Abstract
Hepatitis C virus contains a second open reading frame within the core gene, designated core+1/ARF. Here we demonstrate for the first time expression of core+1/ARF protein in the context of a bicistronic JFH1-based replicon and report the production of two isoforms, core+1/L (long) and core+1/S (short), with different kinetics.
TEXT
Hepatitis C virus (HCV) infection is a major cause of non-A, non-B hepatitis, which frequently leads to chronic liver disease and hepatocellular carcinoma (1); no vaccine is available, while the recent broadly effective therapy is highly expensive (2, 3). The single-stranded positive-sense RNA genome of HCV (9.6 kb) encodes a polyprotein precursor of about 3,000 amino acids flanked by conserved untranslated regions (UTRs) (4, 5). The UTRs are required for RNA translation and replication, and an internal ribosome entry site (IRES) within the 5′ UTR controls translation initiation (6). The polyprotein is processed by cellular and viral proteases to yield at least 10 structural and nonstructural proteins (C, E1, E2, p7, NS2, NS3, NS4A, NS4B, NS5A, and NS5B) (4, 5).
Previous studies from our laboratory and others have shown that HCV possesses a second functional open reading frame (ORF) within the core region of the polyprotein encoding an additional protein designated core+1/alternative reading frame protein (ARFP) (7–9). This alternate ORF is present in all six HCV genotypes (9) but has been studied only in subtypes 1a and 1b. At first, a 17-kDa protein (p17), initially named ARFP (alternative reading frame protein), F (frameshift), or core+1 (to indicate its position), was shown to be synthesized in rabbit reticulocyte lysate (RRL) from the initiating codon of the polyprotein sequence of a genotype 1a HCV type 1 (HCV-1) strain by a +1 frameshift occurring in an A-rich core-encoding region (codons 8 to 11) (7–10). However, the expression of this form of core+1/ARFP (known as F) is limited to the HCV-1 isolate, which is unique in carrying a stretch of 10 consecutive adenine nucleotides within codons 8 to 11 (7, 11, 12), and its expression in transfected cells is dependent on cytoplasmic transcription (11). Interestingly, other core+1/ARFP forms are expressed independently of the A-rich sequence by an alternative translation mechanism(s). Internal translation initiation at methionine codons 85/87 of HCV-1a and HCV-1b, resulting in a shorter form of core+1/ARFP, has been observed in transfected hepatoma cells (13–15). Furthermore, codon 26 of HCV-1a (12) and HCV-1b (15) was also found to function as an internal translation initiation site in mammalian cells. Additionally, a +1 frameshift at codon 42 of HCV-1b, followed by a rephrasing into the core ORF at stop codon 144, was observed in Escherichia coli (16).
The detection of specific anti-core+1/ARFP antibodies and T-cell responses in HCV-infected patients (especially with hepatocellular carcinoma), as reported by many independent laboratories, provides strong evidence that core+1/ARFP is produced during natural infection (7–9, 17–21). These studies suggest a link between core+1/ARFP and development of liver cancer (22–27). Conversely, core+1/ARFP has no obvious effect on virus replication following electroporation of Huh7-Lunet cells with full-length JFH1 wild-type and mutated viral RNAs or in mice xenografted with human liver tissue (28). Those studies were based on a mutational approach which theoretically abolishes the expression of core+1/ARFP and on comparisons of the replication kinetics of the wild-type and the mutated JFH1 viruses. However, to date, elucidation of core+1/ARFP expression in the context of HCV genome remains elusive.
The aim of this study was to investigate whether JFH1 supports the expression of core+1/ARFP. To this end, we used JFH1-based bicistronic replicons, tagged with the firefly luciferase gene at nucleotide (nt) 630, which would allow us to monitor and compare the levels of translation initiation of core and core+1 ORFs.
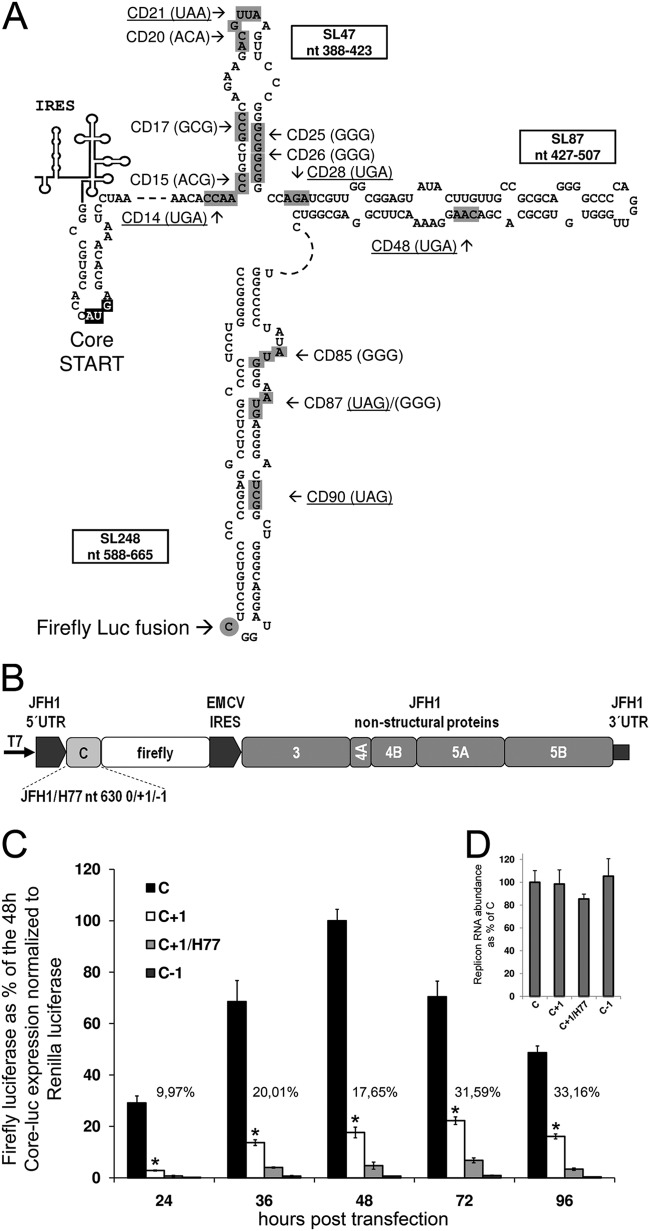
In order to investigate whether the JFH1 isolate expresses core+1/ARFP, we fused the firefly luciferase N-terminally with core+1/ARFP in the context of a bicistronic JFH1-based replicon (Fig. 1A and B). Luciferase tagging stabilizes the highly unstable core+1/ARFP (29) and provides a quantitative measure of the translation efficiency of core and core+1 ORFs (13, 29). Firefly luciferase was fused at nt 630 of the JFH1 replicon in all three reading frames (0/+1/-1 or core/core+1/core-1; Table 1) by PCR amplification of the JFH1 core-coding region (nt 342 to 630) and insertion into the AgeI and XbaI sites of the original pFK-i389LucNS3-3′-JFH1-dg replicon vector (30). Furthermore, to exclude expression from the initial firefly luciferase AUG, the site was mutated to GGG together with an introduction of a BglII site (pFK-i630-BglII-core+1/LucNS3-3′-JFH1-dg) as well. A hybrid luciferase-bearing replicon was constructed using the core-coding region (nt 342 to 630) of the H77 prototypic strain as described for JFH1. The replicons, after linearization with MluI, were used to produce RNA with T7 RNA polymerase (Promega), as described previously (31). The in vitro-transcribed RNAs were introduced into human hepatoma Huh7-Lunet cells by Lipofectamine transfection (2 μl Lipofectamine 2000 [Life Technologies]–0.5 μg replicon RNA on 105 cells plated in a 24-well tissue culture dish), and firefly luciferase expression was measured (dual-luciferase reporter assay; Promega). A capped-polyadenylated Renilla luciferase expressing RNA, produced as previously described (32), was included in the transfection (0.1 μg) as a normalizer and was measured with the dual-luciferase reporter assay (Promega).
FIG 1.
Expression from the core+1 frame within the JFH1-based replicon. (A) Prediction of the secondary RNA structure of stem-loops SL47, SL87, and SL248. Nucleotide positions are illustrated within the boxes of the respective stem-loops according to JFH1 numbering. Mutations introduced at various codons (gray boxes) of core+1 (CD) are shown with an arrow and designated according to codon number, while the mutated codons are noted in parentheses. The introduced nonsense/stop codons are underlined. The gray circle highlights the start of the core+1 fusion with the firefly luciferase (Luc). (B) Schematic representation of the bicistronic subgenomic JFH1-based replicon (pFK-i630-BglII-LucNS3-3′-JFH1-dg). The replicon is composed of the JFH1 5′ and 3′ UTRs and nonstructural protein region, while the core-coding region belongs to either JFH1 (nucleotides 342 to 630) or H77 (nucleotides 342 to 630) fused to the coding sequence of the firefly luciferase gene in the 0, +1, or −1 frame. The encephalomyocarditis virus (EMCV) IRES directs the expression of the nonstructural proteins. (C) Huh7-Lunet cells were transfected with RNA of the pFK-i630-BglII-LucNS3-3′-JFH1-dg-based replicons with the luciferase gene fused in the 0, +1, and −1 (C, C+1, and C-1) frame of the JFH1 core-coding region or the +1 frame of the corresponding H77 core-coding region. The levels of JFH1 core+1 expression are noted as percentages of the respective time point 0 frame expression level. (D) Real-time PCR quantification of replicon RNA abundance 48 h posttransfection. Quantification was achieved by amplification of the 5′ nontranslated region (5′NTR) of the replicon, while the YWHAZ cellular gene was used for normalization. At least three independent repetitions were performed; the error bars represent standard deviations. *, P < 0.02 (Student's t test).
TABLE 1.
List of oligonucleotides used for cloning and real-time quantitative PCRa
| Primer (assay) | Sequence |
|---|---|
| core 630 JFH1 | 5′-GGCGTCTTCCCCAGATCTCCATCCTGCCCAGCC-3′ |
| core+1 630 JFH1 | 5′-GGCGTCTTCCCCAGATCTGCCATCCTGCCCAGCC-3′ |
| core-1 630 JFH1 | 5′-GGCGTCTTCCCCAGATCTTCACCTGCCCAGCC-3′ |
| core+1 630 H77 | 5′-GGCGTCTTCCCCAGATCTGCCATCCCGCCCACCC-3′ |
| N328b | 5′-CTGTCTTCACGCAGAAAGCG-3′ |
| N329c | 5′-CCGAACGGACATTTCGAAG-3′ |
| N323Bd | 5′-GGGGAAGACGCCAAAAAC-3′ |
| CD20 M F | 5′-CCCAGAAGACATTAAGTTCCC-3′ |
| CD20 M R | 5′-GGGAACTTAATGTCTTCTGGG-3′ |
| CD21S F | 5′-CCAGAAGACGTGAAGTTCCCG-3′ |
| CD21S R | 5′-CGGGAACTTCACGTCTTCTGG-3′ |
| CD25/26 M F | 5′-AGTTCCCGGGGGGGGGCCAGATCG −3′ |
| CD25/26 M R | 5′-CGATCTGGCCCCCCCCCGGGAACT −3′ |
| CD14S F | 5′-CCAAAAGAAACATGAACCCGTCGCCCAGAAG-3′ |
| CD14S R | 5′-CTTCTGGGCGACGGTTCATGTTTCTTTTGG-3′ |
| CD28S F | 5′-CGGCGGCCTGATCGTTGGCGGAG-3′ |
| CD28S R | 5′-GCCAACGATCAGGCCGCCGCCCG-3′ |
| CD48S F | 5′-GGTGTGCGCACGATGAGGAAAACTACGG-3′ |
| CD48S R | 5′-CCGAAGTTTTCCTCATCGTGCGCACACC-3′ |
| CD85/87S F | 5′-GGCCCCTATGGGGGAGGGAGGGACTCG-3′ |
| CD85/87S R | 5′-CGAGTCCCTCCCTCCCCCATAGGGGCC-3′ |
| CD87S F | 5′-CTATATGGGATAGAGGGACTCGGC-3′ |
| CD87S R | 5′-AGCCGAGTCCCTCTATCCCATATAG-3′ |
| CD90S F | 5′-GGAATGAGGGACTAGGCTGGGCAGG-3′ |
| CD90S R | 5′-CCTGCCCAGCCTAGTCCCTCATTCC-3′ |
| JFH1-5NTR276F (RT-PCR for IRES) | 5′-GGCCTTGTGGTACTGCCTGATA-3′ |
| JFH1-5NTR354R (RT-PCR for IRES) | 5′-GGATTTGTGCTCATGGTGCA-3′ |
| YWHAZR | 5′-GGATGTGTTGGTTGCATTTCCT-3′ |
| YWHAZF | 5′-GCTGGTGATGACAAGAAAGG-3′ |
Mutated codons are underlined.
Forward for core 630 JFH1, core+1 630 JFH1, core-1 630 JFH1, core+1 630 H77, and all reverse primers (R) of STable2.
Reverse for N323B and all forward primers (F) of STable2.
Forward for N329 in AUG→GGG of luciferase original start.
Expression from the core frame peaked 48 h posttransfection, while core+1 frame expression peaked at 72 h (Fig. 1C). Expression of the core+1 frame relative to core expression increased gradually over time. Expression from the −1 frame was used as a negative control, as no ORF is predicted to exist in that frame. Surprisingly, far less core+1 frame expression was observed with the replicons bearing H77 sequences, suggesting that the insertion of genotype 1 core-coding sequences into the genotype 2 genome may negatively affect core+1/ARFP expression at a mechanistic level (Fig. 1C). All the mutations described above had no effect on replicon RNA abundance (Fig. 1D).
To provide insight into the expression of core+1/ARFP and the isoforms produced in the replicon system, a number of mutations were designed (Fig. 1A and Table 1). Initially, we sought to confirm the specificity of expression of core+1/ARFP in the replicon system, by introducing a nonsense mutation at core+1 ORF codon 90 (CD90S), which was expected to abolish the production of all known forms of core+1/ARFP. As shown in Fig. 2A, the nonsense mutation at core+1 codon 90 abolished the expression of core+1/ARF-luc at all of the time points tested.
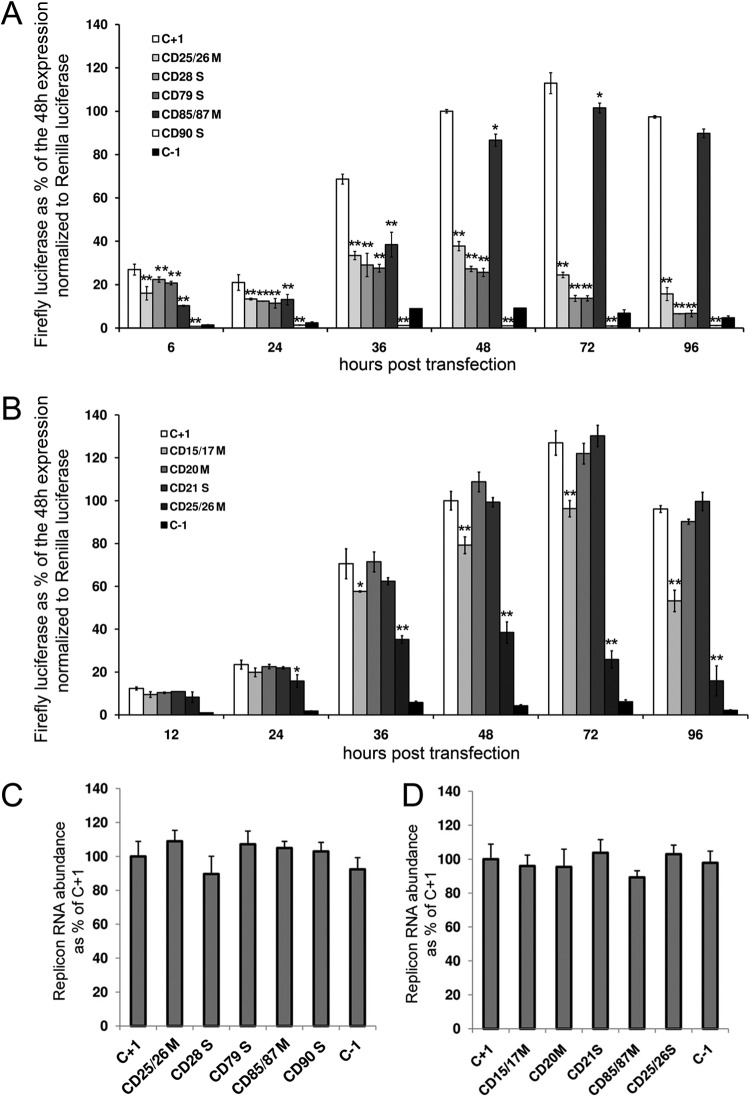
FIG 2.
core+1 frame expression initiates from two distinct sites within the core-coding region. (A and B) Huh7-Lunet cells were transfected with RNA from the pFK-i630-BglII-core+1/LucNS3-3′-JFH1-dg-based replicons bearing mutations at various codons (CD) of JFH1 core+1. Firefly luciferase produced from the mutated replicons at various time points posttransfection is expressed as a percentage of the production from the wild-type replicon (C+1) 48 h posttransfection. The expression is normalized to Renilla luciferase expression from a DNA plasmid and further normalized to replicon RNA levels as measured by reverse transcription-qPCR (RT-qPCR). CDs are designated according to the core+1 codon number. Codons annotated with an S represent introduced nonsense/stop codons and those annotated with an M introduced mutated codons. (C and D) Real-time PCR quantification of replicon RNA abundance 48 h posttransfection. Quantification was achieved by amplification of the 5′NTR of the replicon, while the YWHAZ cellular gene was used for normalization. At least three independent repetitions were performed; the error bars represent standard deviations. *, P < 0.05; **, P < 0.02 (Student's t test).
To determine the starting codon of core+1 frame and the possible existence of more than one isoform, we designed a series of nonsense mutations across the core sequence. Various starting codons/mechanisms have been reported in the past, including the frameshift at codons 8 to 11 (7–9, 33), internal initiation at codon 26 (12, 15), a frameshift at codon 42 (16), and a shorter form initiating at codons 85/87 (13, 15). As shown in Fig. 2, a nonsense mutation at codon 21 (CD21S) had no effect on core+1 expression, indicating that core+1/ARFP is not produced by ribosomal frameshifting at codons 8 to 11 (Fig. 2B). A nonsense mutation at codon 28 (CD28S) was the first that partially hindered core+1 expression, showing an increasing effect over time (Fig. 2A). A similar effect was observed when a nonsense mutation was inserted at codon 79 (CD79S), suggesting the absence of a core+1 start codon within that region (Fig. 2A). The significant difference between CD21S and CD28S in the measured levels and the total abolishment of core+1 expression between CD79S and CD90S indicated the existence of two isoforms of core+1: a short form (core+1/S) initiating between codons 79 and 90 and a long form (core+1/L) initiating between codons 21 and 28. Remarkably, as the reduction of core+1 frame expression in CD28S was minimal at the early stages of the replication, core+1/S seemed to be the predominant form, while at later time points core+1/L took over, reducing core+1/S expression to background levels (Fig. 2A). It must be noted that all the mutations described above had no effect on replicon RNA abundance, as measured by quantitative PCR (qPCR) 48 h posttransfection (Fig. 2C and D and Table 1).
Among the aforementioned putative initiation codons, those that fitted the expression profiles in the replicon system were codons 25/26 for core+1/L and codons 85/87 for core+1/S (12, 13, 15). After changing codons 26, 85, and 87 individually to GGG (which has never been reported to serve as an alternative initiation codon), the expression from the core+1 frame was similar to that seen with the control (data not shown). However, when mutations were introduced into both alternative initiation codons, codons 25 and 26 (CD25/26M), the reduction in the expression from core+1 frame was similar to that observed for CD28S and CD79S with similar kinetics, signifying a major role of codons 25/26 in the initiation of core+1/L (Fig. 2A). As the mutations in codons 25/26 partially disrupt the integrity of stem-loop SL47 (nt 388 to 423), we introduced analogous mutations on the complementary strand of the stem at codons 15 and 17 (CD15/17M) (Fig. 2B). The mutations showed a limited effect on core+1 frame expression, especially late posttransfection, possibly highlighting a mechanistic role of the stem in the core+1/L expression. On the other hand, mutations in both alternative initiation codons, 85/87 (CD85/87M), resulted in a significant early reduction of core+1 expression which was much less apparent 48 h posttransfection (Fig. 2A). The kinetics of CD85/87M were opposite those of CD28S and CD79S, demonstrating an early gene type of expression from codons 85/87.
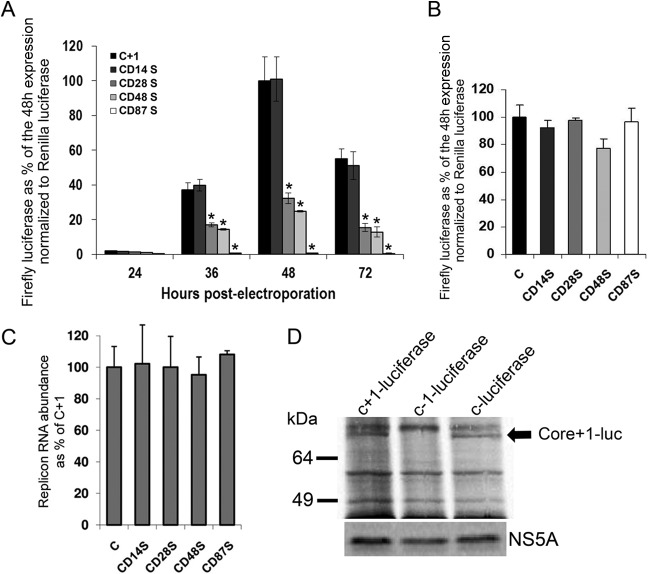
The mutational analysis described above clearly illustrated the synthesis of two isoforms of core+1, an early short form and a late long form, in the context of the HCV replicon. However, for the verification of core+1 expression, we attempted to also detect the expressed protein by biochemical methods. As the amount of core+1 produced with the luciferase replicon is estimated to be at picogram levels (∼10 pg/106 cells, as extrapolated from the activity of recombinant firefly luciferase), we aimed for investigation of the larger-scale replicon system via electroporation of replicon RNA into Huh7-lunet cells. To recapitulate via electroporation the results obtained by lipofection, we assessed the kinetics of replicons encompassing a number of nonsense mutations in the core+1 frame. Electroporation was performed in Huh7-Lunet cells resuspended in RPMI medium or Cytomix as previously described (34, 35). Nonsense mutations at codons 14, 28, 48, and 87 (CD14S, CD28S, CD48S, and CD87S, respectively) resulted in unaffected core+1 frame expression for CD14S, more than 60% inhibition of expression for CD28S/CD48S, and total inhibition for CD87S, as expected (Fig. 3A). All the mutations described above had no effect on replicon RNA abundance (Fig. 3B). CD14S and CD48S further supported the absence of a frameshift at either codons 8 to 11 or codon 42. It must be noted that introduction of the mutations described above in the replicon expressing luciferase in the 0 frame had a minor effect or no effect on luciferase expression (Fig. 3C). Notably, the replication efficiency peaked earlier than in lipofection 48 h postelectroporation. In order to optimize detection of core+1/ARFP, electroporated cells were supplemented with 0.5 mCi of [35S]methionine-cysteine for 24 h in methionine-cysteine-free Dulbecco's modified Eagle's medium (DMEM)–0.5% fetal bovine serum (FBS). Cell extracts (48 h postelectroporation) were subsequently immunoprecipitated using a rabbit polyclonal antibody against firefly luciferase (sc-32896; Santa Cruz Biotechnology), and proteins were separated by SDS-PAGE and visualized by autoradiography. A ca. 70-kDa band was observed in the lanes corresponding only to core+1/ARFP/L-luc (predicted 68 kDa) and core-luc (predicted 72 kDa) and not to core-1-luc (Fig. 3D). A band corresponding to core+1/ARFP/S-luc was not detected, possibly because of its low (∼10%) abundance 48 h postelectroporation. It must also be noted that the luciferase intensity in the gel and its enzymatic activity may not completely correlate due to the variations in the attached peptide corresponding to HCV sequences at the 2 reading frames. Furthermore, the expression of the core+1 ORF was also shown by a replicon carrying a core+1 glutathione S-transferase (GST) fusion (data not shown).
FIG 3.
Immunodetection of core+1 production within the JFH1-based replicon. (A) Huh7-Lunet cells were electroporated with RNA from the pFK-i630-BglII-LucNS3-3′-JFH1-dg) firefly luc-JFH1-based replicons bearing nonsense mutations at various codons (CD) of JFH1 core+1. Firefly luciferase produced from the mutated replicons at various time points postelectroporation is expressed as a percentage of the production from the wild-type replicon (C+1) 48 h postelectroporation. The expression is normalized to Renilla luciferase expression from a DNA plasmid and further normalized to replicon RNA levels as measured by RT-qPCR. (B) Huh7-Lunet cells were electroporated with RNAs representing firefly luc-JFH1-based replicons with firefly fused on a core frame bearing nonsense mutations at various codons (CD) of JFH1 core+1. Firefly luciferase produced from the mutated replicons at various time points postelectroporation is expressed as a percentage of the production from the wild-type replicon (C) 48 h posttransfection. The expression is normalized to Renilla luciferase expressed from a plasmid. (C) Real-time PCR quantification of replicon RNA abundance 48 h postelectroporation. Quantification was achieved by amplification of the 5′NTR of the replicon, while the YWHAZ cellular gene was used for normalization. At least three independent repetitions were performed; the error bars represent standard deviations. *, P < 0.02 (Student's t test). (D) SDS-PAGE autoradiography for immunoprecipitated firefly luciferase and NS5A from lysates 48 h postelectroporation of Huh7-Lunet cells electroporated with RNA from the pFK-i630-BglII-core+1/LucNS3-3′-JFH1-dg-based replicons. The arrow represents the bands for core/core+1-luc proteins. The figure represents the results of one of three independent experiments.
Our report provides evidence supporting the idea of expression of core+1/ARFP in JFH1-based replicons and shows for the first time expression of core+1/ARFP from a 2a subtype strain; all previous studies were limited to studying exclusively genotype 1 isolates. Specifically, we show the expression of two forms of core+1/ARFP, designated core+1/L and core+1/S, with different kinetics of expression during the replication of the JFH1 replicon. Core+1/S was the predominant core+1 form during the early stages of replication, while core+1/L expression increased later in the replication cycle. Conversely, the lack of core+1 expression in the context of the chimeric H77/JFH1 replicon suggests novel translation mechanisms. It should be taken into account that the two isoforms may differ in some of their functions. For example, different isoforms of the hepatitis D virus delta antigen play roles in different stages of the viral replication cycle: the small delta antigen is necessary for replication (36), while the large delta antigen is produced at later stages and is involved in packaging (37). Core+1/ARFP isoforms produced by frameshift have been observed previously in cell-free systems (7, 8). Our results strongly suggest that the core+1/ARFP/F isoform is not produced in the replicon system. Alternatively, our mutational analysis provides strong evidence that, in accordance with previous observations, codons 25/26 serve as translation initiator codons for core+1/ARFP/L, while codons 85/87 serve as translation initiator codons for core+1/S (12, 15).
In conclusion, the results of this study support the idea of expression of core+1 ORF in the context of the HCV replication, confirm the presence of alternative translation initiation mechanisms that are based on internal translation at codons 25/26 and 85/87, and unambiguously demonstrate the lack of frameshift at codons 8 to 11 which has long been speculated.
ACKNOWLEDGMENTS
This work was funded by the Hellenic Secretariat of Research, grant ARISTEIA-1560 (ESPA2007-2013). I.K.-L. and I.K. were financially supported by the Greek State Scholarships Foundation (IKY) and IKY-Siemens, respectively.
Also, we are grateful to Milton A. Typas for helpful discussions, Robert H. A. Coutts for critical reading of the manuscript, and Elina Aslanoglou for technical support.
REFERENCES
- 1.Hoofnagle JH. 2002. Course and outcome of hepatitis C. Hepatology 36(Suppl 1):S21–S29. doi: 10.1053/jhep.2002.36227. [DOI] [PubMed] [Google Scholar]
- 2.Houghton M, Abrignani S. 2005. Prospects for a vaccine against the hepatitis C virus. Nature 436:961–966. doi: 10.1038/nature04081. [DOI] [PubMed] [Google Scholar]
- 3.Brennan T, Shrank W. 13August2014, posting date New expensive treatments for hepatitis C infection. JAMA doi: 10.1001/jama.2014.8897. [DOI] [PubMed] [Google Scholar]
- 4.Poenisch M, Bartenschlager R. 2010. New insights into structure and replication of the hepatitis C virus and clinical implications. Semin Liver Dis 30:333–347. doi: 10.1055/s-0030-1267535. [DOI] [PubMed] [Google Scholar]
- 5.Moradpour D, Penin F, Rice CM. 2007. Replication of hepatitis C virus. Nat Rev Microbiol 5:453–463. doi: 10.1038/nrmicro1645. [DOI] [PubMed] [Google Scholar]
- 6.Pestova TV, Kolupaeva VG, Lomakin IB, Pilipenko EV, Shatsky IN, Agol VI, Hellen CU. 2001. Molecular mechanisms of translation initiation in eukaryotes. Proc Natl Acad Sci U S A 98:7029–7036. doi: 10.1073/pnas.111145798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Varaklioti A, Vassilaki N, Georgopoulou U, Mavromara P. 2002. Alternate translation occurs within the core coding region of the hepatitis C viral genome. J Biol Chem 277:17713–17721. doi: 10.1074/jbc.M201722200. [DOI] [PubMed] [Google Scholar]
- 8.Xu Z, Choi J, Yen TS, Lu W, Strohecker A, Govindarajan S, Chien D, Selby MJ, Ou J. 2001. Synthesis of a novel hepatitis C virus protein by ribosomal frameshift. EMBO J 20:3840–3848. doi: 10.1093/emboj/20.14.3840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Walewski JL, Keller TR, Stump DD, Branch AD. 2001. Evidence for a new hepatitis C virus antigen encoded in an overlapping reading frame. RNA 7:710–721. doi: 10.1017/S1355838201010111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Choi J, Xu Z, Ou JH. 2003. Triple decoding of hepatitis C virus RNA by programmed translational frameshifting. Mol Cell Biol 23:1489–1497. doi: 10.1128/MCB.23.5.1489-1497.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Vassilaki N, Kalliampakou KI, Mavromara P. 2008. Differences in the expression of the hepatitis C virus core+1 open reading frame between a nuclear and a cytoplasmic expression system. J Gen Virol 89:222–231. doi: 10.1099/vir.0.83260-0. [DOI] [PubMed] [Google Scholar]
- 12.Baril M, Brakier-Gingras L. 2005. Translation of the F protein of hepatitis C virus is initiated at a non-AUG codon in a +1 reading frame relative to the polyprotein. Nucleic Acids Res 33:1474–1486. doi: 10.1093/nar/gki292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Vassilaki N, Mavromara P. 2003. Two alternative translation mechanisms are responsible for the expression of the HCV ARFP/F/core+1 coding open reading frame. J Biol Chem 278:40503–40513. doi: 10.1074/jbc.M305504200. [DOI] [PubMed] [Google Scholar]
- 14.Vassilaki N, Boleti H, Mavromara P. 2008. Expression studies of the HCV-1a core+1 open reading frame in mammalian cells. Virus Res 133:123–135. doi: 10.1016/j.virusres.2007.10.019. [DOI] [PubMed] [Google Scholar]
- 15.Boumlic A, Vassilaki N, Dalagiorgou G, Kochlios E, Kakkanas A, Georgopoulou U, Markoulatos P, Orfanoudakis G, Mavromara P. 2011. Internal translation initiation stimulates expression of the ARF/core+1 open reading frame of HCV genotype 1b. Virus Res 155:213–220. doi: 10.1016/j.virusres.2010.10.007. [DOI] [PubMed] [Google Scholar]
- 16.Boulant S, Becchi M, Penin F, Lavergne JP. 2003. Unusual multiple recoding events leading to alternative forms of hepatitis C virus core protein from genotype 1b. J Biol Chem 278:45785–45792. doi: 10.1074/jbc.M307174200. [DOI] [PubMed] [Google Scholar]
- 17.Cohen M, Bachmatov L, Ben-Ari Z, Rotman Y, Tur-Kaspa R, Zemel R. 2007. Development of specific antibodies to an ARF protein in treated patients with chronic HCV infection. Dig Dis Sci 52:2427–2432. doi: 10.1007/s10620-006-9630-2. [DOI] [PubMed] [Google Scholar]
- 18.Bain C, Parroche P, Lavergne JP, Duverger B, Vieux C, Dubois V, Komurian-Pradel F, Trepo C, Gebuhrer L, Paranhos-Baccala G, Penin F, Inchauspe G. 2004. Memory T-cell-mediated immune responses specific to an alternative core protein in hepatitis C virus infection. J Virol 78:10460–10469. doi: 10.1128/JVI.78.19.10460-10469.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Komurian-Pradel F, Rajoharison A, Berland JL, Khouri V, Perret M, Van Roosmalen M, Pol S, Negro F, Paranhos-Baccalà G. 2004. Antigenic relevance of F protein in chronic hepatitis C virus infection. Hepatology 40:900–909. doi: 10.1002/hep.1840400420. [DOI] [PubMed] [Google Scholar]
- 20.Troesch M, Jalbert E, Canobio S, Boulassel MR, Routy JP, Bernard NF, Bruneau J, Lapointe N, Boucher M, Soudeyns H. 2005. Characterization of humoral and cell-mediated immune responses directed against hepatitis C virus F protein in subjects co-infected with hepatitis C virus and HIV-1. AIDS 19:775–784. doi: 10.1097/01.aids.0000168971.57681.6e. [DOI] [PubMed] [Google Scholar]
- 21.Chuang WC, Allain JP. 2008. Differential reactivity of putative genotype 2 hepatitis C virus F protein between chronic and recovered infections. J Gen Virol 89:1890–1900. doi: 10.1099/vir.0.83677-0. [DOI] [PubMed] [Google Scholar]
- 22.Rüster B, Zeuzem S, Roth WK. 1996. Hepatitis C virus sequences encoding truncated core proteins detected in a hepatocellular carcinoma. Biochem Biophys Res Commun 219:911–915. doi: 10.1006/bbrc.1996.0340. [DOI] [PubMed] [Google Scholar]
- 23.Yeh CT, Lu SC, Chu CM, Liaw YF. 1997. Molecular cloning of a defective hepatitis C virus genome from the ascitic fluid of a patient with hepatocellular carcinoma. J Gen Virol 78(Pt 11):2761–2770 http://vir.sgmjournals.org/content/78/11/2761.long. [DOI] [PubMed] [Google Scholar]
- 24.Yeh CT, Lo SY, Dai DI, Tang JH, Chu CM, Liaw YF. 2000. Amino acid substitutions in codons 9–11 of hepatitis C virus core protein lead to the synthesis of a short core protein product. J Gastroenterol Hepatol 15:182–191. doi: 10.1046/j.1440-1746.2000.02066.x. [DOI] [PubMed] [Google Scholar]
- 25.Hayashi J, Furusyo N, Ariyama I, Sawayama Y, Etoh Y, Kashiwagi S. 2000. A relationship between the evolution of hepatitis C virus variants, liver damage, and hepatocellular carcinoma in patients with hepatitis C viremia. J Infect Dis 181:1523–1527. doi: 10.1086/315431. [DOI] [PubMed] [Google Scholar]
- 26.Ogata S, Nagano-Fujii M, Ku Y, Yoon S, Hotta H. 2002. Comparative sequence analysis of the core protein and its frameshift product, the F protein, of hepatitis C virus subtype 1b strains obtained from patients with and without hepatocellular carcinoma. J Clin Microbiol 40:3625–3630. doi: 10.1128/JCM.40.10.3625-3630.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Alam SS, Nakamura T, Naganuma A, Nozaki A, Nouso K, Shimomura H, Kato N. 2002. Hepatitis C virus quasispecies in cancerous and noncancerous hepatic lesions: the core protein-encoding region. Acta Med Okayama 56:141–147. [DOI] [PubMed] [Google Scholar]
- 28.Vassilaki N, Friebe P, Meuleman P, Kallis S, Kaul A, Paranhos-Baccala G, Leroux-Roels G, Mavromara P, Bartenschlager R. 2008. Role of the hepatitis C virus core+1 open reading frame and core cis-acting RNA elements in viral RNA translation and replication. J Virol 82:11503–11515. doi: 10.1128/JVI.01640-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Vassilaki N, Boleti H, Mavromara P. 2007. Expression studies of the core+1 protein of the hepatitis C virus 1a in mammalian cells. The influence of the core protein and proteasomes on the intracellular levels of core+1. FEBS J 274:4057–4074. [DOI] [PubMed] [Google Scholar]
- 30.Wakita T, Pietschmann T, Kato T, Date T, Miyamoto M, Zhao Z, Murthy K, Habermann A, Krausslich HG, Mizokami M, Bartenschlager R, Liang TJ. 2005. Production of infectious hepatitis C virus in tissue culture from a cloned viral genome. Nat Med 11:791–796. doi: 10.1038/nm1268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kaul A, Woerz I, Meuleman P, Leroux-Roels G, Bartenschlager R. 2007. Cell culture adaptation of hepatitis C virus and in vivo viability of an adapted variant. J Virol 81:13168–13179. doi: 10.1128/JVI.01362-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kotta-Loizou I, Vassilaki N, Pissas G, Kakkanas A, Bakiri L, Bartenschlager R, Mavromara P. 2013. Hepatitis C virus core+1/ARF protein decreases hepcidin transcription through an AP1 binding site. J Gen Virol 94:1528–1534. doi: 10.1099/vir.0.050328-0. [DOI] [PubMed] [Google Scholar]
- 33.Ratinier M, Boulant S, Combet C, Targett-Adams P, McLauchlan J, Lavergne JP. 2008. Transcriptional slippage prompts recoding in alternate reading frames in the hepatitis C virus (HCV) core sequence from strain HCV-1. J Gen Virol 89:1569–1578. doi: 10.1099/vir.0.83614-0. [DOI] [PubMed] [Google Scholar]
- 34.van den Hoff MJ, Christoffels VM, Labruyere WT, Moorman AF, Lamers WH. 1995. Electrotransfection with “intracellular” buffer. Methods Mol Biol 48:185–197. [DOI] [PubMed] [Google Scholar]
- 35.Hashemi A, Roohvand F, Ghahremani MH, Aghasadeghi MR, Vahabpour R, Motevau F, Memarnejadian A. 2012. Optimization of transfection methods for Huh-7 and Vero cells: comparative study. Tsitol Genet 46:19–27. doi: 10.3103/S0095452712060035. [DOI] [PubMed] [Google Scholar]
- 36.Chao M, Hsieh SY, Taylor J. 1990. Role of two forms of hepatitis delta virus antigen: evidence for a mechanism of self-limiting genome replication. J Virol 64:5066–5069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Luo GX, Chao M, Hsieh SY, Sureau C, Nishikura K, Taylor J. 1990. A specific base transition occurs on replicating hepatitis delta virus RNA. J Virol 64:1021–1027. [DOI] [PMC free article] [PubMed] [Google Scholar]