ABSTRACT
Campylobacter jejuni is a leading cause of bacterial diarrheal disease and a frequent commensal of the intestinal tract in poultry and other animals. For optimal growth and colonization of hosts, C. jejuni employs two-component regulatory systems (TCSs) to monitor environmental conditions and promote proper expression of specific genes. We analyzed the potential of C. jejuni Cjj81176_1484 (Cjj1484) and Cjj81176_1483 (Cjj1483) to encode proteins of a cognate TCS that influences expression of genes possibly important for C. jejuni growth and colonization. Transcriptome analysis revealed that the regulons of the Cjj81176_1484 (Cjj1484) histidine kinase and the Cjj81176_1483 (Cjj1483) response regulator contain many common genes, suggesting that these proteins likely form a cognate TCS. We found that this TCS generally functions to repress expression of specific proteins with roles in metabolism, iron/heme acquisition, and respiration. Furthermore, the TCS repressed expression of Cjj81176_0438 and Cjj81176_0439, which had previously been found to encode a gluconate dehydrogenase complex required for commensal colonization of the chick intestinal tract. However, the TCS and other specific genes whose expression is repressed by the TCS were not required for colonization of chicks. We observed that the Cjj1483 response regulator binds target promoters in both unphosphorylated and phosphorylated forms and influences expression of some specific genes independently of the Cjj1484 histidine kinase. This work further expands the signaling mechanisms of C. jejuni and provides additional insights regarding the complex and multifactorial regulation of many genes involved in basic metabolism, respiration, and nutrient acquisition that the bacterium requires for optimal growth in different environments.
IMPORTANCE Bacterial two-component regulatory systems (TCSs) link environmental cues to expression of specific genes that enable optimal bacterial growth or colonization of hosts. We found that the Campylobacter jejuni Cjj1484 histidine kinase and Cjj1483 response regulator function as a cognate TCS to largely repress expression of target genes encoding a gluconate dehydrogenase complex required for commensal colonization of the chick intestinal tract, as well as other genes encoding proteins for heme or iron acquisition, metabolism, and respiration. We also discovered different modes by which Cjj1483 may mediate repression with and without Cjj1484. This work provides insight into the signal transduction mechanisms of a leading cause of bacterial diarrheal disease and emphasizes the multifactorial and complex regulation of specific biological processes in C. jejuni.
INTRODUCTION
Despite increasing surveillance and food safety measures, the number of cases of foodborne diarrheal disease caused by Campylobacter jejuni has increased over the last few decades. As such, C. jejuni remains one of the leading causes of diarrheal disease in the United States and many other developed countries throughout the world (1, 2). C. jejuni is a natural commensal bacterium of many wild and agriculturally important animals, particularly poultry (3, 4). In chickens, C. jejuni promotes a persistent colonization of the mucus layer lining the ceca and large intestine without causing disease (5). These zoonotic infections lead to contamination of poultry meats for human consumption (6). In humans, C. jejuni adheres to and invades epithelial cells of the lower intestinal tract and colon, which stimulates inflammation that often leads to watery or bloody diarrhea (7). Infection of humans is typically self-limiting. However, Guillain-Barré syndrome, an autoimmune disorder that results in an acute paralysis of the peripheral nervous system, may develop after diarrheal disease (8, 9).
Many pathogenic bacteria utilize two-component regulatory systems (TCSs) to link environmental cues to transcription of specific genes. TCSs may function by activating and/or repressing transcription of different sets of genes. Typical TCSs are composed of a sensor histidine kinase (HK) and a DNA-binding response regulator (RR) (10, 11). Upon sensing a specific signal, the HK autophosphorylates at a conserved histidine residue (12). This phosphohistidine then serves as the substrate for the RR, which catalyzes the transfer of the phosphate to an aspartic acid on its receiver domain. Once phosphorylated, the RR can influence gene expression by binding to promoter regions of target genes to mediate either transcriptional activation or repression. In some instances, the HK also exhibits phosphatase activity that can dephosphorylate the response regulator (10). The latter mechanism is a means to provide finite control of the response regulator in the absence of a stimulus.
Relative to many bacterial species, C. jejuni contains few TCSs, with only up to seven cognate TCSs predicted to be encoded in the genome (13, 14). Through studies by many groups, five cognate TCSs of C. jejuni have been characterized, including FlgSR, DccRS, PhosSR, CprRS, and RacRS. Many of these have been implicated in controlling expression of genes required for host interactions. For instance, the FlgSR TCS monitors formation of the flagellar type III secretion system, the MS ring, and rotor structures to activate expression of σ54-dependent flagellar genes required for motility and commensal colonization (15–21). Furthermore, DccRS, CprRS, and RacRS are required for wild-type (WT) levels of commensal colonization of the chick ceca (22–26). Whereas DccRS directly influences expression of genes encoding colonization determinants (22), CprRS is required for expression of genes for biofilm formation (24). RacRS is required for expression of genes encoding proteins for the heat shock response and fumarate metabolism, depending on the strain and growth conditions used for analysis (25–27). Additionally, the PhosSR TCS is required for expression of 12 genes, 5 of which encode proteins required for phosphate acquisition (28). However, this TCS is not required for in vivo colonization under the parameters tested.
Another cognate TCS may be encoded by Cjj81176_1484 (Cjj1484; Cj1492c as annotated in the C. jejuni NCTC11168 genome [14]) and the immediately downstream gene, Cjj81176_1483 (Cjj1483; Cj1491c as annotated in the C. jejuni NCTC11168 genome [14]). Cjj1484 is predicted to encode a putative sensor HK (Cjj1484), whereas Cjj1483 appears to encode a putative DNA-binding RR (Cjj1483). This TCS appears to be conserved in a wide variety of C. jejuni strains. In this study, we investigated the potential for these genes to encode a cognate TCS that may influence transcription of genes required for host interactions. Our findings indicate that Cjj1484 and Cjj1483 appear to function as a cognate TCS mainly involved in transcriptional repression of genes encoding proteins involved in commensal colonization of chicks and metabolic processes related to redox potential and iron or heme acquisition when C. jejuni is grown in vitro. In addition, the TCS is modestly required for transcription of an invasion gene. Furthermore, we provide evidence that the Cjj1483 RR may function independently of the Cjj1484 HK to modulate transcription of some genes. Finally, we show that Cjj1483 can be phosphorylated and that specific DNA binding to target promoters occurs with and without phosphorylation. We propose that the previously uncharacterized Cjj1484-Cjj1483 TCS of C. jejuni represses expression of some colonization factors outside the host and likely functions in combination with other transcription factors to fine-tune expression of specific genes involved in various processes that are important for the biology of C. jejuni.
MATERIALS AND METHODS
Bacterial strains and plasmids.
All strains and plasmids used in this study are listed in Tables S1 and S2 in the supplemental material. C. jejuni strains were routinely grown from freezer stocks under microaerobic conditions (10% CO2, 5% O2, and 85% N2) on Mueller-Hinton (MH) agar containing 10 μg/ml trimethoprim at 37°C for 48 h. Strains were then restreaked onto appropriate media and grown for an additional 16 h. When appropriate, antibiotics were added to media at the following concentrations: 20 μg/ml chloramphenicol, 100 μg/ml kanamycin, 30 μg/ml cefoperazone, and 0.1, 0.5, 1, 2, or 5 mg/ml streptomycin. Escherichia coli DH5α and BL21 strains were grown on LB agar or in LB broth containing 100 μg/ml ampicillin, 100 μg/ml kanamycin, 12.5 μg/ml tetracycline, or 20 μg/ml chloramphenicol, as necessary. C. jejuni strains were stored at −80°C in a mixture of 85% MH broth and 15% glycerol. E. coli strains were stored at −80°C in a mixture of 80% LB broth and 20% glycerol.
Construction of C. jejuni mutants.
C. jejuni mutants were constructed through electroporation by previously published methods (29). Briefly, the Cjj1484-Cjj1483 locus was amplified via PCR from the C. jejuni 81-176 chromosome by using primers containing 5′ BamHI sites located approximately 750 bp upstream and downstream of the locus. The PCR product was cloned into the BamHI site of pUC19 to create pDAR812 (pUC19::Cjj1484-1483). An HpaI site was created in Cjj1484 on pDAR812 through PCR-mediated mutagenesis, resulting in pPML101 (30, 31). A cat-rpsL resistance cassette was removed from pDRH265 by SmaI digestion and ligated into the HpaI site within Cjj1484 or the EcoRV site within Cjj1483 (29). Resultant plasmids contained the cat-rpsL cassette within Cjj1484 (pPML107) or Cjj1483 (pPML102).
Plasmids pPML107 and pPML102 were subsequently electroporated into C. jejuni 81-176 Smr (DRH212) or C. jejuni 81-176 Smr ΔastA (DRH461), and transformants were recovered on MH agar plates containing chloramphenicol to obtain PML321 (81-176 Smr Cjj1484::cat-rpsL), PML322 (81-176 Smr Cjj1483::cat-rpsL), and PML324 (81-176 Smr ΔastA Cjj1483::cat-rpsL) (15, 29).
In-frame deletions of Cjj1484 and Cjj1483 were constructed by PCR-mediated mutagenesis using pDAR812 (30). The mutations were verified by DNA sequencing and resulted in plasmids pPML113 (pUC19::ΔCjj1483) and pPML334 (pUC19::ΔCjj1484). These plasmids were then electroporated into C. jejuni strains containing cat-rpsL interruptions in the respective genes on the chromosome, and mutants were recovered on MH agar containing 0.5 to 5 mg/ml streptomycin. Deletion of Cjj1484 and Cjj1483 was verified by colony PCR, resulting in the creation of PML335 (81-176 Smr ΔCjj1483), PML337 (81-176 Smr ΔastA ΔCjj1483), and PML360 (81-176 Smr ΔCjj1484).
To create point mutations in the predicted catalytic residues of Cjj1484 and Cjj1483, PCR-mediated mutagenesis was performed using pDAR812 as the template to create pPML239 (pUC19::Cjj1483D58A), pPML340 (pUC19::Cjj1483D58E), pPML732 (pUC19::Cjj1483D58N), and pPML817 (pUC19::Cjj1484H195A) (30). pPML239 was electroporated into PML322, and pPML340 and pPML732 were electroporated into PML324 to replace Cjj1483::cat-rpsL on the chromosome with the respective genes containing point mutations. Mutants were recovered on MH agar containing 0.5 to 5 mg/ml streptomycin and were verified by colony PCR and DNA sequencing to result in PML242 (81-176 Smr Cjj1483D58A), PML739 (81-176 Smr ΔastA Cjj1483D58E), and PML769 (81-176 Smr ΔastA Cjj1483D58N).
The Cjj81176_0063-Cjj81176_0064 (Cjj0063-Cjj0064), Cjj81176_1386-Cjj81176_1385 (Cjj1386-Cjj1385), Cjj81176_1619-Cjj81176_1620 (Cjj1619-Cjj1620), and chuCD loci were amplified from the C. jejuni 81-176 chromosome via PCR, using primers containing 5′ BamHI sites located approximately 750 bp upstream and downstream of each locus. The PCR products were cloned into the BamHI site of pUC19 to create pLKB156 (pUC19::chuCD), pPML770 (pUC19::Cjj0063-0064), pPML771 (pUC19::Cjj1386-1385), and pPML772 (pUC19::Cjj1619-1620). An HpaI site was created in Cjj1619 (on pPML772) through PCR-mediated mutagenesis to create pPML1247 (30, 31). A cat-rpsL resistance cassette was removed from pDRH265 by SmaI digestion and was ligated into the ClaI site within chuC, the EcoRV site within Cjj0064, or the HpaI site within Cjj1386 or Cjj1619, resulting in pPML1249, pPML1253, pPML1254, and pPML1261, respectively (29).
Plasmids pPML1249, pPML1253, pPML1254, and pPML1261 were subsequently electroporated into C. jejuni 81-176 Smr (DRH212) and were recovered on MH agar plates containing chloramphenicol. The following mutants were obtained and confirmed by colony PCR: PML1262 (81-176 Smr Cjj0064::cat-rpsL), PML1274 (81-176 Smr chuC::cat-rpsL), PML1280 (81-176 Smr Cjj1386::cat-rpsL), and PML1305 (81-176 Smr Cjj1619::cat-rpsL).
Construction of plasmids for complementation of C. jejuni in trans.
To create plasmids to be used for complementation in trans, a fragment beginning 400 bp upstream of the Cjj1483 translational start site and ending at the Cjj1483 stop codon was amplified from pDAR812 by a PCR using primers with 5′ BamHI sites. Following restriction digestion, the fragment was ligated into the BamHI site of pRY112 to create pPML533 (pRY112::Cjj1483) (32). A fragment containing the 203 bp upstream of the start codon of flaA followed by an N-terminal FLAG tag was amplified by a PCR using primers with a 5′ XbaI site and a 3′ BamHI site. Following restriction digestion, the fragment was inserted into the corresponding sites in pRY108 to create pDAR1604 (pRY108::PflaA-N FLAG). This plasmid allowed for expression of N-terminally FLAG-tagged proteins from the strong flaA promoter of C. jejuni. To create a Cjj1484 complementation plasmid, the region from codon 2 to the stop codon of Cjj1484 was amplified from pDAR812 by use of primers containing 5′ BamHI sites. This fragment was digested with BamHI and then ligated into the BamHI site of pDAR1604 to create pPML968 (pDAR1604::Cjj1484). In addition, we amplified Cjj1484H195A from pPML817 in a manner similar to that described above and cloned the gene into the BamHI site of pDAR1604 to create pPML1119 (pDAR1604::Cjj1484H195A).
Plasmids pRY112, pPML533, pPML968, pPML1119, and pDAR1604 were transformed into chemically competent E. coli DH5α/pRK212.1, which contains the conjugation transfer element (33). These plasmids were then conjugated into appropriate C. jejuni strains as previously described (34). Transconjugants containing pPML533 and pRY112 were recovered on MH agar containing trimethoprim, streptomycin, and chloramphenicol. Transconjugants containing pPML968, pPML1119, and pDAR1604 were recovered on MH agar containing trimethoprim, streptomycin, and kanamycin. The presence of the correct plasmid was confirmed by colony PCR.
Primer extension analysis.
WT C. jejuni was grown from a freezer stock on MH agar containing trimethoprim for 48 h at 37°C under microaerobic conditions and then restreaked on MH agar and grown for another 16 h. Total RNA was extracted with RiboZol (Amresco). To identify the transcriptional start site of Cjj1483, a primer (5′-ATCAATGATTCTCTAGCTT-3′) that bound 50 bases downstream of the start codon was used. The primer was end labeled with [γ-32P]ATP by using a polynucleotide kinase from an Excel cycle sequencing kit (Epicentre Tech) and then mixed with RNA and Superscript II reverse transcriptase (Invitrogen) to create labeled cDNA. A sequencing ladder was generated using the end-labeled primer, with pDAR812 as the template. The cDNA and sequencing ladder were run in a 6% acrylamide gel, dried, and analyzed using a Storm 820 phosphorimager according to the manufacturer's instructions (Amersham Biosciences).
Chick colonization assays.
All use of animals in experimentation has been approved by the Institutional Animal Care and Use Committee at the University of Texas Southwestern Medical Center. The ability of WT or isogenic mutant C. jejuni 81-176 Smr strains to colonize the ceca of chicks after oral inoculation was determined as previously described (35). Briefly, fertilized chicken eggs (SPAFAS) were incubated for 21 days at 37.8°C with appropriate humidity and rotation in a Sportsman II model 1502 incubator (Georgia Quail Farms Manufacturing Company). One day after hatching, chicks were orally inoculated with 100 μl of MH broth containing approximately 102 CFU of either a WT or mutant strain. C. jejuni strains were prepared for infection by being suspended from MH agar plates after 16 h of growth at 37°C under microaerobic conditions in MH broth and then diluted to achieve the appropriate inoculum for oral gavage of chicks. Dilutions of the inoculum were plated on MH agar to assess the number of bacteria in each inoculum. At 7 days postinfection, chicks were sacrificed, the cecal contents were removed and suspended in phosphate-buffered saline (PBS), and serial dilutions were plated on MH agar containing trimethoprim and cefoperazone. Following 72 h of growth at 37°C under microaerobic conditions, bacteria were counted to determine the number of CFU per gram of cecal content. Recovered colonies were analyzed by colony PCR to verify that WT and mutant strains were isolated from respectively infected chicks.
Collection of RNA for microarray analysis.
WT C. jejuni 81-176 Smr (DRH212) and isogenic ΔCjj1484 (PML360) and ΔCjj1483 (PML335) mutants were grown from freezer stocks on MH agar containing trimethoprim at 37°C under microaerobic conditions for 48 h. Strains were then restreaked on MH agar and grown for an additional 16 h. C. jejuni growth from the plates was suspended in MH broth and diluted in 25 ml of MH broth to an optical density at 600 nm (OD600) of approximately 0.05. Strains were then grown statically at 37°C under microaerobic conditions for 8 h to achieve mid-log-phase growth. Following growth, strains were suspended in 10× stop solution (95% ethanol plus 5% phenol), incubated on ice for 20 min, pelleted by centrifugation, and stored at −80°C (36). Pellets were suspended in 1 ml of RiboZol (Amresco), and RNA was removed by chloroform extraction. Total RNA (60 μg) was treated with DNase I (Invitrogen) and then purified through an RNeasy minicolumn (Qiagen). Reverse transcriptase PCR (RT-PCR) was performed to confirm the absence of DNA from the RNA samples.
Transcriptome analysis with DNA microarrays.
The DNA microarray-based transcriptome analysis was performed as previously described (37). Briefly, gene expression comparisons were performed indirectly by comparing the transcriptome profile of WT C. jejuni 81-176 Smr with the profile of C. jejuni 81-176 Smr ΔCjj1484 (PML360) or 81-176 Smr ΔCjj1483 (PML335). Total RNA samples (20 μg) for each strain were labeled with Cy3-dUTP during cDNA production by reverse transcriptase and were mixed with Cy5-dUTP-labeled reference genomic DNA from WT C. jejuni 81-176 Smr before being hybridized separately to custom cDNA microarrays (Gene Expression Omnibus [GEO] platform GPL6315 on Corning UltraGAPS slides). Microarrays were scanned using an Axon GenePix 4000B microarray laser scanner (Axon Instruments, Union City, CA). The microarray experiments were performed with two technical replicates per array, with two replicate features for each coding sequence (CDS) per array. GenePix 4.0 software was used to process the spot and background intensities, and data normalization was performed to compensate for differences in the amount of template or unequal Cy3 or Cy5 dye incorporation. GeneSpring 7.3 software (Silicon Genetics, Palo Alto, CA) was used to analyze the normalized data, and the parametric statistical t test was used to determine the significance of the centered data (P values of <0.05 indicate significance), with adjustment of the individual P values by use of the Benjamini-Hochberg false discovery rate multiple-test correction in the GeneSpring analysis package.
qRT-PCR analysis.
C. jejuni strains were grown from freezer stocks on MH agar containing trimethoprim at 37°C under microaerobic conditions for 48 h. Strains were then restreaked on MH agar and grown for an additional 16 h. C. jejuni growth from the plates was suspended in MH broth and diluted in 25 ml of MH broth to an OD600 of approximately 0.1. Strains were then grown statically at 37°C under microaerobic conditions for 8 h to achieve mid-log-phase growth. Total RNA was extracted with RiboZol (Amresco), and RNA was treated with DNase I (Invitrogen). RNA was diluted to a concentration of 50 ng/μl before analysis. Semiquantitative real-time RT-PCR (qRT-PCR) was performed using a model 7500 real-time PCR system (Applied Biosystems) with secD mRNA detection as an endogenous control. mRNA transcript levels in strains DRH212 and DRH461 served as WT controls for determinations of relative gene expression in isogenic mutants.
Analysis of growth of C. jejuni strains.
WT C. jejuni 81-176 Smr (DRH212) and isogenic ΔCjj1484 (PML360) and ΔCjj1483 (PML335) mutants were grown from freezer stocks on MH agar containing trimethoprim at 37°C under microaerobic conditions for 48 h. Strains were then restreaked on MH agar and grown for an additional 16 h. C. jejuni growth from the plates was suspended in MH broth and diluted in 25 ml of MH broth containing trimethoprim to an OD600 of approximately 0.1. Iron-rich growth conditions were created by adding Fe2SO4 to MH broth to a final concentration of 40 μM. Iron-limited growth conditions were created by adding the iron chelator deferoxamine mesylate (DFO) to MH broth to a final concentration of 20 μM (38). Cultures were grown at 37°C under microaerobic conditions without shaking, and growth was measured via spectrometry to determine the OD600. All strains were analyzed in triplicate.
Gluconate dehydrogenase activity in C. jejuni strains.
WT C. jejuni 81-176 Smr (DRH212) and the isogenic ΔCjj1484 mutant (PML360) were grown from freezer stocks on MH agar containing trimethoprim at 37°C under microaerobic conditions for 48 h. Strains were then restreaked on MH agar and grown for an additional 16 h. C. jejuni growth from the plates was suspended in MH broth and diluted in 25 ml of MH broth containing trimethoprim to an OD600 of approximately 0.1. Cultures were grown statically at 37°C under microaerobic conditions until growth reached mid-log phase. Cells were pelleted, washed twice in 20 mM NaPO4 (pH 7.0), disrupted by sonication, and centrifuged at 13,000 rpm for 5 min to remove cell debris. Gluconate dehydrogenase assays were performed as previously described (39). Briefly, each reaction mixture contained 1 ml of 0.1 M KPO4 buffer (pH 6.0), 0.1 ml of 1 mM 2,6-dichlorophenol-indophenol (DCIP), 0.1 ml of 3 mM phenazine methosulfate (PMS), 0.1 ml of 1 M d-gluconate, and 0.2 mg of total protein from the C. jejuni cell lysate in a total volume of 3 ml. Enzyme activity was determined through continuous measurement of DCIP reduction at 600 nm at 25°C. Three separate assays were performed, with each strain analyzed in triplicate.
Motility assays.
WT C. jejuni 81-176 Smr (DRH212) and isogenic ΔCjj1484 (PML360) and ΔCjj1483 (PML335) mutants were grown from freezer stocks on MH agar containing trimethoprim at 37°C under microaerobic conditions for 48 h. Strains were then restreaked on MH agar and grown for an additional 16 h. C. jejuni growth from the plates was suspended in MH broth and diluted in 10 ml of MH broth containing trimethoprim to an OD600 of approximately 0.8. Strains were stabbed into MH motility agar (MH broth plus 0.4% agar) by use of an inoculation needle and incubated for 30 h at 37°C under microaerobic conditions. The size of the motility zone was measured for each strain.
Purification of proteins and generation of antisera.
Primers containing in-frame 5′ BamHI restriction sites at codon 2 and the stop codon were used to amplify Cjj1483 from pDAR812, Cjj1483D58A from pPML239, Cjj1483D58E from pPML340, and Cjj1483D58N from pPML732. Following restriction digestion, the fragments were ligated into the BamHI site of pGEX-4T-2, generating pPML165, pPML278, pPML757, and pPML758, respectively. The plasmids were then transformed into E. coli BL21(DE3) and induced with 1 mM IPTG (isopropyl-β-d-thiogalactopyranoside). The N-terminal glutathione S-transferase (GST)-tagged proteins were purified from the soluble fraction by use of glutathione-Sepharose beads (GE Healthcare). Following GST tag removal by thrombin-mediated cleavage, the recombinant proteins were eluted from benzamidine-Sepharose following the manufacturer's instructions (GE Healthcare). Glycerol was added to a final concentration of 10%, and proteins were stored at −80°C. Recombinant WT Cjj1483 was then used to immunize five mice to generate polyclonal antisera (Cocalico Biologicals).
Primers containing in-frame 5′ BamHI restriction sites at codon 2 and the stop codon were used to amplify Cjj1484 from pDAR812. Following restriction digestion, the fragments were ligated into the BamHI site of pET28-6×His-MBP to generate pPML848 (40). The plasmid encoding an N-terminal 6×His–maltose binding protein (MBP) fusion was transformed into E. coli BL21(DE3) and induced with 300 μM IPTG at 16°C. The recombinant protein was purified from the soluble fraction by use of Ni-nitrilotriacetic acid (Ni-NTA)–agarose (Qiagen) as previously described (41). The 6×His-MBP tag was removed following cleavage with tobacco etch virus (TEV) protease, and the recombinant protein was purified from the tag by use of amylose agarose per the manufacturer's instructions (New England BioLabs). Glycerol was added to a final concentration of 10%, and proteins were stored at −80°C. Recombinant Cjj1484 was then used to immunize five mice to generate polyclonal antisera (Cocalico Biologicals).
In vitro phosphorylation of Cjj1483 proteins with AcP.
Radiolabeled acetyl phosphate (AcP) was created as described previously (42). Fifty picomoles of Cjj1483 protein was incubated with 10 μl of AcP-generating reaction mixture for 20 min at 37°C. Proteins were separated by 12.5% SDS-PAGE without boiling, and the gel was analyzed using a Storm 820 phosphorimager according to the manufacturer's instructions (Amersham Biosciences).
EMSAs.
Recombinant Cjj1483 proteins were purified as described above. A 320-bp DNA fragment spanning positions −297 to +20 relative to the transcriptional start site was amplified for the Cjj81176_0438 (Cjj0438) and Cjj1386 promoters. Electrophoretic mobility shift assays (EMSAs) were performed based on a modified protocol (43). Briefly, 0.5 to 2.0 μM WT or Cjj1483 mutant protein in the presence or absence of 50 mM lithium acetyl phosphate (Li-AcP) was incubated with 32P-labeled DNA at 4°C for 20 min. Competition experiments were performed through the addition of unlabeled DNA for the genes mentioned above, with the aphA-3 promoter as a nonspecific control. Unlabeled DNA was added at ratios of 1:1, 2:1, and 5:1 relative to 32P-labeled DNA, and 2 μM Cjj1483, with or without Li-AcP, was used. After electrophoresis, analysis was performed with a Storm 820 phosphorimager according to the manufacturer's instructions (Amersham Biosciences).
Microarray data accession number.
Microarray data were deposited in the GEO database under accession number GSE66942.
RESULTS
Genomic and transcriptional organization of the Cjj1484-Cjj1483 locus.
We analyzed the potential for Cjj1484 and Cjj1483 to encode a TCS that influences expression of specific genes in C. jejuni 81-176. On the C. jejuni 81-176 chromosome, these genes are predicted to be in an operon, with Cjj1484 (encoding a putative HK) at the 5′ end and Cjj1483 (encoding a putative RR) immediately downstream and in the same orientation as Cjj1484 (Fig. 1A). In order to assess the transcriptional organization of this locus, we performed primer extension analysis to identify potential transcriptional start sites and promoters for these genes. We detected a strong primer extension product that begins 180 bases upstream of the start codon of Cjj1483, but we were not able to detect a product representing a transcriptional start site for Cjj1484 (Fig. 1B and data not shown). However, transcriptome sequencing by RNA-seq analysis of C. jejuni 81-176 performed by another group identified transcriptional start sites immediately upstream of both genes (44). We attempted to determine if Cjj1484 and Cjj1483 were cotranscribed from the promoter upstream of Cjj1484, but we were unable to detect a product by reverse transcriptase PCR analysis of a bicistronic mRNA (data not shown). To discern whether Cjj1483 can be translated from transcripts originating upstream of Cjj1484 or Cjj1483, we constructed an 81-176 ΔCjj1484 mutant, which removed the promoter for Cjj1483. In this mutant, we detected production of Cjj1483 at approximately half the level seen with the WT strain (Fig. 1C). These findings combined indicate that Cjj1483 is mostly like cotranscribed with Cjj1484 from the promoter upstream of Cjj1484 and is also independently transcribed from its own promoter.
FIG 1.
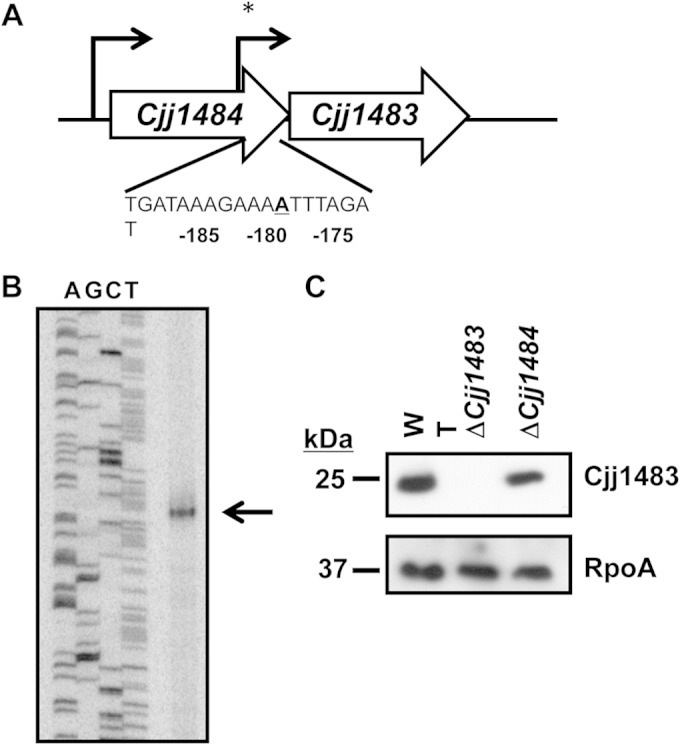
Genetic and transcriptional organization of the Cjj1484-Cjj1483 TCS. (A) Organization of Cjj1484 and Cjj1483 on the C. jejuni 81-176 chromosome. Arrows indicate transcriptional start sites for the genes as determined by RNA-seq analysis (44) or by primer extension (*) (this work), as shown in panel B. The DNA sequence below indicates the transcriptional start site for Cjj1483 that occurs at the adenosine residue 180 bases upstream of the translational start site for the gene, as determined by primer extension. (B) Primer extension analysis of Cjj1483. The product of primer extension analysis was run alongside a sequencing ladder, using the same primer as that for the primer extension reaction. The arrow indicates the product of primer extension. (C) Immunoblot of Cjj1483 production in whole-cell lysates of WT C. jejuni 81-176 Smr and isogenic mutants lacking Cjj1483 or Cjj1484. Cjj1483 was detected with a specific antiserum generated against recombinant Cjj1483. The ΔCjj1484 mutant lacks the region of the gene containing the promoter and transcriptional start site for a monocistronic Cjj1483 transcript depicted in panel A. Detection of the RNA polymerase α subunit (RpoA) served as a loading control for the whole-cell lysates.
Analysis of C. jejuni ΔCjj1484 and ΔCjj1483 mutants in commensal colonization of chicks.
We assessed whether the putative Cjj1484-Cjj1483 TCS is necessary for colonization of a natural host by infecting 1-day-old chicks with an inoculum of 102 CFU of WT C. jejuni 81-176 Smr or the isogenic ΔCjj1484 or ΔCjj1483 mutant. At 7 days postinfection, the levels of C. jejuni in the chick ceca were determined. WT C. jejuni colonized, on average, at a level of 1.55 × 109 CFU per g cecal content (Fig. 2). We did not detect any statistically significant colonization defect due to deletion of Cjj1484 or Cjj1483, as the mutants colonized the chicks at levels similar to that of the WT strain. These data suggest that this putative TCS is likely not required for interaction with the natural avian host.
FIG 2.
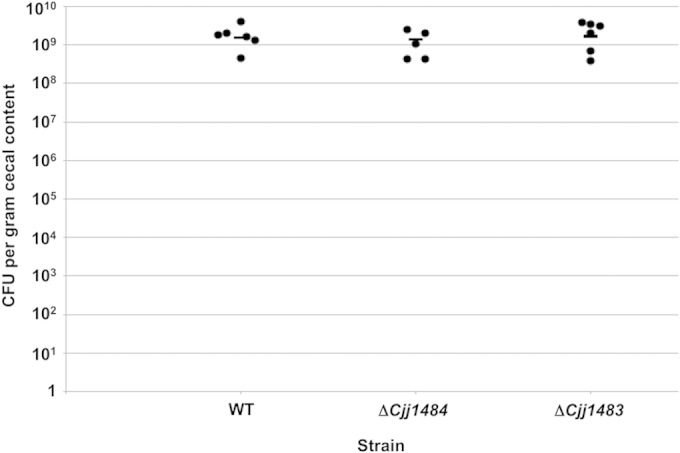
Commensal colonization capacities of wild-type C. jejuni and isogenic mutants lacking Cjj1484 or Cjj1483. One-day-old chicks were orally inoculated with approximately 102 CFU of wild-type C. jejuni 81-176 Smr or isogenic mutant strains. Each dot represents the amount of C. jejuni recovered from the ceca of each chick at day 7 postinfection. The geometric mean for each group is depicted by a horizontal bar. Statistical analysis was performed using the Mann-Whitney U test. Both mutants colonized at levels equal to that of the WT strain and showed no statistically significant differences.
Transcriptome analysis of C. jejuni ΔCjj1484 and ΔCjj1483 mutants.
In order to identify a biological role for the putative Cjj1484-Cjj1483 TCS in C. jejuni, we performed transcriptome analysis using DNA microarrays to identify a potential regulon for Cjj1484 and Cjj1483. WT and mutant C. jejuni strains were grown to mid-log phase at 37°C under microaerobic conditions for isolation of mRNAs to be used in transcriptome analysis. Genes whose expression was increased or decreased at least 2-fold were classified as putative members of the regulon controlled by Cjj1484 or Cjj1483 (Table 1 includes an abbreviated list of genes analyzed in this study; Tables S3 and S4 in the supplemental material include all identified genes). Genes selected for additional analysis met one or more of the following criteria: (i) genes with the highest degrees of expression difference in mutants relative to the WT strain; (ii) genes whose expression was dysregulated in both the ΔCjj1484 and ΔCjj1483 mutants; (iii) genes identified in the analysis that were likely grouped into operons and cotranscribed; and/or (iv) genes encoding C. jejuni proteins whose functions have previously been characterized. Selected genes were analyzed by semiquantitative RT-PCR to validate the results from the DNA microarray analysis (Fig. 3).
TABLE 1.
Condensed list of genes differentially expressed in C. jejuni 81-176 Smr Δ1484 or ΔCjj1483 compared to WT C. jejuni 81-176 Smr by microarray analysisa
| Gene classd and locus tag | Gene name | Putative function | WT/ΔCjj1484 mutant expression ratiob | WT/ΔCjj1483 mutant expression ratioc |
|---|---|---|---|---|
| Class I | ||||
| Cjj81176_0438 | Gluconate dehydrogenase, subunit III | 0.07 | 0.14 | |
| Cjj81176_0439 | Gluconate dehydrogenase, subunit I | 0.08 | 0.13 | |
| Cjj81176_1603e | chuC | Hemin transport ATP-binding protein | 0.09 | |
| Cjj81176_0064e | Putative cytochrome c protein | 0.32 | ||
| Class II | ||||
| Cjj81176_0210 | Possible transferrin transport protein | 2.51 | 0.47 | |
| Cjj81176_0211 | Possible transferrin transport protein | 2.70 | 0.45 | |
| Cjj81176_1619 | exbB2 | Ferric enterobactin transport protein | 3.44 | 0.33 |
| Cjj81176_1620 | exbD2 | Ferric enterobactin transport protein | 2.62 | 0.39 |
| Cjj81176_0063e | Hypothetical protein | 0.27 | ||
| Cjj81176_0315f | peb3 | Glycoprotein; putative adhesion or transport protein | 3.52 | |
| Class III | ||||
| Cjj81176_1385e | Hypothetical protein | 0.19 | ||
| Cjj81176_1386e | Hypothetical protein | 0.11 | ||
| Class IV | ||||
| Cjj81176_1257e | ciaC | Invasion protein | 2.23 |
Shown are a subset of genes identified to be differentially expressed in the mutants and further analyzed in this work. Complete lists of genes that were differentially expressed in the mutants are shown in Tables S3 and S4 in the supplemental material.
Expression of genes was increased or decreased 2-fold in the C. jejuni ΔCjj1484 mutant.
Expression of genes was increased or decreased 2-fold in the C. jejuni ΔCjj1483 mutant.
Genes were divided into one of four classes, as described in the text, based on their expression in the C. jejuni ΔCjj1484 and ΔCjj1483 mutants.
Expression of these genes was not affected in the C. jejuni ΔCjj1484 strain, as determined by microarray analysis.
Expression of this gene was not affected in the C. jejuni ΔCjj1483 strain, as determined by microarray analysis.
FIG 3.
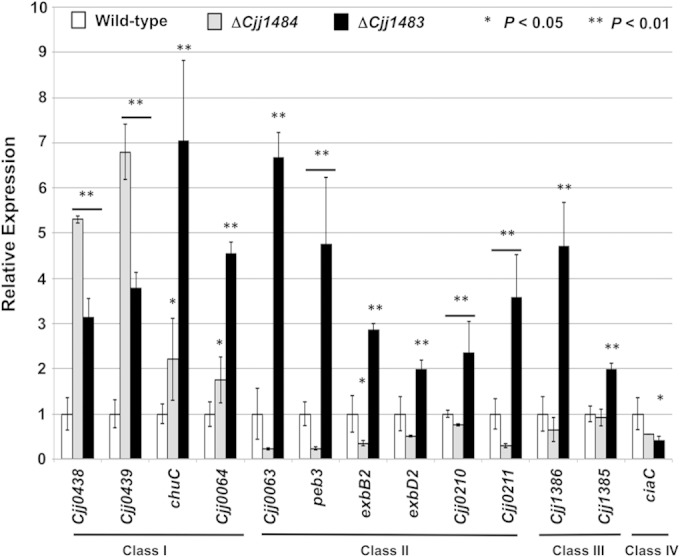
Semiquantitative real-time PCR analysis of transcription of a subset of genes initially identified by microarray analysis of WT and mutant C. jejuni strains. The expression of each gene in the WT strain, as measured by qRT-PCR, was set to 1. Expression of each gene in the ΔCjj1484 and ΔCjj1483 mutants is shown relative to that in WT C. jejuni. All strains were examined in triplicate, and the error bars indicate standard deviations. Statistically significant differences in gene expression between WT C. jejuni and mutant strains are indicated (*, P < 0.05; **, P < 0.01) and were determined by the Student t test. Bars below asterisks indicate instances where gene expression levels in both the ΔCjj1483 and ΔCjj1484 mutants were significantly different from that in WT C. jejuni.
In combining the results from the DNA microarray and qRT-PCR analyses, we discovered that genes could be grouped into four classes based on how mutation of the Cjj1484 HK and the Cjj1483 RR affected transcription of the genes. Generally, the Cjj1483 RR had a repressive effect on transcription of target genes, but the Cjj1484 HK demonstrated both positive and negative effects on transcription. We found that class I genes included chuC (encoding a transport protein for hemin), Cjj81176_0438 (Cjj0438) and Cjj81176_0439 (Cjj0439) (both of which encode subunits of the gluconate dehydrogenase complex that is required for colonization of chicks) (39), and Cjj81176_0064 (encoding a putative cytochrome c protein). Mutation of Cjj1484 and Cjj1483 resulted in 2- to 7-fold increases in expression of these genes as shown by qRT-PCR (Fig. 3).
Class II genes included Cjj81176_0210 (Cjj0210) and Cjj81176_0211 (Cjj0211) (encoding putative transport proteins for transferrin), exbB2 and exbD2 (encoding putative transport proteins for enterobactin) (45), peb3 (encoding a glycoprotein that may function as an adhesin or transport protein) (46–48), and Cjj81176_0063 (encoding a protein of unknown function). Compared to the expression in the WT strain, mutation of the Cjj1484 HK resulted in 2.5- to 5-fold decreases in expression of most of these genes, but mutation of the Cjj1483 RR resulted in 2- to 7-fold increases in expression as shown by qRT-PCR (Fig. 3). These results suggest that the RR has a repressive effect on transcription, whereas the HK may function to remove repression, likely by acting on the Cjj1483 RR.
Class III genes included Cjj81176_1385 and Cjj81176_1386, which encode two hypothetical proteins of unknown function. For these genes, mutation of the Cjj1483 RR resulted in 2- to 5-fold increases in expression, suggesting that the RR functions as a repressor of these genes (Fig. 3). However, mutation of the Cjj1484 HK did not affect transcription of these genes. This finding may indicate that the RR functions independently of the HK to affect expression of these genes.
Class IV genes are exemplified by ciaC, encoding a protein required for invasion (49). For ciaC, transcription was reduced 2- to 2.5-fold by mutation of both the Cjj1484 HK and the Cjj1483 RR, indicating that the TCS had a positive effect on transcription of this gene.
Phenotypic analysis of the C. jejuni ΔCjj1484 and ΔCjj1483 mutants.
In our transcriptome analysis of the C. jejuni ΔCjj1484 and ΔCjj1483 TCS mutants, genes encoding various iron-binding and transport systems were found to be dysregulated in the mutants relative to the WT strain. For example, chuC was expressed at higher levels in both TCS mutants than in the WT, whereas expression of Cjj0210, Cjj0211, exbB2, and exbD2 was reduced in the mutant lacking the Cjj1484 HK but increased in the mutant lacking the Cjj1483 RR. Due to these findings, we investigated whether the TCS mutants demonstrated altered growth in Mueller-Hinton (MH) broth with various levels of iron. In normal MH broth at 37°C under microaerobic conditions, both the ΔCjj1484 and ΔCjj1483 mutants grew as well as the WT C. jejuni strain over the course of 32 h (Fig. 4A). In MH broth supplemented with 40 μM Fe2SO4, we noted a slight enhancement of growth of the Cjj1483 RR mutant relative to the WT strain over the course of the assay, and the difference was statistically significant (Fig. 4B). In contrast, the Cjj1484 HK mutant did not consistently demonstrate increased growth relative to that of the WT strain under this high-iron condition. In MH broth containing the iron chelator deferoxamine mesylate (DFO) (20 μM) to stimulate iron-limiting conditions, neither the ΔCjj1484 mutant nor the ΔCjj1483 mutant consistently demonstrated a growth difference compared to the WT strain (Fig. 4C). Other than a small enhancement of growth in the Cjj1483 RR mutant under high-iron conditions, dysregulation of expression of iron transport systems did not significantly affect the ability of the Cjj1484-Cjj1483 TCS mutants to grow in vitro.
FIG 4.
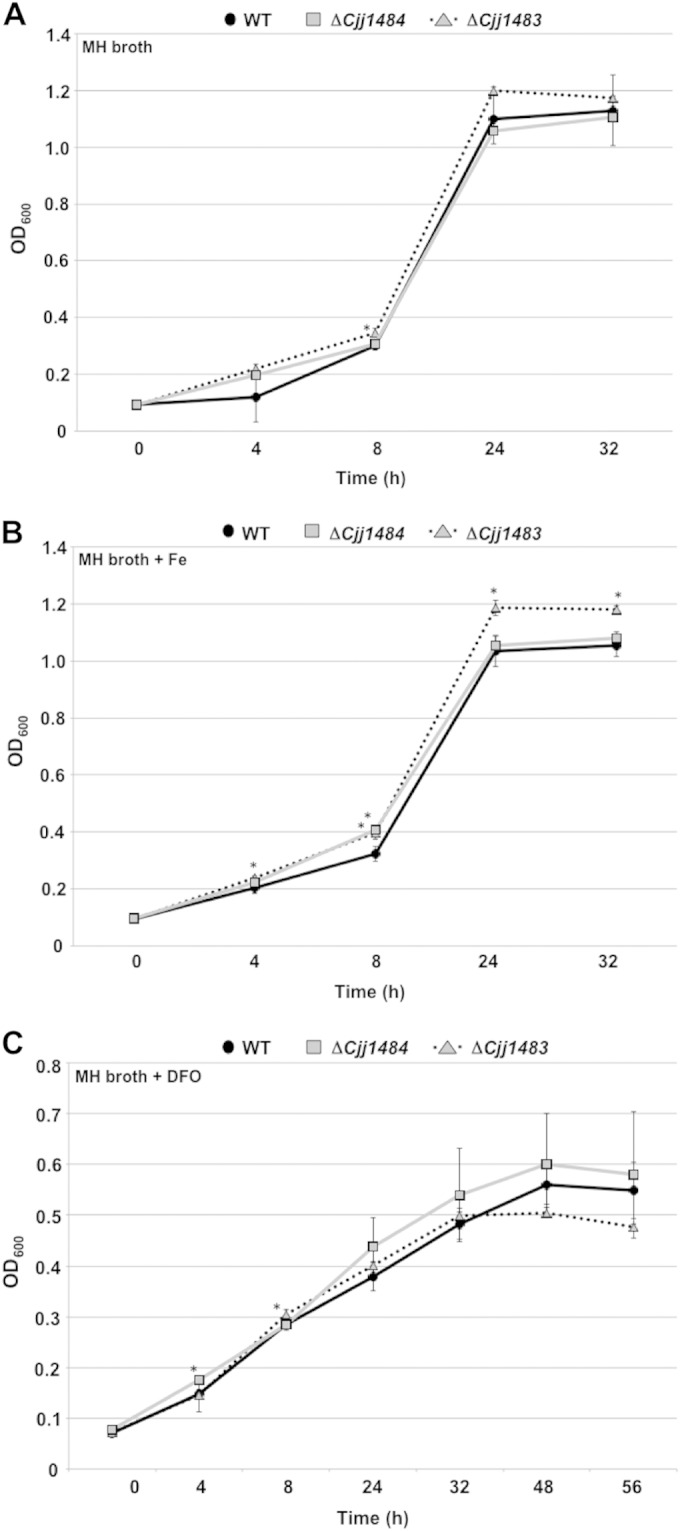
Analysis of growth of WT and mutant C. jejuni strains. C. jejuni strains were grown in MH broth alone (A) or in MH broth containing 40 μM Fe2SO4 to simulate high iron levels (B) or 20 μM deferoxamine mesylate (DFO), an iron chelator, to simulate iron-depleted conditions (C). Strains were grown under microaerobic conditions at 37°C for 32 to 56 h. All strains were analyzed in triplicate, and the data are presented as average OD600 readings for strains at each time point. Error bars indicate standard deviations. Asterisks indicate data points for the mutants that are statistically significantly (P < 0.05) different from those of the WT strain.
Due to the increased in vitro expression of Cjj0438 and Cjj0439, which both encode components of a C. jejuni gluconate dehydrogenase complex (39), in each of our TCS mutants, we analyzed whether this increased transcription translated to increased gluconate dehydrogenase activity in cellular lysates. For this analysis, we compared gluconate dehydrogenase activities of WT C. jejuni and the Cjj1484 HK mutant, which was observed to have approximately 5- to 7-fold-increased transcription of Cjj0438 and Cjj0439. In our assays, we found that the ΔCjj1484 mutant did indeed demonstrate an 80% increase in gluconate dehydrogenase activity (Table 2). Thus, disruption of the Cjj1484-Cjj1483 TCS caused aberrant gluconate dehydrogenase activity in C. jejuni.
TABLE 2.
Gluconate dehydrogenase activities in C. jejuni 81-176 Smr and C. jejuni 81-176 Smr ΔCjj1484
Lysates of strains after microaerobic growth at 37°C were analyzed by a coupled enzyme assay. The activity of gluconate dehydrogenase is reported in nanomoles of DCIP reduced per minute per milligram of protein in the lysates. Data represent the averages and SD for three assays, with each strain analyzed in triplicate.
Statistically significantly different from that of the WT strain (P < 0.05).
We also observed that expression of flaA, encoding the major flagellin required for flagellar motility of C. jejuni, was reduced approximately 3-fold in the ΔCjj1484 mutant (see Table S3 in the supplemental material). However, we did not detect any motility defects in strains lacking the Cjj1484 HK or the Cjj1483 RR in motility agar (data not shown), indicating that disruption of the TCS does not significantly affect the motility of C. jejuni.
Analysis of phosphotransfer through the Cjj1484-Cjj1483 TCS and effects on gene expression.
Bioinformatic analysis indicated that Cjj1484 contains domains typical of many bacterial HKs (Fig. 5A). A PAS9 domain that may function as a specific sensor domain for Cjj1484 is located within the N-terminal 120 residues. Bioinformatic analysis of this domain revealed some conservation of residues for a heme-binding pocket. However, due to an inability to purify sufficient levels of Cjj1484, it is unclear if the PAS domain senses heme or a related compound. This domain is followed by a region that would be expected to form the dimerization and histidine phosphotransfer (DHp) domain in most bacterial HKs. This domain usually contains a conserved histidine residue that is autophosphorylated upon sensing of a signal (10). While this domain in Cjj1484 is only weakly predicted, H195 is the best candidate for a histidine residue to be modified by autophosphorylation. An HATPase domain that typically binds ATP for the autophosphorylation reaction of HKs is present at the C terminus (10). No transmembrane domains are apparent within Cjj1484 to suggest that it is linked to the inner membrane of C. jejuni. Thus, Cjj1484 is most likely a cytoplasmic HK.
FIG 5.
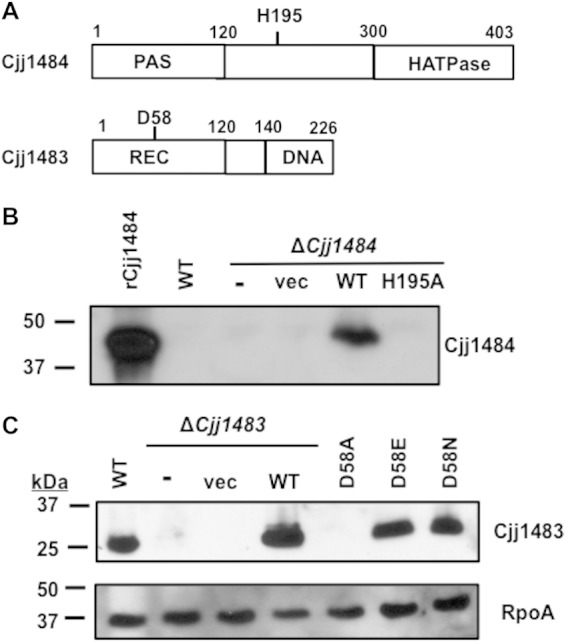
Construction and stability of Cjj1484-Cjj1483 TCS point mutants. (A) Putative domain organization of the Cjj1484 HK and the Cjj1483 RR. Predicted domains were identified by BLAST analysis. Conserved residues that are predicted to participate in phosphotransfer for each protein are indicated. The Cjj1484 HK is predicted to contain a PAS sensory domain and a C-terminal HATPase domain for binding and hydrolysis of ATP. Cjj1484 does not have a strongly predicted dimerization and histidine phosphotransfer (DHp) domain, but H195 is the most likely histidine that is modified by autophosphorylation. The Cjj1483 RR is predicted to contain an N-terminal REC domain common to many response regulators and a C-terminal winged-helix DNA-binding domain. (B) Immunoblot analysis of WT Cjj1484 and Cjj1484H195A in C. jejuni. An antiserum specific to recombinant Cjj1484 (rCjj1484) was generated and then used to analyze production of WT and mutant Cjj1484 proteins in C. jejuni whole-cell lysates. C. jejuni ΔCjj1484 was complemented in trans with a plasmid expressing WT Cjj1484 or Cjj1484H195A (H195A) from the promoter for flaA (encoding the major flagellin of C. jejuni) to overexpress the proteins. In addition, the ΔCjj1484 strain was not complemented (−) or was complemented with vector alone (vec). (C) Immunoblot analysis of WT Cjj1483 or Cjj1483 with various mutations at D58 in C. jejuni whole-cell lysates. C. jejuni ΔCjj1483 was complemented in trans with a plasmid expressing WT Cjj1483 from its native promoter as shown in Fig. 1A. In addition, the ΔCjj1483 strain was not complemented (−) or was complemented with vector alone (vec). For the Cjj1483D58A, Cjj1483D58E, and Cjj1483D58N mutants, the genes encoding the mutations replaced the WT Cjj1483 gene on the chromosome of C. jejuni so that the genes were expressed from native promoters. Detection of RpoA served as a loading control for the whole-cell lysates.
In a prototypical TCS, mutation of the phosphoacceptor histidine of the HK disrupts autophosphorylation and prevents subsequent phosphotransfer to the cognate RR. Considering this mechanism, we mutated H195 to monitor whether this residue is autophosphorylated to serve as the phosphodonor for Cjj1483. However, WT Cjj1484 was largely undetectable in whole-cell lysates, making it difficult to assess whether Cjj1484H195A is stable in C. jejuni (Fig. 5B). Therefore, we analyzed production of WT Cjj1484 and Cjj1484H195A overexpressed in trans from the flaA promoter in C. jejuni ΔCjj1484. Although we were able to detect overexpressed WT Cjj1484, we were unable to detect Cjj1484H195A upon expression from the same flaA promoter (Fig. 5B). We concluded that mutation of H195 likely creates an unstable Cjj1484 protein, and this point mutant was not analyzed further.
Cjj1483 displays features common to many bacterial RRs. The first 120 residues compose a receiver domain, with D58 as the most likely aspartate residue for phosphorylation by phosphotransfer from the Cjj1484 HK or other potential phosphodonors (Fig. 5A). At the C terminus of Cjj1483 is a predicted wing-helix DNA-binding domain that likely facilitates interactions between the RR and target promoters. For many RRs, mutation of the conserved phosphoacceptor aspartic acid to an alanine or asparagine prevents phosphotransfer and alters function. In some RRs, alteration of the aspartate to a glutamate mimics a phosphoaspartate, resulting in RR activities associated with an RR that is naturally phosphorylated by its cognate HK (50). However, in other response regulators, this alteration inactivates the protein (similarly to an alanine mutation), causing the RR to constitutively function as it does in the unphosphorylated state.
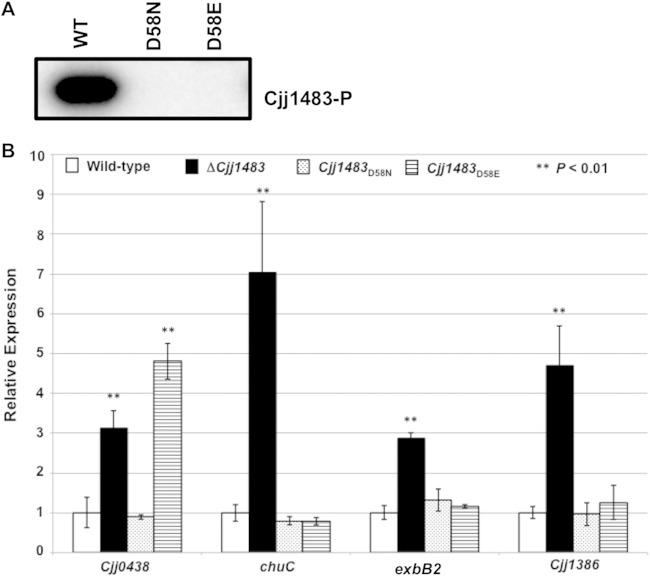
Similar to our observations described above, WT Cjj1483 was easily detected in whole-cell lysates, and its production was restored in a ΔCjj1483 mutant upon expression of the gene in trans from its native promoter (Fig. 5C). However, we were unable to detect Cjj1483D58A when the respective allele replaced WT Cjj1483 at the native locus. In contrast, Cjj1483D58E and Cjj1483D58N were stable and produced at WT levels in C. jejuni (Fig. 5C). As such, the latter mutant Cjj1483 proteins were used in all subsequent studies.
We first attempted to biochemically analyze the potential phosphotransfer mechanism through the Cjj1484-Cjj1483 TCS. We were able to abundantly purify WT Cjj1483, Cjj1483D58E, and Cjj1483D58N as soluble proteins from E. coli. However, we were unable to consistently express and purify a soluble form of the WT Cjj1484 HK. Therefore, we were unable to perform autokinase assays with Cjj1484 and subsequent phosphotransfer assays with Cjj1483. To determine whether D58 was the potential phosphoacceptor residue of Cjj1483, we performed autophosphorylation assays with radiolabeled acetyl phosphate (AcP). WT Cjj1483 was modified by autophosphorylation with AcP, but both Cjj1483D58E and Cjj1483D58N were not (Fig. 6A). These findings strongly suggest that D58 is the phosphoacceptor aspartate of the Cjj1483 RR.
FIG 6.
Analysis of effects of Cjj1483 D58 mutations on expression of target genes. (A) In vitro phosphorylation assay of WT and mutant Cjj1483 RR proteins. Recombinant WT Cjj1483, Cjj1483D58E, and Cjj1483D58N proteins were incubated with radiolabeled AcP for 20 min, separated by SDS-PAGE, and then exposed on a phosphorimager. The signal indicates the ability of the WT protein but not the Cjj1483 D58 mutants to autophosphorylate. (B) Analysis of expression of select genes within the Cjj1483 regulon in WT C. jejuni or isogenic mutants producing Cjj1483D58E or Cjj1483D58N mutant protein. The expression of each gene in the WT strain, as measured by qRT-PCR, was set to 1. Expression levels of each gene in the ΔCjj1483, Cjj1483D58N, and Cjj1483D58E mutants are shown relative to that in WT C. jejuni. All strains were examined in triplicate, and the error bars indicate the standard deviations. Statistically significant differences in gene expression between WT C. jejuni and mutant strains are indicated (**, P < 0.01) and were determined by the Student t test.
With this in mind, we analyzed gene expression in C. jejuni 81-176 Cjj1483D58E and Cjj1483D58N mutants by qRT-PCR. In contrast to the effects of the ΔCjj1483 mutant, Cjj1483D58E and Cjj1483D58N repressed transcription of chuC, Cjj1386, and exbB2 similarly to WT Cjj1483 (Fig. 6B). Since both mutant proteins demonstrated the same activity, we assume that these proteins were both mimicking the unphosphorylated state of Cjj1483 in an activity to repress expression of these genes. Thus, these results together suggest that Cjj1483 likely binds these promoters to repress transcription in C. jejuni in the absence of phosphorylation.
In contrast, Cjj1483D58E and Cjj1483D58N had different effects on expression of Cjj0438. We found that Cjj0438 expression was increased 5-fold in the D58E mutant, similar to the case in the ΔCjj1483 mutant (Fig. 6B). However, the level of expression of Cjj0438 in C. jejuni 81-176 Cjj1483D58N was similar to that of the WT strain. We envision two possible interpretations of these results. In one case, Cjj1483D58E might mimic a phosphorylated version of the protein in C. jejuni that cannot specifically repress transcription from the Cjj0438 promoter. Alternatively, mutation of D58 to glutamate may disrupt the binding activity of the Cjj1483 RR to the Cjj0438 promoter but not to other target promoters.
Colonization ability of mutants lacking genes within the Cjj1484-Cjj1483 TCS regulon.
As we showed above, the Cjj1484-Cjj1483 TCS is required to repress expression of the Cjj0438-Cjj0439 operon, which encodes a gluconate dehydrogenase complex required for WT levels of commensal colonization of the chick ceca (39). Having identified more members of the Cjj1484 regulon, we assessed if any other genes repressed by the RR are required for colonization of chicks. We generated mutants in chuC, exbB2, Cjj0064, and Cjj1386 in a C. jejuni 81-176 Smr strain. Chicks were infected with WT C. jejuni 81-176 Smr or the isogenic mutants at an inoculum of 102 CFU. Chicks were then sacrificed at day 7 postinfection, and the levels of C. jejuni in the ceca of chicks were enumerated. In this assay, WT C. jejuni colonized 2.7 × 107 to 1.17 × 109 CFU per g cecal content, with a geometric mean of 2.95 × 108 CFU per g cecal content (Fig. 7). Mutants lacking exbB2, Cjj0064, or chuC colonized chicks at levels similar to those for WT C. jejuni. Although the colonization levels did not reach statistical significance, we did notice a modest 3.5-fold decrease in colonization for the Cjj1386 mutant.
FIG 7.
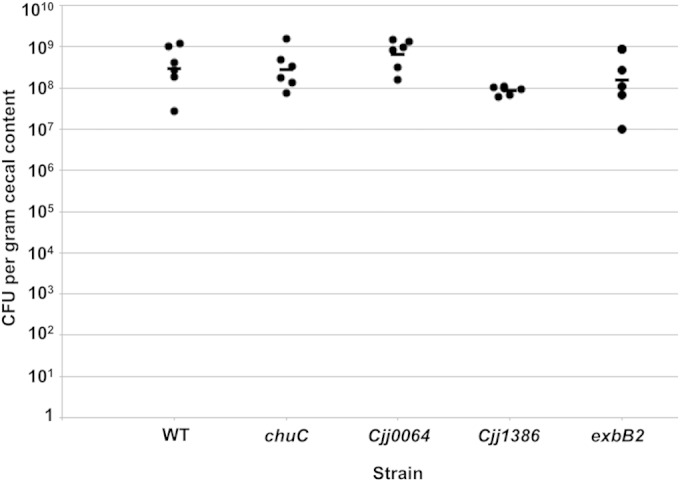
Commensal colonization capacities of wild-type C. jejuni and mutant strains lacking specific genes within the Cjj1483 regulon. One-day-old chicks were orally inoculated with approximately 102 CFU of WT C. jejuni 81-176 Smr or isogenic mutant strains. Each dot represents the amount of C. jejuni recovered from the ceca of each chick at day 7 postinfection. The geometric mean for each group is depicted by a horizontal bar. Statistical analysis was performed using the Mann-Whitney U test. All mutants showed no statistically significant differences.
DNA-binding activity of the Cjj1483 RR to the Cjj0438 promoter.
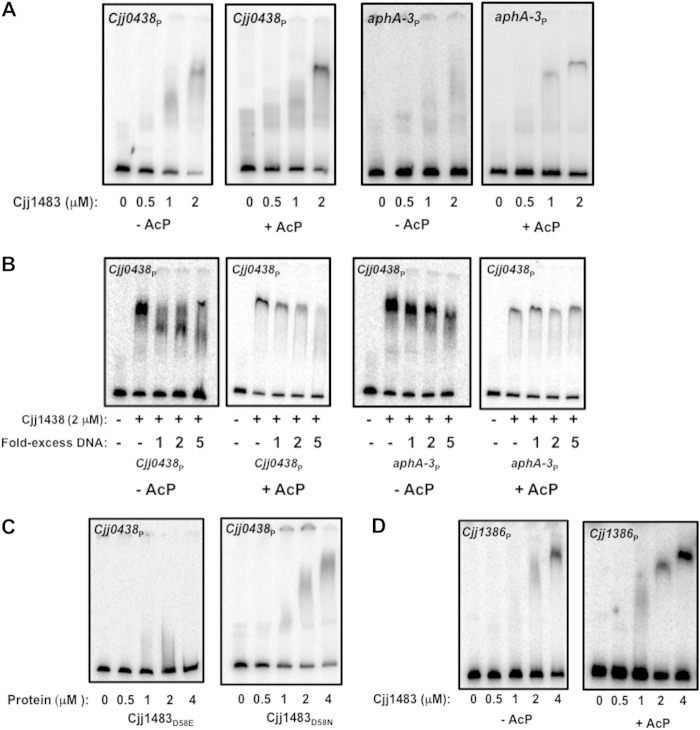
In order to better understand the ability of Cjj1483 to influence gene expression in C. jejuni, we analyzed the potential DNA-binding activity of the RR for target promoters of genes within its regulon. EMSAs with recombinant WT Cjj1483, Cjj1483D58E, and Cjj1483D58N and a DNA fragment encompassing the Cjj0438 promoter were employed. As shown in Fig. 8A, unphosphorylated WT Cjj1483 bound to the Cjj0438 promoter. The amount of DNA bound by Cjj1483 increased as the concentration of the protein increased. We also analyzed the effect of phosphorylation of Cjj1483 on DNA binding. For this analysis, Cjj1483 was pretreated with Li-AcP for 20 min and used in DNA-binding assays. We did notice a slight enhancement in the DNA-binding activity of Cjj1483 relative to the unphosphorylated form, but it is unclear whether this enhanced binding is significant (Fig. 8A, left panels). Unphosphorylated Cjj1483 did not bind to a promoter for aphA-3, encoding kanamycin resistance, but the phosphorylated form did demonstrate a nonspecific, weak binding ability with this promoter (Fig. 8A, right panels). To determine whether binding to the Cjj0438 promoter was specific, we performed DNA-binding assays with an excess of a specific competitor (unlabeled Cjj0438 promoter) or a nonspecific competitor (unlabeled aphA-3 promoter). When Cjj1483 was not phosphorylated, DNA binding could still be observed with a 5-fold excess of unlabeled Cjj0438 promoter (Fig. 8B, left panels). Furthermore, the aphA-3 promoter did not compete with Cjj0438 for binding by unphosphorylated Cjj1483 (Fig. 8B, right panels). When Cjj1483 was pretreated with Li-AcP to stimulate phosphorylation prior to EMSAs, Cjj1483 binding to the Cjj0438 promoter was reduced by a 2-fold excess of unlabeled DNA and completely reduced by a 5-fold excess. Again, the aphA-3 promoter did not effectively compete for binding by phosphorylated Cjj1483 (Fig. 8B, right panels). The results of these competitive binding assays may indicate that the Cjj1483 RR can bind in both the unphosphorylated and phosphorylated forms, but the phosphorylated form may have a specific but reduced binding affinity for target promoters. Note that binding by the phosphorylated Cjj1483 RR occurred with other nonspecific DNA that we analyzed (data not shown). This observation indicates that analysis of in vitro DNA binding by the Cjj1483 RR may be problematic in terms of assessing or interpreting with confidence the degree of binding specificity.
FIG 8.
Electrophoretic mobility shift assays for analysis of DNA-binding activity of the Cjj1483 RR. For panels A and B, purified WT Cjj1483 protein was added to various promoter DNAs at concentrations ranging from 0 to 2 μM. For panels C and D, WT Cjj1483 or the Cjj1483D58E or Cjj1483D58N mutant protein was added at concentrations ranging from 0 to 4 μM. For panels A, B, and D, “−AcP” and “+AcP” indicate whether or not Cjj1483 was pretreated with Li-AcP to autophosphorylate the protein prior to addition to DNA-binding assay mixtures. (A) Binding of Cjj1483 to the radiolabeled promoter for Cjj0438 or aphA-3 in the absence of competition. (B) Binding of Cjj1483 to the radiolabeled promoter for Cjj0438 in the presence of unlabeled competitor DNA (Cjj0438 promoter; left panels) or unlabeled noncompetitor DNA (aphA-3 promoter; right panels). (C) Binding of Cjj1483D58E and Cjj1483D58N mutant proteins to the radiolabeled promoter for Cjj0438 in the absence of competition. (D) Binding of Cjj1483 to the radiolabeled promoter for Cjj1386 in the absence of competition.
We next examined the DNA-binding ability of Cjj1483D58E and Cjj1483D58N with Cjj0438. Compared to WT Cjj1483, Cjj1483D58E was unable to bind the Cjj0438 promoter, but Cjj1483D58N did appear to bind this DNA (Fig. 8C). Considering that Cjj1483D58E was unable to repress expression of Cjj0438 in C. jejuni (Fig. 6B), we propose that this mutation may mimic a constitutively phosphorylated state and that WT Cjj1483 likely has full DNA-binding ability to repress expression of Cjj0438 in the unphosphorylated form. In support of this hypothesis, Cjj1483D58N bound to the Cjj0438 promoter and repressed expression of the gene (Fig. 6B and 8C), suggesting that this protein mimics a constitutively unphosphorylated form that binds DNA specifically to repress gene expression.
Analysis of Cjj1483 binding to the Cjj1386 promoter.
In order to further analyze the biological activity of the Cjj1484-Cjj1483 TCS in C. jejuni, we assayed the ability of the Cjj1483 RR to bind the promoter region for the previously uncharacterized Cjj1386-Cjj1385 operon. As shown in Fig. 3, deletion of Cjj1484 did not have a strong effect on expression of either Cjj1386 or Cjj1385. However, deletion of Cjj1483 resulted in 2- to 5-fold increases in expression of Cjj1385 and Cjj1386. These data suggest that Cjj1483 may function as a repressor for this promoter, independently of the putative cognate Cjj1484 HK.
We performed EMSAs with the promoter for Cjj1386-Cjj1385 and purified Cjj1483 alone or after phosphorylation by Li-AcP. We observed that both the unphosphorylated and phosphorylated forms of the RR could bind to the promoter of Cjj1386, with phosphorylation perhaps promoting Cjj1483 binding to the DNA at a lower protein concentration (Fig. 8D). We attempted to analyze if this binding was specific for the promoter of Cjj1386 by analyzing the binding ability of Cjj1483 in the presence of excess unlabeled Cjj1386 promoter DNA and nonspecific aphA-3 promoter DNA. However, neither DNA in excess efficiently competed for binding (data not shown). Regardless, our collective data are consistent with the Cjj1483 RR repressing transcription from the Cjj1386-Cjj1385 promoter in C. jejuni in either an unphosphorylated or phosphorylated state that is independent of the Cjj1484 HK. It is likely that Cjj1483, which can be expressed independently of Cjj1484 from a monocistronic transcript (Fig. 1B), is periodically produced without the Cjj1483 HK and can influence gene expression in an unphosphorylated state or with phosphorylation originating from a noncognate HK or phosphodonor.
DISCUSSION
In this work, we investigated one of the remaining uncharacterized putative TCSs of C. jejuni, encoded by Cjj1484 and Cjj1483 on the C. jejuni 81-176 genome. Our findings include evidence that the Cjj1484 HK and the Cjj1483 RR likely function as a cognate TCS to influence expression of a common set of genes. In addition, we provide evidence that Cjj1483 influences expression of genes independently of its cognate Cjj1484 HK. In either case, the TCS or Cjj1483 alone appears to largely repress transcription of genes under in vitro growth conditions. Most genes whose expression is controlled by the Cjj1484-Cjj1483 TCS encode proteins that function in various metabolic processes, including heme and/or iron uptake and respiration. In addition, this TCS represses transcription of one known colonization factor, the gluconate dehydrogenase complex (39). Biochemical analysis allowed us to make predictions about the DNA-binding activity of Cjj1483 toward target promoters in relation to its phosphorylation state. Although the Cjj1484-Cjj1483 TCS was not required for commensal colonization of the natural avian host, we suspect that the system may play a role with other transcriptional regulators in finely controlling transcription of metabolic genes that may be important ex vivo and during transmission from one host to another.
The regulons for Cjj1484 and Cjj1483 were found to contain many overlapping genes. Our findings strongly suggest that the Cjj1484 HK and the Cjj1483 RR function as a cognate TCS. Further support for these factors forming a cognate TCS includes their apparent operonic organization on the C. jejuni chromosome. Complete verification would require biochemical analysis to reveal specific phosphotransfer from Cjj1484 to Cjj1483. We attempted to perform such an analysis, but we were unable to purify the Cjj1484 HK in a soluble state in sufficient quantities. We also acquired evidence that transcription of certain genes, such as Cjj1386 and Cjj1385, is specifically influenced by the Cjj1483 RR alone but is unaffected by mutation of the Cjj1484 HK. Additionally, we identified a promoter within the Cjj1484 coding sequence that expresses Cjj1483 without the HK. This finding gives credence to the hypothesis that the Cjj1483 RR may be produced without the HK in certain situations to influence expression of specific genes independently of the Cjj1484 HK. Therefore, while Cjj1483 appears to have a classical function as an RR in a cognate TCS with influence from its cognate HK for regulation of specific genes, the RR appears to be expressed and to function independently of the HK in controlling expression of other genes.
We experienced several difficulties in performing a complete biochemical and genetic analysis of the Cjj1484-Cjj1483 TCS, which may indicate some unusual features of this TCS compared to other systems. First, the WT Cjj1484 HK could be detected in C. jejuni only upon overexpression of the gene in trans, indicating that the HK is either produced at extremely low levels or rapidly turned over. Second, we could not absolutely prove that H195 is the histidine residue that is modified by autophosphorylation upon sensing of a stimulus. Mutation of this residue resulted in a protein that was undetectable in C. jejuni even upon overexpression. For the Cjj1483 RR, we showed that mutation of D58 to an asparagine or glutamate prevented the protein from autophosphorylating by using radiolabeled AcP as a phosphodonor. Curiously, mutation of D58 of Cjj1483 to an alanine resulted in an unstable protein that could not be detected. In many RRs, this mutation does not cause instability. Currently, it is unclear whether this mutational analysis and resultant stability issues for the Cjj1484-Cjj1483 TCS imply a significantly altered biochemistry of signal perception, phosphotransfer, and transduction compared to that of other TCSs.
By combining results from expression analysis and DNA-binding assays using WT and mutant TCS proteins, we are able to propose four putative models for how the Cjj1484-Cjj1483 TCS influences expression of different classes of genes, depending on the phosphorylation state and activities of the Cjj1484 HK and the Cjj1483 RR (Fig. 9). We propose that for the class I genes, which include the Cjj0438 operon, the Cjj0063 operon, and chuC, the Cjj1484 HK and the Cjj1483 RR function together to repress expression of these genes. For these genes, we propose that the Cjj1483 RR in an unphosphorylated state binds promoter DNA to repress gene expression and that the Cjj1484 HK may have a phosphatase activity to maintain Cjj1483 in an unphosphorylated state. This model would explain why mutation of Cjj1484 causes derepression of expression of these genes similarly to the case in a mutant lacking the Cjj1483 RR. Specifically for Cjj0438, we noticed that mutation of the phosphorylated aspartate in Cjj1483 caused different effects on DNA binding to the Cjj0438 promoter and expression of the gene. Cjj1483D58N bound the Cjj0438 promoter DNA effectively, but Cjj1483D58E did not. In addition, Cjj1483D58N repressed expression of Cjj0438, but Cjj1483D58E did not. We interpret these data as suggesting that Cjj1483D58N likely mimics an unphosphorylated state that can still bind DNA to repress gene expression, but Cjj1483D58E may function as a constitutively phosphorylated protein that is unable to bind the Cjj0438 promoter and repress expression. Curiously, we did observe that Cjj1483D58E was still able to repress expression of chuC, which may indicate that alteration of the aspartate residue by certain mutations ultimately affects the ability of the RR to recognize and bind different promoters. Also, as discussed below, other regulators affect expression of chuC, which may have an influence on the ability of Cjj1483D58E to repress expression of this gene.
FIG 9.
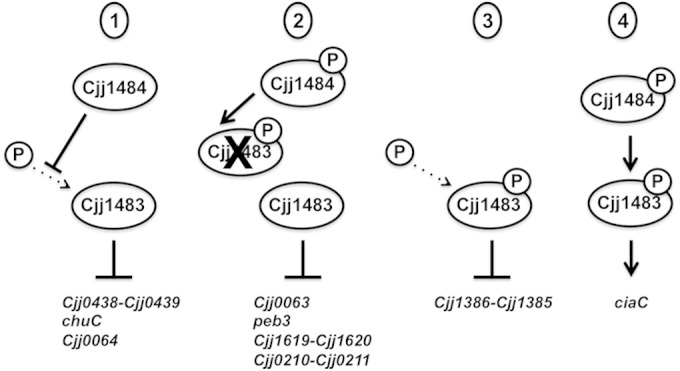
Putative models for how the Cjj1484-Cjj1483 TCS may mediate regulation of different classes of C. jejuni genes. For models 1 to 3, Cjj1483 is proposed to function as a transcriptional repressor, whereas for model 4, Cjj1483 likely functions as a transcriptional activator. In each model, we indicated whether Cjj1483 must be phosphorylated to mediate transcriptional regulation and how phosphorylation or prevention of phosphorylation may occur. Dotted arrows indicate phosphorylation that may occur through noncognate phosphodonors. In model 1, Cjj1484 is predicted to serve as a phosphatase to maintain Cjj1483 in an unphosphorylated state, whereas in model 2, Cjj1484 is predicted to phosphorylate Cjj1483 to inhibit its repressive activity. In model 3, the Cjj1484 HK plays no role in gene expression. In this model, phosphorylation of Cjj1483 via noncognate phosphodonors may mediate repression by the response regulator. In model 4, phosphorylation of the Cjj1483 RR via Cjj1484 appears to be required for Cjj1483 to function as a transcriptional activator. See Discussion for more details.
For the class II genes, which include Cjj0063, peb3, and the exbB2 and Cjj0210 operons, we found that mutation of the Cjj1484 HK resulted in repression of gene expression, but expression of these genes in the presence of the Cjj1483 RR mutant was derepressed. We propose that like the case with Cjj0438 and chuC, Cjj1483 in the unphosphorylated form may mediate repression. This repression may be relieved by Cjj1484 promoting phosphorylation of the RR (Fig. 9). This model would explain how the lack of Cjj1483 phosphorylation in the HK mutant may lead to increased repression of these genes.
For the Cjj1386 operon, which comprises class III genes, we did not find any evidence that the Cjj1484 HK was involved in expression of this operon. However, in our DNA-binding assays, we noted that both the unphosphorylated and phosphorylated Cjj1483 RR forms bound Cjj1386 promoter DNA, with the phosphorylated form of Cjj1483 perhaps binding DNA at a lower protein concentration. Regardless, we propose that the Cjj1483 RR can repress gene expression independently of the HK, with a potential noncognate phosphodonor possibly being used to autophosphorylate the Cjj1483 RR and enhance its DNA binding (Fig. 9). Lastly, we provided evidence that both Cjj1484 and Cjj1483 are required for WT levels of expression of the class IV gene ciaC, suggesting that phosphotransfer through the TCS resulting in phosphorylation of Cjj1483 is likely a transcriptional activator for ciaC (Fig. 9). In all, our data suggest various modalities for how the Cjj1484-Cjj1483 TCS can influence expression of different sets of genes.
In this work, we identified genes within the Cjj1484-Cjj1483 TCS regulon and provided some evidence for direct regulation by the TCS. Genes within this regulon encode proteins involved in different metabolic activities, such as gluconate respiration (Cjj0438 and Cjj0439), heme uptake (ChuC), ferric enterobactin transport (ExbB2 and ExbD2), and possibly transferrin transport (Cjj0210 and Cjj0211). Although we did not observe any large differences in the ability of C. jejuni mutants lacking the TCS to grow under iron-rich or iron-depleted conditions, we did observe increased gluconate dehydrogenase activity in a mutant lacking the Cjj1484 HK, suggesting at least one physiological consequence of an impaired Cjj1484-Cjj1483 TCS. At present, the functions of two other members, Cjj1385 and Cjj1386, are unknown. In bacteria, it is common for expression of heme and iron acquisition systems or respiration to be moderately influenced by diverse regulatory systems. We propose that regulation of these genes is likely complex in C. jejuni. Indeed, in searching the C. jejuni literature, we found that expression of these genes is influenced by multiple regulatory factors or growth conditions. For instance, previous studies have shown that expression of many genes within the Cjj1484-Cjj1483 regulon is reduced under high-iron conditions or by hyperosmotic shock but increased at an acidic pH (51–53). In addition, the ferric uptake regulator (Fur), which monitors the iron status of the bacterium, largely represses expression of these genes (54). For Cjj0210-Cjj0211, chuC, exbB2, and exbD2, evidence exists that the peroxide regulator (PerR) also influences expression of these genes (55). A recent study of the RacRS TCS of C. jejuni revealed that chuC, Cjj0210-Cjj0211, and Cjj1385-Cjj1386 expression levels were all decreased in a racR mutant (27). However, expression of the Cjj1484-Cjj1483 TCS, which is the subject of this work, was significantly higher in the racR mutant (27). Further investigation was unable to demonstrate that RacR interacted specifically with the promoters for chuC, Cjj0210-Cjj0211, and Cjj1385-Cjj1386, creating some speculation about whether the RacRS TCS may only indirectly regulate expression of these genes. In our current work, we demonstrated that the Cjj1483 RR bound directly to the promoter of Cjj1386, indicating a strong possibility that the RacRS TCS mediates regulation of expression of this gene, and possibly others, through the Cjj1484-Cjj1483 TCS. Considering these other global transcriptome analyses of C. jejuni, it is likely that expression of genes within the Cjj1484-Cjj1483 regulon is multifactorial, with multiple regulators finely controlling expression of these genes to an appropriate level that benefits the biology of C. jejuni in different situations.
We were unable to demonstrate that the Cjj1484-Cjj1483 TCS is required for commensal colonization of the natural avian host. However, this TCS largely represses in vitro expression of genes, such as Cjj0438 and Cjj0439, that are required to be expressed in vivo for WT levels of colonization of chicks. Thus, mutations of Cjj1484 or Cjj1483 cause enhanced transcription of Cjj0438, Cjj0439, and other possible colonization factors that are within the Cjj1484-Cjj1483 regulon. Results from our work suggest that an aberrant increase in expression of genes caused by mutation of the Cjj1484-Cjj1483 TCS does not appear to negatively affect the ability of C. jejuni to promote colonization of chicks. Although the data did not meet statistical significance, we did note that the Cjj1386 mutant had a 3.5-fold decrease in colonization. This finding may indicate that the Cjj1386 operon encodes proteins that modestly affect colonization. Although the Cjj1484-Cjj1483 TCS does not appear to be required for colonization, it is possible that this TCS plays an important role in repressing expression of colonization factors when the bacterium is outside a host. This model suggests that the repressive activity of the TCS in vivo is likely decreased so that expression of Cjj0438 and Cjj0439 is sufficient to promote colonization. Further studies will be required to determine if the TCS has an important role in regulating expression of genes ex vivo that may assist the bacterium in surviving outside a host or prime the bacterium for transmission to a new host. Furthermore, it is possible that this TCS plays an important role in regulating expression of genes required for infection of humans to promote diarrheal disease.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by NIH grants R01AI065539 (D.R.H.) and R21AI103643 (D.R.H.). P.M.L. was supported by NIH training grant T32 AI007520. This work was also funded by the USDA, Agricultural Research Service (CRIS project 5325-42000-230-047).
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/JB.02564-14.
REFERENCES
- 1.Crim SM, Iwamoto M, Huang JY, Griffin PM, Gilliss D, Cronquist AB, Cartter M, Tobin-D'Angelo M, Blythe D, Smith K, Lathrop S, Zansky S, Cieslak PR, Dunn J, Holt KG, Lance S, Tauxe R, Henao OL, Centers for Disease Control and Prevention . 2014. Incidence and trends of infection with pathogens transmitted commonly through food—Foodborne Diseases Active Surveillance Network, 10 U.S. sites, 2006–2013. MMWR Morb Mortal Wkly Rep 63:328–332. [PMC free article] [PubMed] [Google Scholar]
- 2.Olson CK, Ethelberg S, van Pelt W, Tauxe RV. 2008. Epidemiology of Campylobacter jejuni infections in industrialized nations, p 163–189. In Nachamkin I, Szymanski CM, Blaser MJ (ed), Campylobacter, 3rd ed ASM Press, Washington, DC. [Google Scholar]
- 3.Lindblom G-B, Sjorgren E, Kaijser B. 1986. Natural Campylobacter colonization in chickens raised under different environmental conditions. J Hyg (Lond) 96:385–391. doi: 10.1017/S0022172400066146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pokamunski S, Kass N, Borochovich E, Marantz B, Rogol M. 1986. Incidence of Campylobacter spp. in broiler flocks monitored from hatching to slaughter. Avian Pathol 15:83–92. doi: 10.1080/03079458608436268. [DOI] [PubMed] [Google Scholar]
- 5.Beery JT, Hugdahl MB, Doyle MP. 1988. Colonization of gastrointestinal tracts of chicks by Campylobacter jejuni. Appl Environ Microbiol 54:2365–2370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Friedman CR, Hoekstra RM, Samuel M, Marcus R, Bender J, Shiferaw B, Reddy S, Ahuja SD, Helfrick DL, Hardnett F, Carter M, Anderson B, Tauxe RV. 2004. Risk factors for sporadic Campylobacter infection in the United States: a case-control study in FoodNet sites. Clin Infect Dis 38(Suppl 3):S285–S296. doi: 10.1086/381598. [DOI] [PubMed] [Google Scholar]
- 7.van Spreeuwel JP, Duursma GC, Meijer CJ, Bax R, Rosekrans PC, Lindeman J. 1985. Campylobacter colitis: histological immunohistochemical and ultrastructural findings. Gut 26:945–951. doi: 10.1136/gut.26.9.945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hadden RD, Karch H, Hartung HP, Zielasek J, Weissbrich B, Schubert J, Weishaupt A, Cornblath DR, Swan AV, Hughes RA, Toyka KV. 2001. Preceding infections, immune factors, and outcome in Guillain-Barre syndrome. Neurology 56:758–765. doi: 10.1212/WNL.56.6.758. [DOI] [PubMed] [Google Scholar]
- 9.Jacobs BC, Van Belkum A, Endtz HP. 2008. Guillain-Barre syndrome and Campylobacter infection, p 245–262. In Nachamkin I, Szymanski CM, Blaser MJ (ed), Campylobacter, 3rd ed ASM Press, Washington, DC. [Google Scholar]
- 10.Gao R, Stock AM. 2009. Biological insights from structures of two-component proteins. Annu Rev Microbiol 63:133–154. doi: 10.1146/annurev.micro.091208.073214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Stock AM, Robinson VL, Goudreau PN. 2000. Two-component signal transduction. Annu Rev Biochem 69:183–215. doi: 10.1146/annurev.biochem.69.1.183. [DOI] [PubMed] [Google Scholar]
- 12.Krell T, Lacal J, Busch A, Silva-Jimenez H, Guazzaroni ME, Ramos JL. 2010. Bacterial sensor kinases: diversity in the recognition of environmental signals. Annu Rev Microbiol 64:539–559. doi: 10.1146/annurev.micro.112408.134054. [DOI] [PubMed] [Google Scholar]
- 13.Fouts DE, Mongodin EF, Mandrell RE, Miller WG, Rasko DA, Ravel J, Brinkac LM, DeBoy RT, Parker CT, Daugherty SC, Dodson RJ, Durkin AS, Madupu R, Sullivan SA, Shetty JU, Ayodeji MA, Shvartsbeyn A, Schatz MC, Badger JH, Fraser CM, Nelson KE. 2005. Major structural differences and novel potential virulence mechanisms from the genomes of multiple Campylobacter species. PLoS Biol 3:e15. doi: 10.1371/journal.pbio.0030015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Parkhill J, Wren BW, Mungall K, Ketley JM, Churcher C, Basham D, Chillingworth T, Davies RM, Feltwell T, Holroyd S, Jagels K, Karlyshev AV, Moule S, Pallen MJ, Penn CW, Quail MA, Rajandream M-A, Rutherford KM, van Vliet AHM, Whitehead S, Barrell BG. 2000. The genome sequence of the food-borne pathogen Campylobacter jejuni reveals hypervariable sequences. Nature 403:665–668. doi: 10.1038/35001088. [DOI] [PubMed] [Google Scholar]
- 15.Hendrixson DR, DiRita VJ. 2003. Transcription of σ54-dependent but not σ28-dependent flagellar genes in Campylobacter jejuni is associated with formation of the flagellar secretory apparatus. Mol Microbiol 50:687–702. doi: 10.1046/j.1365-2958.2003.03731.x. [DOI] [PubMed] [Google Scholar]
- 16.Joslin SN, Hendrixson DR. 2008. Analysis of the Campylobacter jejuni FlgR response regulator suggests integration of diverse mechanisms to activate an NtrC-like protein. J Bacteriol 190:2422–2433. doi: 10.1128/JB.01827-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Joslin SN, Hendrixson DR. 2009. Activation of the Campylobacter jejuni FlgSR two-component system is linked to the flagellar export apparatus. J Bacteriol 191:2656–2667. doi: 10.1128/JB.01689-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Barrero-Tobon AM, Hendrixson DR. 2012. Identification and analysis of flagellar coexpressed determinants (Feds) of Campylobacter jejuni involved in colonization. Mol Microbiol 84:352–369. doi: 10.1111/j.1365-2958.2012.08027.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Boll JM, Hendrixson DR. 2011. A specificity determinant for phosphorylation in a response regulator prevents in vivo cross-talk and modification by acetyl phosphate. Proc Natl Acad Sci U S A 108:20160–20165. doi: 10.1073/pnas.1113013108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Boll JM, Hendrixson DR. 2013. A regulatory checkpoint during flagellar biogenesis in Campylobacter jejuni initiates signal transduction to activate transcription of flagellar genes. mBio 4:e00432-13. doi: 10.1128/mBio.00432-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wosten MMSM, Wagenaar JA, van Putten JPM. 2004. The FlgS/FlgR two-component signal transduction system regulates the fla regulon in Campylobacter jejuni. J Biol Chem 279:16214–16222. doi: 10.1074/jbc.M400357200. [DOI] [PubMed] [Google Scholar]
- 22.MacKichan JK, Gaynor EC, Chang C, Cawthraw S, Newell DG, Miller JF, Falkow S. 2004. The Campylobacter jejuni dccRS two-component system is required for optimal in vivo colonization but is dispensable for in vitro growth. Mol Microbiol 54:1269–1286. doi: 10.1111/j.1365-2958.2004.04371.x. [DOI] [PubMed] [Google Scholar]
- 23.Wosten MM, van Dijk L, Parker CT, Guilhabert MR, van der Meer-Janssen YP, Wagenaar JA, van Putten JP. 2010. Growth phase-dependent activation of the DccRS regulon of Campylobacter jejuni. J Bacteriol 192:2729–2736. doi: 10.1128/JB.00024-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Svensson SL, Davis LM, MacKichan JK, Allan BJ, Pajaniappan M, Thompson SA, Gaynor EC. 2009. The CprS sensor kinase of the zoonotic pathogen Campylobacter jejuni influences biofilm formation and is required for optimal chick colonization. Mol Microbiol 71:253–272. doi: 10.1111/j.1365-2958.2008.06534.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bras AM, Chatterjee S, Wren BW, Newell DG, Ketley JM. 1999. A novel Campylobacter jejuni two-component regulatory system important for temperature-dependent growth and colonization. J Bacteriol 181:3298–3302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Apel D, Ellermeier J, Pryjma M, Dirita VJ, Gaynor EC. 2012. Characterization of Campylobacter jejuni RacRS reveals roles in the heat shock response, motility, and maintenance of cell length homogeneity. J Bacteriol 194:2342–2354. doi: 10.1128/JB.06041-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.van der Stel AX, van Mourik A, Heijmen-van Dijk L, Parker CT, Kelly DJ, van de Lest CH, van Putten JP, Wosten MM. 8 April 2014. The Campylobacter jejuni RacRS system regulates fumarate utilization in a low oxygen environment. Environ Microbiol doi: 10.1111/1462-2920.12476. [DOI] [PubMed] [Google Scholar]
- 28.Wosten MM, Parker CT, van Mourik A, Guilhabert MR, van Dijk L, van Putten JP. 2006. The Campylobacter jejuni PhosS/PhosR operon represents a non-classical phosphate-sensitive two-component system. Mol Microbiol 62:278–291. doi: 10.1111/j.1365-2958.2006.05372.x. [DOI] [PubMed] [Google Scholar]
- 29.Hendrixson DR, Akerley BJ, DiRita VJ. 2001. Transposon mutagenesis of Campylobacter jejuni identifies a bipartite energy taxis system required for motility. Mol Microbiol 40:214–224. doi: 10.1046/j.1365-2958.2001.02376.x. [DOI] [PubMed] [Google Scholar]
- 30.Makarova O, Kamberov E, Margolis B. 2000. Generation of deletion and point mutations with one primer in a single cloning step. Biotechniques 29:970–972. [DOI] [PubMed] [Google Scholar]
- 31.Higuchi R. 1990. Recombinant PCR, p 77–183. In Innis MA, Gelfand DH, Sninsky JJ, White TJ (ed), PCR protocols: a guide to methods and applications. Academic Press, London, United Kingdom. [Google Scholar]
- 32.Yao R, Alm RA, Trust TJ, Guerry P. 1993. Construction of new Campylobacter cloning vectors and a new mutational cat cassette. Gene 130:127–130. doi: 10.1016/0378-1119(93)90355-7. [DOI] [PubMed] [Google Scholar]
- 33.Figurski DH, Helinski DR. 1979. Replication of an origin-containing derivative of plasmid RK2 dependent on a plasmid function provided in trans. Proc Natl Acad Sci U S A 76:1648–1652. doi: 10.1073/pnas.76.4.1648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Guerry P, Yao R, Alm RA, Burr DH, Trust TJ. 1994. Systems of experimental genetics for Campylobacter species. Methods Enzymol 235:474–481. doi: 10.1016/0076-6879(94)35163-5. [DOI] [PubMed] [Google Scholar]
- 35.Hendrixson DR, DiRita VJ. 2004. Identification of Campylobacter jejuni genes involved in commensal colonization of the chick gastrointestinal tract. Mol Microbiol 52:471–484. doi: 10.1111/j.1365-2958.2004.03988.x. [DOI] [PubMed] [Google Scholar]
- 36.Gaynor EC, Wells DH, MacKichan JK, Falkow S. 2005. The Campylobacter jejuni stringent response controls specific stress survival and virulence-associated phenotypes. Mol Microbiol 56:8–27. doi: 10.1111/j.1365-2958.2005.04525.x. [DOI] [PubMed] [Google Scholar]
- 37.Pryjma M, Apel D, Huynh S, Parker CT, Gaynor EC. 2012. FdhTU-modulated formate dehydrogenase expression and electron donor availability enhance recovery of Campylobacter jejuni following host cell infection. J Bacteriol 194:3803–3813. doi: 10.1128/JB.06665-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Holmes K, Mulholland F, Pearson BM, Pin C, McNicholl-Kennedy J, Ketley JM, Wells JM. 2005. Campylobacter jejuni gene expression in response to iron limitation and the role of Fur. Microbiology 151:243–257. doi: 10.1099/mic.0.27412-0. [DOI] [PubMed] [Google Scholar]
- 39.Pajaniappan M, Hall JE, Cawthraw SA, Newell DG, Gaynor EC, Fields JA, Rathbun KM, Agee WA, Burns CM, Hall SJ, Kelly DJ, Thompson SA. 2008. A temperature-regulated Campylobacter jejuni gluconate dehydrogenase is involved in respiration-dependent energy conservation and chicken colonization. Mol Microbiol 68:474–491. doi: 10.1111/j.1365-2958.2008.06161.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Orchard RC, Kittisopikul M, Altschuler SJ, Wu LF, Suel GM, Alto NM. 2012. Identification of F-actin as the dynamic hub in a microbial-induced GTPase polarity circuit. Cell 148:803–815. doi: 10.1016/j.cell.2011.11.063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Skerker JM, Prasol MS, Perchuk BS, Biondi EG, Laub MT. 2005. Two-component signal transduction pathways regulating growth and cell cycle progression in a bacterium: a system-level analysis. PLoS Biol 3:e334. doi: 10.1371/journal.pbio.0030334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Quon KC, Marczynski GT, Shapiro L. 1996. Cell cycle control by an essential bacterial two-component signal transduction protein. Cell 84:83–93. doi: 10.1016/S0092-8674(00)80995-2. [DOI] [PubMed] [Google Scholar]
- 43.McIver KS, Myles RL. 2002. Two DNA-binding domains of Mga are required for virulence gene activation in the group A streptococcus. Mol Microbiol 43:1591–1601. doi: 10.1046/j.1365-2958.2002.02849.x. [DOI] [PubMed] [Google Scholar]
- 44.Dugar G, Herbig A, Forstner KU, Heidrich N, Reinhardt R, Nieselt K, Sharma CM. 2013. High-resolution transcriptome maps reveal strain-specific regulatory features of multiple Campylobacter jejuni isolates. PLoS Genet 9:e1003495. doi: 10.1371/journal.pgen.1003495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zeng X, Xu F, Lin J. 2013. Specific TonB-ExbB-ExbD energy transduction systems required for ferric enterobactin acquisition in Campylobacter. FEMS Microbiol Lett 347:83–91. doi: 10.1111/1574-6968.12221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Linton D, Allan E, Karlyshev AV, Cronshaw AD, Wren BW. 2002. Identification of N-acetylgalactosamine-containing glycoproteins PEB3 and CgpA in Campylobacter jejuni. Mol Microbiol 43:497–508. doi: 10.1046/j.1365-2958.2002.02762.x. [DOI] [PubMed] [Google Scholar]
- 47.Rangarajan ES, Bhatia S, Watson DC, Munger C, Cygler M, Matte A, Young NM. 2007. Structural context for protein N-glycosylation in bacteria: the structure of PEB3, an adhesin from Campylobacter jejuni. Protein Sci 16:990–995. doi: 10.1110/ps.062737507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Min T, Vedadi M, Watson DC, Wasney GA, Munger C, Cygler M, Matte A, Young NM. 2009. Specificity of Campylobacter jejuni adhesin PEB3 for phosphates and structural differences among its ligand complexes. Biochemistry 48:3057–3067. doi: 10.1021/bi802195d. [DOI] [PubMed] [Google Scholar]
- 49.Christensen JE, Pacheco SA, Konkel ME. 2009. Identification of a Campylobacter jejuni-secreted protein required for maximal invasion of host cells. Mol Microbiol 73:650–662. doi: 10.1111/j.1365-2958.2009.06797.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Klose KE, Weiss DS, Kustu S. 1993. Glutamate at the site of phosphorylation of nitrogen-regulatory protein NTRC mimics aspartyl-phosphate and activates the protein. J Mol Biol 232:67–78. doi: 10.1006/jmbi.1993.1370. [DOI] [PubMed] [Google Scholar]
- 51.Cameron A, Frirdich E, Huynh S, Parker CT, Gaynor EC. 2012. Hyperosmotic stress response of Campylobacter jejuni. J Bacteriol 194:6116–6130. doi: 10.1128/JB.01409-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Reid AN, Pandey R, Palyada K, Naikare H, Stintzi A. 2008. Identification of Campylobacter jejuni genes involved in the response to acidic pH and stomach transit. Appl Environ Microbiol 74:1583–1597. doi: 10.1128/AEM.01507-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Butcher J, Stintzi A. 2013. The transcriptional landscape of Campylobacter jejuni under iron replete and iron limited growth conditions. PLoS One 8:e79475. doi: 10.1371/journal.pone.0079475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Butcher J, Sarvan S, Brunzelle JS, Couture JF, Stintzi A. 2012. Structure and regulon of Campylobacter jejuni ferric uptake regulator Fur define apo-Fur regulation. Proc Natl Acad Sci U S A 109:10047–10052. doi: 10.1073/pnas.1118321109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Palyada K, Sun YQ, Flint A, Butcher J, Naikare H, Stintzi A. 2009. Characterization of the oxidative stress stimulon and PerR regulon of Campylobacter jejuni. BMC Genomics 10:481. doi: 10.1186/1471-2164-10-481. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.