ABSTRACT
Although the complete genome sequences of three strains of Mycoplasma bovis are available, few studies have examined gene function in this important pathogen. Mycoplasmas lack the biosynthetic machinery for the de novo synthesis of nucleic acid precursors, so nucleases are likely to be essential for them to acquire nucleotide precursors. Three putative membrane nucleases have been annotated in the genome of M. bovis strain PG45, MBOVPG45_0089 and MBOVPG45_0310, both of which have the thermonuclease (TNASE_3) functional domain, and MBOVPG45_0215 (mnuA), which has an exonuclease/endonuclease/phosphatase domain. While previous studies have demonstrated the function of TNASE_3 domain nucleases in several mycoplasmas, quantitative comparisons of the contributions of different nucleases to cellular nuclease activity have been lacking. Mapping of a library of 319 transposon mutants of M. bovis PG45 by direct genome sequencing identified mutants with insertions in MBOVPG45_0310 (the Δ0310 mutant) and MBOVPG45_0215 (the Δ0215 mutant). In this study, the detection of the product of MBOVPG45_0215 in the Triton X-114 fraction of M. bovis cell lysates, its cell surface exposure, and its predicted signal peptide suggested that it is a surface-exposed lipoprotein nuclease. Comparison of a ΔmnuA mutant with wild-type M. bovis on native and denatured DNA gels and in digestion assays using double-stranded phage λ DNA and closed circular plasmid DNA demonstrated that inactivation of this gene abolishes most of the cellular exonuclease and endonuclease activity of M. bovis. This activity could be fully restored by complementation with the wild-type mnuA gene, demonstrating that MnuA is the major cellular nuclease of M. bovis.
IMPORTANCE Nucleases are thought to be important contributors to virulence and crucial for the maintenance of a nutritional supply of nucleotides in mycoplasmas that are pathogenic in animals. This study demonstrates for the first time that of the three annotated cell surface nuclease genes in an important pathogenic mycoplasma, the homologue of the thermostable nuclease identified in Gram-positive bacteria is responsible for the majority of the nuclease activity detectable in vitro.
INTRODUCTION
Although the complete genomes of many Mycoplasma species are now available, the scarcity of tools for exploring their molecular biology has resulted in an understanding of the function of their genes more rudimentary than that which is available for many other prokaryotes. The genomes of three strains of the important bovine pathogen Mycoplasma bovis, the type strain PG45, the Hubei-1 strain, and the HB0801 strain, have been sequenced and published recently (1–3), but apart from several reports on phase switching in M. bovis (4–7), few studies have examined the molecular mechanisms underlying their pathogenicity and virulence or the metabolic functions of the products of their genes.
Mycoplasmas evolved from Clostridium-like bacteria by a process of reductive evolution and are believed to have retained only the regulatory and metabolic systems essential for their survival, as there is no evidence of residual pseudogenes like those seen in the genomes of some other pathogens that have evolved in a similar manner. They lack a cell wall, and thus, their cytoplasmic membrane is in direct contact with the surrounding milieu. As the genomes of mycoplasmas mainly encode genes involved in catabolism rather than genes involved in biosynthetic pathways, procurement of nutrients and passage of them into the cytoplasm are critical functions. Catabolic enzymes on the cell surface are likely to play a role in liberating complex nutrients for transport into the cell and may also play a role in pathogenesis by damaging host cells. Mycoplasmas lack the biosynthetic machinery for the de novo synthesis of nucleic acid precursors (8), so their intrinsic nuclease activity is likely to be essential for acquisition of nucleotides and thus for their replication and persistence.
Membrane nuclease activity in mycoplasmas was first reported for M. pulmonis, a respiratory and genital tract pathogen of rodents (9). Later, external membrane-associated nuclease activity was found in all 20 Mycoplasma species tested (10). Examination of a recombinant λ phage library containing fragments of the M. pulmonis genome identified the mnuA gene to be responsible for nuclease activity (11). Nuclease activity has also been demonstrated in vitro following cloning, expression, and purification of predicted membrane nucleases from M. hyopneumoniae (12), M. gallisepticum (13), M. genitalium (14), M. pneumoniae (15), and M. agalactiae (16). However, most studies on membrane nucleases of mycoplasmas have been based on an examination of in vitro profiles of activity following protein purification (12–16). Genome sequencing has identified genes encoding many additional putative nucleases in mycoplasmas that are predicted to be located either on the cell surface or within the cytoplasm, but these studies cannot assess the relative significance of a particular nuclease in the cell. Comparison of knockout mutants with wild-type cells is needed to assess the quantitative significance of a particular protein and provide a greater understanding of the range of different nucleases within a species of mycoplasma.
In the study reported here, the nuclease activity of membrane nucleases against various substrates was assessed by using gene-knockout mutants, and comparison of them with wild-type M. bovis was used to establish the relative significance of the nuclease encoded by the mnuA gene (MBOVPG45_0215) in M. bovis.
MATERIALS AND METHODS
Bacterial strains and culture conditions.
M. bovis strain PG45 (ATCC 25523) was cultured at 37°C in modified Frey's broth or on mycoplasma agar plates (modified Frey's broth without phenol red with 1% agar added), while the mutants were selected in the presence of 50 μg gentamicin/ml (17). Escherichia coli DH5α cells (Life Technologies) were used for cloning and the development of constructs for use in complementation studies.
Identification of transposon mutants with disrupted putative nuclease genes.
Following the generation of an M. bovis strain PG45 mutant library with a series of transposons based on Tn4001, 319 insertion sites were characterized by direct genome sequencing (17). The nucleotide sequences were matched with the M. bovis strain PG45 genome sequence (3) using the BLAST program and the NCBI nucleotide sequence database (www.ncbi.nlm.nih.gov). Two independent clones that had insertions in different lipoprotein genes predicted to encode membrane nucleases, MBOVPG45_0215 (MnuA) and MBOVPG45_0310 (lipoprotein-nuclease), were identified.
Nuclease zymograms.
The nuclease zymogram method used was based on the inhibition of nuclease activity by sodium dodecyl sulfate (SDS) and the capacity of most nucleases to renature following removal of SDS by diffusion (10). The M. bovis P45 parent and the two mutants were cultured in 10-ml volumes at 37°C to late log phase. Cells were pelleted by centrifugation at 11,000 × g for 20 min at 4°C, washed twice with phosphate-buffered saline (PBS), suspended in 40% glycerol, and stored at −20°C. Protein concentrations were determined with a protein assay dye reagent (Bio-Rad) using bovine serum albumin as a reference standard. Samples for nuclease zymograms were prepared by mixing cells 1:1 with 2× SDS-PAGE lysis buffer (18) and heating at 100°C for 5 min.
The nuclease zymograms used to compare the nuclease activity in the wild type and the transposon mutants were generated as described previously (10). The stacking and resolving gels were prepared as described previously (18), with the exception that sheared salmon sperm DNA (Sigma) was incorporated into the resolving gel at a concentration of 10 μg/ml. For preparation of denatured DNA gels, sheared salmon sperm DNA was heated at 100°C for 15 min and then immediately cooled by putting it on ice for 10 min, and this was incorporated into the polyacrylamide gel as described above. Total cell proteins were separated by electrophoresis in 10 or 12.5% polyacrylamide resolving gels in SDS-PAGE buffer (25 mM Tris base, 192 mM glycine, 0.1% [wt/vol] SDS). Broad-range prestained protein markers (Fermentas) were used as molecular mass standards.
For nuclease renaturation, gels were washed on a rocking platform for 15 min in 10 to 100 volumes of incubation buffer (0.04 M Tris, pH 7.5, 1% skim milk powder, 0.04% β-mercaptoethanol). This was repeated a further 3 times, and finally, the gels were incubated overnight at room temperature in this buffer. The gel was then incubated statically at 37°C in 10 to 100 volumes of incubation buffer supplemented with 2 mM CaCl2 and 2 mM MgCl2. To examine the activity of MnuA in the presence of calcium or magnesium ions, renaturation was also performed in buffers containing either CaCl2 or MgCl2.The gels were then stained with ethidium bromide (0.5 mg/ml), destained in water for 20 to 30 min, examined using UV transillumination, and imaged. Nuclease activity was detected as zones of clearance on the gels, indicating hydrolysis of the DNA in the gel. As the presence of ethidium bromide in the gel did not interfere with the nuclease action, the gels could be repeatedly incubated in renaturation buffer and reimaged after further ethidium bromide staining.
2-D electrophoresis.
For two-dimensional (2-D) gel electrophoresis, 20-ml mycoplasma cultures were grown to late log phase and cells were harvested by centrifugation at 11,000 × g for 20 min at 4°C. The pellet was washed thrice with PBS, with centrifugation at 16,000 × g for 5 min at room temperature being performed between each wash. After the final wash, the pellet was suspended in 500 μl sample preparation solution {8 M urea, 4% 3-[(3-cholamidopropyl)-dimethylammonio]-1-propanesulfonate (CHAPS), 2% Pharmalyte–immobilized pH gradient (IPG) buffer, pH 3 to 10, 40 mM dithiothreitol (DTT), double-distilled water to 25 ml} using a 25-gauge needle. The sample was sonicated four times for 15 s each time on ice, with a 5-min interval being used between each sonication. The sample was then centrifuged at 70,000 × g for 30 min at 15°C. The supernatant was collected, divided into 100-μl aliquots, and stored at −80°C. Protein concentrations were determined with the protein assay dye reagent (Bio-Rad) using known concentrations of bovine serum albumin as a reference.
The sample was processed using a 2-D cleanup kit (GE Healthcare) according to the manufacturer's recommendations, and the pellet was suspended in rehydration solution (8 M urea, 2% CHAPS, 0.5% Pharmalyte–IPG buffer, 3 to 10 pH, 2.8 mg DTT/ml, double-distilled water), vortexed for 30 s until fully dissolved, and stored at −80°C.
For the first-dimension isoelectric focusing (IEF), 125 μl of the protein solution was loaded onto a 7-cm pH 3 to 10 linear-gradient IPG strip (GE Healthcare). The gel was rehydrated for 6 h at 30 V and another 6 h at 60 V, and then focusing was performed at 200 V for 1 h, 500 V for 1 h, 1,000 V for 1 h, 1,000 to 8,000 V for 1 h, and 8,000 V for 1.5 h (for a total of 14,742 V · h over 17 h) using an IEF unit (Bio-Rad). After IEF, the IPG strip was equilibrated for 15 min in equilibration buffer (75 mM Tris-HCl, pH 8.8, 6 M urea, 30% glycerol, 2% sodium dodecyl sulfate, 0.002% bromophenol blue, 1% [wt/vol] DTT) and then treated for 15 min with the same buffer, except that the buffer contained 2.5% iodoacetamide and DTT was omitted. The equilibrated strips were then loaded onto SDS-polyacrylamide gels, the proteins were separated by SDS-PAGE using the same conditions described above for one-dimensional electrophoresis, and the resultant gel was used to generate nuclease zymograms.
Triton X-114 phase partitioning.
Phase partitioning of proteins was performed as described previously (19, 20). Mycoplasma cells were pelleted from 20 ml of a late-log-phase broth culture by centrifugation at 11,000 × g for 20 min at 4°C, washed thrice with PBS, and then suspended in 0.5 ml PBS containing 0.5% Triton X-114 (Sigma). The solution was incubated on ice for 60 min, with the solution being mixed every 15 min. The solution was centrifuged at 12,000 × g for 30 min at 4°C, and the supernatant was carefully loaded onto a 1-ml 6% (wt/vol) sucrose cushion in PBS containing 0.06% (vol/vol) Triton X-114 and incubated at 37°C for 9 min to achieve phase separation. The sample was then centrifuged at 400 × g for 7 min at 37°C. The supernatant was aspirated, leaving an oily pellet containing the hydrophobic fraction. The pellet was suspended in 0.5 ml of ice-cold PBS, and the proteins were precipitated by adding 0.5 ml methanol-chloroform (4:1), followed by centrifugation of the suspension at 10,000 × g for 1 min at 4°C. The supernatant was removed, and the pellet was partially air dried and then suspended in 100 μl 8 M urea in PBS and used to generate nuclease zymograms.
Trypsin treatment.
Intact M. bovis cells were treated with trypsin to partially digest cell surface proteins, as described previously (12, 21). M. bovis cells were cultured, and the cell pellet was washed in 50 mM Tris, 0.145 M NaCl, pH 7.4 (Tris-salt [TS] buffer). This was repeated twice, and the cells were finally suspended in 600 μl TS buffer and then divided into 7 equal aliquots. A dilution series of trypsin (Sigma) at 500, 250, 125, 62, 31, and 15 μg/ml was made in TS buffer; 100 μl of each dilution, as well as a control without any trypsin, was added to separate aliquots of cells; and the suspension was incubated at 37°C for 30 min. Digestion was stopped by the addition of 200 μl of 0.125% (wt/vol) trypsin inhibitor (Sigma). The trypsin-treated cells were collected by centrifugation and suspended in TS buffer, and the proteins in the sample were used to generate nuclease zymograms.
Assays for nuclease activity.
The nuclease activity of wild-type M. bovis and the putative nuclease mutants was analyzed by 1% agarose gel electrophoresis as described previously (12). The exonuclease and endonuclease activities of the cells were determined by incubating proteins from cells from late-log-phase cultures suspended in 50 μl of nuclease reaction buffer (25 mM Tris-HCl, pH 8.8, 10 mM CaCl2, 10 mM MgCl2) at 37°C with 500 ng of double-stranded phage λ DNA (New England BioLabs) or 2.0 μg of closed circular plasmid DNA (plasmid pRecAGKIRRPG2, which was generated to disrupt a specific gene target by homologous recombination in M. bovis) as the substrates. Aliquots (10 μl) were removed after different time intervals, the reactions were stopped by the addition of EDTA to a final concentration of 20 mM, and the aliquots were mixed 4:1 with 5× loading buffer (Bioline) and then immediately loaded onto an agarose gel made in 0.5× TPE buffer (1× TPE is 36 mM Tris, 30 mM NaH2PO4, and 1 mM EDTA) containing 0.1 μg ethidium bromide/ml and electrophoresed in 0.5× TPE buffer.
Gene complementation studies.
To confirm that the MBOVPG45_0215 (mnuA) locus was responsible for most of the nuclease activity observed in wild-type M. bovis, the complete mnuA gene, along with its upstream region and terminator sequence, was amplified from M. bovis strain PG45 using the primers MBOV0215 for (AGAAATCAAAAAAAGCAGGGT) and MBOV0215 rev (TTGTCCAAGTCTTCATATTGTGC), both of which contained NotI cleavage sites. The PCR mixtures were incubated through one cycle at 94°C for 5 min, followed by 35 cycles of 94°C for 30 s, 50°C for 30 s, and 68°C for 1.5 min and a final extension at 68°C for 5 min. The amplified product was ligated into the T/A site of the pGEM-T vector (Promega) and, after cleavage with NotI, subcloned into dephosphorylated pIRR45, an M. bovis oriC plasmid (22) that carried the tetM resistance marker, to generate the complementation plasmid pMnuAIRR45. Approximately 5 μg of pMnuAIRR45 was introduced into the Δ0215 (mnuA) M. bovis mutant. Five transformant clones were picked and passaged once at a 1:9 dilution and analyzed by performing nuclease zymograms.
Sequence analysis.
The ProtParam tool (http://web.expasy.org/protparam/) was used to assess the physicochemical properties of membrane nucleases (23). The Metagene program (http://metagene.cb.k.u-tokyo.ac.jp/) (24) was used to identify ribosomal binding sites. Searches of the Molligen database (http://services.cbib.u-bordeaux2.fr/molligen/) were performed with the amino acid sequences of the putative membrane nucleases and the BLASTp program, and the MUSCLE program, implemented in the Molligen website, was used to align similar amino acid sequences (25). The Prosite (http://prosite.expasy.org/) (26) and NCBI (27) databases were used to predict the conserved domains and binding sites. The PSORTb (v3.02) tool (http://www.psort.org/psortb/) was used to predict the cellular localization of the proteins (28).
RESULTS
Identification of disruptions in nuclease genes.
The M. bovis strain PG45 genome contains three annotated putative membrane nuclease genes, MBOVPG45_0089, MBOVPG45_0215, and MBOVPG45_0310. The predicted molecular mass of the product of MBOVPG45_0215 is 44.13 kDa, that of the product of MBOVPG45_0310 is 41.56 kDa, and that of the product of MBOVPG45_0089 is 20.51 kDa. Mapping of a library of 319 transposon mutants of the M. bovis PG45 genome identified 1 mutant (the Δ0310 mutant) with a disruption in MBOVPG45_0310, which codes for the nuclease of a putative nucleotide transporter operon (12, 13, 29, 30). Another mutant (the Δ0215 mutant) had a disruption in MBOVPG45_0215, which encodes a membrane nuclease, MnuA. Mutant Δ0215 had an insertion in the mnuA gene 31 bp from the predicted start codon, while mutant Δ0310 had an insertion 823 bp into the gene. There were no observable differences between the in vitro growth of these two putative nuclease mutants and wild-type M. bovis in mycoplasma medium.
SDS-PAGE nuclease assays.
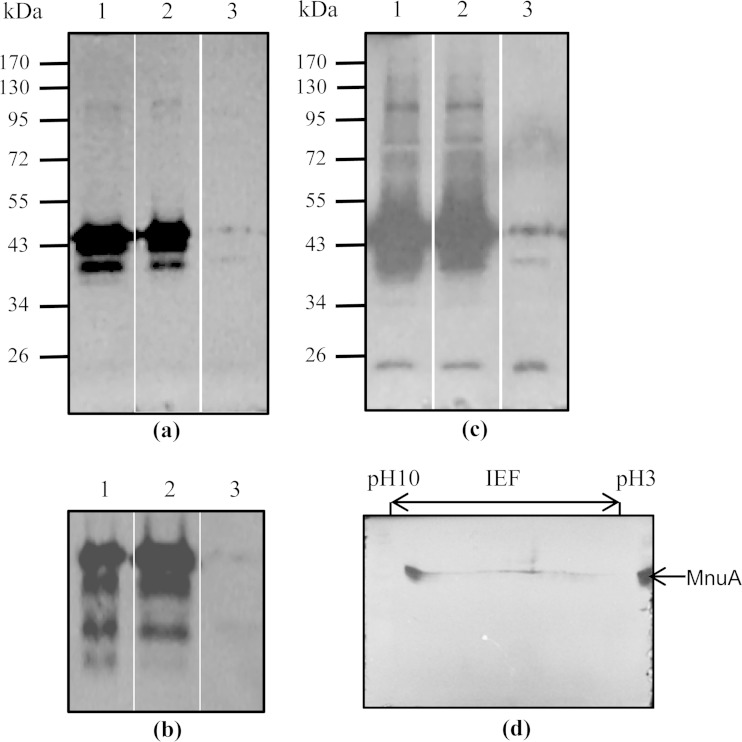
Zymogram analysis revealed nuclease activity as bands of hydrolyzed DNA (zones of clearance) that did not fluoresce when gels were stained with ethidium bromide. Initially, the nuclease activity of the Δ0310 mutant was compared with that of wild-type M. bovis strain PG45 in gels containing double-stranded DNA (dsDNA). There was no detectable difference between the Δ0310 mutant and wild-type M. bovis in SDS-PAGE nuclease gels. Zones of clearance, corresponding in estimated mass to the predicted mass of MnuA, were detected in zymograms of both strains within 2 h of renaturation (Fig. 1a), and the intensity of these zones continued to increase with time. After 3 h of renaturation, four distinct zones could be seen (Fig. 1b), and after 8 h of renaturation, another zone of clearance could be seen (Fig. 1c). The M. bovis PG45 MBOVPG45_0089 open reading frame encodes a putative nuclease with an estimated molecular mass of 20.51 kDa (Table 1), which could be the nuclease with a predicted molecular mass of approximately 21 kDa that was detected after 8 h of renaturation.
FIG 1.
Nuclease zymograms of whole-cell proteins of wild-type and mutant M. bovis in SDS-polyacrylamide gels containing sheared salmon sperm DNA. Nuclease activity causes hydrolysis of the DNA, resulting in zones of clearance that are apparent as black bands after staining with ethidium bromide. Lane 1, wild-type M. bovis; lane 2, Δ0310 mutant; lane 3, Δ0215 mutant. (a) After renaturation for 3 h; (b) after 3 h of renaturation in a gel electrophoresed for a longer period of time, revealing four distinct bands; (c) after 24 h of renaturation; (d) whole-cell proteins of wild-type M. bovis after IEF and separation by SDS-PAGE in a gel containing sheared salmon sperm DNA, demonstrating a single zone of clearance, corresponding to MnuA in size and isoelectric point, after renaturation for 12 h, with the right-hand lane containing whole-cell proteins separated only in the second dimension (MnuA).
TABLE 1.
Membrane nucleases of M. bovis PG45 and their orthologs in other mycoplasmasa
| Species | GenBank accession number of reference sequence | MBOVPG45_0215 |
MBOVPG45_0310 |
MBOVPG45_0089 |
|||
|---|---|---|---|---|---|---|---|
| ORF | No. of amino acids (% identity) | ORF | No. of amino acids (% identity) | ORF | No. of amino acids (% identity) | ||
| M. bovis PG45 | NC_014760.1 | MBOVPG45_0215 | 409 | MBOVPG45_0310 | 389 | MBOVPG45_0089 | 190 |
| M. bovis Hubei-1 | NC_015725.1 | MMB_0635 | 422 (92) | MMB_0542 | 287 (99) | MMB_0081 | 180 (98) |
| M. bovis HB0801 | CP002058.1 | Mbov_0674 | 422 (92) | Mbov_0580 | 389 (99) | Mbov_0087 | 190 (98) |
| M. agalactiae PG2 | NC_009497.1 | MAG_5900 | 404 (73) | MAG_5040 | 390 (74) | MAG_0790 | 164 (67) |
| M. agalactiae 5632 | FP671138.1 | MAG_6550 | 404 (74) | MAG_5510 | 390 (75) | MAG_0850 | 190 (67) |
| M. fermentans JER | NC_014552.1 | MFE_05240 | 647 (36) | MFE_03090 | 392 (43) | MFE_01650 | 193 (39) |
| M. pulmonis UAB CTIP | NC_002771.1 | MYPU_6940 | 394 (34) | MYPU_0250 | 256 (31) | MYPU_1390 | 190 (29) |
| M. pulmonis UAB CTIP | NC_002771.1 | MYPU_6930b | 1,125 (37) | ||||
| M. arthritidis 158L3-1 | NC_011025.1 | MARTH_orf075 | 466 (34) | MARTH_orf834 | 311 (35) | MARTH_orf871 | 234 (37) |
| M. arthritidis 158L3-1 | NC_011025.1 | MARTH_orf723 | 346 (31) | ||||
| M. crocodyli MP145 | NC_014014.1 | MCRO_0442 | 687 (30) | MCRO_0706 | 301 (36) | MCRO_0604 | 211 (36) |
| M. alligatoris A21JP2 | Not completed | MALL_0151 | 431 (30) | MALL_0678c | 207 (28) | MALL_0678c | 207 (41) |
| M. mobile 163K | NC_006908.1 | MMOB1270c | 200 (28) | MMOB1270c | 200 (31) | ||
| M. hyorhinis HUB-1 | NC_014448.1 | MHR_0549 | 445 (29) | MHR_0063 | 319 (35) | MHR_0255 | 136 (28) |
| M. hyorhinis HUB-1 | NC_014448.1 | MHR_0206 | 434 (28) | ||||
| M. hominis ATCC 23114 | NC_013511.1 | MHO_0310 | 489 (31) | MHO_0730 | 321 (29) | MHO_0660 | 317 (36) |
| M. conjunctivae HRC/581 | NC_012806.1 | MCJ_001570 | 391 (27) | MCJ_000150 | 305 (31) | MCJ_005730 | 215 (29) |
| M. hyopneumoniae 232 | NC_006360.1 | mhp597 | 377 (34) | mhp379 | 310 (34) | ||
| M. hyopneumoniae 7448 | NC_007332.1 | MHP7448_0580 | 377 (33) | MHP7448_0368 | 310 (34) | MHP7448_0270 | 208 (27) |
| M. hyopneumoniae J | NC_007295.1 | MHJ_0581 | 377 (34) | MHJ_0364 | 310 (34) | MHJ_0262 | 208 (28) |
| M. synoviae 53 | NC_007294.1 | MS53_0284 | 452 (38) | MS53_0110 | 233 (38) | ||
| M. gallisepticum R low | NC_004829.2 | MGA_0637 | 402 (26) | MGA_0676 | 276 (29) | ||
| M. gallisepticum R(high) | CP001872.1 | MGAH_0637 | 389 (26) | MGAH_0676 | 276 (29) | ||
| M. gallisepticum F | CP001873.1 | MGF_0074 | 389 (27) | MGF_0241 | 276 (29) | ||
| M. genitalium G37 | NC_000908.2 | MG186c | 250 (27) | MG186c | 250 (30) | ||
| M. pneumoniae M129 | NC_000912.1 | MPN491 | 474 (33) | MPN133 | 301 (30) | ||
| M. penetrans HF-2 | NC_004432.1 | MYPE4380 | 452 (36) | ||||
| Ureaplasma parvum serovar 3 ATCC 700970 | NC_002162.1 | UU486 | 434 (33) | ||||
| Ureaplasma parvum serovar 3 ATCC 700970 | NC_002162.1 | UU055d | 1,138 (31) | ||||
MBOVPG45_0215 and its orthologs belong to the EEP domain superfamily. MBOVPG45_0310 and MBOVPG45_0089 and their orthologs have TNASE_3 domains. ORF, open reading frame.
M. pulmonis MYPU_6930 has two EEP domains flanking an N-acetylmuramoyl-l-alanine amidase domain region.
Orthologs have identity with both the MBOVPG45_0310 and MBOVPG45_0089 M. bovis genes.
U. parvum UU055 has an EEP domain as well as a 5′-nucleotidase domain within COG0737.
The nuclease activity of the Δ0215 mutant was also compared with that of wild-type M. bovis strain PG45 on gels containing dsDNA. There was no detectable nuclease activity in the zymogram of the Δ0215 mutant after 2 h of incubation, and after 8 h of incubation only a single cleared zone was seen at about 21 kDa. This clearly indicated that disruption of MBOVPG45_0215, a predicted membrane nuclease, abolished most nuclease activity in M. bovis. In addition to this, two-dimensional gel electrophoresis of the membrane proteins of M. bovis strain PG45 was performed, with the second dimension being performed in gels containing dsDNA. Only a single intense zone of clearance was seen on the gel at a position predicted to correspond to that of MnuA (Fig. 1d).
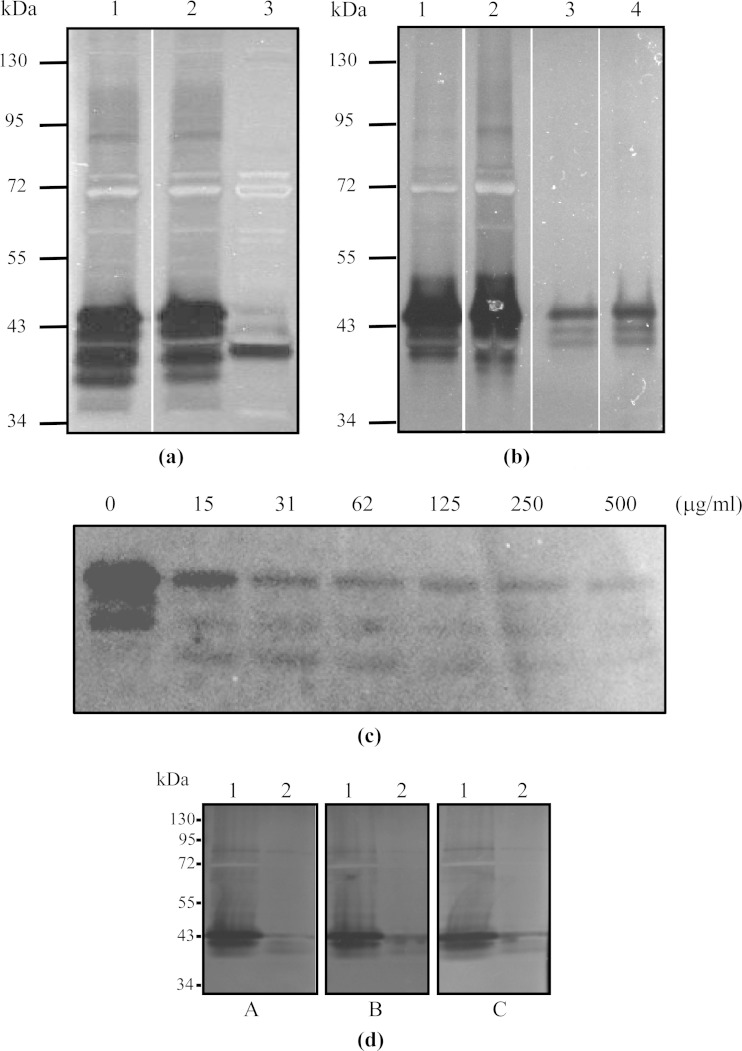
We further examined the nuclease activity of wild-type M. bovis and the Δ0215 mutant against denatured DNA (Fig. 2a). Four zones of clearance of almost equal intensity were observed in the wild type, while the Δ0215 mutant produced a single band with an intensity similar to that observed in the wild type, indicating that the Δ0215 mutant had some nuclease activity against denatured DNA. Triton X-114 partitioning of M. bovis proteins revealed that although there was some nuclease activity within the hydrophilic fraction (Fig. 2b), most was within the hydrophobic fraction. The nuclease activity was also shown to be susceptible to proteolytic cleavage after treatment with increasing concentrations of trypsin (Fig. 2c). To examine the effect of different divalent cations on the nuclease activity, renaturation was performed in buffers containing only Mg2+ or only Ca2+. No differences were seen after renaturation in buffers containing different divalent cations, suggesting that the MnuA nuclease can utilize either calcium or magnesium ions (Fig. 2d).
FIG 2.
(a) Nuclease zymograms of whole-cell proteins of wild-type and mutant M. bovis strains in gels containing denatured DNA. Lane 1, wild-type M. bovis; lane 2, the Δ0310 mutant; lane 3, the Δ0215 mutant. (b) Nuclease zymogram of Triton X-114-fractionated proteins of M. bovis and the Δ0310 mutant on a gel containing denatured DNA. Lane 1, hydrophobic fraction of M. bovis PG45; lane 2, hydrophobic fraction of the Δ0310 mutant; lane 3, hydrophilic fraction of M. bovis PG45; lane 4, hydrophilic fraction of the Δ0310 mutant. (c) Nuclease zymogram of trypsin-treated M. bovis strain PG45 cells. Trypsin concentrations (μg/ml) are indicated above each lane. Increased degradation of MnuA-associated nuclease activity is apparent with increasing concentrations of trypsin. (d) Nuclease zymogram of whole-cell proteins of wild-type M. bovis and the Δ0215 mutant in renaturation buffers containing MgCl2 (A), CaCl2 (B), or both MgCl2 and CaCl2 (C). Lanes 1, wild-type M. bovis; lanes 2, the Δ0215 mutant. No differences were observed, suggesting that the MnuA nuclease can utilize either calcium or magnesium ions.
Whole-cell nuclease assays.
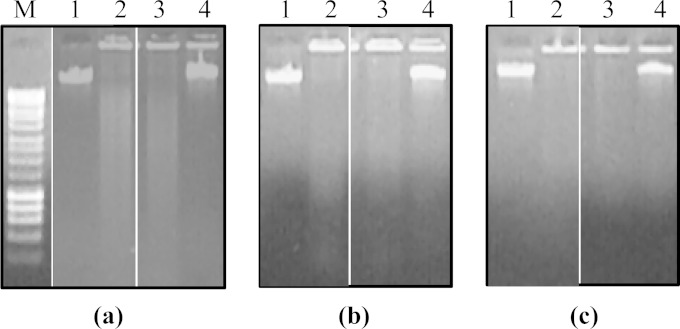
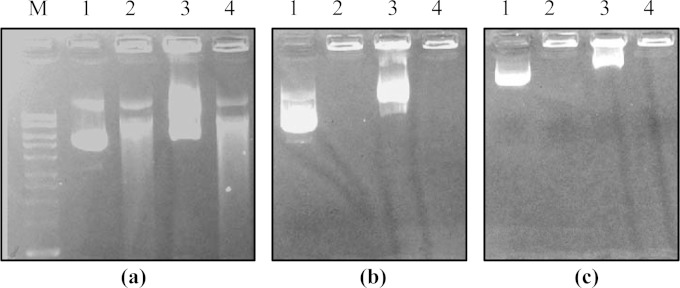
The nuclease activity of wild-type M. bovis and the putative nuclease gene mutants against different substrates was compared in 1% agarose gels. In the presence of M. bovis, the degradation of the linear dsDNA substrate resulted in the appearance of a DNA smear that increased in intensity with an increase in the duration of incubation. Wild-type M. bovis substantially degraded 500 ng of double-stranded phage λ DNA within 15 min, while the phage λ DNA appeared to be unaffected after 60 min when incubated with the Δ0215 mutant (Fig. 3). These data suggest that MnuA of M. bovis has exonuclease activity and that disruption of mnuA greatly reduces the cellular nuclease activity of M. bovis. The endonuclease activity of MnuA was demonstrated using closed circular DNA as the substrate. In the presence of wild-type M. bovis, the plasmid DNA was cleaved into an open circular form within 15 min and was subsequently digested over time with the concurrent appearance of a DNA smear, while the plasmid remained intact even after 90 min of incubation with the Δ0215 mutant (Fig. 4). This clearly suggested that MnuA has endonuclease activity against closed circular plasmid DNA. The low levels of residual nuclease activity in the Δ0215 mutant may originate from the product of MBOVPG45_0310 or MBOVPG45_0089.
FIG 3.
Digestion of phage λ DNA by wild-type and mutant M. bovis cells. Sampling time points were 15 min (a), 30 min (b), and 60 min (c). Lane M, molecular mass markers (Bioline Hyperladder I); lanes 1, nuclease-free water (negative control); lanes 2, wild-type M. bovis; lanes 3, the Δ0310 mutant; lanes 4, the Δ0215 (mnuA) mutant. There was complete digestion of 500 ng phage λ DNA within 15 min of incubation with wild-type M. bovis and the Δ0310 mutant, but the Δ0215 mutant induced only a minor degradation of phage λ DNA after 60 min.
FIG 4.
Digestion of closed circular plasmid DNA by wild-type and mutant M. bovis cells. Sampling time points were 15 min (a), 45 min (b), and 90 min (c). Lane M, molecular mass markers (Bioline Hyperladder I); lanes 1, nuclease-free water (negative control); lanes 2, wild-type M. bovis; lanes 3, the Δ0215 (mnuA) mutant; lanes 4, the Δ0310 mutant. Within 15 min of incubation with wild-type M. bovis or the Δ0310 mutant there was some degradation of the plasmid, while plasmid incubated with the Δ0215 mutant remained intact after 90 min. Note that samples were loaded onto the gel at different times, and thus, the plasmid migrated different distances in each lane.
Restoration of nuclease activity by complementation with wild-type mnuA.
Plasmid pIRR45 was used as a shuttle vector for gene complementation studies in M. bovis, as it has been shown to persist extrachromosomally for at least 20 passages in vitro. In order to confirm that mnuA was indeed essential for nuclease activity, a complete mnuA gene with its upstream sequence was introduced into the Δ0215 mutant of M. bovis on this plasmid. Complementation of the Δ0215 mutant fully restored the nuclease activity of M. bovis (Fig. 5), proving that mnuA encodes most of the cellular nuclease activity in M. bovis.
FIG 5.
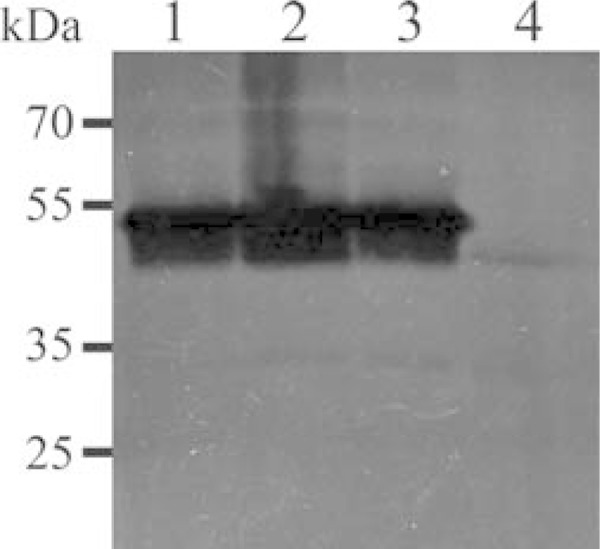
Gene complementation studies. Nuclease zymograms of whole-cell proteins of clones of the Δ0215 mutant complemented with the pMnuAIRR45 plasmid, M. bovis strain PG45, and the Δ0215 mutant. Lanes 1 and 2, the Δ0215 mutant complemented with pMnuAIRR45 expressing MnuA; lane 3 wild-type M. bovis strain PG45; lane 4, the Δ0215 mutant. Complementation with mnuA fully restored nuclease activity.
Membrane nucleases in M. bovis and their homologues.
Orthologues of the three annotated putative membrane nucleases in the genome of M. bovis strain PG45, MBOVPG45_0089, MBOVPG45_0215, and MBOVPG45_0310, were identified in a number of phylogenetically distant mycoplasmas belonging to the M. hominis and M. pneumoniae clusters and are listed in Table 1, along with their size and identity to their respective M. bovis orthologues. Most species had only a single MBOVPG45_0215 orthologue, while M. pulmonis, M. arthritidis, M. hyorhinis, and Ureaplasma parvum each contained two orthologues. However, an orthologue of MBOVPG45_0215 could not be identified in the sequenced genomes of M. synoviae, M. mobile, or M. genitalium. No orthologue of either MBOVPG45_0089 or MBOVPG45_0310 could be identified in M. penetrans or Ureaplasma parvum. Most mycoplasmas contained multiple genes encoding putative nucleases; however, M. genitalium and M. penetrans appeared to contain only a single gene coding for a membrane nuclease. Interestingly, both M. hyopneumoniae strains J and 7448 have orthologues of all three M. bovis nucleases, while M. hyopneumoniae strain 232 is missing the MBOVPG45_0089 orthologue.
Genomic location and features of membrane nucleases in M. bovis.
MBOVPG45_0310 is part of a 7.6-kbp ABC transporter operon that is predicted to be involved in nucleotide transport (12, 13, 16, 29, 30). However, MBOVPG45_0089 and MBOVPG45_0215 do not appear to lie within operons. The region upstream of MBOVPG45_0089 contains a putative fba, predicted to code for fructose-1,6-bisphosphate aldolase, while the region downstream has a putative rplK, coding for the 50S ribosomal protein L11 in most mycoplasmas in the M. hominis cluster.
Although either a 5′-3′ exonuclease gene or a 5′ nucleotidase gene has been annotated in the vicinity of MBOVPG45_0215 orthologues in some mycoplasmas, the locations of these genes vary. The upstream gene (MBOVPG45_0216), which belongs to the peptidase M17 superfamily and contains polypeptide and ion binding sites, is in the opposite orientation, and these two genes lie between two insertion sequences, ISMbov3 and ISMbov6. The region downstream of ISMbov3 contains the 37.4-kbp integrative conjugative element (ICE) B2 (ICEB2 region), one of the two ICEs found in the M. bovis genome (31). The area downstream of the ICEB2 region contains three insertion sequences followed by a truncated 5′-3′ exonuclease gene (MBOVPG45_0179). It appears that MBOVPG45_0215 and MBOVPG45_0179 have been separated by the insertion of the ICEB2 element. This is supported by the fact that the regions downstream of the MBOVPG45_0215 homologues in M. bovis strain Hubei-1 and M. agalactiae strains PG2 and 5632 encode a 5′ nucleotidase on the complementary strand. Similarly, the regions upstream of the MBOVPG45_0215 orthologues in M. hyopneumoniae and M. hyorhinis encode 5′-3′ exonucleases. M. bovis PG45 has additional genes coding for the 5′-3′ exonuclease (MBOVPG45_0688) and the 5′ nucleotidase (MBOVPG45_0690), separated from each other by the ISMbov3 transposase.
The physicochemical and other features of the membrane nucleases of M. bovis were predicted using the ProtParam tool (Table 2). MBOVPG45_0089, MBOVPG45_0215, and MBOVPG45_0310 were estimated to have molecular masses of 20.51 kDa, 44.13 kDa, and 41.56 kDa, respectively, and were predicted to be basic proteins with a high isoelectric constant (pI) and a high instability index, suggesting that they are stable. Neither promoter regions nor Shine-Dalgarno-like sequences could be identified upstream of any of the three genes. All the genes were predicted to encode a hydrophobic amino-terminal signal sequence and prokaryotic lipoprotein cleavage site, and PSORTb (v3.02) predicted that all three are located extracellularly (28).
TABLE 2.
Predicted characteristics of membrane nucleases of M. bovis PG45
| Mnemonic | Genome location (nucleotides) | Size of ORF |
Mass (kDa) of predicted mature protein | pI of predicted mature protein | Lysine content (%) of predicted mature protein | Size of signal sequence (no. of amino acids) | Instability index of predicted mature protein | |
|---|---|---|---|---|---|---|---|---|
| No. of nucleotides | No. of amino acids | |||||||
| MBOVPG45_0089 | 100310–100882 | 573 | 190 | 20.51 | 9.40 | 14.0 | 1–19 | 42.68 |
| MBOVPG45_0215 | 250107–248878 | 1,230 | 409 | 44.13 | 9.03 | 11.2 | 1–25 | 33.36 |
| MBOVPG45_0310 | 348768–347599 | 1,170 | 389 | 41.56 | 6.63 | 12.4 | 1–25 | 23.05 |
Conserved domain structure in membrane nucleases.
The conserved domains in the membrane nucleases of M. bovis are listed in Table 3. MBOVPG45_0215 belongs to subfamily cd10283 and possesses a domain structure (from amino acids 78 to 402) related to DNase 1 (DNase I, EC 3.1.21.1) in a subfamily that also includes M. pulmonis MnuA, a membrane-associated nuclease. Mycoplasma orthologues of MBOVPG45_0215 belong to the cluster of orthologous groups (COG) coding for extracellular nucleases (COG2374). Interestingly, M. pulmonis MYPU_6940 has a single exonuclease/endonuclease/phosphatase (EEP) domain, while MYPU_6930 has two EEP domains on either side of an N-acetylmuramoyl-l-alanine amidase domain. More surprisingly, U. parvum UU055 has a single EEP domain between residues 783 and 1130 as well as a 5′-nucleotidase domain from COG0737 between residues 231 and 624, while in another strain of M. bovis, Hubei-1, and both the sequenced M. agalactiae strains, a 5′-nucleotidase domain is found in the vicinity of the MBOVPG45_0215 homologues. The proximity of the 5′-nucleotidase domain to membrane nucleases containing an EEP domain may suggest complementary action, as 5′-nucleotidases remove 5′-phosphate groups from nucleoside 5′-monophosphates (32).
TABLE 3.
Domain structure and conserved residues within membrane nucleases of M. bovis PG45
| Mnemonic (GenBank accession number) | Length (no. of amino acids) | Domain family | Conserved domain location (amino acids) | Active catalytic site amino acids | Ca2+ binding site amino acids | Mg2+ binding site amino acids | PO43− binding site amino acids | Domain hits |
|---|---|---|---|---|---|---|---|---|
| MBOVPG45_0089a (YP_004055955.1) | 190 | Thermonuclease | 39–181 | R (57), E (65), R (110) | D (43), D (62), T (63) | Micrococcal nuclease (COG1525), staphylococcal nuclease homologues SNase (cd00175, smart00318) | ||
| MBOVPG45_0215b (YP_004056061.1) | 409 | EEP superfamily | 78–402 | N (84), E (119), E (169), H (238), D (289), N (291), D (336), D (398), H (399) | N (84), E (119), D (289), D (336), D (398), H (399) | L (86), H (238), D (289), N (291), H (399) | Predicted extracellular nuclease (COG2374), MnuA_DNase1-like (cd10282, cd10283, cd09075), EEP domain superfamily (cl00490) | |
| MBOVPG45_0310a (YP_004056149.1) | 389 | Thermonuclease | 181–342 | R (219), E (227), R (272) | D (194), D (224), T (225) | Micrococcal nuclease (COG1525), SNase homologues (cd00175, smart00318) |
Prosite.
NCBI.
All the mycoplasma orthologues of MBOVPG45_0089 and MBOVPG45_0310 belong to the micrococcal nuclease (thermonuclease) COG1525 (Table 3). The regions between amino acids 39 and 181 in MBOVPG45_0089 and amino acids 181 and 342 in MBOVPG45_0310 had significant identity with the thermonuclease (TNASE_3) domain and staphylococcal nuclease (SNase) profiles. The TNASE_3 domain, first described for the Nuc thermonuclease of Staphylococcus aureus (33), was conserved in all of the mycoplasma homologues of MBOVPG45_0089 and MBOVPG45_0310.
Conserved amino acid residues in membrane nucleases.
Multiple-sequence alignments indicated that much of the similarity between the mycoplasma orthologues of MBOVPG45_0215 (data not shown) was associated with the EEP domain. For simplicity, the sequences of M. pulmonis MYPU_6930 and U. parvum UU055 were omitted from the alignment, because they each have two domains. Both M. fermentans MFE_05240 and M. crocodyli MCRO_0442 have an amino-terminal extension of more than 300 amino acid residues compared to the sequences of the other orthologues. The residues at positions 84 (N), 119 (E), 169 (E), 238 (H), 289 (D), 291 (N), 336 (D), 398 (D), and 399 (H) in MBOVPG45_0215 (Table 3) are highly conserved and comprise the active catalytic site (34). Residues 84, 119, 289, 336, 398, and 399 can be predicted to be involved in the binding of magnesium ions (34), and those at positions 86 (L), 238, 289, 291, and 399 can be predicted to comprise a putative phosphate binding site (35). While the calcium binding sites conserved in subfamily cd10282 (DNase 1 family) are not conserved in MnuA (subfamily cd10283), suggesting that activity may be dependent on magnesium ions alone, the levels of nuclease activity attributable to MnuA were found to be similar in the presence of only Ca2+, only Mg2+, and both Mg2+ and Ca2+, suggesting that the MnuA nuclease can utilize either calcium or magnesium ions.
Multiple-sequence alignments indicated that much of the similarity between the mycoplasma homologues of MBOVPG45_0089 (data not shown) and MBOVPG45_0310 (data not shown) was associated with the TNASE_3 domain. MBOVPG45_0310 and its orthologues M. agalactiae MAG_5040, M. fermentans MFE_03090, and M. synoviae MS53_0284 have an amino-terminal extension of approximately 200 amino acids compared to the sequences of the orthologues in other mycoplasmas. The residues at positions 194 (D), 224 (D), and 225 (T) in MBOVPG45_0310 and 43 (D), 62 (D), and 63 (T) in MBOVPG45_0089 were strictly conserved (Table 3). These residues are reported to be involved in the binding of calcium ions (36). The residues arginine, glutamic acid, and arginine, located, respectively, at positions 219 (R), 227 (E), and 272 (R) in MBOVPG45_0310 and 57 (R), 65 (E), and 110 (R) in MBOVPG45_0089, are also strictly conserved and can be predicted to comprise the active catalytic site (36). Conservation of these predicted calcium binding and catalytic residues in MBOVPG45_0089 and MBOVPG45_0310 and their orthologues suggests that these are calcium-dependent nucleases.
DISCUSSION
Mycoplasma nucleases were first reported in the mid-1960s (37). Several of these nucleases have been demonstrated to contribute to pathogenicity and cytotoxicity (38, 39). In addition, the M. arthritidis-derived mitogen (MAM) has been demonstrated to exhibit DNase activity (40).
Mollicutes salvage and interconvert but are unable to synthesize de novo purines and pyrimidines (8), with the exception of M. penetrans, which has an orotate-related pathway to convert carbamoyl phosphate into uridine-5′-monophosphate (41). An early study demonstrated the transport of nucleobases and nucleosides into the growing cells of many mollicutes: all transported adenine, guanine, and uracil, while none transported cytosine (42). However, nucleobase or nucleoside transporters were not found in the M. genitalium and M. pneumoniae genomes, and it was suggested that transporters with a wider range of substrates, including the 11 ATP binding cassette (ABC) transporters and the major facilitator superfamily (MFS) primary active transporter, could be involved in nucleic acid precursor import (43). In addition, mollicutes are believed to have a relatively large number of multifunctional or substitute proteins, as exemplified by one protein that performs as both a lactate dehydrogenase and a malate dehydrogenase (44).
It is believed that the nucleic acid metabolism of mollicutes relies on the import of nucleosides and nucleobases and probably of small oligonucleotides, with oligonucleotides and nucleic acids being included in culture media and in host tissues processed by membrane nucleases to facilitate import (45).
MBOVPG45_0310 and its orthologues in most members of the M. hominis and M. pneumoniae phylogenic clusters lie within a putative nucleotide transporter operon (12, 13, 16, 29, 30), and the nuclease activity has been demonstrated for M. agalactiae MAG_5040 (16), M. gallisepticum MGA_0676 (13), M. hyopneumoniae mhp379 (12), M. pneumoniae MPN133 (15), and M. genitalium MG186 (14). It has recently been demonstrated that MslA, the MBOVPG45_0311 homologue in M. gallisepticum, binds single- and double-stranded DNA, suggesting that it may act in concert with these nucleases, with MslA binding and delivering oligonucleotides to the exonuclease, which then processes the oligonucleotides to generate individual nucleotides for transport into the cell via the ABC transporter (13).
Most mycoplasmas contain multiple genes encoding putative membrane nucleases, but BLAST searches with the three putative membrane nucleases of M. bovis found only a single nuclease in M. genitalium MG186 (Table 1). Earlier studies on M. genitalium found only a single nuclease in the genome (14). Similarly, only a single membrane nuclease could be identified in M. penetrans. Searches of the genomes of the M. mycoides cluster of mycoplasmas with the three M. bovis membrane nucleases failed to detect any orthologue, although membrane nuclease activity has previously been demonstrated in some members of this cluster (10).
The MBOVPG45_0310 orthologue in M. pneumoniae, MPN133, has been reported to possess a unique EKS (glutamic acid-lysine-serine)-rich region between residues 72 and 110 (data not shown). This region has been shown to be essential for binding and internalization of the protein into cells, but it is not essential for nuclease activity or immunogenicity (15). Alignment of the sequence of MBOVPG45_0215 with the sequences of its orthologues revealed a unique region in M. pneumoniae MPN491 between residues 175 and 218 with a more than 70% EKS amino acid content (data not shown). Whether it is also involved in binding of this protein by cells needs to be examined.
Transposon mutagenesis has been widely used to disrupt genes in mycoplasmas. Comparison of mutants with wild-type strains can determine the quantitative role of each protein in vivo, which is particularly important when there appears to be functional redundancy. In a previous study, nuclease zymograms were generated for 20 Mycoplasma species (10) and multiple nucleases were predicted to be expressed by many of these species. However, our comparison of the Δ0215 mutant with wild-type M. bovis found that this single mutation eliminated multiple bands of nuclease activity in the zymogram, suggesting that most of these bands may be derived from MnuA.
Comparison of the Δ0215 mutant with wild-type M. bovis on native and denatured DNA gels, as well as in digestion assays using double-stranded phage λ DNA and closed circular plasmid DNA, clearly indicated that disruption of mnuA abolished most of the cellular nuclease activity of M. bovis. Furthermore, complementation of this mutant with the wild-type mnuA gene fully restored nuclease activity, thereby proving that MnuA is the most potent exo- and endonuclease detectable in M. bovis in vitro.
There was no detectable difference between the Δ0310 mutant and wild-type M. bovis in SDS-PAGE nuclease gels and no detectable loss of whole-cell nuclease activity in the mutant, and there was little evidence of a nuclease corresponding in size to the product of MBOVPG45_0310 in the zymograms of the Δ0215 mutant. Nuclease activity probably corresponding to the product of MBOVPG45_0089 could be detected in zymograms after 8 h of renaturation. This protein has a TNASE_3 domain, like the product of MBOVPG45_0310. In a study on M. hyopneumoniae, neither rabbit antiserum against whole M. hyopneumoniae cells nor serum from pigs inoculated with live M. hyopneumoniae bound to recombinant mhp379 (the MBOVPG45_0310 orthologue), even though a monospecific rat antiserum against the recombinant protein reacted with it (12), suggesting that the protein is immunogenic but is expressed at very low levels during culture in vitro and during infection. Similarly, the level of expression of MG186 (another MBOVPG45_0310 orthologue) in M. genitalium cells was found to be low, and the protein was found to be poorly recognized by antibodies in sera from infected patients (14). However, in another study, antibodies against MAG_5040 (another MBOVPG45_0310 orthologue) could be detected for up to 9 months in sheep naturally infected with M. agalactiae (16), even though the levels of expression of protein were very low in vitro (46). The low level of expression of these proteins containing the TNASE_3 domain and the results of the study reported here clearly indicate that nucleases containing the EEP domain are comparatively more potent in most mycoplasmas. The single band of nuclease activity detected in the Δ0215 mutant at a molecular mass of approximately 40 kDa in gels containing denatured DNA may have originated from the MBOVPG45_0310 gene, the product of which has a predicted molecular mass of 41.56 kDa, but the much greater activity of the products of the MBOVPG45_0215 gene precluded unambiguous identification of this band in the Δ0310 mutant. It is also possible that this band contains a cleavage product derived the MBOVPG45_0690 gene, which contains a 5′ nucleotidase domain.
The ability to create genetically modified organisms by eliminating virulence genes is considered a powerful approach for the development of effective vaccines. Although the Δ0215 mutant was able to grow in vitro in mycoplasma medium, which contains high concentrations of nucleic acid precursors, it may have a limited capacity to generate nucleic acid precursors in vivo and may have potential as an attenuated vaccine. During infection, pathogens usually have a limited capacity to obtain these nucleic acid precursors. Yersinia pestis has been rendered avirulent by mutations blocking the purine biosynthesis pathway, even though the mutants could grow well in vitro (47). As mycoplasmas are likely to be dependent on intrinsic nuclease activity to generate nucleic acid precursors, disruption of nuclease activity might be predicted to render them avirulent in vivo.
This is the first study to demonstrate the quantitative role of different membrane nucleases in a mycoplasma. The detection of the product of MBOVPG45_0215 in the Triton X-114 fraction of M. bovis cell lysates, its cell surface exposure, and its predicted signal peptide suggest that it is a lipoprotein nuclease, and inactivation of the mnuA gene that encodes it was shown to abolish most cellular nuclease activity of M. bovis.
ACKNOWLEDGMENT
S.S. was supported by an AusAid scholarship (now Australia Award) provided by the Australian Department of Foreign Affairs and Trade.
REFERENCES
- 1.Li Y, Zheng H, Liu Y, Jiang Y, Xin J, Chen W, Song Z. 2011. The complete genome sequence of Mycoplasma bovis strain Hubei-1. PLoS One 6:e20999. doi: 10.1371/journal.pone.0020999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Qi J, Guo A, Cui P, Chen Y, Mustafa R, Ba X, Hu C, Bai Z, Chen X, Shi L, Chen H. 2012. Comparative geno-plasticity analysis of Mycoplasma bovis HB0801 (Chinese isolate). PLoS One 7:e38239. doi: 10.1371/journal.pone.0038239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wise KS, Calcutt MJ, Foecking MF, Roske K, Madupu R, Methe BA. 2011. Complete genome sequence of Mycoplasma bovis type strain PG45 (ATCC 25523). Infect Immun 79:982–983. doi: 10.1128/IAI.00726-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Behrens A, Heller M, Kirchhoff H, Yogev D, Rosengarten R. 1994. A family of phase- and size-variant membrane surface lipoprotein antigens (Vsps) of Mycoplasma bovis. Infect Immun 62:5075–5084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lysnyansky I, Ron Y, Yogev D. 2001. Juxtaposition of an active promoter to vsp genes via site-specific DNA inversions generates antigenic variation in Mycoplasma bovis. J Bacteriol 183:5698–5708. doi: 10.1128/JB.183.19.5698-5708.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lysnyansky I, Sachse K, Rosenbusch R, Levisohn S, Yogev D. 1999. The vsp locus of Mycoplasma bovis: gene organization and structural features. J Bacteriol 181:5734–5741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ron Y, Flitman-Tene R, Dybvig K, Yogev D. 2002. Identification and characterization of a site-specific tyrosine recombinase within the variable loci of Mycoplasma bovis, Mycoplasma pulmonis and Mycoplasma agalactiae. Gene 292:205–211. doi: 10.1016/S0378-1119(02)00679-0. [DOI] [PubMed] [Google Scholar]
- 8.Paulsen IT, Nguyen L, Sliwinski MK, Rabus R, Saier MH Jr. 2000. Microbial genome analyses: comparative transport capabilities in eighteen prokaryotes. J Mol Biol 301:75–100. doi: 10.1006/jmbi.2000.3961. [DOI] [PubMed] [Google Scholar]
- 9.Minion FC, Goguen JD. 1986. Identification and preliminary characterization of external membrane-bound nuclease activities in Mycoplasma pulmonis. Infect Immun 51:352–354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Minion FC, Jarvill-Taylor KJ, Billings DE, Tigges E. 1993. Membrane-associated nuclease activities in mycoplasmas. J Bacteriol 175:7842–7847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jarvill-Taylor KJ, VanDyk C, Minion FC. 1999. Cloning of mnuA, a membrane nuclease gene of Mycoplasma pulmonis, and analysis of its expression in Escherichia coli. J Bacteriol 181:1853–1860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Schmidt JA, Browning GF, Markham PF. 2007. Mycoplasma hyopneumoniae mhp379 is a Ca2+-dependent, sugar-nonspecific exonuclease exposed on the cell surface. J Bacteriol 189:3414–3424. doi: 10.1128/JB.01835-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Masukagami Y, Tivendale KA, Mardani K, Ben-Barak I, Markham PF, Browning GF. 2013. The Mycoplasma gallisepticum virulence factor lipoprotein MslA is a novel polynucleotide binding protein. Infect Immun 81:3220–3226. doi: 10.1128/IAI.00365-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Li L, Krishnan M, Baseman JB, Kannan TR. 2010. Molecular cloning, expression, and characterization of a Ca2+-dependent, membrane-associated nuclease of Mycoplasma genitalium. J Bacteriol 192:4876–4884. doi: 10.1128/JB.00401-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Somarajan SR, Kannan TR, Baseman JB. 2010. Mycoplasma pneumoniae Mpn133 is a cytotoxic nuclease with a glutamic acid-, lysine- and serine-rich region essential for binding and internalization but not enzymatic activity. Cell Microbiol 12:1821–1831. doi: 10.1111/j.1462-5822.2010.01513.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cacciotto C, Addis MF, Coradduzza E, Carcangiu L, Nuvoli AM, Tore G, Dore GM, Pagnozzi D, Uzzau S, Chessa B, Pittau M, Alberti A. 2013. Mycoplasma agalactiae MAG_5040 is a Mg(2+)-dependent, sugar-nonspecific SNase recognised by the host humoral response during natural infection. PLoS One 8:e57775. doi: 10.1371/journal.pone.0057775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sharma S, Markham PF, Browning GF. 2014. Genes found essential in other mycoplasmas are dispensable in Mycoplasma bovis. PLoS One 9:e97100. doi: 10.1371/journal.pone.0097100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Duffy MF, Noormohammadi AH, Baseggio N, Browning GF, Markham PF. 1998. Polyacrylamide gel-electrophoresis separation of whole-cell proteins, p 279–298. In Miles RJ, Nicholas RAJ (ed), Methods in molecular biology: mycoplasma protocols. Human Press, Totowa, NJ. [DOI] [PubMed] [Google Scholar]
- 19.Duffy MF, Noormohammadi AH, Baseggio N, Browning GF, Markham PF. 1998. Immunological and biochemical characterisation of membrane proteins, p 267–278. In Miles RJ, Nicholas RAJ (ed), Methods in molecular biology: mycoplasma protocols. Human Press, Totowa, NJ. [DOI] [PubMed] [Google Scholar]
- 20.Duffy MF, Walker ID, Browning GF. 1997. The immunoreactive 116 kDa surface protein of Mycoplasma pneumoniae is encoded in an operon. Microbiology 143(Pt 10):3391–3402. doi: 10.1099/00221287-143-10-3391. [DOI] [PubMed] [Google Scholar]
- 21.Panicker IS, Kanci A, Chiu CJ, Veith PD, Glew MD, Browning GF, Markham PF. 2012. A novel transposon construct expressing PhoA with potential for studying protein expression and translocation in Mycoplasma gallisepticum. BMC Microbiol 12:138. doi: 10.1186/1471-2180-12-138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sharma S. 2012. Molecular tools to study pathogenesis of Mycoplasma bovis. School of Veterinary Science, The University of Melbourne, Melbourne, Australia. [Google Scholar]
- 23.Gasteiger E, Hoogland C, Gattiker A, Duvand S, Wilkins MR, Appel RD, Bairoch A. 2005. Protein identification and analysis tools in ExPASy server, p 571–607. In Walker JM. (ed), The proteomics protocols handbook. Humana Press, Totowa, NJ. [Google Scholar]
- 24.Noguchi H, Taniguchi T, Itoh T. 2008. MetaGeneAnnotator: detecting species-specific patterns of ribosomal binding site for precise gene prediction in anonymous prokaryotic and phage genomes. DNA Res 15:387–396. doi: 10.1093/dnares/dsn027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Barre A, de Daruvar A, Blanchard A. 2004. MolliGen, a database dedicated to the comparative genomics of Mollicutes. Nucleic Acids Res 32:D307–D310. doi: 10.1093/nar/gkh114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.de Castro E, Sigrist CJ, Gattiker A, Bulliard V, Langendijk-Genevaux PS, Gasteiger E, Bairoch A, Hulo N. 2006. ScanProsite: detection of PROSITE signature matches and ProRule-associated functional and structural residues in proteins. Nucleic Acids Res 34:W362–W365. doi: 10.1093/nar/gkl124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Marchler-Bauer A, Lu S, Anderson JB, Chitsaz F, Derbyshire MK, DeWeese-Scott C, Fong JH, Geer LY, Geer RC, Gonzales NR, Gwadz M, Hurwitz DI, Jackson JD, Ke Z, Lanczycki CJ, Lu F, Marchler GH, Mullokandov M, Omelchenko MV, Robertson CL, Song JS, Thanki N, Yamashita RA, Zhang D, Zhang N, Zheng C, Bryant SH. 2011. CDD: a conserved domain database for the functional annotation of proteins. Nucleic Acids Res 39:D225–D229. doi: 10.1093/nar/gkq1189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yu NY, Wagner JR, Laird MR, Melli G, Rey S, Lo R, Dao P, Sahinalp SC, Ester M, Foster LJ, Brinkman FS. 2010. PSORTb 3.0: improved protein subcellular localization prediction with refined localization subcategories and predictive capabilities for all prokaryotes. Bioinformatics 26:1608–1615. doi: 10.1093/bioinformatics/btq249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Browning GF, Marenda MS, Noormohammadi AH, Markham PF. 2011. The central role of lipoproteins in the pathogenesis of mycoplasmoses. Vet Microbiol 153:44–50. doi: 10.1016/j.vetmic.2011.05.031. [DOI] [PubMed] [Google Scholar]
- 30.Hallamaa KM, Browning GF, Tang SL. 2006. Lipoprotein multigene families in Mycoplasma pneumoniae. J Bacteriol 188:5393–5399. doi: 10.1128/JB.01819-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Marenda M. 2014. Genomic mosaics. In Browning GF, Citti C (ed), Mollicutes: molecular biology and pathogenesis. Caister Academic Press, Norfolk, United Kingdom. [Google Scholar]
- 32.Johnson S, Pitcher D. 2000. Distribution of ecto 5′-nucleotidase on Mycoplasma species associated with arthritis. FEMS Microbiol Lett 192:59–65. doi: 10.1016/S0378-1097(00)00409-2. [DOI] [PubMed] [Google Scholar]
- 33.Taniuchi H, Anfinsen CB, Sodja A. 1967. The amino acid sequence of an extracellular nuclease of Staphylococcus aureus. 3. Complete amino acid sequence. J Biol Chem 242:4752–4758. [PubMed] [Google Scholar]
- 34.Gueroult M, Picot D, Abi-Ghanem J, Hartmann B, Baaden M. 2010. How cations can assist DNase I in DNA binding and hydrolysis. PLoS Comput Biol 6:e1001000. doi: 10.1371/journal.pcbi.1001000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Whisstock JC, Wiradjaja F, Waters JE, Gurung R. 2002. The structure and function of catalytic domains within inositol polyphosphate 5-phosphatases. IUBMB Life 53:15–23. doi: 10.1080/15216540210814. [DOI] [PubMed] [Google Scholar]
- 36.Hynes TR, Fox RO. 1991. The crystal structure of staphylococcal nuclease refined at 1.7 Å resolution. Proteins 10:92–105. doi: 10.1002/prot.340100203. [DOI] [PubMed] [Google Scholar]
- 37.Razin S, Knyszynski L, Lifshitz Y. 1964. Nucleases of mycoplasmas. J Gen Microbiol 36:323–331. doi: 10.1099/00221287-36-2-323. [DOI] [PubMed] [Google Scholar]
- 38.Bendjennat M, Blanchard A, Loutfi M, Montagnier L, Bahraoui E. 1999. Role of Mycoplasma penetrans endonuclease P40 as a potential pathogenic determinant. Infect Immun 67:4456–4462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Paddenberg R, Wulf S, Weber A, Heimann P, Beck LA, Mannherz HG. 1996. Internucleosomal DNA fragmentation in cultured cells under conditions reported to induce apoptosis may be caused by mycoplasma endonucleases. Eur J Cell Biol 71:105–119. [PubMed] [Google Scholar]
- 40.Diedershagen M, Overbeck S, Arlt S, Plumakers B, Lintges M, Rink L. 2007. Mycoplasma arthritidis-derived superantigen (MAM) displays DNase activity. FEMS Immunol Med Microbiol 49:266–271. doi: 10.1111/j.1574-695X.2006.00189.x. [DOI] [PubMed] [Google Scholar]
- 41.Sasaki Y, Ishikawa J, Yamashita A, Oshima K, Kenri T, Furuya K, Yoshino C, Horino A, Shiba T, Sasaki T, Hattori M. 2002. The complete genomic sequence of Mycoplasma penetrans, an intracellular bacterial pathogen in humans. Nucleic Acids Res 30:5293–5300. doi: 10.1093/nar/gkf667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.McIvor RS, Kenny GE. 1978. Differences in incorporation of nucleic acid bases and nucleosides by various Mycoplasma and Acholeplasma species. J Bacteriol 135:483–489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Pollack JD. 2002. The necessity of combining genomic and enzymatic data to infer metabolic function and pathways in the smallest bacteria: amino acid, purine and pyrimidine metabolism in Mollicutes. Front Biosci 7:d1762–d1781. doi: 10.2741/pollack. [DOI] [PubMed] [Google Scholar]
- 44.Cordwell SJ, Basseal DJ, Pollack JD, Humphery-Smith I. 1997. Malate/lactate dehydrogenase in mollicutes: evidence for a multienzyme protein. Gene 195:113–120. doi: 10.1016/S0378-1119(97)00063-2. [DOI] [PubMed] [Google Scholar]
- 45.Finch LR, Mitchell A. 1992. Sources of nucleotides, p 211–230. In Maniloff J, McElhaney RN, Finch LR, Baseman JB (ed), Mycoplasmas: molecular biology and pathogenesis. American Society for Microbiology, Washington, DC. [Google Scholar]
- 46.Cacciotto C, Addis MF, Pagnozzi D, Chessa B, Coradduzza E, Carcangiu L, Uzzau S, Alberti A, Pittau M. 2010. The liposoluble proteome of Mycoplasma agalactiae: an insight into the minimal protein complement of a bacterial membrane. BMC Microbiol 10:225. doi: 10.1186/1471-2180-10-225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Brubaker RR. 1972. The genus Yersinia: biochemistry and genetics of virulence. Curr Top Microbiol Immunol 57:111–158. doi: 10.1007/978-3-642-65297-4_4. [DOI] [PubMed] [Google Scholar]