ABSTRACT
In order to understand factors that may influence latency-associated transcription and latency-associated transcript (LAT) phenotypes, we studied the expression of the herpes simplex virus 2 (HSV-2) LAT-associated microRNAs (miRNAs). We mapped the transcription initiation sites of all three primary miRNA transcripts and identified the ICP4-binding sequences at the transcription initiation sites of both HSV-2 LAT (pri-miRNA for miR-I and miR-II, which target ICP34.5, and miR-III, which targets ICP0) and L/ST (a pri-miRNA for miR-I and miR-II) but not at that of the primary miR-H6 (for which the target is unknown). We confirmed activity of the putative HSV-2 L/ST promoter and found that ICP4 trans-activates the L/ST promoter when the ICP4-binding site at its transcription initiation site is mutated, suggesting that ICP4 may play a dual role in regulating transcription of L/ST and, consequently, of miR-I and miR-II. LAT exon 1 (containing LAT enhancer sequences), together with the LAT promoter region, comprises a bidirectional promoter required for the expression of both LAT-encoded miRNAs and miR-H6 in latently infected mouse ganglia. The ability of ICP4 to suppress ICP34.5-targeting miRNAs and to activate lytic viral genes suggests that ICP4 could play a key role in the switch between latency and reactivation.
IMPORTANCE The HSV-2 LAT and viral miRNAs expressed in the LAT region are the most abundant viral transcripts during HSV latency. The balance between the expression of LAT and LAT-associated miRNAs and the expression of lytic viral transcripts from the opposite strand appears to influence whether individual HSV-infected neurons will be latently or productively infected. The outcome of neuronal infection may thus depend on regulation of gene expression of the corresponding primary miRNAs. In the present study, we characterize promoter sequences responsible for miRNA expression, including identification of the primary miRNA 5′ ends and evaluation of ICP4 response. These findings provide further insight into the virus' strategy to tightly control expression of lytic cycle genes (especially the neurovirulence factor, ICP34.5) and suggest a mechanism (via ICP4) for the transition from latency to reactivated productive infection.
INTRODUCTION
Herpes simplex virus 1 (HSV-1) and HSV-2 are closely related herpesviruses. HSV-1 typically infects the facial region and establishes a lifelong latent infection in sensory neurons of the trigeminal ganglia (TG), while HSV-2 typically infects the genital region and establishes a lifelong latent infection in sensory neurons of the sacral dorsal root ganglia. Periodically, either virus may reactivate to cause symptomatic or asymptomatic recurrences in the area served by these sensory neurons.
Both HSV-2 and HSV-1 acutely and latently express miRNAs, which are proposed to play an important role in regulating whether an infection will be productive or latent. These miRNAs, which are conserved more in location than in sequence (1, 2, 9, 10), reduce expression of viral genes transcribed from the opposite strand (2, 8–10) and likely also have additional functions (3–5) (see Fig. 1A).
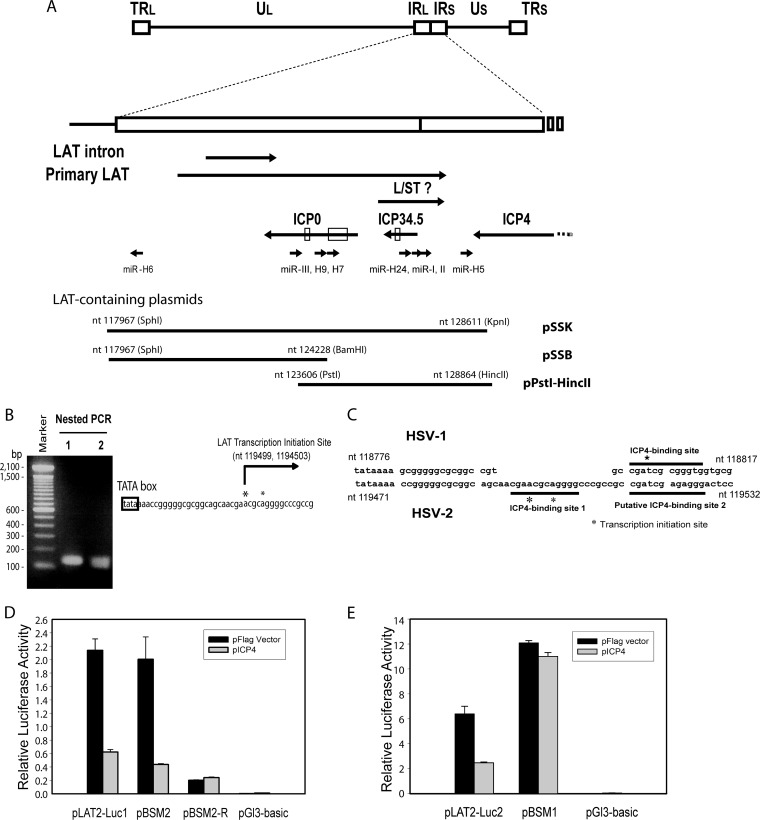
FIG 1.
Identification of HSV-2 LAT ICP4-binding sequence. (A) Schematic diagram of HSV-2 LAT region. The HSV-2 L/ST homolog is labeled. The relative positions of the inserts in LAT-containing plasmids, including pSSK, pSSB, and pPstI-HincII, are shown. (B) Mapping of HSV-2 LAT transcription initiation site. The HSV-2 LAT transcription start sites were mapped to nt 119499 (which was more dominant) and nt 1194503 with a 5′ RACE kit and total RNAs from HSV-2-infected Vero cells. (C) Comparison of the HSV-2 and HSV-1 LAT transcription initiation regions. Two HSV-1 ICP4-binding motifs, including putative ICP4-binding site 1 and putative ICP4-binding site 2 (underlined), were identified near the LAT transcription initiation site. (D) Mutation of putative ICP4-binding site 2 does not affect HSV-2 ICP4-mediated repression of LAT promoter reporter activity in cells transfected with LAT promoter reporters with or without pICP4, indicating that putative ICP4-binding site 2 does not function to limit LAT expression. pLAT2-Luc1 contains LAT sequences ranging from bp −392 to bp +239 relative to the dominant LAT transcription initiation site (nt 119499). pBSM2 contains LAT sequences ranging from bp −392 to bp +239 relative to the dominant LAT transcription initiation site (nt 119499), with substitution mutations in putative ICP4-binding site 2. pBSM2-R contains the same insert as pBSM2, oriented in the reverse direction, in the pGL3-Basic reporter. Firefly luciferase activity was normalized with cotransfected pRL-Ts, a Renilla luciferase reporter. (E) Mutation of putative ICP4-binding site 1, which overlaps the LAT transcription start sites, abolishes inhibition of the LAT promoter reporter activity by HSV-2 ICP4. LAT promoter reporters were cotransfected with or without pICP4 in 293 cells. pLAT2-Luc2 contains LAT promoter and putative ICP4 binding site 1 sequences ranging from bp −392 to bp +8 relative to the dominant LAT transcription initiation site (nt 119499). pBSM1 contains LAT sequences ranging from bp −392 to bp +8 relative to the dominant LAT transcription initiation site (nt 119499), with substitution mutations at putative ICP4-binding site 1.
HSV-2 LAT-encoded miRNAs, miR-I (also called miR-H3) and miR-II (also called miR-H4), as well as the corresponding HSV-1 LAT-encoded miRNAs, are processed from the primary latency-associated transcript (LAT) and from a L/ST (long/short junction-spanning transcripts) that traverses the viral long/short repeat junction, target ICP34.5, an inhibitor of PKR and a major viral neurovirulence factor (2, 6–11). The LAT promoter is highly active during latency, leading to transcription of the stable LAT intron and the unstable primary LAT (12, 13). Deletion of the LAT promoter in both HSV-1 and HSV-2 impairs viral reactivation (14–20). Although the role of the miRNAs in latency and reactivation is unclear, deletion of HSV-1 miR-H3 and miR-LAT-ICP34.5 (also named miR-H4) increases viral virulence by 4- to 10-fold in a neuronal cell culture model (8). HSV-2 miR-III (also named miR-H2) and its HSV-1 homolog are also processed from the primary LAT and target the immediate-early transactivator ICP0 (2, 10, 21–24). miR-H6 (identified in both HSV-1 and HSV-2) is expressed from sequences upstream of the LAT and from the strand opposite LAT and HSV-1 miR-H1 (1, 10, 25). miR-H6 is highly expressed during latency from an unknown primary transcript (1, 10, 26). In HSV-1, miR-H6 was reported to target ICP4, while HSV-2 miR-H6 was reported not to target ICP4 (10, 26).
HSV-1 ICP4, a key viral trans-activator, inhibits both LAT and L/ST expression by binding to specific binding sites near the transcription start sites (27–29). We previously showed that miR-I is expressed by the LAT promoter during latency in the ganglia of infected guinea pigs and mice (9). However, mutation of the HSV-1 ICP4-binding site at the HSV-1 L/ST transcription initiation sites increases the expression of HSV-1 miR-LAT-ICP34.5, which corresponds in location to HSV-2 miR-II, suggesting that the L/ST promoter also contributes to miR-I expression at times when ICP4 inhibition of L/ST expression is relieved. Therefore, ICP4 is likely directly involved in the transcriptional regulation of HSV-2 LAT-encoded miRNAs.
Because miR-I, miR-II, miR-III, and miR-H6 are the most abundantly expressed HSV-2 viral miRNAs during latency and because the LAT-encoded miRNAs contribute to silencing important productive cycle genes, it is of interest to understand the regulation of the primary transcripts from which these miRNAs are processed. In addition to identifying the miR-H6 promoter, we thus undertook to characterize the primary LAT (a precursor for miR-I, miR-II, and miR-III), the L/ST transcript (a precursor for miR-I and miR-II), and the as-yet-unnamed miR-H6 primary transcript by identifying their 5′ transcription start sites and characterizing the interactions of their promoters with the important viral regulator, ICP4 (30–32), which has been shown to reduce transcription of HSV-1 LAT and HSV-1 L/ST (27, 33, 34) through interaction with sequences at their transcription start sites.
MATERIALS AND METHODS
Cells, viruses, miRNA inhibitor and antibodies.
Sequences of HSV-2 strain HG52 (GenBank accession no. NC_001798) and HSV-1 strain 17syn+ (GenBank accession no. NC_001806) were used as reference sequences. Vero, HEK 293, HeLa, and U2OS cell lines were obtained from ATCC. HSV-2 strain 333 was obtained from Gary Hayward (Johns Hopkins University, Baltimore, MD). HSV2 mutant virus ΔLAT (14) has a 624-bp NotI (nucleotides [nt] 119109)-NotI (nt 119732) deletion that removes the LAT promoter, its TATA box, and the adjacent 241 bp of LAT exon 1. HSV2 ΔR is a rescuant virus for HSV2 ΔLAT (14). HSV2 ΔLAT-P2 (35) has a 339-bp deletion in LAT exon 1 sequences between the 5′ end of the primary LAT and the 5′ end of the LAT intron (from nt 119770 to nt 120108) (previously named LAP-2) (see Fig. 5A). Polyclonal rabbit anti-HSV-2 ICP34.5 and ICP0 antibodies were previously described (2, 9). Anti-β-tubulin was obtained from BD Biosciences (San Jose, CA). Detection of HSV-2 ICP34.5 by Western blotting was performed as described previously (9). The miR-III inhibitor, a 2′-O-methyl-modified RNA, was also synthesized by Dharmacon (2). The nonspecific small interfering RNA (NS siRNA) control was also obtained from Dharmacon (9).
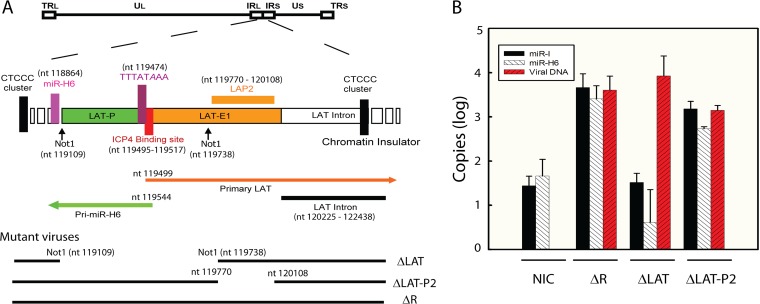
FIG 5.
HSV-2 LAP2 region is not essential for miR-H6 expression during latency. (A) Diagram of HSV-2 LAT region and mutant viruses used in panel B. ΔLAT-P2 is a LAP2 deletion mutant. ΔLAT is a NotI-NotI LAT promoter deletion mutant. (B) Expression of both miR-I and miR-H6 are abolished in mice ganglia latently infected with ΔLAT but not ΔLAT-P2. TG from mice infected with ΔLAT (n = 12), its rescuant ΔR (n = 12), and ΔLAP-P2 (n = 12) were extracted after 21 days postinoculation. Mice (n = 12) inoculated with plain medium were used as no-infection controls (NIC). Six mouse TG from three mice of each group were pooled, and each group thus contained four pooled TG samples. The error bar represents the standard deviations of four pooled samples in each group (each containing six ganglia from three mice). Total RNA and DNA were prepared from these pooled TG. The positive result for miR-I and miR-H6 (at ∼60 copies, with an upper 95% confidence interval of ∼100 copies) reflects the assay background in mouse TG.
Plasmid construction.
pSSK (containing the entire HSV-2 long repeat sequence), pSSB (containing long repeat sequences upstream of LAT and extending to a BamHI site in an ICP0 intron), and pPstI-HincII (containing partial LAT sequences but not the LAT promoter sequences) were described previously (9). The relative positions of the LAT-containing plasmids, including pSSK, pSSB, and pPstI-HincII, are shown in Fig. 1A. pLAT2-Luc1, containing the HSV-2 LAT promoter and a partial LAT exon 1 sequence, was constructed by inserting the NotI fragment (including the HSV-2 LAT promoter) from pSSB into the pFlag vector and then subcloning into the pGL3-Basic luciferase vector (Promega, Fitchburg, WI) BamHI and HindIII sites (2). pLAT2-Luc1R is a reverse direction (opposite strand) version of pLAT2-Luc1.
Luciferase-expressing promoter mutants were produced by overlap PCR, using two sets of oligonucleotide primers. Products were cloned into the NotI site of pLAT2-Luc or into the EcoRI site of pGL3-Basic vector (in some cases after an intermediate cloning step in pCR4-TOPO cloning vector (Life Technologies, Carlsbad, CA). All mutant plasmids were confirmed by both enzymatic digestion and sequencing. pBSM2 and pBSM2-R, containing the LAT promoter and a partial exon 1 sequence (nt 119107 to nt 119738) with mutations of putative ICP4-binding site 2, were constructed by overlap PCR. LAT sequences (nt 119073 to nt 119778) with putative ICP4 binding site mutations were first amplified by PCR using pSSB as the template with the primers oBP3 (GCGAGCCGGGCAGAGTGCGGA) and oMBP (CGCCGCCGatCgagaGGGACTCCGGAGAAGGAAGGCT; substitution mutations are indicated with the corresponding lowercase letters) or with primers oBP3 (GCGAGCCGGGCAGAGTGCGGA) and oMFP (GTCCCgagtGtaCGGCGGCGGGCCCCTGCGTT) to yield overlapping fragments containing the mutant LAT promoter insert for pBSM2. A mixture of these purified PCR fragments were used as the template and reamplified with oMBP and oFP1. The corresponding PCR product was digested with NotI and subsequently cloned into pLAT2-Luc1 using the NotI sites. pBSM2-R contains the same insert as pBSM2, oriented in the reverse direction, in the pGL3-Basic reporter. pLAT2-Luc2 containing the LAT promoter and ICP4-binding site 1 (nt 119073 to nt 119504) was constructed by first cloning the PCR fragment obtained using oBP3 and oST516 (CCCCTGCGTTCGTTGCTGCCGCGCCCCCGGTT) into the pCR4-TOPO cloning vector and then subcloning the EcoRI fragment into the pGL3-Basic vector using the EcoRI site. pBSM1, containing the LAT promoter and putative ICP4-binding site 1 mutant sequence (nt 119073 to nt 119504), was constructed by cloning the LAT sequence, amplified by oBP3 and oST517 (CCCCTGCtcgaGTTGCTGCCGCGCCCCCGGTT; substitution mutations of the ICP4-binding site 1 are indicated in lowercase) into the pCR4-TOPO cloning vector and subcloning the EcoRI fragment into the pGL3-Basic vector using the EcoRI site. pICP4 (including the HSV-2 ICP4 coding region under the control of the CMV-IE promoter) was obtained from Kening Wang and Jeffrey Cohen (National Institutes of Health/National Institute of Allergy and Infectious Disease, Bethesda, MD) (2). pCR432-499 containing HSV-2 ICP34.5 exon 1 sequences was cloned using oST432 (GAGCCCAGCCGCCCGCCATGT) and oST499 (TGTTCGCCCACTCTGCGTCGTCGT). pFlag-miRIII, containing ∼800 bp of HSV-2 ICP0 exon 3 spanning miR-III, was obtained by cloning the PCR product obtained using primers oST537 (GTCGTGCCGAGAGTGGCCTCTCTT) and oST538 (CGGCCTGGGACGACGGAGA) with pSSB as the template into the pCR4-TOPO cloning vector and then subcloning into the pFlag vector (Sigma, St. Louis, MO) at the EcoRI site. p-565+37 and p-428+37 are luciferase reporters that contain the L/ST promoter sequences ranging from bp −565 to bp 37 relative to the dominant LS/T transcription initiation site (nt 125799) as characterized in the manuscript. p-565+37 and p-428+37 contain the wild-type ICP4-binding sequence. p-565−9 and p-428−9 are luciferase reporters that contain L/ST promoter sequences ranging from bp −565 to bp −9 relative to the dominant L/ST transcription initiation site. p-565−9 and p-428−9 do not contain the ICP4-binding sequence. p-565+14M is also a luciferase reporter containing sequences ranging from bp −565 to bp 14 relative to the dominant L/ST transcription initiation site (nt 125799), with mutation of the ICP4-binding sequence from ATCGTCTCTTCG to gaatTCTCTTCG. To obtain these clones, PCR products using HSV-2 333 genomic DNA as the template and oligonucleotide pairs, including oST576 (CGGGTGTACTCCAAGAACCCATTAGCAT) and oST575 (GGAGCCCCGGAGCTCCGAA), oST577 (GGGGAACCAATAGGGGCCGATCA) and oST575, oST576, and oST578 (ACGATGGGAGCCCCGCGTATATATC), oST577 and oST578, along with oST576 and oST579 (TCCGAAGAGAattcGGAGCCCCGCGTATATAC, wherein mutations of the ICP4-binding site are labeled with lowercase letters) were first cloned into a pCR4-TOPO vector and then subcloned into the pGL3-Basic vector. Additional successive deletion constructs for L/ST promoter reporters, including p-254+14M and p-131+14M, contain a mutated ICP4-binding site but range in start position from bp −253 to bp −131 relative to the dominant L/ST transcription initiation site. Similarly, to construct the above two luciferase reporters, PCR products using HSV-2 333 genomic DNA as the template and oligonucleotide pairs, including oST582 (CTGCTAATTACCGCGAGCGGGAA) and oST579 with oST583 (CGGTCGCCGGGGCGGAGT) and oST579, were first cloned into a pCR4-TOPO cloning vector and then subcloned into pGL3-Basic vector (Promega). An additional L/ST core promoter reporter, p-71−12, was constructed using synthetic oligonucleotides, oST854 (AATTCGGGAGCCCCGCGTATATATCCGCGAGGGCCCGGCGCCGCCCCGCCGCTCCGCCCGCCCCAG) and oST855 (AATTCTGGGGCGGGCGGAGCGGCGGGGCGGCGCCGGGCCCTCGCGGATATATACGCGGGGCTCCCG). p-71−12 contains sequences from 71 bp upstream of the dominant L/ST transcription initiation site (nt 125799) to just downstream of the TATAA box, excluding the ICP4-binding sequence. Annealed oST854 and oST855 were directly cloned into the EcoRI site of the pGL3-Basic vector. Illustrations of inserts contained in p-565+37, p-428+37, p-565−9, p-428−9, and p-565+14M are included in Fig. 2C and E. Two luciferase reporters, p-650+74, including nearly the entire LAT exon 1 and 74 bp of sequences downstream of the primary miR-H6 (pri-miR-H6) dominant transcription initiation site (nt 119544), and p-650+106, including nearly the entire LAT exon 1 and 106 bp of sequences downstream of the pri-miR-H6 dominant transcription initiation site (nt 119544), were constructed by subcloning PCR-amplified LAT region sequences using the primer pair oST691 (GCTCTCTCACACGAGACACACGCA) and oST686 (CTGTGGGCATTTCTGCTGCGTCA) or the primer pair oST691 and oST687 (TTTATAAAACCGGGGGCGCGGCA) into a pGL3-Basic luciferase reporter. A diagram illustrating inserts in p-650+74 and p-650+106 is included in Fig. 4A.
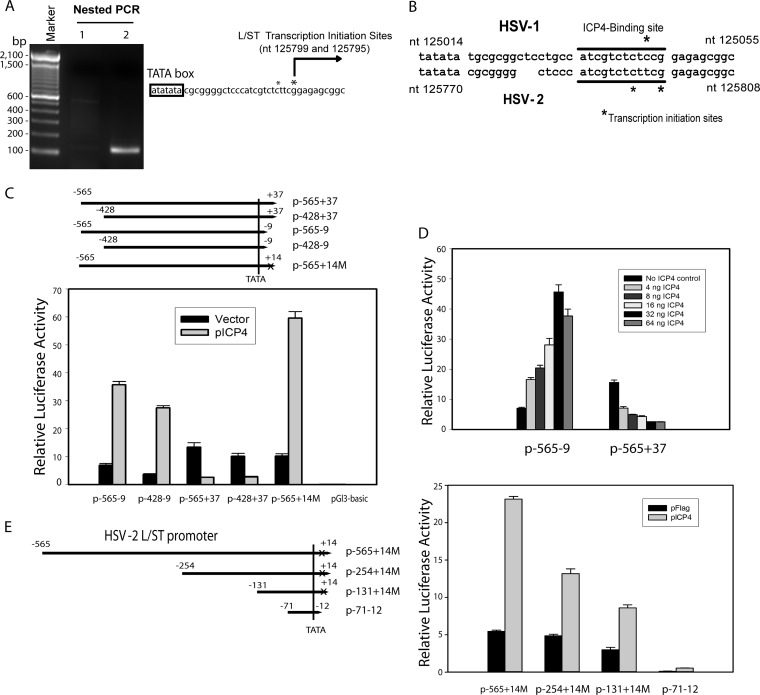
FIG 2.
Characterization of the putative HSV-2 L/ST promoter. (A) Mapping of the HSV-2 L/ST homolog transcription initiation sites. HSV-2 L/ST homolog promoter transcription initiation sites were mapped with a 5′ RACE kit and total RNAs from pPstI-HincII-transfected 293 cells. *, Transcription initiation site. (B) Comparison of the HSV-1 and HSV-2 L/ST transcription initiation regions. The ICP4-binding site is underlined. *, Transcription initiation site. NCBI sequence positions of the TATA boxes are shown. (C) ICP4 not only not inhibits the activity of an HSV-2 L/ST ICP4-binding site mutation or deletion construct but also trans-activates the ICP4-binding site mutant or deleted L/ST promoter reporter. L/ST luciferase reporters were cotransfected with or without ICP4 in 293 cells. p-565−9 and p-428−9 contain upstream L/ST promoter sequences but exclude the ICP4 binding site at the transcription initiation site. p-565+14M contains the upstream L/ST promoter sequence with a mutated L/ST ICP4-binding site. p-565+37 and p-428+37 contain the upstream L/ST promoter sequence and the wild-type L/ST ICP4-binding site. (D) Dose dependence of ICP4 on regulation of L/ST promoter is analyzed by cotransfecting different amounts of HSV-2 ICP4 plasmid. Both the repression of wild-type L/ST promoter reporter (p-565+37) and the activation of the ICP4-binding deletion promoter reporter (p-565−9) is ICP4 dose dependent, suggesting that HSV-2 ICP4 likely plays a double role in regulating L/ST promoter. (E) Activation of the L/ST promoter by ICP4 is not dependent on an ICP4-binding sequence. ICP4 is capable of inhibiting a series of successive deletion promoter reporters (p-565+14M, p-254+14M, and p-131+14M) containing a mutated ICP4-binding site, but ranging in start position from bp 565 to bp 131 upstream of the L/ST dominant transcription initiation site, in transfected 293 cells. An L/ST core promoter reporter (p-71−12) containing ∼71 bp sequences upstream of the dominant transcription initiation site showed drastic reduced promoter activity; however, it is still capable of being activated by ICP4 after normalized with the Renilla luciferase control.
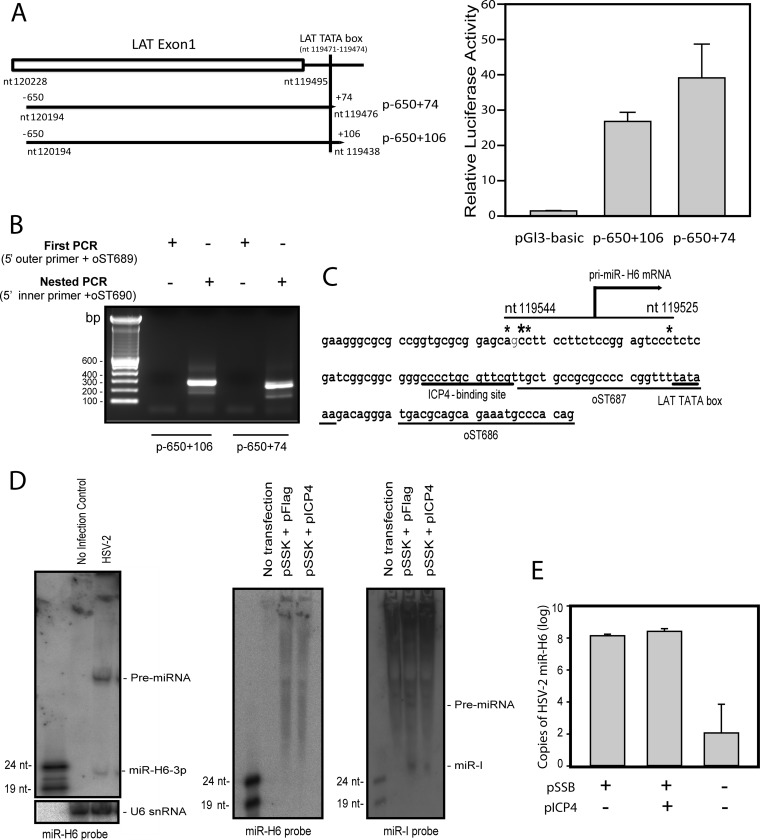
FIG 4.
Investigation of the miR-H6 promoter. (A) Mapping of pri-miR-H6 promoter activity. The left panel is the diagram of two pri-miR-H6 promoter luciferase reporters, p-650+106 that includes most of the LAT exon 1 sequence ranging from bp −650 to bp +106 relative to the dominant pri-miR-H6 transcription initiation site (nt 119544) as determined in the panel B, and p-650+74 that includes LAT exon 1 sequence ranging from bp −650 to bp +74 relative to the dominant pri-miR-H6 transcription initiation site (nt 119544). Both pri-miR-H6 reporters (p-650+106 and p-65+74) show strong promoter activities when transfected in 293 cells compared to pGL3-Basic vector. The luciferase activity is normalized with cotransfected pRL-Ts Renilla luciferase activity. This figure is representative of three independent experiments that yielded similar results. (B) Mapping of the 5′ transcriptional initiation site for miR-H6 primary transcript. Total RNAs from 293 cells transfected with reporter plasmids containing the potential pri-miR-H6 promoter region were used to map the 5′ transcription imitation site. (C) Diagram of miR-H6 primary mRNA transcription site. pri-miR-H6 does not use the LAT TATA box but is transcribed from a TATA-less promoter and initiates upstream of the LAT TATA box. The LAT ICP4-binding site, the LAT TATA box, and the positions for oST687 and oST686 used to clone the p-650+74 and p-650+106 are underlined. *, Transcription initiation site. (D) miR-H6 is below the detection limit of Northern blot in cells transfected with a LAT plasmid (pSSK) with or without pICP4, an HSV-2 ICP4 expression plasmid (middle panel), although it is detectable in infected cell cultures (left panel). In contrast, miR-I is detectable by Northern blotting in the pSSK-transfected cells, and ICP4 reduces miR-I expression in the transfected cells (right panel). pSSK contains the entire LAT promoter and LAT intron, including both miR-I and miR-H6 coding sequence (see Fig. 1A). (E) miR-H6 is detected by real-time PCR in cells transfected with pSSB (containing the entire LAT promoter, the LAT intron, and a partial ICP0 coding sequence; see Fig. 1A); however, ICP4 has no impact on the expression of miR-H6 in the pSSB-transfected cells. The positive result for miR-H6 (at 100 copies, with an upper 95% confidence interval of ∼10,000 copies) reflects the assay background in transfected 293 cells.
Transfection and dual-luciferase assay.
Plasmids and RNA oligonucleotides were transfected into 293 cells using Lipofectamine 2000 (Life Technologies) according to the manufacturer's instructions. The luciferase reporters were cotransfected with pRL-Ts, a Renilla luciferase reporter (36). The dual-luciferase assay was performed with a dual-luciferase assay kit (Promega) as described previously (36). The firefly luciferase activity was normalized to the Renilla luciferase activity. Figure panels comprise a representative of a minimum of three independent transfection experiments that yielded similar results.
Mapping of transcription start sites by 5′ RACE.
The transcription initiation site for the HSV-2 LAT was mapped using Vero cells infected with HSV-2 strain 333 at an MOI of 2. Total RNAs were prepared at 18 h postinfection by TRIzol (Life Technologies). Then, 10 μg of the total RNA was used to determine the transcription start site of HSV-2 LAT by 5′ RACE (rapid amplification of cDNA ends) with a RLM-RACE kit according to the manufacturer's instructions (Applied Biosystems, Carlsbad, CA). Primers (oST556, CCCGAGGCAAGAGGCGGA; oST555, CCTCGGAGGCGCGGAAGA) were used as gene-specific primers. The nest-PCR products were separated by using a 2% agarose gel, and the corresponding band was purified, ligated to a pCR4-TOPO cloning vector, transformed into TOP10 cells (Life Technologies), and sequenced. Similarly, the transcription initiation site for the HSV-2 L/ST was determined using RNA from 293 cells transfected with pSSK by 5′ RACE with a RLM-RACE kit. oST574 (CGGTCTAGGGTTGAACCGGCGA) and oST575 (GGAGCCCCGGAGCTCCGAA) were used as gene-specific primers. The transcription initiation site for the HSV-2 miR-H6 primary miRNA was mapped by 5′ RACE with the RLM-RACE kit using RNA from 293 cells transfected with p-650+106 and p-650+74. Primers (oST689, CTTCATAGCCTTATGCAGTTGCTC; oST690, GTTCCATCTTCCAGCGGATAGAA) specific for the firefly luciferase gene were used as gene-specific primers.
Infection of mice with HSV-2.
Six-week-old female Swiss-Webster mice (Simonsen Laboratories, Gilroy, CA) were anesthetized by intraperitoneal injection with sodium pentobarbital, followed by topical corneal administration of 0.5% proparacaine hydrochloride. The eyes of the mice were inoculated with 10 μl of viral stock (HSV-2 ΔLAT, 106 PFU/ml; HSV-2 ΔR, 106 PFU/ml; or HSV-2 ΔLAT-P2, 106 PFU/ml). Mice were treated with acyclovir starting at 40 h postinfection. The TG were removed at 21 days after inoculation and snap-frozen. Total DNA and RNA were extracted from ganglia in parallel using an AllPrep DNA/RNA kit (Qiagen, Valencia, CA) after homogenization with an Omni rotor-stator homogenizer (Omni International, Marietta, GA). 18S rRNA in these RNA samples was quantified by real-time PCR with a TaqMan ribosomal control kit (Applied Biosystems) and used to normalize RNA loading.
Detection of miRNAs by Northern blotting and real-time PCR.
The oligonucleotide probes for miR-I and U6 snRNA were described previously (9). Oligonucleotide oST592 (GATCCAAGGGCAGAAGAAGATGGG) was used to detect HSV-2 miR-H6-3p. The Northern blot conditions for the detection of HSV-2 miR-I, miR-H6, and U6 snRNA were described previously (9).
Detection of HSV-2 ICP34.5, ICP0, and L/ST using Northern blotting in HSV-2-infected cell cultures.
A total of 5 μg of total RNA from U2OS cells infected with HSV-2 or from uninfected controls were resolved in a formaldehyde denaturing 1% agarose gel (Life Technologies). For DNA probes, after transfer to GeneScreen Plus hybridization transfer membrane (Perkin-Elmer), the membrane was incubated in Hybrisol containing 50% formamide and 6× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate; EMD Millipore) at 60°C overnight with an HSV-2 ICP34.5-specific probe or with an HSV-2 ICP0-specific probe labeled with [α-32P]dCTP using a random priming kit (Promega). The HSV-2 ICP34.5-specific DNA probe was generated using an EcoRI digestion fragment of pCR432-499 containing ICP34.5 exon 1 sequences (nt 423 to nt 713). The membrane was then washed once with 2× SSC–0.5% sodium dodecyl sulfate (SDS) for 30 min at room temperature, once with 0.5× SSC–0.5% SDS, and twice with 0.2× SSC–0.1% SDS for 30 min each time at 60°C. For the ICP34.5-specific or S/LT-specific RNA probes, the temperature for blocking, hybridization, and washing was raised to 78°C. To prepare the ICP34.5-specific RNA probe, pCR432-499 containing HSV-2 ICP34.5 exon 1 sequences was first digested with PstI and then used as the template for in vitro transcription using T7 RNA polymerase and a MAXIscript SP6/T7 transcription kit (Life Technologies). To prepare the L/ST-specific RNA probe, pCR432-499 was first digested with NotI and then used as the template for in vitro transcription using SP6 RNA polymerase using the MAXIscript SP6/T7 transcription kit. To make the ICP0-specific DNA probe, the EcoRI fragment from pFlag-miRIII containing 805 bp (from nt 3839 to nt 4645) of ICP0 exon 3 sequences (not overlapping with the LAT intron) was labeled with [α-32P]dCTP using the random priming kit (Promega).
RESULTS
Mapping of the HSV-2 primary LAT transcription initiation sites and the LAT ICP4-binding sequences.
To determine whether HSV-2 ICP4 functions similarly to HSV-1 ICP4 in controlling LAT promoter activity, we first mapped the HSV-2 LAT transcription initiation site by 5′ RACE PCR using total RNA from Vero cells infected with HSV-2. Two primary LAT transcription initiation sites were mapped: at nt 119499 and at nt 1194503 (Fig. 1B). Transcripts initiating at nt 119499 exceed those initiating at nt 119503 by approximately 5:1, suggesting that nt 119499 is the dominant transcription initiation site for HSV-2 LAT. Sequence analysis revealed two potential ICP4-binding sequences (with core sequences AACGCA and ATCGAG) near the LAT transcription initiation sites, both homologous to the HSV-1 LAT ICP4-binding site (Fig. 1C) (with core sequence ATCGCG). Mutation of each of these sites revealed that HSV-2 ICP4 represses the HSV-2 LAT promoter reporter with mutations at ICP4-binding site 2 (Fig. 1D) but does not repress the HSV-2 LAT promoter reporter with mutations at the ICP4-binding site 1, indicating that the predicted ICP4-binding site 1 at the LAT transcription initiation site is the functional ICP4-binding site.
Mapping of the L/ST transcriptional initiation sites and the L/ST ICP4-binding sequences.
The same cDNA, prepared from infected Vero cells and used to successfully map the LAT transcription initiation site, was used to map the predicted L/ST transcription initiation site using L/ST-specific primers. However, no PCR product was obtained using the same 5′ RACE PCR method, possibly because the predicted HSV-2 L/ST transcript is inhibited too efficiently by ICP4 during lytic infection. Using total RNAs from cells transfected with pSSK, a LAT plasmid containing the potential L/ST promoter and sequences (but no functional ICP4 sequences), the L/ST transcription initiation sites were mapped to nt 125795 and nt 125799 (Fig. 2A). The ratio of transcripts using nt 125795 versus nt 125799 is approximately 3:5. Thus, nt 125799 appears to be the dominant transcription initiation site for L/ST. The putative HSV-2 L/ST ICP4-binding sequence closely resembles the HSV-1 L/ST ICP4-binding sequence (Fig. 2B). HSV-2 ICP4 inhibits L/ST luciferase reporters, including p-565+37 and p-428+37, that contain the putative ICP4-binding sequence (Fig. 2C), but does not inhibit L/ST reporters that do not contain the wild-type putative ICP4-binding sequence (p-565−9, p-428−9, and p-565+14M) (Fig. 2C and D). ICP4 transactivates L/ST promoters that do not contain the wild-type ICP4-binding sequence, in contrast to findings with the LAT ICP4-binding site mutation (in Fig. 1E). Furthermore, activation or inhibition of these L/ST promoter and mutant reporters depends on ICP4 dose (Fig. 2D). Although in the absence of ICP4 binding sites, ICP4 is expected to activate minimal promoters containing only TATA homologies (37), normalization of the L/ST firefly luciferase reporter activity to the activity of cotransfected pRL-Ts (a thromboxane synthase Renilla luciferase reporter (36) likely corrected for any nonspecific ICP-4 induced activation of the basal promoter. Thus, the dose-dependent activation by ICP4 of the ICP4 site-deleted L/ST mutant promoter was unexpected. To further investigate whether, in addition to the ICP4-binding sequence at the L/ST transcription initiation site, there is an ICP4-binding site upstream of the L/ST promoter, a series of successive deletion constructs were made, all containing a mutated ICP4-binding site but ranging in start position from bp 565 to bp 131 upstream of the L/ST dominant transcription initiation site (Fig. 2E). Although deletions from the 5′ end of the L/ST promoter reduced promoter activity in transfected cells, these modified promoters were still activated by ICP4 (Fig. 2E), so specific sequences responsible for HSV-2 ICP4 activation of the L/ST promoter in ICP4 deletion mutants were not identified. This suggests that the mechanism for this activation might be indirect. An additional L/ST core promoter reporter, p-71−12 containing the sequences from 71 bp upstream of the L/ST dominant transcription initiation site (nt 125799) to just downstream of the TATAA box, excluding the ICP4-binding sequence, was constructed by cloning synthetic oligonucleotides into the pGL3-Basic vector. The additional 60-bp deletion in the L/ST promoter from bp −131 to bp −71 dramatically reduced L/ST promoter reporter activity, suggesting that the sequence between bp −131 and bp −71 is required for efficient L/ST promoter activity. However, ICP4 was still capable of activating the minimal promoter contained in p584-585. Since no obvious ICP4-binding sequences were identified within the 130-bp L/ST core promoter region, this suggests that HSV-2 ICP4 likely specifically activates the L/ST core promoter by interacting with other basic transcription factors specific to the L/ST core promoter, rather than directly interacting with an ICP4-binding sequence.
Northern blot analysis of ICP34.5, L/ST and ICP0 transcripts in HSV-2-infected cell culture.
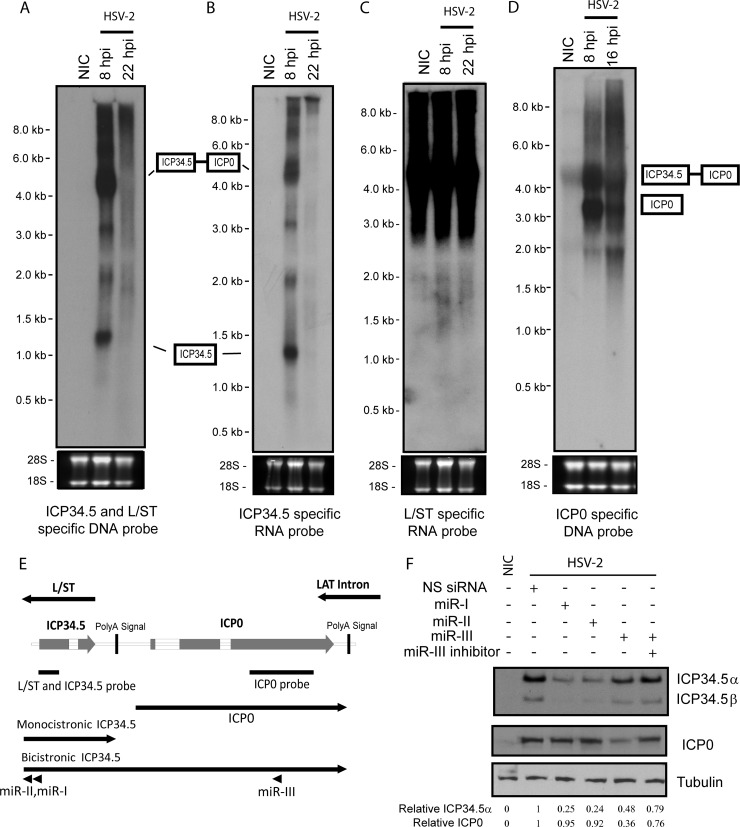
L/ST is transcribed antisense to and completely overlaps ICP34.5. Northern blot analysis of total RNAs from HSV-2-infected Vero cells using an ICP34.5-specific RNA probe, an L/ST-specific RNA probe, and a DNA probe specific to both ICP34.5 and L/ST indicates that in productive infection, HSV-2 L/ST is transcribed at levels too low to be detected by Northern blotting (Fig. 3A to C), a finding consistent with the reported Northern blot results for HSV-1 L/ST (34). This result also suggests that HSV-2 ICP4 efficiently downregulates L/ST expression in infected cell culture. Nonspecific signals were detected using the HSV-2 L/ST-specific RNA probe, likely due to extremely high GC content and repeat sequences in the L/ST-ICP34.5 region.
FIG 3.
Northern hybridization of HSV-2-infected Vero cells, using probes that detect ICP0, L/ST, and ICP34.5. Total RNA from Vero cell infected with HSV-2 at 0, 8, and 22 h postinfection (hpi) was separated by denatured agarose gel, blotted onto a membrane, and detected with an ICP34.5-specific DNA probe (A), an ICP34.5-specific RNA probe (B), an L/ST-specific RNA probe (C), or an ICP0-specific DNA probe (D). (E) Illustrated diagram of the ICP34.5-ICP0 region and probes used in the Northern blot analyses. Monocistronic ICP34.5 and bicistronic ICP34.5/ICP0 are labeled in the diagram. The relative position of miR-I and miR-III are also labeled in the diagram. (F) HSV-2 expresses both a monocistronic ICP34.5 and an ICP34.5/ICP0 bicistronic mRNA. Vero cells pretransfected with 20 nM synthetic miR-I, miR-II, and miR-III, together with or without miR-III inhibitor, at 6 h prior infection with HSV-2 at an MOI of 2. Cells were harvested at 16 hpi, and total proteins were analyzed by Western blotting with the anti-HSV-2 ICP34.5 antibody. The same membrane was stripped prior to incubation with the anti-HSV-2 ICP0 antibody and subsequently with the anti-beta tubulin antibody. NIC, no-infection control. NS siRNA, nonspecific synthetic siRNA. The intensities of ICP34.5, ICP0, and tubulin signals were quantified by using ImageQuant TL8.1 software (GE Healthcare). The intensities of ICP34.5 and ICP0 signals were normalized to the intensity of corresponding tubulin signals and are labeled underneath the panel.
The ICP34.5-specific RNA probe identified 6 discrete RNA transcripts ranging from approximately 1.3 to 9 kb. Based on the previously determined ICP34.5 transcription initiation site (2) and the predicted poly(A) signal site for ICP34.5, the 1.3-kb transcript likely corresponds to the monocistronic ICP34.5 mRNA. Since miR-I is expressed antisense to exon 1 of ICP34.5 (Fig. 3E), miR-I is expected to reduce both the mRNA transcript and ICP34.5 protein expression via miR-I specific RNAi-induced silencing complex (RISC). Indeed, both the 1.3- and the 4.4-kb transcripts were previously observed and was shown to be a specific target of miR-I, which efficiently reduces ICP34.5 expression (9). Similarly, miR-III is completely complementary to the coding sequencing of ICP0, and was shown to reduce expression of ICP0 by reducing the quantity of mRNA transcripts containing the ICP0 sequences and by inhibiting the translation through miR-III-specific RISC (2). The relative positions of miR-I, miR-II, and miR-III are shown in Fig. 1A and Fig. 3E. The 4.4-kb transcript is of a size that would correspond to an ICP34.5-ICP0 bicistronic mRNA. A 4.4-kb band is also observed with the ICP0-specific probe, supporting the hypothesis that this band represents a bicistronic ICP34.5-ICP0 RNA (Fig. 3D and E). Transfecting miR-III, which targets ICP0 exon 3, into the cells prior to infection reduced ICP34.5 expression by ca. 52%, suggesting that any ICP34.5 produced from this bicistronic mRNA is also a target for miR-III and the ICP34.5-ICP0 bicistronic mRNA can produce ICP34.5 (Fig. 3F). The 2- and 3-kb bands may represent alternative splicing products using the 5′ splicing site of ICP34.5 and a 3′ splicing site of ICP0 or another cryptic splice site in ICP0, since a weak 2-kb band is also seen with the ICP0 probe and the 3-kb band would overlap the strong ICP0 signal with the ICP0 probe. However, no corresponding products were obtained by reverse transcription-PCR (RT-PCR) (data not shown), possibly due to the high GC content and repeat sequences in this region.
Identification of the putative miR-H6 promoter and transcription start site.
HSV-2 miR-H6 is expressed in both latently and acutely infected guinea pigs and mice (1, 26) from sequences upstream of the LAT promoter (relative to the LAT) but from the strand opposite the LAT. The miR-H6 primary transcript and promoter have not been described. A primary miR-H6 mRNA was not detectable in total RNAs from HSV-2-infected cells using Northern blot with miR-H6-specific probes (data not shown). We were also unsuccessful at mapping the primary miR-H6 transcription initiation site using the same total RNA sample from infected cells that we used to identify the primary LAT transcription initiation site or the same RNA from pPstI-HincII-transfected cells that we used to identify the L/ST transcription initiation site (data not shown). Previous experiments showed that deleting the LAT promoter and part of the LAT exon 1 region abolished miR-H6 expression in vivo and that the NotI-NotI fragment (which contains the LAT promoter) has bidirectional promoter activity (2), suggesting that the LAT promoter and the LAT exon 1 region might contain promoter sequences for both primary miR-H6 and LAT. The LAT TATA box is the only potential TATA box in this region, so we previously speculated that the LAT TATA box could be bidirectionally active for both the LAT and primary miR-H6.
To test whether the LAT TATA box is required for miR-H6 direction promoter activity, we constructed two luciferase reporters: p-650+106 that includes most of the LAT exon 1 sequence but terminates 33 bp upstream of the LAT TATA box and p-650+74 that includes LAT exon 1 sequence and the LAT TATA box (Fig. 4A). Both reporters showed strong (20- to 30-fold over baseline) luciferase activity, suggesting that the LAT TATA box or sequences immediately upstream of it are likely not required for primary miR-H6 promoter activity. In addition, inclusion of sequence 33 bp upstream of the LAT promoter slightly reduced pri-miR-H6 promoter activity, suggesting that there is a negative correlation between pri-miR-H6 promoter and LAT promoter activities. We next used luciferase-specific primers to map transcription start sites in total RNA from cells transfected with these two luciferase reporters. The 5′ ends of the transcripts of the potential primary miR-H6 transcripts (nt 119544, 119545, 119543, and 119525) mapped using p-650+106 and p-650+74 are identical, with two initiation sites 17 bp apart (Fig. 4B and C). Nucleotide 119544 appears to be the dominant transcription initiation site. Although miR-H6 is readily detectable in infected cell cultures, miR-H6 is below the Northern blot detection limit in cells transfected with pSSK (which contains the promoter and coding sequence of both LAT and primary miR-H6) with or without pICP4 (Fig. 4D). This result suggests that LAT promoter activity antisense to the primary miR-H6 is much stronger than the miR-H6 promoter activity. However, using a miR-H6 specific RT-PCR assay, miR-H6 is detectable in cells transfected with pSSB, a LAT plasmid containing the LAT promoter, LAT exon 1, LAT intron, and partial LAT exon 2, excluding the ICP0 promoter sequence (Fig. 1A and Fig. 4E). Unlike miR-I (Fig. 4D), miR-H6 expression is not affected by the presence of ICP4 (Fig. 4E), likely because the LAT ICP4-binding site does not overlap the miR-H6 initiation site or promoter region, being located approximately 17 to 35 bp downstream of the primary miR-H6 transcription initiation sites.
The LAP2 region is not required for miR-H6 expression in the ganglia of infected guinea pig during latency.
To investigate the role of sequences further upstream (relative to miR-H6) on miR-H6 expression, we used a mutant virus, ΔLAT-P2, in which has a 339-bp deletion in LAT exon 1 sequences between the 5′ end of the primary LAT and the 5′ end of the LAT intron (from nt 119770 to nt 120108) (Fig. 5A). Mice were inoculated corneally with ΔLAT-P2, the LAT promoter/partial exon 1 deletion mutant ΔLAT, and wild-type HSV-2. miR-I, miR-H6, and viral DNA copy number were assayed from TG extracted from these animals 30 days postinoculation. Consistent with our previous observation, miR-I and miR-H6 were not detected in ΔLAT-infected ganglia by real-time PCR (Fig. 5B), suggesting the LAT NotI-NotI sequences (which contains the LAT promoter and 220 bp of the LAT exon 1 sequence) are required for detectable miR-H6 expression during latency in vivo. miR-I and miR-H6 expression in ΔLAT-P2-infected ganglia was only slightly lower than that in wild-type HSV-2-infected ganglia (Fig. 5B), indicating that the LAP2 region is not required for miR-H6 expression. In a cell culture transfection system, we previously showed that LAT promoter sequences are not required for pri-miR-H6 reporter activity. Thus, the first 220 bp of LAT exon 1 likely play a more important role in regulating pri-miR-H6 expression.
DISCUSSION
In the present study, we mapped the transcription initiation sites for the HSV-2 LAT, L/ST, and primary miR-H6 mRNA using RNAs from cells infected with HSV-2, transfected with a plasmid containing LAT sequence, and transfected with a plasmid containing the putative primary miR-H6 promoter, respectively. We found that although ICP4 inhibits the expression of LAT and L/ST through the ICP4-binding site at the transcription initiation site, it had no impact on the expression of the primary miR-H6 in transfected cells. We also found that the LAT promoter and the adjacent 220-bp LAT exon1 sequences, and not the downstream LAP2 sequences, are critical for efficient expression of miR-H6 and the LAT-encoded miRNAs in infected ganglia during latency.
Recent studies showed that both HSV-1- and HSV-2-encoded miRNAs reduce the expression of ICP0 and ICP34.5. This likely reduces viral replication in neurons and contributes to the phenotype attributed to the LAT (2, 8, 9). ICP4, as a key regulator of miRNA expression, thus likely plays an important role in this process. ICP4 expression likely also accounts for reported reduced expression of LAT and LAT-associated miRNAs in an explant reactivation model (38) and soon after induction of reactivation in latently infected ganglia cultured with or without nerve growth factor or epidermal growth factor (39–41). Thus, viral genes on the LAT strand (LAT and L/ST) appear to reduce viral replication in neurons, while genes on the opposite strand (ICP4, ICP34.5, and ICP0) appear to promote viral replication in neurons, with ICP4 as the master regulator of this cassette of genes that influence the outcome of neuronal infection.
ICP4's dual role in regulating L/ST expression is also interesting. As with HSV-1 L/ST (29, 33, 34, 42), HSV-2 L/ST is difficult to detect in cell culture infected with wild-type virus (Fig. 3). Although ICP4 occupation of the HSV-2 L/ST ICP4-binding site during lytic replication was not directly demonstrated, viruses with mutations in the HSV-1 L/ST ICP4-binding site show significant increases in L/ST expression (29, 42) and of HSV-1 miR-LAT-ICP34.5 (also named miR-H4, and a homolog of HSV-2 miR-II) (2), leading to reduced expression of ICP34.5 (29, 42) in infected cell culture, indicating that HSV-1 L/ST is tightly controlled by ICP4 by interaction with the ICP4-binding site during lytic replication. The present finding that ICP4 also influences HSV-2 L/ST expression suggests a similar mechanism for HSV-2. Although ICP4 acts through the ICP4-binding site to reduce L/ST expression, HSV-2 ICP4 dramatically activates the L/ST promoter in a dose-dependent manner when the L/ST ICP4-binding site is disrupted, possibly by interacting with essential transcriptional factors to increase the L/ST promoter activity (43, 44). This implies that under circumstances that reduce ICP4 binding to its site near the L/ST transcription start site, ICP4 may increase L/ST expression and thus lead to further increased expression of miR-I and miR-II.
In infected cell culture, ICP34.5 is transcribed as both a monocistronic and an ICP0-ICP34.5 bicistronic mRNA, likely because polyadenylation at the ICP34.5 poly(A) site is inhibited during late viral infection. Usually, the splicing efficiency of the second ORF in bicistronic transcripts is reduced dramatically due to greater distance to the 5′ cap structure, and the resultant intron-containing transcripts tend to be trapped in the nucleus and subsequently degraded (45). Here we show that miR-III, which targets ICP0, also reduces ICP34.5 expression, suggesting that some bicistronic mRNAs are exported to the cytosol and used as the template for the translation of HSV-2 ICP34.5. Thus, the three most common LAT-strand miRNAs all target ICP34.5, which is consistent with a high level of importance to the virus of maintaining tight control over ICP34.5 expression. It is interesting that HSV-2 possesses many mechanisms for controlling ICP34.5 expression, including the ICP34.5 promoter, the LAT promoter (which produces ICP34.5-inhibiting miRNAs), the L/ST promoter (which produces ICP34.5-inhibiting miRNAs), ICP27 (which inhibits ICP34.5 splicing), and ICP4 (which regulates levels of LAT and L/ST and thus the levels of the miRNAs that they produce). Northern blot analysis of HSV-1 ICP34.5 identified three significant transcripts, of 1.4, 3.5, and 5 kb (46). The 3.5- and 5-kb transcripts were proposed to be ICP34.5 readthrough transcripts that contain ICP0 coding sequences, although no experimental evidence was provided. The similar transcriptional pattern of HSV-1 and HSV-2 ICP34.5 expression suggests a conserved mechanism for ICP0-targeting miRNA in regulating ICP34.5 expression.
In LAT exon 1 sequences, we identified promoter activity that likely is responsible for miR-H6 expression from the strand opposite LAT. The function of LAT exon 1 sequences in latency and reactivation is unclear (19, 35, 47–54), with a possible LAT-independent role in viral recurrence reported in HSV-1. The miR-H6 primary transcript, which starts just downstream of the LAT transcription start site, is likely too unstable to allow direct detection of the primary transcript in infected cells. Because, during latency, levels of HSV-1 and HSV-2 miR-H6 are comparable to those of miR-H2 or miR-III (1, 10, 26, 55), the relative balance of the bidirectional transcription for LAT and primary miR-H6 in vivo during latency is likely regulated differently from that in cell culture. Unlike the other HSV-2 miRNA promoters studied here, this promoter is not ICP4 responsive. Sequences more than 220 bp upstream of this promoter are not essential for miR-H6 expression. We showed previously that, in contrast to a report for HSV-1 miR-H6, HSV-2 miR-H6 does not target its corresponding ICP4 (26). Although the role and target of HSV-2 miR-H6 is unknown, and abrogation of its expression by insertion of a polyadenylation site directly upstream of the miRNA had no influence on acute viral infection or recurrence (26), it is possible that miR-H6 promoter sequences in exon 1 could be important for viral latency and recurrence phenotypes. During latency, these sequences (in HSV-1) are enriched for euchromatin marks (56–59), likely playing a role in the expression of miR-H6 during latency and thus possibly also playing a role in the activity of the nearby LAT promoter.
HSV-1 L/ST encodes two ORFs, ORF-O and ORF-P, which can reduce the synthesis or block the expression of HSV-1 ICP4 in productive infection (33, 60). However, we are unable to find an HSV-2 homolog of ORF-O or ORF-P in HSV-2 L/ST. A hypothetical 130-amino-acid ORF at ∼220 bp downstream of the HSV-2 L/ST start site (nt 126019 to nt 126048) has no homology to HSV-1 ORF-P, suggesting that L/ST more likely functions through miRNAs. A hypothetical 192-amino-acid ORF at ∼300 bp downstream of the HSV-2 primary miR-H6 transcript start site (nt 118257 - nt 119198) has no corresponding ORF in HSV-1. Since the primary miR-H6 transcript is degraded to generate mature miR-H6, pri-miR-H6 likely also functions via miRNA instead of protein, although it is not known whether the hypothetical protein potentially encoded by HSV-2 pri-miR-H6 is expressed during latency or lytic infection.
We identified more than one transcription initiation site for HSV-2 LAT, L/ST, and pri-miR-H6 mRNA in infected or transfected non-neuronal cell cultures. The relevance of the less dominant sites is unknown, and it is conceivable that these sites may be more dominant in some cell types in vivo.
Although the exact function of HSV-2 miRNAs are not known, latently expressed miRNAs are conserved in location between HSV-1 and HSV-2, and there is increasing evidence that these miRNAs may play an important role in the switch between latent and productive infection. Thus, an understanding of how the mRNAs themselves are regulated is likely to provide important clues to their function in different types of cells. The ability of ICP4 to suppress ICP34.5-targeting miRNAs and to (under circumstances of no ICP4 binding to the L/ST start site) increase activity of a promoter directing their expression suggest that ICP4 could play a key role in the switch between latency and reactivation. Of interest, recent studies have focused on the potential function of VP16 as the possible viral inducer of recurrence (61)—the present study suggests that ICP4 should be considered a candidate for this role as well.
ACKNOWLEDGMENTS
We thank Keith Peden and Haruhiko Murata for critical readings of the manuscript. We also thank Jeffrey Cohen and Kening Wang for providing the HSV-2 ICP4 expression plasmid.
This study was supported by the intramural research programs of the Center for Biologics Evaluation and Research. This study was also supported by EY022697 (T.P.M.), EY002687 (Vision Core grant), and an unrestricted grant from Research To Prevent Blindness to the Department of Ophthalmology and Visual Sciences at Washington University.
REFERENCES
- 1.Jurak I, Kramer MF, Mellor JC, van Lint AL, Roth FP, Knipe DM, Coen DM. 2010. Numerous conserved and divergent microRNAs expressed by herpes simplex viruses 1 and 2. J Virol 84:4659–4672. doi: 10.1128/JVI.02725-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Tang S, Patel A, Krause PR. 2009. Novel less-abundant viral microRNAs encoded by herpes simplex virus 2 latency-associated transcript and their roles in regulating ICP34.5 and ICP0 mRNAs. J Virol 83:1433–1442. doi: 10.1128/JVI.01723-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bartel DP, Chen CZ. 2004. Micromanagers of gene expression: the potentially widespread influence of metazoan microRNAs. Nat Rev Genet 5:396–400. doi: 10.1038/nrg1328. [DOI] [PubMed] [Google Scholar]
- 4.Cullen BR. 2006. Viruses and microRNAs. Nat Genet 38(Suppl):S25–S30. doi: 10.1038/ng1793. [DOI] [PubMed] [Google Scholar]
- 5.He L, Hannon GJ. 2004. MicroRNAs: small RNAs with a big role in gene regulation. Nat Rev Genet 5:522–531. doi: 10.1038/nrg1379. [DOI] [PubMed] [Google Scholar]
- 6.Chou J, Kern ER, Whitley RJ, Roizman B. 1990. Mapping of herpes simplex virus-1 neurovirulence to gamma 134.5, a gene nonessential for growth in culture. Science 250:1262–1266. doi: 10.1126/science.2173860. [DOI] [PubMed] [Google Scholar]
- 7.Chou J, Roizman B. 1992. The gamma 1(34.5) gene of herpes simplex virus 1 precludes neuroblastoma cells from triggering total shutoff of protein synthesis characteristic of programmed cell death in neuronal cells. Proc Natl Acad Sci U S A 89:3266–3270. doi: 10.1073/pnas.89.8.3266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Flores O, Nakayama S, Whisnant AW, Javanbakht H, Cullen BR, Bloom DC. 2013. Mutational inactivation of herpes simplex virus 1 microRNAs identifies viral mRNA targets and reveals phenotypic effects in culture. J Virol 87:6589–6603. doi: 10.1128/JVI.00504-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tang S, Bertke AS, Patel A, Wang K, Cohen JI, Krause PR. 2008. An acutely and latently expressed herpes simplex virus 2 viral microRNA inhibits expression of ICP34.5, a viral neurovirulence factor. Proc Natl Acad Sci U S A 105:10931–10936. doi: 10.1073/pnas.0801845105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Umbach JL, Kramer MF, Jurak I, Karnowski HW, Coen DM, Cullen BR. 2008. MicroRNAs expressed by herpes simplex virus 1 during latent infection regulate viral mRNAs. Nature 454:780–783. doi: 10.1038/nature07103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Whitley RJ, Kern ER, Chatterjee S, Chou J, Roizman B. 1993. Replication, establishment of latency, and induced reactivation of herpes simplex virus gamma 1 34.5 deletion mutants in rodent models. J Clin Invest 91:2837–2843. doi: 10.1172/JCI116527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Farrell MJ, Dobson AT, Feldman LT. 1991. Herpes simplex virus latency-associated transcript is a stable intron. Proc Natl Acad Sci U S A 88:790–794. doi: 10.1073/pnas.88.3.790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Stevens JG, Wagner EK, Devi-Rao GB, Cook ML, Feldman LT. 1987. RNA complementary to a herpesvirus alpha gene mRNA is prominent in latently infected neurons. Science 235:1056–1059. doi: 10.1126/science.2434993. [DOI] [PubMed] [Google Scholar]
- 14.Krause PR, Stanberry LR, Bourne N, Connelly B, Kurawadwala JF, Patel A, Straus SE. 1995. Expression of the herpes simplex virus type 2 latency-associated transcript enhances spontaneous reactivation of genital herpes in latently infected guinea pigs. J Exp Med 181:297–306. doi: 10.1084/jem.181.1.297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Leib DA, Bogard CL, Kosz-Vnenchak M, Hicks KA, Coen DM, Knipe DM, Schaffer PA. 1989. A deletion mutant of the latency-associated transcript of herpes simplex virus type 1 reactivates from the latent state with reduced frequency. J Virol 63:2893–2900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sawtell NM, Thompson RL. 1992. Herpes simplex virus type 1 latency-associated transcription unit promotes anatomical site-dependent establishment and reactivation from latency. J Virol 66:2157–2169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Steiner I, Spivack JG, Lirette RP, Brown SM, MacLean AR, Subak-Sharpe JH, Fraser NW. 1989. Herpes simplex virus type 1 latency-associated transcripts are evidently not essential for latent infection. EMBO J 8:505–511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Thompson RL, Sawtell NM. 1997. The herpes simplex virus type 1 latency-associated transcript gene regulates the establishment of latency. J Virol 71:5432–5440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Trousdale MD, Steiner I, Spivack JG, Deshmane SL, Brown SM, MacLean AR, Subak-Sharpe JH, Fraser NW. 1991. In vivo and in vitro reactivation impairment of a herpes simplex virus type 1 latency-associated transcript variant in a rabbit eye model. J Virol 65:6989–6993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yoshikawa T, Hill JM, Stanberry LR, Bourne N, Kurawadwala JF, Krause PR. 1996. The characteristic site-specific reactivation phenotypes of HSV-1 and HSV-2 depend upon the latency-associated transcript region. J Exp Med 184:659–664. doi: 10.1084/jem.184.2.659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cai W, Astor TL, Liptak LM, Cho C, Coen DM, Schaffer PA. 1993. The herpes simplex virus type 1 regulatory protein ICP0 enhances virus replication during acute infection and reactivation from latency. J Virol 67:7501–7512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cai WZ, Schaffer PA. 1989. Herpes simplex virus type 1 ICP0 plays a critical role in the de novo synthesis of infectious virus following transfection of viral DNA. J Virol 63:4579–4589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hagglund R, Roizman B. 2004. Role of ICP0 in the strategy of conquest of the host cell by herpes simplex virus 1. J Virol 78:2169–2178. doi: 10.1128/JVI.78.5.2169-2178.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Halford WP, Schaffer PA. 2001. ICP0 is required for efficient reactivation of herpes simplex virus type 1 from neuronal latency. J Virol 75:3240–3249. doi: 10.1128/JVI.75.7.3240-3249.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cui C, Griffiths A, Li G, Silva LM, Kramer MF, Gaasterland T, Wang XJ, Coen DM. 2006. Prediction and identification of herpes simplex virus 1-encoded microRNAs. J Virol 80:5499–5508. doi: 10.1128/JVI.00200-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tang S, Bertke AS, Patel A, Margolis TP, Krause PR. 2011. Herpes simplex virus 2 microRNA miR-H6 is a novel latency-associated transcript-associated microRNA, but reduction of its expression does not influence the establishment of viral latency or the recurrence phenotype. J Virol 85:4501–4509. doi: 10.1128/JVI.01997-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Farrell MJ, Margolis TP, Gomes WA, Feldman LT. 1994. Effect of the transcription start region of the herpes simplex virus type 1 latency-associated transcript promoter on expression of productively infected neurons in vivo. J Virol 68:5337–5343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lagunoff M, Randall G, Roizman B. 1996. Phenotypic properties of herpes simplex virus 1 containing a derepressed open reading frame P gene. J Virol 70:1810–1817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lee LY, Schaffer PA. 1998. A virus with a mutation in the ICP4-binding site in the L/ST promoter of herpes simplex virus type 1, but not a virus with a mutation in open reading frame P, exhibits cell-type-specific expression of gamma(1)34.5 transcripts and latency-associated transcripts. J Virol 72:4250–4264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Clements JB, Watson RJ, Wilkie NM. 1977. Temporal regulation of herpes simplex virus type 1 transcription: location of transcripts on the viral genome. Cell 12:275–285. doi: 10.1016/0092-8674(77)90205-7. [DOI] [PubMed] [Google Scholar]
- 31.Dixon RA, Schaffer PA. 1980. Fine-structure mapping and functional analysis of temperature-sensitive mutants in the gene encoding the herpes simplex virus type 1 immediate-early protein VP175. J Virol 36:189–203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Knipe DM, Ruyechan WT, Roizman B, Halliburton IW. 1978. Molecular genetics of herpes simplex virus: demonstration of regions of obligatory and nonobligatory identity within diploid regions of the genome by sequence replacement and insertion. Proc Natl Acad Sci U S A 75:3896–3900. doi: 10.1073/pnas.75.8.3896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Randall G, Roizman B. 1997. Transcription of the derepressed open reading frame P of herpes simplex virus 1 precludes the expression of the antisense gamma(1)34.5 gene and may account for the attenuation of the mutant virus. J Virol 71:7750–7757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yeh L, Schaffer PA. 1993. A novel class of transcripts expressed with late kinetics in the absence of ICP4 spans the junction between the long and short segments of the herpes simplex virus type 1 genome. J Virol 67:7373–7382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yoshikawa T, Stanberry LR, Bourne N, Krause PR. 1996. Downstream regulatory elements increase acute and latent herpes simplex virus type 2 latency-associated transcript expression but do not influence recurrence phenotype or establishment of latency. J Virol 70:1535–1541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tang S, Yamanegi K, Zheng ZM. 2004. Requirement of a 12-base-pair TATT-containing sequence and viral lytic DNA replication in activation of the Kaposi's sarcoma-associated herpesvirus K8.1 late promoter. J Virol 78:2609–2614. doi: 10.1128/JVI.78.5.2609-2614.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Smith CA, Bates P, Rivera-Gonzalez R, Gu B, DeLuca NA. 1993. ICP4, the major transcriptional regulatory protein of herpes simplex virus type 1, forms a tripartite complex with TATA-binding protein and TFIIB. J Virol 67:4676–4687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Neumann DM, Bhattacharjee PS, Giordani NV, Bloom DC, Hill JM. 2007. In vivo changes in the patterns of chromatin structure associated with the latent herpes simplex virus type 1 genome in mouse trigeminal ganglia can be detected at early times after butyrate treatment. J Virol 81:13248–13253. doi: 10.1128/JVI.01569-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Du T, Zhou G, Roizman B. 2011. HSV-1 gene expression from reactivated ganglia is disordered and concurrent with suppression of latency-associated transcript and miRNAs. Proc Natl Acad Sci U S A 108:18820–18824. doi: 10.1073/pnas.1117203108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Du T, Zhou G, Roizman B. 2012. Induction of apoptosis accelerates reactivation of latent HSV-1 in ganglionic organ cultures and replication in cell cultures. Proc Natl Acad Sci U S A 109:14616–14621. doi: 10.1073/pnas.1212661109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Du T, Zhou G, Roizman B. 2013. Modulation of reactivation of latent herpes simplex virus 1 in ganglionic organ cultures by p300/CBP and STAT3. Proc Natl Acad Sci U S A 110:E2621–E2628. doi: 10.1073/pnas.1309906110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Randall G, Lagunoff M, Roizman B. 2000. Herpes simplex virus 1 open reading frames O and P are not necessary for establishment of latent infection in mice. J Virol 74:9019–9027. doi: 10.1128/JVI.74.19.9019-9027.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wagner LM, Bayer A, Deluca NA. 2013. Requirement of the N-terminal activation domain of herpes simplex virus ICP4 for viral gene expression. J Virol 87:1010–1018. doi: 10.1128/JVI.02844-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wagner LM, DeLuca NA. 2013. Temporal association of herpes simplex virus ICP4 with cellular complexes functioning at multiple steps in Pol II transcription. PLoS One 8:e78242. doi: 10.1371/journal.pone.0078242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zheng ZM. 2003. Split genes and their expression in Kaposi's sarcoma-associated herpesvirus. Rev Med Virol 13:173–184. doi: 10.1002/rmv.387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Chou J, Roizman B. 1986. The terminal a sequence of the herpes simplex virus genome contains the promoter of a gene located in the repeat sequences of the L component. J Virol 57:629–637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bertke AS, Patel A, Krause PR. 2007. Herpes simplex virus latency-associated transcript sequence downstream of the promoter influences type-specific reactivation and viral neurotropism. J Virol 81:6605–6613. doi: 10.1128/JVI.02701-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bhattacharjee PS, Tran RK, Myles ME, Maruyama K, Mallakin A, Bloom DC, Hill JM. 2003. Overlapping subdeletions within a 348-bp in the 5′ exon of the LAT region that facilitates epinephrine-induced reactivation of HSV-1 in the rabbit ocular model do not further define a functional element. Virology 312:151–158. doi: 10.1016/S0042-6822(03)00174-0. [DOI] [PubMed] [Google Scholar]
- 49.Bloom DC, Hill JM, Devi-Rao G, Wagner EK, Feldman LT, Stevens JG. 1996. A 348-base-pair region in the latency-associated transcript facilitates herpes simplex virus type 1 reactivation. J Virol 70:2449–2459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hill J, Patel A, Bhattacharjee P, Krause P. 2003. An HSV-1 chimeric containing HSV-2 latency associated transcript (LAT) sequences has significantly reduced adrenergic reactivation in the rabbit eye model. Curr Eye Res 26:219–224. doi: 10.1076/ceyr.26.3.219.14896. [DOI] [PubMed] [Google Scholar]
- 51.Hill JM, Garza HH Jr, Su YH, Meegalla R, Hanna LA, Loutsch JM, Thompson HW, Varnell ED, Bloom DC, Block TM. 1997. A 437-base-pair deletion at the beginning of the latency-associated transcript promoter significantly reduced adrenergically induced herpes simplex virus type 1 ocular reactivation in latently infected rabbits. J Virol 71:6555–6559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Jarman RG, Loutsch JM, Devi-Rao GB, Marquart ME, Banaszak MP, Zheng X, Hill JM, Wagner EK, Bloom DC. 2002. The region of the HSV-1 latency-associated transcript required for epinephrine-induced reactivation in the rabbit does not include the 2.0-kb intron. Virology 292:59–69. doi: 10.1006/viro.2001.1265. [DOI] [PubMed] [Google Scholar]
- 53.Jin L, Peng W, Perng GC, Brick DJ, Nesburn AB, Jones C, Wechsler SL. 2003. Identification of herpes simplex virus type 1 latency-associated transcript sequences that both inhibit apoptosis and enhance the spontaneous reactivation phenotype. J Virol 77:6556–6561. doi: 10.1128/JVI.77.11.6556-6561.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Perng GC, Slanina SM, Yukht A, Drolet BS, Keleher W Jr, Ghiasi H, Nesburn AB, Wechsler SL. 1999. A herpes simplex virus type 1 latency-associated transcript mutant with increased virulence and reduced spontaneous reactivation. J Virol 73:920–929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Jurak I, Hackenberg M, Kim JY, Pesola JM, Everett RD, Preston CM, Wilson AC, Coen DM. 2014. Expression of herpes simplex virus 1 microRNAs in cell culture models of quiescent and latent infection. J Virol 88:2337–2339. doi: 10.1128/JVI.03486-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Cliffe AR, Garber DA, Knipe DM. 2009. Transcription of the herpes simplex virus latency-associated transcript promotes the formation of facultative heterochromatin on lytic promoters. J Virol 83:8182–8190. doi: 10.1128/JVI.00712-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Creech CC, Neumann DM. 2010. Changes to euchromatin on LAT and ICP4 following reactivation are more prevalent in an efficiently reactivating strain of HSV-1. PLoS One 5:e15416. doi: 10.1371/journal.pone.0015416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kubat NJ, Amelio AL, Giordani NV, Bloom DC. 2004. The herpes simplex virus type 1 latency-associated transcript (LAT) enhancer/rcr is hyperacetylated during latency independently of LAT transcription. J Virol 78:12508–12518. doi: 10.1128/JVI.78.22.12508-12518.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kubat NJ, Tran RK, McAnany P, Bloom DC. 2004. Specific histone tail modification and not DNA methylation is a determinant of herpes simplex virus type 1 latent gene expression. J Virol 78:1139–1149. doi: 10.1128/JVI.78.3.1139-1149.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Randall G, Lagunoff M, Roizman B. 1997. The product of ORF O located within the domain of herpes simplex virus 1 genome transcribed during latent infection binds to and inhibits in vitro binding of infected cell protein 4 to its cognate DNA site. Proc Natl Acad Sci U S A 94:10379–10384. doi: 10.1073/pnas.94.19.10379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Thompson RL, Preston CM, Sawtell NM. 2009. De novo synthesis of VP16 coordinates the exit from HSV latency in vivo. PLoS Pathog 5:e1000352. doi: 10.1371/journal.ppat.1000352. [DOI] [PMC free article] [PubMed] [Google Scholar]