ABSTRACT
Haloarcula japonica, an extremely halophilic archaeon that requires high concentrations of NaCl for growth, accumulates the C50 carotenoid bacterioruberin (BR). By homology analysis, a gene cluster, including c0507, c0506, and c0505, was found and predicted to be involved in the synthesis of bacterioruberin. To elucidate the function of the encoded enzymes, we constructed Ha. japonica mutants of these genes and analyzed carotenoids produced by the mutants. Our research showed that c0507, c0506, and c0505 encoded a carotenoid 3,4-desaturase (CrtD), a bifunctional lycopene elongase and 1,2-hydratase (LyeJ), and a C50 carotenoid 2″,3″-hydratase (CruF), respectively. The above three carotenoid biosynthetic enzymes catalyze the reactions that convert lycopene to bacterioruberin in Ha. japonica. This is the first identification of functional CrtD and CruF in archaea and elucidation of the complete biosynthetic pathway of bacterioruberin from lycopene.
IMPORTANCE Haloarcula japonica, an extremely halophilic archaeon, accumulates the C50 carotenoid bacterioruberin (BR). In this study, we have identified three BR biosynthetic enzymes and have elucidated their functions. Among them, two enzymes were found in an archaeon for the first time. Our results revealed the biosynthetic pathway responsible for production of BR in Ha. japonica and provide a basis for investigating carotenoid biosynthetic pathways in other extremely halophilic archaea. Elucidation of the carotenoid biosynthetic pathway in Ha. japonica may also prove useful for producing the C50 carotenoid BR efficiently by employing genetically modified haloarchaeal strains.
INTRODUCTION
Carotenoids are natural pigments synthesized by bacteria, archaea, algae, fungi, and plants (1). They are involved in photosynthesis as accessory pigments (2), functioning as antioxidants (3, 4), light protection pigments (5, 6), and membrane stabilizers (7). Thus far, more than 750 carotenoids have been identified and classified as C30, C40, and C50 carotenoids, depending on the number of carbons in their carotene backbones (1). Most of these compounds are based on the symmetric C40 backbone, phytoene, which is formed by condensation of two molecules of geranylgeranyl pyrophosphate (GGPP; C20PP) (8). Only a small number of C30 carotenoids, which arise from the fusion of two molecules of farnesyl pyrophosphate (FPP; C15PP), and even fewer C50 carotenoids, which are derived from the C40 structure by the addition of two 5-carbon (C5) isoprene units, have been discovered to date (1).
In nature, several restricted groups of bacteria, including species belonging to the bacterial genera Corynebacterium, Dietzia, and Micrococcus, and extremely halophilic archaea, such as Halobacterium salinarum, show accumulation of C50 carotenoids (9, 10). To date, only three biosynthetic pathways of cyclic C50 carotenoids, the ε-cyclic C50 carotenoid decaprenoxanthin in Corynebacterium glutamicum (11–13), the β-cyclic C50 carotenoid 2,2′-bis-(4-hydroxy-3-methybut-2-enyl)-β,β-carotene in Dietzia sp. strain CQ4 (14), and the γ-cyclic C50 carotenoid sarcinaxanthin in Micrococcus luteus NCTC2665 (15), have been described, based on their structures. The acyclic C50 carotenoid bacterioruberin (BR) is known to be produced in some extremely halophilic archaea. BR of Haloferax volcanii has been isolated and identified by chromatographic (thin-layer chromatography [TLC] and high-performance liquid chromatography [HPLC]), spectroscopic (mass spectrometry [MS], nuclear magnetic resonance [NMR] analysis, and circular dichroism [CD] spectroscopy), and chemical (silylation and methylation) methods (16). Haloferax mediterranei can also accumulate BR, and a two-stage culture method has been applied to increase production of C50 carotenoids in the laboratory (17). In addition to studies on the antioxidant properties of BR (18), the regulating mechanism of BR biosynthesis in Hb. salinarum has been investigated (19). Hb. salinarum is also known to produce the bacteriorhodopsin light-induced proton pump, comprising retinal and bacterioopsin (20). The carotenoids and retinal biosynthetic pathways of Hb. salinarum have been briefly described previously (10). That report proposed that retinal and BR are synthesized from a common precursor, lycopene, which is generated from phytoene, as occurs in C40 carotenoid biosynthetic pathways. Although the enzyme Lye, which catalyzes the committed step in BR biosynthesis, has been described in Hb. salinarum, the details of the BR biosynthetic pathway remain unclear.
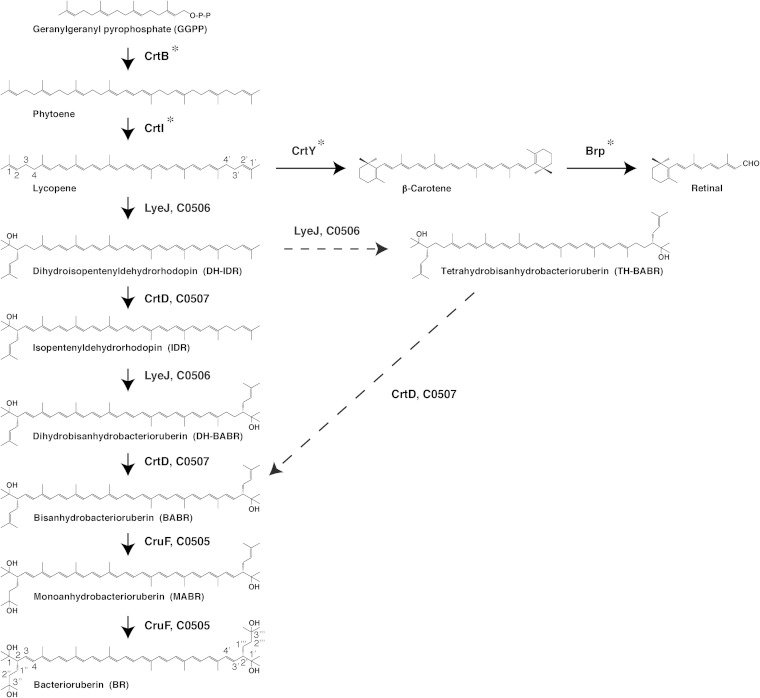
Haloarcula japonica is a predominantly triangular, disc-shaped, extremely halophilic archaeon that requires high concentrations of NaCl for growth (21). In our previous study, we found that Ha. japonica can produce isopentenyldehydrorhodopin (IDR; C45), bisanhydrobacterioruberin (BABR; C50), and monoanhydrobacterioruberin (MABR; C50) as intermediates and that BR is produced as the final product (22). In addition, it has been suggested that Ha. japonica also produces the retinal proteins cruxrhodopsin (23) and cruxhalorhodopsin (24). Thus, retinal could also be synthesized from carotenoids in Ha. japonica. Based on the results of our previous study and the proposed biosynthetic pathways of carotenoids in Hb. salinarum (10), the carotenoid pathways and retinal biosynthetic pathways in Ha. japonica are suggested to be as shown in Fig. 1. Phytoene synthase (CrtB) condenses two GGPP molecules to yield the colorless carotenoid phytoene. Lycopene is generated from phytoene via a series of desaturation reactions that are catalyzed by phytoene desaturase (CrtI). Subsequently, the steps are divided into the retinal biosynthetic pathway and the BR biosynthetic pathway. In retinal synthesis, the cyclization of lycopene to β-carotene is catalyzed by lycopene β-cyclase (CrtY), and β-carotene is then cleaved to form a retinal (C20) by β-carotene dioxygenase (Brp) cleavage. In the BR biosynthetic pathway, lycopene is used as a precursor and is converted to BR by introduction of two C5 isoprene units, two double bonds, and four hydroxyl groups into the lycopene. All of the related enzymes and the complete pathway for biosynthesis of BR from lycopene remain unknown.
FIG 1.
Proposed main steps in the carotenoid and retinal biosynthetic pathways of Haloarcula japonica. Solid arrows signify the main carotenoid biosynthetic steps. The dashed arrows indicate other possible carotenoid biosynthetic steps. Asterisks indicate the unidentified carotenoid biosynthetic enzymes.
In this study, we identified and characterized three main BR biosynthetic enzymes in Ha. japonica, namely, C0506, C0507, and C0505. They function as a bifunctional lycopene elongase and 1,2-hydratase (LyeJ), a carotenoid 3,4-desaturase (CrtD), and a C50 carotenoid 2″,3″-hydratase (CruF), respectively, to generate BR from lycopene. These findings contribute to elucidation of the complete carotenoid biosynthetic pathway in extremely halophilic archaea.
MATERIALS AND METHODS
Microbial strains and growth conditions.
All strains and plasmids used in this study are listed in Table 1. The wild-type and mutant strains of Haloarcula japonica (JCM 7785T) were grown at 37°C in the dark with a complex medium, as described previously (25). The medium was supplemented with 8 μg/ml pravastatin (a gift from Daiichi Sankyo, Tokyo, Japan) instead of mevinoline, when required. Escherichia coli was cultured in LB medium at 37°C (26). Ampicillin (50 μg/ml) was added when required.
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Relevant property or propertiesa | Source or reference |
|---|---|---|
| Strains | ||
| E. coli JM109 | Host for cloning vectors | Laboratory stock |
| E. coli JM110 | Host used for preparing plasmid that is free of Dam or Dcm methylation | Laboratory stock |
| Ha. japonica | Wild type (JCM 7785T) | 21 |
| Ha. japonica Δc0507 | c0507 gene mutant of Ha. japonica | This work |
| Ha. japonica Δc0506 | c0506 gene mutant of Ha. japonica | This work |
| Ha. japonica Δc0505 | c0505 gene mutant of Ha. japonica | This work |
| Ha. japonica Δc0507(pJc0507) | Δc0507 complement with pJc0507 | This work |
| Ha. japonica Δc0506(pJc0506) | Δc0506 complement with pJc0506 | This work |
| Ha. japonica Δc0505(pJc0505) | Δc0505 complement with pJc0505 | This work |
| Plasmids | ||
| pUC119 | E. coli cloning vector; Apr | Laboratory stock |
| pWL102 | E. coli-haloarchaea shuttle vector; Apr Mevr | 29 |
| pDrHj2b | E. coli-haloarchaea shuttle vector; Apr Mevr | Laboratory stock |
| pWL102-Δc0507 | pWL102 derivative containing disrupted fragment of c0507 gene | This work |
| pDrHj2-Δc0506 | pDrHj2 derivative containing disrupted fragment of c0506 gene | This work |
| pDrHj2-Δc0505 | pDrHj2 derivative containing disrupted fragment of c0505 gene | This work |
| pJFZ33 | pWL102 derivative carrying Ha. japonica csg promoter and ftsZ2 gene; Apr Mevr | 27 |
| pJc0507 | pJFZ33 derivative in which ftsZ2 is replaced by the c0507 gene | This work |
| pJc0506 | pJFZ33 derivative in which ftsZ2 is replaced by the c0506 gene | This work |
| pJc0505 | pJFZ33 derivative in which ftsZ2 is replaced by the c0505 gene | This work |
Apr, ampicillin resistance; Mevr, pravastatin resistance.
The pDrHj2 plasmid was constructed as follows. The sequence between SacI and KpnI in pUC119 was replaced with the mevinoline resistance gene, and the sequence between HindIII and XbaI was replaced with the gene of a halophilic β-galactosidase (bgaH) under the control of csg promoter. The bgaH gene was obtained from Haloferax alicantei and was oriented in the direction opposite that in which the mevinoline resistance gene was oriented. The csg promoter is from Haloarcula japonica. Ha. japonica has a large amount of a glycoprotein (cell surface glycoprotein [CSG]) on the cell surface, suggesting that the csg promoter is powerful.
Isolation of genomic DNA and total RNA from Ha. japonica.
Ha. japonica genomic DNA was isolated as described previously (27). Total RNA of Ha. japonica was prepared using Sepasol RNA I (Nacalai Tesque, Kyoto, Japan) according to the manufacturer's instructions. The resulting total RNA was further treated with DNase I (GE Healthcare, Bucks, United Kingdom) to remove trace amounts of contaminating genomic DNA.
RT-PCR and PCR.
Oligonucleotide primers for reverse transcription-PCR (RT-PCR) and PCR were purchased from Operon Biotechnologies (Tokyo, Japan). The oligonucleotide primers used in this study are listed in Table 2. RT-PCR was performed under the following conditions. Total RNA (1.0 μg) was reverse transcribed at 45°C for 60 min in 20 μl of the reaction buffer, which contained 20 pmol of each primer (c0507-A, c0506-A, or c0505-A), 0.3 mM (each) deoxynucleotide triphosphate, 2.5 mM manganese (II) acetate, 20 U RNase inhibitor (Toyobo, Osaka, Japan), and 5 U rTth DNA polymerase (Toyobo). The cDNA generated was then amplified by PCR. PCR was carried out using KOD-Plus-Dash or KOD-Dash (Toyobo) DNA polymerase, according to the manufacturer's instructions. Primer set c0507-S/c0507-A, primer set c0506-S/c0506-A, or primer set c0505-S/c0505-A was used to confirm the transcription of c0507, c0506, or c0505 in the wild-type and mutant strains of Ha. japonica.
TABLE 2.
Sequences of the primers used in this study
| Primer | Sequencea | Usage |
|---|---|---|
| c0507-S | 5′-GGTTGGCCTCCAGCTCATTG-3′ | To confirm the transcription of target genes by RT-PCR |
| c0507-A | 5′-CTCGTGGTCCGGCAGGAGTTC-3′ | |
| c0506-S | 5′-TCGCCCTGTTTCTGTACTTCAC-3′ | |
| c0506-A | 5′-GATATCTGGAATCGCCGAGAAGG-3′ | |
| c0505-S | 5′-CACCGAGAACCGGTTTACCATC-3′ | |
| c0505-A | 5′-ACTGCGGCATCTCGTAGATCC-3′ | |
| c0507-S1 | 5′-ATGTGGCCACGAATGCCATAC-3′ | To amplify the target genes by PCR for the construction of mutants |
| c0507-A1 | 5′-GGTCTGTCGGGAGTTCTGGC-3′ | |
| c0506-S1 | 5′-GGACATCGCCTGAGTAATGCC-3′ | |
| c0506-A1 | 5′-GGTCTTTGCCGCTACCCATAC-3′ | |
| c0506-S3 | 5′-TCTGGGCGATGGGGATGCAC-3′ | |
| c0506-A3 | 5′-TTTCGGATTGTGCTCGTCGATATCGG-3′ | |
| c0505-S1 | 5′-GCTGTATGGGTAGCGGCAAAG-3′ | |
| c0505-A1 | 5′-GAGCTGGCTGATTCCAAACGG-3′ | |
| c0505-S3 | 5′-TGGATCCTGGAGCCGTAGCTATC-3′ | |
| c0505-A3 | 5′-AGCTCGATGCCGTAGGAGTACAG-3′ | |
| c0507-S4 | 5′-GAATTCCATATGAGTGACTTGTCCGGTG-3′ | To amplify the intact target genes by PCR for the complementation study |
| c0507-A4 | 5′-GGGGATCCTCATTAGTGGTGGTGGTGGTGGTGGGCGATGTCCTCGATGAGC-3′ | |
| c0506-S4 | 5′-GAATTCCATATGCCAGAACTCCCGACAG-3′ | |
| c0506-A4 | 5′-GGGGATCCTCATTAGTGGTGGTGGTGGTGGTGCCCATACAGCATCACCCAC-3′ | |
| c0505-S4 | 5′-GAATTCCATATGGGTAGCGGCAAAGACC-3′ | |
| c0505-A4 | 5′-GGCCATGGTCATTAGTGGTGGTGGTGGTGGTGCCGCCAGACGGCCCGACCG-3′ |
The introduced NdeI, BamHI, and NcoI restriction sites are underlined.
Construction of c0507, c0506, and c0505 mutant strains.
Mutants with deletions and frameshift mutations were constructed by homologous recombination (28). The plasmid pWL102-Δc0507, containing a disrupted c0507 gene, was constructed as follows. The targeted gene, c0507, was amplified from Ha. japonica using the primer set c0507-S1/c0507-A1. The amplified PCR fragment was ligated into the SmaI site of pUC119, yielding plasmid pUC119-c0507. A 652-bp DNA fragment, ranging from base 378 to base 1029 of c0507, was excised from pUC119-c0507 by Csp45I and EcoRV digestion, followed by blunting and self-ligation, to yield plasmid pUC119-Δc0507. Plasmid pUC119-Δc0507 was digested with EcoRI and HindIII, releasing a disrupted c0507 gene. Plasmid pWL102 (29) was also digested with EcoRI and HindIII to eliminate the Hf. volcanii ori and was then ligated to the disrupted c0507 gene, yielding plasmid pWL102-Δc0507.
Plasmids pDrHj2-Δc0506, containing a disrupted c0506 gene, and pDrHj2-Δc0505, containing a disrupted c0505 gene, were constructed as follows. The targeted genes, c0506 and c0505, were amplified from Ha. japonica using primer set c0506-S1/c0506-A1 and primer set c0505-S1/c0505-A1, respectively. Amplified PCR fragments were ligated into the SmaI site of pUC119, yielding plasmid pUC119-c0506 and pUC119-c0505, respectively. A 292-bp DNA fragment, ranging from base 271 to base 562 of c0506, and a 221-bp DNA fragment, ranging from base 318 to base 538 of c0505, were removed from the pUC119-c0506 and pUC119-c0505 plasmids by Exsite PCR, using primer set c0506-S3/c0506-A3 and primer set c0505-S3/c0505-A3, respectively. This was followed by self-ligation, yielding plasmids pUC119-Δc0506 and pUC119-Δc0505, respectively. Plasmid pUC119-Δc0506 was digested with EcoRI. After blunting, it was further digested with BamHI, and the fragment containing the disrupted c0506 gene was ligated to pDrHj2 (containing only the ColE1 ori), which had been digested with SmaI and BamHI, to yield plasmid pDrHj2-Δc0506. A fragment containing the disrupted c0505 gene was amplified from pUC119-Δc0505 using the c0505-S1/c0505-A1 primer set, and the fragment was ligated to pDrHj2, which had been digested with SmaI, to yield plasmid pDrHj2-Δc0505.
Plasmids pWL102-Δc0507, pDrHj2-Δc0506, and pDrHj2-Δc0505 were subjected to passage through E. coli JM110 to avoid the restriction barrier formation of extremely halophilic archaea (30). Transformation of Ha. japonica was performed using the polyethylene glycol method (27). Transformants were plated onto agar plates containing pravastatin. Pravastatin-resistant colonies were cultured in liquid medium without pravastatin for 96 h and plated onto agar plates without pravastatin in order to isolate recombinants in which the targeted gene on the genome was replaced with the corresponding disrupted gene. Gene disruption was confirmed by PCR analysis.
Complementation of mutant strains.
Three plasmids, pJc0507, pJc0506, and pJc0505, were constructed as follows. The targeted genes, c0507, c0506, and c0505, were amplified from Ha. japonica using primer set c0507-S4/c0507-A4, primer set c0506-S4/c0506-A4, and primer set c0505-S4/c0505-A4, respectively. Amplified PCR fragments were ligated into the SmaI site of pUC119, yielding plasmids pUCc0507, pUCc0506, and pUCc0505, respectively. Plasmids pUCc0507 and pUCc0506 were digested with NdeI and BamHI. The sequences containing targeted genes were ligated to pJFZ33 (a recombinant plasmid in which the Ha. japonica csg promoter sequence, the NdeI restriction site, the Ha. japonica ftsZ2 gene sequence, and the BamHI and NcoI restriction sites were inserted into the E. coli-haloarchaea pWL102 shuttle vector), which had also been digested with NdeI and BamHI, to yield plasmids pJc0507 and pJc0506, respectively. Plasmid pUCc0505 was digested with NdeI and NcoI. The sequence containing c0505 was ligated to pJFZ33, which had also been digested with NdeI and NcoI, to yield a pJc0505 plasmid.
Plasmids pJc0507, pJc0506, and pJc0505 were subjected to passage through E. coli JM110 and then transformed into the corresponding mutant strains as described above.
Total carotenoid extraction from Ha. japonica.
The wild-type and mutant strains of Ha. japonica were precultured at 37°C. Four milliliters of preinoculum was transferred to a 2-liter Erlenmeyer flask containing 400 ml of liquid medium and cultured for 240 h at 37°C in the dark. Cells were harvested by centrifugation (4°C, 4,400 × g, 20 min). Carotenoids were extracted from Ha. japonica essentially as described previously (22) with the following modifications. Cells were suspended in acetone/methanol (7:2 [vol/vol]) which contained 0.1% (wt/vol) 2,6-di-t-butyl-p-cresol antioxidant to avoid the destruction of carotenoids. After cell disruption by sonication, samples were centrifuged (4°C, 840 × g, 2 min) to obtain the supernatant. The pellets were reextracted until all visible pigments were retained in the liquid phase. Finally, the supernatants were collected and evaporated to dryness in a vacuum. The dried carotenoid extract was used for monitoring the HPLC analysis of total carotenoids and mass spectrometric analysis of the purified carotenoids.
HPLC analysis of total carotenoids.
The dried carotenoid extract was dissolved in a small volume of chloroform-methanol (3:1) and analyzed using an HPLC system (SCL-10A; Shimadzu, Kyoto, Japan) equipped with a μBondapak C18 column (Waters, Milford, MA) (3.9 by 300 mm). The eluent was methanol-water (9:1) for the first 10 min, followed by 100% methanol, at a flow rate of 1.5 ml/min (31). Absorption spectra of the carotenoids were recorded with a photodiode-array detector (SPD-M20A; Shimadzu) attached to the HPLC apparatus.
Mass spectrometric analysis of purified carotenoid.
The dried carotenoid extract was dissolved in 2 ml of acetone-hexane (1:1) and was then applied onto a column of DEAE-Toyopearl 650M (Tosoh, Tokyo, Japan) to separate the polar lipids from the carotenoids (32). The column was washed with acetone-hexane (1:1), and the colored fractions were collected. After evaporation to dryness, the collected fractions were dissolved in 1 ml chloroform-methanol (3:1) and spotted onto high-performance thin-layer chromatography (HPTLC) plates (HPTLC silica gel 60 with concentrating zone; Merck Millipore, Darmstadt, Germany). The HPTLC plates were developed with petroleum ether-acetone (7:1) in the dark, and each separated fraction of carotenoids was collected. The collected silica powder was suspended with 1 ml chloroform-methanol (3:1) and then filtered through a polytetrafluoroethylene (PTFE) 0.2-μm-pore-size filter (Lab Lab, Tokyo, Japan). Carotenoids in each fraction were further separated using the HPLC system described above, with the following modification. Elution was performed with 100% methanol at a flow rate of 1.5 ml/min. Carotenoids in peaks were collected, and their relative molecular masses were measured using an MStation JMS-700 mass spectrometry system (Jeol, Tokyo, Japan) in the fast-atom-bombardment (FAB) mode, with m-nitrobenzyl alcohol as a matrix.
Nucleotide sequence accession numbers.
The DNA sequence data for c0507, c0506, and c0505 were deposited in the DNA Data Bank of Japan (DDBJ), the European Molecular Biology Laboratory (EMBL), and the GenBank nucleotide sequence database. Their GenBank accession numbers are LC008542, LC008543, and LC008544, respectively.
RESULTS
Identification of candidate genes.
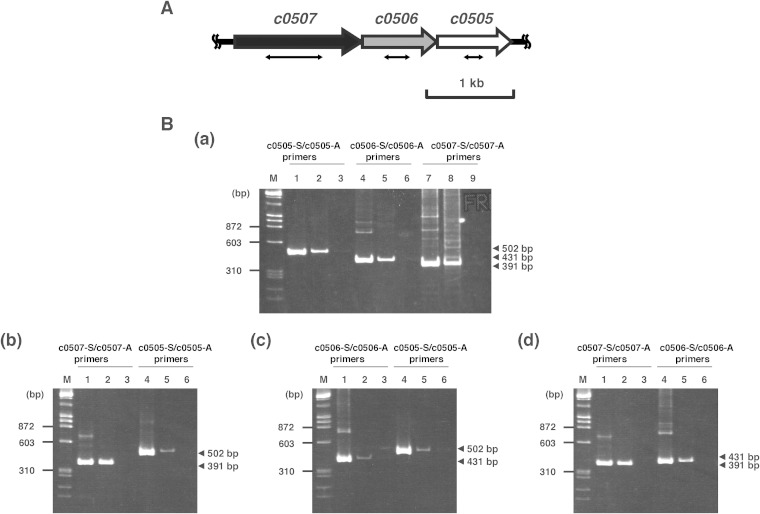
Recently, the draft genome sequence of Ha. japonica was determined (33). By homology analysis, some open reading frames (ORFs) (c0507, c0506, and c0505) were predicted to be candidate genes encoding carotenoid biosynthetic enzymes. C0506 had 31% amino acid sequence identity to lycopene elongase (CrtEb) from Corynebacterium glutamicum and had 60% amino acid sequence identity to a lycopene elongase homolog (Lye) from Hb. salinarum (10). C0507 had 29% amino acid sequence identity to CrtI from Pantoea ananatis (34). Furthermore, C0507 also had 26% and 24% amino acid sequence identity to CrtD from Rhodobacter capsulatus (35) and Deinococcus radiodurans R1 (36), respectively. C0505, the homolog of CruF, had 30% and 31% amino acid identity to those of Synechococcus sp. strain PCC 7002 (37) and Deinococcus radiodurans R1 (38), respectively. In addition, c0507, c0506, and c0505, in that order, cluster on the genome of Ha. japonica (Fig. 2A), and RT-PCR analysis revealed that those genes were cotranscribed. Since carotenoid synthesis-related genes in some bacteria are assembled in clusters or in neighborhoods (34, 39), these genes were predicted to be involved in the carotenoid synthesis of Ha. japonica.
FIG 2.
The c0507-c0506-c0505 gene cluster and agarose gel electrophoresis of the RT-PCR products. (A) The c0507-c0506-c0505 gene cluster. The double-headed arrows show the deleted parts in the corresponding mutants: a 652-bp fragment, ranging from base 378 to base 1029 of c0507, was removed in the Δc0507 strain; a 292-bp fragment, ranging from base 271 to base 562 of c0506, was removed in the Δc0506 strain; and a 221-bp fragment, ranging from base 318 to base 538 of c0505, was removed in the Δc0505 strain. (B) Agarose gel electrophoresis of the RT-PCR product. (Panel a) The genomic DNA and total RNA were isolated from wild-type Ha. japonica. (Panel b) The genomic DNA and total RNA were isolated from the Δc0506 strain. (Panel c) The genomic DNA and total RNA were isolated from the Δc0507 strain. (Panel d) The genomic DNA and total RNA were isolated from the Δc0505 strain. Lanes 1, 4, and 7, the positive-control reaction product using genomic DNA as the template; lanes 2, 5, and 8, the RT-PCR product; lanes 3, 6, and 9, the negative-control reaction product with total RNA not subjected to reverse transcription.
Analysis of c0506 and c0507.
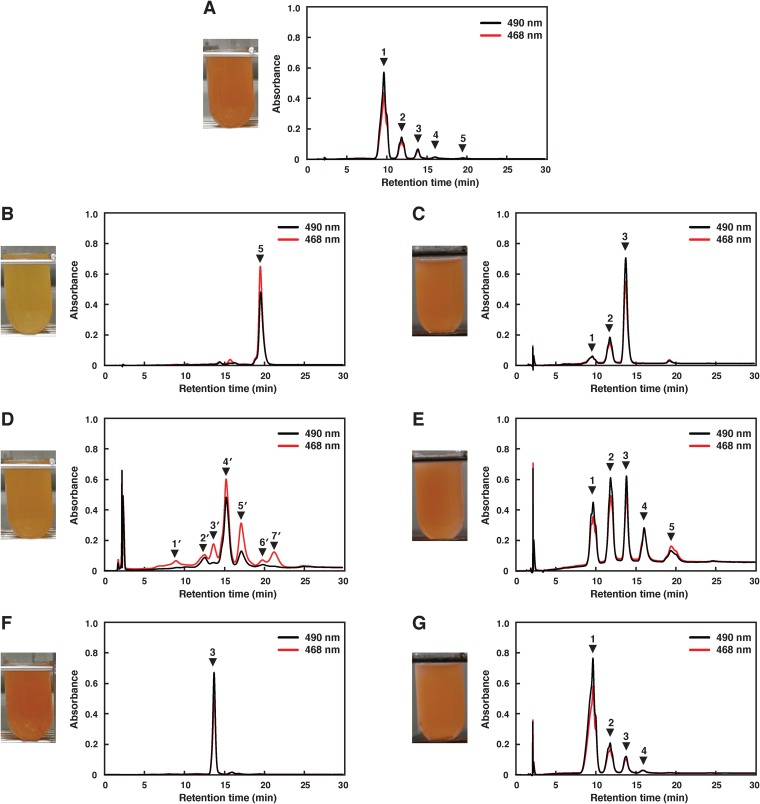
To characterize c0506, a mutant of this gene was constructed by homologous recombination and designated strain Δc0506. The Δc0506 strain showed the same growth rate as wild-type Ha. japonica. The transcription of c0507 and c0505 in the Δc0506 strain were confirmed by RT-PCR (Fig. 2B, panel b). The disruption of c0506 did not affect the expression of the upstream c0507 gene, although the transcription level of the downstream c0505 gene was reduced to less than half that in wild-type Ha. japonica. The cell suspension of the Δc0506 strain was yellow in color, while the cell suspension of wild-type Ha. japonica was red in color (Fig. 3). Thus, the carotenoids produced were likely to be different. The carotenoids produced by the Δc0506 strain were analyzed by HPLC and compared with those of the wild-type Ha. japonica. Five peaks were detected in wild-type Ha. japonica and had been identified as BR (Fig. 3A, peak 1), MABR (peak 2), BABR (peak 3), IDR (peak 4), and lycopene (peak 5), respectively, in our previous study (Fig. 3 and Table 3) (22). In contrast, only one carotenoid (Fig. 3B, peak 5) was detected in the Δc0506 strain. This carotenoid exhibited a retention time of 19.4 min and had λmax values of 443, 469, and 500 nm, which corresponded to an acyclic carotenoid having 11 conjugated double bonds (n = 11). This was similar to the results seen with peak 5 of wild-type Ha. japonica (representing lycopene, with a retention time of 19.4 min and λmax values of 442, 468, and 499 nm [n = 11]). Mass spectrometric analysis showed that peak 5 of the Δc0506 strain had an m/z value of 536. Based on the retention time, absorbance spectrum, and mass spectrometric results, peak 5 of the Δc0506 strain was identified as representing lycopene.
FIG 3.
Cell suspension color of the wild type and of the mutants and their genetically modified Ha. japonica strains, and HPLC elution profiles of carotenoids extracted from them. (A) Wild type. (B) Δc0506 strain. (C) Δc0506(pJc0506) strain. (D) Δc0507 strain. (E) Δc0507(pJc0507) strain. (F) Δc0505 strain. (G) Δc0505(pJc0505) strain. Peak 1, bacterioruberin (BR); peak 2, monoanhydrobacterioruberin (MABR); peak 3, bisanhydrobacterioruberin (BABR); peak 4, isopentenyldehydrorhodopin (IDR); peak 5, lycopene; peak 4′, tetrahydrobisanhydrobacterioruberin (TH-BABR); peak 5′, dihydroisopentenyldehydrorhodopin (DH-IDR). Peaks 1′, 2′, 3′, 6′, and 7′ might be derivatives of lycopene precursors. The eluent was methanol-water (9:1 [vol/vol]) for the first 10 min and then 100% methanol (1.5 ml/min).
TABLE 3.
Characteristics of carotenoids extracted from the wild type and the mutants and the corresponding genetically modified Ha. japonica strains
| Strain | Peak no. | Retention time (min) | λmax (nm) in HPLC eluent | Conjugated double bonds (n) | Molecular mass (m/z) |
Carotenoida | |
|---|---|---|---|---|---|---|---|
| Measured | Calculated | ||||||
| Ha. japonica WT | 1 | 9.6 | 469, 492, 525 | 13 | 740 | 740 | BR |
| 2 | 11.8 | 469, 491, 522 | 13 | 722 | 722 | MABR | |
| 3 | 13.8 | 467, 490, 521 | 13 | 704 | 704 | BABR | |
| 4 | 16.0 | 453, 479, 509 | 12 | 620 | 620 | IDR | |
| 5 | 19.4 | 442, 468, 499 | 11 | 536 | 536 | Lycopene | |
| Ha. japonica Δc0506 | 5 | 19.4 | 443, 469, 500 | 11 | 536 | 536 | Lycopene |
| Ha. japonica Δc0507 | 4′ | 14.8 | 441, 466, 498 | 11 | 708 | 708 | TH-BABR |
| 5′ | 16.7 | 440, 465, 495 | 11 | 622 | 622 | DH-IDR | |
| Ha. japonica Δc0505 | 3 | 13.7 | 468, 491, 523 | 13 | 704 | 704 | BABR |
| Ha. japonica Δc0506(pJc0506) | 1 | 9.6 | 469, 491, 523 | 13 | ND | 740 | BR |
| 2 | 11.8 | 468, 491, 523 | 13 | ND | 722 | MABR | |
| 3 | 13.8 | 466, 490, 522 | 13 | ND | 704 | BABR | |
| Ha. japonica Δc0507(pJc0507) | 1 | 9.6 | 468, 492, 524 | 13 | ND | 740 | BR |
| 2 | 11.8 | 469, 491, 522 | 13 | ND | 722 | MABR | |
| 3 | 13.8 | 466, 490, 521 | 13 | ND | 704 | BABR | |
| 4 | 16.0 | 454, 480, 510 | 12 | ND | 620 | IDR | |
| 5 | 19.4 | 443, 468, 499 | 11 | ND | 536 | Lycopene | |
| Ha. japonica Δc0505(pJc0505) | 1 | 9.6 | 468, 492, 525 | 13 | ND | 740 | BR |
| 2 | 11.8 | 467, 492, 522 | 13 | ND | 722 | MABR | |
| 3 | 13.8 | 466, 490, 520 | 13 | ND | 704 | BABR | |
| 4 | 16.0 | 454, 479, 509 | 12 | ND | 620 | IDR | |
BR, bacterioruberin; MABR, monoanhydrobacterioruberin; BABR, bisanhydrobacterioruberin; IDR, isopentenyldehydrorhodopin; TH-BABR, tetrahydrobisanhydrobacterioruberin; DH-IDR, dihydroisopentenyldehydrorhodopin. Carotenoids were analyzed using an HPLC system equipped with a μBondapak C18 column (Waters) (3.9 by 300 mm) and were eluted with methanol-water (9:1 [vol/vol]) for the first 10 min and then with 100% methanol (1.5 ml/min). Absorption spectra of the carotenoids were recorded with a photodiode array detector attached to the HPLC apparatus (Fig. 3). The relative molecular masses of the purified carotenoids were measured using an MStation JMS-700 mass spectrometry system (Jeol) in the FAB mode with m-nitrobenzyl alcohol as a matrix. ND, not determined.
In the Δc0506 strain, no C45 and C50 carotenoids were detected, suggesting that the biosynthetic pathway for carotenoids was discontinued at lycopene. Therefore, we deduced that c0506 of Ha. japonica encodes a functional enzyme that catalyzes the conversion of lycopene to a BR precursor.
There seem to be two types of desaturation reactions in the carotenoid biosynthetic pathway of Ha. japonica. In the first reaction, CrtI converts phytoene into lycopene by the introduction of four double bonds. In the second reaction, CrtD forms double bonds at C-3,4 and C-3′,4′ of the lycopene derivatives. According to homology analysis results, C0507 shows amino acid sequence homology to both CrtI and CrtD. To investigate the function of C0507, a mutant of c0507, designated strain Δc0507, was constructed by homologous recombination. The Δc0507 strain showed the same growth rate as wild-type Ha. japonica. The transcripts of c0506 and c0505 in the Δc0507 strain were confirmed by RT-PCR, although their transcription levels were reduced to less than half those seen in wild-type Ha. japonica (Fig. 2B, panel c). The cell suspension of the Δc0507 strain was orange in color (Fig. 3D).
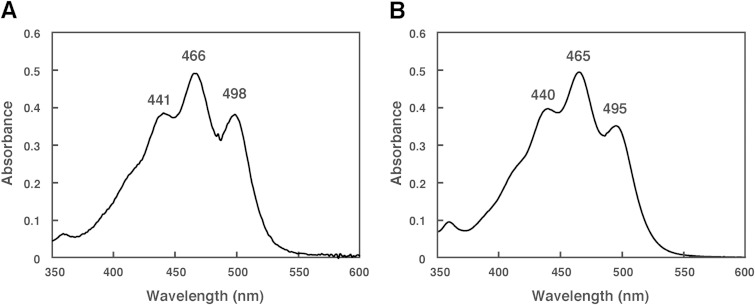
The HPLC elution profile of carotenoids produced by the Δc0507 strain is shown in Fig. 3D. The absorption spectrum indicated that peak 1′, peak 2′, peak 3′, peak 4′, peak 5′, peak 6′, and peak7′ had 9, 10, 9, 11, 11, 7, and 9 conjugated double bonds, respectively. Among these, peak 4′ and peak 5′, representing the main carotenoids in the Δc0507 strain, possessed the most abundant conjugated double bonds, suggesting that peak 4′ and peak 5′ represent the final products or their derivatives. The absorption spectra of peak 4′ and peak 5′ are shown in Fig. 4. Mass spectrometric analysis also showed that peak 4′ and peak 5′ had m/z values of 708 (4 mass units more than BABR) and 622 (2 mass units more than IDR), respectively (Table 3). Based on these data, peak 4′ and peak 5′ were identified as representing TH-BABR and dihydroisopentenyldehydrorhodopin (DH-IDR; C45), respectively. Since phytoene did not accumulate in the Δc0507 strain, the desaturation reactions that convert phytoene into lycopene were not catalyzed by C0507, or the desaturation reactions were catalyzed cooperatively by C0507 and other phytoene desaturases. We reached that conclusion because, in a Bradyrhizobium strain, two distinct crt gene clusters are involved in the synthesis of its carotenoids (spirilloxanthin and canthaxanthin). Each cluster contains the genes crtE (GGPP synthase), crtB, and crtI, leading to the common precursor lycopene (39). In addition, the main products of the Δc0507 strain, TH-BABR and DH-IDR, lacked the double bonds at C-3,4 and C-3′,4′. Therefore, we deduced that C0507 functions as a CrtD, involved in the desaturation reactions that form double bonds at C-3,4 of DH-IDR and C-3′,4′ of DH-BABR. This is the first time that a CrtD has been identified functionally in archaea. Peak 1′, peak 2′, peak 3′, peak 6′, and peak 7′ remain unexplained. Because the number of conjugated double bonds in these compounds was less than 11 and because the number of conjugated double bonds in lycopene was 11, these peaks might represent derivatives of lycopene precursors.
FIG 4.
Absorption spectra of the carotenoids extracted from the Δc0507 strain. (A) Peak 4′ (tetrahydrobisanhydrobacterioruberin [TH-BABR]; λmax = 441, 466, and 498 nm). (B) Peak 5′ (dihydroisopentenyldehydrorhodopin [DH-IDR]; λmax = 440, 465, and 495 nm). Spectra were measured using a photodiode-array detector attached to the HPLC apparatus, with a mobile phase of 100% methanol.
The biosynthetic pathway for carotenoids in the Δc0506 strain was discontinued at lycopene, and the Δc0507 strain accumulated DH-IDR and TH-BABR. C0506 was suggested to catalyze the conversion of lycopene to DH-IDR. The conversion contains two reactions, viz., introduction of a C5 isoprene unit at C-2 and hydration at C-1,2 of the terminal of lycopene (Fig. 1). Therefore, C0506 in Ha. japonica is likely to be a bifunctional lycopene elongase and 1,2-hydratase. The lye gene of Hb. salinarum, a homolog of c0506, was expressed in E. coli, and the gene product catalyzes the conversion of lycopene to TH-BABR (10). In this conversion, introduction of a C5 isoprene unit at C-2 and hydration at C-1,2 of the terminal of lycopene were both catalyzed by Lye. This result supports our prediction that C0506 would be a bifunctional enzyme. So we redefined Lye as a bifunctional enzyme and designated C0506 from Ha. japonica LyeJ.
In order to confirm the function of C0506 and C0507, an in vivo complementation study was performed. Plasmid pJc0506, containing the intact c0506 gene, was introduced into the Δc0506 strain, and the transformant was named the Δc0506(pJc0506) strain. HPLC analysis revealed that the biosynthesis of the final BR product was restored (Fig. 3C and Table 3). However, the main products were BABR and MABR. That was probably a result of the fact that the transcription level of the downstream c0505 gene is still low in the complemented strain. Plasmid pJc0507, containing the intact c0507 gene, was introduced into the Δc0507 strain. The transformant Δc0507(pJc0507) restored the production of BR (Fig. 3E and Table 3). Since the transcription levels of the downstream genes, c0506 and c0505, are probably still low in the complemented strain, the intermediates (MABR, BABR, IDR, and lycopene) accumulated and the proportion of final product diminished in the Δc0507(pJc0507) transformant.
Analysis of c0505.
The hydroxyl groups at C1 and C1′ of lycopene were introduced by C0506. However, the enzyme that introduces hydroxyl groups to the C3″ and C3″′ of BABR had not been identified. C0505, the homolog of CruF, was selected as a candidate enzyme for this reaction. Its mutant was constructed by homologous recombination and designated strain Δc0505. The Δc0505 strain also showed the same growth rate as wild-type Ha. japonica. The transcriptions of c0507 and c0506 in the Δc0505 strain were confirmed by RT-PCR, and the disruption of c0505 did not affect the expression of the upstream genes in the same cluster (Fig. 2B, panel d).
The cell suspension of the Δc0505 strain was red in color. In HPLC, only one peak (Fig. 3F, peak 3) was detected in the Δc0505 strain, and the peak exhibited a retention time of 13.7 min and λmax values of 468, 491, and 523 nm (n = 13) (Table 3). Mass spectrometric analysis showed that peak 3 of the Δc0505 strain had an m/z value of 704, compatible with that of BABR. Based on these data, peak 3 of the Δc0505 strain was identified as representing BABR. These data demonstrated that the carotenoid biosynthetic pathway of the Δc0505 strain was discontinued at BABR and indicated that C0505 catalyzes the reaction that introduces hydroxyl groups to C3″ and C3″′ of BABR to generate BR (Fig. 1). Therefore, C0505 in Ha. japonica is a C50 carotenoid 2″,3″-hydratase.
The in vivo complementation study was also assessed using the intact c0505 gene-introduced transformant Δc0505(pJc0505). HPLC analysis revealed that the carotenoid composition of the Δc0505(pJc0505) transformant was restored and was similar to that of wild-type Ha. japonica (Fig. 3G and Table 3).
DISCUSSION
The C50 carotenoid BR is known to accumulate in several extremely halophilic archaea (15–19), but the identity of the responsible biosynthetic pathway remains unclear. In this study, we have identified three main genes encoding enzymes responsible for the biosynthesis of BR from lycopene for the first time.
The biosynthetic pathway for carotenoids in the Δc0506 strain was discontinued at lycopene, and the Δc0507 strain accumulated DH-IDR and TH-BABR; hence, C0506 was suggested to be a bifunctional lycopene elongase and 1,2-hydratase, catalyzing the reaction that introduces a C5 isoprene unit and a hydroxyl group to the terminal of lycopene. In strain Δc0507, TH-BABR and DH-IDR were detected. Therefore, C0507 was proposed to be a carotenoid 3,4-desaturase and to be involved in the desaturation reactions that form double bonds at C-3,4 of DH-IDR and C-3′,4′ of DH-BABR. It seems that two types of possible biosynthetic pathways from lycopene to BABR exist in wild-type Ha. japonica. The first type of pathway involves immediate conversion of DH-IDR, generated from lycopene by C0506 to IDR by C0507. Subsequently, C0506 further introduces a C5 isoprene unit and a hydroxyl group to the other terminal of IDR to generate the C50 carotenoid DH-BABR, which is then desaturated by C0507 to form BABR. The second type involves C0506-mediated catalysis of reactions that introduce two C5 isoprene units and two hydroxyl groups to each end of the C40 lycopene to generate the C50 TH-BABR, which is then converted to BABR by C0507. IDR was detected as an intermediate, but TH-BABR was not detected in the wild-type Ha. japonica strain. Thus, it can be suggested that the former is the main pathway in wild-type Ha. japonica. On the other hand, it has been proposed that the biosynthetic pathway of BABR from lycopene in Hb. salinarum involves the latter pathway (10); therefore, the carotenoid biosynthetic pathway in Ha. japonica is possibly different from that in Hb. salinarum.
Based on the proposed carotenoid and retinal biosynthetic pathways in Ha. japonica (Fig. 1), the Δc0506 strain was expected to accumulate β-carotene or retinal in addition to lycopene. However, only lycopene was detected when the Δc0506 strain was grown in the dark. Our previous study showed that the transcription of the cruxrhodopsin (a retinal protein) gene is regulated by high light intensity (23), and β-carotene was detected when Ha. japonica was grown in light (unpublished data). So it is possible that β-carotene cannot be accumulated when the Δc0506 strain is grown in the dark.
The carotenoid 1,2-hydratase catalyzes the synthesis of some carotenoids that contain hydroxyl groups by hydration at the C-1,2 or C-1′,2′ double bond. Two types of carotenoid 1,2-hydratases (CrtC and CruF) have been discovered to date. The first type is the CrtC-type carotenoid 1,2-hydratase that has been primarily found in purple bacteria, such as Rhodobacter capsulatus, which produces spheroidene (35, 40). The second type is the CruF-type carotenoid 1,2-hydratase that has been found in bacteria such as Synechococcus sp. strain PCC 7002, which produces myxol-2′ fucoside (41), and two species of Deinococcus (38). C0505, the C50 carotenoid 2″,3″-hydratase from Ha. japonica, is a homolog of the CruF-type carotenoid 1,2-hydratase. This is the first time that an example of CruF has been functionally identified in archaea, although C0505 catalyzes hydration at the C-2″,3″ and C-2″′,3″′ double bond of BABR instead of at the C-1,2 or C-1′,2′ double bond of lycopene. The carotenoid hydratases of the same CruF type from different bacteria would have their own substrate specificities.
In a previous genomic sequence analysis of Ha. japonica, c0507 revealed its sequence homology to crtI of P. ananatis (34). However, our results demonstrated that C0507, the CrtD equivalent, is involved in desaturation reactions that form double bonds at C-3,4 of DH-IDR and C-3′,4′ of DH-BABR. Thus, the CrtI that converts phytoene to lycopene has not been identified. In addition, C0184, C1220, C1219, and C1158 from Ha. japonica also show amino acid sequence homology to GGPP synthase (CrtE), phytoene synthase (CrtB), lycopene β-cyclase (CrtY), and β-carotene cleavage dioxygenase (Brp), respectively. Additionally, further studies are necessary to investigate the remaining unidentified carotenoid biosynthetic enzymes in order to elucidate the complete carotenoid biosynthetic pathway in Ha. japonica.
It has been suggested that the antioxidant capacity of carotenoids is linked to the number of conjugated double bonds and hydroxyl groups (3, 4, 42). BR, which contains 13 conjugated double bonds and four hydroxyl groups, is regarded to be an effective free-radical scavenger. Our previous study has shown that the free-radical scavenging capacity of BR is much higher than that of β-carotene, which contains 11 conjugated double bonds (22). Moreover, BR is a dipolar C50 carotenoid, and it was suggested to act as a “rivet” in the cell membrane, to increase its rigidity and mechanical strength (7).
In summary, we have identified three carotenoid biosynthetic enzymes and have elucidated their functions. Among them, CrtD and CruF were found in an archaeon for the first time. Our results revealed the biosynthetic pathway responsible for production of BR from lycopene in Ha. japonica and provide a basis for investigating carotenoid biosynthetic pathways in other extremely halophilic archaea. Elucidation of the carotenoid biosynthetic pathway in Ha. japonica may also prove useful for producing the C50 carotenoid BR efficiently by employing genetically modified haloarchaeal strains.
ACKNOWLEDGMENTS
We thank M. Ishikawa (Material Analysis Suzukake-dai Center, Technical Department, Tokyo Institute of Technology) for mass spectrometric analysis and Y. Yamamoto, A. Takada, and N. Shinohara (Biomaterial Analysis Center, Technical Department, Tokyo Institute of Technology) for the DNA sequence analysis.
This study was partially supported by Grant-in-Aid for Scientific Research (C) from Japan Society for the Promotion of Science and Takahashi Industrial and Economic Research Foundation (to R.Y.).
REFERENCES
- 1.Britton G, Liaaen-Jensen S, Pfander H. 2004. Carotenoids handbook. Birkhäuser Verlag, Basel, Switzerland. [Google Scholar]
- 2.Holt NE, Zigmantas D, Valkunas L, Li XP, Niyogi KK, Fleming GR. 2005. Carotenoid cation formation and the regulation of photosynthetic light harvesting. Science 307:433–436. doi: 10.1126/science.1105833. [DOI] [PubMed] [Google Scholar]
- 3.Miller NJ, Sampson J, Candeias LP, Brameley PM, Rice-Evans CA. 1996. Antioxidant activities of carotenes and xanthophylls. FEBS Lett 384:240–242. doi: 10.1016/0014-5793(96)00323-7. [DOI] [PubMed] [Google Scholar]
- 4.Naguib YM. 2000. Antioxidant activities of astaxanthin and related carotenoids. J Agric Food Chem 48:1150–1154. doi: 10.1021/jf991106k. [DOI] [PubMed] [Google Scholar]
- 5.Dundas ID, Larsen H. 1963. A study on the killing by light of photosensitized cells of Halobacterium salinarium. Arch Mikrobiol 46:19–28. doi: 10.1007/BF00406383. [DOI] [PubMed] [Google Scholar]
- 6.Shahmohammadi HR, Asgarani E, Terato H, Saito T, Ohyama Y, Gekko K, Yamamoto O, Ide H. 1998. Protective roles of bacterioruberin and intracellular KCl in the resistance of Halobacterium salinarium against DNA damaging agents. J Radiat Res 39:251–262. doi: 10.1269/jrr.39.251. [DOI] [PubMed] [Google Scholar]
- 7.Lazrak T, Milon A, Wolff G, Albrecht AM, Miehe M, Ourisson G, Nakatani Y. 1987. Comparison of the effects of inserted C40- and C50-terminally dihydroxylated carotenoids on the mechanical properties of various phospholipid vesicles. Biochim Biophys Acta 903:132–141. doi: 10.1016/0005-2736(87)90163-5. [DOI] [PubMed] [Google Scholar]
- 8.Dogbo O, Laferriere A, D'Harlingue A, Camara B. 1988. Carotenoid biosynthesis: isolation and characterization of a bifunctional enzyme catalyzing the synthesis of phytoene. Proc Natl Acad Sci U S A 85:7054–7058. doi: 10.1073/pnas.85.19.7054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Goodwin TW. 1980. The biochemistry of the carotenoids: plants, 2nd ed, vol 1 Chapman and Hall, New York, NY. [Google Scholar]
- 10.Dummer AM, Bonsall JC, Cihla JB, Lawry SM, Johnson GC, Peck RF. 2011. Bacterioopsin-mediated regulation of bacterioruberin biosynthesis in Halobacterium salinarum. J Bacteriol 193:5658–5667. doi: 10.1128/JB.05376-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Krubasik P, Kobayashi M, Sandmann G. 2001. Expression and functional analysis of a gene cluster involved in the synthesis of decaprenoxanthin reveals the mechanisms for C50 carotenoid formation. Eur J Biochem 268:3702–3708. doi: 10.1046/j.1432-1327.2001.02275.x. [DOI] [PubMed] [Google Scholar]
- 12.Heider SA, Peters-Wendisch P, Wendisch VF. 10 September 2012, posting date Carotenoid biosynthesis and overproduction in Corynebacterium glutamicum. BMC Microbiol doi: 10.1186/1471-2180-12-198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Heider SA, Peters-Wendisch P, Netzer R, Stafnes M, Brautaset T, Wendisch VF. 2014. Production and glucosylation of C50 and C40 carotenoids by metabolically engineered Corynebacterium glutamicum. Appl Microbiol Biotechnol 98:1223–1235. [DOI] [PubMed] [Google Scholar]
- 14.Tao L, Yao H, Cheng Q. 2007. Genes from a Dietzia sp. for synthesis of C40 and C50 β-cyclic carotenoids. Gene 386:90–97. doi: 10.1016/j.gene.2006.08.006. [DOI] [PubMed] [Google Scholar]
- 15.Netzer R, Stafsnes MH, Andreassen T, Goksoyr A, Bruheim P, Brautaset T. 2010. Biosynthetic pathway for γ-cyclic sarcinaxanthin in Micrococcus luteus: heterologous expression and evidence for diverse and multiple catalytic functions of C50 carotenoid cyclases. J Bacteriol 192:5688–5699. doi: 10.1128/JB.00724-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ronnekleiv M, Liaaen-Jensen S. 1995. Bacterial carotenoids 53* C50-carotenoids 23; carotenoids of Haloferax volcanii versus other halophilic bacteria. Biochem Syst Ecol 23:627–634. doi: 10.1016/0305-1978(95)00047-X. [DOI] [Google Scholar]
- 17.Fang CJ, Ku KL, Lee MH, Su NW. 2010. Influence of nutritive factors on C50 carotenoids production by Haloferax mediterranei ATCC 33500 with two-stage cultivation. Bioresour Technol 101:6487–6493. doi: 10.1016/j.biortech.2010.03.044. [DOI] [PubMed] [Google Scholar]
- 18.Mandelli F, Miranda VS, Rodrigues E, Mercadante AZ. 2012. Identification of carotenoids with high antioxidant capacity produced by extremophile microorganisms. World J Microbiol Biotechnol 28:1781–1790. doi: 10.1007/s11274-011-0993-y. [DOI] [PubMed] [Google Scholar]
- 19.El-Sayed WSM, Takaichi S, Saida H, Kamekura M, Abu-Shady M, Seki H, Kuwabara T. 2002. Effects of light and low oxygen tension on pigment biosynthesis in Halobacterium salinarum, revealed by a novel method to quantify both retinal and carotenoids. Plant Cell Physiol 43:379–383. doi: 10.1093/pcp/pcf044. [DOI] [PubMed] [Google Scholar]
- 20.Oesterhelt D, Stoeckenius W. 1971. Rhodopsin-like protein from the purple membrane of Halobacterium halobium. Nat New Biol 233:149–152. [DOI] [PubMed] [Google Scholar]
- 21.Horikoshi K, Aono R, Nakamura S. 1993. The triangular halophilic archaebacterium Haloarcula japonica strain TR-1. Experientia 49:497–502. [Google Scholar]
- 22.Yatsunami R, Ando A, Yang Y, Takaichi S, Kohno M, Matsumura Y, Ikeda H, Fukui T, Nakasone K, Fujita N, Sekine M, Takashina T, Nakamura S. 17 March 2014, posting date Identification of carotenoids from the extremely halophilic archaeon Haloarcula japonica. Front Microbiol doi: 10.3389/fmicb.2014.00100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yatsunami R, Kawakami T, Ohtani H, Nakamura S. 1999. Transcriptional regulation of cruxrhodopsin gene from extremely halophilic archaeon Haloarcula japonica strain TR-1. Nucleic Acids Symp Ser 42:73–74. doi: 10.1093/nass/42.1.73. [DOI] [PubMed] [Google Scholar]
- 24.Yatsunami R, Aono S, Nakamura S. 2000. The gene encoding a novel halorhodopsin-like protein of extremely halophilic archaeon Haloarcula japonica strain TR-1. Nucleic Acids Symp Ser 44:1–2. doi: 10.1093/nass/44.1.1. [DOI] [PubMed] [Google Scholar]
- 25.Robb FT, DasSarma S, Fleischmann EM. 1995. Archaea: a laboratory manual. Cold Springer Harbor Laboratory Press, Cold Spring Harbor, NY. [Google Scholar]
- 26.Sambrook J, Fritsch EF, Maniatis T. 1989. Molecular cloning: a laboratory manual, 2nd ed Cold Springer Harbor Laboratory Press, Cold Spring Harbor, NY. [Google Scholar]
- 27.Onodera M, Yatsunami R, Tsukimura W, Fukui T, Nakasone K, Takashina T, Nakamura S. 2013. Gene analysis, expression, and characterization of an intracellular α-amylase from the extremely halophilic archaeon Haloarcula japonica. Biosci Biotechnol Biochem 77:281–288. [DOI] [PubMed] [Google Scholar]
- 28.Peck RF, DasSarma S, Krebs MP. 2000. Homologous gene knockout in the archaeon Halobacterium salinarum with ura3 as a counterselectable maker. Mol Microbiol 35:667–676. [DOI] [PubMed] [Google Scholar]
- 29.Lam WL, Doolittle WF. 1989. Shuttle vectors for the archaebacterium Halobacterium volcanii. Proc Natl Acad Sci U S A 86:5478–5482. doi: 10.1073/pnas.86.14.5478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Holmes ML, Nuttall SD, Dyall-Smith ML. 1991. Construction and use of halobacterial shuttle vectors and further studies on Haloferax DNA gyrase. J Bacteriol 173:3807–3813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Takaichi S, Mochimaru M, Maoka T, Katoh H. 2005. Myxol and 4-ketomyxol 2′-fucosides, not rhamnosides, from Anabaena sp. PCC 7120 and Nostoc punctiforme PCC 73102, and proposal for the biosynthetic pathway of carotenoids. Plant Cell Physiol 46:497–504. [DOI] [PubMed] [Google Scholar]
- 32.Takaichi S, Maoka T, Masamoto K. 2001. Myxoxanthophyll in Synechocystis sp. PCC 6803 is myxol 2′-dimethyl-fucoside, (3R,2′S)-myxol 2′-(2,4-di-O-methyl-α-l-fucoside), not rhamnoside. Plant Cell Physiol 42:756–762. [DOI] [PubMed] [Google Scholar]
- 33.Nakamura S, Nakasone K, Takashina T. 2011. Genetics and genomics of triangular disc-shaped halophilic archaeon Haloarcula japonica TR-1, p 364–381. In Horikoshi K, Antranikian G, Bull AT, Robb FT, Stetter KO (ed), Extremophiles handbook, vol 2 Springer, Heidelberg, Germany. [Google Scholar]
- 34.Misawa N, Nakagawa M, Kobayashi K, Yamano S, Izawa Y, Nakamura K, Harashima K. 1990. Elucidation of the Erwinia uredovora carotenoid biosynthetic pathway by functional analysis of gene products expressed in Escherichia coli. J Bacteriol 172:6704–6712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Armstrong GA, Alberti M, Leach F, Hearst JE. 1989. Nucleotide sequence, organization and nature of the protein products of the carotenoid biosynthesis gene cluster of Rhodobacter capsulatus. Mol Gen Genet 216:254–268. doi: 10.1007/BF00334364. [DOI] [PubMed] [Google Scholar]
- 36.Tian B, Sun ZT, Xu ZJ, Shen SC, Wang H, Hua YJ. 2008. Carotenoid 3′,4′-desaturase is involved in carotenoid biosynthesis in the radioresistant bacterium Deinococcus radiodurans. Microbiology 154:3697–3706. doi: 10.1099/mic.0.2008/021071-0. [DOI] [PubMed] [Google Scholar]
- 37.Maresca JA, Graham JE, Bryant DA. 2008. The biochemical basis for structural diversity in the carotenoids of chlorophototrophic bacteria. Photosynth Res 97:121–140. doi: 10.1007/s11120-008-9312-3. [DOI] [PubMed] [Google Scholar]
- 38.Sun ZT, Shen SC, Wang C, Wang H, Hu YP, Jiao JD, Ma TT, Hua YJ. 2009. A novel carotenoid 1,2-hydratase (CruF) from two species of the non-photosynthetic bacterium Deinococcus. Microbiology 155:2775–2783. doi: 10.1099/mic.0.027623-0. [DOI] [PubMed] [Google Scholar]
- 39.Giraud E, Hannibal L, Fardoux J, Jaubert M, Jourand P, Dreyfus B, Sturgis JN, Vermeglio A. 2004. Two distinct crt gene clusters for two different functional classes of carotenoid in Bradyrhizobium. J Biol Chem 279:15076–15083. doi: 10.1074/jbc.M312113200. [DOI] [PubMed] [Google Scholar]
- 40.Takaichi S. 2009. Distribution and biosynthesis of carotenoids, p 97–117. In Hunter CN, Daldal F, Thurnauer MC, Beatty JT (ed), The purple phototrophic bacteria, vol 28 Springer, Dordrecht, the Netherlands. [Google Scholar]
- 41.Graham JE, Bryant DA. 2009. The biosynthetic pathway for myxol-2′ fucoside (myxoxanthophyll) in the cyanobacterium Synechococcus sp. strain PCC 7002. J Bacteriol 191:3292–3300. doi: 10.1128/JB.00050-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Albrecht M, Takaichi S, Steiger S, Wang ZY, Sandmann G. 2000. Novel hydroxycarotenoids with improved antioxidative properties produced by gene combination in Escherichia coli. Nat Biotechnol 18:843–846. doi: 10.1038/78443. [DOI] [PubMed] [Google Scholar]