Abstract
Background:
Coccidiosis is a serious protozoal disease of poultry. The identification of Eimeria species has important implications for diagnosis and control as well as for epidemiology. The molecular characterization of Eimeria species infecting Egyptian baladi chickens was investigated.
Methods:
Eimeria species oocysts were harvested from intestines of naturally infected Egyptian baldi chickens. The morphometry characterization of oocysts along with COCCIMORPH software was done. The DNA was extracted initially by freezing and thawing then the prepared samples was subjected to commercial DNA kits. The DNA products were analyzed through conventional polymerase chain reaction by using amplified region (SCAR) marker.
Results:
The PCR results confirmed the presence of 7 Eimeria species in the examined fecal samples of Egyptian baldi breed with their specific ampilicon sizes being E. acervulina (811bp), E. brunette (626bp), E. tenella (539bp), E. maxima (272bp), E. necatrix (200bp), E. mitis (327bp) and E. praecopx (354bp). A sequencing of the two most predominant species of Eimeria was done, on E. tenella and E. máxima. Analysis of the obtained sequences revealed high identities 99% between Egyptian isolates and the reference one. Similarly, E. maxima isolated from Egyptian baldi chickens showed 98% nucleotide identities with the reference strain. Only single nucleotide substitution was observed among the Egyptian E. tenella isolates (A181G) when compared to the reference one. The Egyptian isolates acquired 4 unique mutations (A68T, C164T, G190A and C227G) in compared with the reference sequence.
Conclusion:
This is the first time to identify the 7 species of Eimeria from Egyptian baladi chickens.
Keywords: Chickens, DNA, Eimeria species, PCR, Nucleotide, Sequencing
Introduction
Coccidiosis is a serious protozoal disease of poultry; characterized by damage to the intestinal epithelium, leading to inefficient feed conversion, and reduction in weight gain resulting in economic loss (1, 2). It is caused by protozoa of the genus Eimeria. Infection with one or several of the 7 Eimeria species infecting chickens including E. acervulina, E. brunetti, E. maxima, E. mitis, E. necatrix, E. praecox, and E. tenella, leads to variable signs from subclinical enteric infection to sub-acute mortality (3).
The baladi breeds became hybrid breeds, most of which are raised by growers for two or six weeks before being sold to village women. Village households raise the chickens as a source of eggs and meat or small income (4).
The identification of Eimeria species has important implications for diagnosis and control as well as for studying their epidemiology and population biology. Traditionally, species of Eimeria have been identified by morphometry and morphological features of the sporulated oocysts (using a microscope) and the specific host from which they originate (5). But, these criteria can be unreliable. For example, individual oocysts of E. brunetti and E. maxima can be the same or very similar in size and shape, thus preventing unequivocal identification. Biochemical, immunological and molecular techniques can overcome the limitations of traditional approaches for parasite identification (6, 7). In particular, DNA approaches, such as arbitrarily-primed PCR (AP-PCR), sequencing and specific PCR approaches, have proven useful for the identification, detection or characterization of Eimeria species. The PCR has been utilized to ‘fingerprint’ avian Eimeria species (8, 9).
Therefore, the aim of this study was molecular identification of Eimeria species infecting Egyptian baladi chicken in Beni-Suef province.
Materials and Methods
Samples collection
A total of 700 gut samples of Egyptian baladi chickens suspected for coccidiosis from different farms of baladi chickens and small holders rearing system located in Beni-Suef province were microscopically examined for Eimeria species oocysts. A total of 140 positive fecal samples were collected from gut samples of the examined intestine to undergo the study.
Morphological identification
Oocyst morphology and size were determined by measuring length and width of 50 oocysts having similar morphological features using ocular micrometer (10, 11). Furthermore, Coccimorph identification of Eimeria species was identified with COCCIMORPH soft-ware (http://www.coccidia.icb.usp.br/coccimorph/). The software was downloaded from the Internet and the oocyst images (400× magnification) were uploaded for species identification as described online (12).
Molecular identification of Eimeria species by polymerase chain reaction (PCR)
The molecular identification of Eimeria species was carried out on pooled fecal samples. Fourteen pools of Eimeria species, each pool represented 10 fecal samples (a total of 140 samples). Coccivac D a living non attenuated vaccine containing 7 Eimeria species was used as control positive.
DNA extraction from fecal samples
DNA extraction was done according to Guven et al. (22) with slight modification. Fifty cycles of freezing, using of liquid nitrogen and thawing, in a shaking water path at 50Cº, were carried out for complete rupturing of oocysts walls without adding sodium hypochlorite or use of glass beads. During this process, 10ul was taken and were examined under the microscope (40 X) to ensure complete oocyst wall destruction. DNA extraction kit was used for genomic DNA extraction from tissue. DNA extracted according to the kits instructions (Biobasic, Inc. Canada, Cat. No. BS427).
Polymerase chain reaction (PCR)
Samples were analyzed by PCR with the SCAR primers for E. tenella, E. acervulina, E. necatrix, E. maxima, E. brunette and E. praecox (9). In addition, ITS1 gene was performed for E. mitis (13, 14); this was showed in Table 1. PCR amplifications were individually made for each primer pair using 40 pmol of each primer, 1 U of platinum Taq DNA polymerase, dNTPs, MgCl2 (Invitrogen®) and 5μL of target DNA in a 25 μL reaction volume (9). The PCR reaction conditions were as follow: initial denaturation cycle at 95°C for five minutes, 30 cycles of 94°C for 45 sec., annealing at 57°C: 63°C for 30 sec for each species primer (as mentioned at Table 1) and 72°C for one and half minutes. The final extension step was at 72°C for seven minutes. Amplifications were carried out in 0.2 mL polypropylene tubes using a Labnet International, Inc software v3.3.4c, Multigene model: Tc9600-G.
Table 1.
The primers used in PCR running
| Species | Primer Name | Primer sequences | Amplicon size (bp) | Annealing temperature |
|---|---|---|---|---|
| (5′→□3′) | ||||
| E. acervulina | Ac-01F | AGTCAGCCACACAATAATGGCAAACATG | 811 | 60°C |
| Ac-01R | AGTCAGCCACAGCGAAAGACGTATGTG | |||
| E. brunetti | Br-01F | TGGTCGCAGAACCTACAGGGCTGT | 626 | 63°C |
| Br01R | TGGTCGCAGACGTATATTAGGGGTCTG | |||
| E. tenella | Tn-01F | CCGCCCAAACCAGGTGTCACG | 539 | 60°C |
| Tn-01R | CCGCCCAAACATGCAAGATGGC | |||
| E. praecox | Pr-01F | AGTCAGCCACCACCAAATAGAACCTTGG | 354 | 58°C |
| Pr-01R | GCCTGCTTACTACAAACTTGCAAGCCCT | |||
| E. mitis | Mt-01F | TATTTCCTGTCGTCGTCTCGC | 327 | 57°C |
| Mt-01R | GTATGCAAGAGAGAATCGGGA | |||
| E. maxima | Mx-01F | GGGTAACGCCAACTGCCGGGTATG | 272 | 58°C |
| Mx-01R | AGCAAACCGTAAAGGCCGAAGTCCTAGA | |||
| E. necatrix | Nc-01F | TTCATTTCGCTTAACAATATTTGGCCTCA | 200 | 57°C |
| Nc-01R | ACAACGCCTCATAACCCCAAGAAATTTTG |
F = foward primer; R = reverse primer.
Sequencing of 2 isolated Eimeria species from the Egyptian baladi chickens for confirmation of PCR results
The gel extraction and purification of PCR products was done. Confirmation of the results of PCR was applied by direct sequencing of 539 bp and 272 bp of SCAR marker gel purified PCR products of the selected most prevalent Eimeria species (E. tenella and E. maxima, respectively) isolated from Egyptian baladi. Briefly, PCR-products were excised from gel and were purified with Wizard® SV gel and PCR clean up system according to the Manufacturer’s instructions. The DNA was dried and shipped for direct sequencing; which was performed by Macrogen Inc. (908 World Meridian Venture Center #60–24, Gasan-dong Geumchun-gu, Seoul 153–781, Korea) in both forward and reverse directions using the same primer sets that have been used for amplification of each PCR product. Sequencing was done on an Applied Biosystems 310 automated DNA sequencer using cycle sequencing ABI prism Big Dye terminator chemistry (a terminator cycle sequencing ready reaction kit) (Perkin-Elmer/Applied Biosystems, Foster City, CA USA). A BLAST analysis was initially performed to establish sequence identity to Gen Bank accessions (15). Comparative sequences analyses were performed using CLUSTAL W Multiple Sequence Alignment Program, version 1.83 (http://www.genome.jp/tools/clustalw/).
Results
Identification of Eimeria species infecting Egyptian baldi chickens Morphological identification
The morphological features of the isolated Eimeria species revealed that 7 species were suspected from the examined Egyptain baldi chickens (E. acervulina, E. maxima, E. mitis, E. necatrix, E. brunetti ,E. tenella and E. praecox) Table 2.
Table 2.
Morphological and molecular identification of Eimeria species isolated from Egyptian baladi chickens
| Pool No | Identified Eimeria species by morphology | Identified Eimeria species by PCR | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Ac | Br | Te | Pr | Mi | Ma | Ne | Ac | Br | Te | Pr | Mi | Ma | Ne | |
| 1 | + | + | + | + | + | + | + | + | + | |||||
| 2 | + | + | + | + | + | + | + | |||||||
| 3 | + | + | + | + | + | + | + | + | + | + | ||||
| 4 | + | + | + | + | + | + | + | + | ||||||
| 5 | + | + | + | + | ||||||||||
| 6 | + | + | + | + | + | |||||||||
| 7 | + | + | + | |||||||||||
| 8 | + | + | + | + | + | + | + | |||||||
| 9 | + | + | + | + | + | + | ||||||||
| 10 | + | + | + | |||||||||||
| 11 | + | + | ||||||||||||
| 12 | + | + | + | + | ||||||||||
| 13 | + | + | + | + | ||||||||||
| 14 | + | + | + | + | + | |||||||||
Pools from 1–14 are fecal samples of Egyptian baldi chickens./ Ac. E. acervulina, Br. E. brunette, Te. E. tenella, Ma. E. maxima, Ne. E. necatrix, Mi. E. mitis, Pr. E. praecopx (354bp)
COCCIMORPH identification
Identification of Eimeria spp. using COCCIMORPH software revealed suspected 7 species of Eimeria.
Molecular identification
The results of molecular identification proved the presence of 7 Eimeria species in the examined Egyptian baldi chicken fecal samples. The primers were sufficiently sensitive and specific enabling the discrimination of seven Eimeria species. The amplified fragments presented different sizes: E. acervulina (811 bp), E. brunette (626 bp), E. tenella (539 bp), E. mitis (310 bp), E. praecox (354 bp), E. maxima (272 bp) and E. necatrix (200 bp) (Fig. 1 & Fig. 2).
Fig. 1:
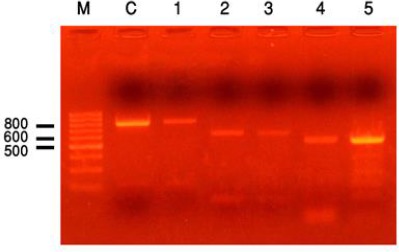
Amplicone of E. acervulina(811bp) Amplicone of E. burnetti (626bp)/ Amplicone of E. tenella (539bp)/ Lane M molecular weight marker 100bp, lane C control positive for E. acervulina 811bp, lane 1 pool No 3 positive for (E. acervulina 811bp), lane 2 control positive for E. brunetti 626bp, lane 3 pool No 3 positive for (E. brunetti 626bp), lane 4 control positive for E. tenella 539bp and lane 5 pool No positive for (E. tenella 539bp)
Fig. 2:
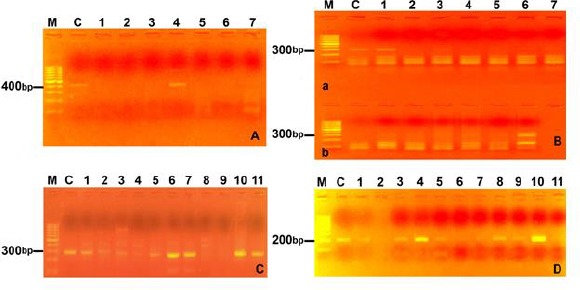
A. Amplicone of E. praecox (354bp). Lane M molecular weight marker 100bp, lane C control positive for E. praecox. Lane 4 represents the pool No 4, the only positive pool for E. praecox from 14 pool. B. Amplicone of E. mitis (327bp). Lane M molecular weight marker 100bp, lane C control positive for E. mitis. Lane 1a represent pool No 1, lane 6 b represent pool No 13 C. Amplicone of E. maxima (272bp). Lane M molecular weight marker 100bp, lane C control positive for E. maxima. Pools No 1, 2, 3, 5, 6, 7, 10 and 11 were positive for E. maxima. D. Amplicone of E. necatrix (200 bp). Lane M molecular weight marker 100bp, lane C control positive for E. necatrix. Pools No 1, 3, 4, 5, 8, 9 and 10 were positive for E. necatrix
Sequence analysis of RAPD-SCAR marker gene
Four PCR products were sequenced including isolates obtained from native Egyptian baladi chickens infected with the most prevalent Eimeria species (E. tenella and E. maxima). Sequencing was done using Eimeria species-specific primers. Sequences E. tennela revealed 99% overall identities between Egyptian isolates and the reference one (Gen Bank accession No. AY5716 34.1). Similarly, E. maxima isolated from baldi breed showed 99% nucleotide nucleotide identities with the reference strain (Gen Bank accession No. AY571588.1). Only single nucleotide substitution was observed among the Egyptian E. tenella isolates (A181G) when compared to the reference one. On the other hand Egyptian E. maxima isolates acquired 4 unique mutations (A68T, C164T, G190A and C227G) when analyzed with the reference sequence (Fig. 3).
Fig. 3.
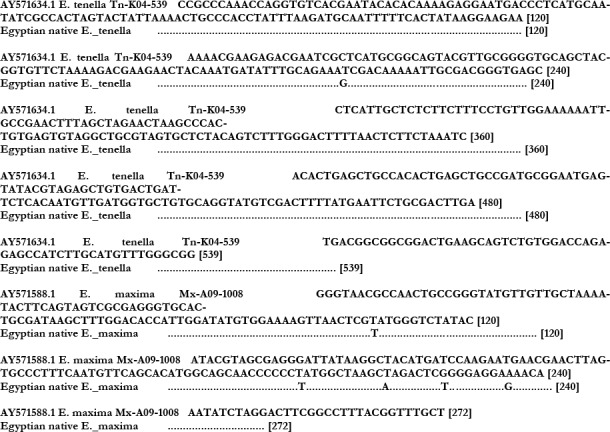
: Deduced nucleotide sequences of the 539bp of E. tenella and 272bp of E. maxima RAPD-SCAR marker gene
*Egyptian native means Egyptian baladi **Dots indicates identical nucleotides
Discussion
Initially, 7 Eimeria species were recorded morphologically and molecularly in baldi chickens in Beni Suef province. Eimeria species identified were E. tenella, E. acervulina, E. necatrix, E. maxima, E. mitis, E. brunette and E. praecox. In Egypt, no study reported the 7 species of Eimeria species but numerous studies revealed the morphological features of Eimeria species; 5 Eimeria species (E. necatrix, E. tenella, E. acervulina, E. mitis and E. maxima) were recorded among the examined Egyptian native chicks (16, 17). Moreover, 6 Eimeria species were reported from 4 Egyptian governorates (Qalubeia, Sharkeia, Fayoum and Giza) which were E. necatrix, E. acervulina, E. pracox, E. maxima, E. mitis and E. tenella (18). This variation with the present work may be attributed to the area of the study. Worldwide, many studies estimated the 7 Eimeria species in different breeds and localities (19–22).
Traditionally, identification of Eimeria spp. has been based on the morphological characteristics of oocysts, parasite biology, clinical signs of the affected animals, and the typical macroscopic lesions assessed during necropsy. However, in a natural setting mixed infections of different Eimeria spp. are commonly encountered and morphological characteristics and pathological changes may overlap, hindering accurate diagnosis and undermining detection of subclinical disease (23). COCCIMORPH tool is, an innovative approach developed for identification of eimerian oocysts of poultry and rabbits through which digital images of unidentified sporulated Eimeria ocysts are uploaded for species identification based on sporulated oocyst morphology (12). COCCIMORPH tool recorded the same findings of microscopical examination in both of baldi chickens.
Due to the thick and resistant oocyst wall of Eimeria spp. (24), several means of breaking down the oocyst wall have been described, including hot phenol incubation (25), repeated freezing and thawing (26), enzyme digestion after sodium hypochlorite incubation (27), passage through a high pressure cell (28), grinding in liquid nitrogen (29) and grinding by glass beads (9) and they mentioned that the most commonly used protocol for DNA extraction from Eimeria species is based on glass bead grinding of oocysts combined with classic phenol-chloroform DNA extraction. In the present study, we overcame this problem by freezing in liquid nitrogen and thawing in a water bath at 50 °C. This caused breaking of the oocysts wall and liberation of oocysts content facilitate DNA extraction by using of tissue DNA extraction kits under our laboratory conditions. This method for DNA extraction was like that of Guven et al. (22) except freezing and thawing for 50 times and without using sodium hypochlorite, glass beads or stool DNA extraction kits.
The measurements of the oocysts can vary due to changes in metabolism of parasites or birds, and even in the value of the shape morphometric indices that may overlap and lead to misleading conclusions regarding the species (30). Consequently, application of molecular tools for identification and characterization of these parasites has been carried out to ensure isolation of 7 Eimeria spp. in Beni-Suef province by PCR technique.
The results of molecular diagnosis by conventional PCR technique using amplified region (SCAR) marker proved the presence of 7 Eimeria species in the examined fecal samples of baldi chickens with their specific ampilicon sizes (E. acervulina (811bp), E. brunette (626bp), E. tenella (539bp), E. maxima (272bp), E. necatrix (200bp), E. mitis (327bp), E. praecopx (354bp). PCR findings coincide with the morphological findings of baldi chicken Eimeria species.
PCR confirmed the presence of 7 Eimeria species in Beni Suef province for the first time. In Egypt, this result more or less as of other study (9), even they used the same primers but they reported Eimeria species in broilers chickens by use of multiplex PCR (E. necatrix, E. acervulina, E. pracox, E. maxima, E. mitis and E. tenella). Moreover, using of the same primers (SCAR) detected 7 Eimeria species by multiplex PCR (31, 11). Besides, 7 Eimeria species was recorded by PCR amplification with species-specific primers for the internal transcribed spacer (ITS) sequence or the small RNA subunit sequence of each of the seven species of Eimeria (30, 32, 33). On other hand, the PCR results of Eimeria species shouldn’t detect 7 species but may give 2 species only; Eimeria praecox and Eimeria mitis (34). This variation of PCR results may be because of locality, used primers, methods of DNA extraction and DNA quantity in the used samples. All these studies were investigated Eimeria species other breeds of chickens. Therefore, the present study was unique in investigation of Eimeria species in Egyptian baladi chickens.
Sequencing of the excised DNA bands obtained after amplification of DNA of the two most prevalent Eimeria species, E. tenella and E. maxima, using Eimeria species-specific primers was done. Blast analysis of the obtained sequences confirmed the species specificity of the used primers as previously reported (9); E. tenella multisequence alignment indicated high nucleotide identities (99%) between Egyptian isolates and the reference one. Similarly, E. maxima isolated from Egyptian baladi breed showed 98% nucleotide identities with the reference strain. Only single nucleotide substitution was observed among the Egyptian E. tenella isolates (A181G) when compared to the reference one. On the other hand Egyptian E. maxima isolates acquired 4 unique mutations (A68T, C164T, G190A and C227G) when analyzed with the reference sequence. The significance of these mutations cannot be assessed specifically with the non-translatable nature of the target SCAR marker into functional amino acids. Further studies would be done using different primer sets for amplification of other genes.
Conclusion
Baldi chickens were found infected by the 7 species of Eimeria first time in Egypt. The identification of Eimeria species was confirmed by PCR. Mutations and differences of Eimeria spp genomes between Eimeria species isolates compared with the reference strains have to be studied further.
Acknowledgment
The authors appreciated many greats for Dr. Salama Abohama for continuous helping in PCR and sequencing. The authors declare that there is no conflict of interests.
References
- 1.Shirley MW, Smith AL, Tomley FM.The biology of avian Eimeria with an emphasis on their control by vaccination. Adv Parasitol. 2005; 60, 285– 330. [DOI] [PubMed] [Google Scholar]
- 2.Morris GM, Gasser RB.Biotechnological advances in the diagnosis of avian coccidiosis and the analysis of genetic variation in Eimeria. Biotechnol Adv. 2006; 24, 590– 603. [DOI] [PubMed] [Google Scholar]
- 3.McDougald LR.Protozoal infections. In: Saif YM, Barnes H.J., Glisson J.R., Fadly A.M., McDougald L.R., Swayne D.E. (Eds.), Diseases of Poultry. Iowa State Press, Ames, IA, USA: 2003: 974– 985. [Google Scholar]
- 4.FAO Mapping traditional poultry hatcheries in Egypt. Prepared by M. Ali Abd-Elhakim, Olaf Thieme, Karin Schwabenbauer and Zahra S. Ahmed. AHBL - Promoting strategies for prevention and control of HPAI 2009, Rome.
- 5.McDougald LR, Fuller L, Mattiello R.A Survey of Coccidia on 43 Poultry Farms in Argentina. Avian The first sentence should be a short brief of what you did and what you got. 1997; 41( 4), 923– 929. [PubMed] [Google Scholar]
- 6.Andrews RH, Chilton NB.Multilocus enzyme electrophoresis: a valuable technique for providing answers to problems in parasite systematics. Int J Parasitol. 1999; 29, 213– 53. [DOI] [PubMed] [Google Scholar]
- 7.Gasser RB.PCR-based technology in veterinary parasitology. Vet Parasitol. 1999; 84, 229– 58. [DOI] [PubMed] [Google Scholar]
- 8.Tsuji N, Kawazu S, Ohta M, Kamio T, Isobe T, Shimura K, Fujisaki K.Discrimination of eight chicken Eimeria species using the two-step polymerase chain reaction. J Parasitol. 1997; 83, 966– 970. [PubMed] [Google Scholar]
- 9.Fernandez S, Pagotto AH, Furtado MM, Katsuyama AM, Madeira AM, Gruber A.A multiplex PCR assay for the simultaneous detection and discrimination of the seven Eimeria species that infect domestic fowl. Parasito. Res. 2003; 127, 317– 325. [DOI] [PubMed] [Google Scholar]
- 10.Reid WM, Long PL.A diagnostic chart for nine species of fowl coccidia. Georgia Uni. Re. Rep. 1979; 335. [Google Scholar]
- 11.Carvalho FS, Wenceslau AA, Teixeira M, Carneiro JAM, Melo ADB, Albuquerque GR.Diagnosis of Eimeria species using traditional and molecular methods in field studies. Ve. Parasitol. 2011b; 176, 95– 100. doi: 10.1016/j.vetpar.2010.11.015. Epub 2010 Nov. [DOI] [PubMed] [Google Scholar]
- 12.Castãnón C, Fraga J, Fernandez S, Gruber A, Costa L.Biological shape characterization for automatic image recognition and diagnosis of protozoan parasites of the genus Eimeria. Pattern Recognitio. 2007; 40, 1899– 1910. [Google Scholar]
- 13.Schnitzler BE, Thebo PL, Tomley FM, Uggala A, Shirley MW.PCR identification of chicken Eimeria: a simplified read-out. Avian Pathol. 1999; 28, 89– 93. [DOI] [PubMed] [Google Scholar]
- 14.Jenkins MC, Miska K, Klopp S.Application of polymerase chain reaction based on ITS1 rDNA to speciate Eimeria. Avian Dis. 2006; 50, 110– 114. [DOI] [PubMed] [Google Scholar]
- 15.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ.Basic local alignment search tool. J Mol Biol. 1990; 15, 403– 410. [DOI] [PubMed] [Google Scholar]
- 16.Ahmed NE, NegmEldin MM, El Akabawy LM, El Medawy RS.Incidences of some protozoan parasites in Birds. KafrElsheikh Vet Med J. 2003; 1 ( 1), 235– 251. [Google Scholar]
- 17.Al-Gawad AA, Mahdy OA, El-Massry AA, Al-Aziz MSA.Studies on Coccidia of Egyptian Balady Breed Chickens. Li Sci J. 2012; 9 ( 3), 568– 576. [Google Scholar]
- 18.Kutkat MA, Shalaby HA, EL.Khateeb RM, Abu ELezz NM, Zayed AA, Abdel Razik AB, Nasef SA, Amer MM.Molecular diagnosis of Eimeria and Clostridia in simultaneously infected chickens. Global Vet. 2009; 3 ( 1), 26– 31. [Google Scholar]
- 19.Lee HA, Hong S, Chunge Y, Kim O.Sensitive and specific identification by polymerase chain reaction of Eimeriatenella and Eimeria maxima, important protozoan pathogens in laboratory avian facilities. Lab Anim Res. 2011; 27 ( 3), 255– 258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jatau ID, Sulaiman NH, Musa IW, Lawal AI, Okubanjo OO, Isah I, Magaji Y.Prevalence of Coccidia Infection and Preponderance Eimeria Species in Free Range Indigenous and Intensively Managed Exotic Chickens during Hotwet Season, in Zaria, Nigeria Asian J Poult Sc. 2012; 6( 3), 79– 88. [Google Scholar]
- 21.Nikam SV, Kanse VS, Jadhav BN, Jaid EL.Comparative study of seasonal incidence of chicken coccidiosis in eight districts of Marathwada region, Maharashtra state, India. Trends in Parasitol Res. 2012; 1 ( 1), 7– 9. [Google Scholar]
- 22.Guven E, Beckstead RB, Kar S, Vatansever Z, Karaer Z.Molecular identification of Eimeria species of broiler chickens in Turkey. Ankara Üniv. Vet Fak Derg. 2013; 60, 245– 250. [Google Scholar]
- 23.Long PL, Joyner LP.Problems in identification of species of Eimeria. J Protozool. 1984; 31, 535– 541. [DOI] [PubMed] [Google Scholar]
- 24.Ryley JF.Cytochemistry, physiology, and biochemistry. In: Hammond D.M., Long P.L. (Eds.), The Coccidia. University Park Press, Baltimore, MD, USA: 1973; 151– 154. [Google Scholar]
- 25.Stucki U, Braun R, Roditi I. Eimeria tenella: characterization of a 5S ribosomal RNA repeat unit and its use as a species-specific probe. Exp Parasitol. 1993; 76, 68– 75. [DOI] [PubMed] [Google Scholar]
- 26.Jinneman KC, Wetherington JH, Hill WE, Adams AM, Johnson JM, Tenge BJ, Dang NL, Manger RL, Wekell MM.Template preparation for PCR and RFLP of amplification products for the detection and identification of Cyclospora sp. And Eimeria spp. oocysts directly from raspberries. J Food Prot. 1998; 61, 1497– 1503. [DOI] [PubMed] [Google Scholar]
- 27.Zhao X, Duszynski DW, Loker ES.A simple method of DNA extraction for Eimeria species. J Microbiol Methods. 2001; 44, 131– 137. [DOI] [PubMed] [Google Scholar]
- 28.Abrahamsen MS, Clark TG, White MW.An improved method for isolating RNA from coccidian oocysts. J Parasitol. 1995; 81, 107– 109. [PubMed] [Google Scholar]
- 29.Tsuji N, Ohta M, Kawazu S, Kamio T, Isobe T, Shimura K, Fujisaki K.DNA polymorphism of srRNA gene among Eimeria tenella strains isolated in Japan. J Vet Med Sci. 1999; 61, 1331– 1333. [DOI] [PubMed] [Google Scholar]
- 30.Sun XM, Pang W, Jia T, Yan WC, He G, Hao LL, Bentue M, Suo X.Prevalence of Eimeria Species in Broilers with Subclinical Signs from Fifty Farms. Avian Dis. 2009; 53( 2), 301– 305. [DOI] [PubMed] [Google Scholar]
- 31.Carvalho FS, Wenceslau AA, Teixeira M, Albuquerque GR.Molecular diagnosis of Eimeria species affecting naturally infected Gallus gallus. Genet Mol Res. 2011a; 10 ( 2), 996– 1005. doi: 10.4238/vol10-2gmr1043. [DOI] [PubMed] [Google Scholar]
- 32.Morris GM, Woods WG, Richards DG, Gasser RB.The application of a polymerase chain reaction (PCR)-based capillary electrophoretic technique provides detailed insights into Eimeria populations in intensive poultry establishments. Mol Cell Probes. 2007; 21: 288– 294. [DOI] [PubMed] [Google Scholar]
- 33.Aarthi S, Raj GD, Raman M, Gomathinayagam S, Kumanan K.Molecular prevalence and preponderance of Eimeria spp. among chickens in Tamil Nadu, India. Parasitol Res. 2010; 107( 4), 1013– 1017. doi: 10.1007/s00436-010-1971-2. Epub 2010 Jul 6. [DOI] [PubMed] [Google Scholar]
- 34.Meireles MV, Roberto LO, Riera RF.Identification of Eimeria mitis and Eimeria praecox in Broiler Feces Using Polymerase Chain Reaction. Brazilian J Poult Sci. 2004; 6 ( 4), 249– 252. [Google Scholar]


