ABSTRACT
Uukuniemi virus (UUKV) is a tick-borne member of the Phlebovirus genus (family Bunyaviridae) and has been widely used as a safe laboratory model to study aspects of bunyavirus replication. Recently, a number of new tick-borne phleboviruses have been discovered, some of which, like severe fever with thrombocytopenia syndrome virus and Heartland virus, are highly pathogenic in humans. UUKV could now serve as a useful comparator to understand the molecular basis for the different pathogenicities of these related viruses. We established a reverse-genetics system to recover UUKV entirely from cDNA clones. We generated two recombinant viruses, one in which the nonstructural protein NSs open reading frame was deleted from the S segment and one in which the NSs gene was replaced with green fluorescent protein (GFP), allowing convenient visualization of viral infection. We show that the UUKV NSs protein acts as a weak interferon antagonist in human cells but that it is unable to completely counteract the interferon response, which could serve as an explanation for its inability to cause disease in humans.
IMPORTANCE Uukuniemi virus (UUKV) is a tick-borne phlebovirus that is apathogenic for humans and has been used as a convenient model to investigate aspects of phlebovirus replication. Recently, new tick-borne phleboviruses have emerged, such as severe fever with thrombocytopenia syndrome virus in China and Heartland virus in the United States, that are highly pathogenic, and UUKV will now serve as a comparator to aid in the understanding of the molecular basis for the virulence of these new viruses. To help such investigations, we have developed a reverse-genetics system for UUKV that permits manipulation of the viral genome. We generated viruses lacking the nonstructural protein NSs and show that UUKV NSs is a weak interferon antagonist. In addition, we created a virus that expresses GFP and thus allows convenient monitoring of virus replication. These new tools represent a significant advance in the study of tick-borne phleboviruses.
INTRODUCTION
The Bunyaviridae are the largest grouping of RNA viruses, comprising >350 members. The family includes a wide range of enveloped, single-stranded RNA viruses with tripartite genomes of negative-sense or ambisense polarity. The Phlebovirus genus contains 70 viruses divided into two groups: the sandfly fever group, transmitted mainly by phlebotomines and mosquitoes, and the Uukuniemi-like virus group, transmitted by ticks. Uukuniemi virus (UUKV) itself was originally isolated in 1964 from an Ixodes ricinus tick in Finland (1). UUKV has served as a prototype tick-borne phlebovirus for many years and has aided in advances in bunyavirus structural and architectural determination (2–4), bunyavirus glycoprotein studies (5–9), and studies of bunyavirus entry into mammalian cells (10–12).
Like other phleboviruses, the three UUKV genome segments, designated small (S), medium (M), and large (L), are found in the form of ribonucleoprotein (RNP) complexes in association with the nucleocapsid (N) protein and the RNA-dependent RNA polymerase (or L protein). The L segment codes for the L protein, the M segment encodes the two envelope glycoproteins Gn and Gc, and the S segment encodes the N protein in the negative-sense orientation and the nonstructural NSs protein in the positive-sense orientation in the genomic RNA, employing an ambisense coding strategy (13, 14). Notably, UUKV does not encode a nonstructural NSm protein on the M segment, and this seems to be a distinguishing feature between the tick-borne and dipteran-borne phleboviruses (15).
Recently, a number of new tick-borne phleboviruses have been reported (16), including severe fever with thrombocytopenia syndrome virus (SFTSV) and Heartland virus (HRTV), which can cause fatal disease in humans. Originally described in the Henan and Hubei provinces in China (17), SFTSV has now been found to be more widespread in China (18) and has also been isolated in Japan and South Korea, suggesting a more widespread distribution (19, 20). HRTV isolates have so far been restricted to Missouri and Tennessee in the United States, with 10 cases and 2 fatalities (21–23), but one serological survey suggested a widespread occurrence of antigenically similar viruses in farm animals throughout Minnesota (24). These novel viruses also lack evidence for an NSm gene.
Antibodies to UUKV (or a very similar virus[es]) have been detected in humans, birds, rodents, and cows (25, 26), but there is no evidence of disease in these species (27). Thus, UUKV would be a useful comparator to include in studies on the molecular basis of the pathogenesis of STSFV and HRTV in humans. Such investigations will be aided by the availability of reverse-genetics systems that allow specific manipulation of the viral genome, and we have recently established such a system for SFTSV (28). Here, we describe a reverse-genetics system for the recovery of infectious UUKV entirely from cDNA copies of the genome. We created UUKV mutants where the NSs open reading frame (ORF) is deleted in the S segment and where the NSs ORF is replaced by that of enhanced green fluorescent protein (eGFP), allowing rapid visualization of infection. We also show that UUKV NSs serves as a weak interferon (IFN) antagonist in human cells.
MATERIALS AND METHODS
Cells and virus.
BSR cells were grown in Glasgow's minimal essential medium (GMEM) supplemented with 10% tryptose phosphate broth (TPB) and 10% fetal calf serum (FCS). BSR-T7/5 cells, which express T7 RNA polymerase (29), were grown in a similar medium supplemented with 1 mg/ml G418 (Promega). BHK-21 cells were grown in GMEM supplemented with 10% TPB and 10% newborn calf serum (NCS). A549 cells were grown in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% FCS, and A549/BVDV-Npro (30) and A549/PIV5-V (31) cells were grown in a similar medium supplemented with 2 μg/ml puromycin (Melford Laboratories Ltd.). Cells were maintained at 37°C with 5% CO2. The UUKV strain used in this study is derived from the prototype tick isolate S-23 (1), which was plaque purified in chicken embryo fibroblasts (32). The virus was initially grown in BHK-21 cells, and working stocks for use in this paper were then grown in BSR cells.
Plasmids.
Full-length cDNA clones of the S, M, and L segments of UUKV were amplified by PCR (primers are listed in Table 1) and subcloned into plasmid TVT7R(0,0) (33), using approaches described previously (34), so that T7 transcripts would be in the antigenome sense. The plasmids were named pT7UUKS(+), pT7UUKM(+), and pT7UUKL(+). pT7UUKSdelNSs was generated from pT7UUKS(+) by excision PCR to delete nucleotides (nt) 874 to 1695, thus removing the NSs ORF. pT7UUKSdelNSsGFP contained the enhanced green fluorescent protein gene exactly in place of the NSs sequence. The L and N ORFs were amplified from the respective full-length plasmids and subcloned into the expression vector pTM1 to generate pTMUUKL and pTMUUKN. Cloning was done by employing the In-Fusion HD cloning kit (Clontech), and the primers used are shown in Table 1.
TABLE 1.
Oligonucleotide primers used in this study
| Primer | Sequence (5′→3′)a | Purpose |
|---|---|---|
| ST7_Fwd | TACGACTCACTATAGacacaaagacctcca | Amplification of full-length segments for insertion into plasmid TVT7R(0,0) in the antigenome sense |
| ST7_Rev | ATGCCATGCCGACCCacacaaagaccctcc | |
| MT7_Fwd | TACGACTCACTATAGacacaaagacggcta | |
| MT7_Rev | ATGCCATGCCGACCCacacaaagacacggc | |
| LT7_Fwd | TACGACTCACTATAGacacaaagacgccaag | |
| LT7_Rev | ATGCCATGCCGACCCacacaaagtccgcca | |
| SdelNSs:eGFPT7exi1 | tttgggccgaagcccttttagagtcc | Amplification of full-length pT7UUKS(+) lacking the NSs sequence for eGFP insertion |
| SdelNSs:eGFPT7exi2 | gcttaatgttggagggtctttgtgtGG | |
| GFPexi1 | ggcttcggcccaaattacttgtacagc | Amplification of eGFP for insertion into pTUUKVS(+) lacking NSs |
| GFPexi2 | ccaacattaagcatggtgagcaagg | |
| SdelNSsexi1 | tttgggccgaagcccttttagagtcc | Amplification of full-length pT7UUKS(+) to delete NSs, followed by religation |
| SdelNSsexi2 | cggcccaaagcttaatgttggaggg | |
| Sfwd1 | acacaaagaccctccaacattaagc | RT-PCR amplification of the full-length S segment |
| Srev1 | acacaaagacctccaacttagc | |
| NpTM1_Fwd | GAAAAACACGATAATACCatggctatgccggag | Amplification of N and L ORFs for insertion into expression plasmid pTM1 |
| NpTM1_Rev | ATTAGGCCTCTCGAGtcagatcaatgatct | |
| LpTM1_Fwd | GAAAAACACGATAATACCatgcttttagcgatt | |
| LpTM1_Rev | ATTAGGCCTCTCGAGtcaaccaaacatgtc |
Plasmid sequences are indicated in uppercase type, and UUKV sequences are indicated in boldface type. For the primers used for amplification and religation of pT7UUKS(+) to delete NSs, the sticky ends are underlined.
Generation of recombinant UUKV from cDNA.
Subconfluent BSR-T7/5 cells (2 × 105 cells in a 35-mm dish) were transfected by using 5 μl Lipofectamine-2000 (Invitrogen) in 500 μl Opti-MEM (Invitrogen), with 1 μg each of pT7UUKL(+) and pT7UUKM(+) and 1 μg of either pT7UUKS(+), pT7UUKSdelNSs, or pT7UUKSdelNSsGFP. After 7 days of incubation at 37°C, the supernatants were collected, clarified, and used for infection of BSR cells to prepare working stocks. The rescue supernatant from BSR-T7/5 cells was also titrated by immunofocus assays to determine virus yields from transfection. Recombinant viruses were designated rUUKV, rUUKVdelNSs, and rUUKVdelNSsGFP, as appropriate.
RT-PCR analysis.
Viral RNA was extracted from BSR cells infected with wild-type UUKV (wtUUKV), rUUKV, rUUKdelNSs, or rUUKdelNSsGFP by using TRIzol (Life Technologies) according to the manufacturer's instructions. Reverse transcription-PCR (RT-PCR) of the full S segment was performed by using GoScript reverse transcriptase (Promega) and either KOD polymerase (Novagen) or GoTaq polymerase (Promega), using the primers listed in Table 1.
Immunofocus assays.
Immunofocus assays were performed on confluent monolayers of BSR or BHK-21 cells. Serial dilutions of virus, made in phosphate-buffered saline (PBS) containing 2% NCS or FCS as appropriate, were added to the cells and incubated for 1 h at 37°C, followed by the addition of a GMEM overlay supplemented with 2% NCS or FCS and 0.6% Avicel (FMC BioPolymer). The cells were incubated at 37°C for 6 days and then fixed for 1 h with 8% formaldehyde in PBS. After washing with PBS, the cells were permeabilized by treatment for 30 min with PBS containing 0.5% Triton X-100 and 20 mM glycine. The cells were incubated for 1 h with an anti-UUKV N monoclonal antibody (8B11A3) (35) in PBS containing 5% dried milk and 0.1% Tween 20, washed, and incubated for 1 h with anti-mouse horseradish peroxidase (HRP)-linked antibody (catalogue no. A4416; Sigma). Foci were detected by using the TrueBlue peroxidase substrate (KPL).
Western blotting.
BSR cells were infected with recombinant viruses at a multiplicity of infection (MOI) of 5 focus-forming units (FFU) per cell, and cell lysates were prepared at 18 h postinfection (p.i.) by the addition of lysis buffer (20% glycerol, 100 mM Tris-HCl [pH 6.8], 4% SDS, 200 mM dithiothreitol [DTT], 0.2% bromophenol blue). Proteins were separated on an SDS–4-to-12% gradient polyacrylamide gel (Nu-PAGE; Life Technologies) and transferred onto a Hybond ECL nitrocellulose membrane (GE Healthcare Life Sciences). The membrane was blocked by incubation in PBS containing 5% dry milk and 0.1% Tween 20 for 1 h, followed by incubation with anti-UUKV N monoclonal antibody 8B11A3 (35), a rabbit polyclonal antibody against UUKV NSs (A. K. Överby, unpublished data), and a mouse monoclonal antibody against tubulin (catalogue no. T5168; Sigma) for 1.5 h. The membrane was washed and incubated for 1 h with HRP-coupled secondary anti-rabbit (catalogue no. A0545; Sigma) or anti-mouse antibodies. Proteins were detected by using the SuperSignal WestPico chemiluminescent substrate (Pierce), followed by exposure on a Bio-Rad ChemiDoc imager.
Interferon assays.
A biological assay for interferon (IFN) production was performed according to a previously described protocol (34). Briefly, IFN-competent A549 cells were infected with recombinant viruses at an MOI of 3 FFU/cell, or with control Bunyamwera viruses at an MOI of 3 PFU/cell, and incubated at 37°C. The supernatant was collected at 24 h p.i., and any released virus was inactivated by exposure to UV irradiation (8 W; 254 nm at a distance of 2 cm for 4 min with occasional shaking). Twofold serial dilutions of the UV-inactivated supernatant were used to pretreat IFN-incompetent A549/BVDV-Npro cells for 24 h, after which encephalomyocarditis virus (EMCV) (0.03 PFU/cell) was added. Four days later, the cells were stained with crystal violet to monitor EMCV-induced destruction of the monolayers. Relative IFN units (RIU) were calculated as 2N, where N is the number of 2-fold dilutions giving protection of A549/BVDV-Npro cells.
RESULTS
Rescue of wild-type UUKV.
Full-length cDNAs to the genome segments of UUKV were cloned into a T7 transcription plasmid, with an extra G residue at the 5′ end for efficient transcription, such that the antigenome sense RNA would be transcribed by T7 RNA polymerase. The nucleotide sequences of the clones were determined and confirmed to match those in the GenBank database (GenBank accession no. M33551.1, M17417.1, and D10759.1), except for a silent mutation introduced as a genetic marker in the S segment at position 963. This creates an XhoI restriction site in the S segment cDNA. BSR-T7/5 cells (that stably express T7 RNA polymerase) were transfected with a mixture of pT7UUKS(+), pT7UUKM(+), and pT7UUKL(+), and the supernatant was harvested at 7 days posttransfection. The titer of infectious virus was determined by an immunofocus assay and averaged 4.9 × 104 (±1.9 × 104) FFU/ml in 4 experiments. The recombinant virus was designated rUUKV. Working stocks of the virus were prepared by infecting BSR cells with the supernatant from the transfected cells, and titers of 108 FFU/ml were achieved. We also rescued virus by using a 5-plasmid system (36), by additionally transfecting 1 μg each of pTMUUKL and pTMUUKN, and titers of rescued virus averaged 2.9 × 104 (±2.7 × 104) FFU/ml in 4 experiments. As the 5-plasmid rescue system offered no increase in efficiency, the 3-plasmid rescue system is routinely used in our laboratory.
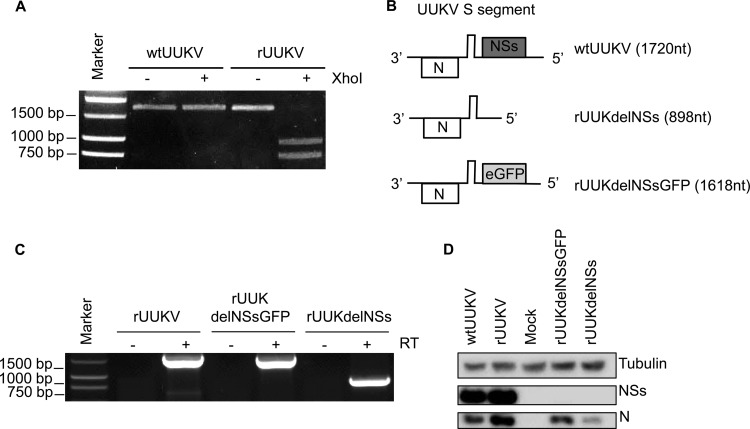
To confirm that rUUKV was indeed generated from cDNA plasmids, total RNA was extracted from BSR cells infected with either authentic UUKV (here referred to as wtUUKV) or rUUKV, and the full-length S segment was amplified by RT-PCR. Digestion of the amplified DNA derived from rUUKV-infected cells with XhoI resulted in two products of the expected sizes, 963 bp and 757 bp, whereas amplified DNA derived from wtUUKV-infected cells was not digested (Fig. 1A).
FIG 1.
Characterization of recombinant viruses. (A) RT-PCR products of the full-length S segment of wtUUKV and rUUKV and digestion with XhoI. Digestion was seen only with the amplified DNA derived from rUUKV, where a silent mutation creating an XhoI site was introduced at position 963. (B) Schematic diagrams of wild-type and mutant S segments lacking the NSs gene. (C) RT-PCR products of full-length S segments from rUUKV, rUUKdelNSsGFP, and rUUKdelNSs showing the size differences corresponding to the structures shown in panel B. (D) Western blot detection of UUKV N and NSs proteins in BSR cells infected with wtUUKV, rUUKV, rUUKdelNSsGFP, or rUUKdelNSs. Cells were infected at an MOI of 5 or mock infected, and cell lysates were prepared at 18 h p.i. Western blots were probed with antitubulin (dilution, 1:5,000), anti-UUKV NSs (dilution, 1:1,000), and anti-UUKV N (dilution, 1:100) antibodies.
Generation of mutant UUKV.
We next used the rescue system to generate two mutant recombinant viruses that lack the NSs open reading frame. First, the NSs ORF was deleted from pT7UUKS(+) such that the intergenic region in the S segment is immediately followed by the genomic 5′ untranslated region (UTR). This results in a short genome segment of 898 nt compared to the 1,720-nt wild-type S segment. Second, the NSs ORF was replaced by the sequences for GFP, in an ambisense orientation, resulting in a GFP-expressing S segment of 1,618 nt. Schematics of the resulting RNAs are shown in Fig. 1B.
The modified S segment cDNA clones were used in place of pT7UUKS(+) in the rescue protocol by transfecting BSR-T7/5 cells as outlined above, and mutant recombinant viruses, named rUUKdelNSs and rUUKdelNSsGFP, were successfully recovered. The titers of virus in the supernatants from transfected cells averaged 3.75 × 104 (±1.2 × 104) FFU/ml for rUUKdelNSs in 4 experiments and 1.0 × 104 (±5.2 × 103) FFU/ml for rUUKdelNSsGFP in 4 experiments. Again, working stocks of the recombinant virus were prepared in BSR cells, and titers of ∼107 FFU/ml were achieved. The sizes of the S segments of the viruses were compared by amplification of infected-cell RNA by specific RT-PCR, as shown in Fig. 1C. Nucleotide sequencing confirmed that no additional mutations were present in these genomes (data not shown). To validate the lack of NSs expression at the protein level, BSR cells were infected with wtUUKV or each of the recombinant viruses (MOI of 5) or were mock infected, and cell lysates were prepared at 18 h p.i. Western blots were probed for N and NSs proteins (Fig. 1D). While N and NSs were detected in BSR cells infected with wtUUKV and rUUKV, only N was detected in cells infected with the NSs-deleted mutants. The signal for N protein in rUUKdelNSs-infected cells was weaker than that in rUUKdelNSsGFP-infected cells.
Phenotypic characterization of rescued viruses in cell culture.
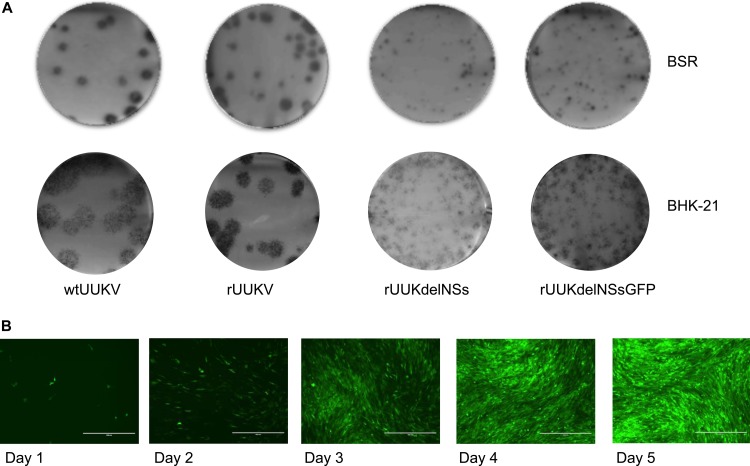
Recombinant viruses were assayed in both BSR and BHK-21 cells by immunofocus assays (Fig. 2A and B). wtUUKV and rUUKV foci were of a similar size (4 to 6 mm in diameter) within one cell type but interestingly were larger in BHK-21 than in BSR cells, considering that the latter are a subclone of BHK-21 cells (37). The NSs-deleted viruses gave smaller foci (<2 mm in diameter) than did the equivalent wild-type viruses.
FIG 2.
Phenotypic characterization of rescued viruses in cell culture. (A) Focus assay for wtUUKV, rUUKV, rUUKdelNSs, or rUUKdelNSsGFP on BSR cells. Foci were stained with anti-UUKV N antibody after 6 days of incubation at 37°C. (B) Focus assay for wtUUKV, rUUKV, rUUKdelNSs, or rUUKdelNSsGFP on BHK-21 cells. Foci were stained with anti-UUKV N antibody after 6 days of incubation at 37°C. (C) Course of infection of the UUKV rUUKdelNSsGFP in BHK-21 cells at 37°C over 5 days. Cells were infected at an MOI of 0.01 FFU/cell.
Figure 2C shows a time course of GFP expression in BHK-21 cells infected with rUUKdelNSsGFP at a very low MOI (0.01 FFU/cell). A few cells were seen to express GFP after 24 h, and the number increased as the virus spread through the monolayer. By 5 days p.i., all the cells were infected. This indicates that rUUKdelNSsGFP will be a useful tool to visualize the replication cycle of UUKV.
Growth properties of recombinant viruses.
The growth properties of wild-type and recombinant viruses in monolayers of BHK-21 cells infected at a low MOI (0.1 FFU/cell) at 37°C were compared. The supernatants of duplicate cultures were harvested at the indicated time points postinfection, and virus titers were determined by an immunofocus assay. Lysates of the cell monolayers were prepared for Western blotting at the same time points. rUUKV and wtUUKV exhibited similar growth properties in BHK cells, reaching titers of 109 FFU/ml (Fig. 3A). The growth kinetics of the mutant viruses lacking NSs were slower, but interestingly, the virus expressing GFP grew better (rUUKdelNSsGFP) (peak titer of ∼108 FFU/ml) than did the virus with no sequences inserted at the NSs locus (rUUKdelNSsGFP) (peak titer of ∼107 FFU/ml). Therefore, NSs is not essential for virus growth in BHK-21 cells but plays a contributory role for efficient replication.
FIG 3.
Growth properties of wtUUKV, rUUKV, rUUKdelNSs, and rUUKdelNSsGFP in BHK-21 cells. Cells were infected at an MOI of 0.1 FFU/ml, and cell culture medium was harvested or cell lysates were prepared at the time points indicated. (A) Growth curves. Infections were carried out in duplicate, and error bars represent the standard errors of the means for a representative experiment. (B) Viral protein synthesis. Cell lysates from the growth curve samples were harvested, and Western blots were probed with antitubulin, anti-UUKV NSs, and anti-UUKV N antibodies, as indicated.
Analysis of viral protein expression (Fig. 3B) showed that at this MOI and with the dilution of antibodies used, NSs protein was first detected in wtUUKV- and rUUKV-infected cells at 24 h p.i. and accumulated thereafter, whereas N was not detected until 48 h p.i. No NSs protein was detected in mutant virus-infected cells, as expected. N was detected earlier in rUUKdelNSsGFP-infected cells than in rUUKdelNSs-infected cells, reflecting the difference in virus growth between these viruses, as shown in Fig. 3A.
UUKV NSs has weak IFN-antagonistic activity.
We investigated the growth of the viruses in an IFN-competent cell line, A549 (a human lung cell line), and a derivative cell line, A549/PIV5-V (A549/V), which expresses the V protein of parainfluenza virus type 5 to block IFN signaling (31). Both wtUUKV and rUUKV grew to titers of ∼106 FFU/ml by 72 h p.i. in A549 cells, but titers were ∼10-fold higher in A549/V cells (Fig. 4). The NSs-deleted viruses achieved titers of ∼105 FFU/ml by 72 h p.i. in A549 cells, and titers were slightly higher in A549/V cells. As noted above for BHK-21 cells, rUUKdelNSsGFP grew better than rUUKdelNSs. These results indicate that although UUKV was able to replicate in an IFN-competent cell line and hence overcome the IFN response, replication was enhanced in A549V cells, where the IFN response is lacking.
FIG 4.
Growth properties of wtUUKV, rUUKV, rUUKdelNSs, and rUUKdelNSsGFP in A549 and A549/V cells. A549 and A549/V cells were infected at an MOI of 2 FFU/cell. The medium was harvested at 72 h p.i. and used for titrations. Infections were done in triplicate, and error bars indicate the standard errors of the means.
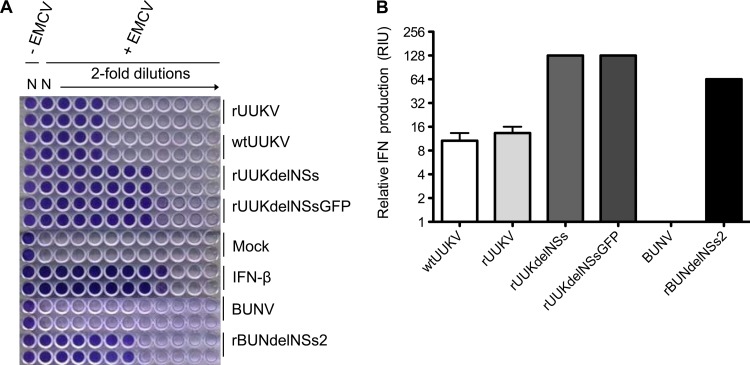
We next carried out a biological assay to measure the production of IFN in the supernatant of virus-infected cells. Briefly, supernatants from A549 cells infected with wtUUKV, rUUKV, rUUKdelNS, or rUUKdelNSsGFP were collected at 24 h p.i. As controls, cells were infected with wild-type Bunyamwera virus (BUNV) or rBUNdelNSs2, a recombinant virus lacking its NSs protein (38), as it is well known that BUNV NSs acts as a strong IFN antagonist (39, 40). Serial dilutions of UV-inactivated supernatants were used to treat IFN-responsive A549/BVDV-Npro cells (41) for 24 h, and the cells were then infected with IFN-sensitive encephalomyocarditis virus (EMCV). (A549/BVDV-Npro cells express the bovine viral diarrhea virus IFN antagonist N-Pro and thus do not produce IFN but remain responsive to exogenously applied IFN [30, 41].) Figure 5A shows that medium from mock-infected or BUNV-infected cells did not confer protection from EMCV infection, indicating that no IFN was present. In contrast, A549/BVDV-Npro cells treated with medium from rBUNdelNSs2-infected cells were protected, similarly to cells treated with recombinant IFN-β. Medium from cells infected with rUUKV or wtUUKV also afforded a degree of protection from EMCV infection, and even more protection was given by medium from cells infected with the recombinant UUKV lacking NSs. The results are quantified in Fig. 5B in terms of relative IFN production from the initially infected A549 cells. These results indicate that unlike BUNV NSs, which is able to fully antagonize the IFN response in A549 cells, UUKV NSs acts as a weak IFN antagonist, as the deletion of NSs results in a 10-fold increase in IFN production, but is unable to fully ablate the IFN response, as infection with wtUUKV and rUUKV also induced IFN production.
FIG 5.
Biological interferon assay. A549 cells were infected with wtUUKV, rUUKV, rUUKdelNSs, rUUKdelNSsGFP, BUNV, or BUNdelNSs2 at an MOI of 3 or mock infected. Media were harvested at 24 h p.i. Serial dilutions of the UV-inactivated supernatant, or dilutions of IFN-β, were used to pretreat A549-Npro cells before infection with EMCV. Cells were stained with crystal violet at 4 days p.i. (A) Assay plates. (B) Relative IFN units (RIU) produced, calculated as RIU = 2N, where N represents the number of wells that are protected from EMCV infection. The mean RIU value was calculated from data from triplicate experiments. Error bars indicate the standard errors of the means.
DISCUSSION
UUKV has served as a model phlebovirus in many aspects of bunyavirus research, as described in the introduction. In addition, minigenome and virus-like particle assays, based on cDNAs cloned into RNA polymerase I vectors, have been described (42, 43). In this paper, we report the successful rescue of UUKV entirely from cloned cDNA copies using our favored T7 RNA polymerase-based approach (36). Furthermore, we were also able to recover recombinant viruses lacking the NSs gene. We show that rUUKV exhibits properties similar to those of the authentic wild-type UUKV, with indistinguishable focus sizes and morphologies, similar growth kinetics in BHK-21 cells, and similar interferon responses in A549 cells.
The growth of the mutant viruses was slower than that of the wild-type virus in BHK-21 cells, and the deletion of NSs from the UUKV genome resulted in a significantly smaller focus size (Fig. 2A and B). This attenuation of growth due to the lack of NSs appears separate from the role of NSs in IFN antagonism (see below), as BHK-21 cells do not produce IFN-α/β (44). For other bunyavirus-cell culture combinations, the lack of NSs expression also results in a degree of growth attenuation independent of the IFN status of the cell, such as BUNV (39), Rift Valley fever virus (RVFV) (45), and Schmallenberg virus (SBV) (34, 46), but interestingly not for La Crosse virus in Vero cells (47). This presumably reflects some other role of NSs, perhaps involving an interaction with cellular partners, for efficient virus replication.
Another observation was that the mutant expressing GFP in place of NSs grew slightly better than did the NSs-deleted virus rUUKdelNSs. At face value, it seems unlikely that GFP itself aids virus replication. It is possible that for rUUKdelNSs, where the S segment intergenic region directly abuts the 5′ UTR (Fig. 1B), antigenome RNA synthesis is impaired. In contrast, the structure of the rUUKdelNSGFP S segment more closely resembles that of the authentic virus, and antigenome synthesis is not compromised. It would be of interest to insert other sequences in place of NSs and determine the growth properties of such recombinant viruses and also to examine RNA synthesis directly; these experiments are in progress. As it stands, the rescue of rUUKdelNSsGFP allows rapid visualization and easy monitoring of viral infection (Fig. 2C).
No IFN induction can be detected in cells following infection with RVFV or SFTSV (48, 49), and IFN is not detected in sera from SFTSV-infected patients (50). In contrast, two other phleboviruses, Punta Toro virus (PTV) and Toscana virus (TOSV), induce various levels of IFN in a strain-dependent manner (51, 52), which has been correlated with the relatively low pathogenicity of the latter viruses in humans compared to that of either RVFV or SFTSV. Our observations indicate that UUKV infection also induces IFN (Fig. 5). When expressed separately from their cognate viruses, both PTV and TOSV NSs proteins act as reasonably potent IFN antagonists (53, 54), as also observed for the NSs protein of sandfly fever Sicilian virus (53, 55). We observed a 10-fold increase in IFN production when A549 cells were infected with the NSs-deleted viruses, suggesting that UUKV NSs also plays a role in antagonizing the host cell IFN response. We also noted a 10-fold increase in the yield of virus from A549/V cells, where IFN signaling is blocked (Fig. 4). Therefore, we conclude that UUKV NSs can act as a weak IFN antagonist in human cells.
RVFV NSs blocks IFN production directly at the transcriptional level, aided because the protein localizes to the nucleus (56). As UUKV NSs localizes in the cytoplasm (57), like other phlebovirus NSs proteins, its mode of antagonizing the IFN response is expected to be different from that of RVFV. TOSV NSs inhibits the dimerization of IFN regulatory factor 3 (IRF-3), hence preventing its localization to the nucleus and the subsequent induction of the IFN-β promoter (52), and this is most likely because the protein inhibits the induction of type I IFN by targeting retinoic acid-inducible gene I (RIG-I) for proteasomal degradation (54). In SFTSV-infected cells, NSs forms cytoplasmic structures termed viroplasm (58), which are believed to redistribute RIG-I signaling components in order to suppress IFN responses (49, 59, 60). Studies to determine whether UUKV NSs uses a mechanism similar to those of TOSV or SFTSV are in progress.
To conclude, we have developed a T7-driven reverse-genetics system for UUKV that enabled the rescue of wild-type virus and the generation of NSs-lacking mutants. In addition, we have shown that NSs is not essential for virus replication but that its absence results in a degree of growth attenuation in cell culture. Recently, we described a reverse-genetics system for pathogenic SFTSV (28). The developed reverse-genetics systems will be useful tools to elucidate the molecular determinants of virulence of the more pathogenic tick-borne phleboviruses. Armed with these two systems, we can now test the hypothesis that pathogenicity in humans is associated with the IFN antagonist ability of the NSs proteins. For instance, chimeric viruses carrying different NSs proteins could be created, under appropriate containment, and their ability to replicate in human cells could be tested. In addition, the potential of reassortment between these tick-borne viruses could be tested by exploiting reverse-genetics systems, since there is evidence for a high degree of reassortment among the dipteran-transmitted Candiru antigenic complex of phleboviruses (61).
ACKNOWLEDGMENTS
We thank Ben Brennan, Stephen Welch, and Klaus Conzelmann for helpful advice and discussions.
This work was funded by the Wellcome Trust. A.K.Ö. is funded by the Laboratory for Molecular Medicine Sweden (MIMS), and R.M.E. is a Wellcome Trust senior investigator.
REFERENCES
- 1.Oker-Blom N, Salminen A, Brummer-Korvenkontio M, Kaeaeriaeinen L, Weckstroem P. 1964. Isolation of some viruses other than typical tick-borne encephalitis viruses from Ixodes ricinus ticks in Finland. Ann Med Exp Biol Fenn 42:109–112. [PubMed] [Google Scholar]
- 2.Overby AK, Pettersson RF, Grunewald K, Huiskonen JT. 2008. Insights into bunyavirus architecture from electron cryotomography of Uukuniemi virus. Proc Natl Acad Sci U S A 105:2375–2379. doi: 10.1073/pnas.0708738105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.von Bonsdorff CH, Pettersson R. 1975. Surface structure of Uukuniemi virus. J Virol 16:1296–1307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pettersson RF. 1975. The structure of Uukuniemi virus, a proposed member of the bunyaviruses. Med Biol 53:418–424. [PubMed] [Google Scholar]
- 5.Overby AK, Popov VL, Pettersson RF, Neve EP. 2007. The cytoplasmic tails of Uukuniemi virus (Bunyaviridae) G(N) and G(C) glycoproteins are important for intracellular targeting and the budding of virus-like particles. J Virol 81:11381–11391. doi: 10.1128/JVI.00767-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Melin L, Persson R, Andersson A, Bergstrom A, Ronnholm R, Pettersson RF. 1995. The membrane glycoprotein G1 of Uukuniemi virus contains a signal for localization to the Golgi complex. Virus Res 36:49–66. doi: 10.1016/0168-1702(95)00006-C. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Andersson AM, Pettersson RF. 1998. Targeting of a short peptide derived from the cytoplasmic tail of the G1 membrane glycoprotein of Uukuniemi virus (Bunyaviridae) to the Golgi complex. J Virol 72:9585–9596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Andersson AM, Melin L, Persson R, Raschperger E, Wikstrom L, Pettersson RF. 1997. Processing and membrane topology of the spike proteins G1 and G2 of Uukuniemi virus. J Virol 71:218–225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Crispin M, Harvey DJ, Bitto D, Halldorsson S, Bonomelli C, Edgeworth M, Scrivens JH, Huiskonen JT, Bowden TA. 2014. Uukuniemi phlebovirus assembly and secretion leave a functional imprint on the virion glycome. J Virol 88:10244–10251. doi: 10.1128/JVI.01662-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lozach PY, Mancini R, Bitto D, Meier R, Oestereich L, Overby AK, Pettersson RF, Helenius A. 2010. Entry of bunyaviruses into mammalian cells. Cell Host Microbe 7:488–499. doi: 10.1016/j.chom.2010.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lozach PY, Kuhbacher A, Meier R, Mancini R, Bitto D, Bouloy M, Helenius A. 2011. DC-SIGN as a receptor for phleboviruses. Cell Host Microbe 10:75–88. doi: 10.1016/j.chom.2011.06.007. [DOI] [PubMed] [Google Scholar]
- 12.Meier R, Franceschini A, Horvath P, Tetard M, Mancini R, von Mering C, Helenius A, Lozach PY. 2014. Genome-wide small interfering RNA screens reveal VAMP3 as a novel host factor required for Uukuniemi virus late penetration. J Virol 88:8565–8578. doi: 10.1128/JVI.00388-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Simons JF, Hellman U, Pettersson RF. 1990. Uukuniemi virus S RNA segment: ambisense coding strategy, packaging of complementary strands into virions, and homology to members of the genus Phlebovirus. J Virol 64:247–255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bouloy M. 2011. Molecular biology of phleboviruses, p 95–128 InPlyusnin A, Elliott RM (ed), Bunyaviridae: molecular and cellular biology. Caister Academic Press, Norfolk, United Kingdom. [Google Scholar]
- 15.Palacios G, Savji N, Travassos da Rosa A, Guzman H, Yu X, Desai A, Rosen GE, Hutchison S, Lipkin WI, Tesh R. 2013. Characterization of the Uukuniemi virus group (Phlebovirus: Bunyaviridae): evidence for seven distinct species. J Virol 87:3187–3195. doi: 10.1128/JVI.02719-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Matsuno K, Weisend C, Kajihara M, Matysiak C, Williamson BN, Simuunza M, Mweene AS, Takada A, Tesh RB, Ebihara H. 2015. Comprehensive molecular detection of tick-borne phleboviruses leads to the retrospective identification of taxonomically unassigned bunyaviruses and the discovery of a novel member of the genus Phlebovirus. J Virol 89:594–604. doi: 10.1128/JVI.02704-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yu XJ, Liang MF, Zhang SY, Liu Y, Li JD, Sun YL, Zhang L, Zhang QF, Popov VL, Li C, Qu J, Li Q, Zhang YP, Hai R, Wu W, Wang Q, Zhan FX, Wang XJ, Kan B, Wang SW, Wan KL, Jing HQ, Lu JX, Yin WW, Zhou H, Guan XH, Liu JF, Bi ZQ, Liu GH, Ren J, Wang H, Zhao Z, Song JD, He JR, Wan T, Zhang JS, Fu XP, Sun LN, Dong XP, Feng ZJ, Yang WZ, Hong T, Zhang Y, Walker DH, Wang Y, Li DX. 2011. Fever with thrombocytopenia associated with a novel bunyavirus in China. N Engl J Med 364:1523–1532. doi: 10.1056/NEJMoa1010095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Liu S, Chai C, Wang C, Amer S, Lv H, He H, Sun J, Lin J. 2014. Systematic review of severe fever with thrombocytopenia syndrome: virology, epidemiology, and clinical characteristics. Rev Med Virol 24:90–102. doi: 10.1002/rmv.1776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kim KH, Yi J, Kim G, Choi SJ, Jun KI, Kim NH, Choe PG, Kim NJ, Lee JK, Oh MD. 2013. Severe fever with thrombocytopenia syndrome, South Korea, 2012. Emerg Infect Dis 19:1892–1894. doi: 10.3201/eid1911.130792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Takahashi T, Maeda K, Suzuki T, Ishido A, Shigeoka T, Tominaga T, Kamei T, Honda M, Ninomiya D, Sakai T, Senba T, Kaneyuki S, Sakaguchi S, Satoh A, Hosokawa T, Kawabe Y, Kurihara S, Izumikawa K, Kohno S, Azuma T, Suemori K, Yasukawa M, Mizutani T, Omatsu T, Katayama Y, Miyahara M, Ijuin M, Doi K, Okuda M, Umeki K, Saito T, Fukushima K, Nakajima K, Yoshikawa T, Tani H, Fukushi S, Fukuma A, Ogata M, Shimojima M, Nakajima N, Nagata N, Katano H, Fukumoto H, Sato Y, Hasegawa H, Yamagishi T, Oishi K, Kurane I, Morikawa S, Saijo M. 2014. The first identification and retrospective study of severe fever with thrombocytopenia syndrome in Japan. J Infect Dis 209:816–827. doi: 10.1093/infdis/jit603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.McMullan LK, Folk SM, Kelly AJ, MacNeil A, Goldsmith CS, Metcalfe MG, Batten BC, Albarino CG, Zaki SR, Rollin PE, Nicholson WL, Nichol ST. 2012. A new phlebovirus associated with severe febrile illness in Missouri. N Engl J Med 367:834–841. doi: 10.1056/NEJMoa1203378. [DOI] [PubMed] [Google Scholar]
- 22.Muehlenbachs A, Fata CR, Lambert AJ, Paddock CD, Velez JO, Blau DM, Staples JE, Karlekar MB, Bhatnagar J, Nasci RS, Zaki SR. 2014. Heartland virus-associated death in Tennessee. Clin Infect Dis 59:845–850. doi: 10.1093/cid/ciu434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pastula DM, Turabelidze G, Yates KF, Jones TF, Lambert AJ, Panella AJ, Kosoy OI, Velez JO, Fisher M, Staples E. 2014. Notes from the field: Heartland virus disease—United States, 2012-2013. MMWR Morb Mortal Wkly Rep 63:270–271. [PMC free article] [PubMed] [Google Scholar]
- 24.Xing Z, Schefers J, Schwabenlander M, Jiao Y, Liang M, Qi X, Li C, Goyal S, Cardona CJ, Wu X, Zhang Z, Li D, Collins J, Murtaugh MP. 2013. Novel bunyavirus in domestic and captive farmed animals, Minnesota, USA. Emerg Infect Dis 19:1487–1489. doi: 10.3201/eid1909.130165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kozuch O, Rajcani J, Sekeyova M, Nosek J. 1970. Uukuniemi virus in small rodents. Acta Virol 14:163–166. [PubMed] [Google Scholar]
- 26.Saikku P, Brummer-Korvenkontio M. 1973. Arboviruses in Finland. II. Isolation and characterization of Uukuniemi virus, a virus associated with ticks and birds. Am J Trop Med Hyg 22:390–399. [DOI] [PubMed] [Google Scholar]
- 27.Hubalek Z, Rudolf I. 2012. Tick-borne viruses in Europe. Parasitol Res 111:9–36. doi: 10.1007/s00436-012-2910-1. [DOI] [PubMed] [Google Scholar]
- 28.Brennan B, Li P, Zhang S, Li A, Liang M, Li D, Elliott RM. 2015. A reverse genetics system for severe fever with thrombocytopenia syndrome virus. J Virol 89:3026–3037. doi: 10.1128/JVI.03432-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Buchholz UJ, Finke S, Conzelmann KK. 1999. Generation of bovine respiratory syncytial virus (BRSV) from cDNA: BRSV NS2 is not essential for virus replication in tissue culture, and the human RSV leader region acts as a functional BRSV genome promoter. J Virol 73:251–259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hilton L, Moganeradj K, Zhang G, Chen YH, Randall RE, McCauley JW, Goodbourn S. 2006. The NPro product of bovine viral diarrhea virus inhibits DNA binding by interferon regulatory factor 3 and targets it for proteasomal degradation. J Virol 80:11723–11732. doi: 10.1128/JVI.01145-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Killip MJ, Young DF, Gatherer D, Ross CS, Short JA, Davison AJ, Goodbourn S, Randall RE. 2013. Deep sequencing analysis of defective genomes of parainfluenza virus 5 and their role in interferon induction. J Virol 87:4798–4807. doi: 10.1128/JVI.03383-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pettersson R, Kaariainen L. 1973. The ribonucleic acids of Uukuniemi virus, a noncubical tick-borne arbovirus. Virology 56:608–619. doi: 10.1016/0042-6822(73)90062-7. [DOI] [PubMed] [Google Scholar]
- 33.Johnson KN, Zeddam JL, Ball LA. 2000. Characterization and construction of functional cDNA clones of Pariacoto virus, the first Alphanodavirus isolated outside Australasia. J Virol 74:5123–5132. doi: 10.1128/JVI.74.11.5123-5132.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Elliott RM, Blakqori G, van Knippenberg IC, Koudriakova E, Li P, McLees A, Shi X, Szemiel AM. 2013. Establishment of a reverse genetics system for Schmallenberg virus, a newly emerged orthobunyavirus in Europe. J Gen Virol 94:851–859. doi: 10.1099/vir.0.049981-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Katz A, Freiberg AN, Backstrom V, Schulz AR, Mateos A, Holm L, Pettersson RF, Vaheri A, Flick R, Plyusnin A. 2010. Oligomerization of Uukuniemi virus nucleocapsid protein. Virol J 7:187. doi: 10.1186/1743-422X-7-187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lowen AC, Noonan C, McLees A, Elliott RM. 2004. Efficient bunyavirus rescue from cloned cDNA. Virology 330:493–500. doi: 10.1016/j.virol.2004.10.009. [DOI] [PubMed] [Google Scholar]
- 37.Sato M, Maeda N, Yoshida H, Urade M, Saito S. 1977. Plaque formation of herpes virus hominis type 2 and rubella virus in variants isolated from the colonies of BHK21/WI-2 cells formed in soft agar. Arch Virol 53:269–273. doi: 10.1007/BF01314672. [DOI] [PubMed] [Google Scholar]
- 38.van Knippenberg I, Carlton-Smith C, Elliott RM. 2010. The N-terminus of Bunyamwera orthobunyavirus NSs protein is essential for interferon antagonism. J Gen Virol 91:2002–2006. doi: 10.1099/vir.0.021774-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bridgen A, Weber F, Fazakerley JK, Elliott RM. 2001. Bunyamwera bunyavirus nonstructural protein NSs is a nonessential gene product that contributes to viral pathogenesis. Proc Natl Acad Sci U S A 98:664–669. doi: 10.1073/pnas.98.2.664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Weber F, Bridgen A, Fazakerley JK, Streitenfeld H, Randall RE, Elliott RM. 2002. Bunyamwera bunyavirus nonstructural protein NSs counteracts the induction of alpha/beta interferon. J Virol 76:7949–7955. doi: 10.1128/JVI.76.16.7949-7955.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hale BG, Knebel A, Botting CH, Galloway CS, Precious BL, Jackson D, Elliott RM, Randall RE. 2009. CDK/ERK-mediated phosphorylation of the human influenza A virus NS1 protein at threonine-215. Virology 383:6–11. doi: 10.1016/j.virol.2008.10.002. [DOI] [PubMed] [Google Scholar]
- 42.Flick R, Pettersson RF. 2001. Reverse genetics system for Uukuniemi virus (Bunyaviridae): RNA polymerase I-catalyzed expression of chimeric viral RNAs. J Virol 75:1643–1655. doi: 10.1128/JVI.75.4.1643-1655.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Overby AK, Popov V, Neve EP, Pettersson RF. 2006. Generation and analysis of infectious virus-like particles of Uukuniemi virus (Bunyaviridae): a useful system for studying bunyaviral packaging and budding. J Virol 80:10428–10435. doi: 10.1128/JVI.01362-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Otsuki K, Maeda J, Yamamoto H, Tsubokura M. 1979. Studies on avian infectious bronchitis virus (IBV). III Interferon induction by and sensitivity to interferon of IBV. Arch Virol 60:249–255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ikegami T, Won S, Peters CJ, Makino S. 2006. Rescue of infectious Rift Valley fever virus entirely from cDNA, analysis of virus lacking the NSs gene, and expression of a foreign gene. J Virol 80:2933–2940. doi: 10.1128/JVI.80.6.2933-2940.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Varela M, Schnettler E, Caporale M, Murgia C, Barry G, McFarlane M, McGregor E, Piras IM, Shaw A, Lamm C, Janowicz A, Beer M, Glass M, Herder V, Hahn K, Baumgartner W, Kohl A, Palmarini M. 2013. Schmallenberg virus pathogenesis, tropism and interaction with the innate immune system of the host. PLoS Pathog 9:e1003133. doi: 10.1371/journal.ppat.1003133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Blakqori G, Weber F. 2005. Efficient cDNA-based rescue of La Crosse bunyaviruses expressing or lacking the nonstructural protein NSs. J Virol 79:10420–10428. doi: 10.1128/JVI.79.16.10420-10428.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Le May N, Mansuroglu Z, Leger P, Josse T, Blot G, Billecocq A, Flick R, Jacob Y, Bonnefoy E, Bouloy M. 2008. A SAP30 complex inhibits IFN-beta expression in Rift Valley fever virus infected cells. PLoS Pathog 4:e13. doi: 10.1371/journal.ppat.0040013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Santiago FW, Covaleda LM, Sanchez-Aparicio MT, Silvas JA, Diaz-Vizarreta AC, Patel JR, Popov V, Yu XJ, Garcia-Sastre A, Aguilar PV. 2014. Hijacking of RIG-I signaling proteins into virus-induced cytoplasmic structures correlates with the inhibition of type I interferon responses. J Virol 88:4572–4585. doi: 10.1128/JVI.03021-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Li DX. 2011. Fever with thrombocytopenia associated with a novel bunyavirus in China. Zhonghua Shi Yan He Lin Chuang Bing Du Xue Za Zhi 25:81–84 (In Chinese.) [PubMed] [Google Scholar]
- 51.Perrone LA, Narayanan K, Worthy M, Peters CJ. 2007. The S segment of Punta Toro virus (Bunyaviridae, Phlebovirus) is a major determinant of lethality in the Syrian hamster and codes for a type I interferon antagonist. J Virol 81:884–892. doi: 10.1128/JVI.01074-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Gori Savellini G, Weber F, Terrosi C, Habjan M, Martorelli B, Cusi MG. 2011. Toscana virus induces interferon although its NSs protein reveals antagonistic activity. J Gen Virol 92:71–79. doi: 10.1099/vir.0.025999-0. [DOI] [PubMed] [Google Scholar]
- 53.Lihoradova OA, Indran SV, Kalveram B, Lokugamage N, Head JA, Gong B, Tigabu B, Juelich TL, Freiberg AN, Ikegami T. 2013. Characterization of Rift Valley fever virus MP-12 strain encoding NSs of Punta Toro virus or sandfly fever Sicilian virus. PLoS Negl Trop Dis 7:e2181. doi: 10.1371/journal.pntd.0002181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Gori-Savellini G, Valentini M, Cusi MG. 2013. Toscana virus NSs protein inhibits the induction of type I interferon by interacting with RIG-I. J Virol 87:6660–6667. doi: 10.1128/JVI.03129-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Habjan M, Pichlmair A, Elliott RM, Overby AK, Glatter T, Gstaiger M, Superti-Furga G, Unger H, Weber F. 2009. NSs protein of Rift Valley fever virus induces the specific degradation of the double-stranded RNA-dependent protein kinase. J Virol 83:4365–4375. doi: 10.1128/JVI.02148-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Billecocq A, Spiegel M, Vialat P, Kohl A, Weber F, Bouloy M, Haller O. 2004. NSs protein of Rift Valley fever virus blocks interferon production by inhibiting host gene transcription. J Virol 78:9798–9806. doi: 10.1128/JVI.78.18.9798-9806.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Simons JF, Persson R, Pettersson RF. 1992. Association of the nonstructural protein NSs of Uukuniemi virus with the 40S ribosomal subunit. J Virol 66:4233–4241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wu X, Qi X, Liang M, Li C, Cardona CJ, Li D, Xing Z. 2014. Roles of viroplasm-like structures formed by nonstructural protein NSs in infection with severe fever with thrombocytopenia syndrome virus. FASEB J 28:2504–2516. doi: 10.1096/fj.13-243857. [DOI] [PubMed] [Google Scholar]
- 59.Wu X, Qi X, Qu B, Zhang Z, Liang M, Li C, Cardona CJ, Li D, Xing Z. 2014. Evasion of antiviral immunity through sequestering of TBK1/IKKepsilon/IRF3 into viral inclusion bodies. J Virol 88:3067–3076. doi: 10.1128/JVI.03510-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Ning YJ, Wang M, Deng M, Shen S, Liu W, Cao WC, Deng F, Wang YY, Hu Z, Wang H. 2014. Viral suppression of innate immunity via spatial isolation of TBK1/IKKepsilon from mitochondrial antiviral platform. J Mol Cell Biol 6:324–337. doi: 10.1093/jmcb/mju015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Palacios G, Tesh R, Travassos da Rosa A, Savji N, Sze W, Jain K, Serge R, Guzman H, Guevara C, Nunes MR, Nunes-Neto JP, Kochel T, Hutchison S, Vasconcelos PF, Lipkin WI. 2011. Characterization of the Candiru antigenic complex (Bunyaviridae: Phlebovirus), a highly diverse and reassorting group of viruses affecting humans in tropical America. J Virol 85:3811–3820. doi: 10.1128/JVI.02275-10. [DOI] [PMC free article] [PubMed] [Google Scholar]