ABSTRACT
Angiopoietin-1 (ANGPT-1) is a secreted glycoprotein that was first characterized as a ligand of the Tie2 receptor. In a previous study using microarray analysis, we found that the expression of ANGPT-1 was upregulated in Kaposi's sarcoma-associated herpesvirus (KSHV)-infected primary effusion lymphoma (PEL) cell lines compared with that in uninfected Burkitt and other leukemia cell lines. Other authors have also reported focal expression of ANGPT-1 mRNA in biopsy specimens of Kaposi's sarcoma (KS) tissue from patients with AIDS. Here, to confirm these findings, we examined the expression and secretion levels of ANGPT-1 in KSHV-infected PEL cell lines and address the mechanisms of ANGPT-1 transcriptional regulation. We also showed that ANGPT-1 was expressed and localized in the cytoplasm and secreted into the supernatant of KSHV-infected PEL cells. Deletion studies of the regulatory region revealed that the region encompassing nucleotides −143 to −125 of the ANGPT-1-regulating sequence was responsible for this upregulation. Moreover, an electrophoretic mobility shift assay and chromatin immunoprecipitation, followed by quantitative PCR, suggested that some KSHV-infected PEL cell line-specific DNA-binding factors, such as OCT-1, should be involved in the upregulation of ANGPT-1 in a sequence-dependent manner.
IMPORTANCE We confirmed that ANGPT-1 was expressed in and secreted from KSHV-infected PEL cells and that the transcriptional activity of ANGPT-1 was upregulated. A 19-bp fragment was identified as the region responsible for ANGPT-1 upregulation through binding with OCT-1 as a core factor in PEL cells. This study suggests that ANGPT-1 is overproduced in KSHV-infected PEL cells, which could affect the pathophysiology of AIDS patients with PEL.
INTRODUCTION
Kaposi's sarcoma (KS)-associated herpesvirus (KSHV), also known as human herpesvirus 8, belongs to the gamma-2 herpesvirus family, which was first identified in KS lesions (1). Epstein-Barr virus (EBV), which also belongs to the gamma-2 herpesvirus family, is frequently associated with malignancies such as Burkitt lymphoma (BL) and nasopharyngeal carcinoma (NPC) (2). KSHV is also associated with several malignancies, i.e., two lymphoproliferative disorders, primary effusion lymphoma (PEL) (3) and multicentric Castleman's disease, as well as KS (4, 5).
It has been reported that KSHV infects various cell types, such as B cells, blood vessel endothelial cells (BECs), lymphatic endothelial cells (LECs), Vero cells, and HEK293 cells (6–9). After infection, KSHV utilizes latency as a default pathway of replication (1, 7). Though viral gene expression profiles might differ between BECs and LECs (10), KSHV is predominantly in latency with its genome binding to the host cell chromosome (10, 11) and governs host gene expression profiles (12) as other viruses do (13, 14). Most KSHV-infected cells are latently infected, and only a limited number of viral genes are expressed in latency: latency-associated nuclear antigen (LANA), viral cyclin (vCYC), viral FLICE inhibitory protein (vFLIP), kaposin (10, 11, 15–18), and viral interferon regulatory factor 3 (vIRF3) (12).
Several viral products of KSHV have been reported to have pivotal effects that contribute to the proliferation of endothelial cells, the viral life cycle, and the secretion of cytokines associated with angiogenic and inflammatory properties; these products include LANA, vIL6, vGPCR, K15, and vIRF3 (12, 19–24). These latency-related viral products may also be involved in enhancement of the expression of various cytokines and growth factors, such as angiopoietin-1 (ANGPT-1), ANGPT-2, vascular endothelial growth factor (VEGF), interleukin-6 (IL-6), IL-8, and tumor necrosis factor alpha (6, 25–29). The angiogenic and inflammatory cytokines regulated by viral proteins or KSHV infection could lead to the induction of lymphangiogenesis, angiogenesis, and antiapoptosis and likely play an important role in KSHV pathogenesis (12, 26, 30–33).
In a previous study, we compared the gene expression profiles of KSHV-infected BC1, BCBL1, and BC3 cells with those of uninfected Daudi, AKATA, Raji, Ramos, and Namalwa cells and MT4, SupT1, Jurkat, and Molt3 leukemia cells. We found that ANGPT-1, a proangiogenic and proinflammatory cytokine, was expressed at significantly higher levels only in KSHV-infected PEL cells (6). ANGPT-1, isolated as a ligand for Tie2, is a glycoprotein secreted from subendothelial stromal cells and hepatic stellate cells (34, 35) and is involved in vascular remodeling, lymphangiogenesis, angiogenesis, and extravasation through ANGPT-1–Tie2 signaling (35, 36). These functions are convincing associations with various oncologic diseases.
Here, we found that ANGPT-1 was expressed in the cytoplasm of KSHV-infected PEL cell lines and actually secreted into the culture medium. Further, we identified a regulatory region affecting ANGPT-1 transcription activity and found that OCT-1 could bind to this region in vitro, as well as in vivo. These findings suggest that the cellular environment established by KSHV infection and/or the PEL cell environment should be involved in the facilitation of ANGPT-1 expression and should affect the pathophysiology of AIDS patients with PEL.
MATERIALS AND METHODS
Cells.
BCBL1, TY1, BC3, BC1, Raji, Namalwa, and BJAB cells were maintained in RPMI 1640 medium (Nacalai Tesque, Kyoto, Japan) supplemented with 20% heat-inactivated fetal bovine serum (FBS), 10 IU/ml penicillin G, and 10 μg/ml streptomycin in a 5% CO2 atmosphere. HEK293 (or just 293) and GP2 cells (TaKaRa-Clontech, Tokyo, Japan), which express a murine leukemia virus gag-pol protein, were maintained in Dulbecco's modified Eagle's medium (DMEM)-high glucose (Nacalai Tesque) supplemented with 10% heat-inactivated FBS, 10 IU/ml penicillin G, and 10 μg/ml streptomycin (Nacalai Tesque). LacZ-VH/BJAB and ANGPT-1-VH/BJAB cells were maintained in RPMI 1640 medium (Nacalai Tesque) supplemented with 20% heat-inactivated FBS, 10 IU/ml penicillin G, 10 μg/ml streptomycin, and 500 μg/ml hygromycin B under a 5% CO2 atmosphere. LacZ-VH/293 and ANGPT-1-VH/293 cells were maintained in DMEM (Nacalai Tesque) supplemented with 10% heat-inactivated FBS, 10 IU/ml penicillin G, 10 μg/ml streptomycin, and 500 μg/ml hygromycin B in a 5% CO2 atmosphere.
Plasmids and retrovirus production.
The gene information of the ANGPT-1 transcription regulatory region was obtained, and the region was amplified (e!Ensemble Human GeneSeqView ENSG00000154188). We initially cloned the region encompassing nucleotides (nt) −2000 to +490 (where +1 is the transcription start site) by PCR with synthetic primers (Greiner, Tokyo, Japan) ANGPT1-ups Fw1 (5′-GGAAGCTTCAAATCAAGCATTATTGGAAAG-3′) and ANGPT1-ups RV1 (5′-AAAAGCTTCACACTCCTTCCGTGCCTCTCG-3′). We cloned the region encompassing nt −898 to +490 with primers ANGPT1-ups Fw4 (5′-GGAAGCTTTATACGCTGCCTGTGGGAAATC-3′) and ANGPT1-ups RV1 by using 100 ng of genomic DNA from BCBL1 as the template. The regulatory sequence and the deletion-containing versions of the regulatory sequence (described below) were cloned into the pGL-3b vector (Promega, Madison, WI). We then verified that the regions extending to nt −2000 and −898 upstream did not show clear differences in transcription activity in BCBL1 cells. Therefore, in this experiment, the region extending to nt −898 upstream was used as the starting reporter construct to determine the element responsible for ANGPT-1 upregulation in KSHV-infected PEL cell lines.
Each deletion mutant construct was obtained by PCR amplification with the −898-nt reporter plasmid used as the template. For the −579-, −178-, −109-, −84-, and nt −58 reporter constructs, ANGPT1-ups Fw5.5 (5′-GGAAGCTTCAGAACATGAAGGGTTGCATTC-3′), ANGPT1-ups Fw8 ((5′-GGAAGCTTTGCTATATTTTAGTAGGTCAGC-3′), ANGPT1-ups Fw8.55 (5′-AAAAGCTTTGCCATGAATCTGCTAAAGGC-3′), ANGPT1-ups Fw9 (GGAAGCTTAGGCAATTGTCTGTGGAAAG-3′), and ANGPT1-ups Fw9.5 (5′-GGAAGCTTAGGGCCATACATGATCGAGGTC-3′) were used, respectively, and ANGPT1-ups RV1 was used as the downstream primer. The D1 fragment (nt −143 to −110) was generated by annealing two synthetic oligonucleotides, ANGPT1-ups FW8.5 SmaI (5′-AACCCGGGAGTGTATTAAGGTGGACTGCTC-3′) and ANGPT1-ups RV8.5 SmaI (5′-AACCCGGGATCAATAAATAGAGCAGTCCAC-3′), followed by a repair reaction with a Klenow fragment (TaKaRa-Clontech), digestion with SmaI, and insertion of a SmaI site just upstream of the nt −58 reporter construct. The D2 fragment (nt −130 to −102) was generated in the same way but with synthetic primers ANGPT1-ups FW8.5 SmaI-2 (5′-AACCCGGGTGGACTGCTCTATTTATTGATT-3′) and ANGPT1-ups RV8.5 SmaI-2 (5′-AACCCGGGTTCATGGCAAATCAATAAATAG-3′). The D1A (nt −143 to −125) and D1B (nt −128 to −110) fragments were generated by annealing phosphorylated synthetic DNA, ANGPT1-ups D1A S (5′-GGGAAGTGTATTAAGGTGGACTCCC-3′) and ANGPT1-ups D1A AS (5′-GGGAGTCCACCTTAATACACTTCCC-3′) for D1A, ANGPT1-ups D1B S (5′-GGGGACTGCTCTATTTATTGATCCC-3′) and ANGPT1-ups D1B AS (5′-GGGATCAATAAATAGAGCAGTCCCC-3′) for D1B, respectively, and insertion of a SmaI site from the −58 reporter construct.
D1A mutant plasmids (Mut1 to -6) were constructed by inserting each of the annealed phosphorylated synthetic oligonucleotides (Mut1, 5′-GGGAAGTGTATTAAGGTGGAAACCC-3′ and 5′-GGGTTTCCACCTTAATACACTTCCC-3′; Mut2, 5′-GGGCCCCGTATTAAGGTGGACTCCC-3′ and 5′-GGGAGTCCACCTTAATACGGGGCCC-3′; Mut3, 5′-GGGAAGTCCCCTAAGGTGGACTCCC-3′ and 5′-GGGAGTCCACCTTAGGGGACTTCCC-3′; Mut4, 5′-GGGAAGTGTATCCCCGTGGACTCCC-3′ and 5′-GGGAGTCCACGGGGATACACTTCCC-3′; Mut5, 5′-GGGAAGTGTATTAAGCCCCACTCCC-3′ and 5′-GGGAGTGGGGCTTAATACACTTCCC-3′; Mut6, 5′-GGGAAGTGTATTAAGGTGGCCCCCC-3′ and 5′-GGGGGGCCACCTTAATACACTTCCC-3′) into a SmaI site of the nt −58 reporter plasmid as described above.
The ANGPT-1 open reading frame (ORF) clone was purchased from the Kazusa ORFeome Project (Product ID FHC0948; Promega). The ORF SgfI-PmeI fragment was cloned into the EcoRV site of the pMT V5His B expression vector (Invitrogen, Carlsbad, CA) with some modifications to generate pMT ANGPT-1-V5His. The ANGPT-1-V5His fragment was recloned into retroviral expression vector pQC XIH (Clontech, Palo Alto, CA), and pQC XIH ANGPT-1-V5His was generated. For retrovirus production, GP2 cells were cotransfected with pQC XIH ANGPT-1V5His and pVSV-G (TaKaRa-Clontech) with LT1 reagent (Mirus, Madison, WI) for 12 h. At 72 h posttransfection, the supernatant was collected to infect BJAB and 293 cells. Hygromycin B (500 μg/ml; Wako Pure Chemicals, Tokyo, Japan) was used to select virus-transduced cells.
The OCT-1 expression vector (pCGN OCT-1) and its parental vector (pCGN) were the kind gifts of W. Herr as described in reference 43.
Transfection and luciferase assay.
For the luciferase assay, 1.5 × 105 PEL cells per 0.5 ml of medium were plated, incubated overnight, and then transfected with 1 μg of each of the reporter plasmids and 0.1 μg of the cytomegalovirus β-galactosidase LacZ expression plasmid by using TransFectin Lipofectin Reagent (Bio-Rad, Hercules, CA). The cells were harvested at 48 h posttransfection. After being washed twice with phosphate-buffered saline (PBS), the cells were lysed in 50 μl of Glo lysis buffer (Promega). Luciferase activity was measured with a Bright-Glo luciferase assay system (Promega) according to the manufacturer's instructions and normalized by β-galactosidase activity (12.5 μg/ml chlorophenol red-β-d-galactopyranoside, Z buffer [0.1 M NaPO4 at pH 7.5, 10 mM KCl, 1 mM MgSO4, 50 mM 2-mercaptoethanol]). To test OCT-1 transactivation activity, either pCGN OCT-1 or the parental vector with either the D1A reporter or the nt −58 reporter plasmid was transfected into 5 × 105 BCBL1 cells. The effector plasmid (pCGN OCT-1) amounts used were 0, 0.3, 0.9, and 1.5 μg.
Immunofluorescence assay (IFA).
Cells were fixed for 1 h with 4% paraformaldehyde (Nacalai Tesque)–PBS and permeabilized in 0.1% Triton X-100 (Nacalai Tesque)–PBS for 30 min. Proteins were detected with the following primary antibodies: a mouse monoclonal antibody against V5 (1:1,000 dilution; Nacalai Tesque) for LacZ-VH/BJAB and ANGPT-1-VH/BJAB and a mouse monoclonal antibody against ANGPT-1 (1:500 dilution; R&D Systems, Minneapolis, MN) for LacZ-VH-/BJAB, ANGPT-1-VH/BJAB, BJAB, BC1, BC3, TY1, and BCBL1. Images were taken with an Olympus FV1000D confocal microscope (Olympus).
ELISA, IP, and Western blotting.
The culture supernatant of the PEL cells (106/ml) was tested with a human ANGPT-1 enzyme-linked immunosorbent assay (ELISA) kit (MyBioSource, San Diego, CA) according to the manufacturer's instructions. ANGPT-2 was measured likewise with a Quantikine ELISA Human ANGPT-2 Immunoassay (R&D Systems) according to the manufacturer's directions. For immunoprecipitation (IP), 30 μl of protein G Sepharose 4 Fast Flow (GE Healthcare, Buckinghamshire, United Kingdom) was washed with IP reaction solution (25 mM HEPES-KOH [pH 7.9], 200 mM NaCl, 5 mM MgCl2, 0.2% NP-40, 10% glycerol, 1 mM dithiothreitol [DTT]), and mixed with culture supernatants from LacZ-VH/BJAB, ANGPT-1-VH/BJAB, BC1 and TY1 cells. After 1 h of incubation with protein G Sepharose to remove nonspecific binding proteins, the Sepharose was excluded and then 10 μl of protein G Sepharose and 2 μg of an ANGPT-1 antibody (R&D Systems) were added to the supernatant for incubation overnight with rotation. The Sepharose was harvested, washed three times with 1 ml of IP reaction solution, suspended in 50 μl of SDS-PAGE sampling buffer, and boiled. For Western blotting, each sample (20 μl) was separated by SDS-PAGE (8% polyacrylamide) and transferred onto a polyvinylidene difluoride (PVDF) membrane (Immun-Blot PVDF membrane for protein blotting; Bio-Rad). The membrane was blocked with 5% dry milk in Tris-buffered saline–Triton X-100 (TBS-T). A specific antibody against ANGPT-1 (1:1,000 dilution; R&D Systems) was added, followed by secondary antibodies conjugated with horseradish peroxidase for detection. The membrane was washed with TBS-T three times, and the chemiluminescence image was obtained with an imaging system (ChemiDoc MP; Bio-Rad).
Electrophoretic mobility shift assay (EMSA).
Nuclear extracts were obtained from BJAB and BCBL1 cells. Cy5-labeled double-stranded D1A (nt −143 to −125;19 nt) oligonucleotide (5′-Cy5-AAGTGTATTAAGGTGGACT-3′ and 5′-Cy5-AGTCCACCTTAATACACTT-3′) was prepared (Greiner) and nonlabeled D1A (5′-AAGTGTATTAAGGTGGACT-3′ and 5′-AGTCCACCTTAATACACTT-3′) and mutant constructs (Mut1, 5′-AAGTGTATTAAGGTGGAAA-3′ and 5′-TTTCCACCTTAATACACTT-3′); Mut2, 5′-CCCCGTATTAAGGTGGACT-3′ and 5′-AGTCCACCTTAATACGGGG-3′; Mut3, 5′-AAGTCCCCTAAGGTGGACT-3′ and 5′-AGTCCACCTTAGGGGACTT-3′; Mut4, 5′-AAGTGTATCCCCGTGGACT-3′ and 5′-AGTCCACGGGGATACACTT-3′; Mut5, 5′-AAGTGTATTAAGCCCCACT-3′ and 5′-AGTGGGGCTTAATACACTT-3′; Mut6, 5′-AAGTGTATTAAGGTGGCCC-3′ and 5′-GGGCCACCTTAATACACTT-3′) were prepared. For the gel shift assay, 10 μg of nuclear extract was mixed with 1.25 pmol of the Cy5 fluorescent tag probe and incubated for 30 min at 25°C in a binding buffer (100 mM HEPES [pH 7.9], 5 mM MgCl2, 2.5 mM DTT, 250 μg/ml bovine serum albumin, 20% Ficoll). Unlabeled D1A probe (20×) or mutant probes (20×) were used for competition experiments. In the case of supershift analysis, 1 μg of each specific antibody against IRF4 (Sigma, St. Louis, MO) or RFX-5 (Sigma) or OCT-1 (MBL Life Science, Japan) was added to the respective reaction mixture. Normal mouse IgG was used for a control reaction mixture. The reacted mixtures were separated on a 0.25× TBE–4% acrylamide gel. The fluorescent image was obtained with a ChemiDoc MP imaging system (Bio-Rad).
Chromatin IP (ChIP).
BJAB or BCBL1 cells (1 × 107) were cross-linked with 1% formaldehyde at room temperature for 10 min, and then glycine was added to a final concentration of 0.125 M. After incubation for 5 min, the cells were washed in PBS and harvested by centrifugation. The cells were lysed in 1 ml of ice-cold low-salt lysis buffer (10 mM Tris-HCl [pH 7.8], 10 mM NaCl, 0.1 mM EDTA) with protease inhibitors. The nuclei were centrifuged, harvested, and resuspended in 300 μl of high-salt buffer (50 mM Tris-HCl [pH 7.8], 500 mM NaCl, 0.1 mM EDTA, 5 mM MgCl2, 20% glycerol) with protease inhibitors. After incubation on ice for 15 min, 700 μl of low-salt buffer with protease inhibitors was added to the samples. Chromatin was sheared into about 500-bp fragments 200 times on ice with a Tomy Ultrasonic Disruptor (UD-201) set to an output of 2 and a 60% duty cycle. The chromatin from 107 cells was incubated with 2 μg of normal mouse IgG and 20 μl of protein G Sepharose (GE Healthcare) and incubated for 0.5 h at room temperature to remove nonspecific binding proteins. The samples with nonspecific binding proteins removed were then incubated with 5 μg of a primary antibody (either normal mouse IgG or a mouse anti-OCT-1 monoclonal antibody [MBL]) and 20 μl of protein G Sepharose at room temperature for 1 h on a rotator. The Sepharose was washed six times with wash buffer (150 mM NaCl, 50 mM Tris-HCl [pH 7.5], 5 mM EDTA, 0.5% [vol/vol] NP-40, 1.0% [vol/vol] Triton X-100). The chromatin-immune complexes were incubated in 200 μl of reversal buffer (10 mM Tris-HCl [pH 8.0], 1 mM EDTA, 300 mM NaCl) at 37°C with 20 μg of RNase A (Roche) for 30 min, 10 μl of proteinase K (Roche) was added, and the mixture was incubated at 65°C overnight. DNA was extracted once with a MonoFas DNA purification kit (GL Sciences) according to the manufacturer's directions. The DNA was then amplified with primers ANGPT1-ups Fw8 (5′-GGAAGCTTTGCTATATTTTAGTAGGTCAGC-3′) and ANGPT1-ups Fw10 RV1 (5′-CAATTTGTAAGACGATCCCGCC-3′), which included the D1A region. The amplification was quantified by quantitative PCR (qPCR) with a LightCycler (Roche Diagnostics).
RESULTS
ANGPT-1 expression in KSHV-infected PEL cells and secretion into culture medium.
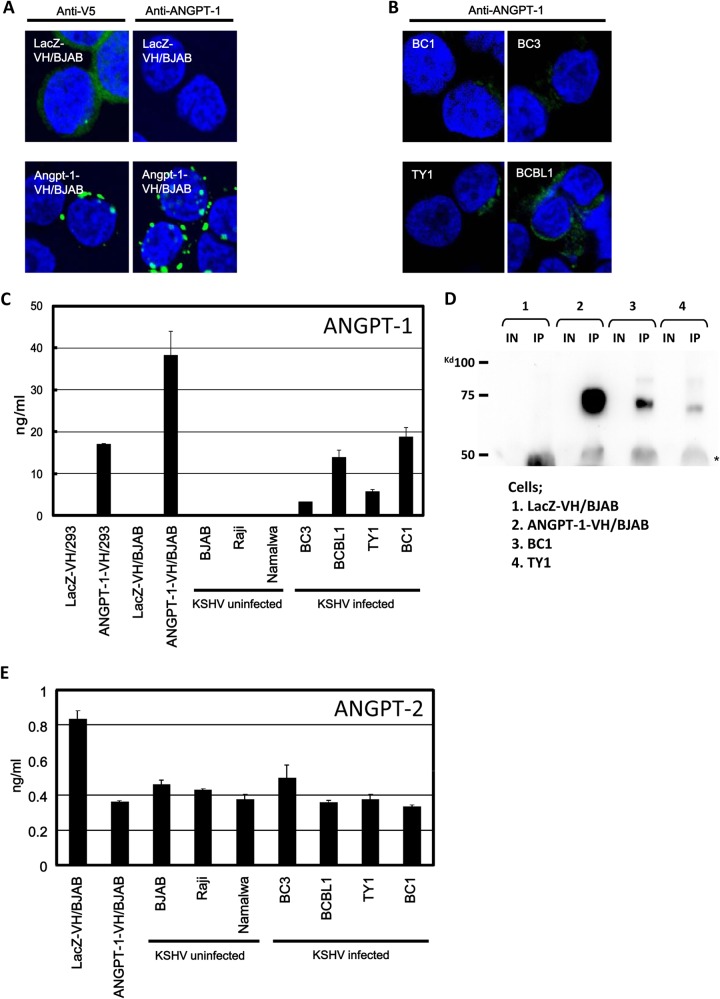
A previous clinical research study revealed the expression of ANGPT-1 mRNA in AIDS-associated KS biopsy specimens (29), and we have shown that the expression of ANGPT-1 in PEL cells appeared to be much higher than that in the T-cell lymphoma and BL cells (6). To investigate whether ANGPT-1 was really produced in the PEL cell lines, we measured the level of ANGPT-1 secretion into the culture supernatant of PEL cells by ELISA and observed its cellular expression and localization by IFA. We generated stably ANGPT-1-expressing ANGPT-1-VH/BJAB and ANGPT-1-VH/293 cells, which were used as positive controls; concomitantly, we used Lac-Z-VH-expressing LacZ-VH/BJAB and LacZ-VH/293 cells as negative controls. Since ANGPT-1-VH and LacZ-VH were tagged with a V5-histidine hexamer (VH) at the C terminus, we assessed whether a mouse anti-V5 antibody, as well as a mouse anti-ANGPT-1 antibody, was valuable for IFA. Compared to the case with no signal upon the staining of LacZ-VH/BJAB cells with anti-ANGPT-1 antibody, diffuse patterns were detected by anti-V5 antibody staining (Fig. 1A). In ANGPT-1-VH/BJAB cells, both anti-V5 and anti-ANGPT-1 antibodies could detect dotted spots. ANGPT-1 was stained diffusely in the cytoplasm of KSHV-infected PEL cells (Fig. 1B). The bigger dot patterns seen in ANGPT-1-VH/BJAB cells may have been due to the massive expression of ANGPT-1-VH in those cells.
FIG 1.
ANGPT-1 expression in KSHV-infected PEL cells and ANGPT-1 and ANGPT-2 secretion into medium. (A and B) ANGPT-1-VH/BJAB and LacZ-VH/BJAB cells were stained with an anti-V5 antibody (A, left, green staining) and an anti-ANGPT-1 antibody (A, right, green staining), followed by anti-mouse IgG conjugated with Alexa 488. KSHV-infected BC1, BC3, BCBL1, and TY1 cells were stained with an anti-ANGPT-1 antibody, followed by anti-mouse IgG conjugated with Alexa 488. (C) ELISA of ANGPT-1 secreted into medium. KSHV-infected BC1, BC3, BCBL1, and TY1 cells and KSHV-negative BJAB, Raji, and Namalwa cells were cultured until they reached a density of 106/ml, and then the supernatant was harvested and ANGPT-1 was measured by ELISA (28). ANGPT-1-VH/BJAB and ANGPT-1-VH/293 cells were used as positive controls, and LacZ-VH/BJAB and LacZ-VH/293 cells were used as negative controls. (D) Detection of secreted ANGPT-1 by Western blotting. ANGPT-1 was immunoprecipitated from the supernatant with an anti-ANGPT-1 antibody and protein G Sepharose and subjected to Western blotting. IN, input; IP, immunoprecipitated. 1, LacZ-VH/BJAB; 2, ANGPT-1-VH/BJAB; 3, BC1; 4, TY1. (E) Detection of secreted ANGPT-2 with an ELISA kit. KSHV-infected PEL cells and EBV-infected or uninfected BL cells were tested as described for ANGPT-1 in panel C.
To examine whether ANGPT-1 was really secreted into the culture medium of PEL cells, we measured ANGPT-1 levels in the culture supernatant by ELISA. We could detect secreted ANGPT-1 in ANGPT-1-VH/293 and ANGPT-1-VH/BJAB cells, which served as positive controls (Fig. 1C). Likewise, we measured secreted ANGPT-1 levels in the culture supernatant of PEL and BL cells, and these levels were clearly higher in the culture medium of KSHV-infected cell lines such as BC1, BC3, BCBL1, and TY1 than in KSHV-uninfected cell lines such as BJAB, Raji, and Namalwa. On the other hand, ANGPT-2 was detected at the same level in these cell lines; the levels were relatively low and did not reach 1 ng/ml (Fig. 1E). Collectively, these data confirmed that KSHV-infected PEL cells expressed a high level of ANGPT-1, but KSHV-uninfected cells—including EBV-infected BL cells (Fig. 1C)—did not express a high level of ANGPT-1, as shown by gene expression profile analysis and the data obtained by reverse transcription-PCR (6). In addition, we detected ANGPT-1 in the culture supernatant of KSHV-infected cells by IP followed by immunoblot analysis, in which ANGPT-1 in the medium was immunoprecipitated with a mouse anti-ANGPT-1 antibody and then identified by Western blotting with the same antibody (Fig. 1D). Together, these results strongly suggested that ANGPT-1 expression should be upregulated in KSHV-infected PEL cells.
The luciferase activity of the regulatory region of ANGPT-1 is enhanced in KSHV-infected PEL cells.
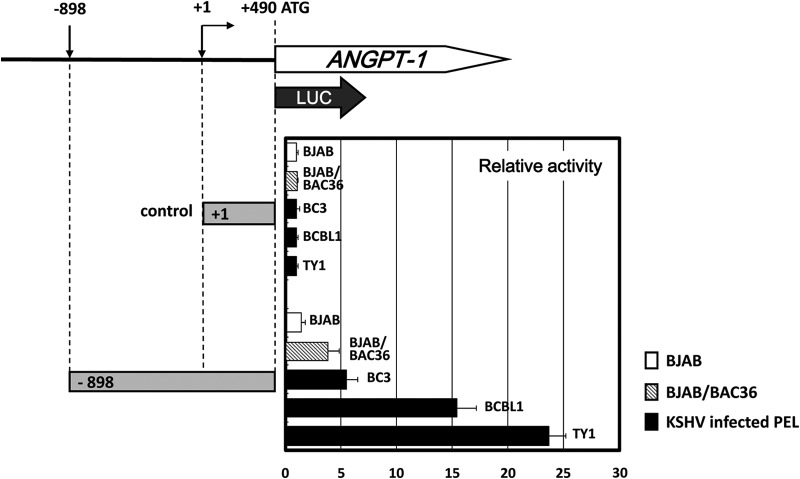
It has been reported that KSHV regulates host gene expression by expressing several viral genes, including vIRF3, vIRF7, and the gene for a viral protein kinase (ORF36) (12, 33). To understand how ANGPT-1 was upregulated in KSHV-infected PEL cells and how KSHV contributed to ANGPT-1 gene expression, we constructed ANGPT-1 promoter-luciferase reporter constructs (−898 to +490 of Luc, where +1 is the transcription start site and +490 is the A in ATG, the ANGPT-1 translation initiation codon) and measured their transcriptional activity. These constructs were tested in KSHV-infected or uninfected cells and BJAB-BAC36 cells, which contained the full KSHV genome in a bacterial artificial chromosome (BAC) (26, 37). The activation of the reporter gene containing the regulatory region of ANGPT-1 was greater than that in the control, i.e., 5.1-fold greater in BC3 cells, 15.1-fold greater in BCBL1 cells, and 23.5-fold greater in TY1 cells, and its activation was 4.1-fold greater in BJAB-BAC36 cells but not in BJAB cells (Fig. 2). These data suggested that ANGPT-1 should be specifically upregulated in KSHV-infected PEL cells.
FIG 2.
Transcriptional activity of the ANGPT-1 regulatory region in KSHV-infected PEL cells. PEL cells, KSHV-negative BJAB cells, and BAC36/BJAB cells were transfected with a luciferase reporter plasmid that contained bp −898 to +490 of ANGPT-1, and a luciferase reporter plasmid that contained bp +1 to +490 was used as a control. Forty-eight hours after transfection, the lysate was prepared and luciferase activity was measured as described in Materials and Methods. Data are shown as fold activity differences from the activity of the basal reporter; the activity of a +1 construct was set at 1-fold.
A 19-nt element is responsible for ANGPT-1 upregulation.
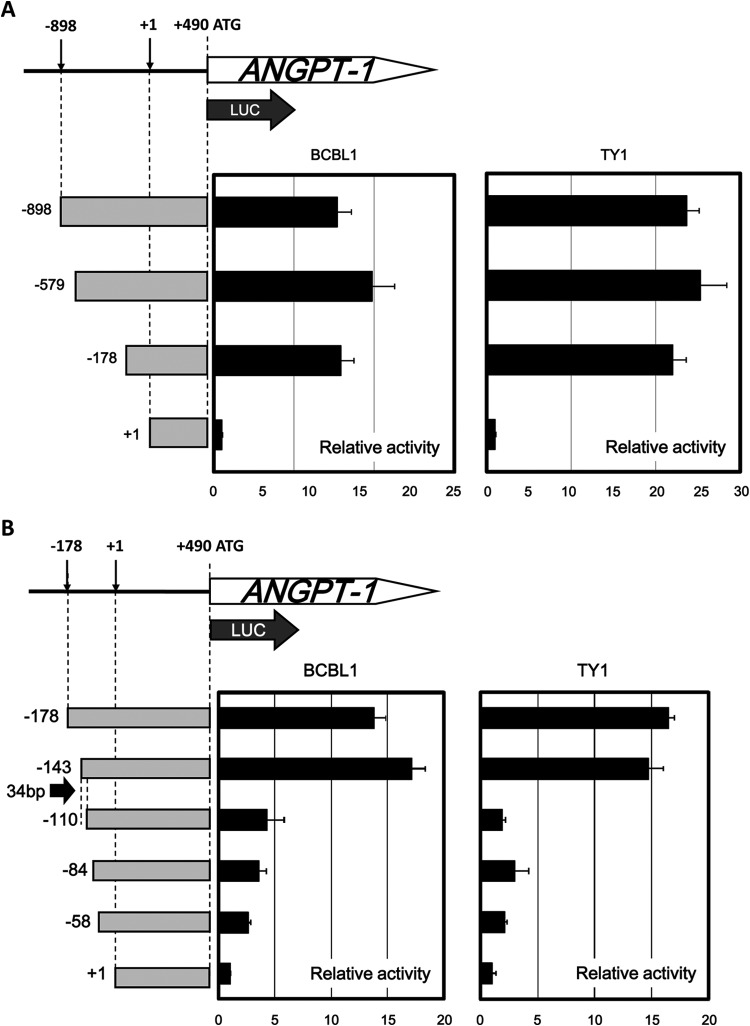
In the experiments described here, it was shown that ANGPT-1 expression was enhanced in KSHV-infected PEL cells, where KSHV could govern the gene expression profile. Therefore, in order to determine the regulatory region responsible for ANGPT-1 transcription in KSHV-infected PEL cells, we made several deletion mutant luciferase reporters (−898, −579, −178, and +1 Luc as a control) and tested them in KSHV-infected BCBL1 and TY1 cells. Two of the deletion reporters (−579 and −178) showed transcriptional activity comparable to that of the full-deletion reporter (−898) in both BCBL1 and TY1 cells (Fig. 3A).
FIG 3.
Deletion analyses of the transcriptional activity of the ANGPT-1 regulatory region in KSHV-infected PEL cells. (A) BCBL1 and TY1 cells were transfected with luciferase reporter plasmids containing parts of the upstream regulatory region of ANGPT-1 as indicated (bp −898, −579, or −178 to +490) and a control plasmid (bp +1 to +490). Luciferase activity was measured 48 h later as described in the text. (B) BCBL1 and TY1 cells were transfected with luciferase reporter plasmids containing parts of the upstream ANGPT-1 regulatory region as shown (bp −178, −143, −109, −84, or −58 to +490) and a control reporter plasmid (bp +1 to +490), and luciferase activity was measured 48 h later. The data shown are fold activity differences from the activity of the basal reporter; the activity of a +1 construct was set at 1-fold.
We next prepared four further deletion mutant luciferase reporters: −143, −110, −84, and −58. Among them, we found a profound decrease in activity between nt −143 and −110 (Fig. 3B), and thus, we focused on this segment and examined whether it contained a responsive element that was sufficiently activated in KSHV-infected PEL cells.
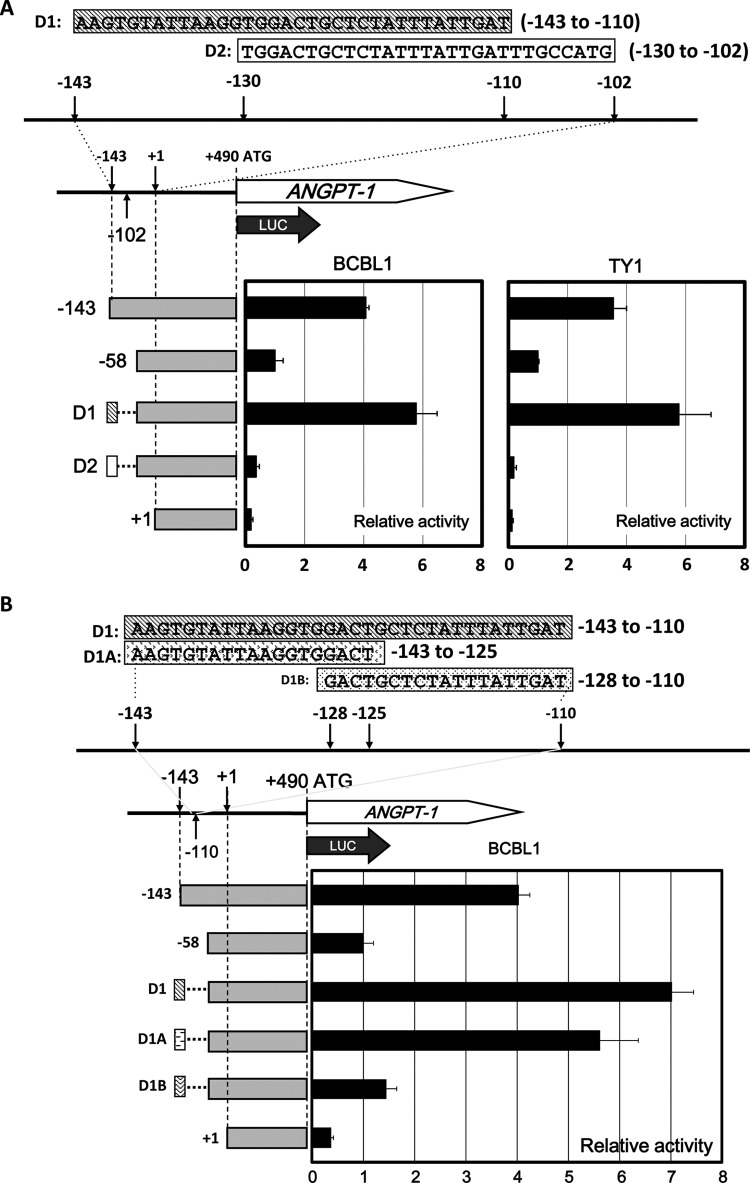
We divided the 34-nt segment into two regions named D1 (the first half; −143 to −110) and D2 (the second half; −130 to −102) and put them in front of the −58 reporter, which showed very low activity close to the basal level of only the proximal promoter (Fig. 4A). Compared to the control luciferase reporter (−58), the transcriptional activity of D1 was clearly higher than that of D2 (Fig. 4A).
FIG 4.
Identification of the minimal enhancing element of ANGPT-1 in KSHV-infected PEL cells. (A) Schematic illustration of two DNA fragments, D1 (bp −143 to −110, oblique shaded rectangle) and D2 (bp −130 to −102, white rectangle), and reporter analysis of the D1 and D2 fragments. Fragments D1 and D2 were inserted into the region upstream of the bp −58 to +490 reporter in the sense orientation, and luciferase activity was measured. (B) Schematic drawing of the deletion constructs with the D1, D2, D1A (bp −143 to −125), and D1B (bp −128 to −110) regions. Fragments D1A and D1B were inserted into the region upstream of the bp −58 to +490 reporter and transfected into BCBL1 cells. Luciferase activity was measured. Data are shown as fold activity differences from the activity of the basal reporter; the activity of the −58 construct was set at 1-fold.
We further designed two reporter plasmids named D1A (the first half of D1; −143 to −125) and D1B (the second half of D1; −128 to −110) by dividing the D1 segment and put them in front of the −58 reporter (Fig. 4B). Luciferase assay showed that D1A had high transcriptional activity similar to that of the positive control (−143) (Fig. 4B). Collectively, these analyses suggested that the 19-nt element (−143 to −125) could be responsible for ANGPT-1 expression regulation in KSHV-infected PEL cell lines.
Nuclear proteins in PEL cells bind to the responsive element (D1A; −143 to −125) for ANGPT-1 transcriptional regulation.
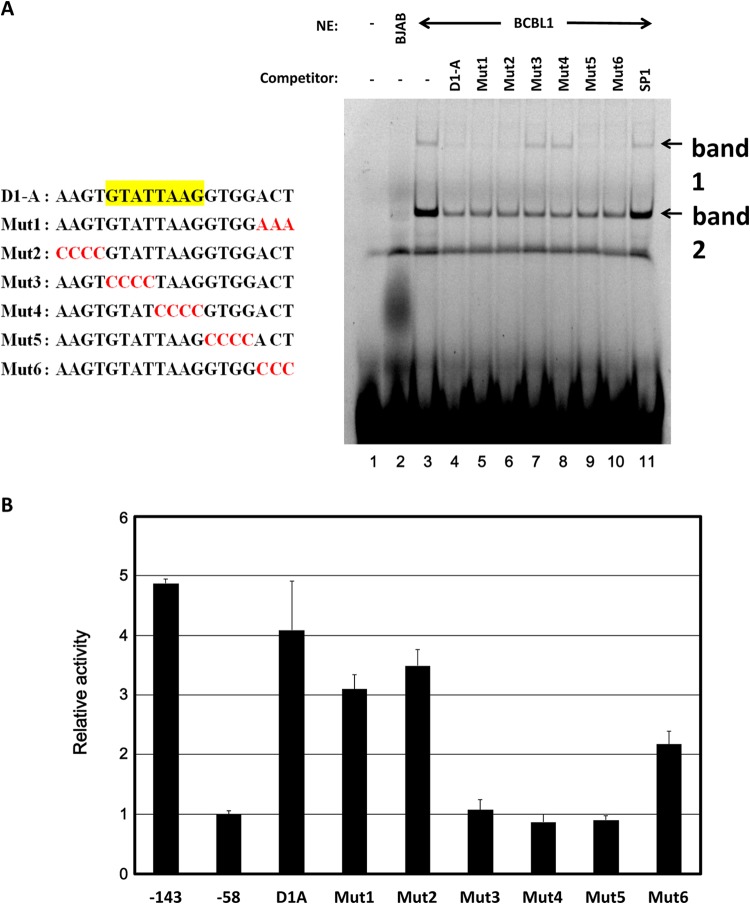
The results described above suggested that the D1A element (−143 to −125) is necessary and sufficient for ANGPT-1 expression in KSHV-infected PEL cell lines. To investigate whether any nuclear factor of KSHV-infected PEL cells is involved in the upregulation of ANGPT-1, we performed an EMSA to identify the factors binding to the 19-bp element. We prepared a Cy5-labeled 19-bp probe and attempted to react it with nuclear proteins of KSHV-infected or uninfected cells. As shown in Fig. 5A, the specific binding factor contained in the nuclear extracts from KSHV-infected (BCBL1) cells could bind to the 19-nt element specifically (lane 3). In fact, there appeared to be two specific shifted bands (1 and 2 in Fig. 5A). These two bands were competed with a nonlabeled 19-bp probe (D1A) to some extent, compared to the bands seen in an SP1-binding consensus competitor (lanes 4 and 11). The following experiment was done with various kinds of mutated competitors (lanes 5 to 10). In the initial experiments, we were careful of the overlapping region between D1A and D1B and prepared a nonlabeled Mut1 probe and thereafter a series of mutant probes, Mut2 to Mut6. Using these probes, we performed a competition EMSA (Fig. 5A, lanes 5 to 10, and B). As shown in Fig. 5A, D1A, Mut1, Mut2, Mut5, and Mut6 competed but Mut3 and Mut4 did not. These data suggested that a core nuclear-factor-binding site in D1A should be the sequence mutated in Mut3 and Mut4, and thus, the sequence should be 5′-GTATTAAG-3′. On the other hand, band 2 competed uniformly with these mutated competitors, except the SP1-binding consensus. Therefore, we focused our attention on band 1, which exhibited more specific competition.
FIG 5.
EMSA of the D1A element. (A, left) Schematic illustration of the D1A region and mutant forms of the D1A region. (A, right) Nuclear extracts (NE) of KSHV-infected PEL (BCBL1) cells were mixed with D1A labeled with Cy5. A 20-fold concentration of each unlabeled mutant probe was used for the competition experiment. A minus sign indicates no nuclear extract or no competitor, respectively. In lane 2, 10 μg of BJAB nuclear extract was the input; in lanes 3 to 11, 10 μg of BCBL1 nuclear extract was used. (B) Mutant plasmids were constructed by inserting each mutant element into the region upstream of the bp −58 to +490 reporter, which contains the ANGPT-1 promoter. Data are shown as fold activity differences from the activity of the basal reporter; the activity of a bp −58 construct was set at 1-fold.
In addition, we prepared reporter constructs with these mutant constructs and checked their luciferase activities (Fig. 5B). The results showed that Mut3 and Mut4 lost their luciferase activity, which further confirmed that the mutated region in Mut3 and Mut4 should be important for the nuclear factor binding that promoted ANGPT-1 expression in KSHV-infected PEL cell lines. Since Mut5 also lost its luciferase activity, some sequences mutated in Mut5 could be involved in this activity without affecting the potential nuclear factor binding.
OCT-1 binds to the responsive element of ANGPT-1.
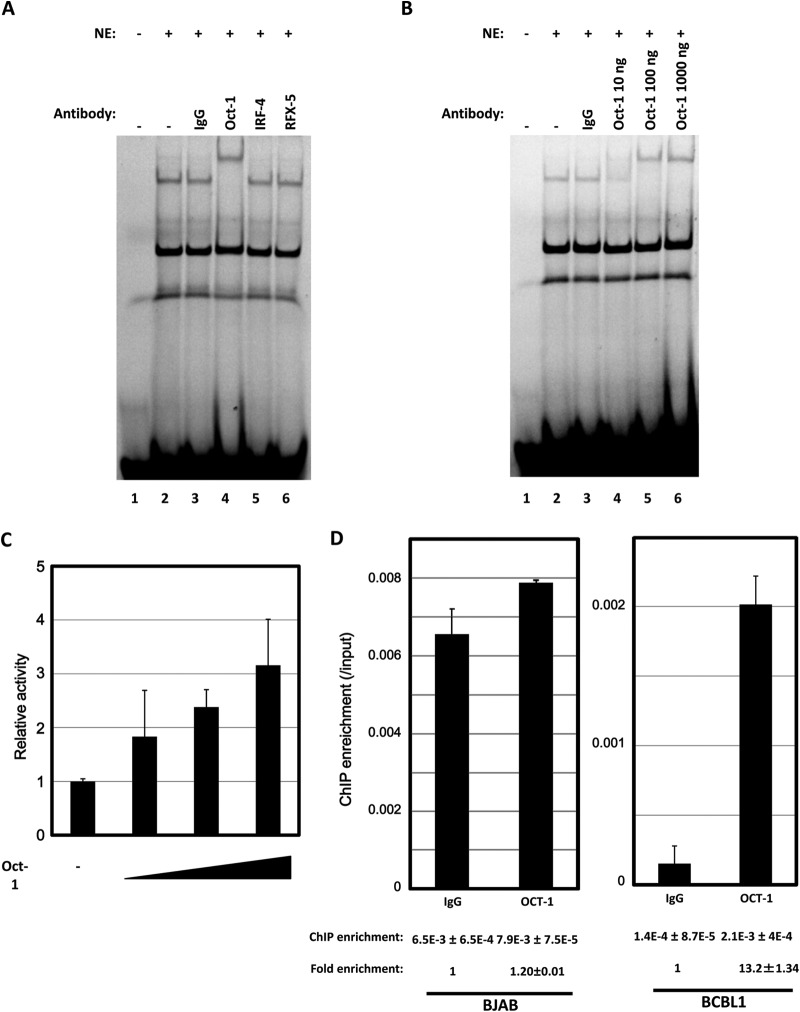
We also tried to identify which factor bound with the D1A element by cutting out shifted band 1 in an EMSA followed by mass spectrometry (MS). Since octamer-binding proteins (OCT-1), interferon regulatory factor 4 (IRF4), and RFX-5 were among the nominated candidates, a supershift EMSA was performed with these antibodies. Compared with the anti-IRF4 and the anti-RFX-5 antibodies, respectively, the anti-OCT-1 antibody showed a supershift (Fig. 6A), even in a dose-dependent manner (Fig. 6B). Thus, it appears that OCT-1 could be involved in the upregulation of ANGPT-1. We then checked whether the expression of OCT-1 could upregulate the luciferase activity of the −58 reporter through the D1A element. Cotransfection of OCT-1 with the D1A reporter resulted in higher luciferase activity than did cotransfection with the −58 reporter, in a dose-dependent manner (Fig. 6C).
FIG 6.
Supershift EMSA with antibodies. (A) Nuclear extracts (NE) of KSHV-infected PEL (BCBL1) cells were mixed with D1A labeled with Cy5. A 1,000-ng sample of each antibody was used for the supershift experiment. Anti-IgG antibody (1,000 ng) was used as a negative control. (B) Samples (10, 100, and 1,000 ng) of anti-OCT-1 were prepared for a dose dependence experiment. (C) pCGN OCT-1 (0, 0.3, 0.9, or 1.5 μg) was transfected with a D1A reporter or a bp −58 reporter (0.3 μg) into BCBL1 cells; the ratio of D1A to −58 luciferase activities of each combination was calculated, and the ratio of D1A to −58 luciferase activities without pCGN OCT-1 was set at 1-fold. (D) qPCR assays were performed with ChIP DNA to test enrichments of the D1A region in the ANGPT-1 promoter. DNA by ChIP with either normal mouse IgG or an anti-OCT-1 antibody was amplified with primers around the D1A region. The amount of the amplified product was quantified by qPCR. The quantified ratios of the ChIP samples to the input DNA were calculated. Thus, each value under the graph is the ratio of the amplified amount obtained with each IgG to the input amount (ChIP enrichment). Fold enrichment values was calculated by setting the normal IgG values at 1.
To confirm that OCT-1 is involved in the upregulation of ANGPT-1 in vivo, we performed ChIP followed by qPCR with mouse normal IgG or an anti-OCT-1 antibody. As shown in Fig. 6D, ChIP enrichment of the region including D1A was about 10-fold higher with the OCT-1 antibody than with normal IgG in the case of BCBL1 cells (Fig. 6D, right). This result suggested that OCT-1 is involved in the upregulation of ANGPT-1 in vivo by binding with this region, though a relatively high background level was observed, but no ChIP enrichment was revealed with the OCT-1 antibody in the case of BJAB cells (Fig. 6D, left).
Taken together, these results show that ANGPT-1 could be upregulated in KSHV-infected cells through OCT-1 binding to the D1A element.
DISCUSSION
KS is recognized as a highly angiogenic and inflammatory tumor (25). It is well understood that KSHV infection could affect the expression and secretion of several cytokines and growth factors, including VEGF and its receptor, matrix metalloproteinases, and angiopoietins, associated with KS (6, 25–28). The angiopoietin gene family encodes four members (ANGPT-1 to -4), and they bind to a tyrosine kinase receptor, Tie1 or Tie2 (34, 35, 38), expressed in endothelial cells. ANGPT-1 is secreted predominantly from mesenchymal cells and acts as an agonist of Tie2 signaling, whereas Angpt-2 acts as an antagonist (34, 35). Activation of Tie2 affects several signaling pathways, including phosphoinositol 3-kinase, endothelial nitric oxide synthase, and growth factor receptor-bound protein 2. These could regulate the proliferation, migration, and survival interaction of endothelial cells (39). Both ANGPT-1 and ANGPT-2 could lead to the development of lymphangiogenesis around islets of Langerhans and pancreatic β-cell tumors in transgenic mice, although tumor angiogenesis was detected only in ANGPT-2 transgenic mice and not in ANGPT-1 transgenic ones (40). It is suggested that ANGPT-1 and ANGPT-2 collaborate in regulating pathological lymphangiogenesis and angiogenesis via different pathways (39, 40).
In human umbilical vein endothelial cells, KSHV infection could upregulate the expression of ANGPT-2, with activation of the promoter via AP-1 and ETS-1 transcriptional factors, which involves the ERK, JNK, and p38 mitogen-activated protein kinase (MAPK) pathways (26). KSHV induced rapid release of ANGPT-2 from the Weibel-Palade bodies of endothelial cells (27). In addition, in KSHV-infected lymphatic endothelial cells, ANGPT-2 was also upregulated by two KSHV lytic genes, vIL-6 and vGPCR, through the MAPK pathway (28). ANGPT-1 could not be detected in KSHV-infected endothelial cells, though it was reported to be expressed at a low level in KS tumor tissue (29). In KSHV-infected PEL cell lines, ANGPT-1 expression was clearly higher than in the other lymphatic cell lines at the RNA level (6), and thus, KSHV and/or the KSHV-infected PEL cell environment could evoke some advantageous effects for the ANGPT-1 expression. On the other hand, an ANGPT-1 receptor, Tie2, was not expressed in KSHV-infected PEL cell lines, and thus, overproduced ANGPT-1 would have an autocrine effect not on these cells but probably on endothelial cells, a putative origin of KS.
Here we provide quantitative evidence that in KSHV-infected PEL cells, ANGPT-1 is localized in the cytoplasm and secreted into the culture medium (Fig. 1), as we showed in our previous DNA array analysis (6). ANGPT-1 overexpression was unique in KSHV infected PEL cell lines, since KSHV-uninfected BL cell lines originated from a B cell lineage, as PEL cells did not show high expression of ANGPT-1. As for ANGPT-2, which was highly expressed in KSHV-infected endothelial cells (26), there was no difference in its production among these cell lines. In order to understand how ANGPT-1 was upregulated, reporter constructs were prepared with the regulatory sequence. The regulatory region of ANGPT-1 up to nt −898 showed high activity in all KSHV-infected PEL cell lines but not in BJAB cells. There was no significant enhancement of transcriptional activity in BJAB cells, but the presence of the full KSHV genome in a BAC (BJAB-BAC36) indeed enhanced transcriptional activity. Consistently, a −898 Luc reporter showed high activity in BJAB cells containing the full-length KSHV genome in the BAC (37, 41).
Analysis of a series of deletion mutant constructs containing the ANGPT-1 regulatory region showed that the D1A region (−143 to −125; AAGTGTATTAAGGTGGACT) should be necessary and sufficient for upregulation of ANGPT-1 transcriptional activity in KSHV-infected PEL cell lines (Fig. 4). In addition, we demonstrated that some nuclear proteins of KSHV-infected cells, but not uninfected cells, could bind the D1A element (Fig. 5A). Mutation analysis of the D1A sequence further revealed that the core element should be 5′-GTATTAAG-3′ and some 5′-GTGG-3′ sequences that were mutated in Mut5 might be involved, since the Mut5 reporter plasmid did not show upregulated activity (Fig. 5B), although the fragment competed with a factor binding to the D1A probe in an EMSA (Fig. 5A).
Among the factors nominated (IRF4, RFX-5, and OCT-1) on the basis of the MS results, IRF4 and RFX-5 are well expressed in the KSHV-infected PEL cell lines (6, 42). While studies of IRF4 and RFX-5 overexpression and supershift analyses with specific antibodies against IRF4 and RFX-5, respectively, did not show that these factors are involved in ANGPT-1 upregulation in KSHV-infected PEL cell lines, an anti-OCT-1 antibody caused a supershift (Fig. 6A and B). The 5′-GTATTAAGG-3′ sequence is partially contained in 5′-GYATGNTAATGARATTCY-3′, which is bound with OCT-1 interacting with the HSV VP16 transactivator (the underlined nucleotides match). Furthermore, ChIP with an OCT-1 antibody followed by qPCR enriched the D1A region compared with the same experiment with normal mouse IgG. Collectively, these data suggested that OCT-1 should be involved in ANGPT-1 upregulation in KSHV-infected PEL cell lines in vitro and in vivo.
As noted, KSHV infection and/or the PEL cell environment could affect ANGPT-1 upregulation. We tested whether several well-known latent genes of KSHV, such as LANA, vCYC, vFLIP, and vIRF3, were involved in the upregulation. However, none of the genes tested showed any activation on the D1A reporter construct (data not shown). Thus, we do not know how KSHV is involved in this upregulation, though kaposin has not been tested because of cloning failure. Such an environment governing PEL might be achieved epigenetically in the long course of PEL establishment. Namely, in PEL cells, the ANGPT-1 regulatory region could be more accessible to OCT-1 than in KSHV-uninfected BL or other cells.
Since the ANGPT-1 receptor Tie2 is not expressed on PEL cell lines (data not shown), ANGPT-1 overproduction from PEL cells should not promote PEL cell growth itself but affect pathophysiology such as exudation/effusion of body fluid and angiogenesis in AIDS patients. In order to know what ANGPT-1 does in AIDS patients, it will be interesting to measure ANGPT-1 in PEL harboring AIDS patients with or without KS and to evaluate their prognosis.
Taking these results together, we showed that ANGPT-1 is activated through an element termed D1A, in which the core is 5′-GTATTAAG-3′, and this core is bound with OCT-1 in KSHV-infected PEL cell lines. Though further investigation must be done to understand the mechanism, ANGPT-1 overproduction may profoundly affect the pathophysiology of AIDS patients with PEL.
REFERENCES
- 1.Chang Y, Cesarman E, Pessin MS, Lee F, Culpepper J, Knowles DM, Moore PS. 1994. Identification of herpesvirus-like DNA sequences in AIDS-associated Kaposi's sarcoma. Science 266:1865–1869. doi: 10.1126/science.7997879. [DOI] [PubMed] [Google Scholar]
- 2.Kingma DW, Weiss WB, Jaffe ES, Kumar S, Frekko K, Raffeld M. 1996. Epstein-Barr virus latent membrane protein-1 oncogene deletions: correlations with malignancy in Epstein-Barr virus–associated lymphoproliferative disorders and malignant lymphomas. Blood 88:242–251. [PubMed] [Google Scholar]
- 3.Bchini R, Capel F, Dauguet C, Dubanchet S, Petit MA. 1990. In vitro infection of human hepatoma (HepG2) cells with hepatitis B virus. J Virol 64:3025–3032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cesarman E, Chang Y, Moore PS, Said JW, Knowles DM. 1995. Kaposi's sarcoma-associated herpesvirus-like DNA sequences in AIDS-related body-cavity-based lymphomas. N Engl J Med 332:1186–1191. doi: 10.1056/NEJM199505043321802. [DOI] [PubMed] [Google Scholar]
- 5.Soulier J, Grollet L, Oksenhendler E, Cacoub P, Cazals-Hatem D, Babinet P, d'Agay MF, Clauvel JP, Raphael M, Degos L, Sigaux F. 1995. Kaposi's sarcoma-associated herpesvirus-like DNA sequences in multicentric Castleman's disease. Blood 86:1276–1280. [PubMed] [Google Scholar]
- 6.Ueda K, Ito E, Karayama M, Ohsaki E, Nakano K, Watanabe S. 2010. KSHV-infected PEL cell lines exhibit a distinct gene expression profile. Biochem Biophys Res Commun 394:482–487. doi: 10.1016/j.bbrc.2010.02.122. [DOI] [PubMed] [Google Scholar]
- 7.Chang HH, Ganem D. 2013. A unique herpesviral transcriptional program in KSHV-infected lymphatic endothelial cells leads to mTORC1 activation and rapamycin sensitivity. Cell Host Microbe 13:429–440. doi: 10.1016/j.chom.2013.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Matteoli B, Broccolo F, Scaccino A, Cottoni F, Angeloni A, Faggioni A, Ceccherini-Nelli L. 2012. In vivo and in vitro evidence for an association between the route-specific transmission of HHV-8 and the virus genotype. J Med Virol 84:786–791. doi: 10.1002/jmv.23246. [DOI] [PubMed] [Google Scholar]
- 9.Vieira J, O'Hearn PM. 2004. Use of the red fluorescent protein as a marker of Kaposi's sarcoma-associated herpesvirus lytic gene expression. Virology 325:225–240. doi: 10.1016/j.virol.2004.03.049. [DOI] [PubMed] [Google Scholar]
- 10.Grundhoff A, Ganem D. 2003. The latency-associated nuclear antigen of Kaposi's sarcoma-associated herpesvirus permits replication of terminal repeat-containing plasmids. J Virol 77:2779–2783. doi: 10.1128/JVI.77.4.2779-2783.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ohsaki E, Ueda K, Sakakibara S, Do E, Yada K, Yamanishi K. 2004. Poly(ADP-ribose) polymerase 1 binds to Kaposi's sarcoma-associated herpesvirus (KSHV) terminal repeat sequence and modulates KSHV replication in latency. J Virol 78:9936–9946. doi: 10.1128/JVI.78.18.9936-9946.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Baresova P, Musilova J, Pitha PM, Lubyova B. 2014. p53 tumor suppressor protein stability and transcriptional activity are targeted by Kaposi's sarcoma-associated herpesvirus-encoded viral interferon regulatory factor 3. Mol Cell Biol 34:386–399. doi: 10.1128/MCB.01011-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chang LS, Wang JT, Doong SL, Lee CP, Chang CW, Tsai CH, Yeh SW, Hsieh CY, Chen MR. 2012. Epstein-Barr virus BGLF4 kinase downregulates NF-kappaB transactivation through phosphorylation of coactivator UXT. J Virol 86:12176–12186. doi: 10.1128/JVI.01918-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rangaswamy US, Speck SH. 2014. Murine gammaherpesvirus M2 protein induction of IRF4 via the NFAT pathway leads to IL-10 expression in B cells. PLoS Pathog 10:e1003858. doi: 10.1371/journal.ppat.1003858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bieleski L, Talbot SJ. 2001. Kaposi's sarcoma-associated herpesvirus vCyclin open reading frame contains an internal ribosome entry site. J Virol 75:1864–1869. doi: 10.1128/JVI.75.4.1864-1869.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sarid R, Wiezorek JS, Moore PS, Chang Y. 1999. Characterization and cell cycle regulation of the major Kaposi's sarcoma-associated herpesvirus (human herpesvirus 8) latent genes and their promoter. J Virol 73:1438–1446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Muralidhar S, Pumfery AM, Hassani M, Sadaie MR, Kishishita M, Brady JN, Doniger J, Medveczky P, Rosenthal LJ. 1998. Identification of kaposin (open reading frame K12) as a human herpesvirus 8 (Kaposi's sarcoma-associated herpesvirus) transforming gene. J Virol 72:4980–4988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ohsaki E, Suzuki T, Karayama M, Ueda K. 2009. Accumulation of LANA at nuclear matrix fraction is important for Kaposi's sarcoma-associated herpesvirus replication in latency. Virus Res 139:74–84. doi: 10.1016/j.virusres.2008.10.011. [DOI] [PubMed] [Google Scholar]
- 19.Watanabe T, Sugaya M, Atkins AM, Aquilino EA, Yang A, Borris DL, Brady J, Blauvelt A. 2003. Kaposi's sarcoma-associated herpesvirus latency-associated nuclear antigen prolongs the life span of primary human umbilical vein endothelial cells. J Virol 77:6188–6196. doi: 10.1128/JVI.77.11.6188-6196.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cousins E, Nicholas J. 2013. Role of human herpesvirus 8 interleukin-6-activated gp130 signal transducer in primary effusion lymphoma cell growth and viability. J Virol 87:10816–10827. doi: 10.1128/JVI.02047-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bais C, Santomasso B, Coso O, Arvanitakis L, Raaka EG, Gutkind JS, Asch AS, Cesarman E, Gershengorn MC, Mesri EA. 1998. G-protein-coupled receptor of Kaposi's sarcoma-associated herpesvirus is a viral oncogene and angiogenesis activator. Nature 391:86–89. doi: 10.1038/34193. [DOI] [PubMed] [Google Scholar]
- 22.Poole LJ, Zong JC, Ciufo DM, Alcendor DJ, Cannon JS, Ambinder R, Orenstein JM, Reitz MS, Hayward GS. 1999. Comparison of genetic variability at multiple loci across the genomes of the major subtypes of Kaposi's sarcoma-associated herpesvirus reveals evidence for recombination and for two distinct types of open reading frame K15 alleles at the right-hand end. J Virol 73:6646–6660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bala K, Bosco R, Gramolelli S, Haas DA, Kati S, Pietrek M, Havemeier A, Yakushko Y, Singh VV, Dittrich-Breiholz O, Kracht M, Schulz TF. 2012. Kaposi's sarcoma herpesvirus K15 protein contributes to virus-induced angiogenesis by recruiting PLCgamma1 and activating NFAT1-dependent RCAN1 expression. PLoS Pathog 8:e1002927. doi: 10.1371/journal.ppat.1002927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ueda K, Ishikawa K, Nishimura K, Sakakibara S, Do E, Yamanishi K. 2002. Kaposi's sarcoma-associated herpesvirus (human herpesvirus 8) replication and transcription factor activates the K9 (vIRF) gene through two distinct cis elements by a non-DNA-binding mechanism. J Virol 76:12044–12054. doi: 10.1128/JVI.76.23.12044-12054.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ensoli B, Sturzl M, Monini P. 2000. Cytokine-mediated growth promotion of Kaposi's sarcoma and primary effusion lymphoma. Semin Cancer Biol 10:367–381. doi: 10.1006/scbi.2000.0329. [DOI] [PubMed] [Google Scholar]
- 26.Ye FC, Blackbourn DJ, Mengel M, Xie JP, Qian LW, Greene W, Yeh IT, Graham D, Gao SJ. 2007. Kaposi's sarcoma-associated herpesvirus promotes angiogenesis by inducing angiopoietin-2 expression via AP-1 and Ets1. J Virol 81:3980–3991. doi: 10.1128/JVI.02089-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ye FC, Zhou FC, Nithianantham S, Chandran B, Yu XL, Weinberg A, Gao SJ. 2013. Kaposi's sarcoma-associated herpesvirus induces rapid release of angiopoietin-2 from endothelial cells. J Virol 87:6326–6335. doi: 10.1128/JVI.03303-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Vart RJ, Nikitenko LL, Lagos D, Trotter MW, Cannon M, Bourboulia D, Gratrix F, Takeuchi Y, Boshoff C. 2007. Kaposi's sarcoma-associated herpesvirus-encoded interleukin-6 and G-protein-coupled receptor regulate angiopoietin-2 expression in lymphatic endothelial cells. Cancer Res 67:4042–4051. doi: 10.1158/0008-5472.CAN-06-3321. [DOI] [PubMed] [Google Scholar]
- 29.Brown LF, Dezube BJ, Tognazzi K, Dvorak HF, Yancopoulos GD. 2000. Expression of Tie1, Tie2, and angiopoietins 1, 2, and 4 in Kaposi's sarcoma and cutaneous angiosarcoma. Am J Pathol 156:2179–2183. doi: 10.1016/S0002-9440(10)65088-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wang L, Damania B. 2008. Kaposi's sarcoma-associated herpesvirus confers a survival advantage to endothelial cells. Cancer Res 68:4640–4648. doi: 10.1158/0008-5472.CAN-07-5988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nishimura K, Ueda K, Sakakibara S, Do E, Ohsaki E, Okuno T, Yamanishi K. 2003. A viral transcriptional activator of Kaposi's sarcoma-associated herpesvirus (KSHV) induces apoptosis, which is blocked in KSHV-infected cells. Virology 316:64–74. doi: 10.1016/S0042-6822(03)00582-8. [DOI] [PubMed] [Google Scholar]
- 32.Guilluy C, Zhang Z, Bhende PM, Sharek L, Wang L, Burridge K, Damania B. 2011. Latent KSHV infection increases the vascular permeability of human endothelial cells. Blood 118:5344–5354. doi: 10.1182/blood-2011-03-341552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jong JE, Park J, Kim S, Seo T. 2010. Kaposi's sarcoma-associated herpesvirus viral protein kinase interacts with RNA helicase a and regulates host gene expression. J Microbiol 48:206–212. doi: 10.1007/s12275-010-0021-1. [DOI] [PubMed] [Google Scholar]
- 34.Davis S, Aldrich TH, Jones PF, Acheson A, Compton DL, Jain V, Ryan TE, Bruno J, Radziejewski C, Maisonpierre PC, Yancopoulos GD. 1996. Isolation of angiopoietin-1, a ligand for the TIE2 receptor, by secretion-trap expression cloning. Cell 87:1161–1169. doi: 10.1016/S0092-8674(00)81812-7. [DOI] [PubMed] [Google Scholar]
- 35.Koh GY. 2013. Orchestral actions of angiopoietin-1 in vascular regeneration. Trends Mol Med 19:31–39. doi: 10.1016/j.molmed.2012.10.010. [DOI] [PubMed] [Google Scholar]
- 36.Brown C, Gaspar J, Pettit A, Lee R, Gu X, Wang H, Manning C, Voland C, Goldring SR, Goldring MB, Libermann TA, Gravallese EM, Oettgen P. 2004. ESE-1 is a novel transcriptional mediator of angiopoietin-1 expression in the setting of inflammation. J Biol Chem 279:12794–12803. doi: 10.1074/jbc.M308593200. [DOI] [PubMed] [Google Scholar]
- 37.Ueda K, Sakakibara S, Ohsaki E, Yada K. 2006. Lack of a mechanism for faithful partition and maintenance of the KSHV genome. Virus Res 122:85–94. doi: 10.1016/j.virusres.2006.07.002. [DOI] [PubMed] [Google Scholar]
- 38.Augustin HG, Koh GY, Thurston G, Alitalo K. 2009. Control of vascular morphogenesis and homeostasis through the angiopoietin-Tie system. Nat Rev Mol Cell Biol 10:165–177. doi: 10.1038/nrm2639. [DOI] [PubMed] [Google Scholar]
- 39.Huang H, Bhat A, Woodnutt G, Lappe R. 2010. Targeting the ANGPT-TIE2 pathway in malignancy. Nat Rev Cancer 10:575–585. doi: 10.1038/nrc2894. [DOI] [PubMed] [Google Scholar]
- 40.Fagiani E, Lorentz P, Kopfstein L, Christofori G. 2011. Angiopoietin-1 and -2 exert antagonistic functions in tumor angiogenesis, yet both induce lymphangiogenesis. Cancer Res 71:5717–5727. doi: 10.1158/0008-5472.CAN-10-4635. [DOI] [PubMed] [Google Scholar]
- 41.Zhou FC, Zhang YJ, Deng JH, Wang XP, Pan HY, Hettler E, Gao SJ. 2002. Efficient infection by a recombinant Kaposi's sarcoma-associated herpesvirus cloned in a bacterial artificial chromosome: application for genetic analysis. J Virol 76:6185–6196. doi: 10.1128/JVI.76.12.6185-6196.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Forero A, Moore PS, Sarkar SN. 2013. Role of IRF4 in IFN-stimulated gene induction and maintenance of Kaposi sarcoma-associated herpesvirus latency in primary effusion lymphoma cells. J Immunol 191:1476–1485. doi: 10.4049/jimmunol.1202514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tanaka M, Herr W. 1990. Differential transcriptional activation by Oct-1 and Oct-2: interdependent activation domains induce Oct-2 phosphorylation. Cell 60:375–386. [DOI] [PubMed] [Google Scholar]