ABSTRACT
Ubiquitin-like domains (Ubls) now are recognized as common elements adjacent to viral and cellular proteases; however, their function is unclear. Structural studies of the papain-like protease (PLP) domains of coronaviruses (CoVs) revealed an adjacent Ubl domain in severe acute respiratory syndrome CoV, Middle East respiratory syndrome CoV, and the murine CoV, mouse hepatitis virus (MHV). Here, we tested the effect of altering the Ubl adjacent to PLP2 of MHV on enzyme activity, viral replication, and pathogenesis. Using deletion and substitution approaches, we identified sites within the Ubl domain, residues 785 to 787 of nonstructural protein 3, which negatively affect protease activity, and valine residues 785 and 787, which negatively affect deubiquitinating activity. Using reverse genetics, we engineered Ubl mutant viruses and found that AM2 (V787S) and AM3 (V785S) viruses replicate efficiently at 37°C but generate smaller plaques than wild-type (WT) virus, and AM2 is defective for replication at higher temperatures. To evaluate the effect of the mutation on protease activity, we purified WT and Ubl mutant PLP2 and found that the proteases exhibit similar specific activities at 25°C. However, the thermal stability of the Ubl mutant PLP2 was significantly reduced at 30°C, thereby reducing the total enzymatic activity. To determine if the destabilizing mutation affects viral pathogenesis, we infected C57BL/6 mice with WT or AM2 virus and found that the mutant virus is highly attenuated, yet it replicates sufficiently to elicit protective immunity. These studies revealed that modulating the Ubl domain adjacent to the PLP reduces protease stability and viral pathogenesis, revealing a novel approach to coronavirus attenuation.
IMPORTANCE Introducing mutations into a protein or virus can have either direct or indirect effects on function. We asked if changes in the Ubl domain, a conserved domain adjacent to the coronavirus papain-like protease, altered the viral protease activity or affected viral replication or pathogenesis. Our studies using purified wild-type and Ubl mutant proteases revealed that mutations in the viral Ubl domain destabilize and inactivate the adjacent viral protease. Furthermore, we show that a CoV encoding the mutant Ubl domain is unable to replicate at high temperature or cause lethal disease in mice. Our results identify the coronavirus Ubl domain as a novel modulator of viral protease stability and reveal manipulating the Ubl domain as a new approach for attenuating coronavirus replication and pathogenesis.
INTRODUCTION
Coronaviruses are emerging human pathogens. Severe acute respiratory syndrome coronavirus (SARS-CoV) caused the epidemic of 2002 to 2003, with a ∼10% case fatality ratio (1). Middle East respiratory syndrome coronavirus (MERS-CoV) is a pathogenic virus that was first identified in humans in 2012 (2). As of 3 February 2015, there have been 965 confirmed cases and 357 deaths (http://www.who.int/csr/don/03-february-2015-mers/en/). For SARS-CoV, the virus emerged from a reservoir in bats, replicated in an intermediate host (civet cats), and spread to humans. The epidemic strain of SARS-CoV evolved for efficient human-to-human spread (3–5). Public health measures of isolation of infected individuals led to the cessation of the epidemic in humans; however, SARS-like viruses remain in bat reservoirs (6–8). For MERS-CoV, dromedary camels now are suspected as the zoonotic source for transmission to humans, since MERS-CoV sequences with 99% nucleotide identity to human MERS-CoV isolates have been detected in respiratory samples from camels (9). Although there are reports of human-to-human transmission of MERS-CoV (10, 11), current strains seem to cause mostly lower respiratory tract disease and are not as highly transmissible as SARS-CoV (12). Other human coronaviruses (HCoV-229E, HCoV-OC43, HCoV-NL63, and HCoV-HKU1) are common in the human population and are the causative agents of upper and lower respiratory tract disease and croup (13–16). To date, there are no FDA-approved antiviral drugs or vaccines to fight human coronavirus-induced disease. In addition, the potential exists for coronaviruses to emerge into the human population from reservoirs in bats or other animals (17). Identifying viral components critical for efficient replication and manifestation of disease will facilitate antiviral drug and vaccine development.
Coronaviruses are RNA viruses that encode a replicase polyprotein at the 5′ end of the positive-strand genome. Upon virus entry, the genomic RNA is translated to produce replicase polyproteins (pp1a and pp1ab), which are processed by virally encoded proteases. Depending on the virus species, the pp1a encodes one or two papain-like proteases (PLPs) and one 3C-like proteinase (3CLpro or Mpro). These proteases process the replicase polyproteins into nonstructural proteins (nsps) that assemble with cellular membranes and facilitate virus replication (reviewed in reference 18). Mouse hepatitis virus (MHV), the murine coronavirus used in this report, is commonly used as a model system to study the replication and pathogenesis of coronaviruses (19).
The MHV replicase product nsp3 consists of multiple domains, including two PLP domains (PLP1 and PLP2), two predicted ubiquitin-like domains (Ubl), an acidic region (Ac), ADP-ribose-1″-phosphatase (ADRP), nucleic acid-binding domain (NAB), coronavirus group 2 marker domain (G2M), transmembrane segment (TM), and coronavirus highly conserved domain (Y) (reviewed in reference 20 and shown in Fig. 1A). The Ubl-1 domain has been shown to interact with nucleocapsid (N) protein, which is important for virus replication (20, 21). Previous studies revealed that MHV PLP1 activity is required for processing the polyprotein at the nsp1/nsp2 and nsp2/nsp3 sites (22). Further, the catalytic activity of PLP1 is required for efficient virus replication (23). PLP2 was shown to recognize and process an LXGG motif and cleave the replicase polyprotein at the nsp3/nsp4 junction (24). The LXGG recognition site is similar to the RLRGG recognition site of cellular deubiquitinating enzymes. Lindner et al. were the first to predict that CoV PLPs could be multifunctional enzymes with both protease and deubiquitinating (DUB) activity (25). Indeed, further studies revealed that SARS-CoV PLpro, MERS-CoV PLpro, HCoV-NL63 PLP2, porcine epidemic diarrhea virus (PEDV) PLP2, and MHV PLP2 are multifunctional enzymes exhibiting protease, DUB, and deISGylating (the ability to deconjugate interferon-stimulated gene 15 protein, ISG15, from substrates [26–33]) activities. CoV PLP activity is required for processing the replicase polyprotein and predicted to modulate the innate immune response by deubiquitination of signaling molecules activated by pattern recognition receptors, such as RIG-I and MDA5 (27–29). Structural studies of the papain-like protease domain of SARS-CoV (35, 36) and, more recently, of MERS-CoV (37) and MHV (Y. Chen, S. N. Savinov, A. M. Mielech, T. Cao, S. C. Baker, and A. D. Mesecar, unpublished data) show that these enzymes belong to the ubiquitin-specific protein (USP) family of deubiquitinating enzymes and reveal the presence of a ubiquitin-like domain (Ubl) located upstream of the protease domain (36). However, the role of the Ubl domain in modulating the enzyme activity of MHV PLP2 is unknown.
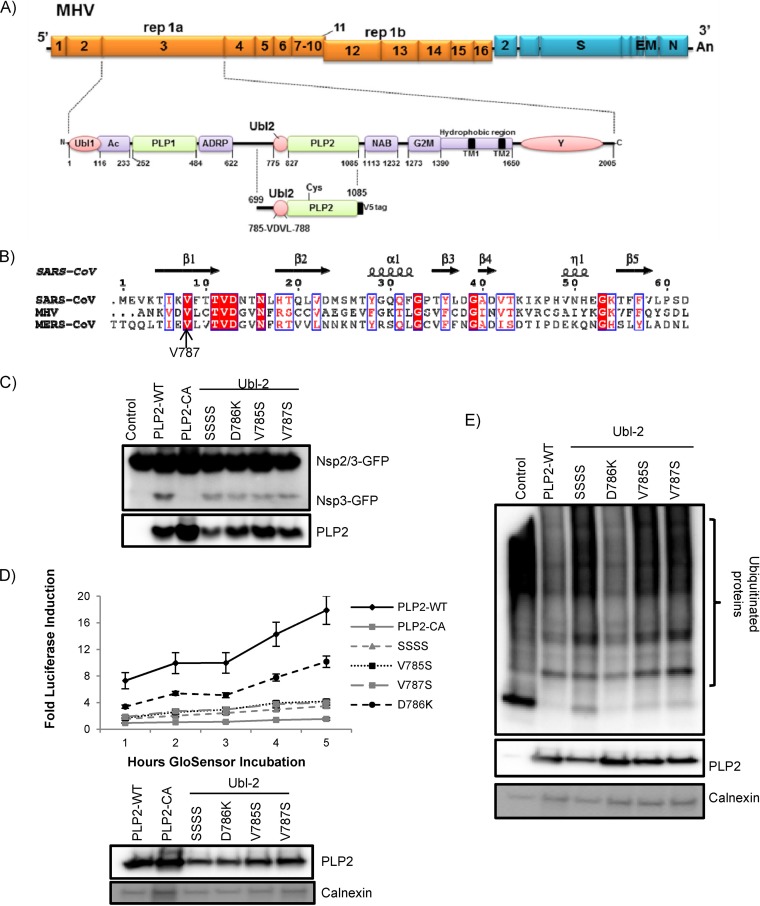
FIG 1.
MHV-A59 Ubl-2 domain influences papain-like protease activity. (A) Schematic diagram of MHV-A59 ORFs and the Ubl-2 and papain-like protease (PLP) domains within nonstructural protein 3. Expression plasmid PLP2 and the VDVL region (aa 785 to 788) are indicated. (B) Alignment of the predicted ubiquitin-like domains from MHV and MERS-CoV with the structural information from the Ubl domain of SARS-CoV (PDB code 2FE8). (C) HEK293T cells were transfected with plasmids expressing the PLP2 wild type, PLP2 catalytic mutant CA, and VDV Ubl-2 domain mutants in the presence of plasmid expressing the nsp2/3-GFP substrate. At 24 h posttransfection, cells were lysed and analyzed by Western blotting. (D) PLP2 activity in live-cell assay. HEK293T cells were transfected with pGlo-RLKGG and the indicated PLP2 expression plasmids. At 14 h postinfection, GloSensor was added and luminescence was assessed hourly. (E) HEK293T cells were transfected with Flag-Ub expression plasmid and the wild type (PLP2-WT) or indicated Ubl-2 mutants. Cells were lysed 24 h posttransfection and analyzed by Western blotting. The figure shows representative data from at least two independent experiments.
Here, we investigated the role of the Ubl domain (designated Ubl-2) adjacent to PLP2 for its function in MHV replication and pathogenesis. We generated proteases and viruses containing mutations within the Ubl domain and found that these mutations decreased PLP2 activity and stability. Further, we found that these mutations resulted in decreased virus replication and a marked attenuation of virulence. Immunization with the Ubl mutant virus protected mice against challenge with wild-type virus. Overall, our data demonstrate, for the first time, that manipulation of the Ubl domain adjacent to a viral protease can ameliorate viral pathogenicity in vivo.
MATERIALS AND METHODS
Cells.
HEK293T cells were cultured in Dulbecco's modified Eagle medium (DMEM) with 10% fetal bovine serum (FBS) and 2% l-glutamine. DBT cells were cultured in minimal essential medium (MEM) with 5% FBS, 2% l-glutamine, and 10% tryptose phosphate broth (TPB). BHK-R cells were kindly provided by Mark Denison (Vanderbilt University Medical Center) and maintained in DMEM supplemented with 10% FBS, 2% l-glutamine, and 0.8 mg/ml G418.
Plasmids and mutagenesis.
PLP2 (amino acids 1525 to 1911) sequence in frame with a V5 epitope tag was codon optimized and synthesized by Gene Script (Piscataway, NJ) (sequence available upon request). Codon-optimized synthetic PLP2 sequence was cloned into pCAGGS-MCS vector. For mutagenesis, an overlapping PCR strategy was used with the primers described in Table 1. The introduced mutations were verified by sequencing. The nsp2/3-green fluorescent protein (GFP) expression plasmid was kindly provided Ralph Baric (University of North Carolina). The Flag-Ub plasmid was kindly provided by Adriano Marchese (Loyola University Chicago).
TABLE 1.
Primers
| Name | Primer sequence |
|
|---|---|---|
| Forward | Reverse | |
| PLP2-CA | 5′ G TCA AAC AAC AAC GCC TAC ATC AAC G | 5′ CGT TGA TGT AGG CGT TGT TGT TTG AC |
| SSSS | 5′ GGC CAA TAA GAG CAG TAG TAG TTG CAC CGT CGA CGG | 5′ CCG TCG ACG GTG CAA CTA CTA CTG CTC TTA TTG GCC |
| V785S | 5′ GGC CAA TAA G AGT GAT GTG CTG TG CAC CGT CGA CG | 5′ CGT CGA CGG TGC ACA GCA CAT CAC TCT TAT TGG CC |
| D786K | 5′ GGC CAA TAA G GTC AAA GTG CTG TG CAC CGT CGA CG | 5′ CGT CGA CGG TGC ACA GCA CTT TGA CCT TAT TGG CC |
| V787S | 5′ GGC CAA TAA G GTC GAT TCG CTG TG CAC CGT CGA CG | 5′ CGT CGA CGG TGC ACA GCG AAT CGA CCT TAT TGG CC |
| AM1 | 5′ CTC GCT AAT AAG AGT TCG TCC TCG TGT ACT GTT GAT GG | 5′ CCA TCA ACA GTA CAC GAG GAC GAA CTC TTA TTA GCG AG |
| AM2 | 5′ CTC GCT AAT AAG GTT GAT TCC TTG TGT ACT GTT GAT GG | 5′ CCA TCA ACA GTA CAC AAG GAA TCA ACC TTA TTA GCG AG |
| AM3 | 5′ CTC GCT AAT AAG TCT GAT GTC TTG TGT ACT GTT GAT GG | 5′ CCA TCA ACA GTA CAC AAG ACA TCA GAC TTA TTA GCG AG |
| Replicase | 5′ GAA TGC CCG GGC GGG ATT TTT GTA TCC | 5′ CCA CAA GAT CTG CCT CGG ACA AAT C |
Protease and deubiquitinase (DUB) activity assays.
To determine the catalytic activity of the PLP2 constructs, 70% confluent HEK293T cells in 12-well CellBIND plates (Corning) were transfected using TransIT-LT1 reagent (Mirus) according to the manufacturer's protocol. For the protease assay, the cells were transfected with 25 ng nsp2/3-GFP plasmid and 300 ng pCAGGS-PLP2-V5 expression plasmids (wild-type PLP2 and various PLP2 mutants). To assess DUB activity, the cells were transfected with 600 ng Flag-Ub plasmid and pCAGGS-PLP2-V5 expression plasmids (wild type and various PLP2 mutants). At 24 h posttransfection, cells were lysed with 300 μl of lysis buffer A containing 4% SDS and 3% dithiothreitol (DTT). Proteins were separated by SDS-PAGE and transferred to a polyvinylidene difluoride (PVDF) membrane in transfer buffer (0.025 M Tris, 0.192 M glycine, 20% methanol) for 1 h at 55 V at 4°C. Following this, the membrane was blocked using 5% dried skim milk in TBST buffer (0.9% NaCl, 10 mM Tris-HCl, pH 7.5, 0.1% Tween 20) overnight at 4°C. The membrane was incubated with polyclonal rabbit anti-GFP antibody (Life Technologies) at a dilution of 1:2,000 for the protease assay or mouse anti-flag (Sigma) at a dilution of 1:2,000 for the DUB assay. The membrane was washed 3 times for 15 min in TBST buffer, followed by incubation with secondary donkey-anti-rabbit-horseradish peroxidase (HRP) antibody at a dilution of 1:2,000 (Amersham) for protease assay or goat anti-mouse-HRP antibody at a dilution of 1:5,000 (Amersham). The membrane then was washed 3 times for 15 min in TBST buffer. Detection was performed using Western lightning chemiluminescence reagent plus (PerkinElmer) and visualized using a FluoroChemE imager (Protein Simple). To verify the expression of the PLP2 constructs, membranes were probed with mouse anti-V5 (Life Technologies) antibody at a dilution of 1:5,000. Mouse anti-calnexin (Cell Signaling) antibody at a dilution of 1:2,000 was used as a loading standard.
Biosensor live cell assay.
To determine the protease activity of the Ubl mutants, the previously described protocol was used (39). Briefly, HEK293T cells were transfected with 37.5 ng pGlo-RLKGG construct and 50 ng of PLP2 expression plasmids. At 18 h posttransfection, GloSensor (Promega) reagent diluted 1:50 in DMEM (10% FBS) was added. The luminescence was measured using a luminometer (Veritas) every hour for 5 h.
Generating Ubl-2 mutant viruses.
To introduce mutations into MHV A59, we used a previously described method (38). Briefly, plasmid encoding the MHV B subclone was mutagenized using primers described in Table 1. Plasmids encoding the complete virus genome were digested with restriction enzymes, gel purified, and ligated using T4 ligase at 16°C overnight. The ligation reaction was isopropanol precipitated, and in vitro RNA transcription was performed using a mMESSAGE mMACHINE kit (Ambion) according to the following protocol: 40.5°C for 25 min, 37.5°C for 50 min, and 40.5°C for 25 min. RNA was electroporated into BHK-R cells, and the electroporated cells were seeded onto DBT cells. The supernatant was harvested 36 h postelectroporation, and plaque assay was performed as described previously (23). RNA was extracted from infected DBT cells at 8 h postinfection using RNeasy Mini (Qiagen), and cDNA was generated using an RT2 first-strand kit (Qiagen). PCR was performed using replicase primers (Table 1), and purified PCR product was sequenced (amino acids 747 to 848). AM2 and AM3 were plaque purified, and AM2 was subjected to deep sequencing.
Temperature shift experiment.
DBT cells in 6-well plates were infected at a multiplicity of infection (MOI) of 0.1 with wild-type MHV or AM2 at 37°C. At 2, 4, or 6 h postinfection, cells were moved to 39.5°C until 14 h postinfection, when supernatant was harvested and virus titer was determined by plaque assay at 37°C as described previously (24).
Expression and purification of wild-type and V787S mutant proteins.
The PLP2 sequence was cloned from pCAGGS-MCS-PLP2 (described above) into ligation-independent cloning (LIC) vector pEV-L8, which is a modified pET-30 plasmid. The expression of wild-type PLP2 and the V787S mutant was performed using Escherichia coli strain BL21(DE3). Cultures were grown in LB medium supplemented with kanamycin (50 μg/ml) at 37°C until the optical density at 600 nm (OD600) reached 0.6. PLP2 expression then was induced with 0.1 mM isopropyl-β-d-thiogalactopyranoside (IPTG) at 25°C for 6 h. Wild-type PLP2 was induced at 25°C for 6 h, while the V787S mutant was induced at 18°C overnight. Cells were harvested by centrifugation at 4,690 × g for 20 min at 4°C and stored at −80°C until use. Cell pellets from 1 liter of culture were resuspended in 40 ml buffer A (25 mM Tris, pH 7.0, 500 mM NaCl, 20 mM imidazole, 5 mM β-mercaptoethanol [βME]) supplemented with dissolved flakes of lysozyme and DNase, lysed through sonication, and centrifuged at 28,880 × g for 30 min (4°C). The supernatant was filtered through a 0.45-μm membrane (Millipore) and loaded onto a 5-ml Ni HiTrap HP column (GE Health Care) preequilibrated with buffer A. The column then was washed with buffer A supplemented with 5% buffer B (25 mM Tris, pH 7.0, 500 mM NaCl, 500 mM imidazole, 5 mM βME) until the UV level was back to the baseline. The protein was eluted through a gradient of 5% to 100% buffer B in 30 column volumes (CV). Fractions were collected and then pooled after enzymatic activity and purity assessment. The N-terminal (His)8 tag then was removed by tobacco etch virus (TEV) protease (His tagged) cleavage by incubating PLP2 and TEV protease together overnight at 4°C while dialyzing into buffer C (25 mM Tris, pH 7.0, 100 mM NaCl, 10 mM βME). Free His tag, TEV protease, and uncleaved PLP2 then were separated from cleaved PLP2 by running the sample over a Ni2+-charged HiTrap column. The flowthrough was collected and then concentrated using Millipore Micron concentrators to a volume of less than 2 ml. The concentrated sample was loaded onto a Superdex-75 HiLoad 26/60 column (GE Healthcare) preequilibrated with buffer D (50 mM HEPES, pH 7.0, 100 mM NaCl, 10 mM dithiothreitol [DTT]) and eluted at a flow rate of 2 ml/min. Fractions containing active enzyme at high purity, as judged by SDS-PAGE, were pooled, concentrated, and flash-frozen in 2% glycerol using liquid nitrogen for storage at −80°C.
Temperature inactivation of wild-type and V787S purified protein.
Wild-type PLP2 and the V787S mutant protein were incubated at 25°C for different time periods (0 to 50 min). At each time point, the specific activity of both enzymes was measured at 25°C using a Synergy multimode microplate reader (BioTek) with 50 μM RLRGG-AMC as the substrate and 3 μM each enzyme. The assay buffer used contained 50 mM HEPES, pH 7.0, 0.1 mg/ml bovine serum albumin (BSA), and 2 mM DTT. The experiments were performed in triplicate in a final volume of 100 μl using 96-well Corning Costar black microplates. Similar experiments were carried out when the enzymes were incubated at 30°C. To analyze the kinetic data, the ratio of the reaction rate at time t to the rate at time 0 was plotted on a logarithmic scale against incubation time. Kinetic data of the V787S PLP2 incubated at 30°C were fitted to a first-order exponential decay model (Ratet/Rate0 = e−kt), from which the inactivation rate constant, kinact, and half-life, t1/2, were determined. The wild-type data were fit to a line, since no significant temperature inactivation was observed.
Thermal Tm analysis using CD.
Thermal melting analyses of the wild-type PLP2 and V787S mutant was carried out with a Chirascan circular dichroism (CD) spectrometer (Applied Photophysics) equipped with a temperature control system (Quantum Northwest Inc.) by monitoring the CD signal at 220 nm while increasing the temperature at a step interval of 0.4°C and at a rate 0.5°C/min. Two ml of protein samples at 1 μM in buffer with 0.1 M potassium phosphate (pH 7.5) was contained in a 10-mm quartz cell (Starna Cells) with magnetic stirring. Thermal scans were performed in three independent experiments for both wild-type and V787S mutant MHV PLP2. The melting temperatures (Tm) were calculated as the first derivative peak using the program SigmaPlot.
Generation of bone marrow-derived macrophages and virus infection.
Femur and tibia from 8-week-old C57BL/6 female mice were prepared, and bone marrow was isolated and cultured in Iscove's modified Dulbecco's medium (IMDM) with 10% FCS, 1% penicillin-streptomycin (pen-strep), 0.1% beta-mercaptoethanol, and 5% macrophage colony-stimulating factor (M-CSF) obtained from L929 supernatants. Additional medium was added at day 3 postisolation, and cells were seeded at day 6 at a density of 2 × 105 cells/well. The following day, cells were infected with wild-type infectious clone MHV (icMHV) A59 and AM2 at an MOI of 1. Medium was changed after 2 h, and supernatant was harvested at various time points postinfection for titration. Supernatant was titrated on murine fibroblast L929 cells (in MEM containing 10% FCS and 1% pen-strep).
Mouse studies.
C57BL/6 mice were purchased from the Jackson Laboratory. The mice were maintained and experiments performed at Loyola University Chicago in accordance with all federal and university guidelines. Four-week-old C57BL/6 male mice were anesthetized with ketamine-xylazine prior to intracranial injection with 600 PFU of wild-type icMHV A59 or AM2 mutant. Weight loss and survival were monitored on a daily basis. Mice were sacrificed when they lost 25% of their initial body weight. In other experiments, mice infected with AM2 at 4 weeks of age and age-matched controls were challenged with 6,000 PFU of wild-type icMHV-A59 at 9 weeks after primary infection. Survival and weight loss were monitored daily. Mice were sacrificed when they lost 25% of their initial body weight. For histology, livers were fixed in 4% formalin and paraffin embedded. Sections were stained with hematoxylin and eosin and analyzed by microscopy.
qRT-PCR analysis.
Brains and livers of 4-week-old male mice infected with wild-type icMHV or AM2 were harvested and homogenized at days 3 and 5 postinfection, and RNA was extracted using RNeasy Mini (Qiagen). Reverse transcription was performed using 2 μg of total RNA and the RT2 first-strand kit (Qiagen) according to the manufacturer's protocol. One μl of cDNA was used to set up the quantitative reverse transcription-PCR (qRT-PCR) according to the manufacturer's protocol using single-primer assay for interleukin-6 (IL-6) (SABiosciences) or replicase primers (Table 1). Threshold cycle (CT) values were normalized to the housekeeping gene (actin).
Nucleotide sequence accession number.
The engineered AM2 strain was subjected to deep sequencing, and the sequence was deposited in GenBank under accession number KP887098.
RESULTS
Identifying the ubiquitin-like domain 2 (Ubl-2) as a modulator of MHV papain-like protease activity.
To identify domains and residues within nsp3 that modulate protease activity, we generated a synthetic, codon-optimized parent construct encoding the protease (PLP2, residues 699 to 1087) in frame with a V5 epitope tag (Fig. 1A) and introduced a series of mutations into the predicted ubiquitin-like domain 2 (Fig. 1B). Protease activity was evaluated using an established transcleavage assay (Fig. 1C). Our goal was to identify a domain that modulated but did not inactivate protease activity. As a negative control, we generated a PLP2 catalytic mutant (C890A) that was inactive in the transcleavage assay. We generated a quadruple mutant (amino acids [aa] 785 to 788; termed VDVL/SSSS) and specific single mutants: V785S, D786K, and V787S. We found that substitution of specific residues within the ubiquitin-like domain 2 adjacent to PLP2 consistently affected protease activity and reduced the abundance of cleavage product detected in the transcleavage assay compared to the activity of PLP2-WT (Fig. 1C). To determine the degree to which mutants' protease activity declined, we used the recently described GloSensor assay (39). We found that, consistent with transcleavage assay results, all Ubl-2 mutants had decreased protease activity. However, VDVL/SSSS, V785S, and V787S showed the highest defect in protease activity, while D786K showed an intermediate phenotype (Fig. 1D). In these experiments, similar levels of wild-type PLP2 and mutants were expressed in each sample. These results implicated the Ubl-2 domain as a modulator of viral protease activity in MHV. Further, we determined the deubiquitinating (DUB) activity of the Ubl-2 mutants. We found that the quadruple mutant and the single mutants V785S and V787S had reduced DUB activity compared to that of wild-type PLP2. In contrast, the D786K mutant had DUB activity similar to that of wild-type PLP2 (Fig. 1E). Therefore, we focused on the Ubl-2 V785S and V787S mutants, since these Ubl mutants exhibited the most significant defects in the cell-based assays.
Ubl-2 mutant virus MHV-AM2 is temperature sensitive.
To determine if substitutions in the Ubl-2 domain affected virus replication, we used reverse genetics (38, 40) to introduce specific mutations into the viral genome and assessed viral replication. Since the phenotype of the mutations in the PLP2 assay (Fig. 1) can be different from the phenotype of the mutant in the context of replicating virus, we generated viruses encoding a quadruple mutation (VDVL/SSSS), designated MHV-AM1, a single mutation (V787S), designated MHV-AM2, and another single mutation (V785S), designated MHV-AM3 (Fig. 2A). The presence of the mutations was confirmed by sequencing within the PLP2 domain. MHV-AM1 was highly impaired, generated pinpoint-sized plaques, and grew to low titer (1 × 103 PFU/ml). The low titer and pinpoint plaques generated by AM1 likely are due to the quadruple mutation in PLP2; however, we cannot rule out the possibility that additional mutations outside PLP2 contribute to this phenotype.
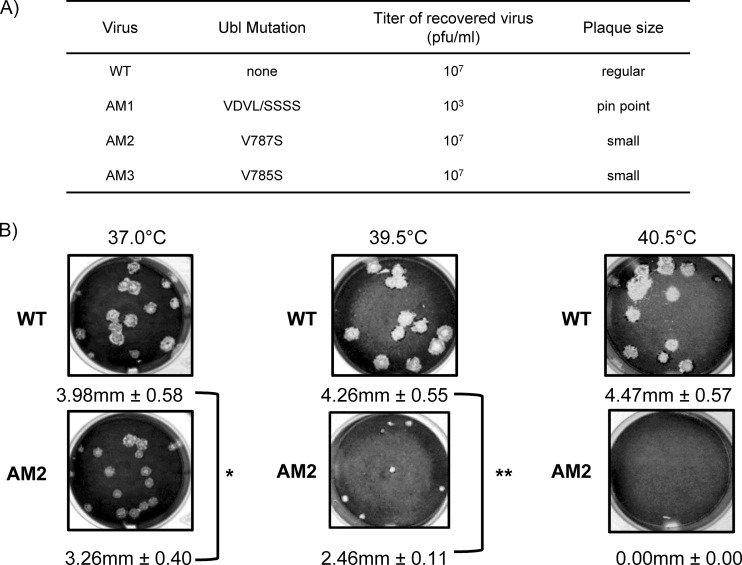
FIG 2.
Murine coronavirus AM2 is temperature sensitive for replication. (A) Summary of the characteristics of Ubl mutant viruses recovered by reverse genetics. (B) Representative plaques generated by wild-type MHV (WT) or AM2 during incubation at the indicated temperature. DBT cells were inoculated with either the WT or AM2, incubated at the indicated temperature, and fixed and stained 48 h postinfection. The diameter of each plaque was measured, and average diameters and standard deviations are indicated for each virus. *, P < 0.05; **, P < 0.001. Representative data from at least two independent experiments are shown.
In contrast, AM2 and AM3 viruses with single-amino-acid substitutions within the Ubl-2 domain replicated to titers similar to that of the parent infectious clone. We found that both AM2 and AM3 viruses generated slightly smaller plaques than those generated by wild-type MHV A59. The small-plaque phenotype can be associated with temperature sensitivity, so we evaluated relative plaque size of the wild type and AM2 by performing plaque assays at 39.5°C and 40.5°C. We found that AM2 produced small plaques at 39.5°C (about 50% reduction compared to the level for the wild type) but was unable to generate plaques at 40.5°C (Fig. 2B), consistent with a temperature-sensitive defect. AM3 also produced plaques that were slightly smaller than those of the wild type but bigger than AM2 plaques at 39.5°C (about 20% reduction compared to the wild type) (data not shown). Because the temperature-sensitive phenotype was more severe for AM2 and because V787 is conserved in other betacoronaviruses, such as SARS-CoV and MERS-CoV (Fig. 1B), we focused our studies on AM2. We verified the engineered sequence of AM2 by deep sequencing of the viral genomic RNA. The 2-nucleotide substitution (GTC to CCC) of V787 was confirmed, and no additional changes were detected.
To determine the replication kinetics of AM2, we infected DBT cells with isogenic wild-type MHV-A59 or AM2 virus at an MOI of 0.1. The cells were incubated at 37°C, supernatant was harvested, and virus titer was determined by plaque assay. We found that at 37°C, AM2 replicated with kinetics similar to those of wild-type virus (Fig. 3A). In addition, no replication defect was observed in bone marrow-derived macrophages, which are important primary target cells during MHV infection (Fig. 3B). However, if infected DBT cells were shifted to a higher temperature (39.5°C), there was a progressive reduction in viral titer with increased time at the higher temperature (Fig. 3C). When the cells were incubated at 37°C for 6 h and then shifted to the higher temperature, the difference between the titers of the WT and AM2 was about half a log. In contrast, when the infected cells were incubated at 37°C for 2 h and then shifted to 39.5°C, the titer of AM2 was two logs lower than that of wild-type virus. Taken together, these data show that AM2 encoding V787S has a temperature-sensitive defect in viral replication.
FIG 3.
Replication kinetics of AM2. (A) AM2 replication is similar to that of MHV-WT at 37°C. DBT cells were infected with AM2 or the WT at an MOI of 0.1. (B) Bone marrow-derived macrophages were infected with AM2 or the WT at an MOI of 1. At the indicated time points, supernatant was harvested and virus titer determined by plaque assay at 37°C. (C) Temperature shift reduces yield of AM2. DBT cells were infected at an MOI of 0.1 at 37°C with AM2 or the WT. At the indicated time point postinfection, cells were moved to 39.5°C until 14 hpi, when supernatant was harvested and virus titer was determined by plaque assay at 37°C. The error bars represent standard deviations within single experiments. Shown are representative data from at least two independent experiments.
nsp3-V787S substitution impairs PLP2 thermal stability.
To investigate the temperature-sensitive phenotype conferred by the V787S substitution, we expressed the wild type and the V787S mutant of MHV PLP2 in E. coli and then purified the enzymes to homogeneity using a multistep chromatography procedure (Fig. 4A). The thermal stability of the wild-type and V787S-purified enzymes then was determined at 25°C and at 30°C by measuring the specific activity of the enzymes as a function of incubation time using the peptide RLRGG-AMC as a substrate. We found that when wild-type and V787S mutant PLP2s were incubated at 25°C, both enzymes were stable over the incubation time course and had similar specific activities (Fig. 4B). The activities shown in Fig. 4B have been normalized to 100%. In contrast, when wild-type and V787S mutant PLP2s were incubated at a higher temperature (30°C), the specific activity of V787S PLP2 decreased rapidly over time, whereas the wild-type protein maintained full activity (Fig. 4B). These results indicate that the V787S enzyme is thermally unstable at 30°C, with a half-life of 27.7 ± 1.1 min.
FIG 4.
Ubl-2 domain is important for MHV PLP2 thermal stability. (A) SDS-PAGE analysis of the final purified PLP2-V787S enzyme. Wild-type and PLP2-V787S mutant PLP2 were expressed and purified from E. coli as described in Materials and Methods. (B) Temperature-dependent inactivation of wild-type PLP2 and PLP2-V787S mutant. The specific activities of the WT and the V787S mutant PLP2 enzymes were measured after incubation at 25°C and 30°C for different time periods. The specific activities then were normalized to the activity at 0 min (Ratet/Rate0 indicates the rate at time t over the initial rate). Kinetic data for the V787S mutant incubated at 30°C were fit to a first-order exponential decay model (Ratet/Rate0 = e−kt). The calculated inactivation rate constant (kinact) for the V787S mutant at 30°C is 0.025 ± 0.001 min−1, and the half-life (t1/2) is 27.7 ± 1.1 min. (C) CD melting curves of WT and PLP2-V787S mutant PLP2. The thermal stability of WT and V787S mutant PLP2 was determined by measuring the CD signal at 220 nm as a function of temperature at a step interval of 0.4°C and at a rate of 0.5°C/min. Three independent experiments were performed for both WT PLP2 (gray) and the V787S mutant (black).
To determine the relative structural stability of the purified proteins over a wider temperature range, we performed thermal denaturation experiments using circular dichroism (CD) analysis to determine the melting temperatures (Tm) of each enzyme. Consistent with results shown in Fig. 4B, the V787S mutant exhibited diminished thermostability (Fig. 4C). The determined melting temperature was 6.8°C lower for the V787S mutant protein than for wild-type PLP2, suggesting that the V787S mutant PLP2 is less stable and unfolds at a lower temperature than wild-type PLP2. Taken together, our data indicate that the V787S substitution diminished the overall protein thermal stability, resulting in decreased enzymatic activity at elevated temperatures.
AM2 exhibits reduced pathogenesis and elicits a protective response in mice.
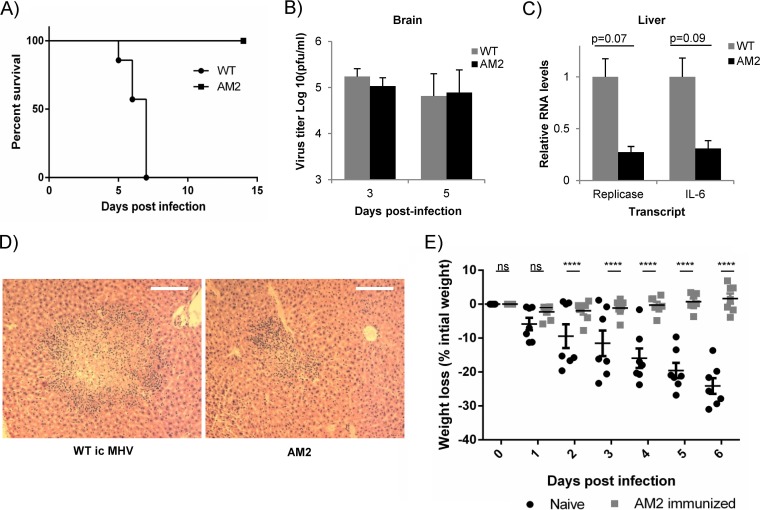
To determine the effect of the Ubl-2 domain mutation on virus pathogenesis, we infected 4-week-old male C57B/L6 mice with 600 PFU of wild-type or AM2 virus by intracranial injection. We monitored weight loss over the course of infection, and mice were humanely euthanized if they lost greater than 25% of their initial body weight. We observed that while 100% of mice infected with wild-type virus succumbed to infection by day 7, all mice infected with AM2 survived the infection (Fig. 5A). We confirmed that the mutation in AM2 was maintained upon mouse infection by sequencing within the PLP2 domain of recovered AM2.
FIG 5.
MHV AM2 is attenuated and generated protective immunity in mice. (A) C57BL/6 mice were infected with 600 PFU WT icMHV or AM2 mutant intracranially and monitored for survival over time (n = 7 for each group). (B) Virus titers in brains of mice infected with 600 PFU WT icMHV or AM2 were determined at the indicated time points p.i. by plaque assay (n = 4 to 5). Error bars represent standard deviations. (C) RNA levels for viral replicase gene and cellular IL-6 were determined using qRT-PCR in livers at day 5 postinfection (n = 6). RNA levels were normalized to actin, and the amount of transcript in WT-infected mice was set to 1. Error bars represent standard errors of the means (SEM). Statistical analysis performed using Student's t test. (D) Representative samples of hematoxylin and eosin staining of formalin-fixed liver samples at day 5 postinfection from mice infected with WT icMHV or AM2 (n = 6 for each group; magnification, ×10). (E) C57BL/6 mice immunized with AM2 mutant and naive age-matched controls were challenged with 6,000 PFU icMHV intracranially 9 weeks after primary infection. The mice were monitored for body weight loss. Error bars represent SEM. ****, P < 0.0001 by two-way analysis of variance (ANOVA). ns, not significant.
We detected similar viral titers in the brains of wild-type- or AM2-infected mice at day 3 or day 5 postinfection (Fig. 5B). In contrast, we found levels of viral replicase and IL-6 were reduced in the livers of AM2-infected mice at day 5 postinfection compared to those of wild-type MHV-infected mice (Fig. 5C). There was no difference in CCL2 or alpha interferon (IFN-α) (data not shown). Furthermore, we analyzed livers for changes in pathology and observed a reduction in the size of the lesions, hepatocyte necrosis, and inflammation in AM2-infected mice compared to results for wild-type-infected mice (Fig. 5D). These data indicate that AM2 is attenuated compared to wild-type MHV.
To determine if primary infection with AM2 mutant virus protected mice from challenge with wild-type virus, we infected AM2-immunized and age-matched naive C57BL/6 mice intracranially with 6,000 PFU of wild-type virus at 9 weeks after primary infection. We monitored body weight loss over the course of infection and found that naive mice lost significantly more weight starting at day 2 postinfection (Fig. 5E). In contrast, AM2-immunized mice did not lose weight and did not exhibit other symptoms of disease upon challenge. Thirteen-week-old C57BL/6 mice are much more resistant to infection than 4-week-old mice, so as expected, no wild type- or AM2-infected mice succumbed to the infection. These results indicate that the immunization with AM2 generated a sufficient immune response to protect the mice from subsequent wild-type challenge.
DISCUSSION
Modulating viral enzyme activity is a powerful approach for generating attenuated viruses. Coronaviruses encode a very large replicase polyprotein that contains multiple enzymatic activities, such as proteases, ADP-ribose-1″-phosphatase (ADRP), methyl-transferase, exonuclease, and RNA-dependent RNA polymerase (Fig. 1). Previous studies have shown that inactivation of the coronavirus papain-like protease by mutagenesis of the required catalytic cysteine residue or inhibition with a protease inhibitor blocks viral replication (23, 34, 35, 41). These approaches demonstrate the requirement for protease activity but represent all-or-nothing approaches and do not allow for the study of reduced or impaired protease activity. Here, we describe an approach involving the mutation of the domain adjacent to the papain-like protease domain, termed the ubiquitin-like domain (Ubl). Structural studies revealed that this domain is highly conserved in coronaviruses, although the function of this domain is unknown. We found that mutation of the Ubl domain destabilizes the adjacent protease domain, leading to the loss of enzymatic activity over time (half-life of ∼30 min at 30°C). When this mutation is introduced into the virus, the mutant virus replicates efficiently in cell culture at 37°C but is temperature sensitive and unable to replicate at high temperature (40.5°C). Indeed, we found that this virus is highly attenuated in infected mice but does replicate sufficiently to induce a protective immune response against wild-type virus. Overall, our studies revealed a new domain that can be exploited for attenuating the pathogenesis of a coronavirus.
Ubl domains have been identified as adjacent to many viral and cellular proteases, particularly the ubiquitin-specific proteases (USPs), a family of deubiquitinating enzymes (DUBs) (42). Ubl domains can be present at the C-terminal or N-terminal end of USP catalytic domains or even can be inserted into the catalytic domain. In addition, USPs may contain one or multiple Ubl domains (43). The Ubl domain of USP14 is required for targeting this cellular DUB to the proteasome. At the proteasome, USP14 removes ubiquitin molecules from proteins that are modified with K48-linked ubiquitin, which serves as a signal for proteosomal degradation (44). Alternatively, the Ubl may regulate function of DUB activity by interacting with other cellular proteins, such as GMP synthetase, which enhances the activity of USP7 (45).
Previous studies have shown that the presence of the Ubl domain of SARS-CoV PLpro is necessary for efficient expression and purification of the enzyme (35). To understand the mechanism by which mutations in the Ubl domain of MHV PLP2 affected function, we mapped the mutated residues V785 and V787 into the recently solved crystal structure of MHV PLP2 (Fig. 6) (Chen et al., unpublished). Interestingly, the overall fold of the protease and Ubl-2 domain is conserved between SARS-CoV PLpro and MHV PLP2. Valine 787, which is the focus of this study, is conserved between MHV and highly pathogenic coronaviruses, such as SARS-CoV and MERS-CoV (Fig. 1B). Valine 787 resides within a β-sheet that helps to maintain the structure of the Ubl. The side chains of valine 785 and valine 787 participate in the formation of a hydrophobic core of the Ubl, which stabilizes this domain. By mutating valine 787 to serine, we introduced a polar side chain that is energetically unable to pack into the hydrophobic core because of significant desolvation penalties. Therefore, mutation of valine 787 or valine 785 likely disrupts the structure of the Ubl domain, thereby leading to the overall destabilization of the PLP2 catalytic domain at elevated temperature. As a consequence, the Ubl mutant enzyme has decreased thermal stability that results in a loss of the specific activity of the enzyme over time in vitro. Furthermore, the destabilization of Ubl-2-PLP2 has an important impact on virus pathogenesis, since we found that MHV-AM2 is attenuated in vivo.
FIG 6.
X-ray structures of the MHV and SARS-CoV Ubl-2 domains. A superposition of the Ubl-2 domains from MHV PLP2 (blue) and SARS-CoV PLpro (orange; PDB code 2FE8) is shown as a stereo view and resulted in an root mean square deviation of 0.74 Å. Side chains of MHV Ubl-2 residues V785 and V787 and their corresponding residues in SARS-CoV are shown as sticks.
Interestingly, our studies revealed that the most dramatic reduction in AM2 viral titer in the mice was in the liver, not the brain, which was the site of inoculation. Since MHV-A59 is hepatotropic, it rapidly disseminates to the liver of infected animals (18). The reduction in viral titer in the liver and the decreased size of lesions in the liver (Fig. 5) could be due to either defective viral replication or a more rapid immune response to clear the virus from the liver. A similar phenotype of virus attenuated in the liver but not the brain was described for the MHV-A59 ns2 mutant (46, 47). In addition, a recent study by Zhang and colleagues showed that an nsp1 mutant virus is attenuated in the liver but replicates efficiently in the brain (48). The attenuated phenotype of the nsp1 mutant virus is hypothesized to be due to a more robust immune response to the virus.
Recent studies on MERS-CoV showed that PLpro has DUB activity and that this activity is important for antagonism of the innate immune response in transfected cells (30, 49). In addition, the arterivirus equine arteritis virus (EAV) PLP DUB activity has been shown to be involved in inhibition of innate immune response in infected cells, suggesting that PLP DUB activity is important for virus pathogenesis in vivo (50). Further studies are needed to determine the role of DUB activity in viral replication and pathogenesis and to determine if the attenuation of AM2 is primarily immune mediated or due to defective replication and dissemination of the virus.
Coronaviruses encode multiple novel enzymatic activities, and previous studies support the concept of modifying/mutating nonstructural proteins to generate attenuated viruses and to identify the role of the protein in virus replication and in causing disease in mice. Mutation of the N-terminal replicase product nsp1 in MHV results in an attenuated infection in mice (51, 52). In addition, mutation of nsp1 in SARS-CoV results in a decrease in antiviral signaling and virus titer in infected cells (53). Mutation of the catalytic residues of the ADP-ribose-1″-phosphatase (ADRP) domain does not diminish virus replication in mice but reduces production of the cytokine IL-6, an important proinflammatory molecule (54–56). Reduction in inflammatory response upon infection with the ADRP mutant may contribute to decreased liver pathology (54). Genetically inactivating the enzyme ExoN, which has proofreading function, results in a hypermutation phenotype virus that is highly attenuated, even in aged, immunocompromised animals (57). Mutation of the 2′-O-methyl-transferase activity in nsp16 revealed that this activity is required to evade viral mRNA detection by the pattern recognition receptor MDA5 (58, 59). Further, deletion or introduction of an inactivating mutation in ns2 in MHV results in the activation of the RNase L pathway, which induces an antiviral state in liver macrophages (46). All of the aforementioned studies reveal the critical roles of viral nonstructural proteins in virus replication and escape from immune surveillance. Our study reveals a role for the Ubl-2 domain in stabilization of the PLP2 catalytic domain of MHV PLP2 and provides a new target for viral attenuation and potentially for antiviral drug development.
ACKNOWLEDGMENTS
This work was supported by NIH grants RO1 AI085089 (to S.C.B. and A.D.M.) and RO1 NS36592 (to S.P.). A.D.M. is also supported by a grant from the Walther Cancer Foundation. A.M.M. was supported in part by an Arthur J. Schmitt Dissertation Fellowship from Loyola University Chicago. E.K. and V.T. were supported by the Swiss National Science Foundation (project 149784).
REFERENCES
- 1.Peiris JS, Guan Y, Yuen KY. 2004. Severe acute respiratory syndrome. Nat Med 10:S88–S97. doi: 10.1038/nm1143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zaki AM, van Boheemen S, Bestebroer TM, Osterhaus AD, Fouchier RA. 2012. Isolation of a novel coronavirus from a man with pneumonia in Saudi Arabia. N Engl J Med 367:1814–1820. doi: 10.1056/NEJMoa1211721. [DOI] [PubMed] [Google Scholar]
- 3.Lau SK, Woo PC, Li KS, Huang Y, Tsoi HW, Wong BH, Wong SS, Leung SY, Chan KH, Yuen KY. 2005. Severe acute respiratory syndrome coronavirus-like virus in Chinese horseshoe bats. Proc Natl Acad Sci U S A 102:14040–14045. doi: 10.1073/pnas.0506735102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Poon LL, Chu DK, Chan KH, Wong OK, Ellis TM, Leung YH, Lau SK, Woo PC, Suen KY, Yuen KY, Guan Y, Peiris JS. 2005. Identification of a novel coronavirus in bats. J Virol 79:2001–2009. doi: 10.1128/JVI.79.4.2001-2009.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Perlman S, Netland J. 2009. Coronaviruses post-SARS: update on replication and pathogenesis. Nat Rev 7:439–450. doi: 10.1038/nrmicro2147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dominguez SR, O'Shea TJ, Oko LM, Holmes KV. 2007. Detection of group 1 coronaviruses in bats in North America. Emerg Infect Dis 13:1295–1300. doi: 10.3201/eid1309.070491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Woo PC, Lau SK, Huang Y, Yuen KY. 2009. Coronavirus diversity, phylogeny and interspecies jumping. Exp Biol Med 234:1117–1127. doi: 10.3181/0903-MR-94. [DOI] [PubMed] [Google Scholar]
- 8.Ge X-Y, Li J-L, Yang X-L, Chmura AA, Zhu G, Epstein JH, Mazet JK, Hu B, Zhang W, Peng C, Zhang Y-J, Luo C-M, Tan B, Wang N, Zhu Y, Crameri G, Zhang S-Y, Wang L-F, Daszak P, Shi Z-L. 2013. Isolation and characterization of a bat SARS-like coronavirus that uses the ACE2 receptor. Nature 503:535–538. doi: 10.1038/nature12711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Briese T, Mishra N, Jain K, Zalmout IS, Jabado OJ, Karesh WB, Daszak P, Mohammed OB, Alagaili AN, Lipkin WI. 2014. Middle East respiratory syndrome coronavirus quasispecies that include homologues of human isolates revealed through whole-genome analysis and virus cultured from dromedary camels in Saudi Arabia. mBio 5:e01146-14. doi: 10.1128/mBio.01146-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Assiri A, McGeer A, Perl TM, Price CS, Al Rabeeah AA, Cummings DA, Alabdullatif ZN, Assad M, Almulhim A, Makhdoom H, Madani H, Alhakeem R, Al-Tawfiq JA, Cotten M, Watson SJ, Kellam P, Zumla AI, Memish ZA, KSA MERS-CoV Investigation Team. 2013. Hospital outbreak of Middle East respiratory syndrome coronavirus. N Engl J Med 369(5):407–416. doi: 10.1056/NEJMoa1306742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Guery B, Poissy J, El Mansouf L, Sejourne C, Ettahar N, Lemaire X, Vuotto F, Goffard A, Behillil S, Enouf V, Caro V, Mailles A, Che D, Manuguerra JC, Mathieu D, Fontanet A, van der Werf S, MERS-CoV Study Group. 2013. Clinical features and viral diagnosis of two cases of infection with Middle East respiratory syndrome coronavirus: a report of nosocomial transmission. Lancet 381:2265–2272. doi: 10.1016/S0140-6736(13)60982-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cotten M, Watson SJ, Zumla AI, Makhdoom HQ, Palser AL, Ong SH, Al Rabeeah AA, Alhakeem RF, Assiri A, Al-Tawfiq JA, Albarrak A, Barry M, Shibl A, Alrabiah FA, Hajjar S, Balkhy HH, Flemban H, Rambaut A, Kellam P, Memish ZA. 2014. Spread, circulation, and evolution of the Middle East respiratory syndrome coronavirus. mBio 5:e01062-13. doi: 10.1128/mBio.01062-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chiu SS, Chan KH, Chu KW, Kwan SW, Guan Y, Poon LL, Peiris JS. 2005. Human coronavirus NL63 infection and other coronavirus infections in children hospitalized with acute respiratory disease in Hong Kong, China. Clin Infect Dis 40:1721–1729. doi: 10.1086/430301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Esper F, Weibel C, Ferguson D, Landry ML, Kahn JS. 2006. Coronavirus HKU1 infection in the United States. Emerg Infect Dis 12:775–779. doi: 10.3201/eid1205.051316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Van der Hoek L, Pyrc K, Jebbink MF, Vermeulen-Oost W, Berkhout RJ, Wolthers KC, Wertheim-van Dillen PM, Kaandorp J, Spaargaren J, Berkhout B. 2004. Identification of a new human coronavirus. Nat Med 10:368–373. doi: 10.1038/nm1024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Vabret A, Mourez T, Gouarin S, Petitjean J, Freymuth F. 2003. An outbreak of coronavirus OC43 respiratory infection in Normandy, France. Clin Infect Dis 36:985–989. doi: 10.1086/374222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lau SK, Li KS, Tsang AK, Lam CS, Ahmed S, Chen H, Chan KH, Woo PC, Yuen KY. 2013. Genetic characterization of betacoronavirus lineage C viruses in bats reveals marked sequence divergence in the spike protein of pipistrellus bat coronavirus HKU5 in Japanese pipistrelle: implications for the origin of the novel Middle East respiratory syndrome coronavirus. J Virol 87:8638–8650. doi: 10.1128/JVI.01055-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mielech AM, Chen Y, Mesecar AD, Baker SC. 2014. Nidovirus papain-like proteases: multifunctional enzymes with protease, deubiquitinating and deISGylating activities. Virus Res 194:184–190. doi: 10.1016/j.virusres.2014.01.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Weiss SR, Leibowitz JL. 2011. Coronavirus pathogenesis. Adv Virus Res 81:85–164. doi: 10.1016/B978-0-12-385885-6.00009-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hurst KR, Ye R, Goebel SJ, Jayaraman P, Masters PS. 2010. An interaction between the nucleocapsid protein and a component of the replicase-transcriptase complex is crucial for the infectivity of coronavirus genomic RNA. J Virol 84:10276–10288. doi: 10.1128/JVI.01287-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hurst KR, Koetzner CA, Masters PS. 2013. Characterization of a critical interaction between the coronavirus nucleocapsid protein and nonstructural protein 3 of the viral replicase-transcriptase complex. J Virol 87:9159–9172. doi: 10.1128/JVI.01275-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ziebuhr J, Thiel V, Gorbalenya AE. 2001. The autocatalytic release of a putative RNA virus transcription factor from its polyprotein precursor involves two paralogous papain-like proteases that cleave the same peptide bond. J Biol Chem 276:33220–33232. doi: 10.1074/jbc.M104097200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Graham RL, Denison MR. 2006. Replication of murine hepatitis virus is regulated by papain-like proteinase 1 processing of nonstructural proteins 1, 2, and 3. J Virol 80:11610–11620. doi: 10.1128/JVI.01428-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kanjanahaluethai A, Baker SC. 2000. Identification of mouse hepatitis virus papain-like proteinase 2 activity. J Virol 74:7911–7921. doi: 10.1128/JVI.74.17.7911-7921.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lindner HA, Fotouhi-Ardakani N, Lytvyn V, Lachance P, Sulea T, Menard R. 2005. The papain-like protease from the severe acute respiratory syndrome coronavirus is a deubiquitinating enzyme. J Virol 79:15199–15208. doi: 10.1128/JVI.79.24.15199-15208.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Barretto N, Jukneliene D, Ratia K, Chen Z, Mesecar AD, Baker SC. 2005. The papain-like protease of severe acute respiratory syndrome coronavirus has deubiquitinating activity. J Virol 79:15189–15198. doi: 10.1128/JVI.79.24.15189-15198.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Clementz MA, Chen Z, Banach BS, Wang Y, Sun L, Ratia K, Baez-Santos YM, Wang J, Takayama J, Ghosh AK, Li K, Mesecar AD, Baker SC. 2010. Deubiquitinating and interferon antagonism activities of coronavirus papain-like proteases. J Virol 84:4619–4629. doi: 10.1128/JVI.02406-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Devaraj SG, Wang N, Chen Z, Tseng M, Barretto N, Lin R, Peters CJ, Tseng CT, Baker SC, Li K. 2007. Regulation of IRF-3-dependent innate immunity by the papain-like protease domain of the severe acute respiratory syndrome coronavirus. J Biol Chem 282:32208–32221. doi: 10.1074/jbc.M704870200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Frieman M, Ratia K, Johnston RE, Mesecar AD, Baric RS. 2009. Severe acute respiratory syndrome coronavirus papain-like protease ubiquitin-like domain and catalytic domain regulate antagonism of IRF3 and NF-kappaB signaling. J Virol 83:6689–6705. doi: 10.1128/JVI.02220-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mielech AM, Kilianski A, Baez-Santos YM, Mesecar AD, Baker SC. 2014. MERS-CoV papain-like protease has deISGylating and deubiquitinating activities. Virology 450-451:64–70. doi: 10.1016/j.virol.2013.11.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Xing Y, Chen J, Tu J, Zhang B, Chen X, Shi H, Baker SC, Feng L, Chen Z. 2013. The papain-like protease of porcine epidemic diarrhea virus negatively regulates type I interferon pathway by acting as a viral deubiquitinase. J Gen Virol 94:1554–1567. doi: 10.1099/vir.0.051169-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yang H, Yang M, Ding Y, Liu Y, Lou Z, Zhou Z, Sun L, Mo L, Ye S, Pang H, Gao GF, Anand K, Bartlam M, Hilgenfeld R, Rao Z. 2003. The crystal structures of severe acute respiratory syndrome virus main protease and its complex with an inhibitor. Proc Natl Acad Sci U S A 100:13190–13195. doi: 10.1073/pnas.1835675100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zheng D, Chen G, Guo B, Cheng G, Tang H. 2008. PLP2, a potent deubiquitinase from murine hepatitis virus, strongly inhibits cellular type I interferon production. Cell Res 18:1105–1113. doi: 10.1038/cr.2008.294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Frieman M, Basu D, Matthews K, Taylor J, Jones G, Pickles R, Baric R, Engel DA. 2011. Yeast based small molecule screen for inhibitors of SARS-CoV. PLoS One 6:e28479. doi: 10.1371/journal.pone.0028479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ratia K, Pegan S, Takayama J, Sleeman K, Coughlin M, Baliji S, Chaudhuri R, Fu W, Prabhakar BS, Johnson ME, Baker SC, Ghosh AK, Mesecar AD. 2008. A noncovalent class of papain-like protease/deubiquitinase inhibitors blocks SARS virus replication. Proc Natl Acad Sci U S A 105:16119–16124. doi: 10.1073/pnas.0805240105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ratia K, Saikatendu KS, Santarsiero BD, Barretto N, Baker SC, Stevens RC, Mesecar AD. 2006. Severe acute respiratory syndrome coronavirus papain-like protease: structure of a viral deubiquitinating enzyme. Proc Natl Acad Sci U S A 103:5717–5722. doi: 10.1073/pnas.0510851103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lei J, Mesters JR, Drosten C, Anemüller S, Ma Q, Hilgenfeld R. 2014. Crystal structure of the papain-like protease of MERS coronavirus reveals unusual, potentially druggable active-site features. Antiviral Res 109:72–82. doi: 10.1016/j.antiviral.2014.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yount B, Denison MR, Weiss SR, Baric RS. 2002. Systematic assembly of a full-length infectious cDNA of mouse hepatitis virus strain A59. J Virol 76:11065–11078. doi: 10.1128/JVI.76.21.11065-11078.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kilianski A, Mielech AM, Deng X, Baker SC. 2013. Assessing activity and inhibition of Middle East respiratory syndrome coronavirus papain-like and 3C-like proteases using luciferase-based biosensors. J Virol 87:11955–11962. doi: 10.1128/JVI.02105-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yount B, Curtis KM, Fritz EA, Hensley LE, Jahrling PB, Prentice E, Denison MR, Geisbert TW, Baric RS. 2003. Reverse genetics with a full-length infectious cDNA of severe acute respiratory syndrome coronavirus. Proc Natl Acad Sci U S A 100:12995–13000. doi: 10.1073/pnas.1735582100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Báez-Santos YM, Barraza SJ, Wilson MW, Agius MP, Mielech AM, Davis NM, Baker SC, Larsen SD, Mesecar AD. 2014. X-ray structural and biological evaluation of a series of potent and highly selective inhibitors of human coronavirus papain-like proteases. J Med Chem 57:2393–2412. doi: 10.1021/jm401712t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhu X, Ménard R, Sulea T. 2007. High incidence of ubiquitin-like domains in human ubiquitin-specific proteases. Proteins 69:1–7. doi: 10.1002/prot.21546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Faesen AC, Luna-Vargas MPA, Sixma TK. 2012. The role of UBL domains in ubiquitin-specific proteases. Biochem Soc Trans 40:539–545. doi: 10.1042/BST20120004. [DOI] [PubMed] [Google Scholar]
- 44.Hu M, Li P, Song L, Jeffrey PD, Chenova TA, Wilkinson KD, Cohen RE, Shi Y. 2005. Structure and mechanisms of the proteasome-associated deubiquitinating enzyme USP14. EMBO J 24:3747–3756. doi: 10.1038/sj.emboj.7600832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Faesen AC, Dirac AMG, Shanmugham A, Ovaa H, Perrakis A, Sixma TK. 2011. Mechanism of USP7/HAUSP activation by its C-terminal ubiquitin-like domain and allosteric regulation by GMP-synthetase. Mol Cell 44:147–159. doi: 10.1016/j.molcel.2011.06.034. [DOI] [PubMed] [Google Scholar]
- 46.Zhao L, Jha BK, Wu A, Elliott R, Ziebuhr J, Gorbalenya AE, Silverman RH, Weiss SR. 2012. Antagonism of the interferon-induced OAS-RNase L pathway by murine coronavirus ns2 protein is required for virus replication and liver pathology. Cell Host Microbe 11:607–616. doi: 10.1016/j.chom.2012.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zhao L, Rose KM, Elliott R, Van Rooijen N, Weiss SR. 2011. Cell-type-specific type I interferon antagonism influences organ tropism of murine coronavirus. J Virol 85:10058–10068. doi: 10.1128/JVI.05075-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zhang R, Li Y, Cowley TJ, Steinbrenner A, Phillips JM, Yount BL, Baric RS, Weiss SR. 14January2015. The nsp1, nsp13, and M proteins contribute to the hepatotropism of murine coronavirus JHM.WU. J Virol doi: 10.1128/JVI.03535-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bailey-Elkin BA, Knaap RCM, Johnson GG, Dalebout TJ, Ninaber DK, van Kasteren PB, Bredenbeek PJ, Snijder EJ, Kikkert M, Mark BL. 2014. Crystal structure of the Middle East respiratory syndrome coronavirus (MERS-CoV) papain-like protease bound to ubiquitin facilitates targeted disruption of deubiquitinating activity to demonstrate its role in innate immune suppression. J Biol Chem 289:34667–34682. doi: 10.1074/jbc.M114.609644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Van Kasteren PB, Bailey-Elkin BA, James TW, Ninaber DK, Beugeling C, Khajehpour M, Snijder EJ, Mark BL, Kikkert M. 2013. Deubiquitinase function of arterivirus papain-like protease 2 suppresses the innate immune response in infected host cells. Proc Natl Acad Sci U S A 110:E838–E847. doi: 10.1073/pnas.1218464110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lei L, Ying S, Baojun L, Yi Y, Xiang H, Wenli S, Zounan S, Deyin G, Qingyu Z, Jingmei L, Guohui C. 2013. Attenuation of mouse hepatitis virus by deletion of the LLRKxGxKG region of Nsp1. PLoS One 8:e61166. doi: 10.1371/journal.pone.0061166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zust R, Cervantes-Barragan L, Kuri T, Blakqori G, Weber F, Ludewig B, Thiel V. 2007. Coronavirus non-structural protein 1 is a major pathogenicity factor: implications for the rational design of coronavirus vaccines. PLoS Pathog 3:e109. doi: 10.1371/journal.ppat.0030109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wathelet MG, Orr M, Frieman MB, Baric RS. 2007. Severe acute respiratory syndrome coronavirus evades antiviral signaling: role of nsp1 and rational design of an attenuated strain. J Virol 81:11620–11633. doi: 10.1128/JVI.00702-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Eriksson KK, Cervantes-Barragán L, Ludewig B, Thiel V. 2008. Mouse hepatitis virus liver pathology is dependent on ADP-ribose-1″-phosphatase, a viral function conserved in the alpha-like supergroup. J Virol 82:12325–12334. doi: 10.1128/JVI.02082-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kuri T, Eriksson KK, Putics A, Züst R, Snijder EJ, Davidson AD, Siddell SG, Thiel V, Ziebuhr J, Weber F. 2011. The ADP-ribose-1″-monophosphatase domains of severe acute respiratory syndrome coronavirus and human coronavirus 229E mediate resistance to antiviral interferon responses. J Gen Virol 92:1899–1905. doi: 10.1099/vir.0.031856-0. [DOI] [PubMed] [Google Scholar]
- 56.Fehr AR, Athmer J, Channappanavar R, Phillips JM, Meyerholz DK, Perlman S. 2015. The NSP3 macrodomain promotes virulence in mice with coronavirus-induced encephalitis. J Virol 89:1523–1536. doi: 10.1128/JVI.02596-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Graham RL, Becker MM, Eckerle LD, Bolles M, Denison MR, Baric RS. 2012. A live, impaired-fidelity coronavirus vaccine protects in an aged, immunocompromised mouse model of lethal disease. Nat Med 18:1820–1826. doi: 10.1038/nm.2972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zust R, Cervantes-Barragan L, Habjan M, Maier R, Neuman BW, Ziebuhr J, Szretter KJ, Baker SC, Barchet W, Diamond MS, Siddell SG, Ludewig B, Thiel V. 2011. Ribose 2′-O-methylation provides a molecular signature for the distinction of self and non-self mRNA dependent on the RNA sensor Mda5. Nat Immunol 12:137–143. doi: 10.1038/ni.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Menachery VD, Yount BL, Josset L, Gralinski LE, Scobey T, Agnihothram S, Katze MG, Baric RS. 2014. Attenuation and restoration of SARS-CoV mutant lacking 2′ O methyltransferase activity. J Virol 88:4251–4264. doi: 10.1128/JVI.03571-13. [DOI] [PMC free article] [PubMed] [Google Scholar]