ABSTRACT
Metal homeostasis in bacterial cells is a highly regulated process requiring intricately coordinated import and export, as well as precise sensing of intracellular metal concentrations. The uptake of zinc (Zn) has been linked to the virulence of Brucella abortus; however, the capacity of Brucella strains to sense Zn levels and subsequently coordinate Zn homeostasis has not been described. Here, we show that expression of the genes encoding the zinc uptake system ZnuABC is negatively regulated by the Zn-sensing Fur family transcriptional regulator, Zur, by direct interactions between Zur and the promoter region of znuABC. Moreover, the MerR-type regulator, ZntR, controls the expression of the gene encoding the Zn exporter ZntA by binding directly to its promoter. Deletion of zur or zntR alone did not result in increased zinc toxicity in the corresponding mutants; however, deletion of zntA led to increased sensitivity to Zn but not to other metals, such as Cu and Ni, suggesting that ZntA is a Zn-specific exporter. Strikingly, deletion of zntR resulted in significant attenuation of B. abortus in a mouse model of chronic infection, and subsequent experiments revealed that overexpression of zntA in the zntR mutant is the molecular basis for its decreased virulence.
IMPORTANCE The importance of zinc uptake for Brucella pathogenesis has been demonstrated previously, but to date, there has been no description of how overall zinc homeostasis is maintained and genetically controlled in the brucellae. The present work defines the predominant zinc export system, as well as the key genetic regulators of both zinc uptake and export in Brucella abortus. Moreover, the data show the importance of precise coordination of the zinc homeostasis systems as disregulation of some elements of these systems leads to the attenuation of Brucella virulence in a mouse model. Overall, this study advances our understanding of the essential role of zinc in the pathogenesis of intracellular bacteria.
INTRODUCTION
The pathogenic alphaproteobacterium Brucella abortus preferentially infects cattle, bison, and elk (1), but the bacteria are also highly efficient at infecting humans. In order to establish a chronic infection in these hosts, the brucellae must survive and replicate within host macrophages (2). While the macrophage serves as the niche for Brucella during a chronic infection, the intracellular environment of these phagocytic immune cells is inhospitable as the bacteria are bombarded with a variety of environmental stresses, including exposure to reactive oxygen species (ROS), low pH, limited oxygen availability, and nutrient deprivation (3). Notwithstanding, the brucellae have evolved multiple strategies to cope with the harsh intramacrophagic environment and ultimately establish a replicative niche in these cells.
With regard to the nutrient limitation experienced by the brucellae within macrophages, metal cations are likely found in extremely low concentrations, and, in fact, macrophages produce transporters, such as the NRAMP family of transporters, that actively remove metal cations from the phagosomal compartment (4). Metal ions are essential micronutrients for all living organisms because these elements serve as important structural and/or enzymatic cofactors of cellular proteins (5, 6). However, while metals are required and beneficial for life, metal ions represent a double-edged sword for the cell as they can also cause significant cellular damage if present in excess of cellular needs. For example, free iron (Fe) and copper (Cu) cations can react with H2O2 and O2− via the Fenton reaction to generate DNA-damaging hydroxyl radicals (7, 8). Additionally, metal ions, including copper (Cu), zinc (Zn), and nickel (Ni), can also lead to equally adverse cellular effects in another way. These metal ions have extremely high binding affinities for generic divalent cation binding sites in proteins, and the binding of these cations to inappropriate sites, such as sites requiring Fe or Mn for proper protein function, can inactivate the proteins, leading to toxicity and cell death (6). Thus, it is not surprising that organisms from single-celled bacteria to multicellular mammals have evolved cellular mechanisms to stringently control the uptake, export, utilization, and storage of metal ions.
It has been estimated that >5% of bacterial proteins bind Zn (9), and several Zn-containing proteins that are important for the basic physiology and virulence of Brucella strains have been identified (10, 11, 12, 13, 14, 15). Moreover, the Zn uptake system protein ZnuA is required for Brucella virulence (16, 17), and Brucella znuA mutants are capable of inducing protective immunity in mice against a subsequent challenge with wild-type Brucella strains (17, 18). While it is clear that high-affinity Zn acquisition is necessary for Brucella virulence, there is very little known about how Zn homeostasis is controlled in these bacteria.
In many bacteria, two well-characterized regulatory systems are used to ensure Zn homeostasis, and these genetic circuits function by cooperatively controlling the expression of membrane-bound transport systems that either import or export Zn cations (19). As alluded to in the previous paragraph, the Zn uptake system, Znu, is employed by numerous Gram-negative bacteria to import Zn cations, and this system is composed of an ABC-type transporter, where ZnuA is the periplasmic-binding protein, ZnuB is the membrane permease, and ZnuC is the ATPase protein (20). Additionally, the expression of the znuABC genes is often controlled by a Zn-responsive transcriptional regulator of the Fur family, called Zur (21). The export of Zn from bacterial cells is often accomplished using the ZntA protein, which is an ATP-dependent transporter employed when cellular Zn concentrations are at toxic levels (19), and the transcription of zntA is regulated by the MerR family transcriptional regulator ZntR (22).
In the present study, we have identified the Zn uptake regulator, Zur, as the primary regulator of the znu system in B. abortus 2308, and we also define the Zn exporter ZntA and its transcriptional regulator ZntR in this bacterium. The experiments described herein were designed (i) to assess the regulation of Zn homeostasis systems by Zur and ZntR in Brucella, (ii) to confirm the function of ZntA as a Zn exporter, and (iii) to characterize the role of the Zn homeostasis regulators in Brucella pathogenesis.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
Brucella abortus 2308 and derivative strains were routinely grown on Schaedler blood agar (SBA), which is composed of Schaedler agar (BD, Franklin Lakes, NJ) containing 5% defibrinated bovine blood (Quad Five, Ryegate, MT), or in brucella broth (BD). For some experiments, Brucella strains were grown in Gerhardt's minimal medium (GMM) (23). In order to cultivate Brucella strains in a zinc-depleted environment for subsequent gene expression analyses, the bacteria were grown in brucella broth to early stationary phase and then subsequently diluted to a concentration of ∼109 CFU/ml in fresh brucella broth or in brucella broth supplemented with the zinc chelator N,N,N′,N′-tetrakis(2-pyridylmethyl)ethylenediamine (TPEN; Sigma, St. Louis, MO) at a final concentration of 100 μM. The cells were then incubated for 30 min at 37°C with constant shaking before RNA was isolated (described below). For other experiments with TPEN, the bacteria were inoculated in brucella broth with or without 10 μM TPEN at a cell concentration of 104 CFU/ml, and growth was measured for viable CFU at 12-h time points by the plating of serial dilutions from the cultures. For cloning and recombinant protein expression, Escherichia coli strains (DH5α and BL21) were grown on tryptic soy agar (BD) or in Luria-Bertani (LB) broth. When appropriate, growth medium was supplemented with ampicillin (100 μg/μl) or kanamycin (45 μg/μl).
Construction of Brucella abortus mutants and genetic complementation.
The zur gene (bab2_1082) in Brucella abortus 2308 was mutated using a nonpolar, unmarked gene excision strategy as described previously (24). Briefly, an approximately 1-kb fragment of the upstream region of bab2_1082 to the second codon of the coding region was amplified by PCR using primers zur-Up-For and zur-Up-Rev and genomic DNA from Brucella abortus 2308 as a template. Similarly, a fragment containing the last two codons of the coding region to approximately 1 kb downstream of each open reading frame (ORF) was amplified with primers zur-Down-For and zur-Down-Rev. The sequences of all oligonucleotide primers used in this study can be found in Table 1. The upstream fragment was digested with BamHI, while the downstream fragment was digested with PstI, and both fragments were treated with polynucleotide kinase in the presence of ATP. Both of the DNA fragments were included in a single ligation mix with BamHI/PstI-digested pNTPS138 (M. R. K. Alley, unpublished data) and T4 DNA ligase (Monserate Biotechnology Group, San Diego, CA). Deletion constructs for zntR (bab1_2018) and zntA (bab1_2019) were generated similarly as described above for the zur deletion construct with appropriate oligonucleotide primers, specifically, primers zntR-Up-For, zntR-Up-Rev, zntR-Down-For, and zntR-Down-Rev for the ΔzntR mutant and primers zntA-Up-For, zntA-Up-Rev, zntA-Down-For, and zntA-Down-Rev for the ΔzntA mutant. The resulting plasmids [pC3035 (Δzur), pC3036 (ΔzntR), and pC3037 (ΔzntA)] (Table 2) were introduced into B. abortus 2308, and merodiploid transformants were obtained by selection on SBA supplemented with kanamycin. Kanamycin-resistant transformants were grown for 6 to 8 h in brucella broth and then plated onto SBA containing 10% sucrose. Genomic DNA from sucrose-resistant, kanamycin-sensitive colonies was isolated and screened by PCR for loss of the zur, zntR, and zntA genes. The isogenic zur, zntR, and zntA mutants derived from B. abortus 2308 were named CC124, CC128, and CC130, respectively. The genotypes of all mutant strains were verified by DNA sequence analyses and confirmed by Southern hybridizations.
TABLE 1.
Oligonucleotide primers used in this study
| Primer name | Sequence (5′ → 3′)a |
|---|---|
| zur-Up-For | CAGGATCCACAATGGGACCA |
| zur-Up-Rev | GTGCGTCGTCATTGTCCAGC |
| zur-Down-For | GTGTGAAACGGTTTTCGGAAAA |
| zur-Down-Rev | AACTGCAGGATCCGCATTATCGTCATCG |
| zntA-Up-For | GCGGATCCAATGTCAGCGATCCGCTGAG |
| zntA-Up-Rev | GTTCATTGGCTCACTCACGGTC |
| zntA-Down-For | AGACAACCTGTACGCTAGAGCGC |
| zntA-Down-Rev | GCCTGCAGAATGGTCGGAACGACATCATCC |
| zntA-Down-Rev2 | TACTGCAGAAATTACTCCGGGACGCAGA |
| zntR-Up-For | GCGGATCCGGAAGCGATCGATCCGCTA |
| zntR-Up-Rev | GAGATACTCGGCAATCACGATG |
| zntR-Up-For4 | TAGGATCCTTTTTGGCGGTGCAATCC G |
| zntR-Up-Rev3 | GAGATACTCGGCAATCACGATG |
| zntR-Down-For | GACCATGATTTTTCGCTTGCTCCTC |
| zntR-Down-Rev | GCCTGCAGCCTTCCAGCAATTCGCCGA |
| rZur-For | GCGGTCTCAGCGCATGACGACGCACCACCATCA |
| rZur-Rev | GCGGTCTCATATCATCACACTTTTCGGGATGCGC |
| rZntR-For | GCGGTCTCAGCGCATGGTCAGCATCCCGATAGGC |
| rZntR-Rev | GCGGTCTCATATCACTAATGCGTGGGGTGCAGG |
| znuAC-IG-For | GAGCAATCGGGGAGGAACAT |
| znuAC-IG-Rev | CTCTGCCTGCATAGACTCTTGGA |
| znt-IG-For | CCTTGGAGGCCTCGCCTAT |
| znt-IG-Rev | CGTCCACACGAAAGCTGATCTG |
| znuA-RT-For | CCCAGCGTATCAATGGGCTT |
| znuA-RT-Rev | TCGATCAAGCCGCTTCACTC |
| znuB-RT-For | CGATACCATGGCCCATTCCG |
| znuB-RT-Rev | CCATGAGAACAAGGCCGAGC |
| znuC-RT-For | GCTGGTGCGCAATGTCGAT |
| znuC-RT-Rev | AAAGCGGCAAGGTGCGGTC |
| zur-RT-For | GTTACGGGACGATGGCTTTCG |
| zur-RT-Rev | AAAGCAACGAGGCCCTGC |
| zntA-RT-For | GCGACGGAAGAGGCGGCAA |
| zntA-RT-Rev | CCTGGTCGTTCCATTGCGCT |
| zntR-RT-For | AATGCGAAGCTCCACTTCTGTCAG |
| zntR-RT-Rev | CGGCTTTTGTTCATCCGCCATG |
Underlined sequences indicate restriction endonuclease recognition sites.
TABLE 2.
Plasmids used in this study
| Plasmid name | Description | Reference or source |
|---|---|---|
| pNPTS138 | Cloning vector, contains sacB gene; Kanr | 41 |
| pASK-IBA6 | Recombinant protein expression vector; Ampr | IBA |
| pC3035 | In-frame deletion of zur plus 1 kb of each flanking region in pNPTS138 | This study |
| pC3036 | In-frame deletion of zntR plus 1 kb of each flanking region in pNPTS138 | This study |
| pC3037 | In-frame deletion of zntA plus 1 kb of each flanking region in pNPTS138 | This study |
| pJB003 | Deletion of zntR and zntA plus 1 kb of each flanking region in pNPTS138 | This study |
| pLS008 | Intact zntA gene plus 1 kb of each flanking region in pNPTS138 | This study |
| pLS009 | Intact zntR gene plus 1 kb of each flanking region in pNPTS138 | This study |
| prZur | Coding region of zur in pASK-IBA6 for recombinant protein purification | This study |
| prZntR | Coding region of zntR in pASK-IBA6 for recombinant protein purification | This study |
Double-mutant strains were also constructed in which zntR and zur or zntR and zntA were deleted from the chromosome. For the zntR zur double mutant, pC3036 was transformed into CC124 (i.e., the Δzur mutant strain), and selection for a strain carrying the zntR deletion was carried out as described above. The resulting ΔzntR zur double mutant strain was named CC127. To generate a zntR zntA double mutant, an approximately 1-kb fragment of the downstream region of zntR to the stop codon of the coding region was amplified by PCR using primers zntR-Up-For4 and zntR-Up-Rev3 and genomic DNA from Brucella abortus 2308 as a template. Similarly, a fragment containing the last two codons of the coding region to approximately 1 kb downstream of zntA was amplified with primers zntA-Down-For and zntA-Down-Rev2. The zntR fragment was digested with BamHI, while the zntA fragment was digested with PstI, and both fragments were treated with polynucleotide kinase in the presence of ATP. Both of the DNA fragments were included in a single ligation mix with BamHI/PstI-digested pNTPS138. The resulting plasmid (pJB003) was transformed into B. abortus 2308, and following the selection of a ΔzntR zntA double mutant strain using the protocol described above, the mutant strain was named JB005.
Genetic complementation was carried out by reconstruction of the deleted locus on the Brucella abortus chromosome. To reconstruct the zntR deletion, a construct was generated containing the wild-type zntR gene with approximately 1 kb of each flanking region. For this, the entire region encompassing zntR and the flanking regions was amplified by PCR using the oligonucleotide primers zntR-Up-For and zntR-Down-Rev, and the amplified fragment was digested with BamHI and PstI. The digested fragment was cloned into BamHI/PstI-digested pNTPS138, and the resulting plasmid construct was named pLS009. pLS009 was transformed into CC128 (i.e., the ΔzntR mutant strain), and selection was carried out as described above. The ΔzntR::zntR complemented strain was named LS004. The ΔzntA deletion strain was similarly complemented, but here the wild-type zntA gene and 1-kb flanking regions were amplified using the oligonucleotide primers zntA-Up-For and zntA-Down-Rev2. The zntA reconstruction plasmid construct was named pLS008, and the complemented ΔzntA mutant strain was named LS005.
Real-time reverse transcription-PCR (RT-PCR).
Brucella abortus 2308 was grown in brucella broth with constant shaking at 37°C to late exponential phase (∼109 CFU/ml). Total RNA was isolated from the cultures, and genomic DNA was removed as described previously (24). cDNA was generated from the final RNA preparation using a SuperScript III cDNA synthesis system (Invitrogen, Carlsbad, CA) according to the manufacturer's protocol, and this cDNA was used for real-time PCR employing a SYBR green PCR supermix (Roche, Mannheim, Germany). For these experiments, primers for 16S rRNA were used as a control, while gene-specific primers were used for evaluating relative levels of zur, znuA, znuB, znuC, zntA, and zntR mRNAs (Table 1). Parameters for PCR included a single denaturing step for 5 min at 95°C, followed by 40 cycles (denature for 15 s at 95°C, anneal for 15 s at 51°C, and extend for 15 s at 72°C) of amplification. Fluorescence from SYBR green incorporation into double-stranded DNA was measured with an iCycler machine (Bio-Rad), and the relative abundance of mRNA was determined using the Pfaffl equation (25).
Production and purification of recombinant Brucella proteins.
A Strep-tag II system (IBA, Göttingen, Germany) was used to produce a recombinant version of the Brucella Zur and ZntR proteins in E. coli strain BL21. The coding region of the zur gene (bab2_1082) was amplified using the primers rZur-For and rZur-Rev (Table 1), B. abortus 2308 chromosomal DNA as a template, and Taq polymerase (Monserate Biotechnology Group, San Diego, CA). Similarly, the zntR (bab1_2018) coding region was amplified using the primers rZntR-For and rZntR-Rev. The amplified DNA fragment was digested with BsaI and ligated into BsaI-digested pASK-IBA6, which is designed to produce an amino-terminal Strep-tag II-tagged version of the protein of interest. The resulting plasmids, prZur and prZntR, were transformed into E. coli strain BL21, and the strains harboring the recombinant protein expression plasmids were grown to an optical density at 600 nm (OD600) of approximately 0.6 in LB broth before production of the recombinant protein was initiated by the addition of anhydrotetracycline (200 μg/μl final concentration) to the growth medium. Following 2 h of further incubation at 37°C, the bacterial cells were collected by centrifugation (4,200 × g for 10 min at 4°C) and lysed by treatment with CelLyticB (Sigma, St. Louis, MO) in the presence of the protease inhibitor phenylmethanesulfonyl fluoride. The supernatant from the suspension of lysed cells was cleared by centrifugation (14,000 × g for 10 min at 4°C), and the clarified supernatant was passed through an affinity column packed with Strep-Tactin–Sepharose. The column was washed extensively with buffer W (100 mM Tris-HCl, 300 mM NaCl, pH 8.0), and the recombinant protein was eluted with 2.5 mM desthiobiotin in buffer W. The degree of purity of recombinant Zur and ZntR was high, as judged by visualization of a single major band on SDS-PAGE.
EMSAs.
All recombinant Zur (rZur) and rZntR electrophoretic mobility shift assays (EMSAs) were carried out in a 20-μl total reaction volume containing binding buffer composed of 10 mM Tris-HCl (pH 7.4), 50 mM KCl, 1 mM dithiothreitol, 6% glycerol, 50 μg/ml bovine serum albumin, and 50 μg/ml salmon sperm DNA. A 200-bp DNA fragment corresponding to the znuA-znuC intergenic region was amplified by PCR from Brucella abortus 2308 chromosomal DNA using primers znuAC-IG-For and znuAC-IG-Rev (Table 1) and chromosomal DNA from B. abortus 2308 as a template. Similarly, a 150-bp DNA fragment corresponding to the zntR-zntA intergenic region was amplified by PCR from Brucella abortus 2308 chromosomal DNA using primers znt-IG-For and znt-IG-Rev (Table 1). The amplified DNA fragments were purified by agarose gel electrophoresis, and the fragments were end labeled with [γ-32P]ATP (PerkinElmer, San Jose, CA) and polynucleotide kinase (Monserate Biotechnology Group, San Diego, CA). Increasing amounts of rZur protein were mixed with the radiolabeled znuA-znuC intergenic region DNA in binding buffer, and the reaction mixtures were incubated at room temperature for 20 min. As controls, a 50× molar concentration of nonradiolabeled znuA-znuC intergenic region DNA (specific competitor) or nonradiolabeled zntR-zntA intergenic region DNA (nonspecific competitor) was added to some reaction mixtures. For experiments involving rZntR, increasing amounts of rZntR protein were mixed with the radiolabeled zntR-zntA intergenic region DNA in binding buffer, and the reaction mixtures were incubated at room temperature for 20 min. As controls, a 50× molar concentration of nonradiolabeled zntR-zntA intergenic region DNA (specific competitor) or nonradiolabeled cueO promoter region DNA (nonspecific competitor) was added to some reaction mixtures. All binding reaction mixtures were subjected to electrophoresis on 6% native polyacrylamide gels in 0.5× Tris-borate (TB) running buffer for approximately 1 h at 4°C. Following electrophoresis, gels were dried onto 3-mm Whatman paper using a vacuum gel dryer system and visualized by autoradiography.
In some experiments, the effects of EDTA and various divalent metal cations on rZur binding to DNA were assessed. For this, binding reactions were prepared as described above, but 250 μM EDTA was also included in the reaction mixtures. To some samples, an additional 25 μM metal cation (Zn2+, Fe2+, Mn2+, Ni2+, or Cu2+) was included in the binding reaction mixtures. All binding reaction mixtures were incubated at room temperature for 20 min and then subjected to electrophoresis and autoradiography as described above.
Assessment of sensitivities of Brucella strains to metal toxicity by disk diffusion assay.
Brucella strains were grown on Schaedler blood agar at 37°C under 5% CO2 for 48 to 72 h, and the bacterial cells were harvested into phosphate-buffered saline (PBS) and suspended at a concentration of ∼3.33 × 107 CFU/ml in brucella broth containing 0.6% agar (maintained at 55°C). Four milliliters of this suspension was overlaid onto brucella agar plates, and after solidification of the overlay, a sterile 7-mm Whatman disk was placed in the center of each plate. Seven microliters of a 1 M solution of either MgCl2, MnCl2, FeCl3, NiCl2, ZnCl2, or CuCl2 was applied to each filter disk, and the plates were incubated at 37°C with 5% CO2 for 72 h. Zones of inhibition around each disk were then measured in millimeters.
Virulence of Brucella strains in cultured murine macrophages and experimentally infected mice.
Experiments to test the virulence of Brucella strains in primary, murine peritoneal macrophages were carried out as described previously (11). Briefly, resident peritoneal macrophages were isolated from BALB/c mice and seeded in 96-well plates in Dulbecco's modified Eagle's medium with 5% fetal bovine serum, and the following day, the macrophages were infected with opsonized brucellae at a multiplicity of infection (MOI) of 50:1. After 2 h of infection, extracellular bacteria were killed by treatment with gentamicin (50 μg/ml). For the 2-h time point, the macrophages were then lysed with 0.1% deoxycholate in PBS, and serial dilutions were plated on Schaedler blood agar (SBA). For the 24- and 48-h time points, the cells were washed with PBS following gentamicin treatment, and fresh cell culture medium containing gentamicin (20 μg/ml) was added to the monolayer. At the time points specified in the figure legends, the macrophages were lysed, and serial dilutions were plated on SBA. Triplicate wells were used for each Brucella strain tested.
The infection and colonization of mice by Brucella strains were as described previously by Gee et al. (11). BALB/c mice (5 per Brucella strain) were infected intraperitoneally with ∼5 × 104 CFU of each Brucella strain in sterile PBS. The mice were sacrificed at 1, 4, and 8 weeks postinfection, and serial dilutions of spleen homogenates were plated on SBA. All experiments involving animals were approved by the Institutional Animal Care and Use Committee (IACUC) at Virginia Tech.
RESULTS
Specific transcriptional regulators control the expression of the zinc import and export systems in Brucella abortus 2308.
Experimental evidence indicates that zinc (Zn) uptake is mediated by the Znu system (16, 17) in B. abortus 2308, which is encoded on chromosome II (Fig. 1A). An ortholog of the prokaryotic Zn exporter ZntA is also encoded on chromosome I of this strain (Fig. 1B), but the participation of this protein in Zn homeostasis has not been experimentally examined. Within these genetic loci, transcriptional regulatory proteins are also encoded. As part of the znu locus, the gene designated bab2_1082 encodes a Fur family protein that is predicted to function as a Zn uptake regulator (Zur). Likewise, the gene bab1_2018, which is divergently transcribed from zntA, is predicted to encode a MerR family transcriptional regulator, and this protein, called ZntR, is expected to control the expression of zntA. In order to determine the roles of Zur and ZntR in the expression of the Zn uptake and export systems, we constructed nonpolar, isogenic, in-frame deletions of the genes encoding each regulator, and subsequently real-time RT-PCR was used to assess the mRNA levels of the various genes of the Zn uptake and export systems in these deletion strains.
FIG 1.
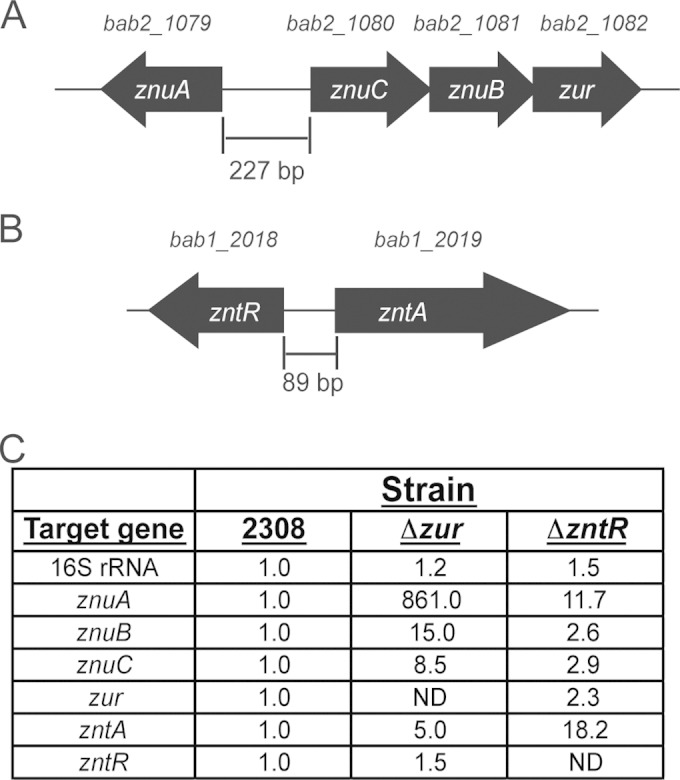
Organization and regulation of the genes encoding the zinc uptake and export systems in Brucella abortus 2308. (A) The zinc uptake (Znu) system is encoded on chromosome II in B. abortus 2308 with znuA being divergently transcribed from the remainder of the uptake system, including znuC, znuB, and zur. (B) The zinc exporter ZntA is encoded on chromosome I in B. abortus 2308, and its potential transcriptional regulator, ZntR, is encoded divergently. (C) Quantitative RT-PCR analysis of zinc uptake and export genes in targeted regulatory mutant strains. The mRNA levels from specific genes encoding elements of the zinc uptake or export system were assessed in B. abortus 2308, an isogenic zur mutant strain, and an isogenic zntR mutant strain. The data are depicted as fold increases in mRNA levels in the mutant strains compared to normalized mRNA levels in the parental strain 2308, and 16S rRNA was evaluated as a control. ND indicates that expression of the gene was not determined. The data represent the average changes in mRNA levels from two biological replicate experiments, each analyzed in triplicate.
In the B. abortus zur mutant, znuA mRNA was extremely elevated compared to the levels found in the parental strain 2308, and likewise, znuB and znuC mRNA levels were higher in the zur mutant strain than in strain 2308 (Fig. 1C). These data indicate that Zur serves as a transcriptional repressor, and this is in line with what other investigators have observed, with Zur negatively controlling znu expression in several diverse bacteria, including Bacillus subtilis (26), Escherichia coli (27), and Pseudomonas aeruginosa (28). Of interest, mRNA levels for zntA, which encodes the putative Zn exporter, were also significantly elevated in the zur mutant. Elevated znuABC expression in the B. abortus zur mutant would be expected to result in an influx of Zn into the cell that would predictably be counteracted by a corresponding increase in zntA expression, which would prevent intracellular Zn levels from reaching toxic levels in the zur mutant.
We observed a similar compensatory situation for the B. abortus zntR mutant (Fig. 1C). zntA mRNA levels were significantly increased in the zntR mutant strain compared to the level in parental strain 2308, but additionally, znuABC transcript levels were also elevated in the zntR mutant strain compared to levels in 2308. Elevated levels of zntA expression would be expected to lead to inappropriate purging of Zn from the cell, and increased expression of the znuABC system likely is required to reestablish an adequate level of intracellular Zn in the zntR mutant.
ZntR is a transcriptional regulator of zntA expression.
The quantitative RT-PCR data in Fig. 1 demonstrated that ZntR is involved in the expression of zntA as deletion of zntR resulted in elevated zntA levels (Fig. 1C). The MerR family of transcriptional regulators, to which ZntR belongs, are known to bind to DNA, and upon interacting with the appropriate divalent metal cation, MerR-type proteins bend the DNA, leading to the activation of gene expression (29). To gain insight into the zinc responsiveness of ZntR in B. abortus, we cultured Brucella strains in the presence and absence of the zinc-specific chelator N,N,N′,N′-tetrakis(2-pyridylmethyl)ethylenediamine (TPEN). After a 60-min incubation, total RNA was isolated from the cultures, and quantitative RT-PCR was performed to assess the levels of zntA and znuA (as a control) mRNA in the cells (Fig. 2). In the parental strain 2308, zntA levels were significantly decreased when the bacteria were exposed to zinc chelation; however, the deletion of zntR ablated the zinc-responsive expression of zntA (Fig. 2A), demonstrating that ZntR functions to silence the constitutive expression of zntA in a zinc-responsive manner. As expected, the depletion of zinc resulted in highly elevated levels of znuA expression in both the parental strain 2308 and the ΔzntR mutant strain compared to levels in these strains when they were grown in zinc-replete medium (Fig. 2B). These results indicate that ZntR is not the predominant regulator of znuA expression and also that znuA expression is exquisitely sensitive to zinc levels.
FIG 2.
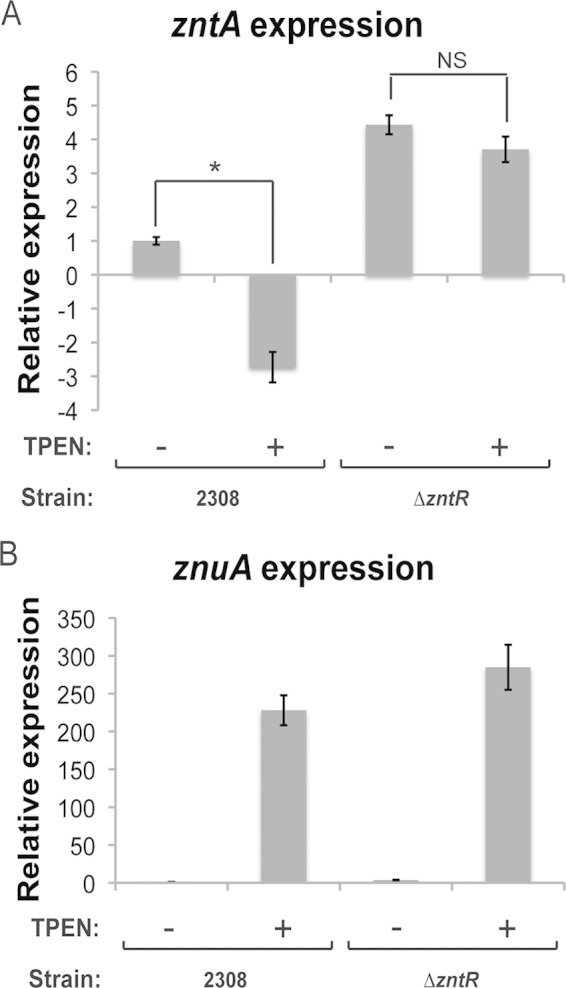
ZntR controls the expression of zntA in a zinc-responsive manner. Quantitative RT-PCR was used to assess the amount of znuA and zntA mRNA produced by B. abortus 2308 or B. abortus ΔzntR incubated in zinc-replete medium, as well as when the same strains were incubated in medium containing the zinc chelator N,N,N′,N′-tetrakis(2-pyridylmethyl)ethylenediamine (TPEN) at a final concentration of 100 μM for 30 min. 16S rRNA levels were assessed as a control, and all mRNA levels were normalized to the level of the parental strain 2308 grown in the absence of TPEN. The graphic bars depict the average level of specific mRNA for each strain and condition listed from two biological replicate experiments, each analyzed in triplicate, and the error bars represent the standard deviations. (A) Relative expression levels of zntA in B. abortus 2308 or the B. abortus ΔzntR mutant strain grown in the absence (−) or presence (+) of TPEN. Statistical significance between the levels of zntA mRNA in a given strain grown with and without TPEN was determined using a t test (*, P < 0.05; NS, not significant). (B) Relative expression levels of znuA in B. abortus 2308 or the B. abortus ΔzntR mutant strain grown in the absence (−) or presence (+) of TPEN.
Zur binds directly to the znuA-znuC intergenic region, and interactions between Zur and DNA require Zn2+.
It is known that Zur binds to the znu promoter regions in other bacteria (21), and therefore, given the role demonstrated for Zur in regulating the expression of the znu genes in Brucella (Fig. 1C), it was hypothesized that Brucella Zur interacts directly with the znu promoters. To test this hypothesis, Brucella Zur was produced in E. coli and purified by affinity chromatography (Materials and Methods), and the recombinant Zur (rZur) was assessed for DNA binding using electrophoretic mobility shift assays (EMSAs). The Brucella rZur bound to the znuA-znuC intergenic region (i.e., a region comprising the znuA promoter and the promoter for znuCB-zur) in a concentration-dependent manner, with binding occurring at concentrations as low as 10 nM rZur (Fig. 3A, upper panel). Importantly, binding between rZur and the radiolabeled znuA-znuC DNA could be inhibited with the addition of unlabeled znuA-znuC competitor DNA but not with the addition of unlabeled DNA corresponding to the zntA-zntR intergenic region. Moreover, rZur did not bind to the Brucella virB promoter (Fig. 3A, lower panel). Taken together, these experiments show that Brucella Zur binds directly to the znuA-znuC promoter regions with high affinity in order to repress expression of znuABC.
FIG 3.
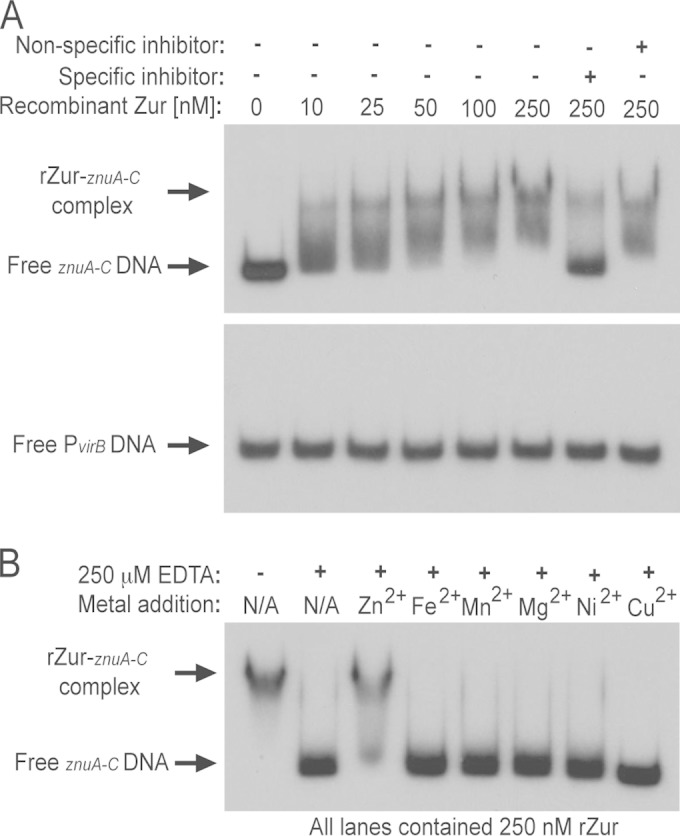
Zur binds directly to the znu (zinc uptake) promoter regions and requires zinc for interactions with DNA. (A) Recombinant Zur (rZur) protein was assessed for binding to the znuA-znuC intergenic region using electrophoretic mobility shift assays (EMSAs). Increasing concentrations of rZur were incubated with radiolabeled znuA-znuC intergenic region DNA, and in some binding reactions, unlabeled specific (znuA-znuC intergenic region DNA) or nonspecific (zntA-zntR intergenic region DNA) competitor DNA fragments were included as controls. The presence (+) and absence (−) of components in the binding reaction mixture are indicated. In addition to testing the binding of rZur to the znuA-znuC intergenic region, interactions between rZur and DNA corresponding to the virB promoter were also assessed as a negative control. (B) EMSAs performed with the znuA-znuC intergenic region DNA and rZur protein in the presence of EDTA and various divalent metal cations. All binding reaction mixtures contained 250 nM rZur and 50 μM ZnCl2, and, where indicated, 250 μM EDTA was included in some samples. To assess the metal specificity of rZur binding to DNA, an additional divalent metal cation (Zn2+, Fe2+, Mn2+, Ni2+, or Cu2+) was added to the binding reaction mixtures at a final concentration of 25 μM in the presence of EDTA.
Fur family proteins typically require metal ions in order to bind to DNA and exert their transcriptional regulatory activities (30). To determine if Brucella Zur requires metal ions, EMSAs were performed with the chelator EDTA added to the binding reaction mixtures (Fig. 3B). The addition of EDTA abolished the binding of rZur to the znuA-znuC intergenic region DNA, indicating that metal ions are necessary for the binding of Brucella Zur to DNA. Importantly, the addition of excess Zn2+ to a binding reaction mixture containing EDTA was able to rescue rZur binding to the znuA-znuC intergenic region DNA, but the addition of other divalent metal cations, such as Fe2+, Mn2+, Mg2+, Ni2+, and Cu2+, was not capable of restoring interactions between rZur and the DNA. These data support the proposition that Zur is a Zn-responsive transcriptional repressor that directly regulates the expression of znuA and znuBC-zur in B. abortus 2308.
ZntR binds directly to the zntA promoter region to control gene expression.
The putative ZntR protein encoded by bab1_2018 in B. abortus 2308 is a MerR family transcriptional regulator. These proteins usually function as transcriptional activators by binding to and bending DNA to expose a functional promoter (29, 31). Given the link between ZntR and zntA expression in Brucella (Fig. 1C and 2), it was hypothesized that ZntR binds to the zntA promoter region, and to test this hypothesis the Brucella ZntR protein was produced in E. coli and purified by affinity chromatography. The purified recombinant ZntR (rZntR) was then used in EMSAs with radiolabeled zntA promoter DNA, and as expected, rZntR bound with high affinity to the zntA promoter region DNA (Fig. 4A). Importantly, the addition of excess unlabeled zntA promoter DNA inhibited binding between rZntR and the labeled zntA DNA, and, moreover, inclusion of excess cueO promoter region DNA did not affect binding of rZntR to the zntA promoter. Additional negative controls were also included (Fig. 4B), and here it was shown that rZntR does not interact with DNA corresponding to the znuA-znuC intergenic region or the promoter of virB.
FIG 4.
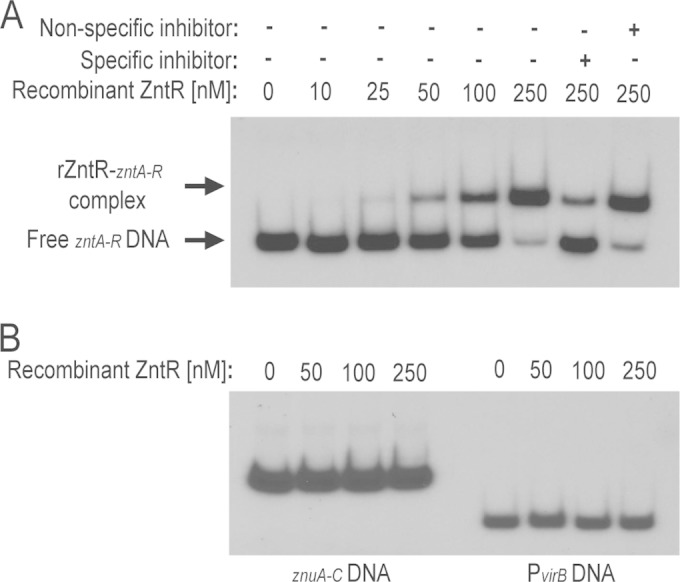
ZntR binds with high affinity to the Brucella zntR-zntA promoter region. (A) Recombinant ZntR (rZntR) protein was assessed for binding to the zntA-zntR intergenic region using electrophoretic mobility shift assays (EMSAs). Increasing concentrations of rZntR were incubated with radiolabeled zntA-zntR intergenic region DNA, and in some binding reactions, unlabeled specific (zntA-zntR intergenic region DNA) or nonspecific (cueO promoter region DNA) competitor DNA fragments were included as controls. The presence (+) and absence (−) of components in the binding reaction mixture are indicated. (B) Specificity of rZntR binding to the zntA-zntR promoter regions. EMSAs were performed as described in panel A, but here, increasing concentrations of rZntR were incubated with radiolabeled znuA-znuC intergenic region DNA or radiolabeled virB promoter (PvirB) region DNA.
In contrast to the situation with Zur (Fig. 3), EDTA did not abolish interactions between rZntR and its target DNA, and, additionally, we did not observe any difference in binding based on the divalent metal cation that was included in the binding reaction mixture (data not shown). These data are not surprising, given that MerR family proteins are known to bind target DNA sequences with high affinity in both the presence and absence of metal cofactors as the inclusion of the metal cation in the protein serves to relax the bent DNA and allow transcription to proceed (31). Altogether, these experiments demonstrate that ZntR from Brucella binds directly to the zntA promoter with high affinity in vitro, suggesting that ZntR interacts with this promoter in vivo in order to control expression of zntA.
The ZntA exporter is required for resistance to Zn toxicity in Brucella abortus 2308.
While the zinc uptake system in Brucella has been characterized by other groups (16, 17), zinc detoxification has not been defined in Brucella. The gene bab1_2019 is predicted to encode an ortholog to the Escherichia coli protein ZntA, which is P-type ATPase that translocates Zn out of the cell (32, 33). Indeed, the Brucella ZntA is 47% identical (64% similar) at the amino acid level to the ZntA protein of E. coli. Therefore, a Brucella abortus zntA mutant was constructed, and this strain, along with the B. abortus zur and zntR mutant strains, was tested for sensitivity to metal toxicity using a disk diffusion assay (Fig. 5). The zur and zntR deletion strains did not exhibit any significant difference in their sensitivities to Zn compared to the sensitivity of the parental strain 2308; however, the zntA mutant was significantly more sensitive to Zn than the other B. abortus strains tested (Fig. 5A). To assess whether the ZntA protein is involved specifically in Zn resistance, the zntA mutant was also assessed for its sensitivity to Ni and Cu. As in the previous experiments, the B. abortus zntA mutant exhibited significantly increased toxicity to Zn compared to that of strain 2308, but there was no difference between strain 2308 and the isogenic zntA mutant in terms of their sensitivities to Ni or Cu (Fig. 5B). The sensitivity of the ΔzntA deletion strain to other divalent metal cations, including magnesium (Mg), manganese (Mn), and iron (Fe), was also accessed; however, we did not observe growth inhibition of the parental strain 2308 or the ΔzntA deletion strain when either was exposed to these metals in the disk diffusion assay (data not shown). Importantly, reconstruction of the zntA locus in the ΔzntA deletion strain resulted in the restoration of the same level of Zn resistance in this strain as that observed in the parental 2308 strain (Fig. 5C). These data support the proposed role of ZntA as a Zn-specific exporter that prevents the toxicity of this metal in B. abortus 2308.
FIG 5.
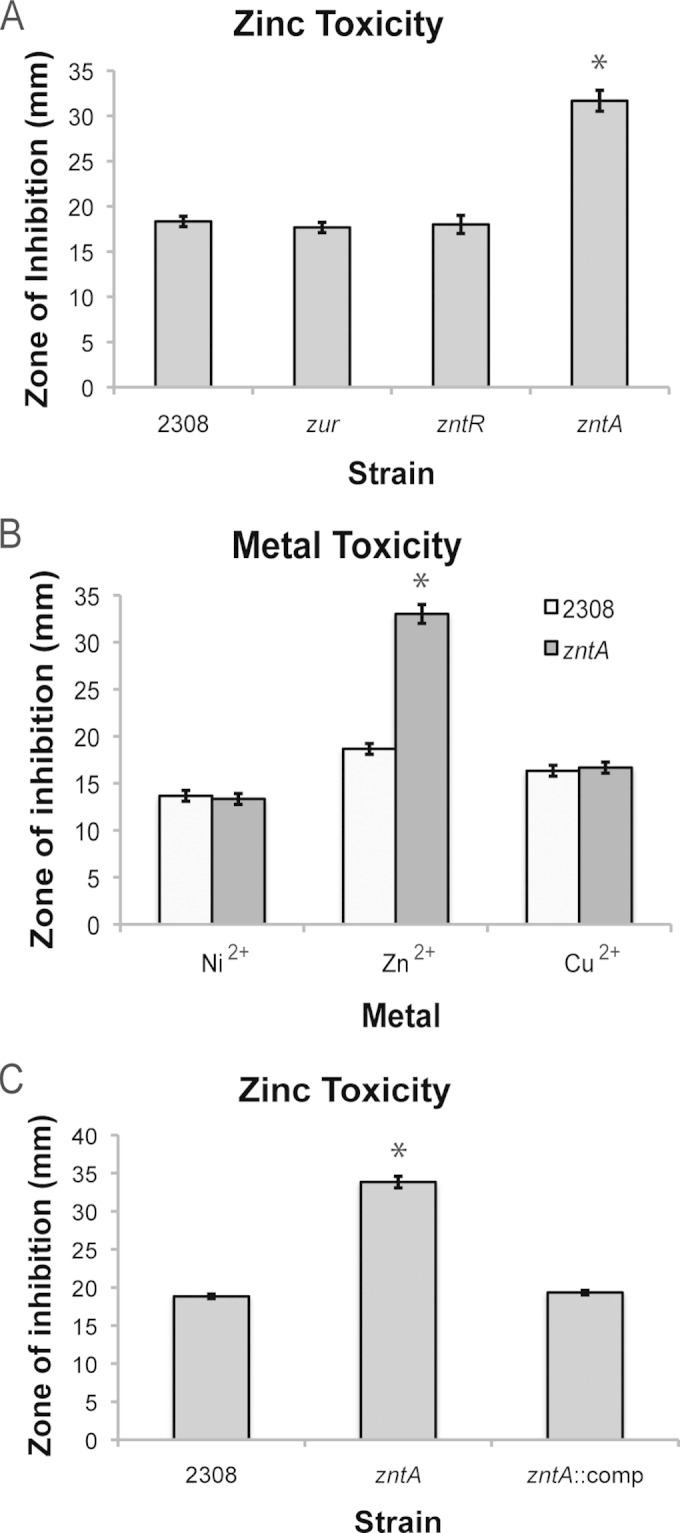
Mutation of zntA, encoding a high-affinity zinc exporter, results in elevated sensitivity to zinc toxicity. (A) Isogenic zur, zntR, and zntA mutants were constructed in Brucella abortus 2308, and these strains were tested in a disk diffusion assay for their comparative susceptibilities to toxic levels of zinc. The results are plotted as the average diameter (±standard deviation) of the zone of inhibition around a disk containing ZnCl2, and the results are from a representative experiment that was performed independently at least three times with triplicate samples for each individual experiment. Statistical significance between results for a given strain and those for the parental strain 2308 was determined by a t test (*, P < 0.05). (B) The zntA mutant is uniquely sensitive to zinc concentrations. A disk diffusion assay was performed to compare the susceptibilities of B. abortus 2308 and the zntA mutant strain to toxic levels of divalent cations, including Ni2+, Zn2+, and Cu2+. The results are shown as the average diameter (±standard deviation) of the zone of inhibition around a disk containing NiCl2, ZnCl2, or CuCl2, and the results are from a single representative experiment that was performed three times independently using triplicate samples for each experiment. Statistical significance between results for the zntA mutant strain and those for the parental strain 2308 was determined by a t test (*, P < 0.05). (C) Genetic complementation of zntA expression restores resistance to zinc toxicity. The wild-type zntA locus was reconstructed on the chromosome of the zntA mutant strain, and this complemented strain (zntA::comp) was assessed for its sensitivity to zinc toxicity using a disk diffusion assay. Data are from a representative experiment performed independently three times using triplicate samples for each experiment. Statistical significance between results for the zntA mutant strain and those for the parental strain 2308 was determined by a t test (*, P < 0.05).
Contribution of zinc homeostasis to Brucella virulence.
The Zn uptake system, ZnuABC, has previously been shown to be required for Brucella virulence (16, 17), and a Brucella znuA mutant was shown to protect mice against subsequent challenge with virulent Brucella strains (17, 18). Moreover, there is evidence that a Brucella znuC mutant is also attenuated in macrophages and HeLa cells (34). To better understand the role of the Zn homeostasis regulators (i.e., Zur and ZntR) and Zn exporter (i.e., ZntA) in Brucella pathogenesis, we assessed the Brucella abortus Δzur, ΔzntR, and ΔzntA mutants for their ability to survive and replicate in cultured murine macrophages and for their virulence in experimentally infected BALB/c mice (Fig. 6). All of the B. abortus strains examined displayed similar intracellular survival and replication patterns in the macrophage cultures (Fig. 6A). Likewise, the isogenic Δzur and ΔzntA mutants exhibited the same capacity to produce chronic spleen infections in mice through 8 weeks postinfection as the parental 2308 strain, but the ΔzntR mutant strain exhibited significant attenuation compared to growth of strain 2308 at 8 weeks postinfection but not at 1 or 4 weeks postinfection (Fig. 6B).
FIG 6.
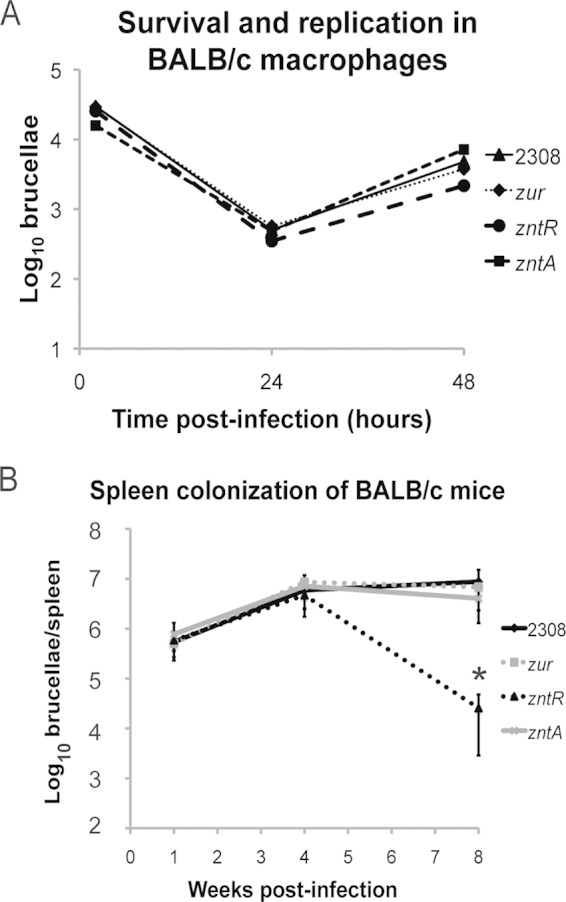
Virulence of B. abortus 2308 and the isogenic zur, zntR, and zntA mutants in murine peritoneal macrophages and experimentally infected mice. (A) Macrophage survival and replication experiments. Cultured resident peritoneal macrophages from BALB/c mice were infected with B. abortus 2308 and the isogenic zur, zntR, and zntA mutant strains. At the indicated times postinfection, the macrophages were lysed, and the number of intracellular brucellae present in these phagocytes was determined by serial dilution, plating, and bacteriologic culture. The results are from a representative experiment that was performed independently four times. (B) Mouse infection experiments. BALB/c mice were infected intraperitoneally with B. abortus 2308 and the isogenic zur, zntR, and zntA mutant strains. Mice were sacrificed at weeks 1, 4, and 8 postinfection, and the number of brucellae colonizing the spleens was determined. The data are presented as the average number of brucellae ± the standard deviation from 5 mice colonized with a specific Brucella strain at a specific time point. These data are from a single experiment using 5 mice per bacterial strain (i.e., n = 5). Statistical significance between results for the zntR mutant strain and those for the parental strain 2308 at 8 weeks postinfection was determined by a t test (*, P < 0.05).
Given that deletion of zntR results in elevated levels of zntA (Fig. 1C) and based on the results of previous studies suggesting that maintaining proper Zn levels in the bacterial cell is important for Brucella virulence (16, 17), we hypothesized that the overexpression of zntA in the ΔzntR mutant is the reason for its attenuation. To test this hypothesis, a ΔzntR zntA double mutant was constructed, and the virulence of this strain, as well as that of a genetically complemented zntR mutant, was assessed in experimentally infected mice. Similar to the first mouse experiment, the ΔzntR mutant strain exhibited significant attenuation at 8 weeks postinfection (Fig. 7A). In contrast, the ΔzntR zntA double mutant and the derivative of the zntR mutant with a reconstructed zntR gene displayed wild-type virulence in mice at this experimental time point. These experimental findings not only confirm the genetic link between the zntR mutation and the attenuation exhibited by the corresponding mutant in mice but also provide evidence that ZntR plays an important role in Brucella virulence by regulating zntA expression.
FIG 7.
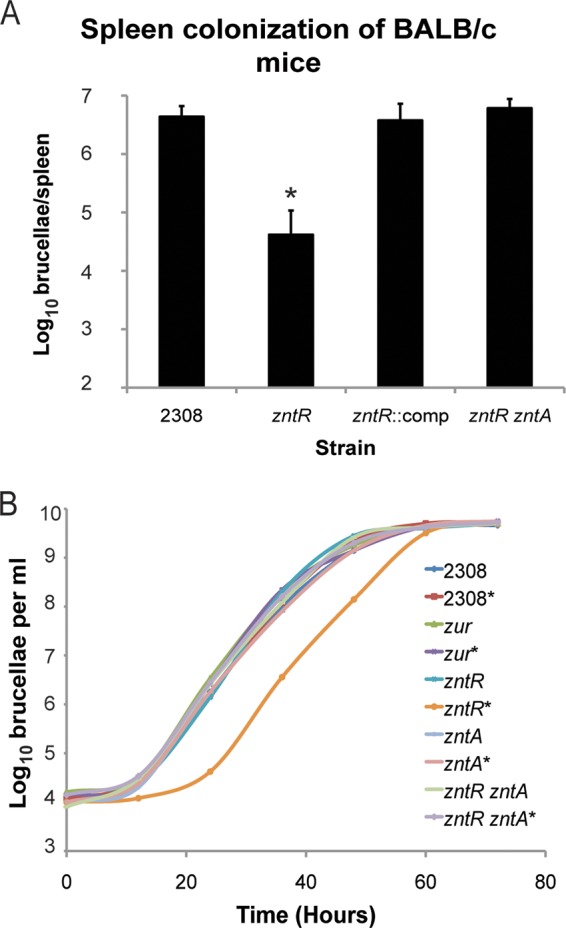
Unregulated expression of zntA in the B. abortus zntR mutant strain leads to attenuation. (A) Mouse infection experiments. BALB/c mice were infected intraperitoneally with Brucella strains, including the parental strain 2308, the isogenic zntR mutant strain, the zntR complemented strain (zntR::comp), and the zntR zntA double mutant strain. The mice were euthanized after 8 weeks, and the bacterial loads in the spleens were assessed. The bars depict the average number of brucellae ± the standard deviation from the 5 mice colonized with a specific Brucella strain. These data are from a single experiment using 5 mice per bacterial strain (i.e., n = 5). Statistical significance between results for the zntR mutant strain and those for the parental strain 2308 at 8 weeks postinfection was determined by a t test (*, P < 0.05). (B) Growth of Brucella strains in zinc-replete and zinc-chelated medium. Brucella strains were grown in rich medium in the presence (indicated by an asterisk) or absence of a 10 μM concentration of the zinc-specific chelator TPEN, and at the specified time points, counts of viable CFU per ml of culture were determined. The graph depicts a representative experiment from three independent trials.
In light of the mouse experiments demonstrating that zntA overexpression is the genetic basis for the attenuation of the zntR mutant (Fig. 7A), the Brucella zinc homeostasis mutant strains were tested for the ability to grow in zinc-chelated medium to determine the effects of zinc limitation on the growth of these mutants (Fig. 7B). For these experiments, Brucella strains were grown in rich medium in the absence or presence of 10 μM N,N,N′,N′-tetrakis(2-pyridylmethyl)ethylenediamine (TPEN), which is a zinc-specific chelator. The parental strain B. abortus 2308 exhibited similar growth kinetics in both the absence and presence of TPEN, as did the zur and zntA mutant strains. Conversely, the zntR mutant strain displayed a growth defect in the zinc-limiting medium, but ultimately the zntR mutant was able to reach the same cell density as the other strains. As expected, growth of a zntR mutant strain with a deletion of the gene encoding the zinc exporter, zntA (i.e., the zntR zntA double mutant strain), was similar to that of the parental strain in both the zinc-replete and zinc-limited environments. These data indicate that loss of some components of the zinc homeostasis regulatory system, specifically zur and zntA, does not significantly affect the ability of B. abortus to grow in zinc-deprived environments; however, zntR is required for optimal growth in zinc-limiting situations due to the capacity of ZntR to curtail the expression of zntA.
DISCUSSION
Given the dichotomy of divalent metal cations being essential yet often toxic to cells, it is not surprising that systems have evolved to coordinate the import, export, and storage of specific metal cations in bacteria. The present study demonstrates that Brucella abortus possesses the systems responsible for Zn uptake (i.e., the ZnuABC system) and Zn export (the ZntA system) and, moreover, that B. abortus employs specific transcriptional regulatory proteins to synchronize the expression of these systems. The ZnuABC system was previously reported to function as a Zn uptake system, and, importantly, mutation in the znu genes leads to attenuation of Brucella in macrophages and mice (16, 17). The function of ZntA as a zinc exporter has been predicted based on its similarity to other Zn export systems (35), but the present study is the first to experimentally validate this previous prediction (Fig. 5). Similarly, the Zur and ZntR proteins were previously hypothesized to function as Zn-responsive regulators of the znuABC and zntA genes, respectively (35), and our data confirm the roles of the proteins as Zn-responsive transcriptional regulators of these Zn transporters (Fig. 1, 2, 3, and 4).
As mentioned in the previous paragraph, the Znu zinc uptake system is required for the wild-type virulence of Brucella (16, 17). These data indicate that zinc is a limiting micronutrient for the brucellae during infection of the host, and, moreover, these experiments underscore the critical role of Zn sequestration in the host defense against invading microorganisms (36). It is important to note that while the Znu system apparently serves as the major high-affinity Zn import system in Brucella, there must be a low-affinity Zn import system in Brucella as well or the construction of znu mutation strains would not have been possible. Nonetheless, the low-affinity Zn transporter is not capable of compensating for the loss of ZnuABC activity in vivo.
In contrast to the situation with the Znu system, the ZntA zinc exporter is not essential for Brucella virulence (Fig. 6). This is interesting in light of recent work demonstrating the requirement of the CtpC zinc exporter for the survival of Mycobacterium tuberculosis in host macrophages during infection (37). The intramacrophagic vacuole containing M. tuberculosis is flooded with Zn in an attempt to kill the bacteria, but M. tuberculosis neutralizes this potentially toxic influx of Zn by elevating production of CtpC (37, 38). These data, along with the data presented in this study, suggest that macrophages employ opposing Zn-based strategies to combat infections by Brucella and M. tuberculosis while also highlighting the evolutionary adaptations that each bacterium has acquired to cope with the various intravacuolar Zn stresses (i.e., elevated toxic levels of Zn versus growth-inhibiting low concentrations of Zn). In terms of the intravacuolar environment, these data suggest that the Brucella-containing vacuole is not comprised of the same divalent metal cation importers that the Mycobacterium-containing vacuole employs to combat the bacteria during an intracellular infection. Additionally, the fact that zntA is dispensable for Brucella virulence further suggests that Zn levels are severely diminished in the Brucella-containing vacuole and that Zn acquisition, but not Zn detoxification via ZntA, is paramount for the survival of Brucella in the host.
While the deletion of zur or zntA does not result in reduced Brucella virulence, the loss of ZntR leads to attenuation in the mouse model late in the infection (i.e., at 8 weeks postinfection) (Fig. 6). We hypothesized that unrepressed expression of zntA in the zntR mutant strain was the basis for the attenuation because the uncontrolled export of Zn from the brucellae in an already Zn-deficient environment would potentially starve the cells, leading to death. Indeed, deletion of zntA in the zntR mutant background alleviated the attenuation observed for the ΔzntR strain (Fig. 7A). These data further indicate that Brucella strains encounter an intramacrophagic environment in which Zn is a limiting nutrient for the bacteria.
Proper zinc homeostasis is clearly required for the pathogenesis of Brucella (Fig. 6 and 7) (16, 17), but the molecular requirements for zinc in Brucella are not fully understood. Nonetheless, several zinc-containing proteins and enzymes are important for Brucella, including the superoxide dismutase C (SodC), which is required for Brucella virulence (10, 11), the carbonic anhydrases I and II, and the histidinol dehydrogenase (12). Additionally, MucR is a zinc finger transcriptional regulatory protein that controls the expression of numerous virulence-associated genetic systems, and MucR is required for Brucella pathogenesis (13, 14, 39, 40). Therefore, the attenuation of the ΔzntR mutant strain may result from a combination of deficiencies related to the lack of sufficient zinc, including the failure of the bacteria to mount an appropriate oxidative stress response, multiple metabolic aberrations, and the inability to coordinate gene expression required for survival within the host. Together, these results indicate that Zn plays wide-ranging and essential roles in the molecular activities of several important proteins and enzymes in Brucella, and the inactivation of these systems through inadequate zinc availability reduces the capacity of Brucella to cause a chronic infection.
ACKNOWLEDGMENTS
We thank Evan Menscher and Ahmed Elhassanny for helpful discussions regarding experimental design and constructive criticism.
This study was supported by funds from the Virginia-Maryland College of Veterinary Medicine and the Fralin Life Science Institute at Virginia Tech to C.C.C. and by grants (AI48499 and AI63516) from NIAID to R.M.R.
REFERENCES
- 1.Pappas G. 2010. The changing Brucella ecology: novel reservoirs, new threats. Int J Antimicrob Agents 36(Suppl 1):S8–S11. doi: 10.1016/j.ijantimicag.2010.06.013. [DOI] [PubMed] [Google Scholar]
- 2.Celli J. 2006. Surviving inside a macrophage: the many ways of Brucella. Res Microbiol 157:93–98. doi: 10.1016/j.resmic.2005.10.002. [DOI] [PubMed] [Google Scholar]
- 3.Roop RM II, Gaines JM, Anderson ES, Caswell CC, Martin DW. 2009. Survival of the fittest: how Brucella strains adapt to their intracellular niche in the host. Med Microbiol Immunol 198:221–238. doi: 10.1007/s00430-009-0123-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Forbes JR, Gros P. 2001. Divalent-metal transport by NRAMP proteins at the interface of host-pathogen interactions. Trends Microbiol 9:397–403. doi: 10.1016/S0966-842X(01)02098-4. [DOI] [PubMed] [Google Scholar]
- 5.Summers AO. 2009. Damage control: regulating defenses against toxic metals and metalloids. Curr Opin Microbiol 12:138–144. doi: 10.1016/j.mib.2009.02.003. [DOI] [PubMed] [Google Scholar]
- 6.Waldron KJ, Robinson NJ. 2009. How do bacterial cells ensure that metalloproteins get the correct metal? Nat Rev Microbiol 7:25–35. doi: 10.1038/nrmicro2057. [DOI] [PubMed] [Google Scholar]
- 7.Imlay JA, Linn S. 1988. DNA damage and oxygen radical toxicity. Science 240:1302–1309. doi: 10.1126/science.3287616. [DOI] [PubMed] [Google Scholar]
- 8.Imlay JA, Chin SM, Linn S. 1988. Toxic DNA damage by hydrogen peroxide through the Fenton reaction in vivo and in vitro. Science 240:640–642. doi: 10.1126/science.2834821. [DOI] [PubMed] [Google Scholar]
- 9.Andreini C, Banci L, Bertini I, Rosato A. 2006. Zinc through the three domains of life. J Proteome Res 5:3173–3178. doi: 10.1021/pr0603699. [DOI] [PubMed] [Google Scholar]
- 10.Tatum FM, Detilleux PG, Sacks JM, Halling SM. 1992. Construction of Cu-Zn superoxide dismutase deletion mutants of Brucella abortus: analysis of survival in vitro in epithelial and phagocytic cells and in vivo in mice. Infect Immun 60:2863–2869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gee JM, Valderas MW, Kovach ME, Grippe VK, Robertson GT, Ng WL, Richardson JM, Winkler ME, Roop RM II. 2005. The Brucella abortus Cu,Zn superoxide dismutase is required for optimal resistance to oxidative killing by murine macrophages and wild-type virulence in experimentally infected mice. Infect Immun 73:2873–2880. doi: 10.1128/IAI.73.5.2873-2880.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lopez M, Köhler S, Winum JY. 2012. Zinc metalloenzymes as new targets against the bacterial pathogen Brucella. J Inorg Biochem 111:138–145. doi: 10.1016/j.jinorgbio.2011.10.019. [DOI] [PubMed] [Google Scholar]
- 13.Mirabella A, Terwagne M, Zygmunt MS, Cloeckaert A, De Bolle X, Letesson JJ. 2013. Brucella melitensis MucR, an orthologue of Sinorhizobium meliloti MucR, is involved in resistance to oxidative, detergent, and saline stresses and cell envelope modifications. J Bacteriol 195:453–465. doi: 10.1128/JB.01336-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Caswell CC, Elhassanny AE, Planchin EE, Roux CM, Weeks-Gorospe JN, Ficht TA, Dunman PM, Roop RM II. 2013. Diverse genetic regulon of the virulence-associated transcriptional regulator MucR in Brucella abortus 2308. Infect Immun 81:1040–1051. doi: 10.1128/IAI.01097-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Herrou J, Crosson S. 2013. Molecular structure of the Brucella abortus metalloprotein RicA, a Rab2-binding virulence effector. Biochemistry 52:9020–9028. doi: 10.1021/bi401373r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kim S, Watanabe K, Shirahata T, Watarai M. 2004. Zinc uptake system (znuA locus) of Brucella abortus is essential for intracellular survival and virulence in mice. J Vet Med Sci 66:1059–1063. doi: 10.1292/jvms.66.1059. [DOI] [PubMed] [Google Scholar]
- 17.Yang X, Becker T, Walters N, Pascual DW. 2006. Deletion of znuA virulence factor attenuates Brucella abortus and confers protection against wild-type challenge. Infect Immun 74:3874–3879. doi: 10.1128/IAI.01957-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Clapp B, Skyberg JA, Yang X, Thornburg T, Walters N, Pascual DW. 2011. Protective live oral brucellosis vaccines stimulate Th1 and th17 cell responses. Infect Immun 79:4165–4174. doi: 10.1128/IAI.05080-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hantke K. 2001. Bacterial zinc transporters and regulators. Biometals 14:239–249. doi: 10.1023/A:1012984713391. [DOI] [PubMed] [Google Scholar]
- 20.Chandra BR, Yogavel M, Sharma A. 2007. Structural analysis of ABC-family periplasmic zinc binding protein provides new insights into mechanism of ligand uptake and release. J Mol Biol 367:970–982. doi: 10.1016/j.jmb.2007.01.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hantke K. 2005. Bacterial zinc uptake and regulators. Curr Opin Microbiol 8:196–202. doi: 10.1016/j.mib.2005.02.001. [DOI] [PubMed] [Google Scholar]
- 22.Hobman JL, Wilkie J, Brown NL. 2005. A design for life: prokaryotic metal-binding MerR family regulators. Biometals 18:429–436. doi: 10.1007/s10534-005-3717-7. [DOI] [PubMed] [Google Scholar]
- 23.Gerhardt P. 1958. The nutrition of brucellae. Bacteriol Rev 22:81–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Caswell CC, Gaines JM, Roop RM II. 2012. The RNA chaperone Hfq independently coordinates expression of the VirB type IV secretion system and the LuxR-type regulator BabR in Brucella abortus 2308. J Bacteriol 194:3–14. doi: 10.1128/JB.05623-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pfaffl MW. 2001. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res 29:e45. doi: 10.1093/nar/29.9.e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gaballa A, Helmann JD. 1998. Identification of a zinc-specific metalloregulatory protein, Zur, controlling zinc transport operons in Bacillus subtilis. J Bacteriol 180:5815–5821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Patzer SI, Hantke K. 2000. The zinc-responsive regulator Zur and its control of the znu gene cluster encoding the ZnuABC zinc uptake system in Escherichia coli. J Biol Chem 275:24321–24332. doi: 10.1074/jbc.M001775200. [DOI] [PubMed] [Google Scholar]
- 28.Ellison ML, Farrow JM, Parrish W, Danell AS, Pesci EC. 2013. The transcriptional regulator Np20 is the zinc uptake regulator in Pseudomonas aeruginosa. PLoS One 8:e75389. doi: 10.1371/journal.pone.0075389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ansari AZ, Bradner JE, O'Halloran TV. 1995. DNA-bend modulation in a repressor-to-activator switching mechanism. Nature 374:371–375. [DOI] [PubMed] [Google Scholar]
- 30.Troxell B, Hassan HM. 2013. Transcriptional regulation by ferric uptake regulator (Fur) in pathogenic bacteria. Front Cell Infect Microbiol 3:59. doi: 10.3389/fcimb.2013.00059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Outten CE, Outten FW, O'Halloran TV. 1999. DNA distortion mechanism for transcriptional activation by ZntR, a Zn(II)-responsive MerR homologue in Escherichia coli. J Biol Chem 274:37517–37524. doi: 10.1074/jbc.274.53.37517. [DOI] [PubMed] [Google Scholar]
- 32.Beard SJ, Hashim R, Membrillo-Hernández J, Hughes MN, Poole RK. 1997. Zinc(II) tolerance in Escherichia coli K-12: evidence that the zntA gene (o732) encodes a cation transport ATPase. Mol Microbiol 25:883–891. doi: 10.1111/j.1365-2958.1997.mmi518.x. [DOI] [PubMed] [Google Scholar]
- 33.Rensing C, Mitra B, Rosen BP. 1997. The zntA gene of Escherichia coli encodes a Zn(II)-translocating P-type ATPase. Proc Natl Acad Sci U S A 94:14326–14331. doi: 10.1073/pnas.94.26.14326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Delrue RM, Lestrate P, Tibor A, Letesson JJ, De Bolle X. 2004. Brucella pathogenesis, genes identified from random large-scale screens. FEMS Microbiol Lett 231:1–12. doi: 10.1016/S0378-1097(03)00963-7. [DOI] [PubMed] [Google Scholar]
- 35.Roop RM II, Anderson E, Ojeda J, Martinson D, Menscher E, Martin DW. 2012. Metal acquisition by Brucella strains, p 179–199. In López-G̃oni I, O'Callaghan D (ed), Brucella: molecular microbiology and genomics. Caister Academic Press, Norfolk, United Kingdom. [Google Scholar]
- 36.Kehl-Fie TE, Skaar EP. 2010. Nutritional immunity beyond iron: a role for manganese and zinc. Curr Opin Chem Biol 14:218–224. doi: 10.1016/j.cbpa.2009.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Botella H, Peyron P, Levillain F, Poincloux R, Poquet Y, Brandli I, Wang C, Tailleux L, Tilleul S, Charrière GM, Waddell SJ, Foti M, Lugo-Villarino G, Gao Q, Maridonneau-Parini I, Butcher PD, Castagnoli PR, Gicquel B, de Chastellier C, Neyrolles O. 2011. Mycobacterial p(1)-type ATPases mediate resistance to zinc poisoning in human macrophages. Cell Host Microbe 10:248–259. doi: 10.1016/j.chom.2011.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Russell DG. 2011. The galvanizing of Mycobacterium tuberculosis: an antimicrobial mechanism. Cell Host Microbe 10:181–183. doi: 10.1016/j.chom.2011.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Arenas-Gamboa AM, Rice-Ficht AC, Kahl-McDonagh MM, Ficht TA. 2011. Protective efficacy and safety of Brucella melitensis 16MΔmucR against intraperitoneal and aerosol challenge in BALB/c mice. Infect Immun 79:3653–3658. doi: 10.1128/IAI.05330-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Dong H, Liu W, Peng X, Jing Z, Wu Q. 2013. The effects of MucR on expression of type IV secretion system, quorum sensing system and stress responses in Brucella melitensis. Vet Microbiol 166:535–542. doi: 10.1016/j.vetmic.2013.06.023. [DOI] [PubMed] [Google Scholar]
- 41.Jacobs C, Domian IJ, Maddock JR, Shapiro L. 1999. Cell cycle-dependent polar localization of an essential bacterial histidine kinase that controls DNA replication and cell division. Cell 97:111–120. [DOI] [PubMed] [Google Scholar]


