ABSTRACT
After the resolution of the acute phase of infection, otherwise quiescent antigen-experienced CD8+ T cells confer rapid protection upon reinfection with viral pathogens or, in the case of persistent viruses, help to maintain control of the infection. Depending on the type of virus, antigen-specific CD8+ T cells have distinct traits, ranging from typical memory cell properties in the case of rapidly cleared viruses to immediate effector functions for persistent viruses. We here show that both the differentiation stage, defined by the expression of cell surface markers, such as CD45RA, CCR7, CD28, and CD27, and distinct expression levels of T-bet and eomesodermin (Eomes) predict the functional profile of antigen-experienced CD8+ T cells. Furthermore, virus-specific CD8+ T cells targeting different respiratory syncytial virus-, influenza A virus-, Epstein-Barr virus (EBV)-, human cytomegalovirus (hCMV)-, and HIV-1-specific epitopes adopt distinct T-bet and Eomes expression patterns that appear to be installed early during the primary response. Importantly, the associations between surface phenotype, T-bet/Eomes expression levels, and the expression of markers that predict CD8+ T-cell function change according to viral infection history, particularly against the background of HIV-1 and, to lesser extent, of human cytomegalovirus and/or Epstein-Barr virus infection. Thus, the functionality of human antigen-experienced CD8+ T cells follows at least two dimensions, one outlined by the surface phenotype and another by the T-bet/Eomes expression levels, which are determined by previous or persistent viral challenges.
IMPORTANCE Functional human CD8+ T-cell subsets have been defined using surface markers like CD45RA, CCR7, CD28, and CD27. However, the induction of function-defining traits, like granzyme B expression, is controlled by transcription factors like T-bet and Eomes. Here, we describe how T-bet and Eomes levels distinctly relate to the expression of molecules predictive for CD8+ T-cell function in a surface phenotype-independent manner. Importantly, we found that central memory and effector memory CD8+ T-cell subsets differentially express T-bet, Eomes, and molecules predictive for function according to viral infection history, particularly so in the context of HIV-1 infection and, to lesser extent, of latent EBV- and/or hCMV-infected, otherwise healthy adults. Finally, we show that the distinct phenotypes and T-bet/Eomes levels of different virus-specific CD8+ T-cell populations are imprinted early during the acute phase of primary infection in vivo. These findings broaden our understanding of CD8+ T-cell differentiation.
INTRODUCTION
The process of viral and host coevolvement has selected for antiviral CD8+ T-cell responses that are successful in ensuring survival of the host. As an apparent consequence, human circulating virus-specific CD8+ T cells display distinct phenotypic and functional properties according to the virus or viral proteins they target (1–3). Acute viruses, such as respiratory syncytial virus (RSV) and influenza A virus, and also some persisting viruses, such as polyomavirus BK, induce CD8+ T cells that predominantly display a CD45RA− CCR7+ CD28+ CD27+ (central memory) or CD45RA− CCR7− CD28+ CD27+ (early differentiated) phenotype (1, 4–10). These and the CD45RA− CCR7− CD28+ CD27− (early-like) subset highly express CD127 (interleukin 7Rα [IL-7Rα]), proliferate vigorously upon stimulation with cognate antigen, and produce several cytokines, among which is IL-2 (11–13). In contrast, Epstein-Barr virus (EBV) and human cytomegalovirus (hCMV) can induce CD8+ T cells with a CD45RA− CCR7− CD28− CD27+ (intermediately differentiated), CD45RA− CCR7− CD28− CD27− (RA− effector-type), or CD45RA+ CCR7− CD28− CD27− (RA+ effector-type) phenotype (1, 3, 5, 14). These subsets less often express IL-7Rα, have stringent proliferation requirements, frequently express granzyme B, and execute immediate cytotoxicity (11, 13, 15).
In contrast to naive cells, antigen-primed T cells rapidly mount protective responses owing to their superior ability to produce molecules that are crucial for the immunological defense. Although the associations between the expression of CD45RA, CCR7, CD27, CD28, and function are well established (1, 4, 11, 12), the actual expression of genes and proteins that ultimately determine T-cell functionality is regulated by transcription factors (TFs). For example, RORγt, FOXP3, and GATA3 drive the generation of type 17, regulatory, and type 2 T cells, respectively, whereas T-bet and eomesodermin (Eomes) are responsible for governing a type 1 cytotoxic differentiation program (16–19). Despite the substantial overlap in DNA-binding sequences and similarity in functions of T-bet and Eomes, such as inducing the expression of gamma interferon (IFN-γ), granzyme B, and IL-2Rβ, the acute-phase CD8+ effector T-cell response in mice is impaired, particularly in the absence of T-bet (16–18, 20–23), whereas Eomes is important for the formation of memory cells and secondary responses in the setting of reinfection (24). Thus, T-bet and Eomes control distinct differentiation programs and different protective functions in CD8+ T cells.
This raises the question of whether T-bet and Eomes expression can be used to distinguish human virus-specific CD8+ T cells with distinct functional programs. We and others have recently shown that T-bet and Eomes expression varies in human CD8+ T-cell subsets and virus-specific populations (6, 25–28). Furthermore, their expression levels were also shown to distinctly correspond to the expression of IL-7Rα, granzymes, and various coinhibitory receptors expressed by exhausted CD8+ T cells (26, 27, 29). Here, we show that the T-bet and Eomes expression state of an individual CD8+ T-cell corresponds to specific functional traits in a manner that is irrespective of the differentiation status defined by the CD45RA/CCR7/CD28/CD27 subdivision. Importantly, the associations between surface phenotype, T-bet/Eomes expression levels, and the expression of IL-7Rα, granzyme K, killer cell lectin-like receptor G1 (KLRG1), and granzyme B were found to vary strongly according to infection history, particularly against the background of HIV-1 infection and, to lesser extent, of EBV and/or hCMV infection. Therefore, the level of T-bet/Eomes expression forms an essential second dimension in comprehending the specific differentiation state of individual human CD8+ T cells.
MATERIALS AND METHODS
Study subjects.
Peripheral blood mononuclear cells (PBMCs) were obtained from the buffy coats of 29 healthy blood donors between the ages of 18 and 64 years (Table 1 and 2). PBMC samples from 13 HIV-1-infected participants of the Amsterdam Cohort studies on the natural history of HIV-1 infection were selected based on being HLA-B8 positive (Table 3). These patients were positive for HIV-1 antibodies at entry into the cohort between October 1984 and April 1985. In previous epidemiological studies, the time since seroconversion of these prevalent cases has been estimated based on the incidence of HIV-1 infection among homosexual participants of the Amsterdam Cohort and was on average 1.5 years before entry into the cohort studies (30). None of these patients had received antiretroviral therapy at analysis (Table 3). Cord blood samples were collected as rest material. PBMCs from two initially hCMV- and EBV-seronegative kidney transplant recipients were longitudinally obtained (Table 4).
TABLE 1.
Virus-specific analysis of healthy subjects
| Subject no. | Sex | Age (in yrs) | Tetramer-specific T cell populations detected |
|---|---|---|---|
| 1 | Male | 36 | RSV NP, influenza MP1, EBV EBNA3a (RLR), EBV BMLF-1 |
| 2 | Male | 40 | RSV NP, influenza MP1, EBV EBNA3a (RPP), EBV BMLF-1 |
| 3 | Female | 37 | Influenza MP1, EBV BMLF-1, hCMV pp65 (NLV) |
| 4 | Female | 24 | Influenza MP1, EBV BMFL-1 (HPV), EBV BMLF-1 |
| 5 | Male | 49 | RSV NP, influenza MP1, EBV EBNA3a (RPP), EBV BMLF-1, hCMV pp65 (NLV and TPR) |
| 6 | Unknown | Unknowna | hCMV pp65 (NLV) |
| 7 | Unknown | Unknowna | hCMV pp65 (NLV) |
| 8 | Unknown | Unknowna | RSV NP, EBV EBNA3a (RLR and RPP) |
| 9 | Female | 37 | RSV NP, EBV EBNA3a (RPP and FLR) |
| 10 | Male | 24 | hCMV pp65 (NLV) |
| 11 | Female | 37 | hCMV pp65 (NLV) |
In between 18 and 64 years of age.
TABLE 2.
Age comparison study groups
| Group (no. of subjects) | Median age (in yrs) [interquartile range] |
|---|---|
| Cord blood (n = 5) | |
| hCMV negative/EBV negative (n = 6) | 28.7 [23.3–32.4]a |
| hCMV positive/EBV negative (n = 3) | 31.1 |
| hCMV negative/EBV positive (n = 5) | 35.7 [30–38.5] |
| hCMV positive/EBV positive (n = 6) | 43.2 [43.2–56.6] |
| HIV infected | 38 [33.5–42] |
Younger than hCMV/EBV double- and HIV-infected individuals (P = 0.04 and P < 0.01, respectively).
TABLE 3.
HIV-infected individualsa
| Subject no. | Gender | Age (in yrs) | Sample moment (no. of yrs after SC) | No. of CD4 cells/μl | No. of CD8 cells/μl | HIV viral load (copies/ml plasma) | cART (no. of yrs after SC) | AIDS | AIDS diagnosis (no. of yrs after SC) | EBV serostatus | hCMV serostatus |
|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | Male | 41 | 3 | 380 | 690 | 32,000 | No | Yes | 5 | ND | + |
| 2 | Male | 51 | 3 | 590 | 590 | 1,000 | No | Yes | 9 | ND | + |
| 3 | Male | 31 | 3 | 710 | 860 | 6,700 | Yes (14) | No | ND | + | |
| 4 | Male | 31 | 3 | 270 | 710 | 360,000 | No | Yes | 6 | ND | + |
| 5 | Male | 38 | 4 | 1,140 | 1,000 | 1,300 | No | No | ND | + | |
| 6 | Male | 44 | 4 | 950 | 790 | 1,000 | Yes (14) | No | ND | + | |
| 7 | Male | 36 | 4 | 1,220 | 910 | 1,000 | No | Yes | 7 | ND | + |
| 8 | Male | 37 | 4 | 460 | 440 | 24,000 | No | Yes | 12 | ND | − |
| 9 | Male | 43 | 4 | 770 | 1,910 | 1,000 | No | Yes | 8 | ND | + |
| 10 | Male | 40 | 4 | 400 | 680 | 1,700 | Yes (15) | Yes | 8 | ND | + |
| 11 | Male | 39 | 4 | 920 | 680 | 1,000 | Yes (15) | Yes | 6 | ND | + |
| 12 | Male | 38 | 4 | 290 | 850 | 13,000 | No | Yes | 5 | ND | + |
| 13 | Male | 30 | 4 | 1,060 | 1,270 | 1,000 | No | Yes | 9 | ND | − |
SC, seroconversion; ND, not determined; cART, combination antiretroviral therapy; +, positive; −, negative.
TABLE 4.
Kidney transplant recipients
| Subject no. | Sex | Age (in yrs) | Immunosuppressive treatment after transplantation |
|---|---|---|---|
| 1 | Male | 25 | Prednisolone, cyclosporine, mycophenolate mofetil |
| 2 | Male | 25 | Prednisolone, cyclosporine, mycophenolate mofetil |
Ethics statement.
The inclusion of patients has been conducted in accordance with the ethical principles set out in the Declaration of Helsinki. Both patient inclusion and blood sample collection were done with approval of the Amsterdam Medical Center Medical Ethical Committee. Written informed consent was obtained prior to data collection.
Isolation of PBMCs.
PBMCs were isolated using standard density gradient centrifugation, after which they were cryopreserved until the day of analysis.
Tetrameric complexes.
All tetrameric complexes were obtained from Sanquin, Amsterdam, the Netherlands. See Table 5 for a list of all tetrameric complexes used.
TABLE 5.
Tetrameric complexes used
| Tetramer specificity | HLA restriction | Virus | Protein | Sequence | Amino acid positions |
|---|---|---|---|---|---|
| hCMV-pp65 | HLA-A*0101 | hCMV | pp65 | YSEHPTFTSQY | 363–373 |
| HLA-A*0201 | hCMV | pp66 | NLVPMVATV | 495–504 | |
| HLA-B*0702 | hCMV | pp65 | TPRVTGGGAM | 417–426 | |
| HLA-B*3501 | hCMV | pp65 | IPSINVHHY | 123–131 | |
| EBV-BZLF1 | HLA-B*3501 | EBV | BZLF-1 | EPLPQGQLTAY | 64–65 |
| EBV-BMLF1 | HLA-A*0201 | EBV | BMLF-1 | GLCTLVAML | 259–267 |
| EBV-EBNA1 | HLA-B*3501 | EBV | EBNA1 | HPVGEADYFEY | 407–417 |
| EBV-EBNA3a | HLA-B*0702 | EBV | EBNA3a | RPPIFIRRL | 247–255 |
| HLA-B*0802 | EBV | EBNA3a | FLRGRAYGL | 193–201 | |
| HLA-A*0301 | EBV | EBNA3a | RLRAEAQVK | 603–611 | |
| FLU-MP | HLA-A*0201 | Influenza A virus | Matrix protein 1 | GILGFVFTL | 58–66 |
| RSV-NP | HLA-B*0702 | RSV | Nucleoprotein | NPKASLLSL | 306–314 |
| HIV-gag | HLA-B*0802 | HIV | Gag (p24) | EIYKRWII | 260–267 |
| HIV-nef | HLA-B*0802 | HIV | Nef | FLKEKGGL | 90–97 |
Tetramer staining and phenotyping of CD8+ T cells.
PBMCs were incubated with the allophycocyanin (APC)-labeled tetrameric complex (see Table 5) for 30 min at 4°C in the dark, after which the surface staining was done under the same conditions. A LIVE/DEAD fixable red cell stain kit (Life Technologies Europe BV, Bleiswijk, Netherlands) was used to exclude dead cells. For the detection of intracellular molecules, cells were fixed and permeabilized using the Foxp3/transcription factor staining set (eBioscience Inc., San Diego, CA, USA). PBMCs were subsequently stained with fluorescently labeled monoclonal antibodies specific for intracellular markers. Monoclonal antibodies used include anti-CD3 V500, anti-CD28 phycoerythrin (PE), anti-CCR7 PE-Cy7, anti-granzyme B Alexa Fluor 700, anti-CD27 APC eFluor 780, anti-Eomes PerCP-eFluor 710 (eBioscience Inc.), anti-KLRG1 Alexa Fluor 488 (31), anti-KLRG1 Alexa Fluor 647 (31), anti-granzyme K PE (Immunotools, Friesoythe, Germany), anti-CD8 brilliant violet 711 (BV711), anti-CD127 (IL-7Rα) BV711, anti-CD45RA BV650, anti-CD8 BV785, anti-Ki-67 BV711, anti-T-bet BV421 (BioLegend, San Diego, CA, USA), and anti-CD28 fluorescein isothiocyanate (FITC) (Sanquin, Amsterdam, the Netherlands). Flow cytometry measurements were performed on an LSRFortessa flow cytometer (BD Biosciences) that was calibrated on a daily basis. Analyses were done using FlowJo version 9.7.5 (FlowJo, Ashland, OR, USA).
Gating strategy.
Lymphocytes were gated using forward/sideward scatter properties (see Fig. S1b in the supplemental material). Duplets were excluded using forward scatter width/height and sideward scatter (SSC) width/height characteristics (see Fig. S1b). Dead cells were excluded using LIVE/DEAD fixable red cell fluorescence intensity (FI) (see Fig. S1b). CD3+ CD8+ tetramer+ events were gated as shown in Fig. S1b. CD8+ T-cell subsets were gated first as CD45RA+ CD27+, CD45RA− CD27+, CD45RA− CD27−, and CD45RA+ CD27− populations (see Fig. S1b). CCR7-, CD27-, and CD28-positive and -negative events were then gated and dragged into the CD45RA/CD27 populations, ultimately yielding 16 different gates that held events with a distinct CD45RA, CD27, CCR7, and CD28 expression profile (see Fig. 1a). T-bet and Eomes populations were gated as shown in Fig. S1a using the SSC plotted against T-bet, the SSC plotted against Eomes, and T-bet plotted against Eomes to determine the final gating. IL-7Rα, granzyme K, KLRG1, and/or granzyme B gates, identifying positive or negative events, were then inserted in the CD45RA/CD27/CCR7/CD28 gates and/or in the T-bet/Eomes gates.
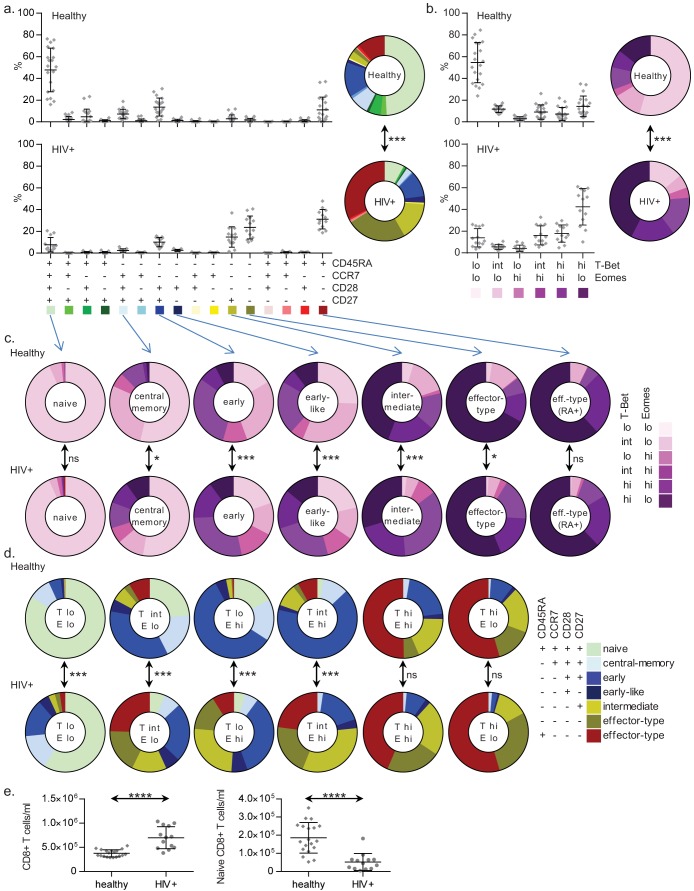
FIG 1.
Shifting associations between the surface phenotype and the T-bet/Eomes expression levels. (a) The distribution of all possible CD45RA/CCR7/CD28/CD27 phenotypes among the total CD8+ T-cell pools of 20 healthy (top) or 13 HIV-1-infected (bottom) individuals; (b) in the same groups, the distribution of the T-bet/Eomes expression states over the total CD8+ T-cell pools, with means and standard deviations (SD) shown; (c) T-bet/Eomes expression states per surface marker-defined subset; (d) reciprocally, expression states of the surface marker-defined subsets per T-bet/Eomes population, where “T” (denoting T-bet) and “E” (denoting Eomes) are followed by either lo, int, or hi, indicating a low, intermediate, or high level of expression of the TFs, respectively (mean percentages are shown; see Fig. S1b and c in the supplemental material for statistical dispersion); (e) absolute numbers of overall (left) and naive (right) CD8+ T cells in healthy and HIV-1-infected individuals. ns, not significant; *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001.
Virological analyses.
Quantitative PCR (qPCR) measurements to determine viral loads and serological assays to determine the presence of antiviral antibodies were done as described previously (32, 33).
Statistical analyses.
The differences in age between study groups were calculated using the Mann-Whitney test in IBM SPSS Statistics v22. For the statistical comparison of CD8+ T-cell properties from healthy and HIV-1-infected individuals, we used repeated-measurement analysis of variance (ANOVA) testing. This was only possible for the two-dimensional analyses (Fig. 1a to d, 2, 3a and b, and 5), as data points for the three-dimensional analyses (see Fig. 4; see also Fig. S4 and S9 in the supplemental material) were sometimes unavailable due to too few or no events in certain CD45RA/CCR7/CD28/CD27/T-bet/Eomes gates in CD8+ T-cell populations from some individuals. Therefore, for the three-dimensional analyses, we used a mixed linear model test. Furthermore, owing to the small population sizes of the EBV/hCMV-serotyped adults (see Fig. 5; see also Fig. S5 in the supplemental material), we were unable to compare individual groups to one another. Here, we analyzed all four groups at once in order to find out whether they were similar or not. Statistical differences between the absolute numbers of overall and naive CD8+ T cells (Fig. 1e) and the expression of IL-7Rα, granzyme K, KLRG1, and granzyme B by hCMV pp65-specific CD8+ T cells in healthy and HIV-1-infected individuals (see Fig. 3c) were assessed with unpaired Student's t tests. Statistical differences between the expression of IL-7Rα, granzyme K, KLRG1, and granzyme B by the surface marker-defined or T-bet/Eomes expression level-defined subsets from groups of healthy adults with different EBV/hCMV infection histories were determined using one-way ANOVA tests (Fig. 5d; see also Fig. S7 and S8 in the supplemental material). Results were considered statistically significant when P values were lower than 0.05.
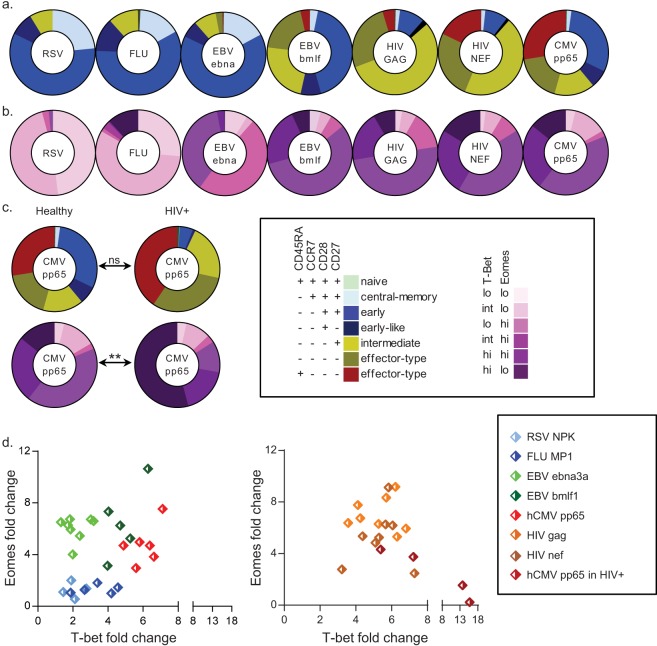
FIG 2.
Virus-specific CD8+ T cells show distinct CD45RA/CCR7/CD28/CD27 and T-bet/Eomes expression levels. The distribution of the CD45RA/CCR7/CD28/CD27 phenotypes (a) and the T-bet/Eomes expression states (b) found among RSV NP (n = 5 individuals)-, influenza A virus (Flu) MP1 (n = 5)-, EBV EBNA3a (n = 8)-, EBV BMLF-1 (n = 5)-, HIV-1 gag (n = 12)-, HIV-1 Nef (n = 11)-, and hCMV pp65 (n = 7)-specific CD8+ T-cell populations; (c) comparison of hCMV pp65-specific CD8+ T cells circulating in healthy and HIV-1-infected individuals (n = 4), with mean percentages shown (see also Fig. S2 in the supplemental material); (d) representative scatter plots depicting the locations of different virus-specific populations in healthy subjects (left) or HIV-1 Gag-, HIV-1 Nef-, and hCMV pp65-specific CD8+ T cells in HIV-1-infected subjects (right) on the T-bet/Eomes plot, shown as geometric mean fluorescence intensity (GMFI) fold changes (overall population GMFI divided by the double-negative population GMFI). ns, not significant; **, P < 0.01.
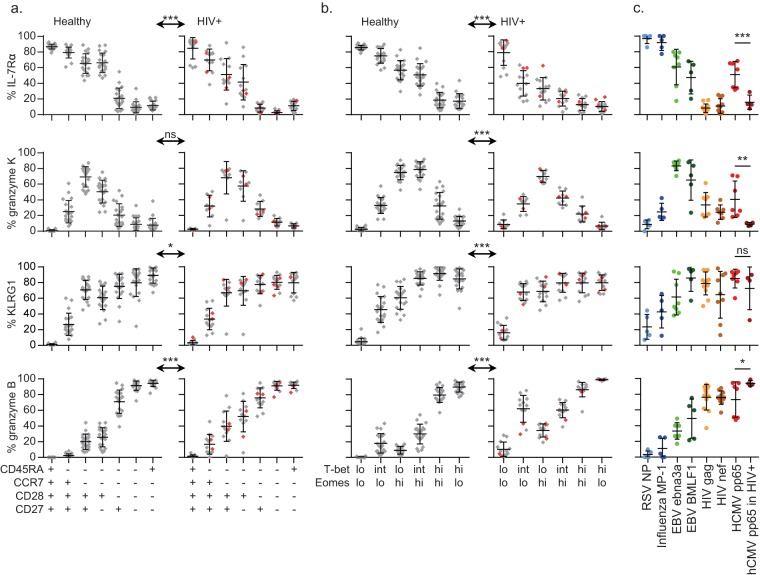
FIG 3.
T-bet and Eomes expression levels are indicators of the functional potential of CD8+ T cells. Expression frequencies of IL-7Rα (first row), granzyme K (second row), KLRG1 (third row), and granzyme B (fourth row) per surface marker-defined subset in 20 healthy (left) or 13 HIV-1-infected (right) individuals (n = 8 for the expression of granzyme K by CD8+ T cells in HIV-1-infected individuals), with means and SD shown (a), per T-bet/Eomes population, with means and SD shown (b), and per virus-specific memory/latency population: RSV NP-specific cells (light blue, n = 5 individuals), influenza A virus MP1-specific cells (dark blue, n = 5), EBV EBNA-3a-specific cells (light green, n = 8), EBV BMLF-1-specific cells (dark green, n = 5), HIV-1 Gag-specific cells (yellow, n = 12), HIV-1 Nef-specific cells (light brown, n = 11), hCMV pp65-specific cells (red, n = 7), and hCMV pp65 circulating in HIV-1-infected individuals (dark red, n = 4), with means and SD shown (c). The two HIV-1-infected patients not coinfected with hCMV are shown in red. ns, not significant; *, P < 0.05; **, P < 0.01; ***, P < 0.001.
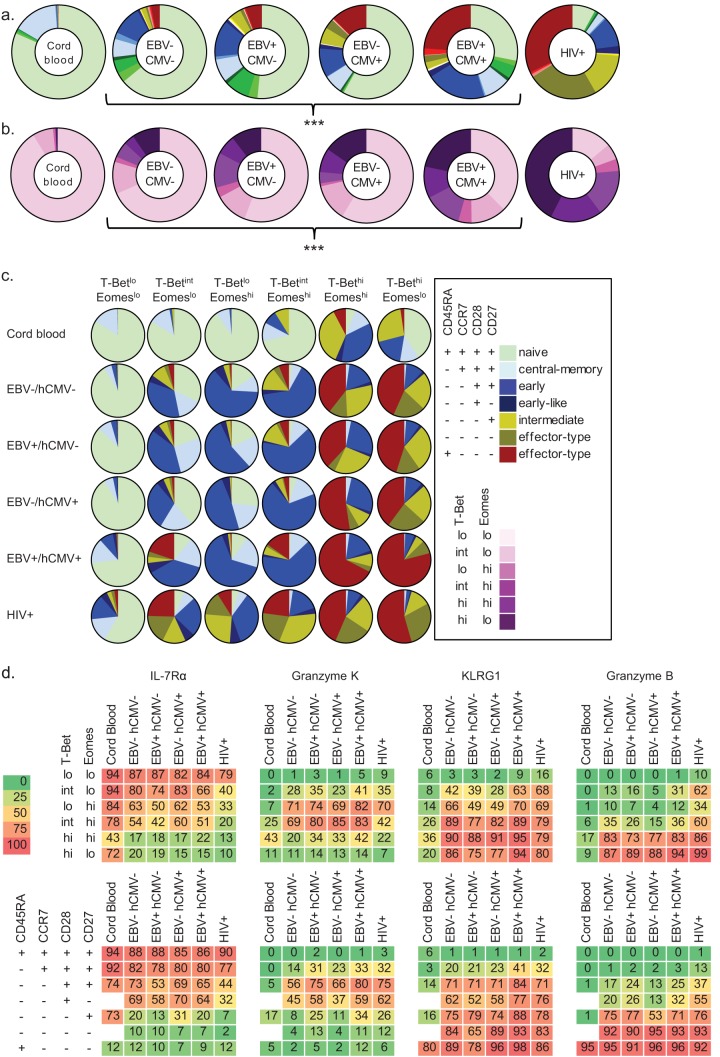
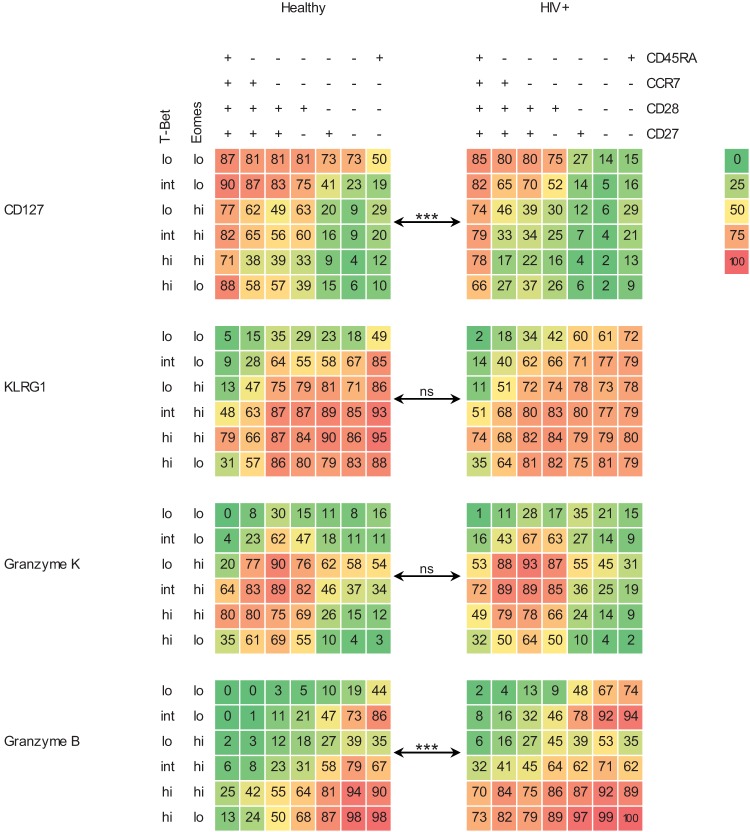
FIG 5.
Infection history influences the associations between surface phenotype, T-bet/Eomes expression levels, and functional potential. The distribution of CD45RA/CCR7/CD28/CD27 subsets (a) and of T-bet/Eomes populations (b) in cord blood samples (n = 5) and in individuals that are EBV/hCMV seronegative (n = 6), EBV monoinfected (n = 5), hCMV monoinfected (n = 3), EBV/hCMV double infected (n = 6), and HIV-1 infected (n = 13), with median percentages shown (see also Fig. S5a and b in the supplemental material); (c) the surface marker-defined subsets per T-bet/Eomes population (mean percentages shown; see also Fig. S6 in the supplemental material); (d) heat maps showing the associations between T-bet/Eomes expression state (y axis, first row), CD45RA/CCR7/CD28/CD27 phenotype (y axis, second row), and expression of IL-7Rα (first column), granzyme K (second column), KLRG1 (third column), and granzyme B (last column), per study group (x axis, both rows). The degree of expression intensity is indicated by increments in color intensity and by the mean percentages of positive cells denoted in the boxes (see Fig. S9 in the supplemental material for statistical dispersion). ***, P < 0.001.
FIG 4.
Combining the CD45RA/CCR7/CD28/CD27 dimension and the T-bet/Eomes dimension more accurately predicts CD8+ T-cell functional potential. Heat maps depicting the degree of expression of IL-7Rα (first row), granzyme K (second row), KLRG1 (third row), and granzyme B (last row) per combined surface marker- and T-bet/Eomes-defined CD8+ T-cell subset plotted on the x and y axes, respectively, circulating in 20 healthy (left) or 13 HIV-1-infected (right) individuals. The degree of expression intensity is indicated by increments in color intensity from green to red and by the mean percentage of positive cells denoted in the boxes (see Fig. S4 in the supplemental material for statistical dispersion). ns, not significant; ***, P < 0.001.
RESULTS
T-bet and Eomes expression levels vary independently during CD8+ T-cell differentiation.
First, we determined the distribution of the CD45RA/CCR7/CD28/CD27 phenotypes among CD8+ T cells circulating in the peripheral blood compartments of healthy donors. Here, the putatively naive, early differentiated, and RA+ effector type form the most substantial subsets (Fig. 1a). As expected, HIV-1 infection strongly influences the characteristics of the CD8+ T-cell compartment (34), in that the naive population was smaller in size, whereas the intermediately differentiated and the RA–/+ effector-type populations were much larger (Fig. 1a). This was also true when comparing absolute numbers of putatively naive CD8+ T cells from healthy and HIV-1-infected individuals. Whereas the absolute overall numbers of CD8+ T cells were increased in the untreated HIV-1-infected individuals, their absolute numbers of CD45RA+/CCR7+/CD28+/CD27+ CD8+ T cells were significantly reduced compared to those of healthy adults (Fig. 1e). Concordant with data from McLane et al. (25), T-bet was expressed in ranges stretching from negative to low (lo), low to intermediate (int), and intermediate to high (hi), whereas Eomes was expressed either in a negative-to-low or intermediate-to-high fashion, ultimately yielding six distinct populations when plotted against each other (see Fig. S1a in the supplemental material). The majority of cells in the healthy individuals displayed a T-betlo Eomeslo expression state, with smaller representations of the other expression states (Fig. 1b). In the HIV-1-infected individuals, most cells showed a T-bethi Eomeslo expression pattern, with only modest proportions of cells in the T-betlo Eomeslo or other states (Fig. 1b).
We then investigated how T-bet and Eomes are expressed by the CD45RA/CCR7/CD28/CD27-defined subsets. As expected, in both healthy and HIV-1-infected subjects, each surface marker-defined subset was found to contain multiple T-bet/Eomes expression states (25), however, to various extents and in healthy individuals seemingly in a restricted range. The putatively naive and central memory populations contained mainly T-betlo Eomeslo cells, with only minor or small representations of the other expression states. The heterogeneity increased among the early differentiated, the early-like, and the intermediately differentiated subsets, whereas the distribution became skewed toward T-betint-hi Eomeslo-hi states among the RA−/+ effector-type subsets (Fig. 1c; see also Fig. S1c in the supplemental material). Remarkably, in the HIV-1-infected individuals, the CD45RA/CCR7/CD28/CD27-defined subsets expressed T-bet and/or Eomes at higher frequencies than what was seen in the healthy adults, nearly irrespective of the CD45RA/CCR7/CD28/CD27 phenotypes (Fig. 1c; see also Fig. S1c). Reciprocally, each of the six T-bet/Eomes populations in the total CD8+ T-cell pool comprised its own restricted range of surface marker-defined T-cell subsets, a distribution that was again different when comparing HIV-1-infected to healthy individuals (Fig. 1d; see also Fig. S1d).
In conclusion, the different CD45RA/CCR7/CD28/CD27-defined CD8+ T-cell subsets are each associated with a specific range of T-bet/Eomes expression patterns in healthy individuals. However, HIV-1 infection changes these relations profoundly.
Virus-specific memory populations display distinct T-bet/Eomes expression levels.
Similar analyses were done for circulating virus-specific CD8+ memory T-cell populations. As expected, RSV nuclear protein (NP)-, influenza A virus matrix protein 1 (MP1)-, and EBV nuclear protein 3a (EBNA3a; a latent cycle protein)-specific cells were phenotypically mainly central memory, early differentiated, and early-like cells (Fig. 2a; see also Fig. S2 in the supplemental material) (5, 7–10). However, the T-bet/Eomes expression states differed considerably: influenza A virus-specific cells held a substantial proportion of T-bethi Eomeslo cells, and EBV EBNA3a-specific cells comprised unique T-betlo Eomeshi and T-betint Eomeshi populations (Fig. 2b; see also Fig. S2). Instead, EBV BamHI-M leftward reading frame 1 (BMLF-1; a lytic cycle protein)-, hCMV phosphoprotein 65 (pp65)-, and HIV-1 group antigen (Gag)- and negative-factor (Nef)-specific cells displayed more advanced surface phenotypes than the other virus-specific CD8 T cell populations, and hCMV-specific cells comprised a sizeable RA+ effector-type population (Fig. 2a; see also Fig. S2), which is in line with previous publications (1, 5, 14, 35). However, the differences between the CD45RA/CCR7/CD28/CD27 phenotypes of hCMV- and HIV-1-specific cells were not necessarily reflected by the T-bet/Eomes expression states. These were quite similar, all comprising large populations of T-betint-hi Eomeslo-hi cells (Fig. 2b; see also Fig. S2). Previously, it was reported that hCMV pp65-specific CD8+ T cells in HIV-1-infected individuals more often displayed a further-differentiated surface phenotype than their counterparts found in healthy individuals (34). Although we were unable to confirm the latter on a statistical level, we did find that the hCMV pp65-specific cells in HIV-1-infected individuals were displaying a different range of T-bet/Eomes expression levels than their counterparts in healthy subjects, particularly concerning an increased number of cells in a T-bethi Eomeslo expression state (Fig. 2c; see also Fig. S2).
In conclusion, virus-specific populations display distinct T-bet/Eomes expression patterns (Fig. 2d). However, even when similar in surface phenotype, virus-specific CD8+ T-cell populations can differ substantially in regard to their T-bet and Eomes expression levels, and vice versa.
T-bet and Eomes expression levels are indicators of the functional potential of CD8+ T cells.
Surface marker-defined differentiation states are linked to specific functional properties of CD8+ T cells. Nevertheless, a substantial degree of functional heterogeneity has been observed within these subsets (11, 12). Therefore, we wanted to know whether T-bet/Eomes expression levels can be used to more accurately define T-cell functionality. The process of intranuclear staining for T-bet and Eomes, and the kinetics of these TFs after stimulation in vitro (25), hampers cell sorting and stimulation assays. Therefore, we defined the functional profiles of the surface marker- or TF-defined subsets by determining the expression of a number of key molecules predictive for functional potential: IL-7Rα (memory marker, mediator of homeostatic proliferation) (13, 36), granzyme K (effector memory marker, mediator of apoptosis, triggers cytokine release and functions through noncytotoxic inhibition of viral replication) (37–39), KLRG1 (coinhibitory receptor, negative regulator of fully differentiated T cells) (40, 41), and granzyme B (effector marker, mediator of apoptosis) (37, 38).
First, we determined the associations between the expression of these molecules and the CD45RA/CCR7/CD28/CD27 phenotypes in both healthy individuals and untreated HIV-1-infected individuals. Early differentiated, early-like, intermediately differentiated, and RA− effector-type cells in HIV-1-infected subjects showed a lower expression of IL-7Rα than their healthy counterparts (Fig. 3a). On the other hand, granzyme B was expressed in the HIV-1-infected subjects earlier than the surface phenotype initially suggested and was already found in substantial amounts in early differentiated and early-like cells (Fig. 3a).
We next identified associations between the expression of these functional markers and that of T-bet and/or Eomes. IL-7Rα was expressed most often by cells in a T-betlo Eomeslo state and declined progressively as the expression of T-bet and/or Eomes increases. Interestingly, for cells in HIV-1-infected individuals, the IL-7Rα expression rates dropped faster than those in healthy subjects (Fig. 3b). Also, granzyme K was expressed particularly by cells in the T-betlo-int Eomeshi expression states and infrequently by cells not expressing Eomes or by cells highly expressing T-bet (Fig. 3b). Furthermore, KLRG1 and granzyme B were found to be expressed in larger amounts by T-betlo-int Eomeslo-hi cells in HIV-1-infected individuals than in healthy subjects (Fig. 3b).
When examining virus-specific populations, the RSV- and influenza A virus-specific CD8+ T cells displayed the highest expression of IL-7Rα. This was lower for EBV- and hCMV-specific cells, until it was nearly absent from the HIV-1-specific cells (Fig. 3c). Granzyme K was expressed most often by both EBV-specific populations and much less frequently by the other virus-specific populations (Fig. 3c). KLRG1 and granzyme B were expressed mainly by EBV BMLF-1- and in particular by hCMV- and HIV-1-specific populations (Fig. 3c). Interestingly, hCMV pp65-specific cells circulating in HIV-1-infected individuals expressed significantly less IL-7Rα and granzyme K, while more often expressing granzyme B, than the same population in healthy subjects (Fig. 3c).
In conclusion, the expression levels of T-bet and Eomes predict for differences in the functional potential of CD8+ T-cell populations. Markedly, the associations between the T-bet/Eomes levels, the CD45RA/CCR7/CD28/CD28 phenotypes, and the expression of IL-7Rα, granzyme K, KLRG1, and granzyme B differ strongly between healthy and HIV-1-infected individuals.
Combining the CD45RA/CCR7/CD28/CD27 dimension and the T-bet/Eomes dimension more accurately predicts CD8+ T-cell functional potential.
We then merged the CD45RA/CCR7/CD28/CD27 dimension with the T-bet/Eomes dimension to study their combined relations to the expression of IL-7Rα, granzyme K, KLRG1, and granzyme B. This analysis further emphasized that cells with an identical surface phenotype can differ substantially regarding their T-bet/Eomes expression levels and their expression of IL-7Rα, granzyme K, KLRG1, and/or granzyme B (see Fig. S3 and S4 in the supplemental material). For example, the few putatively naive cells that were in a T-betlo-int Eomeshi state differed substantially from the naive cells in a T-betlo Eomeslo state with regard to their expression of granzyme K and KLRG1 (Fig. 4; see also Fig. S4). Interestingly, the associations between the T-bet/Eomes expression levels and IL-7Rα, granzyme K, KLRG1, and granzyme B expression varied per CD45RA/CCR7/CD28/CD27-defined subset. Thus, naive T-betlo Eomeslo cells expressed IL-7Rα at a much higher rate than that expressed by the RA+ effector-type cells in a T-betlo Eomeslo state (Fig. 4; see also Fig. S4). In HIV-1-infected subjects, these associations were entirely different, particularly concerning a generally lower expression of IL-7Rα and a higher expression of granzyme B (Fig. 4; see Fig. S4).
Therefore, the combined analysis of the CD45RA/CCR7/CD28/CD27 phenotypes and the TF dimension reveals a more accurate image of the functional potential of an individual CD8+ T cell. Importantly, all these associations, and thus the makeup of the total CD8+ T-cell pool, change significantly against the background of untreated HIV-1 infection, in particular when it concerns the overall expression of IL-7Rα and granzyme B.
Infection history influences the associations between surface phenotype, T-bet/Eomes expression levels, and the functional potential.
These findings urged us to determine whether EBV and/or hCMV infection also influenced the associations between surface phenotype, T-bet/Eomes expression, and the expression of molecules predictive for T-cell function. However, we first examined cord blood samples, which were comprised of mainly naive CD8+ T cells and a modest amount of central memory cells. Remarkably, these samples also were comprised of intermediately differentiated and RA+ effector-type cells, although in very low numbers. The early-like and RA− effector-type subsets were not detected (Fig. 5a; see also Fig. S5a in the supplemental material). Furthermore, the associations between surface phenotype, T-bet/Eomes expression levels, and expression of molecules predictive for function were completely different from those in adult CD8+ T cells. In contrast, whereas healthy adults with different EBV/hCMV infection histories were displaying significantly altered distributions of the surface marker-defined and T-bet/Eomes expression level-defined CD8+ T-cell subsets (Fig. 5a and b; see also Fig. S5), the T-bet/Eomes expression level-defined populations all comprised a similar distribution of the surface marker-defined subsets and vice versa (Fig. 5c; see also Fig. S6 in the supplemental material). Also, when examining the associations between either the CD45RA/CCR7/CD28/CD27 phenotypes or the T-bet/Eomes expression levels and the expression of molecules predictive for function, EBV/hCMV infection history did not appear to alter these. However, if we look at the three dimensions combined, we did find that the overall makeup of the CD8+ T-cell pool, as well as the differentiation state of individual CD8+ T cells, was different between healthy individuals with different EBV/HCMV infection histories. These differences concerned mainly the expression of granzyme K and KLRG1 by the CD45RA/CCR7/CD28/CD27/T-bet/Eomes-defined CD8+ T-cell populations. Granzyme K was more often expressed by EBV single- and EBV/HCMV dual-infected individuals than by cells from EBV-uninfected individuals, and KLRG1 levels were generally higher in the three-dimensionally defined CD8+ T-cell populations from EBV/hCMV dual-infected individuals than in single-infected or uninfected individuals (see Fig. S9 in the supplemental material).
It must be noted that the hCMV/EBV double-negative individuals were younger than the hCMV/EBV double- and HIV-1-seropositive individuals and that the more pronounced differences noted specifically between these groups may also be due to age and pathogen exposure (Table 2), all factors that are known to influence the CD8+ T-cell differentiation state (2, 3, 14).
In conclusion, although not as dramatic as HIV-1 infection, EBV and/or hCMV infection also influences the associations between the surface phenotype, the T-bet/Eomes expression levels, and the expression of molecules predictive for T-cell function in otherwise healthy adults.
T-bet and Eomes expression states of virus-specific memory populations are imprinted early during primary infection in vivo.
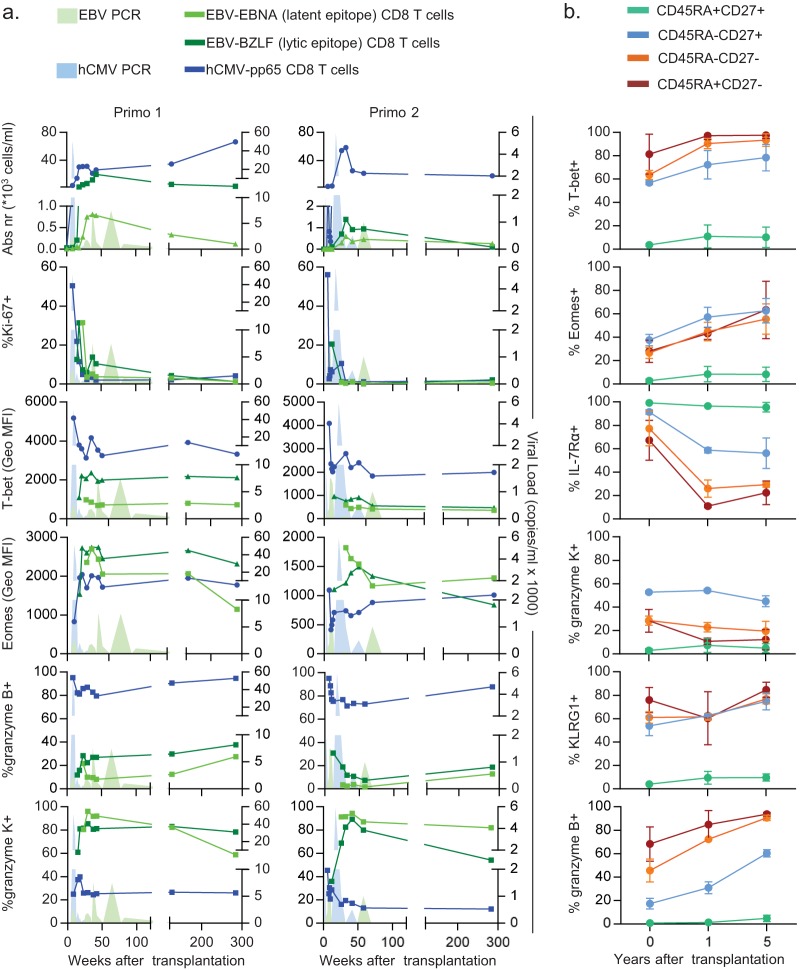
We then wanted to investigate how T-bet and Eomes are expressed by different virus-specific memory populations over the course of primary infection. Therefore, we determined the expression of the TFs by developing hCMV pp65-, EBV EBNA-, and EBV BZLF-1-specific CD8+ T-cell populations in longitudinally obtained samples deriving from two kidney transplant recipients who each nearly concomitantly experienced both a primary hCMV infection and a primary EBV infection.
Similar to the memory populations, acute-phase hCMV-specific cells highly expressed T-bet and much less so Eomes, whereas the reverse was true for the EBV-specific populations (Fig. 6a). Furthermore, acute-phase EBV-specific cells highly expressed granzyme K and only little granzyme B, where again the opposite went for hCMV-specific cells (Fig. 6a). These trends continued until viral loads were undetectable and infections had entered the latency stage (Fig. 6a).
FIG 6.
Early imprinting of T-bet/Eomes expression levels and shifts in the dimensions of CD8+ T cells over the course of primary persistent infections. (a) Characteristics of EBV EBNA- and BZLF-1-specific (light-green and dark-green lines, respectively) and hCMV pp65-specific CD8+ T cells (blue line) circulating in two kidney transplant recipients (left and right columns) who were both EBV and hCMV seronegative prior to receiving kidney allografts from EBV- and hCMV-seropositive donors, followed over the course of a primary EBV and hCMV infection (EBV and hCMV viral loads as determined by qPCR indicated by the light-green and light-blue filled areas, respectively): absolute numbers of tetramer+ events (first row), Ki-67+ tetramer+ events (second row), T-bet GMFI (third row), Eomes GMFI (fourth row), granzyme B+ tetramer+ events (fifth row), and granzyme K+ tetramer+ events (last row). (b) Graphs showing the expression of T-bet (first plot), Eomes (second plot), IL-7Rα (third plot), granzyme K (fourth plot), KLRG1 (fifth plot), and granzyme B (last plot) per CD45RA/CD27-defined subset (lines) over the course of transplantation and EBV/hCMV seroconversion.
Because these patients progressed from an EBV/hCMV-seronegative status to a dually infected status, with a long-term follow-up period during viral latency, this provided us with the unique opportunity to see whether EBV/hCMV infection history indeed changes the associations between the surface marker phenotype, T-bet/Eomes expression levels, and expression of molecules predictive for function, also over time. In all surface marker-defined subsets, an increase in expression of T-bet, Eomes, and granzyme B was evident (Fig. 6b). Interestingly, the expression of IL-7Rα initially declined, only to slightly increase again long after the acute phase of infection (Fig. 6b). The changes were less apparent for granzyme K and KLRG1 (Fig. 6b).
Thus, the differential expression of T-bet, Eomes, granzyme K, and granzyme B by these virus-specific CD8+ memory T-cell populations appears to be imprinted early during the acute phase of primary infection. Furthermore, also long after resolution of the primary infection, changes in the associations between different dimensions that define the differentiation state of CD8+ T cells appear to occur.
DISCUSSION
In the current study, we show that specific T-bet and Eomes expression states relate to distinct expression patterns of IL-7Rα, granzyme K, KLRG1, and granzyme B expression, proteins that are predictive for the functional profile of CD8+ T cells (11, 13, 41, 42). We show that, by combining the surface phenotype with the TF expression pattern dimension, a more accurate image of the differentiation state of an individual CD8+ cell may be obtained. The variable associations between CD45RA/CCR7/CD28/CD27 phenotypes, the T-bet/Eomes expression levels, and the expression of IL-7Rα, granzyme K, KLRG1, and granzyme B, when comparing healthy with untreated HIV-1-infected individuals, emphasize how at a given time point, for example, a central memory T cell circulating in one individual is not necessarily the same functional entity as a central memory T cell circulating in another. Infection history shapes the makeup of the circulatory CD8+ T-cell pool, in some cases also affecting other virus-specific cells, as illustrated by hCMV pp65-specific cells circulating in HIV-1-infected individuals. The reasons for this are unknown but may involve an effect of a more inflammatory microenvironment, which may be reinforced by an increase in hCMV reactivations in these patients. The latter is supported by the finding of Papagno et al. (34), who showed that in HIV-1-infected individuals, unlike hCMV-and EBV-specific CD8+ T cells, influenza A virus-specific CD8+ T cells displayed a differentiation state similar to that of their circulating counterparts in healthy individuals (34). In regard to HIV-1 infection itself, it must be noted that all patients included in the current study were viremic. Jagannathan et al. demonstrated that the presence or absence of HIV-1 viremia indeed also impacts the differentiation state of HIV-1-specific CD8+ T cells (43). Furthermore, Hersperger et al. recently described how HIV-1-specific CD8+ T cells from elite controllers generally display a higher expression of T-bet and cytotoxic effector molecules, like perforin and granzyme B, than chronically infected progressors and highly active antiretroviral therapy-suppressed individuals, arguing also for a strong effect of an individual's immunological makeup on the phenotype of virus-specific CD8+ T cells (44). Because the frequency of virus-specific CD8+ T cells targeting a single virus in the total CD8+ T-cell pool is low, we expect that the changing associations between T-cell function, surface marker expression, and T-bet/Eomes expression levels, according to infection history, may be regulated on several levels other than simply the presence of virus-specific populations. For example, human CMV, EBV, and HIV-1 have been shown to utilize strategies aimed at disrupting T-cell activation on various levels that are likely to impact the overall makeup of the CD8+ T-cell pool as well (45–48). Finally, in the context of viral coinfection, heterologous immunity, the process by which a T cell originally raised against, for example, EBV is also able to react to presented hCMV epitopes, although probably with a different affinity/avidity, could also impact our findings. However, it is very difficult to hypothesize on the frequencies by which such events would occur. Nevertheless, if indeed certain CD8+ T-cell subsets are displaying different traits against the background of HIV-1 infection or EBV/hCMV dual infection, then it would be interesting to see how EBV/hCMV latency (or the absence of these infections) influences T-cell-inducing vaccination responses, for example, those induced with yellow fever 17D vaccination.
It is important to note that T-bet and Eomes are part of a vast and intricately interwoven network of a multitude of other TFs, the complexity of which we are only just beginning to comprehend. As such, looking at T-bet and Eomes, although important, provides us with a rather narrow scope. Indeed, in the current study, we show that the T-bet/Eomes expression levels alone cannot fully account for the infection history-dependent variations in the functional potential of CD8+ T-cell subsets. Kurachi et al. recently showed how the absence of the basic leucine zipper TF (BATF) from murine T cells perturbed the overall balance of other TFs affected by BATF and therewith also the translation of their many direct and indirect targets that ultimately produce the T-cell phenotype and functional profile (49). T-bet and Eomes are also known to interact with other TFs, such as Runt-related (Runx) TF 1 and Runx3, and are thereby in their own ways part of this system of TFs acting in T cells (50, 51). Such interactions may, either directly or indirectly, underlie the different associations between T-bet, Eomes, and the expression of IL-7Rα, granzyme K, KLRG1, and granzyme B noted per CD45RA/CCR7/CD28/CD27-defined subsets. In this regard, the network of TFs acting in central memory cells may very well be different from the network acting in the RA+ effector-type cells. Given the many functions of TFs, such as chromatin remodeling, gene accessibility may indeed vary per individual CD8+ T cell, depending on the (re)activation history (16, 17).
We also provide evidence that the distinct T-bet/Eomes expression states found in the memory phase appear to be the result of early imprinting during the acute phase, which is in line with what we found previously on the mRNA level in acute-phase hCMV-specific CD8+ T cells (52). Indeed, each virus has developed its own specialized mode of infection and survival tactics that in turn requires a specialized CD8+ T-cell response. The latter is therefore the product of distinct signals provided by the virus and/or the immunological environment triggered by the infection. Indeed, IL-12 was shown to differentially regulate CD8+ T-cell differentiation in mice by inducing the expression of T-bet, while repressing the expression of Eomes (23). EBV infection may therefore induce specific signals that promote the expression of Eomes over that of T-bet. For the future, identifying the roles of other specific environmental cues in regulating the balance in TF expression could prove to be essential for the successful development of specific T-cell-inducing vaccination strategies.
Supplementary Material
ACKNOWLEDGMENTS
We thank Giso Brasser, Irma Maurer, Nelly D. van der Bom-Baylon, Kirstin M. Heutinck, Ester M. M. van Leeuwen, Hanneke de Kort, Si La Yong, Gijs van Schijndel, Berend Hooibrink, Nan van Geloven, and Wouter J. Kikkert for their technical advice, statistical advice, and useful discussions. We also thank the physicians of the Renal Transplant Unit for their help and useful discussions. We are indebted to all participants for their participation in this study.
Furthermore, the Amsterdam Cohort Studies on HIV Infection and AIDS, a collaboration between the Amsterdam Health Service, the Academic Medical Center of the University of Amsterdam, Sanquin Blood Supply Foundation, the University Medical Center Utrecht, and the Jan van Goyen Clinic, are part of the Netherlands HIV Monitoring Foundation and financially supported by the Center for Infectious Disease Control of the Netherlands National Institute for Public Health and the Environment. Our research is in part supported by the Dutch Kidney Foundation (IP11.32).
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/JVI.03478-14.
REFERENCES
- 1.Appay V, Dunbar PR, Callan M, Klenerman P, Gillespie GM, Papagno L, Ogg GS, King A, Lechner F, Spina CA, Little S, Havlir DV, Richman DD, Gruener N, Pape G, Waters A, Easterbrook P, Salio M, Cerundolo V, McMichael AJ, Rowland-Jones SL. 2002. Memory CD8+ T cells vary in differentiation phenotype in different persistent virus infections. Nat Med 8:379–385. doi: 10.1038/nm0402-379. [DOI] [PubMed] [Google Scholar]
- 2.van Lier RA, ten Berge IJ, Gamadia LE. 2003. Human CD8(+) T-cell differentiation in response to viruses. Nat Rev Immunol 3:931–939. doi: 10.1038/nri1254. [DOI] [PubMed] [Google Scholar]
- 3.van Aalderen MC, Remmerswaal EB, ten Berge IJ, van Lier RA. 2014. Blood and beyond: properties of circulating and tissue-resident human virus-specific alphabeta CD8(+) T cells. Eur J Immunol 44:934–944. doi: 10.1002/eji.201344269. [DOI] [PubMed] [Google Scholar]
- 4.Sallusto F, Lenig D, Forster R, Lipp M, Lanzavecchia A. 1999. Two subsets of memory T lymphocytes with distinct homing potentials and effector functions. Nature 401:708–712. doi: 10.1038/44385. [DOI] [PubMed] [Google Scholar]
- 5.Hislop AD, Gudgeon NH, Callan MF, Fazou C, Hasegawa H, Salmon M, Rickinson AB. 2001. EBV-specific CD8+ T cell memory: relationships between epitope specificity, cell phenotype, and immediate effector function. J Immunol 167:2019–2029. doi: 10.4049/jimmunol.167.4.2019. [DOI] [PubMed] [Google Scholar]
- 6.van Aalderen MC, Remmerswaal EB, Heutinck KM, ten Brinke A, Pircher H, van Lier RA, ten Berge IJ. 2013. Phenotypic and functional characterization of circulating polyomavirus BK VP1-specific CD8+ T cells in healthy adults. J Virol 87:10263–10272. doi: 10.1128/JVI.01540-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Heidema J, de Bree GJ, De Graaff PM, van Maren WW, Hoogerhout P, Out TA, Kimpen JL, van Bleek GM. 2004. Human CD8(+) T cell responses against five newly identified respiratory syncytial virus-derived epitopes. J Gen Virol 85:2365–2374. doi: 10.1099/vir.0.80131-0. [DOI] [PubMed] [Google Scholar]
- 8.de Bree GJ, Heidema J, van Leeuwen EM, van Bleek GM, Jonkers RE, Jansen HM, van Lier RA, Out TA. 2005. Respiratory syncytial virus-specific CD8+ memory T cell responses in elderly persons. J Infect Dis 191:1710–1718. doi: 10.1086/429695. [DOI] [PubMed] [Google Scholar]
- 9.Hoji A, Rinaldo CR Jr. 2005. Human CD8+ T cells specific for influenza A virus M1 display broad expression of maturation-associated phenotypic markers and chemokine receptors. Immunology 115:239–245. doi: 10.1111/j.1365-2567.2005.02135.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.He XS, Mahmood K, Maecker HT, Holmes TH, Kemble GW, Arvin AM, Greenberg HB. 2003. Analysis of the frequencies and of the memory T cell phenotypes of human CD8+ T cells specific for influenza A viruses. J Infect Dis 187:1075–1084. doi: 10.1086/368218. [DOI] [PubMed] [Google Scholar]
- 11.Hamann D, Baars PA, Rep MH, Hooibrink B, Kerkhof-Garde SR, Klein MR, van Lier RA. 1997. Phenotypic and functional separation of memory and effector human CD8+ T cells. J Exp Med 186:1407–1418. doi: 10.1084/jem.186.9.1407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Romero P, Zippelius A, Kurth I, Pittet MJ, Touvrey C, Iancu EM, Corthesy P, Devevre E, Speiser DE, Rufer N. 2007. Four functionally distinct populations of human effector-memory CD8+ T lymphocytes. J Immunol 178:4112–4119. doi: 10.4049/jimmunol.178.7.4112. [DOI] [PubMed] [Google Scholar]
- 13.van Leeuwen EM, de Bree GJ, Remmerswaal EB, Yong SL, Tesselaar K, ten Berge IJ, van Lier RA. 2005. IL-7 receptor alpha chain expression distinguishes functional subsets of virus-specific human CD8+ T cells. Blood 106:2091–2098. doi: 10.1182/blood-2005-02-0449. [DOI] [PubMed] [Google Scholar]
- 14.Gamadia LE, van Leeuwen EM, Remmerswaal EB, Yong SL, Surachno S, Wertheim-van Dillen PM, ten Berge IJ, van Lier RA. 2004. The size and phenotype of virus-specific T cell populations is determined by repetitive antigenic stimulation and environmental cytokines. J Immunol 172:6107–6114. doi: 10.4049/jimmunol.172.10.6107. [DOI] [PubMed] [Google Scholar]
- 15.van Leeuwen EM, Gamadia LE, Baars PA, Remmerswaal EB, ten Berge IJ, van Lier RA. 2002. Proliferation requirements of cytomegalovirus-specific, effector-type human CD8+ T cells. J Immunol 169:5838–5843. doi: 10.4049/jimmunol.169.10.5838. [DOI] [PubMed] [Google Scholar]
- 16.Szabo SJ, Kim ST, Costa GL, Zhang X, Fathman CG, Glimcher LH. 2000. A novel transcription factor, T-bet, directs Th1 lineage commitment. Cell 100:655–669. doi: 10.1016/S0092-8674(00)80702-3. [DOI] [PubMed] [Google Scholar]
- 17.Szabo SJ, Sullivan BM, Stemmann C, Satoskar AR, Sleckman BP, Glimcher LH. 2002. Distinct effects of T-bet in TH1 lineage commitment and IFN-gamma production in CD4 and CD8 T cells. Science 295:338–342. doi: 10.1126/science.1065543. [DOI] [PubMed] [Google Scholar]
- 18.Pearce EL, Mullen AC, Martins GA, Krawczyk CM, Hutchins AS, Zediak VP, Banica M, DiCioccio CB, Gross DA, Mao CA, Shen H, Cereb N, Yang SY, Lindsten T, Rossant J, Hunter CA, Reiner SL. 2003. Control of effector CD8+ T cell function by the transcription factor eomesodermin. Science 302:1041–1043. doi: 10.1126/science.1090148. [DOI] [PubMed] [Google Scholar]
- 19.Kaech SM, Cui W. 2012. Transcriptional control of effector and memory CD8+ T cell differentiation. Nat Rev Immunol 12:749–761. doi: 10.1038/nri3307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Intlekofer AM, Takemoto N, Kao C, Banerjee A, Schambach F, Northrop JK, Shen H, Wherry EJ, Reiner SL. 2007. Requirement for T-bet in the aberrant differentiation of unhelped memory CD8+ T cells. J Exp Med 204:2015–2021. doi: 10.1084/jem.20070841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Joshi NS, Cui W, Chandele A, Lee HK, Urso DR, Hagman J, Gapin L, Kaech SM. 2007. Inflammation directs memory precursor and short-lived effector CD8(+) T cell fates via the graded expression of T-bet transcription factor. Immunity 27:281–295. doi: 10.1016/j.immuni.2007.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Intlekofer AM, Takemoto N, Wherry EJ, Longworth SA, Northrup JT, Palanivel VR, Mullen AC, Gasink CR, Kaech SM, Miller JD, Gapin L, Ryan K, Russ AP, Lindsten T, Orange JS, Goldrath AW, Ahmed R, Reiner SL. 2005. Effector and memory CD8+ T cell fate coupled by T-bet and eomesodermin. Nat Immunol 6:1236–1244. doi: 10.1038/ni1268. [DOI] [PubMed] [Google Scholar]
- 23.Takemoto N, Intlekofer AM, Northrup JT, Wherry EJ, Reiner SL. 2006. Cutting edge: IL-12 inversely regulates T-bet and eomesodermin expression during pathogen-induced CD8+ T cell differentiation. J Immunol 177:7515–7519. doi: 10.4049/jimmunol.177.11.7515. [DOI] [PubMed] [Google Scholar]
- 24.Banerjee A, Gordon SM, Intlekofer AM, Paley MA, Mooney EC, Lindsten T, Wherry EJ, Reiner SL. 2010. Cutting edge: the transcription factor eomesodermin enables CD8+ T cells to compete for the memory cell niche. J Immunol 185:4988–4992. doi: 10.4049/jimmunol.1002042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.McLane LM, Banerjee PP, Cosma GL, Makedonas G, Wherry EJ, Orange JS, Betts MR. 2013. Differential localization of T-bet and Eomes in CD8 T cell memory populations. J Immunol 190:3207–3215. doi: 10.4049/jimmunol.1201556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Smith C, Elhassen D, Gras S, Wynn KK, Dasari V, Tellam J, Tey SK, Rehan S, Liu YC, Rossjohn J, Burrows SR, Khanna R. 2012. Endogenous antigen presentation impacts on T-box transcription factor expression and functional maturation of CD8+ T cells. Blood 120:3237–3245. doi: 10.1182/blood-2012-03-420182. [DOI] [PubMed] [Google Scholar]
- 27.Knox JJ, Cosma GL, Betts MR, McLane LM. 2014. Characterization of T-bet and Eomes in peripheral human immune cells. Front Immunol 5:217. doi: 10.3389/fimmu.2014.00217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Dolfi DV, Mansfield KD, Polley AM, Doyle SA, Freeman GJ, Pircher H, Schmader KE, Wherry EJ. 2013. Increased T-bet is associated with senescence of influenza virus-specific CD8 T cells in aged humans. J Leukoc Biol 93:825–836. doi: 10.1189/jlb.0912438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Buggert M, Tauriainen J, Yamamoto T, Frederiksen J, Ivarsson MA, Michaelsson J, Lund O, Hejdeman B, Jansson M, Sonnerborg A, Koup RA, Betts MR, Karlsson AC. 2014. T-bet and Eomes are differentially linked to the exhausted phenotype of CD8+ T cells in HIV infection. PLoS Pathog 10:e1004251. doi: 10.1371/journal.ppat.1004251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Geskus RB. 2000. On the inclusion of prevalent cases in HIV/AIDS natural history studies through a marker-based estimate of time since seroconversion. Stat Med 19:1753–1769. doi:. [DOI] [PubMed] [Google Scholar]
- 31.Marcolino I, Przybylski GK, Koschella M, Schmidt CA, Voehringer D, Schlesier M, Pircher H. 2004. Frequent expression of the natural killer cell receptor KLRG1 in human cord blood T cells: correlation with replicative history. Eur J Immunol 34:2672–2680. doi: 10.1002/eji.200425282. [DOI] [PubMed] [Google Scholar]
- 32.Klarenbeek PL, Remmerswaal EB, ten Berge IJ, Doorenspleet ME, van Schaik BD, Esveldt RE, Koch SD, ten Brinke A, van Kampen AH, Bemelman FJ, Tak PP, Baas F, de Vries N, van Lier RA. 2012. Deep sequencing of antiviral T-cell responses to HCMV and EBV in humans reveals a stable repertoire that is maintained for many years. PLoS Pathog 8:e1002889. doi: 10.1371/journal.ppat.1002889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.van Manen D, Delaneau O, Kootstra NA, Boeser-Nunnink BD, Limou S, Bol SM, Burger JA, Zwinderman AH, Moerland PD, van 't Slot R, Zagury JF, van 't Wout AB, Schuitemaker H. 2011. Genome-wide association scan in HIV-1-infected individuals identifying variants influencing disease course. PLoS One 6:e22208. doi: 10.1371/journal.pone.0022208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Papagno L, Spina CA, Marchant A, Salio M, Rufer N, Little S, Dong T, Chesney G, Waters A, Easterbrook P, Dunbar PR, Shepherd D, Cerundolo V, Emery V, Griffiths P, Conlon C, McMichael AJ, Richman DD, Rowland-Jones SL, Appay V. 2004. Immune activation and CD8+ T-cell differentiation towards senescence in HIV-1 infection. PLoS Biol 2:E20. doi: 10.1371/journal.pbio.0020020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gamadia LE, Rentenaar RJ, Baars PA, Remmerswaal EB, Surachno S, Weel JF, Toebes M, Schumacher TN, ten Berge IJ, van Lier RA. 2001. Differentiation of cytomegalovirus-specific CD8(+) T cells in healthy and immunosuppressed virus carriers. Blood 98:754–761. doi: 10.1182/blood.V98.3.754. [DOI] [PubMed] [Google Scholar]
- 36.Kimura MY, Pobezinsky LA, Guinter TI, Thomas J, Adams A, Park JH, Tai X, Singer A. 2013. IL-7 signaling must be intermittent, not continuous, during CD8(+) T cell homeostasis to promote cell survival instead of cell death. Nat Immunol 14:143–151. doi: 10.1038/ni.2494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Susanto O, Trapani JA, Brasacchio D. 2012. Controversies in granzyme biology. Tissue Antigens 80:477–487. doi: 10.1111/tan.12014. [DOI] [PubMed] [Google Scholar]
- 38.Bovenschen N, Kummer JA. 2010. Orphan granzymes find a home. Immunol Rev 235:117–127. doi: 10.1111/j.0105-2896.2010.00889.x. [DOI] [PubMed] [Google Scholar]
- 39.Zhong C, Li C, Wang X, Toyoda T, Gao G, Fan Z. 2012. Granzyme K inhibits replication of influenza virus through cleaving the nuclear transport complex importin alpha1/beta dimer of infected host cells. Cell Death Differ 19:882–890. doi: 10.1038/cdd.2011.178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Henson SM, Akbar AN. 2009. KLRG1—more than a marker for T cell senescence. Age 31:285–291. doi: 10.1007/s11357-009-9100-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Voehringer D, Koschella M, Pircher H. 2002. Lack of proliferative capacity of human effector and memory T cells expressing killer cell lectinlike receptor G1 (KLRG1). Blood 100:3698–3702. doi: 10.1182/blood-2002-02-0657. [DOI] [PubMed] [Google Scholar]
- 42.Bratke K, Kuepper M, Bade B, Virchow JC Jr, Luttmann W. 2005. Differential expression of human granzymes A, B, and K in natural killer cells and during CD8+ T cell differentiation in peripheral blood. Eur J Immunol 35:2608–2616. doi: 10.1002/eji.200526122. [DOI] [PubMed] [Google Scholar]
- 43.Jagannathan P, Osborne CM, Royce C, Manion MM, Tilton JC, Li L, Fischer S, Hallahan CW, Metcalf JA, McLaughlin M, Pipeling M, McDyer JF, Manley TJ, Meier JL, Altman JD, Hertel L, Davey RT Jr, Connors M, Migueles SA. 2009. Comparisons of CD8+ T cells specific for human immunodeficiency virus, hepatitis C virus, and cytomegalovirus reveal differences in frequency, immunodominance, phenotype, and interleukin-2 responsiveness. J Virol 83:2728–2742. doi: 10.1128/JVI.02128-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hersperger AR, Martin JN, Shin LY, Sheth PM, Kovacs CM, Cosma GL, Makedonas G, Pereyra F, Walker BD, Kaul R, Deeks SG, Betts MR. 2011. Increased HIV-specific CD8+ T-cell cytotoxic potential in HIV elite controllers is associated with T-bet expression. Blood 117:3799–3808. doi: 10.1182/blood-2010-12-322727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Miyazaki I, Cheung RK, Dosch HM. 1993. Viral interleukin 10 is critical for the induction of B cell growth transformation by Epstein-Barr virus. J Exp Med 178:439–447. doi: 10.1084/jem.178.2.439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Cheung TC, Humphreys IR, Potter KG, Norris PS, Shumway HM, Tran BR, Patterson G, Jean-Jacques R, Yoon M, Spear PG, Murphy KM, Lurain NS, Benedict CA, Ware CF. 2005. Evolutionarily divergent herpesviruses modulate T cell activation by targeting the herpesvirus entry mediator cosignaling pathway. Proc Natl Acad Sci U S A 102:13218–13223. doi: 10.1073/pnas.0506172102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Avdic S, Cao JZ, Cheung AK, Abendroth A, Slobedman B. 2011. Viral interleukin-10 expressed by human cytomegalovirus during the latent phase of infection modulates latently infected myeloid cell differentiation. J Virol 85:7465–7471. doi: 10.1128/JVI.00088-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Majumder B, Janket ML, Schafer EA, Schaubert K, Huang XL, Kan-Mitchell J, Rinaldo CR Jr, Ayyavoo V. 2005. Human immunodeficiency virus type 1 Vpr impairs dendritic cell maturation and T-cell activation: implications for viral immune escape. J Virol 79:7990–8003. doi: 10.1128/JVI.79.13.7990-8003.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kurachi M, Barnitz RA, Yosef N, Odorizzi PM, DiIorio MA, Lemieux ME, Yates K, Godec J, Klatt MG, Regev A, Wherry EJ, Haining WN. 2014. The transcription factor BATF operates as an essential differentiation checkpoint in early effector CD8+ T cells. Nat Immunol 15:373–383. doi: 10.1038/ni.2834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Cruz-Guilloty F, Pipkin ME, Djuretic IM, Levanon D, Lotem J, Lichtenheld MG, Groner Y, Rao A. 2009. Runx3 and T-box proteins cooperate to establish the transcriptional program of effector CTLs. J Exp Med 206:51–59. doi: 10.1084/jem.20081242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Djuretic IM, Levanon D, Negreanu V, Groner Y, Rao A, Ansel KM. 2007. Transcription factors T-bet and Runx3 cooperate to activate Ifng and silence Il4 in T helper type 1 cells. Nat Immunol 8:145–153. doi: 10.1038/ni1424. [DOI] [PubMed] [Google Scholar]
- 52.Hertoghs KM, Moerland PD, van Stijn A, Remmerswaal EB, Yong SL, van de Berg PJ, van Ham SM, Baas F, ten Berge IJ, van Lier RA. 2010. Molecular profiling of cytomegalovirus-induced human CD8+ T cell differentiation. J Clin Invest 120:4077–4090. doi: 10.1172/JCI42758. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.