Abstract
Purpose of Review
The identification and characterization of essential islet transcription factors have improved our understanding of β cell development, provided insights into many of the cellular dysfunctions related to diabetes, and facilitated the successful generation of β cells from alternative cell sources. Recently, noncoding RNAs have emerged as a novel set of molecules that may represent missing components of the known islet regulatory pathways. The purpose of this review is to highlight studies that have implicated noncoding RNAs as important regulators of pancreas cell development and β cell function.
Recent Findings
Disruption of essential components of the microRNA processing machinery, in addition to misregulation of individual miRNAs, has revealed the importance of microRNAs in pancreas development and β cell function. Furthermore, over 1000 islet-specific long noncoding RNAs have been identified in mouse and human islets, suggesting that this class of noncoding molecules will also play important functional roles in the β cell.
Summary
The analysis of noncoding RNAs in the pancreas will provide important new insights into pancreatic regulatory processes that will improve our ability to understand and treat diabetes and may facilitate the generation of replacement β cells from alternative cell sources.
Keywords: non-coding RNA, microRNA, long non-coding RNA, pancreas, beta cells
INTRODUCTION
Significant research efforts are currently underway to understand and prevent diabetes mellitus. Towards this goal, studies in mice and humans have significantly advanced our understanding of the conserved signaling pathways and regulatory factors required for the development and maintenance of functional beta cells. However, it is evident from the current challenges associated with predicting and treating diabetes, in addition to generating alternative sources of endocrine cells, that we are still missing key molecular components of the regulatory pathways that are required to generate, mature and preserve fully functional beta cells. For decades, a principle tenet of molecular biology was that the primary function of RNA was to code for proteins. As a result, most cellular processes were studied in a protein-centric manner. Recent advances in transcriptome biology have revolutionized this thinking by revealing that the mammalian genome encodes a vast number of functional non-protein-coding RNAs (ncRNAs). Characterization of ncRNAs across species and in different tissues has revised how we think about the functions of RNA in a cell. As discussed in this review, several important studies have now implicated roles for ncRNAs in the pancreas. A greater appreciation of the RNA-based mechanisms of gene regulation in the β cell will enhance our understanding of β cell development, function, and disease, and perhaps identify novel molecular targets for the treatment of diabetes. (Table 1)
Table 1.
Key terminology
| ncRNA (noncoding RNA) | An RNA molecule which is not predicted to encode a protein. |
| miRNA (microRNA) | An endogenously produced, small (~22 nucleotide) noncoding RNA that functions predominantly in RNA silencing through post-transcriptional regulation of gene expression. |
| siRNA (Small interfering RNA) | A small (20–25 nucleotide) noncoding RNA that functions in the RNA interference pathway to promote post-transcriptional gene silencing. |
| lncRNA (Long noncoding RNA) | An RNA molecule longer than 200 nucleotides which is not predicted to encode a protein. |
| Dicer | An endoribonuclease essential for maturation and subsequent function of RNA molecules involved in the RNA interference pathway, such as miRNAs and siRNAs. |
| SNP (Single Nucleotide Polymorphism): | A DNA sequence variation occurring commonly within a population useful for identifying disease-causing variants during GWAS. |
| GWAS (Genome Wide Association Studies) | The examination of genetic variants in different individuals to determine whether a specific DNA variant is associated with a particular trait or disease. |
| Cre Recombinase | A tyrosine recombinase enzyme derived from the P1 bacteriophage. The Cre enzyme mediates a site-specific recombination event between two DNA recognition sites (called LoxP sites). Cre expression can be driven by any promoter enabling tissue specific Cre activity and targeted knockout or expression of a gene in a cell specific manner. |
| CRISPR/Cas9 (Clustered Regularly Interspaced Short Palindromic Repeats/Cas9) | A gene editing system that uses an RNA guide molecule in combination with a Cas9 endonuclease to target specific loci in the genome for gene disruption or replacement. |
PANCREAS DEVELOPMENT, FUNCTION AND DISEASE
The pancreas is a multifunctional organ consisting of an exocrine compartment that aids in digestion and an endocrine compartment that maintains glucose homeostasis. Insulin-producing β cells are the predominant cell type in the islets of Langerhans, and the loss or dysfunction of β cells is associated with the set of diseases known as diabetes mellitus. Specifically, Type 1 diabetes (T1D) is an autoimmune disorder in which targeted destruction of the β cells causes chronic hyperglycemia, whereas Type 2 diabetes (T2D) is characterized by a gradual dysfunction and/or loss of β cells over time due to physiological and metabolic overexertion [1]. Although many studies have clarified the complex genetic and molecular mechanisms contributing to the dysfunction and loss of β cells [2,3], we still do not fully understand the disease etiologies, nor have we developed satisfactory treatments. Intriguingly, genome-wide association studies (GWAS) have shown that a majority of diabetes susceptibility loci fall outside protein coding genes. This suggests a role for enhancer regions and/or ncRNAs in maintaining proper β cell function.
In addition to understanding the genetic regulation of islet function, there are considerable research efforts directed towards understanding how β cells naturally develop. This need is in large part due to a goal of regenerative medicine: to replace lost or damaged β cells from alternative cell sources. While much effort has been spent towards improving differentiation and transdifferentiation protocols, the end product still differs from a bona fide β cell and the long term potential of these cells to retain β cell identity and function remains unclear [4,5]. It is therefore likely that we are missing critical regulators of β cell differentiation, and there is evidence that ncRNAs may fulfill this role in the regulation of pancreas development and β cell function.
NONCODING RNAS
Currently, there are two major classes of ncRNAs: housekeeping and regulatory. Housekeeping ncRNAs, such as tRNAs and rRNAs, are expressed ubiquitously and are required for protein synthesis, while regulatory ncRNAs influence gene expression via transcriptional and post-transcriptional regulatory mechanisms [6]. Regulatory ncRNAs are further subdivided according to size: 1) short ncRNAs (~20 nucleotides), such as micro RNAs (miRNAs) and small interfering RNAs (siRNAs); and 2) long noncoding RNAs (lncRNAs) that are defined as transcripts longer than 200 nucleotides [6]. While categorization by size may seem arbitrary, there are also functional distinctions between the two groups: small ncRNAs alter mRNA stability whereas lncRNAs predominantly function at the level of transcription [6,7]. This review provides a general overview of miRNAs and lncRNAs and discusses some representative studies that have characterized their function in pancreatic β cells. A more comprehensive description of the specific miRNAs and their respective functions in pancreatic β cells is summarized in Table 2 and extensively described in a recent review by Özcan [32].
Table 2.
List of microRNAs that function in β cell biology
| Gene/ncRNA | Genotype/Misexpression | Phenotype | Target pathway | References |
|---|---|---|---|---|
| Dicer | Pdx1:Cre; Dicerfl/fl | Pancreas agenesis | Lynn et al., 2007 [8] | |
| Ngn3:Cre; Dicerfl/fl | Loss of beta cell identity | REST | Kanji et al., 2013 [9] | |
| RIP:Cre; Dicerfl/fl | β cell dysfunction | Kalis et al., 2011 [10]; Mandelbaum et al., 2012 [11] | ||
| RIP:CreER; Dicerfl/fl | Defects in insulin production with no loss of β cell identity | Sox6, Bhlhe22 | Melkman-Zehavi et al., 2011 [12] | |
| Ago2 | Ago2 KD in vitro | Enhanced insulin secretion | miR-375 | Tattikota et al., 2013 [13] |
| Ago2 KO in vivo | Defective β cell expansion induced by insulin resistance | miR-375, Cadm1 | Tattikota et al., 2014 [14] | |
| miR-30 | miR-30 expression | β cell differentiation | Snail1, Vimentin | Joglekar et al., 2009 [15] |
| miR-30d OE in vitro | Increased insulin transcription | Tang et al., 2009 [16] | ||
| miR-30d KD in vitro | Decreased insulin transcription induced by glucose | Tang et al., 2009 [16] | ||
| mir-375 | miR-375 OE in vitro | Suppressed glucose stimulated insulin secretion | Mtpn | Poy et al., 2004 [17] |
| miR-375 KD in vitro | Enhanced insulin secretion | Mtpn | Poy et al., 2004 [17] | |
| miR-375 KO in vivo | Increased α cell numbers, defective β cell proliferation, moderate hyperglycemia | Poy et al., 2009 [18] | ||
| miR-7 | RIP:Cre; Mir7a2fl/fl | Improved glucose tolerance due to enhanced insulin secretion | Vesicle exocytosis | Latreille et al., 2014 [19] |
| Pdx1:Cre; Rosa26:Mir7a1 OE | Defects in endocrine differentiation | Pax6 | Kredo-Russo et al., 2012 [20] | |
| miR-7 KD in vitro | Promotes β cell proliferation | mTOR | Wang et al., 2013 [21] | |
| miR-24, miR-26, miR-148, miR-182 | KD in vitro | Down regulation of insulin expression | Sox6, Bhlhe22 | Melkman-Zehavi et al., 2011 [12] |
| miR-184 | miR-184 OE in ob/ob diabetic mice | β cell expansion mediated by insulin resistance | Ago2 | Tattikota, 2014 [14] |
| Let-7 family | Inducible Let-7 OE (iLet-7) | Reduced body weight and size, impaired glucose intolerance, increased insulin production | Insulin-PI3K-mTOR | Zhu et al., 2011 [22] |
| Global Let-7 OE in vivo | Reduced body weight size, reduced fat mass, impaired glucose tolerance | InsR, Irs2 | Frost and Olson, 2011 [23] | |
| Global Let-7 KD in vivo | Improved blood glucose levels and insulin resistance in obese mice | InsR, Irs2 | Frost and Olson, 2011 [23] | |
| miR-9 | miR-9 KD and OE in vitro | Aberrant glucose-stimulated insulin secretion | Sirt1, Onecut2, Slp4 | Ramachandran et al., 2011 [24]; Plaisance et al., 2006 [25] |
| miR-21, miR-34, miR-146 | KD in vitro | Prevents β cell dysfunction and death after cytokine treatment | Roggli et al., 2010 [26] | |
| miR-124 | miR-124a OE in vitro | Impaired glucose signaling and insulin secretion | Foxa2, Rab27a | Baroukh, 2007 [27]; Lovis, Gattesco, and Regazzi, 2008 [28] |
| miR-29 | miR-29 OE in vitro | Impaired insulin secretion, increased apoptosis | Onecut2, Mcl1 | Roggli et al., 2012 [29] |
| miR-33 | miR-33a OE in vitro | Decreased glucose-stimulated insulin secretion | Abca1 | Wijesekara et al., 2012 [30] |
| miR-200 | miR-200 OE in vitro | Beta cell apoptosis | Zeb1 | Filios et al., 2014 [31] |
List of the known miRNAs that have been implicated in β cell development and function. Included are eight miRNAs (bottom 6 rows) that are not discussed in this review due to space restrictions, but are examined in detail in another review [8]. KD - knockdown, KO - knockout, OE- overexpression.
MICRORNAS IN β CELL DEVELOPMENT AND FUNCTION
MicroRNAs are the most well-characterized class of regulatory ncRNAs. MiRNAs are generally believed to negatively regulate gene expression through mRNA cleavage that is mediated by the Argonaute (Ago) family of proteins as part of the RNA-induced silencing complex (RISC) [33]. The finding that miRNAs only require short “seed” sequences in the 5′ end of their transcript to direct Ago to target mRNAs implies that one miRNA can influence the expression of several target genes [34]. It is therefore not surprising that miRNAs play essential regulatory roles in diverse cellular processes, including proliferation, organogenesis, hormone secretion, and apoptosis [6]. Studies have also shown that the misexpression of miRNAs disrupts gene regulatory networks and contributes to disease states, such as cancer and diabetes [33]. In fact, several groups have identified essential roles for miRNAs in β cell development, function, and disease.
The requirement of miRNAs in β cell development was initially demonstrated by removal of Dicer function during different stages of pancreas development (Figure 1a). Given that Dicer is required for the formation of mature miRNAs, loss of Dicer ablates all miRNA function within a cell [35]. The pan-pancreatic loss of Dicer using the Pdx1:Cre allele resulted in severe pancreatic agenesis and neonatal death [8]. The mutant mice also displayed a dramatic loss of all endocrine cell types, which was attributed to a reduction in the number of Neurogenin3 (Ngn3) positive endocrine progenitor cells [8]. In a more recent study, Ngn3:Cre was used to ablate Dicer function specifically in the endocrine progenitor population. The mutant mice had normal embryonic pancreas development; however, shortly after birth the mice developed hyperglycemia due to progressive loss of β cells [9]. Surprisingly, the authors found that loss of miRNA function in endocrine progenitor cells caused upregulation of neuronal genes normally repressed by RE1-silencing transcription factor (REST) [9]. Thus, it appears that miRNAs maintain islet-cell identity by inhibiting expression of REST, thereby restricting expression of neuronal genes.
Figure 1. The role of microRNAs in β cell development, function, and disease.
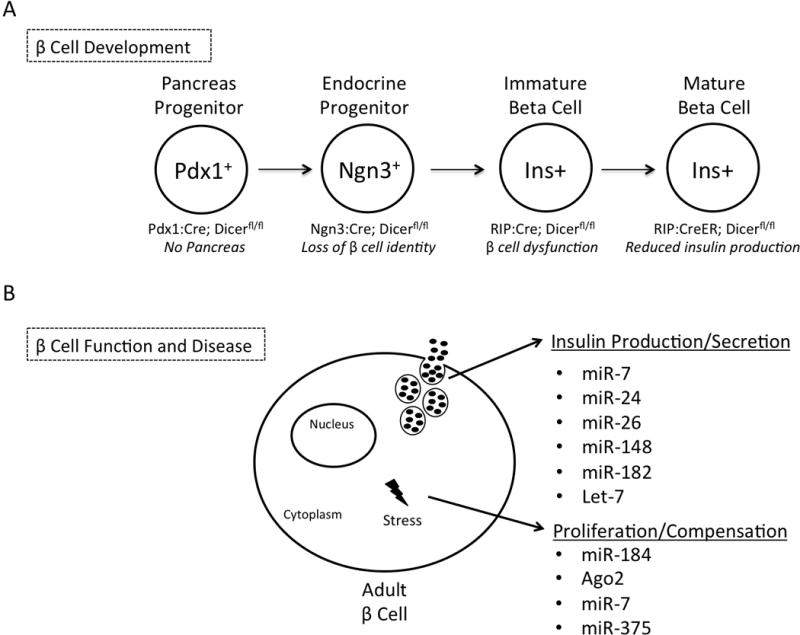
1a) A pictorial summary of the studies that identified a role for global miRNA function during β cell development through temporal ablation of Dicer in the pancreas in vivo. Each major stage of pancreas development is represented along with the genotype of conditional Dicer ablation and corresponding phenotype. 1b) Summary of a subset of individual miRNAs that regulate β cell function. The indicated miRNAs regulate glucose-stimulated insulin secretion and/or play a role in compensatory mechanisms during β cell stress.
While disruption of Dicer function shows that miRNAs are generally required for proper pancreas development and islet cell specification, individual miRNAs have also been implicated in β cell specification (Figure 1b). Kredo-Russo et al. showed that miRNA-7 (miR-7) is expressed during endocrine pancreas development where it directly targets and controls expression levels of the essential pancreatic transcription factor, Pax6 [20]. Additionally, the miR-30 family functions during the epithelial-to-mesenchymal transition by inhibiting mesenchymal mRNAs, such as Vimentin and Snail1, to favor pancreatic epithelial development [15]. Taken together, these studies define a requirement for miRNAs in several layers of β cell development.
Following β cell specification, complex networks of regulatory factors, including miRNAs, are needed to maintain β cell function. Two groups have examined the role of Dicer specifically in the developing β cells by using the Rat Insulin Promoter (RIP):Cre [10,11]. Unlike mice with Dicer deleted throughout the whole pancreas, the β cell specific Dicer mutant mice survived postnatally, but developed a diabetic phenotype around 8 weeks of age due to an overall decrease of β cell number and reduced insulin production and secretion [10,11]. Ultrastructural analysis demonstrated that the Dicer mutant β cells had 50% fewer insulin granules than control β cells [10]. Intriguingly, mutant islets also had a dramatic reduction in insulin transcript levels, suggestive of a defect in insulin gene transcription [10]. Global miRNA function also appears to be required for maintenance of β cell identity. Ablation of Dicer in adult β cells [12] using tamoxifen inducible RIP:CreER;Dicerfl/fl mice, resulted in hyperglycemia, glucose intolerance and a drastic reduction in β cell number [12]. The authors concluded that the loss of miRNA function caused the upregulation of several transcriptional repressors, including Sox6 and Bhlhe22, which in turn triggered decreased insulin expression [12].
Individual miRNAs have also been shown to be essential for many aspects of β cell function (Figure 1b). The most highly expressed miRNA in mouse and human islets, miRNA-375, was first identified in a murine pancreatic β -cell line (Min6) where it was shown to negatively regulate glucose-stimulated insulin secretion [17]. Analyses in mice showed that genetic ablation of miR-375 resulted in hyperglycemia due to reduced β cell mass [18]. This defect was attributed to a significant reduction in β cell proliferation due to the upregulation of several genes that negatively regulate cell growth [18]. Several miRNAs including miR-24, miR-26, miR-148, and miR-182 were also shown in vitro to negatively regulate insulin expression [12]. However, the relatively minimal effect seen with individual miRNA knockdown suggests that a combination of multiple miRNAs maintain insulin expression [12]. Interestingly, the evolutionarily conserved miRNA, miR-7a, was shown to be a negative regulator of both insulin secretion and β cell proliferation through inhibition of mTOR signaling proteins, indicating that a single miRNA can also regulate multiple layers of β cell function [19,21].
Consistent with recent implications that miRNAs maintain tissue homeostasis during a cellular response to stress, several studies have implicated miRNAs in β cell stress and diabetes [33]. Recently, Latreille et al. elucidated the relationship between miR-7a expression levels and two well-known diabetes mouse models [19]. Specifically, they determined that mice with a β-cell specific miR-7a deletion had a similar phenotype to genetically obese (ob/ob) mice; mice had enhanced pancreatic β cell function due to compensatory mechanisms that allowed them overcome insulin resistance [19]. This finding was supported by a corresponding 50% reduction in miR-7a transcript levels in compensating islets from ob/ob mice [19]. Remarkably, they found the opposite defect when they overexpressed miR-7a in vivo: hyperglycemia, reduced plasma insulin levels, and impaired glucose-stimulated insulin secretion [19]. Overexpression of miR-7a also caused decreased Ins1 and Ins2 expression, with a correlating decline in expression of several mature β cell markers [19]. Of note, these phenotypes are strikingly similar to the metabolic phenotype of diabetic db/db mice, which develop hyperglycemia and have reduced plasma insulin levels over time due to β cell dysfunction [19]. Mechanistically, miR-7a was shown to directly regulate genes that control late stages of insulin granule fusion with the plasma membrane, indicating a direct role for miRNAs in regulating β cell function during disease [19]. Similarly, the finding that miR-375 is upregulated in islets isolated from ob/ob mice, combined with an established role for miR-375 in β cell proliferation, suggests a mechanism whereby miR-375 enables β cell proliferation to compensate for metabolic stress [18]. Tattikota et al. further elucidated a mechanistic link between miR-375, Argonaute2 (Ago2), and another miRNA, miR-184 [14]. This study determined that the onset of insulin resistance caused the silencing of miR-184, which then released its constraint on Ago2, a major component of the RNA-induced silencing complex [14]. Ago2 is then able to orchestrate the suppressive function miR-375 thus enhancing β cell proliferation to accommodate the physiological demand for insulin [14]. Taken together, these studies have shown that miRNAs work to maintain β cell function during metabolic stress, and aberrant miRNA expression can be a marker for β cell dysfunction (Figure 1b).
Characterizing miRNAs that regulate β cell function may also reveal novel methods to treat diabetes. For example, the Let-7 miRNA family was shown to negatively regulate glucose-stimulated insulin secretion [22]. A recent study sought to determine if global Let-7 knockdown was sufficient to prevent or rescue impaired glucose intolerance in mice [23]. Researchers put mice on a high fat diet for 10 weeks and then initiated weekly injections with a locked nucleic acid (LNA) modified anti-miR that ablated Let-7 function [23]. After confirming strong reduction of Let-7 transcript in several tissues, researchers found that Let-7 knockdown was sufficient to prevent and treat impaired glucose tolerance brought on by a high fat diet [23]. Overall, miRNAs have been shown to be critical regulators of β cell development and function. Harnessing miRNA therapeutic capabilities will require a more comprehensive understanding of their mechanism of action in the pancreas.
LONG NONCODING RNAS IN β CELL DEVELOPMENT, FUNCTION AND DISEASE
Another abundant species of regulatory ncRNAs identified during the unbiased reconstruction of the transcriptome are the lncRNAs [36]. The grouping of lncRNAs simply by size (>200 nucleotides) and lack of protein-coding potential results in a category of RNAs with diverse properties and functions [37,38]. In general, lncRNAs are post-transcriptionally modified, localized to the nucleus and/or cytoplasm, and often have low expression levels and poor sequence conservation between species [7,39–41]. Thousands of lncRNAs have been identified in numerous cell types and different model organisms; however, with a few important exceptions, their respective functions are largely unknown. LncRNAs that have been carefully analyzed have been shown to regulate processes as varied as imprinting, dosage compensation, pluripotency, and apoptosis [42–45]. These RNAs are predicted to form stable secondary and tertiary structures that are able to interact with proteins and regulate several levels of gene expression [46]. More specifically, lncRNAs can control the expression of genes in cis or in trans by direct binding to transcription factors or by recruiting chromatin-modifying complexes [47–49]. Additionally, rare lncRNAs have been shown to function post-transcriptionally by altering splicing or mRNA stability [50,51].
The identification of several different lncRNA-mediated regulatory mechanisms suggests that we must reassess many biological processes in the context of a role for lncRNAs. This is true for β cell biology, which will benefit from a more comprehensive understanding of the regulatory mechanisms that give rise to a fully specified and functional pancreas. Although lncRNAs have not yet been studied in the embryonic pancreas, their highly specific temporal and spatial expression patterns and ability able to bind chromatin-modifying complexes suggest a mechanism whereby a cell specific lncRNA promotes differentiation towards one lineage over another [48,52]. In fact, there is increasing evidence that lncRNAs are key regulators of differentiation in several cell types including adipocytes, myocytes, and neurons [53]. Deep sequencing combined with novel gene editing techniques, such as cluster regularly interspersed palindromic repeats (CRISPR) [54], will be critical for the identification and characterization of lncRNAs in the embryonic pancreas.
In addition to a putative role during pancreas development, studies suggest that lncRNAs promote β cell function through the regulation of imprinted loci. The lncRNA MEG3 is transcribed downstream of Dlk1, the non-canonical Notch ligand highly expressed in β cells [55,56]. MEG3 acts to silence the imprinted paternal Dlk1 locus by recruitment of polycomb repressive complex 2 (PRC2) [55]. Interestingly, MEG3 is down regulated in human islets from T2D donors, which correlates with hypermethylation and misregulation of the Dlk1 locus [55]. A SNP associated with Type 1 diabetes was also mapped to the MEG3 locus, suggestive of dynamic lncRNA function [57]. Another lncRNA, Kcnq1ot1, is transcribed from an imprinted locus also containing the Kcnq1 gene, which encodes a voltage-gated potassium channel [58]. Interestingly, GWAS mapped a T2D-associated risk variant to the Kcnq1ot1 lncRNA [59]. Further analyses of human fetal samples homozygous for the SNP found increased methylation at the locus; however there was no concomitant change in gene expression of Kcnq1 or Kcnq1ot1 [60]. In contrast, a separate study found elevated Kcnq1ot1 transcript levels in T2D islets [61]. These contradicting results reflect the difficulty of examining complex loci and demonstrate the need for in depth in vivo analysis of lncRNAs.
While these findings are all suggestive of a role for lncRNAs in the β cell biology, characterization of lncRNA function requires tissue-specific analysis. Recently, several groups have catalogued lncRNAs in mouse and human islet cells [61–63]. A study from Moran et al. identified 1128 human islet lncRNAs, a majority of which adhered to previously established lncRNA properties of low expression compared to protein coding genes and a high degree of tissue specificity [7,61]. Remarkably, 55% of identified lncRNAs were islet specific, compared to only 9% of protein-coding genes. Genes in cis to the lncRNAs were also significantly more likely to be islet-specific, suggestive of co-regulation [61]. Additionally, the majority of islet lncRNAs were not expressed in human embryonic pancreas, indicative of an exclusive role in islet maturation and function [61]. Another study identified 1359 lncRNAs in β cells isolated from mouse islets that shared many properties with human islet-specific lncRNAs [61,62]. Interestingly, many of the lncRNAs harbored short spans of conserved sequences that might confer a conserved secondary structure across species [62]. Lastly, Bramswig et al. evaluated the cell-specific epigenetic and transcriptional landscapes of human β and α-cells [63]. This analysis identified 12 β cell-specific and 5 α cell-specific lncRNAs, suggesting that lncRNAs contribute to the unique regulatory environment found in different islet cell types [63]. While these studies lay important groundwork to implicate lncRNAs in β cell function, mechanistic conclusions will be dependent on functional interrogation of individual pancreatic lncRNAs.
CONCLUSIONS
Noncoding RNAs are beginning to be recognized as essential regulators of a wide range of biological processes. MicroRNAs are required for pancreas development and regulate β cell function by controlling expression levels of genes essential for β cell proliferation, insulin production and secretion, and maintenance of β cell identity. Long noncoding RNAs are the most abundant type of ncRNA and a large number of islet-specific lncRNAs have been identified; however, their functional characterization in the pancreas is lacking. Further characterization of pancreatic ncRNAs will extend our understanding of β cell biology and the factors leading to β cell dysfunction and diabetes.
KEY POINTS.
Noncoding RNAs represent a novel set of regulatory molecules that influence pancreatic development and β cell function.
Several miRNAs have been directly implicated in the regulation of β cell development, identity and function
A large number of islet-specific lncRNAs have been identified and potentially represent a novel layer of gene regulation in the pancreatic islet.
Noncoding RNAs could represent novel therapeutic targets for the treatment of diabetes
Acknowledgments
None
Funding:
NIH U01 DK087711
NIH T32 GM008224
Russell Berrie Foundation
Foundation for Diabetes Research
Juvenile Diabetes Foundation
Financial Support and sponsorship:
The work was supported by NIH U01 DK087711 and R01 DK082590 (L.S.), Foundation for Diabetes Research (L.S.), the Russell Berrie Foundation (L.A.) and Juvenile Diabetes Foundation (L.A.). Support for R.A.S. was provided by NIH T32 GM008224.
Footnotes
Financial Support and sponsorship:
None of the authors declare a conflict of interest.
REFERENCES AND RECOMMENDED READING
- 1.Zimmet P, Alberti KG, Shaw J. Global and societal implications of the diabetes epidemic. Nature. 2001;414:782–787. doi: 10.1038/414782a. [DOI] [PubMed] [Google Scholar]
- 2.Talchai C, Xuan S, Lin HV, Sussel L, Accili D. Pancreatic β cell dedifferentiation as a mechanism of diabetic β cell failure. Cell. 2012;150:1223–1234. doi: 10.1016/j.cell.2012.07.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hang Y, Yamamoto T, Benninger RK, et al. The MafA transcription factor becomes essential to islet β-cells soon after birth. Diabetes. 2014;63:1994–2005. doi: 10.2337/db13-1001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pagliuca FW, Millman JR, Gürtler M, et al. Generation of functional human pancreatic β cells in vitro. Cell. 2014;159:428–439. doi: 10.1016/j.cell.2014.09.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rezania A, Bruin JE, Arora P, et al. Reversal of diabetes with insulin-producing cells derived in vitro from human pluripotent stem cells. Nat Biotechnol. 2014;32:1121–1133. doi: 10.1038/nbt.3033. [DOI] [PubMed] [Google Scholar]
- 6*.Eliasson L, Esguerra JL. Role of non-coding RNAs in pancreatic beta-cell development and physiology. Acta Physiol. 2014;211:273–284. doi: 10.1111/apha.12285. This is a comprehensive review of ncRNAs that are known to regulate pancreatic islet function. [DOI] [PubMed] [Google Scholar]
- 7.Ponting CP, Oliver PL, Reik W. Evolution and Functions of Long Noncoding RNAs. Cell. 2009;136:629–641. doi: 10.1016/j.cell.2009.02.006. [DOI] [PubMed] [Google Scholar]
- 8.Lynn FC, Skewes-Cox P, Kosaka Y, et al. MicroRNA expression is required for pancreatic islet cell genesis in the mouse. Diabetes. 2007;56:2938–2945. doi: 10.2337/db07-0175. [DOI] [PubMed] [Google Scholar]
- 9**.Kanji MS, Martin MG, Bhushan A. Dicer1 is required to repress neuronal fate during endocrine cell maturation. Diabetes. 2013;62:1602–1611. doi: 10.2337/db12-0841. The authors explored the role of Dicer in endocrine progenitor cells by ablating Dicer in vivo using the Ngn3:Cre. Mice had normal endocrine cell differentiation but adult islets gradually loss β cells and upregulated neuronal genes. Mechanistic analysis showed that loss of Dicer caused upregulation of REST genes, which promote expression of neuronal genes. Study highlights regulatory mechanisms that maintain endocrine cell identity instead of a neuronal fate. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kalis M, Bolmeson C, Esguerra JL, et al. Beta-cell specific deletion of Dicer1 leads to defective insulin secretion and diabetes mellitus. PLoS One. 2011;6:e29166. doi: 10.1371/journal.pone.0029166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mandelbaum AD, Melkman-Zehavi T, Oren R, et al. Dysregulation of Dicer1 in beta cells impairs islet architecture and glucose metabolism. J Diabetes Res. 2012:e470302. doi: 10.1155/2012/470302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Melkman-Zehavi T, Oren R, Kredo-Russo S, et al. miRNAs control insulin content in pancreatic β cells via downregulation of transcriptional repressors. EMBO J. 2011;30:835–845. doi: 10.1038/emboj.2010.361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tattikota SG, Sury MD, Rathjen T, et al. Argonaute2 regulates the pancreatic β-cell secretome. Mol Cell Proteomics. 2013;12:1214–25. doi: 10.1074/mcp.M112.024786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14**.Tattikota SG, Rathjen T, McAnulty SJ, et al. Argonaute2 mediates compensatory expansion of the pancreatic β cell. Cell Metab. 2014;19:122–134. doi: 10.1016/j.cmet.2013.11.015. This is an elegant study showing the relationship between miR-184, miR-375, and Ago2 during β cell compensation in an insulin resistant mouse model. Reduction of miR-184 during insulin resistance promotes expression of Ago2, which then mediates miR-375 function to inhibit growth suppressor genes. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Joglekar MV, Patil D, Joglekar VM, et al. The miR-30 family microRNAs confer epithelial phenotype to human pancreatic cells. Islets. 2009;1:137–147. doi: 10.4161/isl.1.2.9578. [DOI] [PubMed] [Google Scholar]
- 16.Tang X, Muniappan L, Tang G, Özcan S. Identification of glucose-regulated miRNAs from pancreatic β cells reveals a role for miR-30d in insulin transcription. RNA. 2009;15:287–93. doi: 10.1261/rna.1211209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Poy MN, Eliasson L, Krutzfeldt J, et al. A pancreatic islet-specific microRNA regulates insulin secretion. Nature. 2004;432:226–230. doi: 10.1038/nature03076. [DOI] [PubMed] [Google Scholar]
- 18.Poy MN, Hausser J, Trajkovski M, et al. miR-375 maintains normal pancreatic α- and β-cell mass. Proc Natl Acad Sci. 2009;106:5813–5818. doi: 10.1073/pnas.0810550106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19**.Latreille M, Hausser J, Stützer I, et al. MicroRNA-7a regulates pancreatic β cell function. J Clin Invest. 2014;124:2722–2735. doi: 10.1172/JCI73066. This study showed that miR-7a acts to control β cell function by regulating late stage insulin granule fusion. Mice overexpressing miR-7a developed diabetes due to impaired insulin secretion and β cell dedifferentiation. Intriguingly, miR-7a expression levels are decreased in diabetic mouse models as well as islets from human diabetic patients. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kredo-Russo S, Mandelbaum AD, Ness A, et al. Pancreas-enriched miRNA refines endocrine cell differentiation. Development. 2012;139:3021–3031. doi: 10.1242/dev.080127. [DOI] [PubMed] [Google Scholar]
- 21*.Wang Y, Liu J, Liu C, Naji A, Stoffers DA. microRNA-7 regulates the mTOR pathway and proliferation in adult pancreatic β-cells. Diabetes. 2013;62:887–895. doi: 10.2337/db12-0451. The miRNA miR-7a was found to inhibit proliferation in mouse β cells by targeting five members of the mTOR signaling pathway. This pathway was also found to be conserved in human β cells, representing a potential therapeutic target for diabetes treatment. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhu H, Shyh-Chang N, Segrè AV, et al. The Lin28/let-7 axis regulates glucose metabolism. Cell. 2011;147:81–94. doi: 10.1016/j.cell.2011.08.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Frost RJ, Olson EN. Control of glucose homeostasis and insulin sensitivity by the Let-7 family of microRNAs. Proc Natl Acad Sci. 2011;108:21075–21080. doi: 10.1073/pnas.1118922109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ramachandran D, Roy U, Garg S, Ghosh S, Pathak S, Kolthur-Seetharam U. Sirt1 and mir-9 expression is regulated during glucose-stimulated insulin secretion in pancreatic β-islets. FEBS J. 2011;278:1167–1174. doi: 10.1111/j.1742-4658.2011.08042.x. [DOI] [PubMed] [Google Scholar]
- 25.Plaisance V, Abderrahmani A, Perret-Menoud V, Jacquemin P, Lemaigre F, Regazzi R. MicroRNA-9 Controls the Expression of Granuphilin/Slp4 and the Secretory Response of Insulin-producing Cells. J Biol Chem. 2006;281:26932–26942. doi: 10.1074/jbc.M601225200. [DOI] [PubMed] [Google Scholar]
- 26.Roggli E, Britan A, Gattesco S, et al. Involvement of microRNAs in the cytotoxic effects exerted by proinflammatory cytokines on pancreatic β-cells. Diabetes. 2010;59:978–986. doi: 10.2337/db09-0881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Baroukh N, Ravier MA, Loder MK, et al. MicroRNA-124a regulates Foxa2 expression and intracellular signaling in pancreatic β-cell lines. J Biol Chem. 2007;282:19575–19588. doi: 10.1074/jbc.M611841200. [DOI] [PubMed] [Google Scholar]
- 28.Lovis P, Gattesco S, Regazzi R. Regulation of the expression of components of the exocytotic machinery of insulin-secreting cells by microRNAs. Biol Chem. 2008;389:305–312. doi: 10.1515/BC.2008.026. [DOI] [PubMed] [Google Scholar]
- 29.Roggli E, Gattesco S, Caille D, et al. Changes in microRNA expression contribute to pancreatic β-cell dysfunction in prediabetic NOD mice. Diabetes. 2012;61:1742–1751. doi: 10.2337/db11-1086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wijesekara N, Zhang L, Kang MH, et al. miR-33a modulates ABCA1 expression, cholesterol accumulation, and insulin secretion in pancreatic islets. Diabetes. 2012;61:653–658. doi: 10.2337/db11-0944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Filios SR, Xu G, Chen J, Hong K, Jing G, Shalev A. MicroRNA-200 is induced by thioredoxin-interacting protein and regulates ZEB1 signaling and β cell apoptosis. J Biol Chem. 2014;289:36275–36283. doi: 10.1074/jbc.M114.592360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32*.Özcan S. Minireview: MicroRNA function in pancreatic β cells. Mol Endocrinol. 2014;28:1922–1933. doi: 10.1210/me.2014-1306. A thorough review of all characterized miRNAs that have been associated with pancreatic β cell function. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mendell JT, Olson EN. MicroRNAs in Stress Signaling and Human Disease. Cell. 2012;148:1172–1187. doi: 10.1016/j.cell.2012.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bartel DP. MicroRNAs: Target recognition and regulatory functions. Cell. 2009;136:215–233. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bernstein E, Kim SY, Carmell MA, et al. Dicer is essential for mouse development. Nat Genet. 2003;35:215–217. doi: 10.1038/ng1253. [DOI] [PubMed] [Google Scholar]
- 36.Carninci P, Kasukawa T, Katayama S, et al. The transcriptional landscape of the mammalian genome. Science. 2005;309:1559–1563. doi: 10.1126/science.1112014. [DOI] [PubMed] [Google Scholar]
- 37.Mattick JS, Makunin IV. Non-coding RNA. Hum Mol Genet. 2006;15:R17–R29. doi: 10.1093/hmg/ddl046. [DOI] [PubMed] [Google Scholar]
- 38.Ponjavic J, Ponting CP, Lunter G. Functionality or transcriptional noise? Evidence for selection within long noncoding RNAs. Genome Res. 2007;17:556–565. doi: 10.1101/gr.6036807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Guttman M, Amit I, Garber M, et al. Chromatin signature reveals over a thousand highly conserved large non-coding RNAs in mammals. Nature. 2009;458:223–227. doi: 10.1038/nature07672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Guttman M, Garber M, Levin JZ, et al. Ab initio reconstruction of cell type-specific transcriptomes in mouse reveals the conserved multi-exonic structure of lincRNAs. Nat Biotechnol. 2010;28:503–510. doi: 10.1038/nbt.1633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wang KC, Chang HY. Molecular mechanisms of long noncoding RNAs. Mol Cell. 2011;43:904–914. doi: 10.1016/j.molcel.2011.08.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bartolomei MS, Zemel S, Tilghman SM. Parental imprinting of the mouse H19 gene. Nature. 1991;351:153–155. doi: 10.1038/351153a0. [DOI] [PubMed] [Google Scholar]
- 43.Brockdorff N, Ashworth A, Kay GF, et al. The product of the mouse Xist gene is a 15 kb inactive X-specific transcript containing no conserved ORF and located in the nucleus. Cell. 1992;71:515–526. doi: 10.1016/0092-8674(92)90519-i. [DOI] [PubMed] [Google Scholar]
- 44.Guttman M, Donaghey J, Carey BW, et al. LincRNAs act in the circuitry controlling pluripotency and differentiation. Nature. 2011;477:295–300. doi: 10.1038/nature10398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hung T, Wang Y, Lin MF, et al. Extensive and coordinated transcription of noncoding RNAs within cell-cycle promoters. Nat Genet. 2011;43:621–629. doi: 10.1038/ng.848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Novikova IV, Hennelly SP, Sanbonmatsu KY. Tackling structures of long noncoding RNAs. Int J Mol Sci. 2013;14:23672–23684. doi: 10.3390/ijms141223672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Feng J, Bi C, Clark BS, et al. The Evf-2 noncoding RNA is transcribed from the Dlx-5/6 ultraconserved region and functions as a Dlx-2 transcriptional coactivator. Genes Dev. 2006;20:1470–1484. doi: 10.1101/gad.1416106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zhao J, Sun BK, Erwin JA, Song J-J, Lee JT. Polycomb proteins targeted by a short repeat RNA to the mouse X chromosome. Science. 2008;322:750–756. doi: 10.1126/science.1163045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Rinn JL, Kertesz M, Wang JK, et al. Functional demarcation of active and silent chromatin domains in human HOX loci by noncoding RNAs. Cell. 2007;129:1311–1323. doi: 10.1016/j.cell.2007.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Tripathi V, Ellis JD, Shen Z, et al. The nuclear-retained noncoding RNA MALAT1 regulates alternative splicing by modulating SR splicing factor phosphorylation. Mol Cell. 2010;39:925–938. doi: 10.1016/j.molcel.2010.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51*.Wang Y, Xu Z, Jiang J, et al. Endogenous miRNA sponge lincRNA-RoR regulates Oct4, Nanog, and Sox2 in human embryonic stem cell self-renewal. Dev Cell. 2013;25:69–80. doi: 10.1016/j.devcel.2013.03.002. This study identified the long noncoding RNA, RoR, as a promoter of human embryonic stem cell pluripotency. RoR acts as a miRNA sponge to inhibit suppression of key pluripotency transcription factors. [DOI] [PubMed] [Google Scholar]
- 52.Mercer TR, Dinger ME, Sunkin SM, Mehler MF, Mattick JS. Specific expression of long noncoding RNAs in the mouse brain. Proc Natl Acad Sci. 2008;105:716–721. doi: 10.1073/pnas.0706729105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53*.Fatica A, Bozzoni I. Long non-coding RNAs: new players in cell differentiation and development. Nat Rev Genet. 2014;15:7–21. doi: 10.1038/nrg3606. Excellent review on lncRNA manipulation and resulting phenotypes in model animal systems. [DOI] [PubMed] [Google Scholar]
- 54.Han J, Zhang J, Chen L, et al. Efficient in vivo deletion of a large imprinted lncRNA by CRISPR/Cas9. RNA Biol. 2014;11:829–835. doi: 10.4161/rna.29624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55**.Kameswaran V, Bramswig NC, McKenna LB, et al. Epigenetic regulation of the DLK1-MEG3 microRNA cluster in human type 2 diabetic islets. Cell Metab. 2014;19:135–145. doi: 10.1016/j.cmet.2013.11.016. This study describes the imprinting of the Dlk1-Meg3 locus in human type 2 diabetic islets. Transcriptome analyses showed that a cluster of miRNAs in that locus were epigenetically down regulated in diabetic islets. HITS-CLIP analysis further identified several miRNA targets that are relevant in β cell disease. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Falix FA, Aronson DC, Lamers WH, Gaemers IC. Possible roles of DLK1 in the Notch pathway during development and disease. Biochim Biophys Acta. 2012;1822:988–995. doi: 10.1016/j.bbadis.2012.02.003. [DOI] [PubMed] [Google Scholar]
- 57.Wallace C, Smyth DJ, Maisuria-Armer, et al. The imprinted DLK1-MEG3 gene region on chromosome 14q32.2 alters susceptibility to type 1 diabetes. Nat Genet. 2010;42:68–71. doi: 10.1038/ng.493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58*.Kameswaran V, Kaestner KH. The Missing lnc(RNA) between the pancreatic β-cell and diabetes. Front Genet. 2014;5:200. doi: 10.3389/fgene.2014.00200. Extensive review of long noncoding RNAs that have been implicated in β cell function and disease. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Voight BF, Scott LJ, Steinthorsdottir V, et al. Twelve type 2 diabetes susceptibility loci identified through large-scale association analysis. Nat Genet. 2010;42:579–589. doi: 10.1038/ng.609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60*.Travers ME, Mackay DJ, Dekker Nitert M, et al. Insights into the molecular mechanism for type 2 diabetes susceptibility at the KCNQ1 locus from temporal changes in imprinting status in human islets. Diabetes. 2013;62:987–992. doi: 10.2337/db12-0819. In depth analysis of a large cohort of human islets found that a Type 2 diabetes associated SNP in the imprinted Kcnq1 locus is significantly associated with increased methylation levels. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Morán I, Akerman I, van de Bunt M, et al. Human β cell transcriptome analysis uncovers lncRNAs that are tissue-specific, dynamically regulated, and abnormally expressed in type 2 diabetes. Cell Metab. 2012;16:435–448. doi: 10.1016/j.cmet.2012.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Ku GM, Kim H, Vaughn IW, et al. Research resource: RNA-Seq reveals unique features of the pancreatic β-cell transcriptome. Mol Endocrinol. 2012;26:1783–1792. doi: 10.1210/me.2012-1176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63*.Bramswig NC, Everett LJ, Schug J, et al. Epigenomic plasticity enables human pancreatic α to β cell reprogramming. J Clin Invest. 2013;123:1275–1284. doi: 10.1172/JCI66514. In this paper, ChIP-seq and RNA-seq identified several lncRNAs in human β, α, and exocrine cells. [DOI] [PMC free article] [PubMed] [Google Scholar]


