Abstract
Ebola virus VP35 inhibits alpha/beta interferon production and functions as a viral polymerase cofactor. Previously, the 8-kDa cytoplasmic dynein light chain (LC8) was demonstrated to interact with VP35, but the functional consequences were unclear. Here we demonstrate that the interaction is direct and of high affinity and that binding stabilizes the VP35 N-terminal oligomerization domain and enhances viral RNA synthesis. Mutational analysis demonstrates that VP35 interaction is required for the functional effects of LC8.
TEXT
Ebola virus (EBOV) VP35 is a multifunctional protein critical for both viral innate immune evasion and viral RNA synthesis (1, 2). It contains an N-terminal oligomerization domain and a C-terminal double-stranded RNA (dsRNA) binding domain referred to as the interferon inhibitory domain (IID) (3, 4). VP35 acts as an interferon antagonist, using both dsRNA-binding-dependent and -independent mechanisms, to block the RIG-I-like receptor signaling pathways (5–9). VP35 also participates with the viral large protein (L), the enzymatic component of the RNA-dependent RNA polymerase (RDRP) complex, VP30, and nucleoprotein (NP) in viral RNA synthesis (10, 11). VP35 is critical in this role because it mediates the interaction between L and the RNA template-associated NP (2, 12–14).
VP35 can interact with the highly conserved 8-kDa cytoplasmic dynein light-chain (LC8) protein (15). LC8 is a subunit of the cytoplasmic dynein motor complex, which plays an important role in the microtubule-associated intracellular retrograde transport system (16, 17). It also can exist in a soluble form without association with the dynein motor or microtubule (18). LC8 exists as a dimer containing two identical grooves located at opposite faces of the protein dimer interface (19). These grooves bind to proteins containing consensus motifs of either (K/S)XTQT or G(I/V)QVD (20). VP35 contains an SQTQT motif that is required for LC8 interaction (15). However, LC8 binding was not required for, nor did it inhibit, VP35 interferon (IFN) antagonist function (15, 21). Therefore, the significance of the VP35-LC8 interaction has remained undefined.
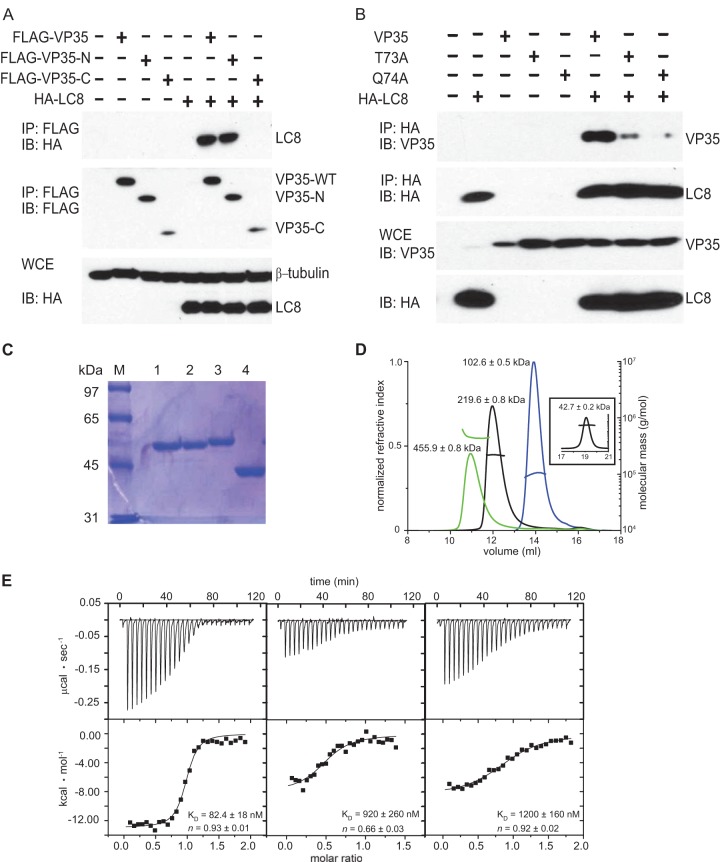
Here we characterized the interaction between LC8 and EBOV VP35 and provide evidence that the interaction stabilizes the VP35 N-terminal oligomerization domain to enhance viral RNA synthesis. The VP35-LC8 interaction was first validated by coimmunoprecipitation (co-IP). Briefly, HEK293T cells were transfected with plasmids expressing hemagglutinin (HA)-tagged LC8 (500 ng) and either full-length FLAG-tagged VP35 (1 μg) (GenBank accession no. AF086833.2) or a VP35 truncation mutant (1 μg) corresponding to the VP35 N-terminal domain (FLAG-VP35-N, residues 1 to 218) or the IID (FLAG-VP35-C, residues 219 to 340). Twenty-four hours posttransfection, anti-FLAG immunoprecipitations were performed. Both wild-type (WT) VP35 and VP35-N coprecipitated with LC8, confirming that VP35 binds LC8 via its N terminus (Fig. 1A). Mutation to alanine of T73 or Q74 within the VP35 71-SQTQT-75 motif disrupted or reduced the interaction, consistent with results of prior studies (15) (Fig. 1B).
FIG 1.
LC8 interacts with EBOV VP35 protein. (A) The N terminus of VP35 is sufficient to interact with LC8. A co-IP experiment was performed with FLAG-tagged WT VP35, VP35-N (containing amino acids [aa] 1 to 218), VP35-C (aa 219 to 340), and HA-tagged LC8 (HA-LC8). VP35 proteins were pulled down by anti-FLAG antibody, and protein expression was detected by Western blotting (immunoblotting [IB]). WCE, whole-cell extracts. (B) VP35 mediates the interaction with LC8 via an LC8 binding consensus motif (K/SXTQT). VP35 mutants (the T73A and Q74A mutants) were utilized in a co-IP experiment with HA-tagged LC8. Protein expression was detected by Western blotting with anti-VP35 and anti-HA antibodies. (C) SDS-PAGE gel of MBP-VP35 50–150 Q74A (lane 1), MBP-VP35 50–150 (lane 2), MBP-LC8 (lane 3), and MBP (lane 4) used in SEC-MALS and ITC experiments. (D) VP35 forms a 1:1 complex with LC8. Elution profiles of MBP-LC8 (blue), MBP-VP35 50–150 (black), MBP-VP35 50–150/MBP-LC8 complex (green), and MBP (inset) from a Superdex 200 column were obtained and analyzed by SEC-MALS. The theoretical monomeric molecular masses for MBP-VP35 50–150 and MBP-LC8 are 55.2 and 54.4 kDa, respectively. (E) ITC data for MBP-LC8 binding to MBP-WT VP35 50–150 (left), mutant MBP-VP35 50–150 Q74A (center), and mutant MBP-VP35 50–150 Q72A Q74A (right). Representative raw data and binding isotherms are shown, and the reported values for KD and n are averages from at least two independent experiments.
To further characterize the interaction, LC8 and VP35 from residues 50 to 150 (VP35 50–150) were expressed in BL21(DE3) Escherichia coli from a modified pET15b vector (Novagen) containing a maltose binding protein (MBP) fusion tag. Cells were lysed using a cell disrupter (Avestin) and centrifuged at 47,000 × g to remove debris. Proteins were purified to homogeneity using a series of chromatographic columns (GE Healthcare), including affinity and ion-exchange columns, prior to a final gel filtration step into buffer containing 10 mM HEPES, pH 7, 150 mM NaCl, and 2 mM tris(2-carboxyethyl)phosphine (TCEP) (Fig. 1C). With a size exclusion chromatography (SEC) column coupled with a Dawn Heleos II multiangle light-scattering (MALS) detector (Wyatt Technologies), MBP-LC8 was demonstrated to behave as a dimer, consistent with results in previous reports (Fig. 1D, blue line) (22, 23), while the MBP-VP35 50–150 behaved as a tetramer (Fig. 1D, black line). Together, MBP-LC8 and MBP-VP35 50–150 form a 1:1 complex, such that two LC8 dimers bind to one VP35 50-150 tetramer (Fig. 1D, green line). Isothermal titration calorimetry (ITC) measurements using a VP-ITC microcalorimeter (Malvern) revealed that VP35 has a high affinity for LC8 (equilibrium dissociation constant [KD] = 82.4 ± 18 nM), while a Q74A single mutant and a Q72A Q74A double mutant that alter the LC8 binding motif in VP35 50–150 show substantially diminished binding to LC8 (KD = 900 ± 260 nM and 1,200 ± 160 nM, respectively) (Fig. 1E, middle and right panels).
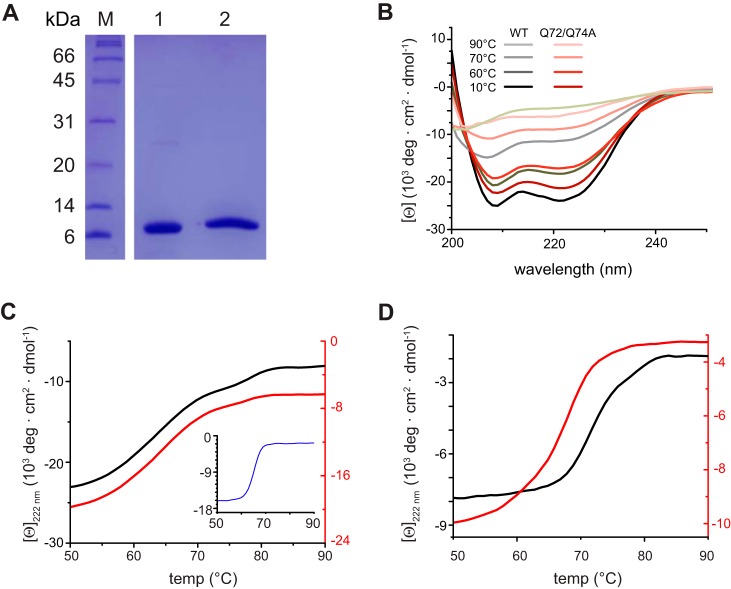
We next assessed the stability of VP35 upon LC8 binding using proteins lacking the MBP fusion tag (Fig. 2A). We performed circular dichroism (CD) wavelength scans, using a Chirascan CD spectrometer (Applied Photophysics), and monitored the change in molar ellipticity of each protein from 10 to 90°C on samples containing 5 to 10 μM VP35 50–150 and the VP35 50–150 Q72A Q74A mutant (Fig. 2B). CD results show that VP35 50–150 has a high alpha-helical content that persists even at higher temperatures (Fig. 2B). Next, we conducted comparative thermal stability analysis between VP35 50–150 and the VP35 50–150 Q72A Q74A mutant in the presence of equimolar LC8. Introduction of the Q72A and Q74A mutations did not significantly affect the stability of VP35 50–150 (Fig. 2B and C). In contrast, addition of LC8 to VP35 50–150, but not to VP35 50–150 Q72A Q74A, differentially enhanced stability. This stability enhancement was predominantly due to LC8-VP35 50–150 binding, as judged by the stability of the mutant control (melting temperature [Tm] = 70.8 ± 1°C for VP35 50–150 versus 66.4 ± 2°C for the VP35 50–150 Q72A Q74A mutant) (Fig. 2D).
FIG 2.
LC8 binding enhances the stability of the N terminus of VP35. (A) SDS-PAGE gel of VP35 50–150 (lane 1) and LC8 (lane 2) used in circular dichroism (CD) experiments. Lane M, molecular mass markers. (B) CD wavelength scans for WT VP35 50–150 (black lines) and the VP35 50–150 Q72A Q74A mutant (red lines) at the temperatures indicated. Mean residue ellipticity is in 103 degrees · cm2 · dmol−1. (C) Thermal denaturation (Tm) of WT VP35 50–150 (black, left axis; Tm = 63.8 ± 0.1°C), the VP35 50–150 Q72A Q74A mutant (red, right axis; Tm = 63.8 ± 1°C), and LC8 (inset, blue; Tm = 66.3 ± 2°C) monitored at 222 nm. (D) Thermal denaturation of the WT VP35 50–150/LC8 complex (black, left axis; Tm = 70.8 ± 1°C) and the VP35 50–150 Q72A Q74A/LC8 mutant complex (red, right axis; Tm = 66.4 ± 2°C) monitored at 222 nm. Reported Tms are averages of results from at least two independent experiments.
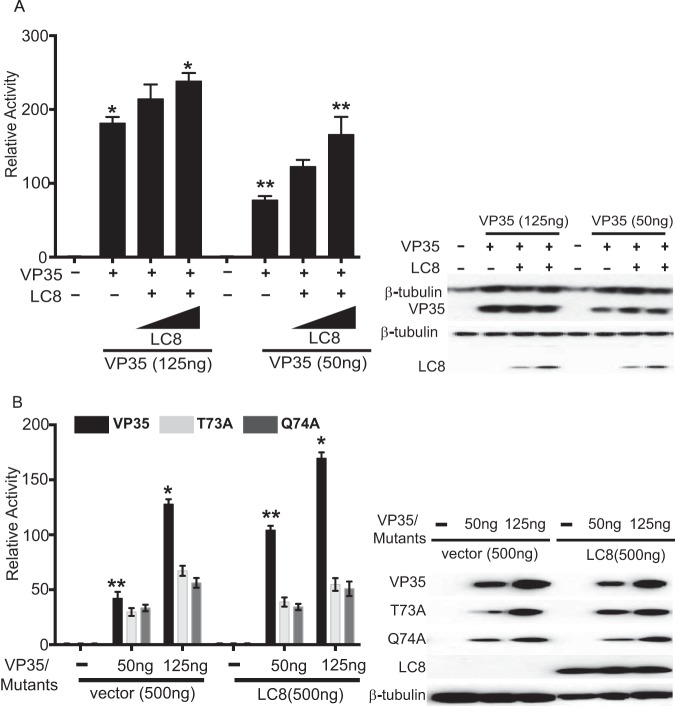
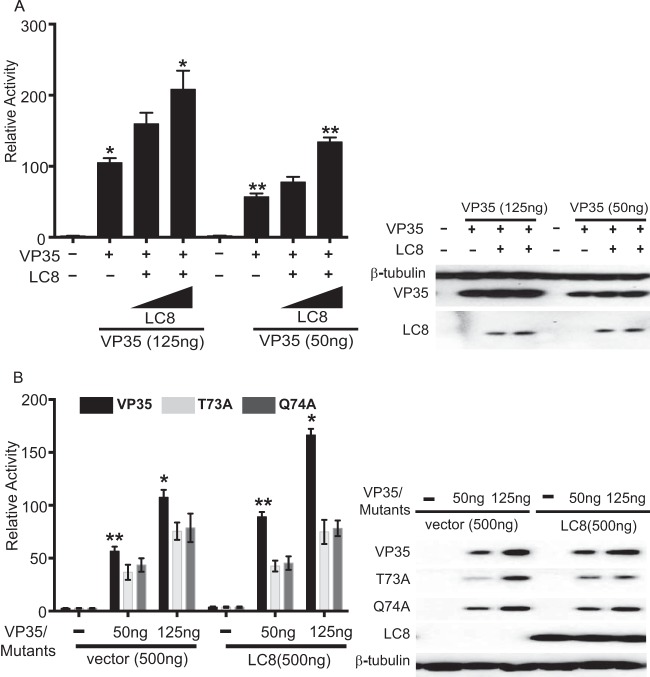
Previously, it was shown that LC8 interaction does not affect VP35 interferon antagonist function, and studies in which we compared levels of inhibition of beta interferon (IFN-β) promoter activation by wild-type VP35 versus T73A or Q74A VP35 confirmed this conclusion (reference 15 and data not shown). However, the effect of LC8 on VP35 viral polymerase cofactor function had not been addressed. We therefore utilized an established EBOV minigenome assay where a model EBOV RNA encoding Renilla luciferase is transcribed and replicated by a viral RDRP complex that is reconstituted by transfection (8, 10). LC8 overexpression enhanced minigenome activity in a dose-dependent manner, with the highest concentration of LC8 plasmid yielding a statistically significant increase. The stimulation was more apparent when lower-than-optimal levels of WT VP35 were used (Fig. 3A). Representative Western blots indicated that VP35 expression levels were similar for each condition and confirmed LC8 expression (Fig. 3A). When VP35 mutants T73A and Q74A were tested, each exhibited reduced activity compared to WT VP35, even when expression levels were accounted for (Fig. 3B; compare the 125-ng conditions for mutant VP35s to the 50-ng conditions for WT VP35). Further, addition of LC8 did not enhance minigenome activity in the mutant VP35 samples (Fig. 3B), suggesting that LC8 binding to VP35 is critical for the LC8-mediated enhancement of minigenome activity.
FIG 3.
LC8 enhances EBOV RNA synthesis in a dose-dependent manner. (A) Increasing amounts of HA-tagged LC8 plasmid (50, 500 ng) were cotransfected with the plasmids required to reconstitute the EBOV RNA polymerase complex (L, NP, VP35, VP30) along with a plasmid encoding the Renilla luciferase minigenome RNA and a firefly luciferase expression plasmid, which served as a control for transfection efficiency. Relative activity was determined by normalizing Renilla luciferase activity to firefly luciferase activity. The error bars indicate standard deviations from three independent replicates. *, P = 0.04; **, P = 0.03, as determined by Student's t test. The Western blot shows expression of β-tubulin, VP35, and LC8 (anti-HA antibody). (B) A minigenome experiment similar to that described for panel A was performed except that WT VP35 or the indicated VP35 mutants were used, and the LC8 plasmid amount was kept constant at 500 ng. The error bars indicate standard deviations from three independent replicates. The Western blot shows the expression of VP35, LC8, and β-tubulin, detected with anti-VP35, anti-HA, and anti-β-tubulin antibodies, respectively. *, P = 0.01; **, P = 0.006, as determined by Student's t test.
We also examined the impact of LC8 on viral transcription by using a minigenome plasmid that lacks the viral antigenomic replication promoter region. This previously described construct lacks replication activity and specifically serves as a measure of viral mRNA synthesis (24). Overexpression of LC8 increased transcription in the presence of WT VP35 but not in the presence of VP35 T73A or VP35 Q74A. Further, VP35 T73A and VP35 Q74A exhibited reduced transcription (Fig. 4A and B).
FIG 4.
LC8 expression enhances EBOV transcription. (A) A minigenome assay was performed as described for Fig. 2A, except that a replication-deficient minigenome construct was used. *, P = 0.01; **, P = 0.0045, as determined by Student's t test. (B) A minigenome experiment similar to that described for panel A was performed except that WT VP35 or the indicated VP35 mutants were included and the LC8 plasmid amount was kept constant at 500 ng. The error bars indicate standard deviations from three independent replicates. The Western blot shows the expression of VP35 and LC8, detected with anti-VP35 and anti-HA antibodies, respectively. *, P = 0.007; **, P = 0.002, as determined by Student's t test.
This work reveals several novel features of relevance to VP35 function. First, we demonstrate that the VP35 oligomerization domain forms tetramers in the context of VP35 50–150. Therefore, full-length VP35 is also likely a tetramer. This is consistent with the ability of paramyxovirus P proteins, which are functionally analogous to filoviral VP35 proteins, to tetramerize (25, 26). Second, we show that VP35 50–150 is sufficient for high-affinity interaction with LC8 in the absence of other viral or cellular factors and that mutation of the LC8-interacting motif 71-SQTQT-75 disrupts interaction, further confirming specificity (Fig. 1 and 2). Third, these data provide the first evidence that LC8 interaction modulates EBOV RNA synthesis. The enhancing activity is specific to the RNA synthesis functions of VP35, as we and others have not seen the effects of the T73A or Q74A mutation on IFN antagonist function (data not shown and reference 15). The N-terminal oligomerization function has previously been implicated as facilitating both filoviral RNA synthesis and IFN antagonism (4, 12, 27). That the enhancement of RNA synthesis requires the interaction of LC8 with VP35 is demonstrated by the inability of LC8 to stimulate activity when the VP35 mutants were used in the minigenome assay. Overall, the data suggest that LC8-dependent stabilization of the oligomerization domain at least in part accounts for the enhanced activity. Given that the impact of LC8 is more dramatic when VP35 levels are limiting, LC8 may function primarily early in infection, when levels of replication complex proteins are low, serving as a means to rapidly establish viral gene expression before effective antiviral responses can be initiated.
LC8 interacts with a variety of viral proteins to facilitate movement of viral components (19, 20, 28–31). The rabies virus P protein, which serves as a rabies virus polymerase cofactor, also interacts with LC8 (32, 33). LC8-P protein interaction was not needed for dynein-mediated viral transport but was required for optimal rabies viral polymerase activity (33). The molecular basis for enhanced rabies virus polymerase activity was not defined, but our data suggest that LC8 stabilizes the P protein oligomer. It will therefore be of interest to determine to what extent other negative-strand RNA viruses, including other filoviruses, use a similar strategy to enhance their replication and transcription machinery.
ACKNOWLEDGMENTS
We thank Christine Schwall for critical reading of the manuscript.
This work was supported by NIH grants R01AI107056 to D.W.L., R01AI059536 to C.F.B., U19AI109945 (C. F. Basler was the principal investigator [PI]) and U19AI109664 (C. F. Basler, PI) to C.F.B. and G.K.A., U19 AI070489 (M. J. Holtzman, PI) to G.K.A., R01AI081914 to G.K.A., and T32-CA09547-37 (P. M. Allen, PI) to D.S.J. and by a Roche Translation and Clinical Research grant in infectious diseases to P.L.
REFERENCES
- 1.Basler CF, Wang X, Mühlberger E, Volchkov V, Paragas J, Klenk HD, García-Sastre A, Palese P. 2000. The Ebola virus VP35 protein functions as a type I IFN antagonist. Proc Natl Acad Sci U S A 97:12289–12294. doi: 10.1073/pnas.220398297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Leung DW, Prins KC, Basler CF, Amarasinghe GK. 2010. Ebolavirus VP35 is a multifunctional virulence factor. Virulence 1:526–531. doi: 10.4161/viru.1.6.12984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Leung DW, Ginder ND, Fulton DB, Nix J, Basler CF, Honzatko RB, Amarasinghe GK. 2009. Structure of the Ebola VP35 interferon inhibitory domain. Proc Natl Acad Sci U S A 106:411–416. doi: 10.1073/pnas.0807854106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Reid SP, Cardenas WB, Basler CF. 2005. Homo-oligomerization facilitates the interferon-antagonist activity of the ebolavirus VP35 protein. Virology 341:179–189. doi: 10.1016/j.virol.2005.06.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Basler CF, Mikulasova A, Martinez-Sobrido L, Paragas J, Mühlberger E, Bray M, Klenk HD, Palese P, Garcia-Sastre A. 2003. The Ebola virus VP35 protein inhibits activation of interferon regulatory factor 3. J Virol 77:7945–7956. doi: 10.1128/JVI.77.14.7945-7956.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hartman AL, Towner JS, Nichol ST. 2004. A C-terminal basic amino acid motif of Zaire ebolavirus VP35 is essential for type I interferon antagonism and displays high identity with the RNA-binding domain of another interferon antagonist, the NS1 protein of influenza A virus. Virology 328:177–184. doi: 10.1016/j.virol.2004.07.006. [DOI] [PubMed] [Google Scholar]
- 7.Cardenas WB, Loo YM, Gale M Jr, Hartman AL, Kimberlin CR, Martinez-Sobrido L, Saphire EO, Basler CF. 2006. Ebola virus VP35 protein binds double-stranded RNA and inhibits alpha/beta interferon production induced by RIG-I signaling. J Virol 80:5168–5178. doi: 10.1128/JVI.02199-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Luthra P, Ramanan P, Mire CE, Weisend C, Tsuda Y, Yen B, Liu G, Leung DW, Geisbert TW, Ebihara H, Amarasinghe GK, Basler CF. 2013. Mutual antagonism between the Ebola virus VP35 protein and the RIG-I activator PACT determines infection outcome. Cell Host Microbe 14:74–84. doi: 10.1016/j.chom.2013.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Prins KC, Cardenas WB, Basler CF. 2009. Ebola virus protein VP35 impairs the function of interferon regulatory factor-activating kinases IKKepsilon and TBK-1. J Virol 83:3069–3077. doi: 10.1128/JVI.01875-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Muhlberger E, Lötfering B, Klenk HD, Becker S. 1998. Three of the four nucleocapsid proteins of Marburg virus, NP, VP35, and L, are sufficient to mediate replication and transcription of Marburg virus-specific monocistronic minigenomes. J Virol 72:8756–8764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mühlberger E, Weik M, Volchkov VE, Klenk HD, Becker S. 1999. Comparison of the transcription and replication strategies of Marburg virus and Ebola virus by using artificial replication systems. J Virol 73:2333–2342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.DiCarlo A, Moller P, Lander A, Kolesnikova L, Becker S. 2007. Nucleocapsid formation and RNA synthesis of Marburg virus is dependent on two coiled coil motifs in the nucleoprotein. Virol J 4:105. doi: 10.1186/1743-422X-4-105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Becker S, Rinne C, Hofsass U, Klenk HD, Mühlberger E. 1998. Interactions of Marburg virus nucleocapsid proteins. Virology 249:406–417. doi: 10.1006/viro.1998.9328. [DOI] [PubMed] [Google Scholar]
- 14.Groseth A, Charton JE, Sauerborn M, Feldmann F, Jones SM, Hoenen T, Feldmann H. 2009. The Ebola virus ribonucleoprotein complex: a novel VP30-L interaction identified. Virus Res 140:8–14. doi: 10.1016/j.virusres.2008.10.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kubota T, Matsuoka M, Chang TH, Bray M, Jones S, Tashiro M, Kato A, Ozato K. 2009. Ebolavirus VP35 interacts with the cytoplasmic dynein light chain 8. J Virol 83:6952–6956. doi: 10.1128/JVI.00480-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.King SM, Barbarese E, Dillman JF III, Benashski SE, Do KT, Patel-King RS, Pfister KK. 1998. Cytoplasmic dynein contains a family of differentially expressed light chains. Biochemistry 37:15033–15041. doi: 10.1021/bi9810813. [DOI] [PubMed] [Google Scholar]
- 17.Rodriguez-Crespo I, Yelamos B, Roncal F, Albar JP, Ortiz de Montellano PR, Gavilanes F. 2001. Identification of novel cellular proteins that bind to the LC8 dynein light chain using a pepscan technique. FEBS Lett 503:135–141. doi: 10.1016/S0014-5793(01)02718-1. [DOI] [PubMed] [Google Scholar]
- 18.Martinez-Moreno M, Navarro-Lerida I, Roncal F, Albar JP, Alonso C, Gavilanes F, Rodriguez-Crespo I. 2003. Recognition of novel viral sequences that associate with the dynein light chain LC8 identified through a pepscan technique. FEBS Lett 544:262–267. doi: 10.1016/S0014-5793(03)00516-7. [DOI] [PubMed] [Google Scholar]
- 19.Garcia-Mayoral MF, Rodriguez-Crespo I, Bruix M. 2011. Structural models of DYNLL1 with interacting partners: African swine fever virus protein p54 and postsynaptic scaffolding protein gephyrin. FEBS Lett 585:53–57. doi: 10.1016/j.febslet.2010.11.027. [DOI] [PubMed] [Google Scholar]
- 20.Lo KW, Naisbitt S, Fan JS, Sheng M, Zhang M. 2001. The 8-kDa dynein light chain binds to its targets via a conserved (K/R)XTQT motif. J Biol Chem 276:14059–14066. doi: 10.1074/jbc.M010320200. [DOI] [PubMed] [Google Scholar]
- 21.Pichlmair A, Kandasamy K, Alvisi G, Mulhern O, Sacco R, Habjan M, Binder M, Stefanovic A, Eberle CA, Goncalves A, Burckstummer T, Muller AC, Fauster A, Holze C, Lindsten K, Goodbourn S, Kochs G, Weber F, Bartenschlager R, Bowie AG, Bennett KL, Colinge J, Superti-Furga G. 2012. Viral immune modulators perturb the human molecular network by common and unique strategies. Nature 487:486–490. doi: 10.1038/nature11289. [DOI] [PubMed] [Google Scholar]
- 22.Benashski SE, Harrison A, Patel-King RS, King SM. 1997. Dimerization of the highly conserved light chain shared by dynein and myosin V J Biol Chem 272:20929–20935. [DOI] [PubMed] [Google Scholar]
- 23.Wang W, Lo KW, Kan HM, Fan JS, Zhang M. 2003. Structure of the monomeric 8-kDa dynein light chain and mechanism of the domain-swapped dimer assembly. J Biol Chem 278:41491–41499. doi: 10.1074/jbc.M307118200. [DOI] [PubMed] [Google Scholar]
- 24.Hoenen T, Jung S, Herwig A, Groseth A, Becker S. 2010. Both matrix proteins of Ebola virus contribute to the regulation of viral genome replication and transcription. Virology 403:56–66. doi: 10.1016/j.virol.2010.04.002. [DOI] [PubMed] [Google Scholar]
- 25.Tarbouriech N, Curran J, Ebel C, Ruigrok RW, Burmeister WP. 2000. On the domain structure and the polymerization state of the Sendai virus P protein. Virology 266:99–109. doi: 10.1006/viro.1999.0066. [DOI] [PubMed] [Google Scholar]
- 26.Tarbouriech N, Curran J, Ruigrok RW, Burmeister WP. 2000. Tetrameric coiled coil domain of Sendai virus phosphoprotein. Nat Struct Biol 7:777–781. doi: 10.1038/79013. [DOI] [PubMed] [Google Scholar]
- 27.Becker S, Mühlberger E. 1999. Co- and posttranslational modifications and functions of Marburg virus proteins. Curr Topics Microbiol Immunol 235:23–34. [DOI] [PubMed] [Google Scholar]
- 28.Merino-Gracia J, Garcia-Mayoral MF, Rodriguez-Crespo I. 2011. The association of viral proteins with host cell dynein components during virus infection. FEBS J 278:2997–3011. doi: 10.1111/j.1742-4658.2011.08252.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Alonso C, Miskin J, Hernaez B, Fernandez-Zapatero P, Soto L, Canto C, Rodriguez-Crespo I, Dixon L, Escribano JM. 2001. African swine fever virus protein p54 interacts with the microtubular motor complex through direct binding to light-chain dynein. J Virol 75:9819–9827. doi: 10.1128/JVI.75.20.9819-9827.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Douglas MW, Diefenbach RJ, Homa FL, Miranda-Saksena M, Rixon FJ, Vittone V, Byth K, Cunningham AL. 2004. Herpes simplex virus type 1 capsid protein VP26 interacts with dynein light chains RP3 and Tctex1 and plays a role in retrograde cellular transport. J Biol Chem 279:28522–28530. doi: 10.1074/jbc.M311671200. [DOI] [PubMed] [Google Scholar]
- 31.Schneider MA, Spoden GA, Florin L, Lambert C. 2011. Identification of the dynein light chains required for human papillomavirus infection. Cell Microbiol 13:32–46. doi: 10.1111/j.1462-5822.2010.01515.x. [DOI] [PubMed] [Google Scholar]
- 32.Raux H, Flamand A, Blondel D. 2000. Interaction of the rabies virus P protein with the LC8 dynein light chain. J Virol 74:10212–10216. doi: 10.1128/JVI.74.21.10212-10216.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tan GS, Preuss MA, Williams JC, Schnell MJ. 2007. The dynein light chain 8 binding motif of rabies virus phosphoprotein promotes efficient viral transcription. Proc Natl Acad Sci U S A 104:7229–7234. doi: 10.1073/pnas.0701397104. [DOI] [PMC free article] [PubMed] [Google Scholar]