2 under the influence of high levels of STAT3; these cells remain refractory, thereby ensuring that a substantial fraction of cells remains latently infected with EBV. Because viral persistence is a result of the balance between lytic and refractory states, this continued resistance to lytic activation signals impacts EBV persistence. Continuing studies at the single-cell level of both STAT3-dependent and -independent mechanisms underlying the refractory state in naturally infected cells will be key to understanding viral persistence and pathogenesis and devising methods to improve viral oncolytic therapies.
Supplementary Material
ACKNOWLEDGMENTS
This study was supported by funds from The Research Foundation for The State University of New York to S.B.-M.
We thank Steven Holland and Alexandra Freeman at the NIAID for providing access to AD-HIES patient samples and the Snyder Laboratory at Stanford University for providing access to LCL ChIP-Seq data via the ENCODE TFBS file database at the UCSC Genome Browser. We also thank Patrick Hearing for his valuable comments during the preparation of the manuscript.
S.B.-M. designed the study; S.K., X.L., C.C., S.C., and E.R.H. carried out the experiments; E.I.C. performed mass spectrometry and its analysis; J.Z. performed bioinformatic analysis; S.K., X.L., E.I.C., J.Z., and S.B.-M. analyzed data and interpreted the findings; and C.C. and S.B.-M. wrote the manuscript.
We have no conflicts of interest to disclose.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/JVI.00121-15.
REFERENCES
- 1.Bhaduri-McIntosh S. 2005. Human herpesvirus-8: clinical features of an emerging viral pathogen. Pediatr Infect Dis J 24:81–82. doi: 10.1097/01.inf.0000151367.14455.9c. [DOI] [PubMed] [Google Scholar]
- 2.Cohen JI. 2000. Epstein-Barr virus infection. N Engl J Med 343:481–492. doi: 10.1056/NEJM200008173430707. [DOI] [PubMed] [Google Scholar]
- 3.Thorley-Lawson DA, Gross A. 2004. Persistence of the Epstein-Barr virus and the origins of associated lymphomas. N Engl J Med 350:1328–1337. doi: 10.1056/NEJMra032015. [DOI] [PubMed] [Google Scholar]
- 4.Hippocrate A, Oussaief L, Joab I. 2011. Possible role of EBV in breast cancer and other unusually EBV-associated cancers. Cancer Lett 305:144–149. doi: 10.1016/j.canlet.2010.11.007. [DOI] [PubMed] [Google Scholar]
- 5.Iizasa H, Nanbo A, Nishikawa J, Jinushi M, Yoshiyama H. 2012. Epstein-Barr virus (EBV)-associated gastric carcinoma. Viruses 4:3420–3439. doi: 10.3390/v4123420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dziurzynski K, Chang SM, Heimberger AB, Kalejta RF, McGregor Dallas SR, Smit M, Soroceanu L, Cobbs CS, HCMV and Gliomas Symposium. 2012. Consensus on the role of human cytomegalovirus in glioblastoma. Neuro Oncol 14:246–255. doi: 10.1093/neuonc/nor227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Perrine SP, Hermine O, Small T, Suarez F, O'Reilly R, Boulad F, Fingeroth J, Askin M, Levy A, Mentzer SJ, Di Nicola M, Gianni AM, Klein C, Horwitz S, Faller DV. 2007. A phase 1/2 trial of arginine butyrate and ganciclovir in patients with Epstein-Barr virus-associated lymphoid malignancies. Blood 109:2571–2578. doi: 10.1182/blood-2006-01-024703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Daigle D, Megyola C, El-Guindy A, Gradoville L, Tuck D, Miller G, Bhaduri-McIntosh S. 2010. Upregulation of STAT3 marks Burkitt lymphoma cells refractory to Epstein-Barr virus lytic cycle induction by HDAC inhibitors. J Virol 84:993–1004. doi: 10.1128/JVI.01745-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hill ER, Koganti S, Zhi J, Megyola C, Freeman AF, Palendira U, Tangye SG, Farrell PJ, Bhaduri-McIntosh S. 2013. Signal transducer and activator of transcription 3 limits Epstein-Barr virus lytic activation in B lymphocytes. J Virol 87:11438–11446. doi: 10.1128/JVI.01762-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bhaduri-McIntosh S, Miller G. 2006. Cells lytically infected with Epstein-Barr virus are detected and separable by immunoglobulins from EBV-seropositive individuals. J Virol Methods 137:103–114. doi: 10.1016/j.jviromet.2006.06.006. [DOI] [PubMed] [Google Scholar]
- 11.Heston L, Rabson M, Brown N, Miller G. 1982. New Epstein-Barr virus variants from cellular subclones of P3J-HR-1 Burkitt lymphoma. Nature 295:160–163. doi: 10.1038/295160a0. [DOI] [PubMed] [Google Scholar]
- 12.Koganti S, Hui-Yuen J, McAllister S, Gardner B, Grasser F, Palendira U, Tangye SG, Freeman AF, Bhaduri-McIntosh S. 2014. STAT3 interrupts ATR-Chk1 signaling to allow oncovirus-mediated cell proliferation. Proc Natl Acad Sci U S A 111:4946–4951. doi: 10.1073/pnas.1400683111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chen EI, Hewel J, Felding-Habermann B, Yates JR III. 2006. Large scale protein profiling by combination of protein fractionation and multidimensional protein identification technology (MudPIT). Mol Cell Proteomics 5:53–56. doi: 10.1074/mcp.T500013-MCP200. [DOI] [PubMed] [Google Scholar]
- 14.Yates JR III, Eng JK, McCormack AL, Schieltz D. 1995. Method to correlate tandem mass spectra of modified peptides to amino acid sequences in the protein database. Anal Chem 67:1426–1436. doi: 10.1021/ac00104a020. [DOI] [PubMed] [Google Scholar]
- 15.Tabb DL, McDonald WH, Yates JR III. 2002. DTASelect and Contrast: tools for assembling and comparing protein identifications from shotgun proteomics. J Proteome Res 1:21–26. doi: 10.1021/pr015504q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Park SK, Yates JR III. 2010. Census for proteome quantification. Curr Protoc Bioinformatics Chapter 13:Unit 13.12.1-11. doi: 10.1002/0471250953.bi1312s29. [DOI] [PubMed] [Google Scholar]
- 17.Li C, Wong WH. 2001. Model-based analysis of oligonucleotide arrays: expression index computation and outlier detection. Proc Natl Acad Sci U S A 98:31–36. doi: 10.1073/pnas.98.1.31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Huang DW, Sherman BT, Lempicki RA. 2009. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc 4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 19.ENCODE Project Consortium. 2011. A user's guide to the encyclopedia of DNA elements (ENCODE). PLoS Biol 9:e1001046. doi: 10.1371/journal.pbio.1001046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Makeyev AV, Liebhaber SA. 2002. The poly(C)-binding proteins: a multiplicity of functions and a search for mechanisms. RNA 8:265–278. doi: 10.1017/S1355838202024627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dinh PX, Beura LK, Panda D, Das A, Pattnaik AK. 2011. Antagonistic effects of cellular poly(C) binding proteins on vesicular stomatitis virus gene expression. J Virol 85:9459–9471. doi: 10.1128/JVI.05179-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ghosh D, Srivastava GP, Xu D, Schulz LC, Roberts RM. 2008. A link between SIN1 (MAPKAP1) and poly(rC) binding protein 2 (PCBP2) in counteracting environmental stress. Proc Natl Acad Sci U S A 105:11673–11678. doi: 10.1073/pnas.0803182105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Vogt DA, Andino R. 2010. An RNA element at the 5′-end of the poliovirus genome functions as a general promoter for RNA synthesis. PLoS Pathog 6:e1000936. doi: 10.1371/journal.ppat.1000936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Xin Z, Han W, Zhao Z, Xia Q, Yin B, Yuan J, Peng X. 2011. PCBP2 enhances the antiviral activity of IFN-alpha against HCV by stabilizing the mRNA of STAT1 and STAT2. PLoS One 6:e25419. doi: 10.1371/journal.pone.0025419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Darnell JE., Jr 2002. Transcription factors as targets for cancer therapy. Nat Rev Cancer 2:740–749. doi: 10.1038/nrc906. [DOI] [PubMed] [Google Scholar]
- 26.Nepomuceno RR, Snow AL, Beatty RP, Krams SM, Martinez OM. 2002. Constitutive activation of Jak/STAT proteins in Epstein-Barr virus-infected B-cell lines from patients with posttransplant lymphoproliferative disorder. Transplantation 74:396–402. doi: 10.1097/00007890-200208150-00017. [DOI] [PubMed] [Google Scholar]
- 27.Yu H, Jove R. 2004. The STATs of cancer—new molecular targets come of age. Nat Rev Cancer 4:97–105. doi: 10.1038/nrc1275. [DOI] [PubMed] [Google Scholar]
- 28.Hu L, Wang J, Liu Y, Zhang Y, Zhang L, Kong R, Zheng Z, Du X, Ke Y. 2011. A small ribosomal subunit (SSU) processome component, the human U3 protein 14A (hUTP14A) binds p53 and promotes p53 degradation. J Biol Chem 286:3119–3128. doi: 10.1074/jbc.M110.157842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kawamura H, Tomozoe Y, Akagi T, Kamei D, Ochiai M, Yamada M. 2002. Identification of the nucleocytoplasmic shuttling sequence of heterogeneous nuclear ribonucleoprotein D-like protein JKTBP and its interaction with mRNA. J Biol Chem 277:2732–2739. doi: 10.1074/jbc.M108477200. [DOI] [PubMed] [Google Scholar]
- 30.Omnus DJ, Mehrtens S, Ritter B, Resch K, Yamada M, Frank R, Nourbakhsh M, Reboll MR. 2011. JKTBP1 is involved in stabilization and IRES-dependent translation of NRF mRNAs by binding to 5′ and 3′ untranslated regions. J Mol Biol 407:492–504. doi: 10.1016/j.jmb.2011.01.050. [DOI] [PubMed] [Google Scholar]
- 31.Zeiner GM, Foldynova S, Sturm NR, Lukes J, Campbell DA. 2004. SmD1 is required for spliced leader RNA biogenesis. Eukaryot Cell 3:241–244. doi: 10.1128/EC.3.1.241-244.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ding Y, Li XR, Yang KY, Huang LH, Hu G, Gao K. 2013. Proteomics analysis of gastric epithelial AGS cells infected with Epstein-Barr virus. Asian Pac J Cancer Prev 14:367–372. doi: 10.7314/APJCP.2013.14.1.367. [DOI] [PubMed] [Google Scholar]
- 33.Malik-Soni N, Frappier L. 2012. Proteomic profiling of EBNA1-host protein interactions in latent and lytic Epstein-Barr virus infections. J Virol 86:6999–7002. doi: 10.1128/JVI.00194-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ivaldi C, Martin BR, Kieffer-Jaquinod S, Chapel A, Levade T, Garin J, Journet A. 2012. Proteomic analysis of S-acylated proteins in human B cells reveals palmitoylation of the immune regulators CD20 and CD23. PLoS One 7:e37187. doi: 10.1371/journal.pone.0037187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lippe R. 2012. Deciphering novel host-herpesvirus interactions by virion proteomics. Front Microbiol 3:181. doi: 10.3389/fmicb.2012.00181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Meckes DG Jr, Gunawardena HP, Dekroon RM, Heaton PR, Edwards RH, Ozgur S, Griffith JD, Damania B, Raab-Traub N. 2013. Modulation of B-cell exosome proteins by gamma herpesvirus infection. Proc Natl Acad Sci U S A 110:E2925–E2933. doi: 10.1073/pnas.1303906110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lundberg E, Fagerberg L, Klevebring D, Matic I, Geiger T, Cox J, Algenas C, Lundeberg J, Mann M, Uhlen M. 2010. Defining the transcriptome and proteome in three functionally different human cell lines. Mol Syst Biol 6:450. doi: 10.1038/msb.2010.106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Schwanhausser B, Busse D, Li N, Dittmar G, Schuchhardt J, Wolf J, Chen W, Selbach M. 2011. Global quantification of mammalian gene expression control. Nature 473:337–342. doi: 10.1038/nature10098. [DOI] [PubMed] [Google Scholar]
- 39.Tian Q, Stepaniants SB, Mao M, Weng L, Feetham MC, Doyle MJ, Yi EC, Dai H, Thorsson V, Eng J, Goodlett D, Berger JP, Gunter B, Linseley PS, Stoughton RB, Aebersold R, Collins SJ, Hanlon WA, Hood LE. 2004. Integrated genomic and proteomic analyses of gene expression in mammalian cells. Mol Cell Proteomics 3:960–969. doi: 10.1074/mcp.M400055-MCP200. [DOI] [PubMed] [Google Scholar]
- 40.Vogel C, de Sousa Abreu R, Ko D, Le SY, Shapiro BA, Burns SC, Sandhu D, Boutz DR, Marcotte EM, Penalva LO. 2010. Sequence signatures and mRNA concentration can explain two-thirds of protein abundance variation in a human cell line. Mol Syst Biol 6:400. doi: 10.1038/msb.2010.59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rowe M, Glaunsinger B, van Leeuwen D, Zuo J, Sweetman D, Ganem D, Middeldorp J, Wiertz EJ, Ressing ME. 2007. Host shutoff during productive Epstein-Barr virus infection is mediated by BGLF5 and may contribute to immune evasion. Proc Natl Acad Sci U S A 104:3366–3371. doi: 10.1073/pnas.0611128104. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.