ABSTRACT
Respiratory syncytial virus (RSV) is the leading cause of lower respiratory tract infections in infants. Despite over 50 years of research, to date no safe and efficacious RSV vaccine has been licensed. Many experimental vaccination strategies failed to induce balanced T-helper (Th) responses and were associated with adverse effects such as hypersensitivity and immunopathology upon challenge. In this study, we explored the well-established recombinant vaccinia virus (rVV) RSV-F/RSV-G vaccination-challenge mouse model to study phenotypically distinct vaccine-mediated host immune responses at the proteome level. In this model, rVV-G priming and not rVV-F priming results in the induction of Th2 skewed host responses upon RSV challenge. Mass spectrometry-based spectral count comparisons enabled us to identify seven host proteins for which expression in lung tissue is associated with an aberrant Th2 skewed response characterized by the influx of eosinophils and neutrophils. These proteins are involved in processes related to the direct influx of eosinophils (eosinophil peroxidase [Epx]) and to chemotaxis and extravasation processes (Chil3 [chitinase-like-protein 3]) as well as to eosinophil and neutrophil homing signals to the lung (Itgam). In addition, the increased levels of Arg1 and Chil3 proteins point to a functional and regulatory role for alternatively activated macrophages and type 2 innate lymphoid cells in Th2 cytokine-driven RSV vaccine-mediated enhanced disease.
IMPORTANCE RSV alone is responsible for 80% of acute bronchiolitis cases in infants worldwide and causes substantial mortality in developing countries. Clinical trials performed with formalin-inactivated RSV vaccine preparations in the 1960s failed to induce protection upon natural RSV infection and even predisposed patients for enhanced disease. Despite the clinical need, to date no safe and efficacious RSV vaccine has been licensed. Since RSV vaccines have a tendency to prime for unbalanced responses associated with an exuberant influx of inflammatory cells and enhanced disease, detailed characterization of primed host responses has become a crucial element in RSV vaccine research. We investigated the lung proteome of mice challenged with RSV upon priming with vaccine preparations known to induce phenotypically distinct host responses. Seven host proteins whose expression levels are associated with vaccine-mediated enhanced disease have been identified. The identified protein biomarkers support the development as well as detailed evaluation of next-generation RSV vaccines.
INTRODUCTION
Respiratory syncytial virus (RSV) is the leading cause of severe lower respiratory tract infections in children, being responsible for up to 80% of the cases of acute bronchiolitis and frequent subsequent hospital admission in industrialized countries (1, 2). It has been estimated that RSV causes 33.8 million acute lower respiratory tract infections globally each year, resulting in 66,000 to 199,000 deaths among children under 5 years of age, 99% of which occur in developing countries (3). In addition to young children, immunocompromised individuals and the elderly are at increased risk for severe RSV disease and hospitalization. A licensed RSV vaccine is currently not available, and the development of a vaccine against RSV infection has proven to be very difficult. Formalin-inactivated RSV (FI-RSV) vaccine trials in the 1960s failed to induce protection upon natural RSV infection and even predisposed patients for enhanced disease, resulting in two deaths and hospitalization of 80% of the vaccinated subjects (4–7). Experimental inactivated and subunit vaccines tend to prime for the induction of unbalanced, type 2 T-helper (Th2) host responses that result in enhanced disease accompanied by an influx of inflammatory cells (8). On the other hand, development of live attenuated RSV vaccines that are more likely to induce balanced host responses appears difficult to tune and these live vaccines tend either to be over- or underattenuated (9, 10). Several RSV infection animal models have been developed, and these are exploited for vaccine research and studies on virus-induced (immuno)pathology. Multiple virological and immunological parameters are assessed in these studies to monitor the virus-host interaction. More recently, “high-resolution” genomics tools have also been applied, in particular, to characterize the host response. Previously, the FI-RSV mouse model had been used to characterize the host lung response in enhanced disease by mRNA profiling (11), and several host gene expression correlates of enhanced disease have been identified. Since it has been shown that immunization with formalin-inactivated mock antigen vaccine, like immunization with FI-RSV itself, also primes for Th2 skewed responses in the FI-RSV mouse model (12), we explored the recombinant vaccinia virus F (rVV-F)/rVV-G vaccination-challenge model of enhanced disease for which the induced disease phenotype depends only on the expression of the RSV F or G protein. For this model, it has been shown that vaccination of mice with rVV-G results in the production of neutralizing antibodies and subsequent reductions in RSV replication upon challenge infection (13, 14). However, these mice develop severe illness and pulmonary eosinophilia. Therefore, this model is ideally suited to the study of vaccine-enhanced disease upon RSV infection. Similarly to rVV-G priming, priming with rVV-F results in the production of neutralizing antibodies but induces a more effective immunity to viral replication than priming with rVV-G (13), without the pulmonary eosinophilia seen upon RSV challenge.
Using this mouse model of vaccine-enhanced disease and applying mass spectrometry (MS)-assisted protein profiling, we studied the molecular mechanism underlying vaccine-induced RSV-specific enhanced disease. With a comparative proteomics approach, we identified seven proteins as biomarkers of enhanced disease. These proteins include eosinophil peroxidase (Epx), chitinase-like-protein 3 (Chil3), and Itgam, representing proteins specific for the infiltrating eosinophils. In addition, proteins that are postulated to play a role in the preceding stages of induction and establishment of skewed vaccine-induced hypersensitivity responses, arginase-I (ArgI), Clca3, purine-rich element-binding protein beta (PurB), and hexokinase III (Hk3), have also been identified. These seven protein biomarkers can be used to support the development, as well as the detailed evaluation, of next-generation RSV vaccines.
MATERIALS AND METHODS
Cells and viruses.
HEp-2 and RK-13 cells were cultured in Dulbecco's modified Eagle's medium (DMEM) and RPMI 1640, respectively (both BioWhittaker, Verviers, Belgium), supplemented with 10% heat-inactivated fetal bovine serum (HI-FBS; Greiner, Frickenhausen, Germany), penicillin (BioWhittaker) (100 U/ml), streptomycin (BioWhittaker) (100 μg/ml), l-glutamine (BioWhittaker) (2 mM), and β-mercaptoethanol (Merck, Darmstadt, Germany) (10 to 5 M) at 37°C with 5% CO2. RSV-A2 (a kind gift from P. J. M. Openshaw, National Heart and Lung Institute, Centre for Respiratory Infection, Imperial College London, London, United Kingdom) was propagated on HEp-2 cells. Recombinant vaccinia viruses (13) were kindly provided by P. L. Collins (National Institutes of Health, Bethesda, MD, USA). Stocks of the parental vaccinia virus (VV-wt) and recombinant vaccinia viruses expressing the RSV fusion protein (rVV-F) and attachment protein (rVV-G) were produced on RK-13 cells. Infectivity of all virus stocks was checked on the corresponding cell line by serial 10-fold dilutions, and titers were calculated using the method of Reed and Muench (15).
Mouse vaccination and challenge studies.
Six-to-8-week-old female BALB/c mice (Netherlands Vaccine Institute, Bilthoven, The Netherlands) were vaccinated by scarification at the tail base with 106 PFU VV-wt, rVV-F, or rVV-G as described before (14, 16). Three weeks after vaccination, the mice were challenged by intranasal inoculation with 5 × 106 50% tissue culture infectious doses (TCID50) of RSV-A2. Weight was measured on a daily basis, and mice were sacrificed by exsanguination under isoflurane anesthesia at 1, 2, 3, 5, and 7 days after RSV challenge (10 animals per group per day). Five animals from each group were subjected to bronchoalveolar lavage (BAL). Subsequently, lungs were inflated with 10% neutral buffered formalin (formalin; Klinipath, Duiven, The Netherlands) and stored in 10% formalin for histopathological examination. The lungs of five other animals per group were dissected. Half of the lungs were transferred to virus transport medium for virus isolation, while the other half were stored in RNAlater (Ambion, Austin, TX, USA) for RNA and protein isolation. To obtain control samples, naive age-matched animals were sacrificed and processed identically. The study was approved by the Animal Ethics Committee and was carried out in accordance with animal experimentation guidelines.
BAL fluid phenotyping.
Cells in BAL fluid were analyzed by flow cytometry as described before (17). In short, BAL fluid samples were centrifuged and residual red blood cells were lysed using red blood cell lysis buffer (Roche Diagnostics GmbH, Mannheim, Germany) for 20 min at room temperature. Cells in BAL fluid were counted and labeled with fluorescent monoclonal antibodies in phosphate-buffered saline (PBS) supplemented with 3% HI-FBS for 60 min on ice. Next, cells were washed once with PBS and subjected to fluorescence-activated cell sorter (FACS) analysis.
Virus isolation.
Virus was isolated from lung tissue by homogenizing half of the lung in 1 ml virus transport medium in a Fastprep-24 instrument (MPbio, Illkirch, France) using ceramic spheres (VWR International, Radnor, PA, USA) (0.25-in. diameter) according to the manufacturer's protocol (20 min and 10 m/s). The homogenate was clarified by centrifugation, and the supernatant was applied on a monolayer of HEp-2 cells in serial 2-fold dilutions. Cells were screened microscopically for cytopathic effect (cpe), and TCID50 values were calculated as described above.
Protein isolation.
After removal of the RNAlater, the lungs were homogenized and lysed in TRIzol and proteins were isolated from the interphase and organic phase that remained after RNA extraction from TRIzol samples. Four volumes of ice-cold (−20°C) acetone was added to these fractions and incubated at −20°C for 1 h. Precipitated proteins were then centrifuged at maximum speed for 5 min. The protein pellet was washed twice with ice-cold 80% acetone. The pellet was air-dried and suspended in 40 μl of lysis buffer containing 30 mM Tris, 7 M urea, 1 M thiourea, and 4% 3-[(3-cholamidopropyl)-dimethylammonio]-1-propanesulfonate (CHAPS). The protein concentration in each sample was determined using a two-dimensional (2-D) Quant kit (GE Healthcare, Amersham, United Kingdom) according to the manufacturer's instructions.
Mass spectrometry.
A total amount of 25 μg protein was loaded onto a 12% SDS-PAGE gel and run at 100 V until the front reached the bottom of the gel. The gel was stained overnight with Coomassie blue silver. Each lane was divided into 5 slices, and each slice was cut into smaller pieces and prepared for protein digestion with trypsin. Peptides were extracted using 2% trifluoroacetic acid (TFA) and buffer B (80% acetonitrile [ACN], 0.5% acetic acid, 1% TFA) and subsequently desalted and concentrated using C18 StageTips and buffer A (0.5% acetic acid, 1% TFA). Peptide mixtures were purified and desalted using C18 StageTips. Peptide separation and sequence determination were performed with a nano-high-performance liquid chromatography system (Agilent 1100 series; Agilent, Amstelveen, The Netherlands) connected to a 7-T linear quadrupole-ion trap-ion cyclotron resonance (ICR) Fourier transform (FT) mass spectrometer (Thermo Electron, Breda, The Netherlands). Peptides were separated on a PicoTip emitter for online electrospray (New Objective, Woburn, MA) (15-cm length, 100-μm inner diameter) packed with 3-μm-diameter C18 beads (Reprosil; Dr Maisch GmbH, Ammerbuch-Entringen, Germany) with a 60-min linear gradient of 2.4% to 40% acetonitrile–0.5% acetic acid at a 300 nl/min flow rate. The four most abundant ions were sequentially isolated and fragmented in the linear ion trap by applying collisionally induced dissociation. Proteins were identified using the MASCOT search engine (Matrix Science, London, United Kingdom) and the human International Protein Index (IPI) database (forward and reverse sequences) and the following search criteria: 20 ppm peptide tolerance, a maximum of 2 missed cleavages, a fixed carbamidomethyl modification of cysteines, and variable oxidation (M) and deamidation (NQ) modification. A UniProt KB mapping table was used to map IPI identifiers (IDs) to UniProt IDs (ftp://ftp.uniprot.org/pub/databases/uniprot/current_release/knowledgebase/idmapping/). Differentially expressed proteins were functionally classified using GO-slim identifiers (18) and Ingenuity pathway analysis (Qiagen, Redwood City, CA, USA).
Validation of peptides and proteins.
Mascot database search files were further processed using MSQuant 1.4.3 to generate first-ranked peptide lists. A validation filter was applied to these lists using the reverse database searches. First, the mass accuracy cutoff value was set at the mass relative error (in parts per million [ppm]) at which 95% of all identified peptides were still included. Then, the reverse database search was used to set the peptide cutoff score at which only 10% of the identified peptides were still included in the reverse database search list. The number of peptide sequences per protein was then calculated, and the list was further validated by the following criteria. Proteins that were identified on the basis of 3 or more peptides were accepted. If 2 peptides were identified, those peptides were not allowed to contain any modifications. If only 1 peptide was identified, no modifications were accepted, peptide scores had to be above 40, and the peptide delta score had to be above 10. This resulted in false-positive rates of 11% for proteins identified with 1 or 2 peptides and 0.9% for proteins identified with 3 or more peptides. After internal calibration of peptides masses by MSQuant and validation as just described, the absolute mass accuracy of all identified peptides was below 15 ppm for all samples, with an average of ∼2 ppm. To remove redundancy at the protein level and to uniquely assign peptides to one protein, the peptides were remapped using Protein Coverage Summarizer (http://ncrr.pnl.gov/software/).
Comparative protein expression analyses.
For each protein in the validated list of proteins, the number of unique parent ions (spectral count) was calculated based on peptide sequences and modifications. The number of spectra matched to peptides from a protein is used as a surrogate measure of protein abundance (19). A negative binomial model that handles overdispersion (20, 21) caused by limited numbers of biological replicates is used to estimate the per-protein counts. To test for differential levels of expression between two conditions, the mean of the normalized per-protein counts under each condition is computed and the means are compared for equality. An exact P value, defined as the probability of a pair of observed counts, is computed as described in reference 22. After computing the normalized mean counts under each set of conditions and the corresponding P values, we controlled for multiple testing by controlling the false-discovery rate (FDR), which was defined as the expected proportion of false rejection among the rejected hypotheses, using the Benjamini and Hochberg (BH) procedure.
RESULTS
Induction of skewed immune responses in an RSV vaccination-challenge mouse model.
BALB/c mice were primed with rVV-F, rVV-G, or VV-wt and challenged with RSV. Body weight, viral titers in the lungs, and the cellular composition of BAL fluid samples were measured, and histopathological examination of the lungs was performed for a period of 7 days. Body weight measurements showed that VV-wt-primed animals remained at the starting weight upon challenge with RSV (Fig. 1A). In contrast, animals that had been primed with the RSV fusion protein (rVV-F) or with the RSV attachment protein (rVV-G) showed a reduction in body weight to approximately 85% of the starting weight in 4 to 5 days. Subsequently, the rVV-F-primed animals returned to the starting weight whereas the weight of the rVV-G-primed animals remained low at 85% of the starting weight until the end of the study at day 7 (Fig. 1A).
FIG 1.
Relative body weights (A) and replication-competent virus titers in the lung (B) of VV wild-type (VV-wt) (black dots)-, rVV-F (white dots)-, and rVV-G (black triangles)-vaccinated mice up to 7 days following RSV challenge. Average numbers with standard errors are indicated.
Virus could be isolated from lung homogenates from all animals in the VV-wt group. Virus titers from VV-wt-primed mice that mounted a primary immune response to RSV peaked at day 3 to a level of 106 TCID50/gram lung tissue. At any time point, the highest titers obtained in rVV-F- or rVV-G-primed mice were at least 10-fold lower than those obtained for the VV-wt-primed mice. In addition, the virus titers in these groups of mice showed an earlier (day 2) and much lower peak than that seen with VV-wt-primed mice, pointing to partial protection from challenge in rVV-F- and rVV-G-primed mice (Fig. 1B).
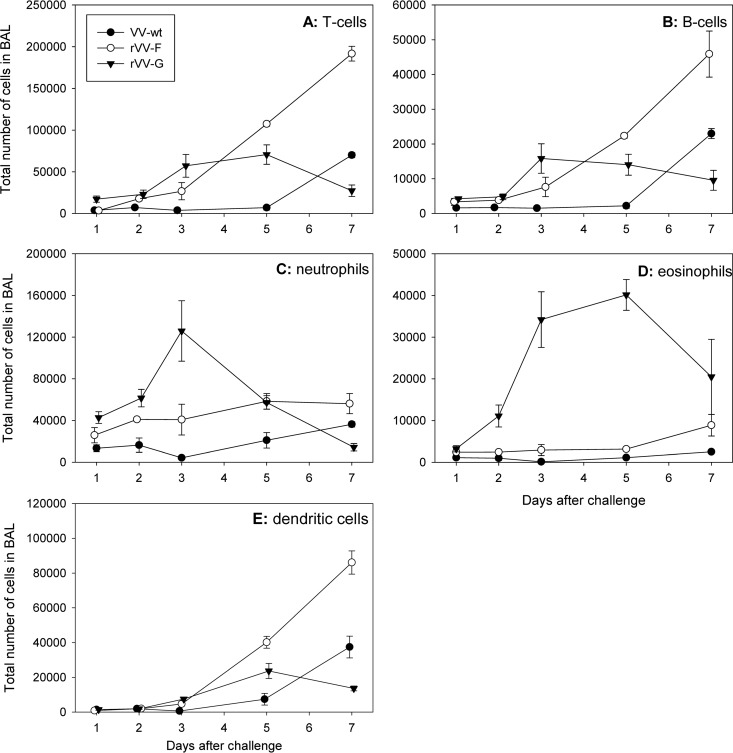
BAL fluid samples were obtained at days 1, 2, 3, 5, and 7 after challenge, and the cellular composition was determined by FACS analysis. Mice primed with VV-wt displayed a typical primary response to RSV infection, with a relatively late and modest influx of cells during the experiment. Mice primed with rVV-F displayed the highest numbers of T cells, B cells, and dendritic cells in the BAL fluid samples, while neutrophils and eosinophils were more abundant in the BAL fluid samples obtained from rVV-G-primed animals (Fig. 2). In contrast, eosinophil numbers in BAL fluid samples were much higher in rVV-G-mediated secondary responses than in rVV-F- or rVV-wt-primed animals (Fig. 2D).
FIG 2.
Cellular composition of BAL fluid. Total numbers of T cells (A), B cells (B), neutrophils (C), eosinophils (D), and dendritic cells (E) in BAL fluid from VV-wt (black dots)-, rVV-F (white dots)-, and rVV-G (black triangles)-vaccinated mice were determined by FACS analyses. Average numbers with standard errors are indicated.
Histopathological examination of slides of lung samples obtained from animals that were sacrificed at day 5 after challenge revealed that both VV-wt- and rVV-F-primed mice experienced interstitial pneumonia characterized by marked peribronchiolar and perivascular infiltrates. In contrast, rVV-G-primed animals experienced a more severe form of bronchointerstitial pneumonia, in some animals even leading to the formation of foci of necrosis associated with the presence of viral antigen. Interestingly, rVV-G-primed animals accounted for the highest level of positive staining of viral antigen of all the groups. While eosinophils were virtually absent in VV-wt- and rVV-F-primed animals, these cells were readily detectable in rVV-G-primed animals. Most eosinophils were located in the perivascular and peribronchiolar infiltrates and, to a lesser degree, in the alveolar walls and lumina (data not shown).
In line with the original observations obtained with this RSV vaccination and challenge model (13, 14), the data collectively demonstrate that the regimens of rVV-F and rVV-G priming indeed mediate the induction of distinct host response phenotypes upon RSV challenge. The collected lung samples are a good substrate to monitor protein expression profiles and to search for differences between the different priming regimens.
Mass spectrometry-assisted protein profiling of phenotypically distinct host responses.
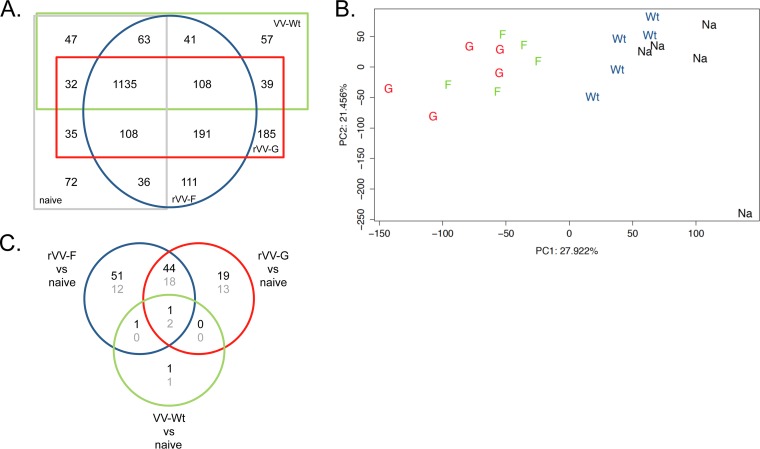
Lungs collected 5 days after RSV challenge were homogenized, and proteins were isolated and subjected to Fourier transform ion cyclotron resonance tandem MS (FT-ICR-MS/MS) analysis. The collected sample set was complemented with identical lung samples obtained from naive mice. Raw FT-ICR-MS/MS data were processed and validated. Between 2,247 and 2,547 distinct proteins were detected for each group of mice. Comparing the different experimental groups of mice, a large overlap in detected proteins was observed (Fig. 3A). Principal component analysis (PCA) of the normalized protein expression data (spectral counts) revealed two major clusters. One cluster consists of VV-wt-primed and naive mouse-derived lung samples, while the other consists of rVV-F and rVV-G lung samples, indicating that protein expression data determined for the two latter groups are similar but are different from those determined for the naive and VV-wt-primed mice (Fig. 3B).
FIG 3.
Proteome analysis of lung samples. (A) Distribution of the number of proteins identified in each group of vaccinated mice upon RSV challenge. (B) PCA of all lung proteome samples on the basis of normalized spectral counts. The first principal component (PC1) and the second principal component (PC2) are depicted on the x axis and the y axis. (C) Numbers of identified and validated proteins that were upregulated (black numbers) or downregulated (gray numbers) upon RSV challenge in response to VV-wt, rVV-F, or rVV-G vaccination compared to the results seen with naive mice.
Next, we applied spectral count analysis to identify proteins differentially expressed between the vaccination groups and naive mice. In this analysis, the number of unique parent ions (spectral counts) is used as a surrogate measure of protein abundance (19). First, spectral counts were compared between VV-wt-, rVV-F-, and rVV-G-vaccinated mice and naive control mice. Lungs from VV-wt-vaccinated mice displayed the smallest changes in protein expression, with only 3 down- and 3 upregulated proteins compared to the results seen with naive mice (Fig. 3C). Vaccination with rVV-F or rVV-G induced higher numbers and more pronounced changes in protein expression (129 and 97 differentially expressed proteins, respectively). The magnitude of these total lung proteome changes reflects a relatively slow-developing primary immune response (VV-wt-primed animals) and a much-faster-developing secondary response in the RSV-specific (rVV-F and rVV-G) primed mice. Only 3 proteins were differentially expressed in all 3 vaccination groups (Fig. 3C). As expected on the basis of the PCA data, a large overlap between the rVV-F-vaccinated mice and rVV-G-vaccinated mice was observed, where 50% to 67% of the proteins identified as differentially expressed with respect to the results seen with the naive mice were also found to be differentially expressed relative to those seen with the naive mice in the other group. Regardless of the priming regimen (rVV-F, rVV-G, or rVV-wt), differentially expressed proteins (relative to naive mice) shared between the experimental groups were always up- or downregulated in the same direction. The rVV-F- and rVV-G-primed responses are indeed highly similar: in a direct comparison of rVV-F- and rVV-G-primed lung tissue samples, only 7 proteins (Epx, Chil3, Itgam, Arg1, Clca3, PurB, and Hk3) are significantly differentially regulated (Table 1). Six of these proteins were more abundantly present in lung tissue of rVV-G-primed mice: 2 proteins (Arg1 and Epx) were upregulated in rVV-G-primed samples relative to rVV-F-primed samples, and 4 (Itgam, Chi3l3, Clca3, and Hk3) were unique to the rVV-G-primed samples. One protein (PurB) was expressed in rVV-F-primed mice only.
TABLE 1.
Differentially expressed proteins in rVV-G- versus rVV-F-primed mice upon challenge with RSV
| UniProt ID | Gene product | Protein name | Spectral count |
rVV-G vs rVV-F |
||
|---|---|---|---|---|---|---|
| rVV-G | rVV-F | Fold change | Adjusted P value | |||
| Q3U1U4 | Itgam | Integrin alpha-M | 3.8 | 0 | Up | 0.002 |
| P49290 | Epx | Eosinophil peroxidase | 12.2 | 1.0 | 12.1 | 0 |
| Q61176 | Arg1 | Arginase 1 | 18.2 | 3.1 | 6.0 | 0.002 |
| Q9D7Z6 | Clca3 | Calcium-activated chloride channel regulator 1 | 6.9 | 0 | Up | 0 |
| O35744 | Chi3l3 | Chitinase-like protein 3 | 5.7 | 0 | Up | 0 |
| Q3TRM8 | Hk3 | Hexokinase 3 | 2.8 | 0 | Up | 0.041 |
| O35295 | Purb | Transcriptional activator protein Pur-beta | 0 | 3.5 | Down | 0.024 |
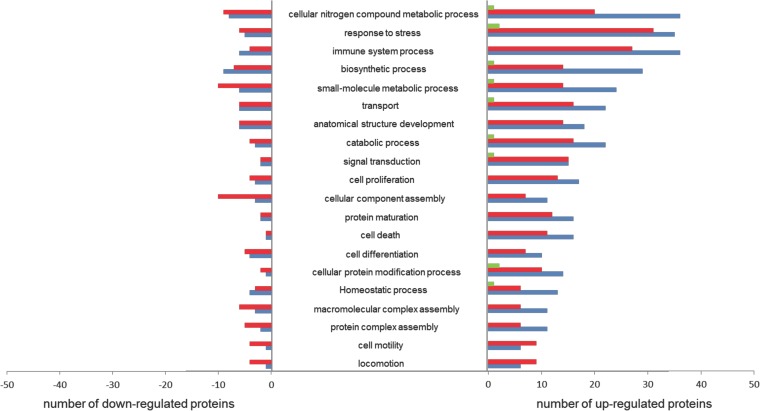
As expected, RSV challenge after VV-wt priming led to only a few differentially expressed proteins in lung tissue at day 5: Eif2s1, Gpd1l, and Armc10 were upregulated compared to naive mice, while Msra, Hba-a1/2, and Krt34 were downregulated. These proteins are involved in responses to stress and metabolic processes (Fig. 4). PCA also showed that the levels of protein expression in lungs from VV-wt and naive mice, both naive to RSV, are highly similar and explained why only 6 differentially expressed proteins were observed (Fig. 3C).
FIG 4.
Functional classification of differentially expressed proteins. Proteins were grouped by functional categories derived from GO slim and UniProt (http://www.uniprot.org/uniprot/). The graph shows the absolute number of differentially expressed proteins that could be assigned to a functional category for VV-wt (green bars)-, rVV-G (red bars)-, and rVV-F (blue bars)-vaccinated mice upon RSV challenge compared to naive mice.
There were many similarities between rVV-F- and rVV-G-primed animals in the alterations of protein expression (Fig. 3). A total of 65 proteins were differentially expressed in the two groups of mice compared to naive mice, and these mainly included proteins involved in metabolic processes and responses to stress (Fig. 4). A total of 63 proteins were differentially expressed only in rVV-F-vaccinated mice and a total of 32 only in rVV-G-vaccinated mice. In both groups, the majority of differentially expressed proteins, involving similar numbers and biological processes, were upregulated (Fig. 3 and 4).
Proteins are known to be involved in multiple pathways and, hence, are likely to have multiple functions. Pathway analysis (Ingenuity) was performed for further functional classification of differentially expressed proteins and to investigate the presumed interactions between these proteins within specific functional categories. A substantial proportion of the proteins differentially expressed in both rVV-F- and rVV-G-primed mice compared to naive mice appeared to play a role in antigen presentation (7.2%), biosynthesis (6.6%), or metabolism (7.8%), but the majority of the proteins were involved in immune and defense responses (16.3%). Within the latter group of differentially expressed proteins, five (Npm1, Snd1, Kars, Krt18, and Eif4a1) have been reported to be involved in host-virus interactions and another eight (BstI, Bst2, Gbp3, Ifit1, Ifit2, Ifit3, Ddx21, and Gbp1) have been shown to be active in antiviral immune responses. Within this group of proteins involved in antiviral immune responses, Gbp3, Ifit2, and Ddx21 were found to be upregulated compared to the levels in naive mice in rVV-F-primed mice only, while Bst2, Ifit1, Ifit3, and Gbp1 were upregulated in both rVV-F- and rVV-G-primed mice. A first line of defense against virus infections is formed by the type I interferon (IFN)-induced proteins. Five proteins in the list of differentially expressed proteins were involved in this defense response. Ifit1, Ifit2, and Ifit3 are members of the family of interferon-induced proteins with tetratricopeptide repeats (IFITs), and Gbp1 and Gbp3 are members of the family of IFN-induced guanylate-binding proteins which are all involved in antiviral responses (23, 24). Ifit1 and Ifit3 were differentially expressed in rVV-F- and rVV-G-primed mice, while Ifit2 was found differentially expressed only in rVV-F-primed mice. All three proteins are expressed in the cytoplasm and mitochondria, but Ifit2 is expressed in microtubules as well and interacts with the cytoskeleton, which may explain the difference observed.
DISCUSSION
Vaccines aim to induce antigen-specific memory responses that mediate safe and effective secondary responses when the host is exposed to the pathogen. Many live-attenuated and inactivated virus vaccines have been developed and have collectively reduced the incidence of disease. However, for RSV and several other viruses, classical formalin-inactivated vaccine production approaches failed, as they prime for unbalanced, skewed Th2-type immune responses upon challenge (7, 25, 26). These hypersensitivity responses are associated with immune pathology and are marked by peribronchiolar monocytic infiltration, including neutrophils and eosinophils (27). In this study, we explored the well-established rVV-RSV vaccination-challenge mouse model to identify protein markers and regulatory mechanisms that are associated with or that underlie vaccine-induced aberrant host responses. Comparing lung tissue samples from rVV-F- and rVV-G-primed animals with control samples from naive mice, we identified many differentially regulated proteins. These proteins are involved in metabolic processes and stress, but a significant proportion were identified as host-immune-response-related proteins (Table 2). By directly comparing vaccine-induced host responses, we were able to uncover subtle but highly relevant lung proteome differences between phenotypically distinct Th1- and Th2-like host responses. Direct comparison of levels of protein expression in lung tissue from RSV-challenged mice that had been vaccinated with rVV-F and rVV-G identified differential expression of only 7 proteins: Epx, Chil3, Itgam, Arg1, Clca3, PurB, and Hk3. These proteins are of particular interest, as their biological functions can be linked to the vaccination-induced host response phenotype and immunopathology.
TABLE 2.
Differentially expressed proteins involved in immune system processes
| UniProt ID | Gene product | Protein name | rVV-F vs naive |
rVV-G vs naive |
rVV-G vs rVV-F |
|||
|---|---|---|---|---|---|---|---|---|
| Fold change | Adjusted P value | Fold change | Adjusted P value | Fold change | Adjusted P value | |||
| P04187 | Gzmb | Granzyme B | Up | 0.000 | Up | 0.033 | ||
| Q02357 | Ank1 | Ankyrin-1 | −5.2 | 0.000 | −5.9 | 0.015 | ||
| P15508 | Sptb | Spectrin beta chain, erythrocytic | −6.2 | 0.000 | −30.7 | 0.011 | ||
| Q8R2Q8 | Bst2 | Bone marrow stromal antigen 2 | 11.7 | 0.000 | 11.2 | 0.004 | ||
| Q9R233 | Tapbp | Tapasin | Up | 0.000 | Up | 0.002 | ||
| P08032 | Spta1 | Spectrin apha chain, erythrocytic 1 | −7.1 | 0.000 | −10.6 | 0.002 | ||
| P36371 | Tap2 | Antigen peptide transporter 2 | Up | 0.001 | Up | 0.001 | ||
| Q64282 | Ifit1 | Interferon-induced protein with tetratricopeptide repeats 1 | Up | 0.001 | Up | 0.001 | ||
| Q60766 | Irgm1 | Immunity-related GTPase family M protein 1 | Up | 0.000 | Up | 0.000 | ||
| Q5I2A0 | Serpina3g | Serine protease inhibitor A3G | Up | 0.000 | Up | 0.000 | ||
| Q64345 | Ifit3 | Interferon-induced protein with tetratricopeptide repeats 3 | Up | 0.000 | Up | 0.000 | ||
| Q9QZ85 | Iigp1 | Interferon-inducible GTPase 1 | 130.7 | 0.000 | 98.4 | 0.000 | ||
| Q01514 | Gbp1 | Interferon-induced guanylate-binding protein 1 | 52.3 | 0.000 | 44.2 | 0.000 | ||
| O35955 | Psmb10 | Proteasome subunit beta type-10 | 35.1 | 0.000 | 37.2 | 0.000 | ||
| P42225 | Stat1 | Signal transducer and activator of transcription 1 | 30.0 | 0.000 | 30.0 | 0.000 | ||
| P21958 | Tap1 | Antigen peptide transporter 1 | Up | 0.000 | ||||
| P01029 | C4b | Complement C4-B | 38.4 | 0.000 | ||||
| P01863 | Ighg | Ig gamma-2A chain C region, A allele | 8.6 | 0.000 | ||||
| P29351 | Ptpn6 | Tyrosine-protein phosphatase nonreceptor type 6 | Up | 0.001 | Up | 0.000 | ||
| P11835 | Itgb2 | Integrin beta-2 | Up | 0.001 | Up | 0.000 | ||
| P47941 | Crkl | Crk-like protein | Up | 0.001 | ||||
| Q61646 | Hp | Haptoglobin | 4.4 | 0.001 | ||||
| Q9Z0E6 | Gbp2 | Interferon-induced guanylate-binding protein 2 | 10.0 | 0.002 | 8.3 | 0.038 | ||
| Q62418 | Dbnl | Drebrin-like protein | 28.5 | 0.002 | ||||
| Q06318 | Scgb1a1 | Uteroglobin | −23.0 | 0.002 | ||||
| Q61107 | Gbp4 | Guanylate-binding protein 4 | Up | 0.004 | ||||
| Q8CFB4 | Gbp5 | Guanylate-binding protein 5 | Up | 0.006 | Up | 0.001 | ||
| P97290 | Serping1 | Plasma protease C1 inhibitor | 9.1 | 0.006 | ||||
| Q64112 | Ifit2 | Interferon-induced protein with tetratricopeptide repeats 2 | Up | 0.007 | ||||
| O89053 | Coro1a | Coronin-1A | 2.8 | 0.012 | 3.2 | 0.012 | ||
| P02089 | Hbb-b2 | Hemoglobin subunit beta-2 | −2.0 | 0.018 | ||||
| P10711 | Tcea1 | Transcription elongation factor A protein 1 | Up | 0.020 | ||||
| P04441 | Cd74 | H-2 class II histocompatibility antigen gamma chain | Up | 0.022 | Up | 0.007 | ||
| Q9EPB4 | Pycard | Apoptosis-associated speck-like protein containing a CARD | 10.2 | 0.023 | 13.5 | 0.005 | ||
| P31230 | Aimp1 | Aminoacyl tRNA synthase complex-interacting multifunctional protein 1 | 16.4 | 0.023 | ||||
| P00493 | Hprt | Hypoxanthine-guanine phosphoribosyltransferase | 3.0 | 0.024 | ||||
| Q60710 | Samhd1 | Deoxynucleoside triphosphate triphosphohydrolase SAMHD1 | 2.2 | 0.027 | 2.3 | 0.030 | ||
| P18468 | H2-Eb1 | H-2 class II histocompatibility antigen, I-A beta chain | Up | 0.028 | Up | 0.018 | ||
| Q99JB2 | Stoml2 | Stomatin-like protein 2, mitochondrial | Up | 0.028 | ||||
| P01027 | C3 | Complement C3 | 2.0 | 0.042 | ||||
| Q64339 | Isg15 | Ubiquitin-like protein ISG15 | Up | 0.045 | ||||
| P09470 | Ace | Angiotensin-converting enzyme | −1.8 | 0.048 | −3.9 | 0.002 | ||
| Q9EQH2 | Erap1 | Endoplasmic reticulum aminopeptidase 1 | 15.7 | 0.044 | ||||
| Q61233 | Lcp1 | Plastin-2 | 2.1 | 0.040 | ||||
| P16110 | Lgals3 | Galectin-3 | 3.5 | 0.037 | ||||
| P01915 | H2-Eb1 | H-2 class II histocompatibility antigen, E-D beta chain | 17.5 | 0.032 | ||||
| P49290 | Epx | Eosinophil peroxidase | 34.0 | 0.007 | 12.0 | 0.000 | ||
| Q3U1U4 | Itgam | Integrin alpha-M | Up | 0.001 | Up | 0.000 | ||
An important feature of RSV-vaccine-primed Th2 skewed host responses is a marked increase in the influx of eosinophils into the lungs. As expected, rVV-G-primed mice indeed accumulated significant numbers of eosinophils in the lungs (BAL fluid) upon RSV challenge (Fig. 2). Eosinophil peroxidase (Epx) is expressed by eosinophils and is localized in the cytoplasmic granules. Against a background of many other cells present in lung tissue samples, we could easily detect increased levels of the Epx protein in the lungs of rVV-G-primed animals (Table 1). Upon RSV challenge, the Epx protein was present at levels that were much (12-fold) lower in the lungs of rVV-F-primed animals. The Epx protein functions as an oxidant and has been shown to be released at sites of infection to mediate lysis of protozoa or parasitic worms. Lung histology from the two fatal cases of the FI-RSV trial in the 1960s revealed a pronounced inflammatory lung infiltrate, including eosinophils and neutrophils (7, 28). This result has subsequently been reproduced in RSV vaccination-challenge animal models (29). The association of lung eosinophilia with enhanced disease in particular has attracted much attention, but it is most likely a secondary event, since rVV-G-primed eosinophil-deficient mice also display enhanced disease upon challenge with RSV (30). Nevertheless, vaccine-primed eosinophilia reflects an unbalanced host response to be avoided, as influx of these cells is regulated by Th2-type cytokines, including interleukin-4 (IL-4), IL-10, and IL-13 (31). Unfortunately, we could not detect these cytokines at the protein level, probably due to the presence of concentrations too low to be detected in a complex mixture of proteins. In the context of RSV infection, the Epx protein can be used for vaccine evaluation purposes. Increased Epx mRNA levels have not been observed in rodent RSV vaccination-challenge models of enhanced disease (11 and unpublished observation).
The Chil3 protein levels are also upregulated in rVV-G-primed animals relative to those primed with the F protein. Chil3 (chitinase-like-protein 3) is a secretory protein (also known as Ym1) with in vitro chemotactic activity for T lymphocytes, bone marrow polymorphonuclear leukocytes, and eosinophils. In addition, it has been demonstrated that Chil3 specifically triggers extravasation of eosinophils. The Chil3 protein shares conserved amino acid residues with the chitinase family of proteins but also harbors a conserved CXC chemokine motif near the NH2 terminus (32). This hybrid type of protein is mainly produced by macrophages and is localized in the lumen of the rough endoplasmic reticulum and the nuclear envelope of these cells and of neutrophils (33). In a murine model of airway hyperresponsiveness (AHR), Chil3 mRNA was also upregulated in the lungs. Interestingly, upregulation preceded the onset of airway inflammation in this model and specific inhibition of Chil3 mRNA expression suppressed the induction of eosinophilia (34), supporting the idea of a critical role for Chil3 in eosinophilic inflammation processes. Upregulation of Chil3 protein in the rVV-G-primed mice upon RSV challenge suggests a role for Chil3 in the eosinophilia observed in these mice.
Homing of effector cells in inflamed tissue is facilitated by many cell adhesion factors. In line with the pronounced influx of different cell types into the lungs of rVV-F- and rVV-G-primed animals upon challenge with RSV, we identified Itgam, Itgb2, Lgals3bp, Lama5, and Tns1 as being differentially expressed relative to naive animals. However, only the Itgam protein expression level was found to be significantly increased in lung tissue obtained from rVV-G-vaccinated mice in direct comparisons to lung tissue obtained from rVV-G- and rVV-F-primed animals. The Itgam gene encodes the CD11b integrin (alphaM) polypeptide, which pairs with the polypeptide integrin beta-2 (Itgb2 = CD18 integrin) to form a functional integrin also known as the complement receptor type 3 (CR3) or macrophage-1 (Mac-I) antigen. Itgb2 was also upregulated in rVV-G-primed mice, which indicates increased expression of the functional integrin. In contrast to Itgam, which was detected only in rVV-G-primed mice, Itgb2 was also found to be upregulated in rVV-F-primed mice compared to naive mice and with no statistical difference in expression results compared to rVV-G-primed mice (Table 2). Indeed, Itgam binds to multiple ligands and is involved in other biological processes in addition to cell adhesion (35). Itgam expression was originally reported to be specific for neutrophils and monocytes/macrophages. However, Itgam expression was also induced by IL-5 and IL-33 in eosinophils (36). In an rVV-G-primed Th2 skewed host response, Itgam expression can thus mediate the recruitment of neutrophils as well as of eosinophils to the lungs. The concerted actions of upregulated Chil3 and Itgam proteins may account for the high level of influx of eosinophils in the lungs of the rVV-G-primed animals.
Arginase-I (ArgI) catalyzes the hydrolysis of arginine to ornithine and urea and is constitutively expressed in hepatocytes (37) but has also been recognized as an enzyme involved in the inducible nitric oxide synthase (iNOS) pathway in myeloid cells, which is predominantly regulated by exogenous stimuli (37, 38). In cells isolated from human blood, Arg1 expression has been found thus far only in neutrophils (39). In mice, Arg1 expression has been reported for alternatively activated macrophages (AAM). AAM play an important role under Th2-driven pathological conditions such as asthma and expression of Arg1 together with Chil3 (40), suggesting that these macrophages help to orchestrate the Th2 skewed responses in rVV-G-primed enhanced disease. In addition, the elevated Arg1 levels in lungs from rVV-G-primed mice compared to those from rVV-F-primed mice may also point to a role for type 2 innate lymphocytes, since these cells constitutively express Arg1 (41). Participation of the latter cell type may also be dependent on the presence of IL-33. This cytokine was not detected in our proteome screen. However, microarray analysis of lung tissue samples obtained from the same experiment showed that the level of IL-33 mRNA was increased for rVV-G-primed samples compared with naive lung samples. In a direct comparison between rVV-G- and rVV-F-primed lung tissue samples, the level of the rVV-G-primed IL-33 lung mRNA signal was higher but the difference did not reach statistical significance (data not shown).
Members of the chloride channel, calcium-activated (CLCA) family of proteins, including the murine Clca3 (mClca3, alias gob-5) protein and its human ortholog hCLCA1, have been identified as relevant molecules in diseases with secretory dysfunctions, including asthma and cystic fibrosis (42). mClca3 is also upregulated in rVV-G-primed animals relative to rVV-F-primed animals. Biochemical analysis of the posttranslational processing and intracellular trafficking of the mClca3 protein showed that it does not form an anion channel as suggested by the protein name but is cleaved into two subunits that are fully secreted as a glycosylated protein complex into the extracellular environment (42). Interestingly, it has been demonstrated that hCLCA1 mRNA levels and protein expression are significantly increased in the airway epithelium of asthmatic patients (43, 44), and overexpression of mClca3 in mice resulted in goblet cell metaplasia and mucin overproduction (45). Targeting the mClca3 protein with antibodies inhibited these processes in asthmatic mice (46). Collectively, the data suggest that CLCA proteins may also play an important role in Th2-driven RSV vaccine-induced enhanced lung disease and that detection of Clca3 protein expression could be used to guide RSV vaccine design and evaluation.
Both hexokinase III (Hk3) and purine-rich element-binding protein beta (PurB) have no obvious function in the host response to infection. Hexokinases catalyze the first step in glucose metabolism. Hk3 expression is relatively low in most tissues, with the highest levels reported in lung, kidney, and liver. Hk3 may play a role in protecting against cell death, and its overexpression decreases the oxidant-induced production of reactive oxygen species (ROS) (47). In addition to Hk3 exhibiting a protein expression level that changes due to the influx of specific immune cells, its metabolic function can be related only indirectly to the observed host responses. PurB is the only protein that is expressed at lower levels in rVV-G-primed animals than in rVV-F-primed animals. PurB is a sequence-specific, single-stranded DNA-binding protein implicated in the control of both DNA replication and transcription. Defects in this gene are associated with the development of acute myelogenous leukemia, in which some hematopoietic precursors are arrested in an early stage (48). A nonequal role for PurB in cell division for the distinct immune cell types present in the RSV-challenged lungs may contribute to the skewed cell composition results observed for the different vaccine priming regimens.
In summary, using proteome analysis, we have identified a set of host proteins that are differentially expressed between two phenotypically distinct secondary immune responses, each of which is evoked upon challenge with RSV after priming with closely related, but distinct, RSV vaccine preparations: rVV-F and rVV-G. Vaccination of mice with rVV-G but not rVV-F prior to RSV challenge induces typical vaccine-mediated enhanced disease that is characterized by a marked influx of eosinophils and neutrophils. The host proteins identified as differentially expressed under the two sets of vaccination-dependent conditions are involved in processes related to the direct influx of eosinophils (Epx) and to chemotaxis and extravasation (Chil3) as well as in the homing to the lungs of these cells and neutrophils (Itgam). In addition, the increased protein levels of Arg1 and Chil3 point to a functional and regulatory role for alternatively activated macrophages and type 2 innate lymphoid cells in Th2 cytokine-driven RSV vaccine-mediated enhanced disease. The identification of those proteins supports research on regulatory aspects of vaccine-induced skewed host responses as well as the evaluation of next-generation RSV vaccines.
ACKNOWLEDGMENTS
We thank R. L. de Swart for critical readings of the manuscript.
This study was financially supported by the VIRGO consortium, which is funded by the Netherlands Genomics Initiative and by the Dutch government (FES0908).
REFERENCES
- 1.Bush A, Thomson AH. 2007. Acute bronchiolitis. BMJ 335:1037–1041. doi: 10.1136/bmj.39374.600081.AD. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Simoes EA. 1999. Respiratory syncytial virus infection. Lancet 354:847–852. [DOI] [PubMed] [Google Scholar]
- 3.Nair H, Nokes DJ, Gessner BD, Dherani M, Madhi SA, Singleton RJ, O'Brien KL, Roca A, Wright PF, Bruce N, Chandran A, Theodoratou E, Sutanto A, Sedyaningsih ER, Ngama M, Munywoki PK, Kartasasmita C, Simões EA, Rudan I, Weber MW, Campbell H. 2010. Global burden of acute lower respiratory infections due to respiratory syncytial virus in young children: a systematic review and meta-analysis. Lancet 375:1545–1555. doi: 10.1016/S0140-6736(10)60206-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chin J, Magoffin RL, Shearer LA, Schleble JH, Lennette EH. 1969. Field evaluation of a respiratory syncytial virus vaccine and a trivalent parainfluenza virus vaccine in a pediatric population. Am J Epidemiol 89:449–463. [DOI] [PubMed] [Google Scholar]
- 5.Fulginiti VA, Eller JJ, Joyner JW, Bier J, Sieber OF, Minamitani M, Meiklejohn G. 1969. Respiratory virus immunization. Am J Epidemiol 89:435–448. [DOI] [PubMed] [Google Scholar]
- 6.Kapikian AZ, Mitchell RH, Chanock RM, Shvedoff RA, Stewart CE. 1969. An epidemiologic study of altered clinical reactivity to respiratory syncytial (RS) virus infection in children previously vaccinated with an inactivated RS virus vaccine. Am J Epidemiol 88:405–421. [DOI] [PubMed] [Google Scholar]
- 7.Kim HW, Canchola JG, Brandt CD, Pyles G, Chanock RM, Jensen K, Parrott RH. 1969. Respiratory syncytial virus disease in infants despite prior administration of antigenic inactivated vaccine. Am J Epidemiol 89:422–434. [DOI] [PubMed] [Google Scholar]
- 8.Openshaw PJM, Culley FJ, Olszewska W. 2001. Immunopathogenesis of vaccine-enhanced RSV disease. Vaccine 20(Suppl 1):S27–S31. doi: 10.1016/S0264-410X(01)00301-2. [DOI] [PubMed] [Google Scholar]
- 9.Wright PF, Karron RA, Belshe RB, Shi JR, Randolph VB, Collins PL, O'Shea AF, Gruber WC, Murphy BR. 2007. The absence of enhanced disease with wild type respiratory syncytial virus infection occurring after receipt of live, attenuated, respiratory syncytial virus vaccines. Vaccine 25:7372–7378. doi: 10.1016/j.vaccine.2007.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Anderson LJ. 2013. Respiratory syncytial virus vaccine development. Semin Immunol 25:160–171. doi: 10.1016/j.smim.2013.04.011. [DOI] [PubMed] [Google Scholar]
- 11.Schuurhof A, Bont L, Pennings JL, Hodemaekers HM, Wester PW, Buisman A, de Rond LC, Widjojoatmodjo MN, Luytjes W, Kimpen JL, Janssen R. 2010. Gene expression differences in lungs of mice during secondary immune responses to respiratory syncytial virus infection. J Virol 84:9584–9594. doi: 10.1128/JVI.00302-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Boelen A, Andeweg A, Kwakkel J, Lokhorst W, Bestebroer T, Dormans J, Kimman T. 2000. Both immunisation with a formalin-inactivated respiratory syncytial virus (RSV) vaccine and a mock antigen vaccine induce severe lung pathology and a Th2 cytokine profile in RSV-challenged mice. Vaccine 19:982–991. [DOI] [PubMed] [Google Scholar]
- 13.Olmsted RA, Elangot N, Prince GA, Murphy BR, Johnson PR, Mosst B, Chanock RM, Collins PL. 1986. Expression of the F glycoprotein of respiratory syncytial virus by a recombinant vaccinia virus: comparison of the individual contributions of the F and G glycoproteins to host immunity. Proc Natl Acad Sci U S A 83:7462–7466. doi: 10.1073/pnas.83.19.7462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Openshaw PJ, Clarke SL, Record FM. 1992. Pulmonary eosinophilic response to respiratory syncytial virus infection in mice sensitized to the major surface glycoprotein G. Int Immunol 4:493–500. [DOI] [PubMed] [Google Scholar]
- 15.Reed LJ, Muench H. 1938. A simple method of estimating fifty percent endpoints. Am J Hyg (Lond) 27:493–497. [Google Scholar]
- 16.Stott EJ, Ball LA, Young KK, Furze J, Wertz GW. 1986. Human respiratory syncytial virus glycoprotein G expressed from a recombinant vaccinia virus vector protects mice against live-virus challenge. J Virol 60:607–613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.van Rijt LS, Kuipers H, Vos N, Hijdra D, Hoogsteden HC, Lambrecht BN. 2004. A rapid flow cytometric method for determining the cellular composition of bronchoalveolar lavage fluid cells in mouse models of asthma. J Immunol Methods 288:111–121. doi: 10.1016/j.jim.2004.03.004. [DOI] [PubMed] [Google Scholar]
- 18.Harris MA, Clark J, Ireland A, Lomax J, Ashburner M, Foulger R, Eilbeck K, Lewis S, Marshall B, Mungall C, Richter J, Rubin GM, Blake JA, Bult C, Dolan M, Drabkin H, Eppig JT, Hill DP, Ni L, Ringwald M, Balakrishnan R, Cherry JM, Christie KR, Costanzo MC, Dwight SS, Engel S, Fisk DG, Hirschman JE, Hong EL, Nash RS, Sethuraman A, Theesfeld CL, Botstein D, Dolinski K, Feierbach B, Berardini T, Mundodi S, Rhee SY, Apweiler R, Barrell D, Camon E, Dimmer E, Lee V, Chisholm R, Gaudet P, Kibbe W, Kishore R, Schwarz EM, Sternberg P, Gwinn M, et al. 2004. The Gene Ontology (GO) database and informatics resource. Nucleic Acids Res 32:D258–D261. doi: 10.1093/nar/gkh036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Choi H, Fermin D, Nesvizhskii AI. 2008. Significance analysis of spectral count data in label-free shotgun proteomics. Mol Cell Proteomics 7:2373–2385. doi: 10.1074/mcp.M800203-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cameron AC, Trived PK. 1998. Regression analysis of count data. Cambridge University Press, Cambridge, United Kingdom. [Google Scholar]
- 21.Whitaker L. 1914. On the Poisson law of small numbers. Biometrika 10:36–71. [Google Scholar]
- 22.Anders S, Huber W. 2010. Differential expression analysis for sequence count data. Genome Biol 11:R106. doi: 10.1186/gb-2010-11-10-r106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhou X, Michal JJ, Zhang L, Ding B, Lunney JK, Liu B, Jiang Z. 2013. Interferon induced IFIT family genes in host antiviral defense. Int J Biol Sci 9:200–208. doi: 10.7150/ijbs.5613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nordmann A, Wixler L, Boergeling Y, Wixler V, Ludwig S. 2012. A new splice variant of the human guanylate-binding protein 3 mediates anti-influenza activity through inhibition of viral transcription and replication. FASEB J 26:1290–1300. doi: 10.1096/fj.11-189886. [DOI] [PubMed] [Google Scholar]
- 25.Griffin DE, Pan CH, Moss WJ. 2008. Measles vaccines. Front Biosci 13:1352–1370. doi: 10.2741/2767. [DOI] [PubMed] [Google Scholar]
- 26.de Swart RL, van den Hoogen BG, Kuiken T, Herfst S, van Amerongen G, Yüksel S, Sprong L, Osterhaus ADME. 2007. Immunization of macaques with formalin-inactivated human metapneumovirus induces hypersensitivity to hMPV infection. Vaccine 25:8518–8528. doi: 10.1016/j.vaccine.2007.10.022. [DOI] [PubMed] [Google Scholar]
- 27.Blanco JCG, Boukhvalova MS, Shirey KA, Prince GA, Vogel SN. 2011. New insights for development of a safe and protective RSV vaccine. Hum Vaccin 6:482–492. doi: 10.4161/hv.6.6.11562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Prince GA, Curtis SJ, Yim KC, Porter DD. 2001. Vaccine-enhanced respiratory syncytial virus disease in cotton rats following immunization with Lot 100 or a newly prepared reference vaccine. J Gen Virol 82(Pt 12):2881–2888. [DOI] [PubMed] [Google Scholar]
- 29.Graham BS, Henderson GS, Tang Y, Xiaotao L, Neuzil KM, Colley DG. 1993. Priming immunization determines T helper cytokine mRNA expression patterns in lungs of mice challenged with respiratory syncytial virus. J Immunol 151:2032–2040. [PubMed] [Google Scholar]
- 30.Castilow EM, Legge KL, Varga SM. 2008. Eosinophils do not contribute to respiratory syncytial virus vaccine-enhanced disease. J Immunol 181:6692–6696. doi: 10.4049/jimmunol.181.10.6692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.De Swart RL, Kuiken T, Timmerman HH, van Amerongen G, Van Den Hoogen BG, Vos HW, Neijens HJ, Andeweg AC, Osterhaus AD. 2002. Immunization of macaques with formalin-inactivated respiratory syncytial virus (RSV) induces interleukin-13-associated hypersensitivity to subsequent RSV infection. J Virol 76:11561–11569. doi: 10.1128/JVI.76.22.11561-11569.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Owhashi M, Arita H, Hayai N. 2000. Identification of a novel eosinophil chemotactic cytokine (ECF-L) as a chitinase family protein. J Biol Chem 275:1279–1286. doi: 10.1074/jbc.275.2.1279. [DOI] [PubMed] [Google Scholar]
- 33.Chang NC, Hung SI, Hwa KY, Kato I, Chen JE, Liu CH, Chang AC. 2001. A macrophage protein, Ym1, transiently expressed during inflammation is a novel mammalian lectin. J Biol Chem 276:17497–17506. doi: 10.1074/jbc.M010417200. [DOI] [PubMed] [Google Scholar]
- 34.Iwashita H, Morita S, Sagiya Y, Nakanishi A. 2006. Role of eosinophil chemotactic factor by T lymphocytes on airway hyperresponsiveness in a murine model of allergic asthma. Am J Respir Cell Mol Biol 35:103–109. doi: 10.1165/rcmb.2005-0134OC. [DOI] [PubMed] [Google Scholar]
- 35.Tan S-M. 2012. The leucocyte β2 (CD18) integrins: the structure, functional regulation and signalling properties. Biosci Rep 32:241–269. doi: 10.1042/BSR20110101. [DOI] [PubMed] [Google Scholar]
- 36.Suzukawa M, Koketsu R, Iikura M, Nakae S, Matsumoto K, Nagase H, Saito H, Matsushima K, Ohta K, Yamamoto K, Yamaguchi M. 2008. Interleukin-33 enhances adhesion, CD11b expression and survival in human eosinophils. Lab Invest 88:1245–1253. doi: 10.1038/labinvest.2008.82. [DOI] [PubMed] [Google Scholar]
- 37.Morris SM. 2007. Arginine metabolism: boundaries of our knowledge. J Nutr 137(Suppl 2):1602S–1609S. [DOI] [PubMed] [Google Scholar]
- 38.Murray PJ, Wynn TA. 2011. Obstacles and opportunities for understanding macrophage polarization. J Leukoc Biol 89:557–563. doi: 10.1189/jlb.0710409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Jacobsen LC, Theilgaard-Mönch K, Christensen EI, Borregaard N. 2007. Arginase 1 is expressed in myelocytes/metamyelocytes and localized in gelatinase granules of human neutrophils. Blood 109:3084–3087. doi: 10.1182/blood-2006-06-032599. [DOI] [PubMed] [Google Scholar]
- 40.Martinez FO, Helming L, Gordon S. 2009. Alternative activation of macrophages: an immunologic functional perspective. Annu Rev Immunol 27:451–483. doi: 10.1146/annurev.immunol.021908.132532. [DOI] [PubMed] [Google Scholar]
- 41.Bando JK, Nussbaum JC, Liang HE, Locksley RM. 2013. Type 2 innate lymphoid cells constitutively express arginase-I in the naive and inflamed lung. J Leukoc Biol 94:877–884. doi: 10.1189/jlb.0213084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mundhenk L, Alfalah M, Elble RC, Pauli BU, Naim HY, Gruber AD. 2006. Both cleavage products of the mCLCA3 protein are secreted soluble proteins. J Biol Chem 281:30072–30080. doi: 10.1074/jbc.M606489200. [DOI] [PubMed] [Google Scholar]
- 43.Toda M, Tulic MK, Levitt RC, Hamid Q. 2002. A calcium-activated chloride channel (HCLCA1) is strongly related to IL-9 expression and mucus production in bronchial epithelium of patients with asthma. J Allergy Clin Immunol 109:246–250. doi: 10.1067/mai.2002.121555. [DOI] [PubMed] [Google Scholar]
- 44.Woodruff PG, Boushey HA, Dolganov GM, Barker CS, Yang YH, Donnelly S, Ellwanger A, Sidhu SS, Dao-Pick TP, Pantoja C, Erle DJ, Yamamoto KR, Fahy JV. 2007. Genome-wide profiling identifies epithelial cell genes associated with asthma and with treatment response to corticosteroids. Proc Natl Acad Sci U S A 104:15858–15863. doi: 10.1073/pnas.0707413104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhang HI, He L. 2010. Overexpression of mclca3 in airway epithelium of asthmatic murine models with airway inflammation. Chin Med J (Engl) 123:1603–1606. doi: 10.3760/cma.j.issn.0366-6999.2010.12.022. [DOI] [PubMed] [Google Scholar]
- 46.Song L, Liu D, Wu C, Wu S, Yang J, Ren F, Li Y. 2013. Antibody to mCLCA3 suppresses symptoms in a mouse model of asthma. PLoS One 8:e82367. doi: 10.1371/journal.pone.0082367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wyatt E, Wu R, Rabeh W, Park H-W, Ghanefar M, Ardehali H. 2010. Regulation and cytoprotective role of hexokinase III. PLoS One 5:e13823. doi: 10.1371/journal.pone.0013823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Johnson EM, Daniel DC, Gordon J. 2014. The Pur protein family: genetic and structural features in development and disease. J Cell Physiol 228:930–937. doi: 10.1002/jcp.24237. [DOI] [PMC free article] [PubMed] [Google Scholar]