Abstract
As critical players in gene regulation, RNA binding proteins are taking center stage in our understanding of cellular function and disease. In our era of bench-top sequencers and unprecedented computational power, biological questions can be addressed in a systematic, genome-wide manner. Development of high-throughput sequencing methodologies provides unparalleled potential to discover new mechanisms of disease-associated perturbations of RNA homeostasis. Complementary to candidate single-gene studies, these innovative technologies may elicit the discovery of unexpected mechanisms, and allow us to determine the widespread influence of the multifunctional RNA binding proteins on their targets. As disruption of RNA processing is increasingly implicated in neurological diseases, these approaches will continue to provide insights into the roles of RNA binding proteins in disease pathogenesis.
Keywords: RNA binding protein, neurodegeneration, high-throughput sequencing, splicing, polyadenylation
RNA binding proteins and RNA processing
If DNA is the blueprint for a cell, then transcribed RNA represents bits of information retrieved from DNA to direct cellular function and promote cell survival. Prior to guiding cell function, these nascent RNAs must first undergo extensive processing and precise localization, both of which are highly dynamic processes that require a complex interplay among proteins interacting with RNA, known as RNA binding proteins (RBPs, see Glossary). As with any multi-step, multi-component procedure, exact homeostatic control of RNA processing is essential for the sustained health and proper function of the eukaryotic cell. RBPs bind to specific sequences or secondary structures within the RNA molecule to modulate co- and post-transcriptional processing steps (depicted in Fig. 1).
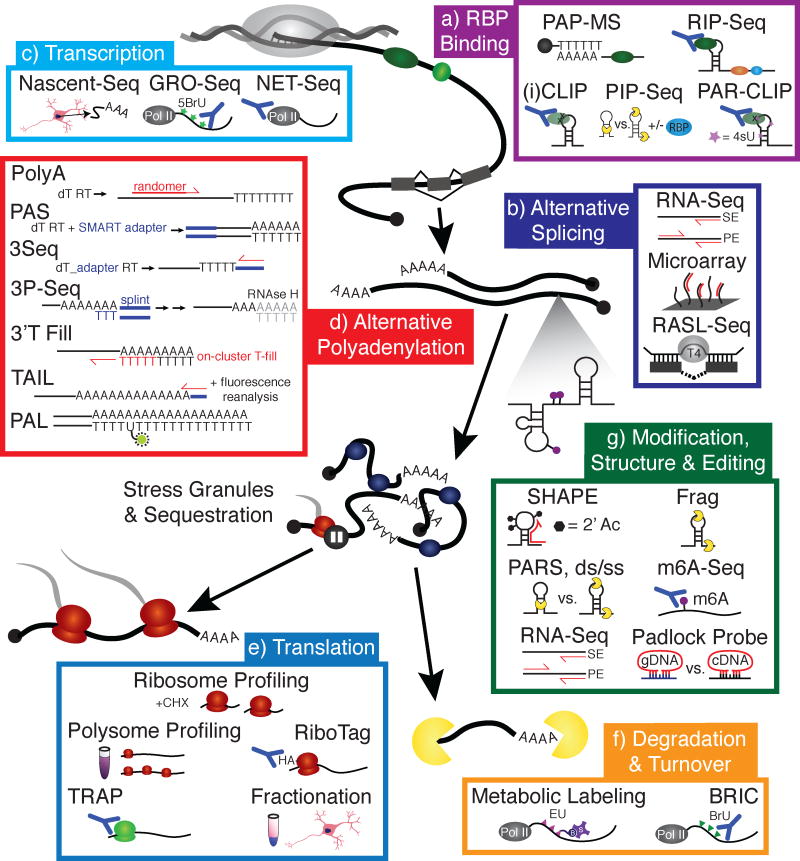
Figure 1. High-throughput sequencing enables quantification of RNA processing steps on a global scale.
a | RBP Binding. Poly(A) purification and liquid chromatography mass spectrometry (PAP-MS) is a method that involves poly(A) selection of RBP-bound RNA followed by proteomic analysis to identify mRNA-bound proteins, enabling the identification of novel RBPs. RNA immunoprecipitation (RIP) is a method for identifying whole transcripts associated with an RBP by immunoprecipitating the RBP with bound RNA, then subjecting the isolated RNA to RNA-seq analysis. Cross-linking immunoprecipitation (CLIP)-seq and individual nucleotide resolution CLIP (iCLIP) improve on the resolution of RIP-Seq by utilizing UV to crosslink RNAs to protein. This enables both more stringent washing to reduce false positives, and a digestion step that reveals specific RBP target regions and motifs. Photoactivatable-ribonucleoside-enhanced crosslinking and immunoprecipitation (PAR-CLIP) is similar to iCLIP, but crosslinking efficiency is increased with the metabolic labeling of RNA by 4-thio-uracil (4sU). Protein interaction profile (PIP)-Seq characterizes the structural dependence of RBP-RNA interactions through the inclusion of proteinase K dependent libraries using proteinase K.
b | Alternative Splicing. Splicing inclusion and exclusion events can be detected by a simple RNA-Seq experiment, either single end (SE) or paired end (PE) reads, provided sufficient read depth at exon-exon and exon-intron junctions, and does not require the splicing event to be annotated. Microarrays can also be used to detect splicing changes, and are more sensitive to detecting events in lowly expressed transcripts compared to RNA-Seq; however, the assay is limited to the number of events on an array, as well as prior knowledge of an inclusion/exclusion event. A less comprehensive but very sensitive technique, RNA-mediated oligonucleotide Annealing, Selection, and Ligation with sequencing (RASL-seq), utilizes a ligation reaction to detect an event based on ligation of oligomers complementary to alternative exon-exon junctions.
c | Transcription. Nascent-Seq involves the sequencing of nascent RNAs isolated from the nucleus using centrifugation and fractionation. Global run-on sequencing (GRO-Seq) involves pausing of the transcription machinery, then reinitiation with the addition of brominated nucleotides which are incorporated into nascent transcripts and facilitate immunopurification of the nascent RNAs by antibodies specific for the 5-bromouridine (5BrU). To avoid transcription pausing, native elongating transcript sequencing (NET-Seq) employs immunopurification of RNA polymerase II (pol II) with its associated transcripts for sequencing.
d | Alternative Polyadenylation. Poly(A) site usage can be determined by several techniques, the majority of which involve positive selection by oligo(d)Ts and sequencing into the poly(A) tail. PolyA-Seq involves oligo(d)T-primed first strand synthesis and randomer-primed second strand synthesis. Poly(A) site sequencing (PAS-Seq) also uses oligo(d)T-primed first strand synthesis, but with the inclusion of a SMART adapter added at the end of first strand synthesis, reducing the need for internal randomer priming. 3Seq takes a somewhat modified approach, with oligo(d)T-primed first strand synthesis that includes an adapter for second strand synthesis. A problem with internal priming is the risk that the PCR will not proceed all the way to the poly(A) site, making it impossible to determine alternative polyadenylation. Furthermore, oligo(d)T priming risks polymerase slippage. 3P-Seq avoids this with the use of a splint ligation to attach adapters, while 3′T-fill fills in the poly(A) tract with d(T)s on the flow cell of the sequencing instrument prior to initiation of sequencing. In addition to alternative poly(A) site identification, poly(A) length can be critical in the case of RBPs that bind the poly(A) tail. Poly(A) size can be determined by TAIL-Seq, which utilizes special software to reanalyze the fluorescence signal on the flow cell to eliminate false ‘T’ base calls due to residual signal from reading the poly(A) tract. Alternatively, poly(A) tail length sequencing (PAL-Seq) utilizes stoichiometric incorporation of a modified uridine during poly(A) sequencing that can be fluorescently tagged, identifying tail length as proportional to fluorescence intensity.
e | Translation. Methods for identifying actively translating transcripts involve purification of ribosomes or polysomes by fractionation and sequencing of the associated RNAs, as in ribosome and polysome profiling. Ribosome labeling has enhanced these techniques. In a method similar to CLIP, tagged ribosomes can be immunoprecipitated and the associated RNAs sequenced, as in RiboTag. Alternatively, lineage-specific promoters driving EGFP-labeled ribosomes allow for cell-specific isolation of polysomes, as in TRAP-Seq, further enhancing the resolution of nascent RNA sequencing. One aspect of translational control is localization of the translation machinery and associated RNAs, which can be probed by compartmental fractionation and either sequencing or proteomic analysis of each fraction.
f | Degradation & Turnover. Measuring rates of global mRNA synthesis and degradation is most commonly carried out with a pulse-chase experiment to label nascent transcripts followed by the observed loss of label as transcripts are degraded. Metabolic labeling can be performed with 5-ethynyl uridine (EU), which can be biotinylated with click chemistry for purification of pulse-labeled transcripts, or with 4-thiouridine which can also be biotinylated. Similarly, RNA can be pulsed with 5-bromo-uridine (BrU) in BrU immunoprecipitation chase-deep sequencing analysis (BRIC-Seq). These techniques require an immense amount of input material, making it a challenging assay for difficult-to-obtain cell types, such as iPS-derived neurons.
g | Modifications, Structure & Editing. High-throughput methods for determining secondary structure include selective 2′-hydroxyl acylation analyzed by primer extension (SHAPE)-Seq, which involves the acetylation of specific bases in single-stranded loops and bulges, blocking cDNA synthesis and enabling single nucleotide resolution of certain secondary structures. To glean information on regions of secondary structure, differential digestion with single and double-strand specific nucleases can be utilized. Fragmentation sequencing (Frag-Seq) utilizes a single-strand-specific nuclease to identify regions of RNA involved in base pairing structures. Parallel analysis of RNA structure (PARS)-Seq compares sequencing libraries of single strand (ss) and double strand (ds)-specific nuclease-treated RNA to identify secondary structure. To identify RNA editing events, such as A-to-I editing, RNA-Seq often suffices. However, to probe a specific site, padlock probes can be designed to compare the identity of a single base between cDNA generated from RNA to the genomic DNA. m6A-Seq enables low-resolution sequencing of m6A modifications through nucleobase IP.
Opportunities for misregulation of RNA processing abound, often caused by mutations within RBP binding sites or in the RBPs themselves, altering RBP-RNA interactions. Such dysfunction has been identified as the culprit in countless human diseases [6, 7] and is increasingly recognized as a central component in neurodegenerative disorders. In mice, more than half of the known or putative RBPs can be detected in brain tissue by in-situ hybridization, and a subset of these are specific to neural cells [8], consistent with neurons being highly susceptible to mutations in RBP binding elements and aberrant RBP interactions. Furthermore, RBPs expressed in the central nervous system are intimately involved in the regulation of alternative splicing, which is also more prevalent in cells of the nervous system than in any other cell-type [9, 10]. Finally, RBPs are required to protect mRNAs from premature translation and degradation during their transport from soma to dendrites and axon, enabling de novo protein synthesis at synapses [11–14]. Genomic approaches have recently provided major insights into the multiple ways RBPs influence the fate of their targets and create vast opportunities to reveal the roles of RBPs in neurological diseases. In this review, we describe genome-wide technologies used to identify and characterize RBPs in the context of RNA processing and neurological disease.
Identifying RBPs and their targets
Although hundreds of RBPs have been predicted based on homology to known RNA binding domains, only a subset of these have been validated and characterized in vivo. To identify novel RBPs, poly(A) affinity purification and mass spectrometry (PAP-MS) is a straightforward technique in which mRNA-protein complexes are purified by poly(A) selection, and bound proteins are identified by mass spectrometry (Fig. 1A). This technique has enabled identification of RBPs lacking a canonical RNA binding domain [15]; however, poly(A) selection misses intron-bound RBPs and RBPs bound to non-polyadenylated species like unprocessed mRNA, microRNAs and their precursors. Putative RBPs can then be validated by techniques utilizing immunoprecipitation and high-throughput sequencing (Seq). The most basic of these is RNA immunoprecipitation (RIP)-Seq [16] (Fig. 1A). This technique identifies RBP-associated transcripts, but does not reveal precise binding sites, and is potentially encumbered by false positives due to low stringency washes in the absence of cross-linking [17]. To determine specific binding sites, cross-linking and immunoprecipitation (CLIP)-Seq (or HITS-CLIP) [18] is now commonly used (Fig. 1A). UV irradiation (inefficiently) induces covalent bonds (crosslinks) between protein and nucleic acid, enabling both stringent washes to remove non-specific and indirect interactions, as well as RNA size-trimming by RNase digestion to hone in on specific binding sites [19–29]. CLIP-Seq has proved invaluable for precise identification of in-vivo RBP binding sites and providing insights into RBP functions in disease and development [30–34]). Variations of CLIP-Seq include metabolic labeling with photoreactive thiolated nucleotides to enhance UV-crosslinking efficiency (Fig. 1A). This technique, termed photoactivatable-ribonucleoside-enhanced CLIP (PAR-CLIP) [35] gets closer to nucleotide-level resolution of binding sites [36], with the caveat that 4-thiouridine at high concentrations was recently shown to inhibit rRNA synthesis and induce a nucleolar stress response [37]. Leveraging the observation that reverse transcription of isolated RNA terminates at the cross-linked nucleotide, iCLIP [38] and cross-link-induced mutations sites (CIMS) [18] (Fig. 1A) pinpoint the exact site of protein-RNA interaction. CLIP data can also be analyzed in conjunction with in-vitro techniques such as RNA SELEX [39], SEQRS [40], RNAcompete [41, 42], RNA Bind-n-Seq [43], or interactome profiling [44].
A disadvantage of the CLIP methods is their inability to identify a binding site composed of multiple motifs that are distal in the RNA primary sequence, but form a single binding site through secondary structure formation, such as a stem-loop. Although RBPs often have primary sequence specificities, it is likely that they recognize these sequences in the context of a particular RNA structure. CLIP methods are limited by their reliance on RNA-protein crosslink formation, whose efficiency is highly dependent on the molecular geometry of the RNA-protein interface [45]. Alternative techniques have been developed, such as protein interaction profile sequencing (PIP-Seq) [46] (Fig. 1A), which utilizes single and double strand-specific RNases, together with or without proteinase to uncover how RNA structure influences RBP binding. Computational approaches may also aid in the prediction of the RNA structure at RBP binding sites, as was done with the amyotrophic lateral sclerosis (ALS)-associated FUS/TLS [47] and Lin28 [48, 49].
CLIP-Seq has also provided valuable insights into the roles of RBPs in neural development and disease. Genome-wide identification of RBP binding sites was achieved for the neuron-specific Nova proteins involved in paraneoplastic opsoclonus-myoclonus ataxia (POMA) [20]. Thousands of binding sites and functional rules underlying Nova regulation of splicing were identified by correlating binding sites with splicing alterations induced by the absence of Nova. Similar efforts in cultured cells [23, 24, 47, 50–52] or mouse and human brains [31, 32, 53, 54] have demonstrated largely different binding patterns for TDP-43 and FUS/TLS, two RBPs linked to ALS/frontotemporal dementia (FTD). CLIP-Seq of the muscleblind proteins involved in myotonic dystrophy (DM) [22, 27, 55] identified mostly 3′UTR and intronic binding, supporting a role in regulation of splicing as well as sub-cellular localization and translation for a subset of targets. CLIP-Seq of the cytoplasmic polyribosome-associated FMRP protein involved in fragile X syndrome also revealed a distinct binding pattern through coding regions of its RNA targets, consistent with a role for FMRP in translational repression by promoting ribosome stalling on mRNAs [25]. Finally, CLIP-Seq has also associated novel RBPs involved in neurodegenerative disease, including a role for Park7 defects in early-onset Parkinson’s disease [56, 57]. Overall, CLIP-Seq combined with appropriate computational analyses has become a powerful tool for elucidating RBP functions and for providing significant insight into the mechanisms by which misregulation of these RBPs leads to neurodegenerative disease.
Exploring alternative splicing co- and post-transcriptionally
Pre-mRNA splicing, the process of intron removal and joining of exons, is tightly regulated by RBPs, several of which, including MBNL1/2, TDP-43, FUS/TLS, TAF15, EWS, hnRNPA1 and hnRNPA2/B1, have been implicated in neurodegenerative diseases such as myotonic dystrophy, multi-system proteinopathy and ALS. The potential impact of alternative splicing, the process whereby multiple isoforms are generated from the same genic locus [58, 59], is also increasingly recognized in neurodegenerative diseases including Alzheimer’s disease and Parkinson’s disease [60] (Fig. 2). In the last decade, microarray and sequencing studies have revealed that >90% of human genes undergo alternative splicing. Expectedly, disrupting the function of a single RBP often results in a dramatic effect on transcript diversity [20, 30–32]. To identify alternatively spliced exons, multiple high-throughput techniques have been developed (Fig. 1B). An early technique utilized microarrays that interrogate exon-exon junctions [61, 62]. However, this method is limited by the number of probes that can fit on an array, and only examines already annotated events. As an alternative approach, both novel and annotated splicing events can be identified by standard RNA-Seq through the analysis of exon-junction reads [59, 63–65], but sensitivity is highly dependent on sequencing depth, i.e. the number of reads that map to a particular genic region. Because sequencing depth is proportional to cost, this approach is not cost-effective but allows for novel isoform discovery. Nevertheless, in the case of lowly expressed alternative splicing events, microarrays can outperform RNA-Seq. Another sequencing-based, targeted approach to measuring alternatively spliced events is RNA-mediated oligonucleotide Annealing, Selection, and Ligation with Next-Gen sequencing (RASL-Seq) [66, 67]. Here, for each exon-exon junction a pair of DNA oligonucleotides is designed to hybridize immediately upstream and downstream of the junction. Following annealing, mRNA is captured on a poly(A)-selective solid support and unbound oligonucleotide probes removed. Treatment with ligase will join the pair of DNA probes to form a PCR-amplifiable product only in the presence of the exon-exon junction. Using pools of DNA probe pairs during ligation and barcoded (sequence-indexed) primers during PCR allows for the simultaneous interrogation of several hundred to thousands of specific splicing events from a large number of samples in a single next-generation sequencing run, thus making this method amenable for large-scale drug screening [67].
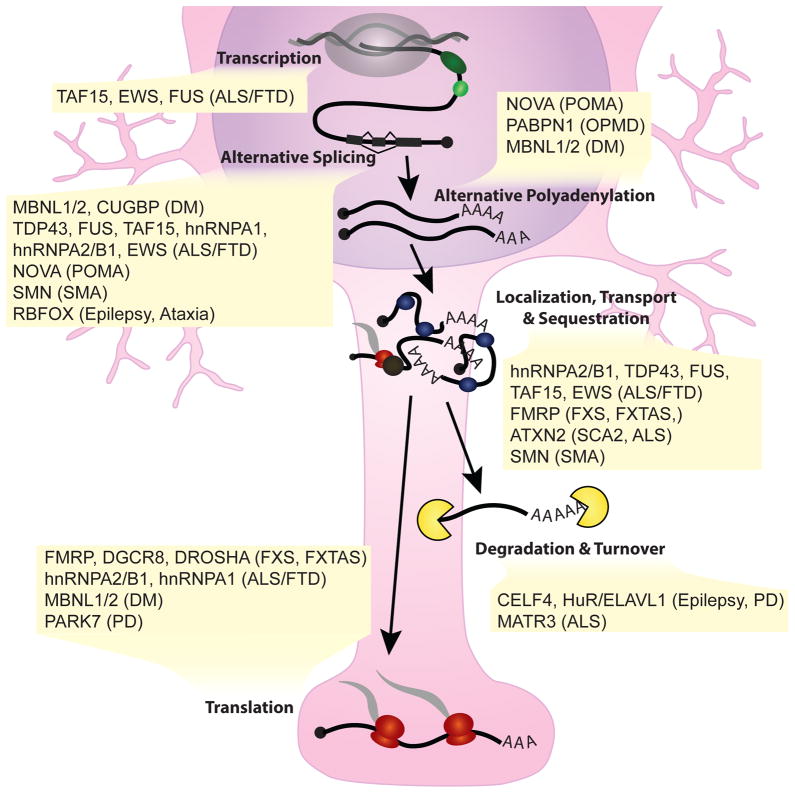
Figure 2. RBPs are implicated in several neurological diseases.
RBPs implicated in neurological diseases play a role in several steps of RNA processing, including transcription, alternative splicing, alternative polyadenylation, localization of transcripts including sequestration into inclusions and stress granules, translation, RNA degradation and turnover. The endogenous function of these RBPs often lends significant insight into the mechanism of pathology. ALS, amyotrophic lateral sclerosis; FTD, frontotemporal dementia; DM, myotonic dystrophy; POMA, paraneoplastic opsoclonus-myoclonus ataxia; SMA, spinal muscular atrophy; FXTAS, fragile X-associated tremor/ataxia syndrome; FXS, fragile X syndrome; PD, Parkinson’s disease; OPMD, oculopharyngeal muscular dystrophy; SCA2, spinocerebellar ataxia type 2.
Splicing of many human genes has previously been shown to be co-transcriptional [68, 69]. Intriguingly, a class of RBPs known as the FET family (consisting of FUS/TLS, EWS and TAF15) has been associated with ALS/FTD and proposed to affect both transcription elongation as well as splicing [23, 31, 53, 70–72] (Fig. 2). To assess co-transcriptional splicing, several methods have been developed. Nascent-Seq (Fig. 1C) utilizes sub-cellular fractionation to isolate nascent transcripts for sequencing [73], enabling the genome-wide study of co-transcriptional regulation mechanisms. Several variations of this technique have emerged (Fig. 1C) including Global Run-On sequencing (GRO-Seq), where transcription initiation and elongation are halted and restarted in the presence of the nucleotide analog 5-bromouridine 5′-triphosphate (BrUTP). Nascent RNA is then isolated by BrUTP immunoprecipitation and sequenced [74], enabling identification of actively transcribed regions, but requiring re-initiation of transcription elongation under artificial conditions. In contrast, Native Elongating Transcript sequencing (NET-Seq) identifies nascent transcripts under physiological conditions [75] via immunoprecipitation of the pol II complex and associated RNAs without crosslinking (Fig. 1C). GRO-Seq and NET-Seq can be directly applied to study in vivo nascent RNA populations. DM, ALS/FTD, spinal muscular atrophy (SMA) and POMA are all conditions accompanied by splicing alterations due to the disruption of different RBPs, yet it is not clear if these RBP-regulated splicing events occur co- or post-transcriptionally. Enlisting these techniques to quantify splicing changes in healthy and disease patient tissue as well as cellular or animal models of disease may uncover interplays between elongation and disease-linked splicing alterations and provide insight into subtle differences between the effects of various mutants that might correlate with therapeutic sensitivity.
Identifying alternative polyadenylation by PolyA sequencing
The selection of the 3′ end cleavage site within pre-mRNA transcripts followed by addition of a polyA tail is mediated by coordinated roles of RBPs. Defects in polyadenylation and tail-length undoubtedly result in aberrant gene expression and neuronal dysfunction; for example, dysfunction of PABPN1, a protein involved in polymerization of the poly(A) tail, is implicated in Oculopharyngeal Muscular Dystrophy (OPMD) (Fig. 2).
Recently, sequencing-based methods have been developed to analyze alternative 3′ cleavage and polyadenylation (Fig. 1D). Most of the early methods entail isolation of poly(A)+ RNA and oligo(d)T-primed cDNA synthesis. In PolyA-Seq [76], first strand synthesis (FSS) is oligo(d)T-primed and second strand synthesis is primed with a randomer. In poly(A) site sequencing (PAS-Seq) [77], FSS is carried out with a (d)T primer followed by ligation of adapters to both cDNA ends. Finally, in 3Seq [78], FSS is primed with an oligo(d)T containing a 3′ adapter. Internal priming is a major concern in these techniques, as sufficient sequence upstream of the poly(A) site must be identified to obtain mappable reads. An additional problem with (d)T priming is the propensity for polymerase slippage due to the repetitive nature of the poly(A) tail. To overcome this, a method called 3P-Seq [79] that experimentally defines 3′ ends independently of isolation of mRNA through poly(A)-stretches has also been used. 3P-Seq utilizes a splint ligation that attaches an adapter to the poly(A) tail as well as a poly(T)-adapter to the mRNA, thus avoiding poly(T) priming and simultaneously adding a biotin moiety to facilitate purification. Reverse transcription creates a stretch of (d)T complementary to the poly(A) tail that is partially digested with RNase H, leaving the sequence immediately adjacent to the site of polyadenylation available for sequencing. Notably, the quantitative ability of direct sequencing is not clear, since 3P-Seq requires multiple enzymatic steps and adapter ligations, increasing chances of ligation-induced biases [80]. Another technique, termed 3′T-Fill [81], incorporates a ‘filling-in’ step of the poly(A) tail with (d)T after libraries are clustered on the flow cell of the sequencing instrument, then starts sequencing at the site of polyadenylation.
Since alternative polyadenylation is important for RNA localization, degradation and translation, poly(A) sequencing has an important role in the study of neuronal function and neurological diseases. Variations on 3Seq and PAS-Seq were recently utilized to identify novel roles of PABPN1 in alternative polyadenylation. Loss of PABPN1 function in an OPMD mouse model, and cells expressing mutant trePABPN1, showed a widespread shift towards utilization of proximal polyadenylation sites resulting in a shorter 3′UTR [82, 83]. Similarly, a role in polyadenylation was recently uncovered for MBNL proteins associated with DM [55]. Although it is not yet determined whether alternative splicing and alternative polyadenylation are coordinately regulated, there is increasing evidence that multiple levels of RNA processing are misregulated in neurological diseases.
In addition to alternative poly(A) site usage, poly(A) length is another relatively unexplored feature of mRNAs that may play a role in neurodegeneration, particularly in the case of PABPN1. Two techniques have recently been developed to determine poly(A) length: TAIL-Seq [84] and Poly(A)-tail length profiling by sequencing (PAL-seq) [85] (Fig. 1D). TAIL-Seq involves the ligation of a biotin-containing adapter to the 3′ end of mRNAs, followed by a partial digestion by RNase T1, which cleaves downstream of guanosine, thus protecting the poly(A) tail. The biotinylated RNA is streptavidin purified, 5′ phosphorylated, gel purified, ligated to a 5′ adapter, reverse-transcribed, amplified and paired-end sequenced. The first read uncovers the identity of the mRNA and the second determines the poly(A) length. However, one of the major obstacles to determining poly(A) length by sequencing is the residual fluorescent ‘T’ signal on the flow cell from sequencing long tracts of ‘T’s, which can drown out the signal of a non-T base and results in overestimating poly(A) tail length. To overcome this, TAIL-Seq incorporates an additional fluorescence re-analysis to determine the actual template base, and thus significantly reduces overestimation of poly(A) length [84]. PAL-Seq, on the other hand, uses stochastic incorporation of a biotinylated uridine base that, when bound to fluorescently tagged streptavidin, gives a fluorescence intensity proportional to poly(A) length, though not with the same resolution as TAIL-Seq [85]. In this method, total RNA is 3′ splint ligated to a biotinylated adapter, partially digested, size selected, and biotin purified. RNAs are ligated to a 5′ adapter, reverse transcribed, and size selected. Next, sequencing clusters are generated on an Illumina flow cell, but prior to sequencing, a primer is hybridized 3′ to the terminal A of the poly(A) sequence and then extended into the poly(A) tail with dTTPs and biotin-conjugated dUTPs. After sequencing into the poly(A) proximal region for mapping purposes, the flow cell is incubated with fluorophore-conjugated streptavidin, which binds the biotin and generates fluorescence proportional in intensity (based on a standard curve generated with synthetic poly(A) tails of known length) to poly(A) length. Performing TAIL-Seq and PAL-Seq on OPMD patient tissues will likely reveal differential poly(A) lengths in mRNA substrates, which may be key to uncovering PABPN1’s role in disease pathogenesis.
Exploring translation
RBPs involved in translational control of mRNA have been associated with neurodegenerative diseases such as fragile X syndrome (FXS) and fragile X-associated tremor/ataxia syndrome (FXTAS) (Fig. 2). Sequencing and microarray analyses have been used to study translational control at the genome-wide level [12] (Fig. 1E). A subset of these techniques utilizes sequencing of ribosome-associated mRNA populations, an approach termed ‘polysome profiling’, which uses the degree of ribosome occupancy of an mRNA as a measure of its translational efficiency. This approach entails isolation, purification, and sequencing of polysome-associated mRNA by sucrose gradient density centrifugation, and has been utilized to elucidate mechanisms of disease caused by FMRP in FXS [86]. Polysome profiling can be used for a variety of cell types but does not allow isolation of cell-type specific polysomes in complex tissues such as brain. Translating ribosome affinity purification (TRAP) utilizes a line of Bacterial Artificial Chromosome (bacTRAP) transgenic mice with lineage specific promoters driving expression of EGFP-tagged ribosomal protein L10 [87, 88], allowing for isolation of polysome-associated RNA from specific cell types by EGFP immunoaffinity purification. Similarly, Ribotag mice harbor a loxP-flanked version of the last exon of the ribosomal protein L22 gene (Rpl22) upstream of a hemagglutinin (HA)-tagged version of that same exon at the endogenous locus. Crossing Ribotag mice with lineage-specific Cre recombinase driver lines results in genomic deletion of the untagged exon and usage of the tagged exon, resulting in expression of HA-tagged L22 protein in the desired cell types [89]. Thus, this approach avoids a separate bacTRAP line for each cell type and employs the endogenous promoter to ensure near physiological levels of the tagged protein.
A similar technique termed ‘ribosome profiling’ yields precise positions of translating ribosomes on mRNAs in a transcriptome-wide manner and thus allows deeper insights into gene-dependent dynamics of translational control [90, 91]. In a manner reminiscent of CLIP, ribosome profiling involves treatment of cells with cycloheximide to prevent ribosome disassembly, followed by limited micrococcal nuclease mediated digestion of RNA. The ribosome-protected mRNA fragments are isolated, sequenced and mapped onto the genome. It will be informative to analyze the translational state in systems that allow recently discovered repeat associated non-ATG translation (RAN) translation, which appears to be a common theme in microsatellite-associated neurological disorders [92, 93].
Mechanisms and alterations of mRNA turnover in neurological diseases
The last phase of RNA processing is degradation, which can occur before or after the processing steps discussed above. RNA degradation is regulated by RBPs, some of which have been implicated in epilepsy and Parkinson’s disease including CELF4 [94] and Hu/ELAV [26] while others have been associated with ALS, including MATR3 [95] (Fig. 2). The current state-of-the-art in quantifying mRNA turnover rates is by metabolic labeling (Fig. 1F). Cells are provided synthetic nucleoside analogs containing a reactive functional group (typically 4-thiouridine (4sU) [96] or 5-ethynyl uridine (EU) [97]), which are incorporated into nascent transcripts by the endogenous transcription machinery. Total RNA is then isolated and the labeled fraction biotinylated using label-specific chemistry. Using a pulse-chase technique, labeled RNA can be sequenced at various time points during the chase to quantify degradation rates of particular RNAs via RNA-Seq or quantitative real-time PCR (qPCR). Labeled RNA can also be quantified during the pulse to determine synthesis rates. An analogous method, 5′-bromo-uridine (BrU) immunoprecipitation chase-deep sequencing analysis (BRIC-Seq) (Fig. 1F), utilizes incorporation of BrU and immunoprecipitation of BrU-containing RNAs [98]. However, the benefit of the 4sU and EU methods is the strength of the streptavidin-biotin interaction, which allows for stringent washing and cDNA synthesis on a solid support, eliminating an elution and precipitation step that can significantly reduce yields for downstream qPCR analyses. While CLIP has facilitated study of decay-associated RBPs involved in neurodegeneration, such as CELF4 [94] and ELAV [26], techniques that globally assay mRNA turnover could provide valuable insight into mechanisms of disease with pathological RNAs, such as those containing repeat expansions.
Secondary structure, post-transcriptional modifications and editing of RNAs modulate RBP binding
Base-pairing, hairpins, bulges, multi-loops, and unstructured regions in RNA play a significant role in the processing of transcripts, in part by mediating RBP binding and function. As mentioned above, current CLIP-Seq protocols do not readily identify structural preferences for RBP binding; however, several high-throughput methods have been developed to literally untangle the structure of RNA and the structural parameters of RBP targets (Fig. 1G). In selective 2′-hydroxyl acylation analyzed by primer extension (SHAPE)-Seq [99], single-stranded loops and bulges are selectively acetylated, halting cDNA synthesis and generating sequencing libraries of various length corresponding to the location of single-stranded secondary structure at single-nucleotide resolution. To identify regions of secondary structure, fragmentation sequencing (FragSeq) utilizes a single-strand specific nuclease, leaving RNA with secondary structure for sequencing [100]. In addition, parallel analysis of RNA structure (PARS)-Seq compares sequencing libraries generated from treating mRNA with either a single strand or double strand specific nuclease to define secondary structure [101]. While these techniques address RNA secondary structure at a genome-wide level, they do not address RBP specificity for a particular structural motif. PIP-Seq may be able to address the RNA structure parameters necessary for RBP binding. In addition to technical advances, there are several computational tools for predicting structures within identified RBP binding sites. RBPmotif is a web server that utilizes such tools to predict secondary structure, including base-pairing, hairpin loops, bulge loops, multi-loops, and unstructured regions [102]. No genome-wide studies have been done to examine the role of RNA structure in neurodegenerative disease; however, PIP-Seq and similar techniques could further elucidate the mechanism of pathology by identifying potentially altered structural requirements of RBP mutants in neurodegenerative disorders, or possibly a role of RNA structure in RBP sequestration and repeat expansion.
RNA also undergoes extensive post-transcriptional modifications, including methylation of the cap structure and the nucleobases of mRNA. Recent approaches have been developed to identify such marks on RNA (Fig. 1G). For example, m6A-Seq [103] also called MeRIP-Seq [104] generate low-resolution genome-wide maps of N6-methyladenosine (m6A), a modification recently shown to play a role in transcript stability [105, 106]. After RNA is fragmented, an antibody against m6A selects for m6A-containing fragments for sequencing. These techniques have not yet been directly applied to the study of neurodegenerative diseases, but given the recently identified role of m6A in transcript processing, doing so could reveal another layer of RNA-mediated pathology.
Finally, RNA is modified at the level of primary structure during a process called RNA editing, in which RNA deaminases convert adenosine to inosine (which is read as guanosine during reverse transcription) or cytosine to uracil [107]. Normal editing of GluR2 was found significantly impaired in motor neurons from ALS patients, which is a potential cause of excessive calcium influx that may contribute to motor neuron death [108–112]. In addition, abnormal editing of the glutamate transporter EAAT2 was identified in ALS patients [113], and ADAR2 was recently proposed to bind expanded RNAs in C9ORF72 ALS/FTD patients [114]. Finally, hnRNPA2/B1 recently linked to ALS/FTD [115] has been proposed as an enhancer of RNA editing [116], further emphasizing the potential role of editing in ALS/FTD. Single end (SE) or paired end (PE) RNA-Seq often proves sufficient to detect sequence changes due to RNA editing. So-called padlock probes can be used to interrogate candidate editing events by detecting single base changes at tens of thousands of specific positions [117] (Fig. 1G). The assay consists of an oligonucleotide designed such that its ends hybridize to isolated RNA to form a nearly circular structure at the target site except for a small gap. This gap is filled enzymatically and resulting sequences are compared between genomic DNA and cDNA, a base change being indicative of an editing event.
Perspectives and Summary
Neurological diseases are increasingly recognized as associated with RNA regulatory dysfunction caused by reduced or aberrant RBP activity. Comprehensive analysis of the functions of these RBPs and identification of their target RNA regulatory networks are necessary to keep pace with the accelerated rate of their discovery in genetic diseases, and to enhance the development of therapeutics. These analyses have been aided by the development of new genome-wide, high-throughput techniques to resolve the role of RBPs in RNA processing as depicted in Figure 1.
Although genome-wide technologies have greatly enhanced our ability to study complex neurological diseases, several major challenges still exist (see Outstanding Questions). One of the greatest hurdles to the study of disease, and cell biology in general, is the identification and isolation of rare cells that exhibit the phenotype of interest. Recent advances in single-cell technology are beginning to address these deficits and resolve cellular heterogeneity (Fig. 3). The ability to identify phenotypes with single cell resolution is particularly relevant for neurological studies, as the brain is naturally highly heterogeneous. Several neurological diseases including DM, C9ORF72 ALS/FTD and FXTAS are characterized by nuclear RNA foci, which occur in only a subset of cells. Thus, when a bulk sample is studied, this small but critical subpopulation may be missed. In addition, repeat expansions can result in somatic mosaicism, and the gradient of phenotypes is lost in the average [118]. For example, repeat expansions as seen in ALS often differ in size between cell types, and this variability can account for variable pathology [119]. Furthermore, in diseases such as ALS/FTD that present with protein inclusions due to mutation or mislocalization of aggregation-prone RBPs, only a small percentage of cells possess the pathological hallmarks. Application of single-cell technology is critical for overcoming issues of bulk sample heterogeneity and identifying such rare and variable cell types. There have been many developments in the single-cell field, which are discussed in the review by Shapiro et al. [120]. It would be ideal to combine single-cell capture with the genome-wide techniques described here, but currently most of these methods require more material than can be obtained from single cells.
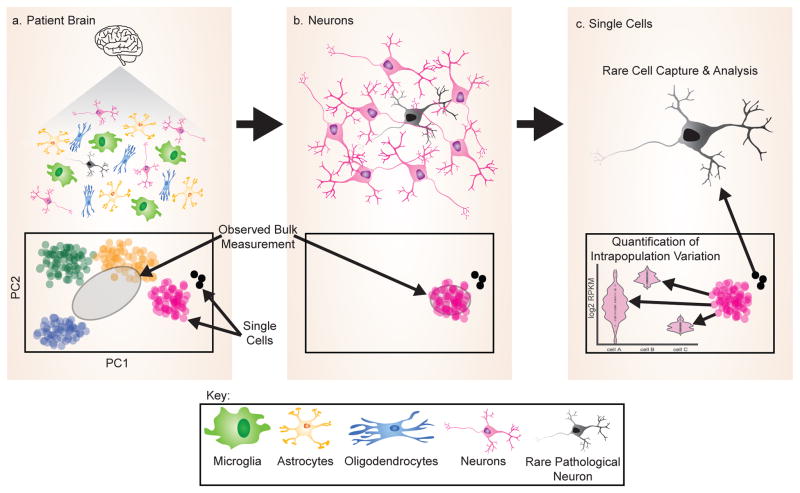
Figure 3. Single cell analysis identifies rare cell types and intra-population variation.
a | Analysis of bulk cell populations, such as brain tissue, prevents the identification of rare cell types, such as neurons with RBP inclusions, in addition to reducing evidence of phenotypic gradients by generating an average.
b | Isolation of a cell type, such as the neuron, may also fail to enable identification of a rare cell.
c | With the isolation and analysis of single cell expression profiles it becomes possible to not only identify rare cell types that differ from the bulk population, but also to identify variation within a bulk population, such as differences in splicing.
As the list of proteins and pathways linked to each neurodegenerative disease grows, directed approaches that focus on individual players are no longer sufficient to fully define the mechanisms of pathology. Rather, it is becoming necessary to adjust experimental approaches to identify characteristic global signatures of neurodegenerative diseases. To accomplish this, genome-wide approaches can identify comprehensive molecular snapshots, which can then be used to generate more accurate experimental models of disease, enhance diagnostic accuracy and identify additional therapeutic targets. By using these techniques to observe the entire molecular “forest” characteristic of the disease state, model systems can be developed to recapitulate the unique disease signature rather than modifying a single target that may not encompass the entire pathology. In addition to the novel and unique insights provided by genome-wide techniques, these methods also produce their share of challenges. Because such a large number of targets and players are identified, there is the inherent problem of prioritizing which candidates to pursue. One aspect of this is determining the rate of false positives and false negatives, some of which can be addressed by utilizing appropriate controls. A second component of prioritization is to determine which targets are causative to the phenotype and which are merely bystanders, changing as the result of a change in a common upstream regulator. A third consideration is the emerging roles of non-coding RNAs that are often overlooked with current methodologies or computational analyses. Indeed, several (l)ncRNAs have been implicated in neurodegenerative disease, as reviewed in [121, 122]. In addition to addressing this issue from a biological standpoint, there is also the issue of sheer computational power needed to wade through this ocean of data. This can be facilitated by the development of robust and efficient computational pipelines, but the ease with which such large datasets can be generated in today’s age of benchtop sequencers requires further consideration to avoid a data processing and analysis bottleneck. Finally, and perhaps most significantly, high-throughput genome-wide analyses are proving that RBPs play a significant role in the pathology of neurodegenerative diseases. As master regulators of several aspect of cellular function, it may prove necessary to target the availability of the normal versus mutant RBP, rather than its downstream targets and effectors. Some therapeutics are being developed to address this [114, 123], including antisense oligonucleotides that directly block RBPs or indirectly block RBP binding by either altering RNA secondary structure or causing RNA degradation.
As these genome-wide techniques become more widely utilized, their application to the complex involvement of RBPs in neurological disease will bring much needed clarity to understanding, modeling and ultimately curing these devastating pathologies.
Outstanding Questions
What is the role of RNA secondary structure, editing, and modification in the pathology of neurodegenerative diseases? Changes in RNA primary and secondary structure have yet to be thoroughly studied in the context of neurodegeneration. Of particular interest given the availability of new reagents and protocols are internal methylation and RNA secondary structure. With the recent evidence for a role of m6A in transcript stability, and potential roles in RNA localization and translation, applying m6A-seq/MeRIP-Seq to a neurodegenerative model could identify misregulation of these processes through changes in RBP binding. Furthermore, for repeat expansion diseases, determining how secondary structure arising from long tracts of repeats influences RBP-mediated pathology is essential for understanding RBP sequestration and/or formation of foci.
How can we address the issue of heterogeneity in tissue and cellular models of neurodegeneration? One of the greatest hurdles to understanding the mechanisms of RBP-mediated neuropathology is the complexity of brain tissue and populations of iPSC/ESC-derived neural cells. Advances in single cell isolation platforms, like the Fluidigm C1 Auto-Prep System or other single-cell capture devices, are aiding in overcoming this hurdle, but there needs to be improvements in sequencing techniques to allow for a smaller amount of starting material. Techniques like CLIP currently require millions of cells.
What will application of these techniques reveal about the mechanisms of neuropathology? Few of the reviewed techniques aside from CLIP and its variations have been applied to the study of RBPs in the context of neuropathology, but doing so could reveal novel mechanisms of disease as well as potential therapeutic targets. As discussed in the review, several steps of RNA processing are misregulated in neurodegeneration, but the mechanisms remain elusive.
How can we study the RBPs and RNA transcripts found in foci and stress granules characteristic of several neurodegenerative diseases? There is a growing need for a high-throughput technique for the study of foci and stress granules, and the associated RNA and RBPs, implicated in neurodegeneration. The inability to isolate these complexes has thus far prevented such analyses, but once an efficient method has been developed for their isolation and purification, the associated RNA can be subjected to several of the reviewed techniques in order to better understand how stress granules form and the mechanism by which they contribute to disease pathology.
Highlights.
RNA binding proteins are implicated in several neurodegenerative diseases
A thorough understanding of the role of RBPs in neurodegeneration is lacking
High-throughput techniques reveal novel mechanisms of RBP-mediated neuropathology
Applying high-throughput techniques will provide novel therapeutic targets
Tissue heterogeneity remains an obstacle to understanding neuronal diseases
Acknowledgments
Funding Sources: This work is supported by grants to G.W.Y. from the National Institute of Health (HG004659, NS075449 and U54HG007005) and California Institute for Regenerative Medicine (RB4-06045 and TR3-05676). G.W.Y. is an Alfred P. Sloan Research Fellow. R.B. is a Myotonic Dystrophy Foundation Fellow. C.L.T. is supported by grants from the National Institute of Health (R01NS087227 and P50AG005131), Target ALS (04827 and 0840), the Frick Foundation and the Amyotrophic Lateral Sclerosis Association (grant 2235A). C.L.T. was a recipient of a career Development Award from the Muscular Dystrophy Association and receives salary support from the Ludwig Institute for Cancer Research.
Glossary
- RBP
RNA binding protein; a protein that interacts with RNA to affect downstream function or processing
- Seq
High-throughput sequencing
- RNA Element
Sequence of RNA that is often conserved and has a particular function, for example as a binding site for an RNA binding protein
- Crosslink
Formation of a covalent bond between two entities. In the context of this review, crosslinking refers to the formation of covalent bonds between protein and nucleic acid that are within close physical proximity (within a few angstroms). They can be chemical and reversible (formaldehyde), or photochemical and irreversible (ultraviolet light)
- UTR
Untranslated region; the regions at the 5′ and 3′ ends of transcripts that do not encode protein, but often harbor cis-regulatory elements that are bound by protein
- Randomer
For a defined length of nucleic acid, the set of oligomers with all possible sequences
- Sequencing Adapter
Defined nucleic acid sequences ligated to the end of the nucleic acid fragments of interest before sequencing; allows for hybridization to a sequencing flow cell as well as recognition by the sequencing primer
- Library
The pooled sample of fragmented nucleic acids possessing the necessary adapters for high-throughput sequencing
- Read-mapping
The process of aligning short sequencing reads to a reference genome or sequence
- Splicing
RNA splicing is the process of excising non protein-coding regions of pre-mRNA, called introns, and the joining of exons. The preferential inclusion or exclusion of an exon is termed alternative splicing and contributes significantly to the diversity of the proteome
- Polyadenylation
The process of adding multiple adenosine residues to the 3′ end of transcripts. The poly(A) tail is necessary for nuclear export as well as protecting the 3′ end of the transcript from exonuclease degradation. Poly(A) sites can be located within introns, exons or the 3′UTR of a transcript; however, poly(A) sites in the 3′UTR are more commonly utilized in vivo. Alternative polyadenylation is a common phenomenon in which one of many potential poly(A) sites available is favored. Use of a poly(A) site depends on the core 3′ processing machinery, the strength of cis-elements, transcription dynamics, and other auxiliary factors [1]
- RNA Turnover
The process of RNA degradation. There are several known mechanisms, all of which involve the recruitment and function of several RBPs [2]. The majority of RNAs are degraded in a deadenylation-independent manner, in which the poly(A) tail is shortened, followed by removal of the 5′ cap enabling exonuclease degradation of the RNA. Transcripts can also be targeted for degradation without deadenylation or decapping via microRNA-mediated recruitment of the RISC complex. Another deadenylation independent mechanism of RNA turnover is nonsense-mediated decay (NMD), where the interaction of RBPs Upf1, Upf3 and Nmd2 with mRNAs that contain premature stop codons results in decapping and degradation by exonucleases [3]. RNAs lacking a stop codon are also rapidly deadenylated and subjected to decapping and exonuclease degradation in a pathway known as non-stop decay [4, 5]
- RNA Splint Ligation
The ligation of two RNA molecules brought together via binding of a third bridging oligonucleotide complementary to the two RNA molecules
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Elkon R, Ugalde AP, Agami R. Alternative cleavage and polyadenylation: extent, regulation and function. Nat Rev Genet. 2013;14(7):496–506. doi: 10.1038/nrg3482. [DOI] [PubMed] [Google Scholar]
- 2.Balagopal V, Fluch L, Nissan T. Ways and means of eukaryotic mRNA decay. Biochim Biophys Acta. 2012;1819(6):593–603. doi: 10.1016/j.bbagrm.2012.01.001. [DOI] [PubMed] [Google Scholar]
- 3.He F, Jacobson A. Upf1p, Nmd2p, and Upf3p regulate the decapping and exonucleolytic degradation of both nonsense-containing mRNAs and wild-type mRNAs. Mol Cell Biol. 2001;21(5):1515–30. doi: 10.1128/MCB.21.5.1515-1530.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Frischmeyer PA, et al. An mRNA surveillance mechanism that eliminates transcripts lacking termination codons. Science. 2002;295(5563):2258–61. doi: 10.1126/science.1067338. [DOI] [PubMed] [Google Scholar]
- 5.van Hoof A, et al. Exosome-mediated recognition and degradation of mRNAs lacking a termination codon. Science. 2002;295(5563):2262–4. doi: 10.1126/science.1067272. [DOI] [PubMed] [Google Scholar]
- 6.Lukong KE, et al. RNA-binding proteins in human genetic disease. Trends Genet. 2008;24(8):416–25. doi: 10.1016/j.tig.2008.05.004. [DOI] [PubMed] [Google Scholar]
- 7.Cooper TA, Wan L, Dreyfuss G. RNA and disease. Cell. 2009;136(4):777–93. doi: 10.1016/j.cell.2009.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.McKee AE, et al. A genome-wide in situ hybridization map of RNA-binding proteins reveals anatomically restricted expression in the developing mouse brain. BMC Dev Biol. 2005;5:14. doi: 10.1186/1471-213X-5-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yeo G, et al. Variation in alternative splicing across human tissues. Genome Biol. 2004;5(10):R74. doi: 10.1186/gb-2004-5-10-r74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Castle JC, et al. Expression of 24,426 human alternative splicing events and predicted cis regulation in 48 tissues and cell lines. Nat Genet. 2008;40(12):1416–25. doi: 10.1038/ng.264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Holt CE, Bullock SL. Subcellular mRNA localization in animal cells and why it matters. Science. 2009;326(5957):1212–6. doi: 10.1126/science.1176488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kapeli K, Yeo GW. Genome-wide approaches to dissect the roles of RNA binding proteins in translational control: implications for neurological diseases. Front Neurosci. 2012;6:144. doi: 10.3389/fnins.2012.00144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Darnell JC, Klann E. The translation of translational control by FMRP: therapeutic targets for FXS. Nat Neurosci. 2013 doi: 10.1038/nn.3379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Alami NH, et al. Axonal transport of TDP-43 mRNA granules is impaired by ALS-causing mutations. Neuron. 2014;81(3):536–43. doi: 10.1016/j.neuron.2013.12.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Baltz AG, et al. The mRNA-bound proteome and its global occupancy profile on protein-coding transcripts. Mol Cell. 2012;46(5):674–90. doi: 10.1016/j.molcel.2012.05.021. [DOI] [PubMed] [Google Scholar]
- 16.Zhao J, et al. Genome-wide identification of polycomb-associated RNAs by RIP-seq. Mol Cell. 2010;40(6):939–53. doi: 10.1016/j.molcel.2010.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mili S, Steitz JA. Evidence for reassociation of RNA-binding proteins after cell lysis: implications for the interpretation of immunoprecipitation analyses. RNA. 2004;10 (11):1692–4. doi: 10.1261/rna.7151404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Moore MJ, et al. Mapping Argonaute and conventional RNA-binding protein interactions with RNA at single-nucleotide resolution using HITS-CLIP and CIMS analysis. Nat Protoc. 2014;9(2):263–93. doi: 10.1038/nprot.2014.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ule J, et al. CLIP identifies Nova-regulated RNA networks in the brain. Science. 2003;302(5648):1212–5. doi: 10.1126/science.1090095. [DOI] [PubMed] [Google Scholar]
- 20.Licatalosi DD, et al. HITS-CLIP yields genome-wide insights into brain alternative RNA processing. Nature. 2008;456(7221):464–9. doi: 10.1038/nature07488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Darnell RB. RNA protein interaction in neurons. Annu Rev Neurosci. 2013;36:243–70. doi: 10.1146/annurev-neuro-062912-114322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wang ET, et al. Transcriptome-wide regulation of pre-mRNA splicing and mRNA localization by muscleblind proteins. Cell. 2012;150(4):710–24. doi: 10.1016/j.cell.2012.06.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nakaya T, et al. FUS regulates genes coding for RNA-binding proteins in neurons by binding to their highly conserved introns. RNA. 2013;19(4):498–509. doi: 10.1261/rna.037804.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ishigaki S, et al. Position-dependent FUS-RNA interactions regulate alternative splicing events and transcriptions. Sci Rep. 2012;2:529. doi: 10.1038/srep00529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Darnell JC, et al. FMRP stalls ribosomal translocation on mRNAs linked to synaptic function and autism. Cell. 2011;146(2):247–61. doi: 10.1016/j.cell.2011.06.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ince-Dunn G, et al. Neuronal Elav-like (Hu) proteins regulate RNA splicing and abundance to control glutamate levels and neuronal excitability. Neuron. 2012;75 (6):1067–80. doi: 10.1016/j.neuron.2012.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Charizanis K, et al. Muscleblind-like 2-mediated alternative splicing in the developing brain and dysregulation in myotonic dystrophy. Neuron. 2012;75(3):437–50. doi: 10.1016/j.neuron.2012.05.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Licatalosi DD, et al. Ptbp2 represses adult-specific splicing to regulate the generation of neuronal precursors in the embryonic brain. Genes Dev. 2012;26(14):1626–42. doi: 10.1101/gad.191338.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Weyn-Vanhentenryck SM, et al. HITS-CLIP and Integrative Modeling Define the Rbfox Splicing-Regulatory Network Linked to Brain Development and Autism. Cell Rep. 2014 doi: 10.1016/j.celrep.2014.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yeo GW, et al. An RNA code for the FOX2 splicing regulator revealed by mapping RNA-protein interactions in stem cells. Nat Struct Mol Biol. 2009;16(2):130–7. doi: 10.1038/nsmb.1545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lagier-Tourenne C, et al. Divergent roles of ALS-linked proteins FUS/TLS and TDP-43 intersect in processing long pre-mRNAs. Nat Neurosci. 2012;15(11):1488–97. doi: 10.1038/nn.3230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Polymenidou M, et al. Long pre-mRNA depletion and RNA missplicing contribute to neuronal vulnerability from loss of TDP-43. Nat Neurosci. 2011;14(4):459–68. doi: 10.1038/nn.2779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sanford JR, et al. Splicing factor SFRS1 recognizes a functionally diverse landscape of RNA transcripts. Genome Res. 2009;19(3):381–94. doi: 10.1101/gr.082503.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Modic M, Ule J, Sibley CR. CLIPing the brain: studies of protein-RNA interactions important for neurodegenerative disorders. Mol Cell Neurosci. 2013;56:429–35. doi: 10.1016/j.mcn.2013.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hafner M, et al. Transcriptome-wide identification of RNA-binding protein and microRNA target sites by PAR-CLIP. Cell. 2010;141(1):129–41. doi: 10.1016/j.cell.2010.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kishore S, et al. A quantitative analysis of CLIP methods for identifying binding sites of RNA-binding proteins. Nat Methods. 2011;8(7):559–64. doi: 10.1038/nmeth.1608. [DOI] [PubMed] [Google Scholar]
- 37.Burger K, et al. 4-thiouridine inhibits rRNA synthesis and causes a nucleolar stress response. RNA Biol. 2013;10(10) doi: 10.4161/rna.26214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Konig J, et al. iCLIP reveals the function of hnRNP particles in splicing at individual nucleotide resolution. Nat Struct Mol Biol. 2010;17(7):909–15. doi: 10.1038/nsmb.1838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ellington AD, Szostak JW. In vitro selection of RNA molecules that bind specific ligands. Nature. 1990;346(6287):818–22. doi: 10.1038/346818a0. [DOI] [PubMed] [Google Scholar]
- 40.Campbell ZT, et al. Cooperativity in RNA-protein interactions: global analysis of RNA binding specificity. Cell Rep. 2012;1(5):570–81. doi: 10.1016/j.celrep.2012.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ray D, et al. Rapid and systematic analysis of the RNA recognition specificities of RNA-binding proteins. Nat Biotechnol. 2009;27(7):667–70. doi: 10.1038/nbt.1550. [DOI] [PubMed] [Google Scholar]
- 42.Ray D, et al. A compendium of RNA-binding motifs for decoding gene regulation. Nature. 2013;499(7457):172–7. doi: 10.1038/nature12311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lambert N, et al. RNA Bind-n-Seq: quantitative assessment of the sequence and structural binding specificity of RNA binding proteins. Mol Cell. 2014;54(5):887–900. doi: 10.1016/j.molcel.2014.04.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Castello A, et al. System-wide identification of RNA-binding proteins by interactome capture. Nat Protoc. 2013;8(3):491–500. doi: 10.1038/nprot.2013.020. [DOI] [PubMed] [Google Scholar]
- 45.Meisenheimer KM, Koch TH. Photocross-linking of nucleic acids to associated proteins. Crit Rev Biochem Mol Biol. 1997;32(2):101–40. doi: 10.3109/10409239709108550. [DOI] [PubMed] [Google Scholar]
- 46.Silverman IM, et al. RNase-mediated protein footprint sequencing reveals protein-binding sites throughout the human transcriptome. Genome Biol. 2014;15(1):R3. doi: 10.1186/gb-2014-15-1-r3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hoell JI, et al. RNA targets of wild-type and mutant FET family proteins. Nat Struct Mol Biol. 2011;18(12):1428–31. doi: 10.1038/nsmb.2163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wilbert ML, et al. LIN28 binds messenger RNAs at GGAGA motifs and regulates splicing factor abundance. Mol Cell. 2012;48(2):195–206. doi: 10.1016/j.molcel.2012.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Cho J, et al. LIN28A is a suppressor of ER-associated translation in embryonic stem cells. Cell. 2012;151(4):765–77. doi: 10.1016/j.cell.2012.10.019. [DOI] [PubMed] [Google Scholar]
- 50.Sephton CF, et al. Identification of neuronal RNA targets of TDP-43-containing ribonucleoprotein complexes. J Biol Chem. 2011;286(2):1204–15. doi: 10.1074/jbc.M110.190884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Xiao S, et al. RNA targets of TDP-43 identified by UV-CLIP are deregulated in ALS. Mol Cell Neurosci. 2011;47(3):167–80. doi: 10.1016/j.mcn.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 52.Colombrita C, et al. TDP-43 and FUS RNA-binding proteins bind distinct sets of cytoplasmic messenger RNAs and differently regulate their post-transcriptional fate in motoneuron-like cells. J Biol Chem. 2012;287(19):15635–47. doi: 10.1074/jbc.M111.333450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Rogelj B, et al. Widespread binding of FUS along nascent RNA regulates alternative splicing in the brain. Sci Rep. 2012;2:603. doi: 10.1038/srep00603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Tollervey JR, et al. Characterizing the RNA targets and position-dependent splicing regulation by TDP-43. Nat Neurosci. 2011;14(4):452–8. doi: 10.1038/nn.2778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Batra R, et al. Loss of MBNL Leads to Disruption of Developmentally Regulated Alternative Polyadenylation in RNA-Mediated Disease. Mol Cell. 2014;56(2):311–322. doi: 10.1016/j.molcel.2014.08.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.van der Brug MP, et al. RNA binding activity of the recessive parkinsonism protein DJ-1 supports involvement in multiple cellular pathways. Proc Natl Acad Sci U S A. 2008;105(29):10244–9. doi: 10.1073/pnas.0708518105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Blackinton J, et al. Post-transcriptional regulation of mRNA associated with DJ-1 in sporadic Parkinson disease. Neurosci Lett. 2009;452(1):8–11. doi: 10.1016/j.neulet.2008.12.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Pan Q, et al. Deep surveying of alternative splicing complexity in the human transcriptome by high-throughput sequencing. Nat Genet. 2008;40(12):1413–5. doi: 10.1038/ng.259. [DOI] [PubMed] [Google Scholar]
- 59.Wang Z, Burge CB. Splicing regulation: from a parts list of regulatory elements to an integrated splicing code. RNA. 2008;14(5):802–13. doi: 10.1261/rna.876308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Mills JD, Janitz M. Alternative splicing of mRNA in the molecular pathology of neurodegenerative diseases. Neurobiol Aging. 2012;33(5):1012 e11–24. doi: 10.1016/j.neurobiolaging.2011.10.030. [DOI] [PubMed] [Google Scholar]
- 61.Johnson JM, et al. Genome-wide survey of human alternative pre-mRNA splicing with exon junction microarrays. Science. 2003;302(5653):2141–4. doi: 10.1126/science.1090100. [DOI] [PubMed] [Google Scholar]
- 62.Pan Q, et al. Revealing global regulatory features of mammalian alternative splicing using a quantitative microarray platform. Mol Cell. 2004;16(6):929–41. doi: 10.1016/j.molcel.2004.12.004. [DOI] [PubMed] [Google Scholar]
- 63.Katz Y, et al. Analysis and design of RNA sequencing experiments for identifying isoform regulation. Nat Methods. 2010;7(12):1009–15. doi: 10.1038/nmeth.1528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Li H, et al. Determination of tag density required for digital transcriptome analysis: application to an androgen-sensitive prostate cancer model. Proc Natl Acad Sci U S A. 2008;105(51):20179–84. doi: 10.1073/pnas.0807121105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Lovci MT, et al. Rbfox proteins regulate alternative mRNA splicing through evolutionarily conserved RNA bridges. Nat Struct Mol Biol. 2013;20(12):1434–42. doi: 10.1038/nsmb.2699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Zhou Z, et al. The Akt-SRPK-SR axis constitutes a major pathway in transducing EGF signaling to regulate alternative splicing in the nucleus. Mol Cell. 2012;47(3):422–33. doi: 10.1016/j.molcel.2012.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Li H, Qiu J, Fu XD. RASL-seq for massively parallel and quantitative analysis of gene expression. Curr Protoc Mol Biol. 2012;Chapter 4(Unit 4 13):1–9. doi: 10.1002/0471142727.mb0413s98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Wuarin J, Schibler U. Physical isolation of nascent RNA chains transcribed by RNA polymerase II: evidence for cotranscriptional splicing. Mol Cell Biol. 1994;14 (11):7219–25. doi: 10.1128/mcb.14.11.7219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Wetterberg I, et al. In situ transcription and splicing in the Balbiani ring 3 gene. EMBO J. 2001;20(10):2564–74. doi: 10.1093/emboj/20.10.2564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Zhou Y, et al. ALS-associated FUS mutations result in compromised FUS alternative splicing and autoregulation. PLoS Genet. 2013;9(10):e1003895. doi: 10.1371/journal.pgen.1003895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Bertolotti A, et al. EWS, but not EWS-FLI-1, is associated with both TFIID and RNA polymerase II: interactions between two members of the TET family, EWS and hTAFII68, and subunits of TFIID and RNA polymerase II complexes. Mol Cell Biol. 1998;18 (3):1489–97. doi: 10.1128/mcb.18.3.1489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Schwartz JC, et al. FUS binds the CTD of RNA polymerase II and regulates its phosphorylation at Ser2. Genes Dev. 2012;26(24):2690–5. doi: 10.1101/gad.204602.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Khodor YL, et al. Nascent-seq indicates widespread cotranscriptional pre-mRNA splicing in Drosophila. Genes Dev. 2011;25(23):2502–12. doi: 10.1101/gad.178962.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Core LJ, Waterfall JJ, Lis JT. Nascent RNA sequencing reveals widespread pausing and divergent initiation at human promoters. Science. 2008;322(5909):1845–8. doi: 10.1126/science.1162228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Churchman LS, Weissman JS. Nascent transcript sequencing visualizes transcription at nucleotide resolution. Nature. 2011;469(7330):368–73. doi: 10.1038/nature09652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Derti A, et al. A quantitative atlas of polyadenylation in five mammals. Genome Res. 2012;22(6):1173–83. doi: 10.1101/gr.132563.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Shepard PJ, et al. Complex and dynamic landscape of RNA polyadenylation revealed by PAS-Seq. RNA. 2011;17(4):761–72. doi: 10.1261/rna.2581711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Beck AH, et al. 3′-end sequencing for expression quantification (3SEQ) from archival tumor samples. PLoS One. 2010;5(1):e8768. doi: 10.1371/journal.pone.0008768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Jan CH, et al. Formation, regulation and evolution of Caenorhabditis elegans 3′UTRs. Nature. 2011;469(7328):97–101. doi: 10.1038/nature09616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Hafner M, et al. RNA-ligase-dependent biases in miRNA representation in deep-sequenced small RNA cDNA libraries. RNA. 2011;17(9):1697–712. doi: 10.1261/rna.2799511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Wilkening S, et al. An efficient method for genome-wide polyadenylation site mapping and RNA quantification. Nucleic Acids Res. 2013;41(5):e65. doi: 10.1093/nar/gks1249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Jenal M, et al. The poly(A)-binding protein nuclear 1 suppresses alternative cleavage and polyadenylation sites. Cell. 2012;149(3):538–53. doi: 10.1016/j.cell.2012.03.022. [DOI] [PubMed] [Google Scholar]
- 83.de Klerk E, et al. Poly(A) binding protein nuclear 1 levels affect alternative polyadenylation. Nucleic Acids Res. 2012;40(18):9089–101. doi: 10.1093/nar/gks655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Chang H, et al. TAIL-seq: Genome-wide Determination of Poly(A) Tail Length and 3′ End Modifications. Mol Cell. 2014;53(6):1044–52. doi: 10.1016/j.molcel.2014.02.007. [DOI] [PubMed] [Google Scholar]
- 85.Subtelny AO, et al. Poly(A)-tail profiling reveals an embryonic switch in translational control. Nature. 2014 doi: 10.1038/nature13007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Stefani G, et al. Fragile X mental retardation protein is associated with translating polyribosomes in neuronal cells. J Neurosci. 2004;24(33):7272–6. doi: 10.1523/JNEUROSCI.2306-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Heiman M, et al. A translational profiling approach for the molecular characterization of CNS cell types. Cell. 2008;135(4):738–48. doi: 10.1016/j.cell.2008.10.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Doyle JP, et al. Application of a translational profiling approach for the comparative analysis of CNS cell types. Cell. 2008;135(4):749–62. doi: 10.1016/j.cell.2008.10.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Sanz E, et al. Cell-type-specific isolation of ribosome-associated mRNA from complex tissues. Proc Natl Acad Sci U S A. 2009;106(33):13939–44. doi: 10.1073/pnas.0907143106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Ingolia NT, et al. Genome-wide analysis in vivo of translation with nucleotide resolution using ribosome profiling. Science. 2009;324(5924):218–23. doi: 10.1126/science.1168978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Ingolia NT, et al. The ribosome profiling strategy for monitoring translation in vivo by deep sequencing of ribosome-protected mRNA fragments. Nat Protoc. 2012;7(8):1534–50. doi: 10.1038/nprot.2012.086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Zu T, et al. Non-ATG-initiated translation directed by microsatellite expansions. Proc Natl Acad Sci U S A. 2011;108(1):260–5. doi: 10.1073/pnas.1013343108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Cleary JD, Ranum LP. Repeat-associated non-ATG (RAN) translation in neurological disease. Hum Mol Genet. 2013;22(R1):R45–51. doi: 10.1093/hmg/ddt371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Wagnon JL, et al. CELF4 regulates translation and local abundance of a vast set of mRNAs, including genes associated with regulation of synaptic function. PLoS Genet. 2012;8(11):e1003067. doi: 10.1371/journal.pgen.1003067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Johnson JO, et al. Mutations in the Matrin 3 gene cause familial amyotrophic lateral sclerosis. Nat Neurosci. 2014;17(5):664–6. doi: 10.1038/nn.3688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Rabani M, et al. Metabolic labeling of RNA uncovers principles of RNA production and degradation dynamics in mammalian cells. Nat Biotechnol. 2011;29(5):436–42. doi: 10.1038/nbt.1861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Jao CY, Salic A. Exploring RNA transcription and turnover in vivo by using click chemistry. Proc Natl Acad Sci U S A. 2008;105(41):15779–84. doi: 10.1073/pnas.0808480105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Imamachi N, et al. BRIC-seq: A genome-wide approach for determining RNA stability in mammalian cells. Methods. 2013 doi: 10.1016/j.ymeth.2013.07.014. [DOI] [PubMed] [Google Scholar]
- 99.Mortimer SA, et al. SHAPE-Seq: High-Throughput RNA Structure Analysis. Curr Protoc Chem Biol. 2012;4(4):275–97. doi: 10.1002/9780470559277.ch120019. [DOI] [PubMed] [Google Scholar]
- 100.Underwood JG, et al. FragSeq: transcriptome-wide RNA structure probing using high-throughput sequencing. Nat Methods. 2010;7(12):995–1001. doi: 10.1038/nmeth.1529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Kertesz M, et al. Genome-wide measurement of RNA secondary structure in yeast. Nature. 2010;467(7311):103–7. doi: 10.1038/nature09322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Kazan H, Morris Q. RBPmotif: a web server for the discovery of sequence and structure preferences of RNA-binding proteins. Nucleic Acids Res. 2013;41(Web Server issue):W180–6. doi: 10.1093/nar/gkt463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Dominissini D, et al. Topology of the human and mouse m6A RNA methylomes revealed by m6A-seq. Nature. 2012;485(7397):201–6. doi: 10.1038/nature11112. [DOI] [PubMed] [Google Scholar]
- 104.Meyer KD, et al. Comprehensive analysis of mRNA methylation reveals enrichment in 3′ UTRs and near stop codons. Cell. 2012;149(7):1635–46. doi: 10.1016/j.cell.2012.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Wang X, et al. N6-methyladenosine-dependent regulation of messenger RNA stability. Nature. 2014;505(7481):117–20. doi: 10.1038/nature12730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Wang Y, et al. N6-methyladenosine modification destabilizes developmental regulators in embryonic stem cells. Nat Cell Biol. 2014;16(2):191–8. doi: 10.1038/ncb2902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Farajollahi S, Maas S. Molecular diversity through RNA editing: a balancing act. Trends Genet. 2010;26(5):221–30. doi: 10.1016/j.tig.2010.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Hideyama T, et al. Induced loss of ADAR2 engenders slow death of motor neurons from Q/R site-unedited GluR2. J Neurosci. 2010;30(36):11917–25. doi: 10.1523/JNEUROSCI.2021-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Kawahara Y, et al. Glutamate receptors: RNA editing and death of motor neurons. Nature. 2004;427(6977):801. doi: 10.1038/427801a. [DOI] [PubMed] [Google Scholar]
- 110.Kwak S, Kawahara Y. Deficient RNA editing of GluR2 and neuronal death in amyotropic lateral sclerosis. J Mol Med (Berl) 2005;83(2):110–20. doi: 10.1007/s00109-004-0599-z. [DOI] [PubMed] [Google Scholar]
- 111.Aizawa H, et al. TDP-43 pathology in sporadic ALS occurs in motor neurons lacking the RNA editing enzyme ADAR2. Acta Neuropathol. 2010;120(1):75–84. doi: 10.1007/s00401-010-0678-x. [DOI] [PubMed] [Google Scholar]
- 112.Hideyama T, et al. Profound downregulation of the RNA editing enzyme ADAR2 in ALS spinal motor neurons. Neurobiol Dis. 2012;45(3):1121–8. doi: 10.1016/j.nbd.2011.12.033. [DOI] [PubMed] [Google Scholar]
- 113.Flomen R, Makoff A. Increased RNA editing in EAAT2 pre-mRNA from amyotrophic lateral sclerosis patients: involvement of a cryptic polyadenylation site. Neurosci Lett. 2011;497(2):139–43. doi: 10.1016/j.neulet.2011.04.047. [DOI] [PubMed] [Google Scholar]
- 114.Donnelly CJ, et al. RNA toxicity from the ALS/FTD C9ORF72 expansion is mitigated by antisense intervention. Neuron. 2013;80(2):415–28. doi: 10.1016/j.neuron.2013.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Kim HJ, et al. Mutations in prion-like domains in hnRNPA2B1 and hnRNPA1 cause multisystem proteinopathy and ALS. Nature. 2013;495(7442):467–73. doi: 10.1038/nature11922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Garncarz W, et al. A high-throughput screen to identify enhancers of ADAR-mediated RNA-editing. RNA Biol. 2013;10(2):192–204. doi: 10.4161/rna.23208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Li JB, et al. Multiplex padlock targeted sequencing reveals human hypermutable CpG variations. Genome Res. 2009;19(9):1606–15. doi: 10.1101/gr.092213.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.McConnell MJ, et al. Mosaic copy number variation in human neurons. Science. 2013;342(6158):632–7. doi: 10.1126/science.1243472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Dols-Icardo O, et al. Characterization of the repeat expansion size in C9orf72 in amyotrophic lateral sclerosis and frontotemporal dementia. Hum Mol Genet. 2014;23 (3):749–54. doi: 10.1093/hmg/ddt460. [DOI] [PubMed] [Google Scholar]
- 120.Shapiro E, Biezuner T, Linnarsson S. Single-cell sequencing-based technologies will revolutionize whole-organism science. Nat Rev Genet. 2013;14(9):618–30. doi: 10.1038/nrg3542. [DOI] [PubMed] [Google Scholar]
- 121.Wu P, et al. Roles of long noncoding RNAs in brain development, functional diversification and neurodegenerative diseases. Brain Res Bull. 2013;97:69–80. doi: 10.1016/j.brainresbull.2013.06.001. [DOI] [PubMed] [Google Scholar]
- 122.Salta E, De Strooper B. Non-coding RNAs with essential roles in neurodegenerative disorders. Lancet Neurol. 2012;11(2):189–200. doi: 10.1016/S1474-4422(11)70286-1. [DOI] [PubMed] [Google Scholar]
- 123.Wheeler TM, et al. Targeting nuclear RNA for in vivo correction of myotonic dystrophy. Nature. 2012;488(7409):111–5. doi: 10.1038/nature11362. [DOI] [PMC free article] [PubMed] [Google Scholar]