Abstract
Improved diagnostic screening has led to earlier detection of many tumors, but screening may still miss many aggressive tumor types. Proteomic and genomic profiling studies of breast cancer samples have identified tumor markers that may help improve screening for more aggressive, rapidly growing breast cancers. To identify potential blood-based biomarkers for the early detection of breast cancer, we assayed serum samples via matrix-assisted laser desorption ionization–time of flight mass spectrometry from a rat model of mammary carcinogenesis. We found elevated levels of a fragment of the protein dermcidin (DCD) to be associated with early progression of N-methylnitrosourea-induced breast cancer, demonstrating significance at weeks 4 (p = 0.045) and 5 (p = 0.004), a time period during which mammary pathologies rapidly progress from ductal hyperplasia to adenocarcinoma. The highest serum concentrations were observed in rats bearing palpable mammary carcinomas. Increased DCD was also detected with immunoblotting methods in 102 serum samples taken from women just prior to breast cancer diagnosis. To validate these findings in a larger population, we applied a 32-gene in vitro DCD response signature to a dataset of 295 breast tumors and assessed correlation with intrinsic breast cancer subtypes and overall survival. The DCD-derived gene signature was significantly associated with subtype (p < 0.001) and poorer overall survival [HR (95 % CI) = 1.60 (1.01–2.51), p = 0.044]. In conclusion, these results present novel evidence that DCD levels may increase in early carcinogenesis, particularly among more aggressive forms of breast cancer.
Keywords: Dermcidin, Breast cancer, Serum, Microenvironment
Introduction
In recent years, improved diagnostic screening has led to decreased breast cancer mortality, particularly among older women with a history of early-stage invasive breast cancer [1]. Routine surveillance includes mammography, ultrasound, and self- or physician-performed physical examination, methods that may be particularly effective at detecting indolent, slow-growing cancers [2, 3]. Thus, new approaches are needed for detecting more aggressive breast cancer in its earliest stages. Breast cancer heterogeneity is reflected in both the tumor and surrounding stroma. Stromal–tumor interaction can put unique pressure on the evolution of the tumor that may dictate its genetic composition [4]. Gene and protein candidate biomarkers have been identified in normal and neoplastic primary breast tissue [5] and in the secretome of nipple aspirate fluid of breast cancer patients [6]—indicating that cancer-specific changes can be measured outside of the tumor itself. Blood-based tumor markers and genomic profiling [7, 8] may be able to detect early biological changes in epithelial cells or in the tumor microenvironment, allowing for improved disease detection and monitoring.
Previously, we developed analytical methods using matrix-assisted laser desorption ionization–time of flight mass spectrometry (MALDI–TOF MS) to identify changes in the serum peptidome [9, 10]. This type of spectrometry-based protein profiling has been used to identify novel diagnostic markers for disease [11, 15]. MALDI–TOF MS can resolve signals from hundreds of peptides/proteins in complex mixtures, such as human serum. With the removal of large abundant proteins, we can collect accurate signals from peptides and proteins that may have otherwise been suppressed due to their lower abundance. The high-throughput and low-cost screening properties of MALDI–TOF MS make it a suitable method for collecting proteomic data from a large sample set. Over the last decade, many proteomic techniques have been evaluated to complement genomic analyses and to better evaluate the molecular biology of disease through tissue-based or circulating diagnostic or prognostic markers.
In this study, we tested whether serum peptide biomarkers reflective of tumor progression and changes in the microenvironment were detectable in humans and in a rat model for breast cancer. MALDI–TOF MS was used to analyze blood samples from a rat model of mammary carcinogenesis [16] to detect serum proteins that changed during tumor progression. We then used a case–control study of human breast cancer to measure the level of one of the identified proteins that was progression-associated in the rat model. Finally, publicly available data were interrogated to evaluate gene expression of a published gene signature associated with our protein of interest to further elucidate its role as a marker in breast cancers with different molecular characteristics.
Methods
Identification of potential marker of mammary carcinogenesis in a rodent study
Ninety-two female Sprague–Dawley rats (Taconic Farms, Germantown, NY) were obtained at 3 weeks of age. The animals had free access to a chow diet and tap water. They were housed three per cage for the duration of the study. Animals were randomized to either a control group (N = 20) or an experimental group (N = 70) using a random number table to determine group assignment. Mammary tumors were induced by N-methylnitrosourea (NMU) (Ash Stevens, Riverview, MI) prepared according to our previously published method [17]. NMU was dissolved immediately before use in 0.9 % NaCl and adjusted with acetic acid to pH value 4. At 3 weeks of age all 70 experimental rats were given NMU intraperitoneally at a dose of 50 mg/kg body weight [17]. The 20 control rats at the same age received sterile saline only. Following carcinogen treatment, rats were divided at random into a control group (N = 20) and an experimental group (N = 70). Rats were weighed weekly. After NMU treatment, rats were palpated twice a week for the presence of mammary tumors, the clinical endpoint used in this animal model for breast cancer. Five control and 15 carcinogentreated rats were euthanized at 2, 3, 4, and 5 weeks post carcinogen. At 9 weeks post carcinogen, 10 rats bearing palpable tumors were sacrificed. All procedures were executed between 9:00 a.m. and 12:00 p.m.. Animals were rendered unconscious by inhalation of gaseous CO2. Blood was obtained by retro-orbital sinus bleeding and gravity fed into red top tubes, allowed to sit for 10 min at room temperature, and then processed to centrifuge for 10 min in portable clinical centrifuge. Serum was aliquoted, put on dry ice, and stored frozen at −80 °C. Euthanasia was completed via cervical dislocation immediately after blood collection. The entire protocol (Supplemental Materials) was approved by the Colorado State University Institutional Animal Care and Use Committee.
An equal volume of 100 % acetonitrile was added to each serum sample to precipitate out large globular proteins as previously described [9, 10]. Samples were shaken for 30 min. Precipitated protein was spun down and supernatant was collected and mixed with 4-hydroxy-3,5-dimethoxycinnamic acid (sinapinic acid) matrix. Samples were spotted onto a hydrophobic 384-well MALDI plate in quintuplicate and MALDI–TOF MS data were collected on a Voyager DE-Pro spectrometer (Applied Biosystems, Foster City, CA). Daily machine calibrations were done using cytochrome c [12361 (+1), 6181 (+2)] to obtain mass accuracy. Plates were analyzed in linear mode using a 337 nm nitrogen laser, under the following settings optimized using cytochrome c: laser intensity 1,784; accelerating voltage 25,000; grid voltage 23,000; guide wire voltage 1,250; delay time 325 ns; low mass gate 1,000; and molecular mass range 1,000–40,000. Final spectra were generated by summing spectra produced from 20 laser hits at up to 20 places per spot (a total of up to 400 laser hits per summed spectra).
The dataset was aligned and processed to formulate a common set of peaks among spectra as previously described [18]. The intensity values of the resulting peaks were transformed and standardized as previously described [19, 20]. In brief, an intensity value, Yijk, for subject i, spectrum j at peak k, was transformed to Tijk = ln(Yijk − ck + 1.0) where ck is the minimum intensity measured at the kth peak among all spectra. The value 1.0 is arbitrary and included to stabilize the logarithm of any extremely small value. A linear mixed model with the quintuplicates of each sample sharing a Gaussian random intercept was used to identify peaks of interest. Specifically, at each m/z value, the logtransformed intensity described above was compared between the treated and untreated rats at each week of measurements, assessing whether the mean difference was significantly different from zero in that week, using all of the quintuplicate intensity measures of each sample. Subsequently, a test for trend was performed on each peak of interest over the 5 weeks for the treated and untreated rats separately.
To identify peptides represented by the statistically significant peaks from the MALDI–TOF MS analysis, albumin and IgG were removed from the sample via PROT-IA spin columns (Sigma-Aldrich, St. Louis, MO). Laemmli sample buffer was added to the eluate and the sample was heated to 100 °C for 5 min. The proteins were separated on a NuPAGE 4–12 % Bis–Tris Gel (Invitrogen Corporation, Carlsbad, CA) using 2-(N-morpholino)ethane sulfonic acid (MES) running buffer (Invitrogen). The gel was silver stained and the band corresponding to 4,209 Da was excised. The gel slice was digested with trypsin in 50 mmol/L ammonium bicarbonate overnight at 37 °C. Peptides were extracted from the gel slice and desalted with a ≥18 ZipTip (Millipore, Billerica, MA). The material was then analyzed by nano LC/MS using a ThermoFisher LTO (Thermo Fisher Scientific, Waltham, MA) at 0–35 % B over 60 min. Peptides were identified using Xtandem. The database search was performed against the Rat IPI database using Protein Prophet. Results were uploaded to Computational Portal and Analysis System (CAPS) database and interrogated using the following filter to look for individual peptides: ion percent ≥ 0.15; +1 raw score ≥ 200; +2 raw score ≥ 300; +3 raw score ≥ 300; unique peptides > 1.
In order to further characterize the 4,209 m/z species, 3.5 mg protein from rat serum that had been acetonitrile depleted as described above was injected onto a POROS R2/10, 4.6 × 100 mm, column (Applied Biosystems, Carlsbad, CA) and fractionated across a gradient of Buffer A (5 % AcN + 0.1 % TFA) and Buffer B (90 % AcN + 0.1 % TFA) at 2.7 ml/min. 192 fractions were collected from RP-HPLC, stored for 24 h at −80 °C, and lyophilized overnight. For dot blot analysis, the collected fractions (1 µL) were spotted in a 96-well format onto nitrocellulose and the membrane was blocked with 5 % milk in PBS at room temperature. The blot was incubated overnight at 4 °C with a DCD antibody (Santa Cruz Bio-technology, Santa Cruz, CA) specific to the N-terminal (sc-33656). Primary antibodies were visualized with a fluorescent-dye-labeled secondary antibody (Alexa Fluor-680 goat anti-rabbit, Molecular Probes, Eugene, OR) and directly quantified using the LI-COR Biosciences Odyssey infrared imaging system (Lincoln, NE) and associated software. Reverse phase high-performance liquid chromatography (RP-HPLC) fractions that interacted with the DCD antibody were analyzed using MALDI-TOF MS as described above. The fraction that consistently produced the original peak of interest (4,209 m/z) was further validated using carboxypeptidase cleavage. C-terminal sequencing of the fragment of interest was carried out using carboxypeptidases Y and A (Sigma-Aldrich Corporation, St. Louis, MO) and MALDI–TOF mass spectrometry as previously described [21].
Analysis of DCD levels in human serum samples
Breast cancer and age-matched control fasting serum samples were collected prior to diagnosis by the Aichi Cancer Center (ACC), Japan, as part of the Hospital-based Epidemiological Research Program II (HERPACC-II). We utilized 102 female incident cases of breast cancer (age range was 30–79 years) and 102 controls who had not been diagnosed as having any cancer prior to the blood draw matched on age frequency. Informed consent and human subjects research protocols were reviewed by Institutional Review Boards (IRB) at the ACC and at the Fred Hutchinson Cancer Research Center (FHCRC). Blood samples were preserved at 4 °C for 2–4 h until serum separation. The separated serum was then stored initially at −40 °C for 2–4 weeks after the separation, then at −80 °C for long-term storage.
Acetonitrile precipitation (as described above) was used to remove large globular proteins prior to analysis. Immunoblotting of all samples in triplicate was carried out with the same N-terminal antibody to DCD that was used in the identification of the candidate marker. Analysis of immunoblotting data used two-tailed t test. In all statistical analyses, p value <0.05 was taken as statistically significant.
Independent validation of marker in genomic dataset
Our goal was to assess how DCD gene expression was manifested in tumor behavior and characteristics. We examined this by projecting a previously published DCD-derived gene signature onto the publically available breast tumor microarray dataset from the Netherlands Cancer Institute (NKI-295, n = 295) [22]. The DCD expression-based signature was identified by Lowrie et al. [22] in HuH7, a hepatic carcinoma cell. These cells do not express DCD, but overexpression of DCD in these cells leads to improved cell survival. Treatment with a synthetic PIF-CP led to the overexpression of DCD and DCD-related genes. From this, Lowrie et al. generated the signature by comparing genomic microarrays from treated and untreated cells.
We classified the tumors as ‘‘DCD-positive’’ or ‘‘DCD-negative’’ by calculating the Pearson correlation coefficient as described in Creighton et al. [23]; we were able to extract 21 genes in the 32-gene DCD signature from the NKI295 dataset. Prior to applying the Creighton method, all probes were collapsed by statistical mean to a unique Entrez ID, and then data were median centered across all samples. Samples whose correlation coefficient was >0 were classified as ‘‘DCD-positive,’’ and those with a correlation coefficient ≤0 were classified as ‘‘DCD-negative.’’ We then evaluated associations between the DCD classification and tumor subtype (Basal-like, luminal A, luminal B, Her2, and Normal-like) (DF = 4), ER status (Positive and Negative) (DF = 1), tumor grade (well differentiated/intermediate/poorly differentiated) (DF = 2), tumor size (<2 cm, ≥2 cm) (DF = 1), age (20–29, 30–39, 40–49, 50–59, and ≥60) (DF = 4), and node status (positive/negative) (DF = 1) using Chi square tests.
We further examined the association between the DCD classification and overall survival in years to further evaluate associations between DCD and tumor aggressiveness. Specifically, Cox proportional hazard models were used to estimate the mortality hazard ratio (HR) and 95 % confidence interval (CI), comparing the two DCD classification groups unadjusted, and adjusted for the following clinically meaningful variables: tumor size (<2 cm/≥2 cm), grade (well differentiated, intermediate, and poorly differentiated), ER status (positive/negative), HER2 status (positive/negative), age (continuous), and node status (present/absent) [24]. All analyses were performed in R 2.14 using Bioconductor packages.
Results
A peak at 4,209 m/z increases with disease progression identified as DCD
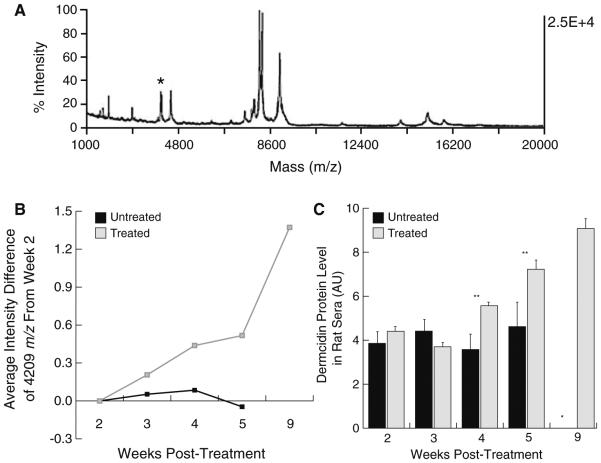
Serum was collected at weekly intervals from 20 control and 70 NMU-treated rats, 2, 3, 4, 5, and 9 weeks after treatment to identify changes in the serum proteome early in mammary carcinogenesis development. MALDI–TOF MS analysis identified four peaks with a week trend that was significant among the treated group when compared to the untreated group (Supplemental Table 1). The peak with the most significant trend was 4,209 m/z (Fig. 1a), so we further evaluated this as a potential marker of disease progression. For the peak 4,209 m/z, the treated group had a highly significant trend over the 5 weeks (p < 0.001) compared to the untreated group (p = 0.9804). Looking at the time points individually, the difference between the two groups reaches statistical significance at week 4 post-NMU treatment (p = 0.045) and becomes highly statistically significant by week 5 (p = 0.004). The intensity level of the peak 4,209 m/z at week 9 showed a 2.7-fold increase over week 5 and a 6.7-fold increase over week 3 (Fig. 1b).
Fig. 1.
a Complete MALDI spectrum of acetonitrile precipitated rat serum, the peak 4209 is in a preceding shoulder of the peak noted with an asterisk above. b Average intensity difference of the 4,209 m/z signal is plotted for the treated and untreated groups at weekly time points through week 5 with an additional time point at week 9 for the treated group (±SD). The difference was statistically significant between the groups at weeks 4 (p = 0.045) and 5 (p = 0.004). c Immunoblotting of treated and untreated groups with a DCD antibody confirmed this trend with significant differences between the two groups at weeks 4 (p = 0.001) and 5 (p = 0.004)
RP-HPLC followed by MALDI-TOF/TOF identified DCD as the most plausible peptide candidate with 22.7 % sequence coverage and two unique peptides (Supplemental Fig. 1A). Carboxypeptidase digestion also identified the N-terminus of DCD as the most plausible peptide candidate (Supplemental Fig. 1B), including both the signal peptide and survival promoting peptide (Supplemental Fig. 1C). Immunoblotting with an N-terminal antibody to DCD confirmed its presence in the fraction containing the maximum amount of 4,209 m/z signal (Data not shown). Immunoblotting of the treated and untreated rat serum samples indicated a statistically significant increase in DCD signal between the treated and untreated groups at weeks 4 (p = 0.001) and 5 (p = 0.004) that was consistent with the intensity difference of the peak 4,209 m/z (Fig. 1c).
DCD Levels are elevated in human sera of breast cancer patients
Comparison of serum samples collected from women just prior to their breast cancer diagnosis with 102 serum samples from age-matched controls who were disease free at serum collection and had no history of cancer showed a trend of elevated levels of DCD via immunoblot [OR (95 % CI) = 1.67 (0.93–3.02), p = 0.093, Fig. 2a].
Fig. 2.
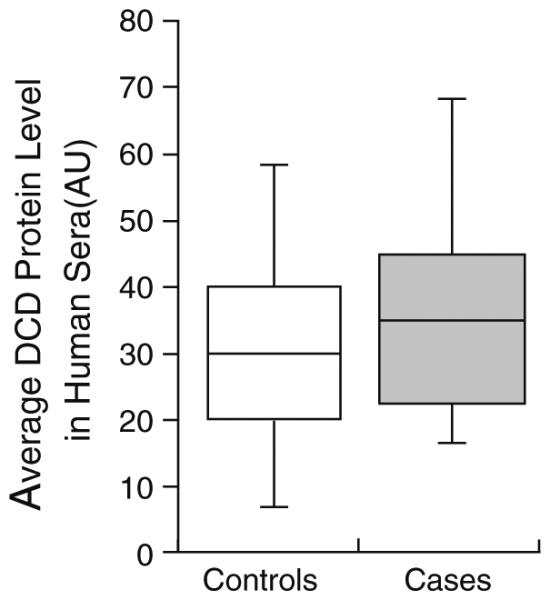
Human sera samples collected at time of breast cancer diagnosis showed significantly elevated levels of DCD compared to a control group (p 0.093)
Tumor characteristics are associated with DCD response
To evaluate whether DCD response is associated with tumor aggressiveness, we utilized the publically available breast tumor microarray data NKI-295 to evaluate a previously published signature of genes up- and downregulated with DCD overexpression in HuH7 cells [22]. Patient characteristics for these 295 patients are described in Table 1. The signature was also significantly associated with the tumor subtype (p < 0.001) and ER status (p < 0.001), but not with grade (p = 0.250), age (p = 0.950), size (p = 0.999), or node status (p = 0.190).
Table 1.
Characteristics of NKI patients by DCD signature correlation (N = 295)
| NKIa
N = 295 (%) |
NKI—negative for DCD signature N = 141 (%) |
NKI—positive for DCD signature N = 154 (%) |
P valuec | |
|---|---|---|---|---|
| Subtypeb | ||||
| Basal-like | 46 (15.6) | 13 (9.2) | 33 (21.4) | <0.01 |
| Her2-overexpressing | 49 (16.6) | 17 (12.0) | 32 (20.8) | |
| Luminal A | 86 (29.2) | 44 (31.2) | 42 (27.3) | |
| Luminal B | 81 (27.5) | 44 (31.2) | 37 (24.0) | |
| Normal | 33 (11.2) | 23 (16.3) | 10 (6.5) | |
| ER status | ||||
| Negative | 69 (23.4) | 17 (12.1) | 52 (33.8) | <0.01 |
| Positive | 226 (76.6) | 124 (87.9) | 102 (66.2) | |
| Grade | ||||
| Well differentiated | 75 (25.4) | 34 (24.1) | 41 (26.6) | 0.25 |
| Intermediate | 101 (34.2) | 55 (39.0) | 46 (29.9) | |
| Poorly differentiated | 119 (40.4) | 52 (36.9) | 67 (43.5) | |
| Size | ||||
| <2 cm | 114 (38.6) | 55 (39.0) | 59 (38.3) | 1.00 |
| ≥2 cm | 181 (61.4) | 86 (61.0) | 95 (61.7) | |
| Node status | ||||
| Absent | 151 (51.2) | 66 (46.8) | 85 (55.2) | 0.19 |
| Present | 144 (48.8) | 75 (53.2) | 69 (44.8) | |
| Age (years) | ||||
| 20–29 | 4 (1.4) | 2 (1.4) | 2 (1.3) | 0.95d |
| 30–39 | 59 (20.0) | 30 (21.3) | 29 (18.8) | |
| 40–49 | 183 (62.0) | 86 (61.0) | 97 (63.0) | |
| ≥50 | 49 (16.6) | 23 (16.3) | 26 (16.9) |
Italicized values indicate statistically significant P value (< 0.05)
DCD dermcidin
NKI—publically available breast cancer dataset from the Dutch Cancer Institute (Amsterdam)
PAM50 subtype classification used [30]
Chi squared used to calculate P values unless otherwise noted
Fisher’s exact test used
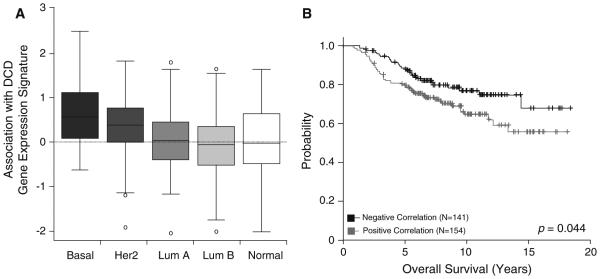
We next assessed the association between expression of the DCD genomic signature and intrinsic subtypes. The signature was more highly expressed in basal-like and HER2 breast cancers compared to other subtypes (Fig. 3a). We found that the DCD signature was more likely to be positively correlated in basal-like breast cancers and negatively correlated in normal-like breast cancers, while luminal cancers had an intermediate phenotype (Fig. 3a). In stratified analyses of all tumors, the DCD genomic signature was a significant predictor of overall breast cancer survival after adjusting for standard clinical variables: tumor size (<2 cm/ ≥2 cm), grade (well differentiated, intermediate, and poorly differentiated), ER status (positive/negative), HER2 status (positive/negative), age (continuous), and node status (present/absent) [HR (95 % CI) = 1.60 (1.01–2.51), p = 0.044, Fig. 3b]. After adjusting for intrinsic subtype the association with overall survival was no longer statistically significant [HR (95 % CI) = 1.40 (0.87–2.27), p = 0.17].
Fig. 3.
b A previously published DCD genomic signature was significantly associated with intrinsic subtype (p < 0.001) and b overall survival (p = 0.044), indicating a positive correlation with more aggressive disease
Discussion
Given that triple-negative tumors are more likely to be interval cancers and are not readily detected by standard screening [25, 26], additional biomarkers for the early detection and progression of this type of breast cancer are needed. Markers that are altered at multiple biological levels, such as the genome and proteome, will provide more complete information on biological signaling pathways that may play a role in these aggressive forms of breast cancer.
Here we identify DCD as a candidate biomarker of early breast cancer in a rat study of mammary carcinogenesis. DCD is a 110 amino acid protein that is characterized by three key peptides: a 47 amino acid DCD-1 peptide, a 19 amino acid signal peptide, and a 30 amino acid proteolysis-inducing factor-core peptide (PIF-CP). Carboxypeptidase digestion was used to confirm the inclusion of the survivalpromoting peptide, PIP-CP, in our DCD fragment. DCD-1 and PIF-CP are the two biologically active peptides and are responsible for antimicrobial activity and cell survival, respectively [27]. PIF-CP is identical to neuronal survival factor Y-P30 and is thought to protect cancer cells in the same manner as it protects neuronal cells from damage [28]. DCD leads to multiple gene expression changes that have been suggested to influence cell proliferation, which in turn influence tumor formation and progression [22].
We evaluated the association between breast cancer and DCD in human sera in a subset of available samples from ACC. Our findings demonstrate higher concentrations in DCD protein expression in the sera of women just prior to a diagnosis of breast cancer. These sera samples and matched controls were taken from Japanese clinic patients. Thus, we cannot exclude the possibility that lifestyle and environmental factors may influence the peptidome of those samples, so generalizability to ethnically diverse populations will need to be assessed. However, we also showed interesting biology of DCD-response genes in a population from the Netherlands. The association between DCD response and tumor aggressiveness is interesting in light of the influence that DCD has on cell proliferation [22] and in protecting neuronal cells from damage [29].
The association we observed between DCD-response gene expression and breast cancer subtype may indicate a biological role for DCD in more aggressive forms of breast cancer. Additionally, across all subtypes, survival was significantly poorer in women who had tumors with a high DCD-response signature. These data suggest that DCD is not only a biomarker of cancer, but it may also act as a progression factor. DCD has already been identified as a candidate survival gene in breast cancer [29], but the subtype-specific association and association of DCD influenced genes with overall survival had not been previously documented. Future studies should consider whether DCD is less sensitive to the detection of luminal breast cancers, given the role it may play in progression of more aggressive breast cancers. Further studies are also necessary to understand the mechanisms of action and to determine if DCD may be a possible target for the treatment of more aggressive breast cancers.
Supplementary Material
Acknowledgments
This work was supported by the National Institutes of Health through the following grants: Women’s Health Initiative (U01 CA152637), Early Detection Research Network (N01 WH 74313), and Cardiovascular Health Study (R01 CA116393). The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Footnotes
Electronic supplementary material The online version of this article (doi:10.1007/s10549-014-2880-3) contains supplementary material, which is available to authorized users.
Conflict of interest The authors declare that they have no conflict of interest or financial relationship with the organizations that sponsored the research.
References
- 1.Buist D, Bosco J, Silliman RA, Gold HT. Long term surveillance mammography and mortality in older women with a history of early stage invasive breast cancer. Breast Cancer Res. 2013;142(1):153–63. doi: 10.1007/s10549-013-2720-x. doi:10.1080/10810730.2013.825673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Anderson WF, Chen BE, Brinton LA, Devesa SS. Qualitative age interactions (or effect modification) suggest different cancer pathways for early onset and late onset breast cancers. Cancer Causes Control. 2007;18:1187–1198. doi: 10.1007/s10552-007-9057-x. doi:10.1007/s10552-007-9057-x. [DOI] [PubMed] [Google Scholar]
- 3.Collett K, Stefansson IM, Eide J, et al. A basal epithelial phenotype is more frequent in interval breast cancers compared with screen detected tumors. Cancer Epidemiol Biomarkers Prev. 2005;14:1108–1112. doi: 10.1158/1055-9965.EPI-04-0394. doi:10.1158/1055 9965.EPI-04-0394. [DOI] [PubMed] [Google Scholar]
- 4.Saunders NA, Simpson F, Thompson EW, et al. Role of intratumoural heterogeneity in cancer drug resistance: molecular and clinical perspectives. EMBO Mol Med. 2012;4:675–684. doi: 10.1002/emmm.201101131. doi:10. 1002/emmm.201101131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Marić P, Ozretić P, Levanat S, et al. Tumor markers in breast cancer evaluation of their clinical usefulness. Coll Antro pol. 2011;35:241–247. [PubMed] [Google Scholar]
- 6.Mannello F, Medda V, Tonti GA. Protein profile analysis of the breast microenvironment to differentiate healthy women from breast cancer patients. Expert Rev Proteomics. 2009;6:43–60. doi: 10.1586/14789450.6.1.43. doi:10.1586/14789450.6.1.43. [DOI] [PubMed] [Google Scholar]
- 7.Ebeling FG, Stieber P, Untch M, et al. Serum CEA and CA 15 3 as prognostic factors in primary breast cancer. Br J Cancer. 2002;86:1217–1222. doi: 10.1038/sj.bjc.6600248. doi:10.1038/sj/bjc/6600248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gion M, Mione R, Leon AE, Dittadi R. Comparison of the diagnostic accuracy of CA27.29 and CA15.3 in primary breast cancer. Clin Chem. 1999;45:630–637. [PubMed] [Google Scholar]
- 9.Brauer HA, Libby TE, Mitchell BL, et al. Cruciferous vegetable supplementation in a controlled diet study alters the serum peptidome in a GSTM1 genotype dependent manner. Nutr J. 2011;10:11. doi: 10.1186/1475-2891-10-11. doi:10.1186/1475-2891-10-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mitchell BL, Yasui Y, Lampe JW, et al. Evaluation of matrix assisted laser desorption/ionization time of flight mass spectrometry proteomic profiling: identification of alpha 2 HS glycoprotein B chain as a biomarker of diet. Proteomics. 2005;5:2238–2246. doi: 10.1002/pmic.200401099. doi:10.1002/pmic.200401099. [DOI] [PubMed] [Google Scholar]
- 11.Bertucci F, Goncalves A. Clinical proteomics and breast cancer: strategies for diagnostic and therapeutic biomarker dis covery. Future Oncol. 2008;4:271–287. doi: 10.2217/14796694.4.2.271. doi:10.2217/14796694.4.2.271. [DOI] [PubMed] [Google Scholar]
- 12.Li J, Orlandi R, White CN, et al. Independent validation of candidate breast cancer serum biomarkers identified by mass spectrometry. Clin Chem. 2005;51:2229–2235. doi: 10.1373/clinchem.2005.052878. doi:10.1373/clinchem.2005.052878. [DOI] [PubMed] [Google Scholar]
- 13.Belluco C, Petricoin EF, Mammano E, et al. Serum pro teomic analysis identifies a highly sensitive and specific dis criminatory pattern in stage 1 breast cancer. Ann Surg Oncol. 2007;14:2470–2476. doi: 10.1245/s10434-007-9354-3. doi:10.1245/s10434 007-9354-3. [DOI] [PubMed] [Google Scholar]
- 14.van Winden AW, Gast MCW, Beijnen JH, et al. Validation of previously identified serum biomarkers for breast cancer with SELDI TOF MS: a case control study. BMC Med Genomics. 2009;2:4. doi: 10.1186/1755-8794-2-4. doi:10.1186/1755-8794-2-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.van Winden AO, Rodenburg W, Pennings JLA, et al. A bead based multiplexed immunoassay to evaluate breast cancer biomarkers for early detection in pre diagnostic serum. IJMS. 2012;13:13587–13604. doi: 10.3390/ijms131013587. doi:10.3390/ijms131013587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Thompson HJ, McGinley JN, Rothhammer K, Singh M. Rapid induction of mammary intraductal proliferations, ductal carcinoma in situ and carcinomas by the injection of sexually immature female rats with 1 methyl 1 nitrosourea. Carcinogen esis. 1995;16:2407–2411. doi: 10.1093/carcin/16.10.2407. [DOI] [PubMed] [Google Scholar]
- 17.Thompson HJ, Adlakha H. Dose responsive induction of mammary gland carcinomas by the intraperitoneal injection of 1 methyl 1 nitrosourea. Cancer Res. 1991;51:3411–3415. [PubMed] [Google Scholar]
- 18.Randolph TW, Yasui Y. Multiscale processing of mass spectrometry data. Biometrics. 2006;62:589–597. doi: 10.1111/j.1541-0420.2005.00504.x. doi:10.1111/j.1541 0420.2005.00504.x. [DOI] [PubMed] [Google Scholar]
- 19.Yasui Y, Pepe M, Thompson ML, et al. A data analytic strategy for protein biomarker discovery: profiling of high dimensional proteomic data for cancer detection. Biostatistics. 2003;4:449–463. doi: 10.1093/biostatistics/4.3.449. doi:10.1093/biostatistics/4.3.449. [DOI] [PubMed] [Google Scholar]
- 20.Yasui Y, McLerran D, Adam BL, et al. An automated peak identification/calibration procedure for high dimensional protein measures from mass spectrometers. J Biomed Biotechnol. 2003;2003:242–248. doi: 10.1155/S111072430320927X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Liska J, Galbavy S, Macejova D, Zlatos J. Histopathology of mammary tumours in female rats treated with 1 methyl 1 nitrosourea. Endocr Regul. 2000;34(2):91–96. [PubMed] [Google Scholar]
- 22.Lowrie AG, Dickinson P, Wheelhouse N, et al. Proteolysis inducing factor core peptide mediates dermcidin induced prolif eration of hepatic cells through multiple signalling networks. Int J Oncol. 2011;39:709–718. doi: 10.3892/ijo.2011.1064. doi:10.3892/ijo-2011.1064. [DOI] [PubMed] [Google Scholar]
- 23.Creighton CJ, Casa A, Lazard Z, et al. Insulin like growth factor I activates gene transcription programs strongly associated with poor breast cancer prognosis. J Clin Oncol. 2008;26:4078–4085. doi: 10.1200/JCO.2007.13.4429. doi:10.1200/JCO.2007.13.4429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fan C, Prat A, Parker JS, et al. Building prognostic models for breast cancer patients using clinical variables and hundreds of gene expression signatures. BMC Med Genomics. 2011;4:3. doi: 10.1186/1755-8794-4-3. doi:10. 1186/1755-8794-4-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Anderson WF, Matsuno R. Breast cancer heterogeneity: a mixture of at least two main types? JNCI J Natl Cancer Inst. 2006;98:948–951. doi: 10.1093/jnci/djj295. doi:10.1093/jnci/djj295. [DOI] [PubMed] [Google Scholar]
- 26.Anders CK, Carey LA. Biology, metastatic patterns, and treatment of patients with triple negative breast cancer. Clin Breast Cancer. 2009;9:S73–S81. doi: 10.3816/CBC.2009.s.008. doi:10.3816/CBC.2009.s.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Stewart GD, Skipworth RJE, Pennington CJ, et al. Varia tion in dermcidin expression in a range of primary human tumours and in hypoxic/oxidatively stressed human cell lines. Br J Cancer. 2008;99:126–132. doi: 10.1038/sj.bjc.6604458. doi:10.1038/sj.bjc.6604458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Stewart GD, Lowrie AG, Riddick ACP, et al. Dermcidin expression confers a survival advantage in prostate cancer cells subjected to oxidative stress or hypoxia. Prostate. 2007;67:1308–1317. doi: 10.1002/pros.20618. doi:10.1002/pros.20618. [DOI] [PubMed] [Google Scholar]
- 29.Porter D, Weremowicz S, Chin K, et al. A neural survival factor is a candidate oncogene in breast cancer. Proc Natl Acad Sci USA. 2003;100:10931–10936. doi: 10.1073/pnas.1932980100. doi:10.1073/pnas.1932980100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Perou CM, Sørlie T, Eisen MB, van de Rijn M, et al. Molecular portraits of human breast tumours. Nature. 2000;406:747–752. doi: 10.1038/35021093. doi:10.1038/35021093. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.