ABSTRACT
Classical swine fever virus (CSFV) is the causative agent of classical swine fever (CSF), a highly contagious, economically important viral disease in many countries. The Erns and E2 envelope glycoproteins are responsible for the binding to and entry into the host cell by CSFV. To date, only one cellular receptor, heparan sulfate (HS), has been identified as being involved in CSFV attachment. HS is also present on the surface of various cells that are nonpermissive to CSFV. Hence, there must be another receptor(s) that has been unidentified to date. In this study, we used a set of small interfering RNAs (siRNAs) against a number of porcine cell membrane protein genes to screen cellular proteins involved in CSFV infection. This approach resulted in the identification of several proteins, and of these, the laminin receptor (LamR) has been demonstrated to be a cellular receptor for several viruses. Confocal analysis showed that LamR is colocalized with CSFV virions on the membrane, and a coimmunoprecipitation assay indicated that LamR interacts with the CSFV Erns protein. In inhibition assays, anti-LamR antibodies, soluble laminin, or LamR protein significantly inhibited CSFV infection in a dose-dependent manner. Transduction of PK-15 cells with a recombinant lentivirus expressing LamR yielded higher viral titers. Moreover, an attachment assay demonstrated that LamR functions during virus attachment. We also demonstrate that LamR acts as an alternative attachment receptor, especially in SK6 cells. These results indicate that LamR is a cellular attachment receptor for CSFV.
IMPORTANCE Classical swine fever virus (CSFV) is the causative agent of classical swine fever (CSF), an economically important viral disease affecting the pig industry in many countries. To date, only heparan sulfate (HS) has been identified to be an attachment receptor for CSFV. Here, using RNA interference screening with small interfering RNAs (siRNAs) against a number of porcine membrane protein genes, we identified the laminin receptor (LamR) to be another attachment receptor. We demonstrate the involvement of LamR together with HS in virus attachment, and we elucidate the relationship between LamR and HS. LamR also serves as an attachment receptor for many viral pathogens, including dengue virus, a fatal human flavivirus. The study will help to enhance our understanding of the life cycle of flaviviruses and the development of antiviral strategies for flaviviruses.
INTRODUCTION
Classical swine fever virus (CSFV) is the causative agent of classical swine fever (CSF), a highly contagious and often fatal viral disease in pigs. The disease leads to significant economic losses in many countries. CSFV is a member of the Pestivirus genus within the Flaviviridae family (1). The virus possesses a single-stranded, positive-sense RNA genome of approximately 12.3 kb (2, 3). Its genome contains a single, large open reading frame that encodes a precursor polyprotein of 3,898 amino acids (aa). The polyprotein is co- and posttranslationally processed by viral and cellular proteases, giving rise to four structural proteins (C, Erns, E1, and E2) and eight nonstructural proteins (Npro, p7, NS2, NS3, NS4A, NS4B, NS5A, and NS5B) (4, 5).
The envelope glycoproteins Erns and E2 are involved in CSFV infection. Erns and E2 are present on the outer surface of the virion (6, 7) and are recognized to be the main targets for neutralizing antibodies (6, 8). They are thus inferred to be responsible for the attachment and entry of CSFV. Soluble Erns and E2 proteins could inhibit CSFV infection and are inferred to interact with different unknown cell surface receptors (9). Similar inhibition was observed with anti-E2 or anti-Erns monoclonal antibodies (MAbs) (10). Analysis of an overlapping peptide library (with the Erns, E1, and E2 proteins displayed on phage surfaces) resulted in the discovery of two peptides (one from Erns and the other from E2) that could bind to host cells with a high affinity and also inhibit the binding of CSFV to cells (11). These findings show that infection with CSFV is highly associated with Erns and E2, which bind with cellular receptors during virus entry.
Viruses rely on the host cell to complete the viral life cycle. Viral replication starts with specific interactions of virion constituents with cellular surface components, i.e., cellular receptors. The interactions between viral attachment proteins and cellular receptors are thought to determine the tissue tropism and host range for viruses. More importantly, antiviral strategies can be designed to prevent virus invasion by blocking the virus-receptor interaction. Thus, the study of cellular receptors can contribute to the understanding of viral entry mechanisms and provide targets for novel antivirals.
Heparan sulfate (HS) is a common nonprotein receptor. It is ubiquitously present on the surface of many cell types and used by various viruses for attachment (12–16). Protein receptors vary greatly, depending on the virus. One virus (such as hepatitis C virus [HCV] [17–23]) may exploit different receptors and entry mechanisms. This strategy is considered to facilitate virus infection. When one receptor is blocked, HCV can still infect the host cell. Some viruses share the same receptors (such as CD46 [24–28]), suggesting that similar entry mechanisms are shared by different viruses. On the basis of this observation, novel cellular receptors may be identified by high-throughput screening in screening assays with the appropriate design.
To date, only HS has been identified to be responsible for the attachment of CSFV to SK6 cells. However, CSFV can still infect SK6 cells even after a single amino acid mutation in Erns removes CSFV's ability to attach to HS (29, 30). In addition, the wide distribution of HS does not match the narrow host range of CSFV. Hence, there must be another unidentified cellular receptor(s) that enables CSFV infection.
The aim of this study was to screen membrane proteins that are critical for CSFV infection using pooled small interfering RNAs (siRNAs) against 29 porcine genes that encode cell membrane proteins. These proteins included defined cellular receptors or proteins that are critical for the infection of members of the Flaviviridae family. We demonstrate here, for the first time, that the laminin receptor (LamR) is a cellular receptor for CSFV and is responsible for its attachment to the cell.
MATERIALS AND METHODS
Cells and viruses.
CSFV-permissive porcine kidney cell lines (PK-15 and SK6) and nonpermissive cell lines (BHK-21, CHO-K1, HEK293T, HEK293, HeLa, Marc-145, MDBK, MDCK, NIH/3T3, and Vero) were cultured in Dulbecco's modified Eagle medium (DMEM; Gibco) supplemented with 10% fetal bovine serum (HyClone) free of bovine viral diarrhea virus (BVDV) and anti-BVDV antibodies.
CSFV-NproFluc, a reporter CSFV strain rescued from the infectious cDNA clone pBRCISM-NproFluc stably expressing the firefly luciferase (Fluc) within the Npro gene (31), was used for siRNA screening. The CSFV Shimen strain (genogroup 1) and the pseudorabies virus (PRV) TJ strain (32) were propagated in PK-15 cells. The CSFV Tianjin/10 strain (genogroup 2) is a tissue-derived virus without a cell culture history.
RNA interference (RNAi) screening.
We designed three siRNA molecules (one for LamR) for 29 membrane protein genes and pooled them for transfection. PK-15 cells were transfected with these siRNAs at a final concentration of 200 nM for 48 h in 48-well plates. Reporter viruses were then infected for another 48 h and assayed for Fluc activity by using a PE Envision system. The screening was run in triplicate. All of the specific siRNAs targeting porcine membrane protein genes were synthesized by Genepharma (sequences will be provided upon request). All of the candidate sequences targeting the genes were searched for in the GenBank database using the BLAST program to avoid homologous sequences in the pig genome.
For testing of siRNAs, PK-15 cells grown to 60% confluence were transfected with the siRNAs indicated below at a final concentration of 200 nM in 48-well plates using the X-tremeGene siRNA transfection reagent (Roche) in accordance with the manufacturer's protocol. After 48 h, the cells were infected with 100 50% tissue culture infective doses (TCID50) CSFV-NproFluc. At 48 h postinfection (hpi), the Fluc activity was assayed as described below.
Preparation of membrane proteins.
Crude membrane fractions of PK-15 cells were prepared as described previously (33). The cells were washed three times with phosphate-buffered saline (PBS) and then incubated for 30 min in 0.01 M triethanolamine hydrochloride, pH 7.5 (TEA buffer), containing 8.5% sucrose, 1 mM phenylmethylsulfonyl fluoride, and 1 mM N-ethylmaleimide and homogenized with 10 strokes in a Dounce homogenizer. Homogenates from the cells were centrifuged at 1,200 × g for 5 min; the supernatants were pelleted at 20,000 × g for 1 h and resuspended in 8.5% sucrose in TEA buffer. After centrifugation at 130,000 × g for 3 h in an SW32 rotor, the cell membranes were collected from the 32%-40% interface and pelleted at 20,000 × g for 1.5 h. For cells transfected with siRNAs against LamR, a ProteoExtract transmembrane protein extraction kit (Merck) was used for membrane extraction. Briefly, the cells were washed with wash buffer and then incubated with extraction buffers I and II (in the Merck kit). The membrane proteins were dissolved in extraction buffer II.
Fluc activity assay.
For time course analysis of Fluc expression, infections of PK-15 cells with the reporter virus were performed in 48-well plates. The supernatant was removed, and the infected cells were collected at the time points indicated below. After washing with cold PBS, the cells in each well were lysed with 100 μl of passive lysis buffer (Promega). This was followed by incubation on a shaker for 20 min at 4°C. Then the lysate was centrifuged for 5 min at 10,000 × g and 4°C. For all other experiments, performed in 96-well plates, 30 μl of passive lysis buffer was added to the cells. The supernatant was assayed for Fluc activity using a dual-luciferase reporter assay system (Promega), and Fluc activity was measured with a TD-20/20 luminometer (Turner Designs) in accordance with the manufacturer's instructions.
Flow cytometry.
PK-15 cells grown to 60% confluence were harvested and fixed in 4% formaldehyde for 30 min at room temperature. After three washes with PBS, the cells were blocked with 5% bovine serum albumin (BSA) in PBS and incubated for 1 h at room temperature. After washing, the cells were resuspended in PBS and incubated with the anti-LamR antibodies (Abcam) or irrelevant rabbit IgG (Abcam) for 2 h at 4°C, followed by fluorescein isothiocyanate (FITC)-labeled goat anti-rabbit secondary antibody (Santa) for 1 h. The cells were subjected to flow cytometry analysis after washing with PBS.
Coimmunoprecipitation (co-IP).
HEK293T cells were transfected with plasmids carrying the Flag-tagged Erns gene or Flag-tagged E2 gene. The transfected cells were harvested at 48 h posttransfection (hpt), washed three times with cold PBS, and lysed with NP-40 buffer (Beyotime) containing 1 mM phenylmethylsulfonyl fluoride (Beyotime) for 1 h at 4°C. Finally, the lysate was centrifuged at 12,000 × g for 10 min at 4°C. The lysate was mixed with the membrane or cytoplasmic extracts of PK-15 cells prepared as described above and then precleared with protein G beads (Roche) plus anti-Flag MAb (Sigma) for 4 h at 4°C. The beads were then washed with NP-40 buffer and boiled in loading buffer (Beyotime). These samples were then subjected to SDS-PAGE, followed by immunoblotting analysis using anti-Flag and anti-LamR antibodies (Abcam).
Confocal imaging.
PK-15 cells grown to 60% confluence were treated with heparinase I (New England BioLabs) for 2 h at room temperature and then inoculated with CSFV at a multiplicity of infection (MOI) of 10 for 2 h on ice. Cells were fixed with 4% paraformaldehyde in PBS for 30 min. The cells were blocked with 5% skimmed milk for 1 h and then incubated with homemade anti-Erns and anti-LamR antibodies (Abcam) for 1 h. Following 1 h of incubation with anti-mouse IgG-FITC antibody (Sigma) and anti-rabbit IgG-tetramethyl rhodamine isocyanate antibody (Sigma), the cells were stained with 4′,6-diamidino-2-phenylindole (DAPI) for 15 min and examined using a Leica SP2 confocal system (Leica Microsystems).
Inhibition assay.
PK-15 or SK6 cells grown in 24-well plates were incubated with polyclonal antibody (PAb) against LamR or soluble laminin (Sigma) at various concentrations at 37°C for 1 h. Following incubation, the cells were washed three times with PBS and inoculated with CSFV at an MOI of 1 for 2 h at 4°C. Unbound virus was removed by washing three times with PBS. The infected cells were cultured for a further 24 h. The culture medium was collected and titrated as described below.
CSFV (5,000 TCID50) was incubated with various concentrations of soluble recombinant LamR (Abcam) for 1 h at 4°C. The viral suspension was then added to PK-15 cells in 24-well plates at 4°C for 2 h. Unbound viruses were removed by washing three times with PBS. The infected cells were cultured for a further 24 h. The culture medium was collected and titrated as described below.
Gene knockdown by shRNAs.
Duplexes of synthesized oligonucleotides containing target sequences were inserted into the lentivirus vector pLVX-shRNA2 (Clontech). To generate lentiviruses for short hairpin RNA (shRNA) delivery, HEK293T cells plated in poly-l-lysine (PLL)-coated 10-cm dishes were transfected with 21 μg of pLVX-LamR-shRNA2 and the packaging plasmids psPAX2 (14 μg) and pMD2.0G (7 μg). At 48 hpt, the culture supernatant was concentrated by centrifugation at 4,500 × g for 20 min at 4°C using an Amicon Ultra-15 centrifugal filter unit with an Ultracel-10 membrane (Millipore). Lentiviral titers on PK-15 or SK6 cells were determined. Serial dilutions of the virus were applied to the cells, and infectivity was determined after 72 h by fluorescence microscopy for green fluorescent protein (GFP) expression. To produce shRNA-expressing cells, PK-15 and SK6 cells were treated with lentivirus at an MOI of 0.1 with culture medium containing 8 μg/ml Polybrene for 48 h. After washing with PBS, the transduced cells were passaged and cultured as described above. LamR expression in PK-15 or SK6 cells was tested by Western blotting using rabbit anti-LamR PAb (1:1,000; Abcam).
Construction of stable cell lines overexpressing LamR.
To construct a stable cell line overexpressing LamR, the LamR gene was cloned and inserted into the pFUGW vector (Addgene) using SfiI to generate the pFUGW-LamR plasmid. HEK293T cells grown in 10-cm dishes were transfected with the recombinant plasmid pFUGW-LamR, the empty vector pFUGW, or the packaging plasmids psPAX2 and pMD2.0G. At 48 hpt, the culture supernatant was concentrated and titrated as described above. The concentrated pseudovirus was transduced into PK-15 or SK6 cells in 6-well plates. Expression of LamR tagged with enhanced GFP (EGFP) in PK-15 or SK6 cells was tested by Western blotting using rabbit anti-LamR PAb (1:1,000; Abcam).
IFA and virus titration.
The titers of CSFV and PRV were determined by an indirect immunofluorescence assay (IFA). Briefly, an IFA-based viral titration assay was performed by infecting PK-15 cells seeded in 96-well plates (approximately 5 × 104 cells/well) with 10-fold serial dilutions and four replicates for each dilution. After a further 48-h incubation, PK-15 cells were washed with PBS, fixed with 4% paraformaldehyde, and permeabilized with 0.1% Triton X-100. The fixed cells were incubated with anti-E2/gB serum for 2 h at 37°C, washed three times with PBS, and then incubated with FITC-labeled goat anti-pig IgG (1:100 dilution in PBS; Sigma) for 1 h at 37°C. After washing three times with PBS, the cells were examined under a fluorescence microscope (TE2000U; Nikon) with a video documentation system. Viral titers were calculated in accordance with the Reed-Muench method and expressed as the number of medium TCID50/ml.
Virus attachment assay and real-time RT-PCR.
The attachment assay was performed as described previously (34–36). Cells in 24-well plates were treated with 1,000 mIU heparinase I (New England BioLabs) for 2 h at room temperature with gentle shaking. The cells were then incubated with CSFV at an MOI of 5 for 2 h at 4°C with gentle agitation. The cells were washed five times with cold PBS to remove the unbound CSFV. The total viral RNA was extracted from CSFV-bound cells using the TRIzol reagent (Invitrogen) and treated with DNase I (TaKaRa) to remove potential genomic DNA contamination. Synthesis of cDNA was performed with 200 ng of total RNA and 20 U of Moloney murine leukemia virus (M-MLV) reverse transcriptase (TaKaRa), 200 μM deoxynucleoside triphosphates (TaKaRa), and 4 μl of 5× M-MLV reverse transcriptase buffer (in the TaKaRa kit) in a total volume of 20 μl. The mixture was incubated for 1 h at 42°C and then for 15 min at 75°C. Quantification of CSFV genomic copies was performed by a previously described real-time reverse transcription-PCR (RT-PCR) assay (37), performed on a Stratgene Mx real-time quantitative PCR system (Agilent Technologies) according to the manufacturer's instructions.
Infection assay in cells treated with heparinase I.
Approximately 104 cells in 96-well plates were washed three times with the binding buffer (PBS containing 0.2% BSA, 0.5 mM CaCl2, and 0.5 mM MgCl2). The cells were then incubated with 100 μl of the binding buffer containing 100 mIU of heparinase I (New England BioLabs) or left untreated. After incubation for 2 h at room temperature with gentle shaking, the enzyme solution was removed and the cells were washed three times with PBS. The cells were then inoculated with 100 TCID50 CSFV-NproFluc on ice. At different time points postinfection (30, 60, 90, and 120 min), the virus was removed and the cells were washed three times, supplied with overlay medium, and incubated for a further 48 h at 37°C. All of the relative infection rates are calculated on the basis of the Fluc activity in triplicate wells.
Statistical analysis.
Correlations within cohorts were evaluated using the Spearman correlation test. Differences between groups were examined for statistical significance using Student's t test. An unadjusted P value of less than 0.05 was considered to be significant.
RESULTS
Membrane proteins required for CSFV infection were screened with siRNAs.
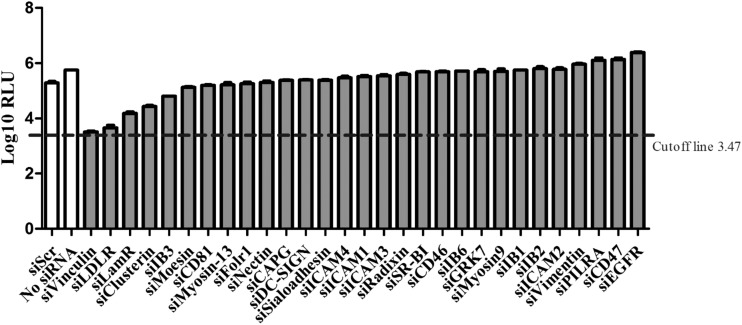
To identify the cellular membrane factors that function during CSFV infection, we selected 29 membrane protein genes for RNAi screening with the reporter virus CSFV-NproFluc. These selected membrane proteins have been identified to be cellular receptors for viruses within the Flaviviridae family, cellular receptors commonly used by various viruses, or virus-host interaction partners of great importance. Therefore, these proteins were selected as having the potential to influence CSFV proliferation. We selected the candidate proteins on the basis of the findings of our previous study (31). Due to the high sensitivity of the Fluc assay, only those candidates with Fluc activity at least 10 times lower than that of the control transfected with scramble siRNA (siScr) were considered to be meaningful.
The siRNA screening suggested that knockdown of the expression of the genes for three proteins had a significant impact on CSFV infection (Fig. 1). These membrane proteins were low-density lipoprotein receptor (LDLr), LamR, and vinculin. Among these, LamR has been identified to be a cellular receptor for different pathogens (38–47). As shown in Fig. 1, siRNA-mediated knockdown of LamR expression reduced Fluc activity by nearly 90%. The results strongly suggested that LamR is critical for CSFV infection.
FIG 1.
Overview of pool of siRNAs against porcine membrane protein genes used in the screening to identify genes required for CSFV infection. The cutoff line represents the Fluc activity of 1 fluorescence focus (31). RLU, relative light units; PILRA, paired immunoglobulin-like type 2 receptor alpha; EGFR, epidermal growth factor receptor.
Confirmation of primary screening.
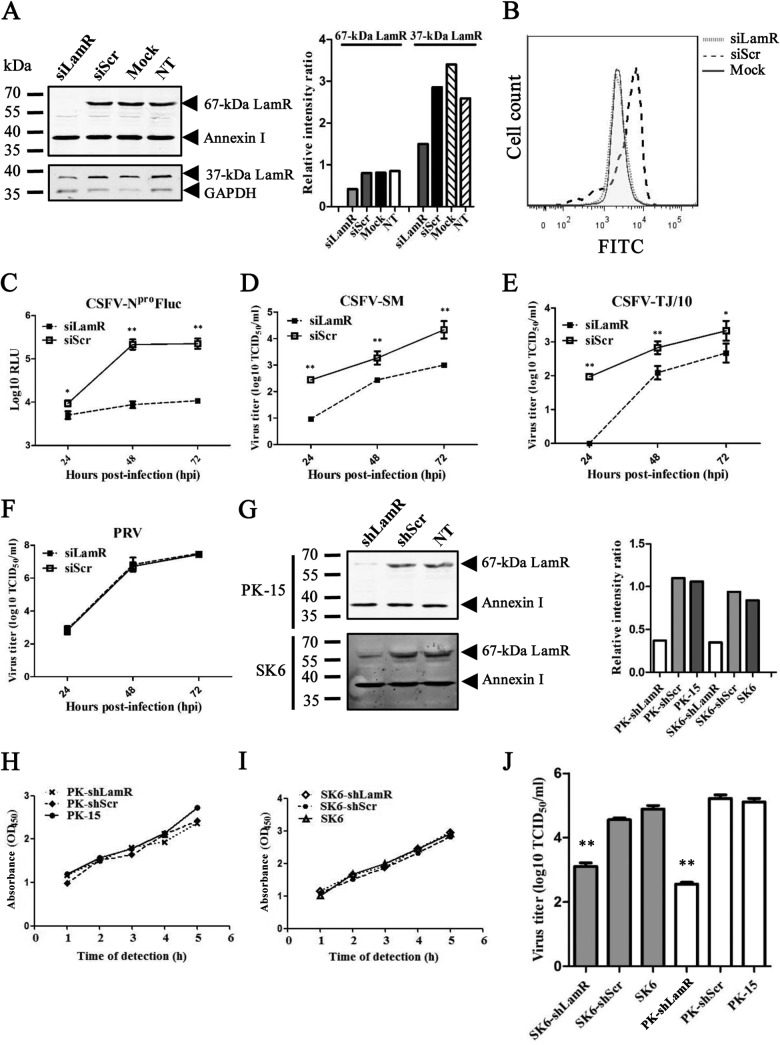
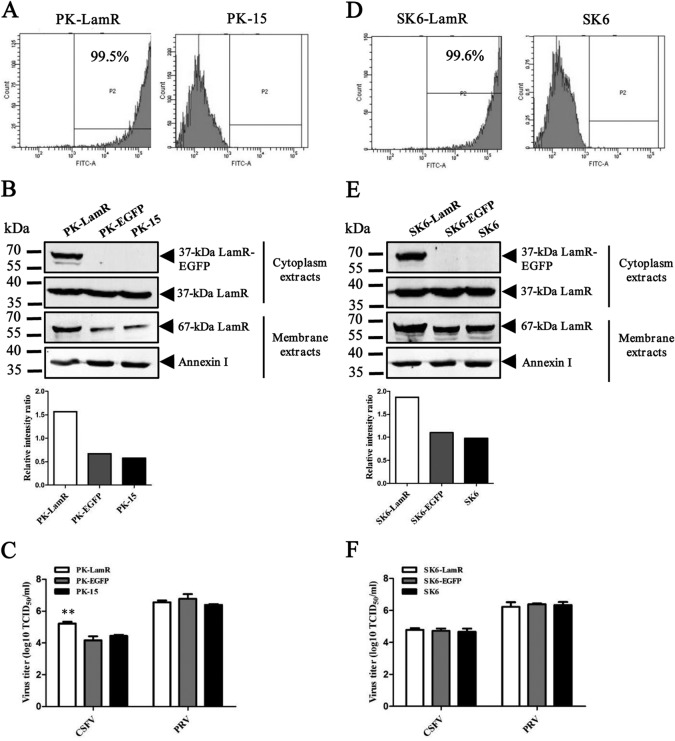
The screening results were first validated using the corresponding siRNAs. The LamR gene can be translated into two different forms: one of approximately 37 kDa located in the cytoplasm and the other of approximately 67 kDa located in the membrane. Western blotting showed that with knockdown of LamR the 67-kDa form was successfully knocked down, but knockdown of LamR showed only a minimal effect on the 37-kDa form. The knockdown effect on the 37-kDa LamR was observed only with a less than 80-ng loading quantity of samples (Fig. 2A). The knockdown effect on the 67-kDa LamR was also confirmed by flow cytometry analysis (Fig. 2B). There was no signal in PK-15 cells transfected with siRNAs against LamR, but there was a strong signal in untreated PK-15 cells, after staining with anti-LamR antibodies (Fig. 2B). The results described above suggest that the 67-kDa LamR correlates with CSFV infection and is likely to be the protein critical for CSFV. Therefore, we targeted the 67-kDa LamR. We then tested the siRNA with wild-type CSFV of different genogroups and PRV at an MOI of 0.01. All three CSFV strains, the reporter virus CSFV-NproFluc, the CSFV Shimen strain, and the CSFV Tianjin/10 strain, shared a similar growth profile in cells with LamR knockdown (Fig. 2C to E). All of the CSFV strains grew poorly in the cells transfected with the siRNA against LamR (siLamR). The viral titer was much lower in cells transfected with siLamR than in cells transfected with siScr at all three time points (24, 48, and 72 hpi). In contrast, PRV exhibited no difference in replication in those cells. The results suggested that only LamR affects CSFV infection (Fig. 2F). Recombinant lentiviruses expressing shRNA against LamR (shLamR) or scramble shRNA (shScr) were also generated and transduced into PK-15 and SK6 cells (another CSFV-permissive swine kidney cell line). Expression of the 67-kDa LamR was knocked down in both cell lines (Fig. 2G), but there was no significant change in cell viability, as indicated by a 3-(4,5-dimethyl-2-thiazolyl)-2,5-diphenyl-2H-tetrazolium bromide (MTT) assay (Fig. 2H and I). The CSFV Shimen strain was used at an MOI of 1 for testing. The culture supernatant was collected at 24 hpi for study of the first cycle of infection. The CSFV titers were significantly reduced after LamR knockdown in both PK-15 and SK6 cells, with higher inhibitory effects being detected in PK-15 cells than in SK6 cells (Fig. 2J).
FIG 2.
LamR is required for CSFV infection. (A) Reduced LamR expression following the knockdown of LamR expression by siRNA. PK-15 cells were transfected with siLamR or siScr, treated only with the transfection reagent (Mock), or left untreated (not treated [NT]). The membrane protein (including the 67-kDa LamR) was extracted by use of a ProteoExtract transmembrane protein extraction kit (Merck) and subjected to Western blot analysis. The remaining cell lysate was used for detection of the 37-kDa LamR. Annexin I (a membrane protein) and GAPDH (glyceraldehyde-3-phosphate dehydrogenase; a cytoplasmic protein) were included as internal references. (B) Detection of LamR expression on the cell surface of PK-15 cells by flow cytometry. siScr-transfected (siScr) or siLamR-transfected (siLamR) PK-15 cells were incubated with anti-LamR antibodies. PK-15 cells incubated with an irrelevant isotype rabbit antibody (Mock) were used as a negative control. (C) Growth curve of CSFV-NproFluc in cells transfected with siLamR or siScr molecules. Cells were infected with virus at an MOI of 0.01. (D) Growth curve of the CSFV Shimen strain (CSFV-SM) in cells transfected with siLamR or siScr molecules. The cells were infected with virus at an MOI of 0.01. (E) Growth curve of the CSFV Tianjin/10 strain (CSFV-TJ/10; genogroup 2) in cells transfected with siLamR or siScr molecules. Cells were infected with virus at an MOI of 0.01. (F) Growth curve of the PRV TJ strain in cells transfected with siLamR or siScr molecules. Cells were infected with virus at an MOI of 0.01. (G) Knockdown effects in cells stably expressing shLamR or shScr. (H) PK-15 cell viability tested with an MTT assay. OD450, optical density at 450 nm. (I) SK6 cell viability tested with an MTT assay. (J) CSFV infection in PK-15 or SK6 cells stably expressing shLamR (PK-shLamR or SK6-shLamR) or shScr (PK-shScr or SK6-shLamR) or in parent cells. Cells were infected with CSFV at an MOI of 1, and the supernatant was collected at 24 hpi. *, P < 0.05; **, P < 0.01.
LamR interacted with the CSFV Erns protein.
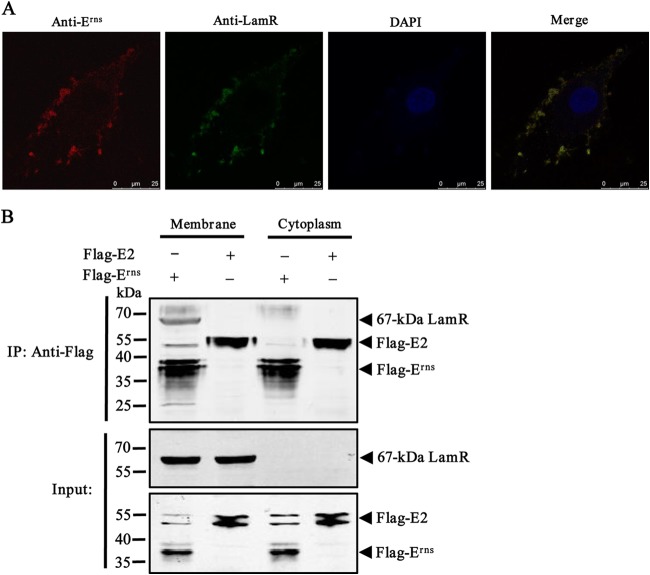
To determine if LamR could act as an interacting partner of CSFV, we verified the interaction between CSFV and LamR by confocal scanning laser microscopy. We treated PK-15 cells with heparinase I and then incubated them with CSFV at an MOI of 10. The results showed that CSFV still attached to the cell surface, and CSFV was highly colocalized with the cell surface LamR that was stained with FITC (Fig. 3A). This binding was specific to LamR but not HS, which is another receptor for CSFV, which had already been removed by heparinase I.
FIG 3.
CSFV envelope protein Erns interacts with LamR. (A) Analysis of the cell surface distribution and colocalization of LamR and cell surface-bound CSFV by indirect immunofluorescence assay in PK-15 cells. (B) Co-IP and Western blotting of the binding of LamR to the CSFV envelope proteins Erns and E2. HEK293T cells were transfected with plasmids expressing either Flag-tagged Erns (Flag-Erns) or Flag-tagged E2 (Flag-E2). The cell lysate was then mixed with membrane or cytoplasm extracts. Co-IP was performed using anti-Flag MAb. The precipitated proteins were analyzed by Western blotting using antibodies against the Flag tag and the LamR protein.
To investigate whether the CSFV envelope protein Erns or E2 mediates the interaction between CSFV and LamR, we transfected HEK293T cells with Flag-Erns or Flag-E2. The cell lysate was mixed with the membrane or cytoplasm extracts and then immobilized on agarose beads conjugated with anti-Flag MAb. The precipitated proteins were analyzed by Western blotting. A mechanical separation process was utilized for membrane protein preparation. The co-IP results showed that LamR interacted with Erns but not E2 (Fig. 3B).
Taken together, these results demonstrate that CSFV binds to LamR and that CSFV Erns mediates the interaction between CSFV and LamR.
CSFV infection was specifically inhibited by soluble LamR, laminin, and antibodies against LamR.
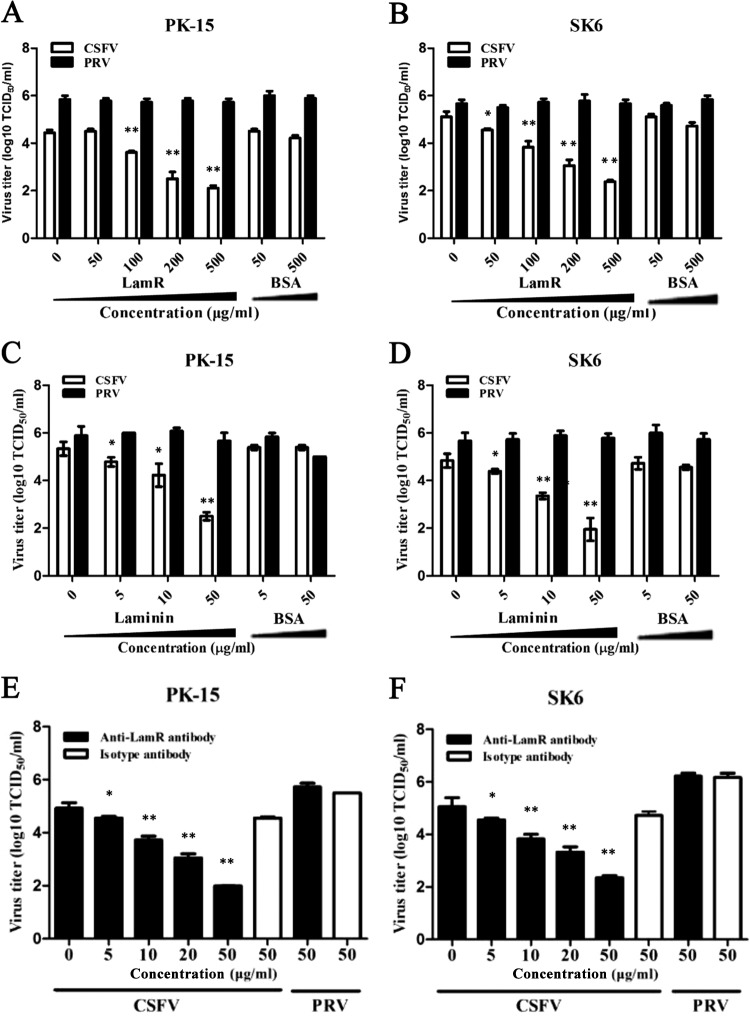
Colocalization of CSFV and LamR suggested that LamR could be a potential cellular receptor for CSFV. We then performed an inhibition assay to investigate if the addition of exogenous LamR would inhibit the binding of CSFV to host cells. Both CSFV and PRV were preincubated with either LamR or BSA before being used to infect PK-15 or SK6 cells. After 24 h, the culture supernatant was collected and titrated. The results from both cell lines indicated that CSFV incubated with LamR showed a decreased infection ability correlated with the increased amounts of soluble LamR in a dose-dependent manner. LamR at 500 μg/ml dramatically reduced CSFV infection, while BSA displayed no significant influence (Fig. 4A and B). The inhibition effects were also found to be specific to CSFV, as exogenous LamR failed to block PRV infection even when it was used at the maximum amount.
FIG 4.
Inhibition of CSFV infection in PK-15 cells by soluble LamR, laminin, or anti-LamR antibodies. (A, B) Influence of LamR on infection of CSFV and PRV in PK-15 cells (A) or SK6 cells (B). CSFV or PRV was incubated with different concentrations of LamR or BSA for 1 h at 4°C, followed by infection of the cells. The infected cell culture supernatant was collected at 48 hpi and titrated. (C, D) Influence of laminin on the invasion of CSFV and PRV in PK-15 cells (C) or SK6 cells (D). Cells were incubated with different concentrations of laminin or BSA for 1 h at 37°C, followed by infection with CSFV or PRV. The infected cell culture supernatant was collected at 48 hpi and titrated. (E, F) Antibody inhibition assay. PK-15 cells (E) or SK6 cells (F) were preincubated with anti-LamR antibodies or irrelevant rabbit IgG, followed by infection with CSFV or PRV. In the assays whose results are presented in panels C to F, the cells were infected with the indicated virus at an MOI of 1 and the supernatant was collected at 24 hpi. Bars represent standard deviations. *, P < 0.05; **, P < 0.01.
LamR is a receptor of laminin, which is a large glycoprotein containing multiple protein-binding domains (48). Therefore, we incubated cells with soluble laminin at different concentrations. Laminin displayed an inhibitory effect on CSFV infection but not PRV infection in a dose-dependent manner. As expected, no inhibition was observed for BSA at the same dose (Fig. 4C and D).
To provide further evidence of the involvement of LamR in CSFV infection, we tested the effects of antibodies against the 67-kDa LamR. PK-15 or SK6 cells were preincubated with anti-LamR PAb and then infected with CSFV or PRV. The results showed that anti-LamR antibodies at a concentration of 5 μg/ml markedly blocked the infectivity of CSFV (Fig. 4E and F). No inhibition was observed with the rabbit IgG control at a concentration of 50 μg/ml, and the anti-LamR antibodies showed no inhibitory effects on PRV infection, suggesting that the inhibition is specific to CSFV.
These results confirmed the interaction between CSFV and LamR on the surface of PK-15 or SK6 cells. When we blocked the virus-host interaction on the cell membrane, CSFV infection was markedly inhibited.
LamR overexpression in PK-15 cells promoted CSFV infection.
An inhibition assay showed that blocking the interaction between LamR and CSFV can inhibit CSFV infection. Therefore, it is likely that LamR is involved in CSFV invasion, possibly as a cellular receptor. CSFV has a relatively restricted host range, with only pigs and wild boar being its natural hosts. This may be due to differences in receptor(s) distribution among different species or because a unique cellular environment is required for CSFV growth. To exclude the second possibility, we first constructed CSFV-permissive cell lines (PK-15 and SK6) overexpressing LamR. Flow cytometry showed that nearly all cells expressed the EGFP-tagged LamR (Fig. 5A and D). The 37-kDa LamR with the EGFP tag was detected in both cell lines, but no 67-kDa LamR with the EGFP tag was observed. We also detected the 67-kDa LamR in the membrane extracts. Western blot analysis showed that the endogenous expression of the 67-kDa LamR was increased. However, no 67-kDa LamR with the EGFP tag was observed (Fig. 5B and E). Interestingly, overexpression of the 67-kDa LamR confers elevated susceptibility to PK-15 cells but not to SK6 cells (Fig. 5C and F). Compared with the parent PK-15 cells and PK-15 cells stably expressing EGFP (PK-EGFP cells), PK-15 cells expressing LamR (PK-LamR cells) seemed to be more suitable for CSFV infection. CSFV replicated more efficiently in PK-LamR cells than in the other two cell lines. In contrast, SK6 cells, SK6 cells expressing EGFP (SK6-EGFP cells), and SK6 cells expressing LamR (SK6-LamR cells) showed similar susceptibilities to CSFV. PK-15 and SK6 cells are both from porcine kidney, but there may be some unidentified differences between them.
FIG 5.
CSFV infection in PK-15 or SK6 cells overexpressing LamR. (A) EGFP-LamR expression in PK-15 cells detected by flow cytometry; (B) LamR expression in PK-15 cells; (C) CSFV titers in PK-LamR, PK-EGFP, and PK-15 cells (**, P < 0.01); (D) EGFP-LamR expression in SK6 cells detected by flow cytometry; (E) LamR expression in SK6 cells; (F) CSFV titers in SK6-LamR, SK6-EGFP, or SK6 cells.
The expression of LamRpig did not confer onto nonpermissive cells susceptibility to CSFV.
Sequence alignment showed that LamR is highly conserved, with only a few amino acid differences being located in the carboxyl terminus of LamRs from different species (Fig. 6). It is of interest to investigate whether the expression of LamR from pigs (LamRpig) correlates with susceptibility to CSFV infection. We transduced cells of various nonpermissive cell lines, including BHK-21, CHO-K1, HeLa, Marc-145, MDBK, MDCK, and Vero cells, with the recombinant lentivirus expressing EGFP-tagged LamRpig. LamRpig was expressed in cells of all of these cell lines. Cells of the stable cell lines were infected with CSFV-NproFluc and passaged eight times. The Fluc activity in these cells was markedly lower than that in PK-15 cells, and no CSFV RNA was detected in the culture supernatant by real-time RT-PCR (data not shown). The results suggest that CSFV replication does not occur in these nonpermissive cell lines stably expressing porcine LamR.
FIG 6.
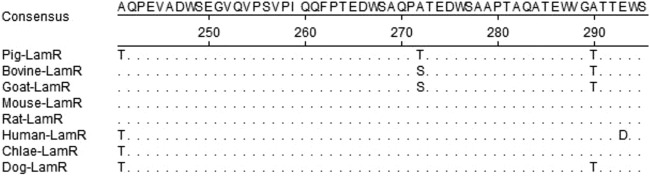
Sequence alignment of LamRs of different species origin. Dots represent identical residues. Chlae, Chlorocelrus aethiops.
LamR overexpression in PK-15 cells promoted CSFV attachment to cells.
The interaction of LamR with CSFV was necessary for CSFV infection, but LamR expression did not confer susceptibility to CSFV onto cells of nonpermissive cell lines. Therefore, we deduced that LamR may be an attachment receptor (which helps the virus attach to the cell surface) but is not involved in entry. To confirm our speculation, we performed attachment assays on cells of two permissive cell lines (PK-15 and SK6) and one nonpermissive cell line (MDBK). The cells were treated with heparinase I and then inoculated with CSFV at an MOI of 5 on ice for 2 h. The unbound virions were removed by washing five times. The level of attached CSFV virions was determined by immediately quantifying the viral RNA by real-time RT-PCR. After treatment with heparinase I, there were over 104 copies of virions binding to different cell lines. This suggests that there is a receptor(s) other than HS that is responsible for virus binding to different cell lines. As predicted, there was no significant difference between parent and EGFP-expressing PK-15 cells in the number of CSFV virions that bound to the cells. According to viral RNA copy numbers, the levels of CSFV binding to LamR-expressing cells were much higher than the levels of CSFV binding to cells of the two other cell lines (Fig. 7). Therefore, we infer that LamR functions in the attachment step of the virus replication cycle.
FIG 7.
Efficiency of CSFV attachment to different cell lines. Cells were incubated with CSFV at an MOI of 5 for 2 h at 4°C. The viral RNA was extracted and quantified by real-time RT-PCR as described previously (33). Bars represent standard deviations. *, P < 0.05; **, P < 0.01.
LamR serves as an alternative attachment receptor for CSFV.
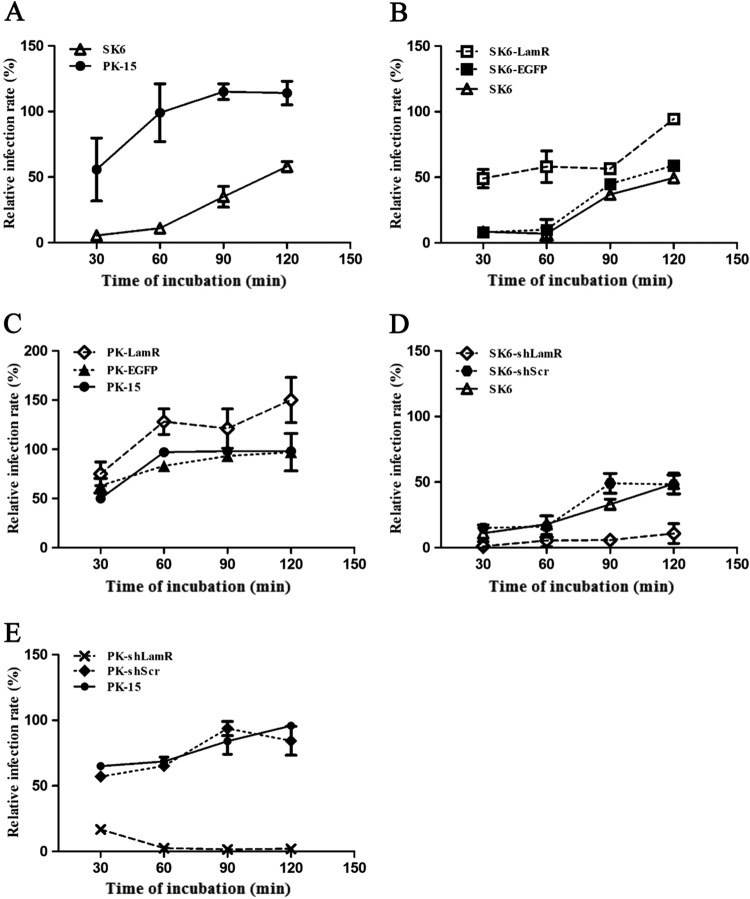
HS has already been identified to be an attachment receptor for CSFV (29). The identification was based on the findings obtained with SK6 cells and not with PK-15 cells. When LamR was overexpressed, CSFV growth seemed to be different in these two cell lines. The results indicate that there are differences between the two cell lines. More importantly, we did not observe complete blocking of CSFV infection by either anti-LamR antibodies or soluble LamR or laminin protein in the two cell lines. Is HS responsible for this CSFV infection without any interaction between LamR and CSFV virions? We further sought to clarify the relationship between the two attachment receptors (LamR and HS), and we analyzed their specific roles. We first tried to clarify the role of HS in PK-15 cells. We incubated PK-15 and SK6 cells with heparinase I or binding buffer and then inoculated them with CSFV-NproFluc on ice for different times (30, 60, 90, and 120 min). Fluc activity was detected at 48 hpi. The results derived from SK6 cells correlated with the results of a previous study (29) (Fig. 8A). After the cells were treated with heparinase I, CSFV displayed poor infectivity in SK6 cells during the 60-min incubation. From then, the infection ability rose with increasing inoculation time. When the cells were inoculated with the virus for 120 min, the relative infection rate reached the peak, up to 50% of the rate for the control. For PK-15 cells, the results seemed to be different. When incubating cells with CSFV for only 30 min, the relative infection rate reached nearly 50%, similar to the results for 120 min of incubation in SK6 cells. This infection efficiency reached a peak at 90 min, almost the same as the time to the peak for the control. The effects of HS seemed to differ in these two cell lines. SK6 cells may use HS as the main attachment receptor. Following the removal of HS, CSFV attachment was significantly inhibited. There may be an alternative attachment pathway which is not as efficient as the HS pathway.
FIG 8.
Cells treated with heparinase I showed distinct susceptibility to CSFV. (A) Relative infection rate of CSFV-NproFluc in PK-15 and SK6 cells treated with heparinase I; (B) relative infection rate of CSFV-NproFluc in SK6-LamR, SK6-EGFP, and SK6 cells treated with heparinase I; (C) relative infection rate of CSFV-NproFluc in PK-LamR, PK-EGFP, and PK-15 cells treated with heparinase I; (D) relative infection rate of CSFV-NproFluc in SK6-shLamR, SK6-shScr, and SK6 cells treated with heparinase I; (E) relative infection rate of CSFV-NproFluc in PK-shLamR, PK-shScr, and PK-15 cells treated with heparinase I. The relative infection rate was defined as the ratio of the rate of infection of cells treated with heparinase I to the rate of infection of cells not treated with heparinase I for the same cell line. Each sample was run in triplicate.
Is LamR the molecule responsible for the alternative pathway? To answer this question, we further performed the test on SK6, SK6-EGFP, and SK6-LamR cells. After treatment as described above, we tested Fluc activity. The results confirmed our prediction (Fig. 8B). The relative rate of infection of SK6 and SK6-EGFP cells showed a high similarity to our earlier results. For SK6-LamR cells, the relative infection rate reached 40% at 30 min and slowly increased to approximately 90% at 120 min. This suggested that LamR plays a supplementary role. However, its affinity may be inferior to that of HS. The same treatment was also performed on PK-15, PK-EGFP, and PK-LamR cells. The relative rate of CSFV infection was enhanced, especially when the incubation time was more than 60 min (Fig. 8C).
To further confirm the role of LamR, we used PK-15 cells expressing shLamR (PK-shLamR cells) and SK6 cells expressing shLamR (SK6-shLamR cells), in which LamR was knocked down by shRNAs. As expected, CSFV could hardly replicate in PK-shLamR or SK6-shLamR cells (Fig. 8D and E).
DISCUSSION
In the study described in this report, we screened pooled siRNAs against porcine membrane protein genes and showed that LamR contributes to CSFV infection. We demonstrated that LamR specifically interacts with the CSFV envelope glycoprotein Erns. We observed decreased CSFV infection in vitro using anti-LamR antibodies or soluble laminin before virus inoculation. CSFV infection was also inhibited in a competition experiment, in which virus was incubated with soluble LamR before inoculation of cells. Transduction of PK-15 cells with LamR resulted in higher viral titers, but LamR transduction did not confer susceptibility to a number of nonpermissive cell lines. An attachment assay proved that LamR functioned during virus attachment, providing an alternative pathway to the HS pathway. Overexpression of LamR resulted in an increase in the binding of CSFV virions to both permissive and nonpermissive cells. All of the evidence suggests that LamR is a cellular receptor for CSFV and that it functions during attachment.
Pooled RNAi screening has been widely used to discover the host genes required for virus infection, especially for screening for virus-host interaction partners (49–53). Several receptors have been identified with the help of genome-wide pooled RNAi screening and indicator viruses (33, 54, 55). Both advantages and disadvantages are obvious. This screening is high throughput and systematic, but it is also costly and labor-intensive. More importantly, only the human genome-wide RNAi pool is currently available. For those viruses which fail to infect humans or primates, the human genome-wide RNAi pool does not appear to be applicable. In this study, we designed several siRNA molecules against selected porcine membrane protein genes and successfully identified LamR to be our target. This strategy could be useful for studies of other animal viruses.
LamR is a cell surface receptor that mediates high-affinity interactions between the cell and laminin (56–58). It was first discovered in 1983 to be a 295-aa peptide with a calculated molecular mass of 32 kDa and an apparent electrophoretic mobility of about 37 kDa in SDS-polyacrylamide gels. It was subsequently processed into a 67-kDa protein (58). Little is known about this process. Both dimerization and modification have been proposed to be possible explanations, but neither has been proven to date (48). The 67-kDa LamR is located in the membrane of cells, while the 37-kDa LamR is located in the cytoplasm. The expression levels of the two different forms of LamR are significantly different. The 37-kDa LamR is abundant in the cell, whereas the 67-kDa LamR is scarce. Interestingly, we found that siRNA molecules against LamR efficiently knock down the 67-kDa form but show only a minimal effect on the 37-kDa form (Fig. 2A). We thought that this may be due to the high level of 37-kDa LamR expression and the self-regulation of the host cells. The 37-kDa LamR seems to be highly stable, whereas the 67-kDa LamR seems to be pretty unstable. Surprisingly, both the transfection and the transduction of LamR into cells resulted in a detectable level of ectopic expression of the 37-kDa but not the 67-kDa LamR. However, we did observe an increased level of endogenous expression of the 67-kDa LamR in the cell membrane (Fig. 5B and D). A possible explanation is that the 67-kDa LamR is the activated, interconvertible, and functional form of the 37-kDa LamR, which is more abundant, inactive, and stable than its 67-kDa counterpart. The two forms of LamR are in a dynamic balance in the cell. LamR has been reported to serve as a binding protein that facilitates the invasion of several pathogens. Using idiotypic antibodies, LamR was identified to be a cellular receptor for tick-borne encephalitis virus (38). Sindbis virus uses LamR as a functional receptor to enter BHK-21 cells (39). Mosquito LamR was found to be a putative receptor for Venezuelan equine encephalitis virus (40) and dengue virus (41–44). In mammalian cells, LamR acts as a cellular receptor for the cellular prion protein (PrP) (45, 46), and it mediates the infection of adeno-associated virus serotypes 8, 2, 3, and 9 (47). On the other hand, the form of LamR that is functional was not determined in the studies described above. In our study, the knockdown results showed that the 67-kDa LamR was successfully knocked down, whereas the 37-kDa LamR was slightly or not affected. Confocal and inhibition assays were performed on intact cells. Thus, the viruses, antibodies, and soluble proteins that were tested had access to the 67-kDa LamR located in the membrane but not to the 37-kDa LamR located in the cytoplasm. These results indicate that the 67-kDa LamR is the functional form of LamR for CSFV infection.
The relationship between HS and LamR is another interesting topic. Our data indicated that HS and LamR work as attachment receptors in both PK-15 and SK6 cells. However, the roles of HS and LamR varied from each other. HS showed a higher affinity to CSFV than LamR did in SK6 cells. If there is abundant HS on the cell surface, increasing the expression of LamR does not enhance the infection rate. The inhibition assay carried out in PK-15 cells showed that LamR did not completely inhibit CSFV infection. However, when both receptors were removed, CSFV could hardly infect the cells. We propose that LamR works as an attachment receptor (as an alternative pathway to the HS pathway). This speculation correlated with the results that CSFV virions bind to different cell lines after cells are treated with heparinase I (Fig. 7). This also explains the observation that the CSFV Erns protein can attach to the surfaces of various cell lines (9, 11).
Dengue virus is among the viruses that use LamR as a cellular receptor, and the virus is another member of the Flaviviridae family. It has been shown that HS acts as a cellular receptor for dengue virus (59). The two molecules also contribute to CSFV infection, which implies that CSFV and dengue virus may use similar mechanisms during the entry stage. We also identified several interacting partners of CSFV that were previously defined to be interacting partners of dengue virus (data not shown). This deserves attention, because understanding the similarities in receptor usage and entry mechanisms among members of the Flaviviridae family can help to enhance our understanding of the infection cycles of these viruses.
The identification of LamR as a CSFV receptor also raises a number of questions with regard to the entry of CSFV. The receptor(s) acting on the pathway of entry remains undefined. LDLr is potentially such a molecule. LDLr is a functional receptor of HCV (17). It is also involved in the entry of various flaviviruses (60). However, there have been paradoxical reports on the role of LDLr in the entry of BVDV: an anti-LDLr MAb was found to inhibit the entry of BVDV in one study but failed to do so in another study, even though the same materials and methods were used (60–62). We observed decreased CSFV growth with LDLr knockdown (Fig. 1). Knockdown of the expression of LDLr was not performed in the study of the role of LDLr during BVDV infection. It would be of interest to determine in future studies whether LDLr is a functional receptor for both BVDV and CSFV.
It is worth mentioning that the ability of a virus to replicate is correlated not only with the availability of host cellular receptors which mediate virus attachment/entry but also with the cell microenvironment that favors virus replication, virus assembly, and release. CSFV is characterized by its limited host range and poor replication ability in vitro. Pigs and wild boar are the only natural hosts of CSFV, which cannot infect ruminants (63). In contrast, BVDV and border disease virus (BDV), two other members of the Pestivirus genus, can infect both ruminants and pigs (63, 64). Moreover, only some types of porcine cell lines support CSFV replication. This poses a question: is the receptor or the cellular environment the major limiting factor for CSFV growth? In this study, we also transfected cells of various nonpermissive cell lines with pBRCISM-NproFluc (the infectious cDNA clone from which CSFV-NproFluc was derived) to screen cell lines for support of CSFV replication. The Fluc activity reached a peak in the first generation and decreased sharply after passage (data not shown). No CSFV protein was detected in the cells by Western blotting, and no CSFV RNA was detected in the supernatant by real-time RT-PCR (data not shown). The results suggest that the cell lines did not support CSFV growth and partially explain why CSFV grows poorly in some porcine cell lines. There may be restriction factors that affect CSFV growth.
In conclusion, we demonstrate for the first time that LamR is an attachment receptor for CSFV. Further study is needed to identify the other receptor(s) responsible for the entry of CSFV.
ACKNOWLEDGMENTS
We thank Xiaolong Fan at Beijing Normal University for revision of the manuscript.
This study was supported by the Heilongjiang Natural Science Foundation (no. ZD201410) and the Natural Science Foundation of China (no. 31201921).
REFERENCES
- 1.Lindenbach BD, Murray CL, Thiel HJ. 2013. Flaviviridae, p 712–746 InKnipe DM, Howley PM, Cohen JI, Griffin DE, Lamb RA, Martin MA, Racaniello VR, Roizman B (ed), Fields virology, 6th ed, vol 2 Lippincott Williams & Wilkins, Philadelphia, PA. [Google Scholar]
- 2.Meyers G, Rümenapf T, Thiel HJ. 1989. Molecular cloning and the nucleotide sequence of the genome of hog cholera virus. Virology 171:555–567. doi: 10.1016/0042-6822(89)90625-9. [DOI] [PubMed] [Google Scholar]
- 3.Moormann RJM, Warmerdam PAM, van der Meer B, Schaaper W, Wensvoort G, Hulst MM. 1990. Molecular cloning and nucleotide sequence of hog cholera virus strain Brescia and mapping of the genomic region encoding envelope protein E1. Virology 177:184–198. doi: 10.1016/0042-6822(90)90472-4. [DOI] [PubMed] [Google Scholar]
- 4.Thiel HJ, Stark R, Weiland E, Rümenapf T, Meyers G. 1991. Hog cholera virus: molecular composition of virions from a pestivirus. J Virol 65:4705–4712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rümenapf T, Unger G, Strauss JH, Thiel HJ. 1993. Processing of the envelope glycoproteins of pestiviruses. J Virol 67:3288–3294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Weiland E, Stark R, Haas B, Rümenapf T, Meyers G, Thiel HJ. 1990. Pestivirus glycoprotein which induces neutralizing antibodies forms part of a disulfide-linked heterodimer. J Virol 64:3563–3569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wensvoort G, Boonstra J, Bodzinga B. 1990. Immunoaffinity purification of envelope protein E1 of hog cholera virus. J Gen Virol 71:531–540. doi: 10.1099/0022-1317-71-3-531. [DOI] [PubMed] [Google Scholar]
- 8.Weiland E, Ahl R, Stark R, Weiland F, Thiel HJ. 1992. A second envelope glycoprotein mediates neutralization of a pestivirus, hog cholera virus. J Virol 66:3677–3682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hulst MM, Moormann RJ. 1997. Inhibition of pestivirus infection in cell culture by envelope proteins E(rns) and E2 of classical swine fever virus: E(rns) and E2 interact with different receptors. J Gen Virol 78:2779–2787. [DOI] [PubMed] [Google Scholar]
- 10.Wang Z, Nie Y, Wang P, Ding M, Deng H. 2004. Characterization of classical swine fever virus entry by using pseudotyped viruses: E1 and E2 are sufficient to mediate viral entry. Virology 330:332–341. doi: 10.1016/j.virol.2004.09.023. [DOI] [PubMed] [Google Scholar]
- 11.Li X, Wang L, Zhao D, Zhang G, Luo J, Deng R, Yang Y. 2011. Identification of host cell binding peptide from an overlapping peptide library for inhibition of classical swine fever virus infection. Virus Genes 43:33–40. doi: 10.1007/s11262-011-0595-7. [DOI] [PubMed] [Google Scholar]
- 12.Sawitzky D, Hampl H, Habermehl KO. 1990. Comparison of heparin-sensitive attachment of pseudorabies virus (PRV) and herpes simplex virus type 1 and identification of heparin-binding PRV glycoproteins. J Gen Virol 71:1221–1225. doi: 10.1099/0022-1317-71-5-1221. [DOI] [PubMed] [Google Scholar]
- 13.Misinzo G, Delputte PL, Meerts P, Lefebvre DJ, Nauwynck HJ. 2006. Porcine circovirus 2 (PCV2) uses heparan sulfate and chondroitin sulfate B glycosaminoglycans as receptors for its attachment to host cells. J Virol 80:3487–3494. doi: 10.1128/JVI.80.7.3487-3494.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Germi R, Crance JM, Garin D, Guimet J, Lortat-Jacob H, Ruigrok RW, Zarski JP, Drouet E. 2002. Heparan sulfate-mediated binding of infectious dengue virus type 2 and yellow fever virus. Virology 292:162–168. doi: 10.1006/viro.2001.1232. [DOI] [PubMed] [Google Scholar]
- 15.Madu IG, Chu VC, Lee H, Regan AD, Bauman BE, Whittaker GR. 2007. Heparan sulfate is a selective attachment factor for the avian coronavirus infectious bronchitis virus Beaudette. Avian Dis 51:45–51. doi: 10.1637/0005-2086(2007)051[0045:HSIASA]2.0.CO;2. [DOI] [PubMed] [Google Scholar]
- 16.Jackson T, Ellard FM, Ghazaleh RA, Brookes SM, Blakemore WE, Corteyn AH, Stuart DI, Newman JW, King AM. 1996. Efficient infection of cells in culture by type O foot-and-mouth disease virus requires binding to cell surface heparan sulfate. J Virol 70:5282–5287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Monazahian M, Böhme I, Bonk S, Koch A, Scholz C, Grethe S, Thomssen R. 1999. Low density lipoprotein receptor as a candidate receptor for hepatitis C virus. J Med Virol 57:223–229. doi:. [DOI] [PubMed] [Google Scholar]
- 18.Pileri P, Uematsu Y, Campagnoli S, Galli G, Falugi F, Petracca R, Weiner AJ, Houghton M, Rosa D, Grandi G, Abrignani S. 1998. Binding of hepatitis C virus to CD81. Science 282:938–941. doi: 10.1126/science.282.5390.938. [DOI] [PubMed] [Google Scholar]
- 19.Blanchard E, Belouzard S, Goueslain L, Wakita T, Dubuisson J, Wychowski C, Rouillé Y. 2006. Hepatitis C virus entry depends on clathrin-mediated endocytosis. J Virol 80:6964–6972. doi: 10.1128/JVI.00024-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Evans MJ, von Hahn T, Tscherne DM, Syder AJ, Panis M, Wölk B, Hatziioannou T, McKeating JA, Bieniasz PD, Rice CM. 2007. Claudin-1 is a hepatitis C virus co-receptor required for a late step in entry. Nature 446:801–805. doi: 10.1038/nature05654. [DOI] [PubMed] [Google Scholar]
- 21.Ploss A, Evans MJ, Gaysinskaya VA, Panis M, You H, de Jong YP, Rice CM. 2009. Human occludin is a hepatitis C virus entry factor required for infection of mouse cells. Nature 457:882–886. doi: 10.1038/nature07684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Scarselli E, Ansuini H, Cerino R, Roccasecca RM, Acali S, Filocamo G, Traboni C, Nicosia A, Cortese R, Vitelli A. 2002. The human scavenger receptor class B type I is a novel candidate receptor for the hepatitis C virus. EMBO J 21:5017–5025. doi: 10.1093/emboj/cdf529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lozach PY, Lortat-Jacob H, de Lacroix de Lavalette A, Staropoli I, Foung S, Amara A, Houles C, Fieschi F, Schwartz O, Virelizier JL, Arenzana-Seisdedos F, Altmeyer R. 2003. DC-SIGN and L-SIGN are high affinity binding receptors for hepatitis C virus glycoprotein E2. J Biol Chem 278:20358–20366. doi: 10.1074/jbc.M301284200. [DOI] [PubMed] [Google Scholar]
- 24.Maurer K, Krey T, Moennig V, Thiel HJ, Rümenapf T. 2004. CD46 is a cellular receptor for bovine viral diarrhea virus. J Virol 78:1792–1799. doi: 10.1128/JVI.78.4.1792-1799.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Naniche D, Varior-Krishnan G, Cervoni F, Wild TF, Rossi B, Rabourdin-Combe C, Gerlier D. 1993. Human membrane cofactor protein (CD46) acts as a cellular receptor for measles virus. J Virol 67:6025–6032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Santoro F, Kennedy PE, Locatelli G, Malnati MS, Berger EA, Lusso P. 1999. CD46 is a cellular receptor for human herpesvirus 6. Cell 99:817–827. doi: 10.1016/S0092-8674(00)81678-5. [DOI] [PubMed] [Google Scholar]
- 27.Gaggar A, Shayakhmetov DM, Lieber A. 2003. CD46 is a cellular receptor for group B adenoviruses. Nat Med 9:1408–1412. doi: 10.1038/nm952. [DOI] [PubMed] [Google Scholar]
- 28.Segerman A, Atkinson JP, Marttila M, Dennerquist V, Wadell G, Arnberg N. 2003. Adenovirus type 11 uses CD46 as a cellular receptor. J Virol 77:9183–9191. doi: 10.1128/JVI.77.17.9183-9191.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hulst MM, van Gennip HG, Moormann RJ. 2000. Passage of classical swine fever virus in cultured swine kidney cells selects virus variants that bind to heparan sulfate due to a single amino acid change in envelope protein E(rns). J Virol 74:9553–9561. doi: 10.1128/JVI.74.20.9553-9561.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hulst MM, van Gennip HG, Vlot AC, Schooten E, de Smit AJ, Moormann RJ. 2001. Interaction of classical swine fever virus with membrane-associated heparan sulfate: role for virus replication in vivo and virulence. J Virol 75:9585–9595. doi: 10.1128/JVI.75.20.9585-9595.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Shen L, Li Y, Chen J, Li C, Huang J, Luo Y, Sun Y, Li S, Qiu HJ. 2014. Generation of a recombinant classical swine fever virus stably expressing the firefly luciferase gene for quantitative antiviral assay. Antiviral Res 109:15–21. doi: 10.1016/j.antiviral.2014.06.006. [DOI] [PubMed] [Google Scholar]
- 32.Luo Y, Li N, Cong X, Wang CH, Du M, Li L, Zhao B, Yuan J, Liu DD, Li S, Li Y, Sun Y, Qiu HJ. 2014. Pathogenicity and genomic characterization of a pseudorabies virus variant isolated from Bartha-K61-vaccinated swine population in China. Vet Microbiol 174:107–115. doi: 10.1016/j.vetmic.2014.09.003. [DOI] [PubMed] [Google Scholar]
- 33.Lesot H, Kühl U, Mark K. 1983. Isolation of a laminin-binding protein from muscle cell membranes. EMBO J 2:861–865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Klimstra WB, Ryman KD, Johnston RE. 1998. Adaptation of Sindbis virus to BHK cells selects for use of heparan sulfate as an attachment receptor. J Virol 72:7357–7366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tamura M, Natori K, Kobayashi M, Miyamura T, Takeda N. 2000. Interaction of recombinant Norwalk virus particles with the 105-kilodalton cellular binding protein, a candidate receptor molecule for virus attachment. J Virol 74:11589–11597. doi: 10.1128/JVI.74.24.11589-11597.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jiang J, Cun W, Wu X, Shi Q, Tang H, Luo G. 2012. Hepatitis C virus attachment mediated by apolipoprotein E binding to cell surface heparan sulfate. J Virol 86:7256–7267. doi: 10.1128/JVI.07222-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhang XJ, Xia H, Everett H, Sosan O, Crooke H, Belák S, Widén F, Qiu HJ, Liu L. 2010. Evaluation of a primer-probe energy transfer real-time PCR assay for detection of classical swine fever virus. J Virol Methods 168:259–261. doi: 10.1016/j.jviromet.2010.05.004. [DOI] [PubMed] [Google Scholar]
- 38.Protopopova EV, Konavalova SN, Loktev VB. 1997. Isolation of a cellular receptor for tick-borne encephalitis virus using anti-idiotypic antibodies. Vopr Virusol 42:264–268. [PubMed] [Google Scholar]
- 39.Wang KS, Kuhn RJ, Strauss EG, Ou S, Strauss JH. 1992. High-affinity laminin receptor is a receptor for Sindbis virus in mammalian cells. J Virol 66:4992–5001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ludwig GV, Kondig JP, Smith JF. 1996. A putative receptor for Venezuelan equine encephalitis virus from mosquito cells. J Virol 70:5592–5599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Thepparit C, Smith DR. 2004. Serotype-specific entry of dengue virus into liver cells: identification of the 37-kilodalton/67-kilodalton high-affinity laminin receptor as a dengue virus serotype 1 receptor. J Virol 78:12647–12656. doi: 10.1128/JVI.78.22.12647-12656.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sakoonwatanyoo P, Boonsanay V, Smith DR. 2006. Growth and production of the dengue virus in C6/36 cells and identification of a laminin-binding protein as a candidate serotype 3 and 4 receptor protein. Intervirology 49:161–172. doi: 10.1159/000089377. [DOI] [PubMed] [Google Scholar]
- 43.Tio PH, Jong WW, Cardosa MJ. 2005. Two dimensional VOPBA reveals laminin receptor (LAMR1) interaction with dengue virus serotypes 1, 2 and 3. Virol J 2:25. doi: 10.1186/1743-422X-2-25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Upanan S, Kuadkitkan A, Smith DR. 2008. Identification of dengue virus binding proteins using affinity chromatography. J Virol Methods 151:325–328. doi: 10.1016/j.jviromet.2008.05.001. [DOI] [PubMed] [Google Scholar]
- 45.Gauczynski S, Peyrin JM, Haïk S, Leucht C, Hundt C, Rieger R, Krasemann S, Deslys JP, Dormont D, Lasmézas CI, Weiss S. 2001. The 37-kDa/67-kDa laminin receptor acts as the cell-surface receptor for the cellular prion protein. EMBO J 20:5863–5875. doi: 10.1093/emboj/20.21.5863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Gauczynski S, Nikles D, El-Gogo S, Papy-Garcia D, Rey C, Alban S, Barritault D, Lasmezas CI, Weiss S. 2006. The 37-kDa/67-kDa laminin receptor acts as a receptor for infectious prions and is inhibited by polysulfated glycanes. J Infect Dis 94:702–709. doi: 10.1086/505914. [DOI] [PubMed] [Google Scholar]
- 47.Akache B, Grimm D, Pandey K, Yant SR, Xu H, Kay MA. 2006. The 37/67-kilodalton laminin receptor is a receptor for adeno-associated virus serotypes 8, 2, 3, and 9. J Virol 80:9831–9836. doi: 10.1128/JVI.00878-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Rea VE, Rossi FW, De Paulis A, Ragno P, Selleri C, Montuori N. 2012. 67 kDa laminin receptor: structure, function and role in cancer and infection. Infez Med 2:8–12. [PubMed] [Google Scholar]
- 49.Karlas A, Machuy N, Shin Y, Pleissner KP, Artarini A, Heuer D, Becker D, Khalil H, Ogilvie LA, Hess S, Mäurer AP, Müller E, Wolff T, Rudel T, Meyer TF. 2010. Genome-wide RNAi screen identifies human host factors crucial for influenza virus replication. Nature 463:818–822. doi: 10.1038/nature08760. [DOI] [PubMed] [Google Scholar]
- 50.Hao L, Sakurai A, Watanabe T, Sorensen E, Nidom CA, Newton MA, Ahlquist P, Kawaoka Y. 2008. Drosophila RNAi screen identifies host genes important for influenza virus replication. Nature 454:890–893. doi: 10.1038/nature07151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Metz P, Dazert E, Ruggieri A, Mazur J, Kaderali L, Kaul A, Zeuge U, Windisch MP, Trippler M, Lohmann V, Binder M, Frese M, Bartenschlager R. 2012. Identification of type I and type II interferon-induced effectors controlling hepatitis C virus replication. Hepatology 56:2082–2093. doi: 10.1002/hep.25908. [DOI] [PubMed] [Google Scholar]
- 52.Zhou H, Xu M, Huang Q, Gates AT, Zhang XD, Castle JC, Stec E, Ferrer M, Strulovici B, Hazuda DJ, Espeseth AS. 2008. Genome-scale RNAi screen for host factors required for HIV replication. Cell Host Microbe 4:495–504. doi: 10.1016/j.chom.2008.10.004. [DOI] [PubMed] [Google Scholar]
- 53.Oh HS, Bryant KF, Nieland TJ, Mazumder A, Bagul M, Bathe M, Root DE, Knipe DM. 2014. A targeted RNA interference screen reveals novel epigenetic factors that regulate herpesviral gene expression. mBio 5(1):e01086-13. doi: 10.1128/mBio.01086-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Panda D, Rose PP, Hanna SL, Gold B, Hopkins KC, Lyde RB, Marks MS, Cherry S. 2013. Genome-wide RNAi screen identifies SEC61A and VCP as conserved regulators of Sindbis virus entry. Cell Rep 5:1737–1748. doi: 10.1016/j.celrep.2013.11.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Lupberger J, Zeisel MB, Xiao F, Thumann C, Fofana I, Zona L, Davis C, Mee CJ, Turek M, Gorke S, Royer C, Fischer B, Zahid MN, Lavillette D, Fresquet J, Cosset FL, Rothenberg SM, Pietschmann T, Patel AH, Pessaux P, Doffoël M, Raffelsberger W, Poch O, McKeating JA, Brino L, Baumert TF. 2011. EGFR and EphA2 are host factors for hepatitis C virus entry and possible targets for antiviral therapy. Nat Med 17:589–595. doi: 10.1038/nm.2341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Rao NC, Barsky SH, Terranova VP, Liotta LA. 1983. Isolation of a tumor cell laminin receptor. Biochem Biophys Res Commun 111:804–808. doi: 10.1016/0006-291X(83)91370-0. [DOI] [PubMed] [Google Scholar]
- 57.Terranova VP, Rao CN, Kalebic T, Margulies IM, Liotta LA. 1983. Laminin receptor on human breast carcinoma cells. Proc Natl Acad Sci U S A 80:444–448. doi: 10.1073/pnas.80.2.444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Malinoff HL, Wicha MS. 1983. Isolation of a cell surface receptor protein for laminin from murine fibrosarcoma cells. J Cell Biol 96:1475–1479. doi: 10.1083/jcb.96.5.1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Chen Y, Maguire T, Hileman RE, Fromm JR, Esko JD, Linhardt RJ, Marks RM. 1997. Dengue virus infectivity depends on envelope protein binding to target cell heparan sulfate. Nat Med 3:866–871. doi: 10.1038/nm0897-866. [DOI] [PubMed] [Google Scholar]
- 60.Agnello V, Abel G, Elfahal M, Knight GB, Zhang QX. 1999. Hepatitis C virus and other Flaviviridae viruses enter cells via low density lipoprotein receptor. Proc Natl Acad Sci U S A 96:12766–12771. doi: 10.1073/pnas.96.22.12766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Flores EF, Donis RO. 1995. Isolation of a mutant MDBK cell line resistant to bovine viral diarrhea virus infection due to a block in viral entry. Virology 208:565–575. doi: 10.1006/viro.1995.1187. [DOI] [PubMed] [Google Scholar]
- 62.Krey T, Moussay E, Thiel HJ, Rümenapf T. 2006. Role of the low-density lipoprotein receptor in entry of bovine viral diarrhea virus. J Virol 80:10862–10867. doi: 10.1128/JVI.01589-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Carbrey EA, Stewart WC, Kresse JL, Snijder ML. 1976. Natural infection of pigs with bovine diarrhoea virus and its differential diagnosis from hog cholera. J Am Vet Med Assoc 169:1217–1219. [PubMed] [Google Scholar]
- 64.Terpstra C, Wensvoort G. 1988. The protective value of vaccine-induced neutralising antibody titres in swine fever. Vet Microbiol 16:123–128. doi: 10.1016/0378-1135(88)90036-3. [DOI] [PubMed] [Google Scholar]