ABSTRACT
MerR metalloregulators alleviate toxicity caused by an excess of metal ions, such as copper, zinc, mercury, lead, cadmium, silver, or gold, by triggering the expression of specific efflux or detoxification systems upon metal detection. The sensor protein binds the inducer metal ion by using two conserved cysteine residues at the C-terminal metal-binding loop (MBL). Divalent metal ion sensors, such as MerR and ZntR, require a third cysteine residue, located at the beginning of the dimerization (α5) helix, for metal coordination, while monovalent metal ion sensors, such as CueR and GolS, have a serine residue at this position. This serine residue was proposed to provide hydrophobic and steric restrictions to privilege the binding of monovalent metal ions. Here we show that the presence of alanine at this position does not modify the activation pattern of monovalent metal sensors. In contrast, GolS or CueR mutant sensors with a substitution of cysteine for the serine residue respond to monovalent metal ions or Hg(II) with high sensitivities. Furthermore, in a mutant deleted of the Zn(II) exporter ZntA, they also trigger the expression of their target genes in response to either Zn(II), Cd(II), Pb(II), or Co(II).
IMPORTANCE Specificity in a stressor's recognition is essential for mounting an appropriate response. MerR metalloregulators trigger the expression of specific resistance systems upon detection of heavy metal ions. Two groups of these metalloregulators can be distinguished, recognizing either +1 or +2 metal ions, depending on the presence of a conserved serine in the former or a cysteine in the latter. Here we demonstrate that the serine residue in monovalent metal ion sensors excludes divalent metal ion detection, as its replacement by cysteine renders a pan-metal ion sensor. Our results indicate that the spectrum of signals detected by these sensors is determined not only by the metal-binding ligand availability but also by the metal-binding cavity flexibility.
INTRODUCTION
A set of bacterial metal ion sensors of the MerR family of transcriptional regulators are essential for controlling toxicity caused by an excess of essential heavy metal ions, such as copper (Cu) or zinc (Zn), or the presence of toxic-only heavy metal ions, such as mercury (Hg), lead (Pb), cadmium (Cd), silver (Ag), or gold (Au) ions (1). Detection of free metal ions in the cytoplasm by these sensors rapidly induces the transcription of sets of genes mostly coding for efflux or detoxification systems (2–5). Two functional regions composed of residues from both monomers can easily be distinguished in the structures of these dimeric regulators, namely, the DNA-binding and metal-binding regions. These regions are connected by two 34-residue antiparallel coiled-coil helices, one from each monomer, which cross over at the dimer interface. According to the current model, the signal detected at the metal-binding site is transmitted through allosteric changes to the DNA-binding domain, resulting in transcriptional activation of the target genes via a DNA distortion mechanism (2, 6).
Metal ions differ in charge and preferred coordination chemistry. In consequence, the ability of a sensor to recognize a particular metal ion depends on the array of putative ligands at the metal-binding site to establish the favorite coordinate-covalent interactions with the metal ion (6). In fact, all characterized metal ion sensors of the MerR family are able to recognize either +1 or +2 metal ions but not both “types.” The crystallographic structures of two members of the family from Escherichia coli, CueR and ZntR, bound to their inducers revealed important aspects of metal coordination and the interactions involved in signal recognition (7). In both regulators, the metal ion is buried in a solvent-inaccessible site at the dimer interface. CueR uses the S atoms of the C112 and C120 cysteine residues from the C-terminal metal-binding loop (MBL) to coordinate one Cu(I) ion in a linear two-coordinated array (7–9). The same ligands are used to bind other +1 ions, such as Ag(I) or Au(I), that also activate CueR (see Fig. 4A). In contrast, the Zn sensor ZntR requires other ligands, besides the C-terminal Cys residues, to coordinate two atoms of Zn(II) at the metal-binding site (7). One of the Zn(II) ions is coordinated by C114 and C124 (equivalent to C112 and C120 of CueR), and the other Zn(II) ion interacts with C115 and H119 from the MBL. The C79 residue, located at the beginning of the dimerization (α5) helix of the other monomer, and a phosphate ion from the buffer used during crystal formation also served as additional ligands to coordinate the Zn(II) atoms in a tetrahedral geometry (7).
FIG 4.
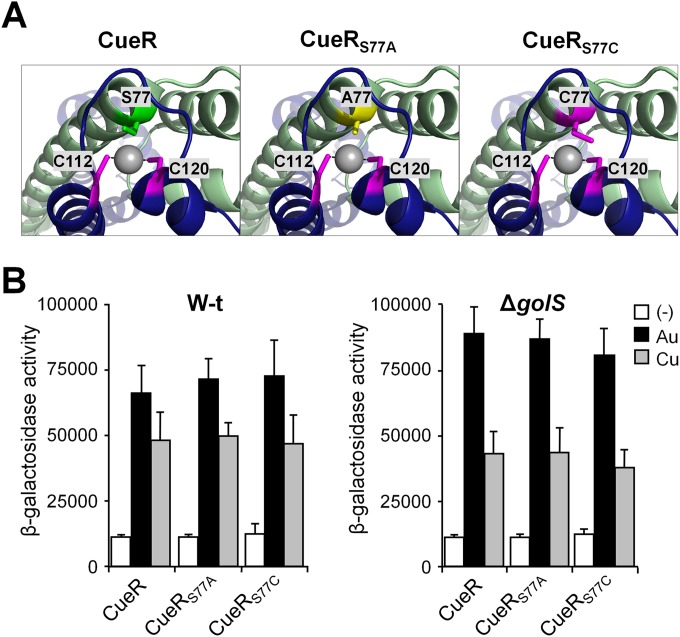
S77A and S77C substitutions do not affect the CueR response to Au or Cu. (A) Structural models of the CueR, CueRS77A, and CueRS77C MBLs. Residues C112 and C120 from one protomer and the residue at position 77 from the other protomer are indicated. (B) β-Galactosidase activities (Miller units) from a plasmid-carried copA::lacZ transcriptional fusion (13) expressed by cells carrying either cueR (CueR) or a mutant allele coding for CueRS77A or CueRS77C, in an otherwise wild-type (W-t) or a ΔgolS genetic background. Cells were grown overnight in LB broth without (−) or with the addition of 40 μM AuHCl4 (Au) or 1 mM CuSO4 (Cu). The data correspond to mean values for four independent experiments performed in duplicate. Error bars correspond to standard deviations.
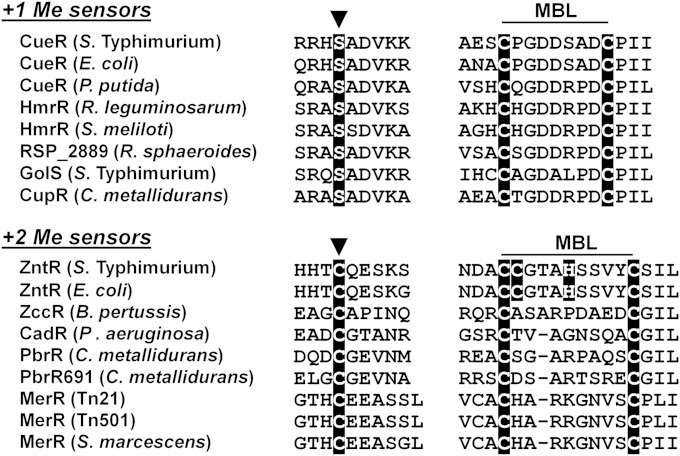
Two homologous monovalent metal ion sensors that coexist in Salmonella enterica were characterized: the nonselective CueR sensor, which responds to Cu(I), Ag(I), and Au(I) with similar sensitivities, and the Au sensor GolS, which discriminates Au(I) from Cu(I) or Ag(I) (10–13). By domain-swapping and site-directed mutagenesis, we established that residues at positions 113 and 118 within the MBL are the main contributors to Au(I) selectivity (12). The presence of a Pro residue at position 113 favors the detection of Cu, while the presence of Pro at position 118 disfavors it. GolS xenologs harbor alanine and proline at positions 113 and 118, respectively, while CueR and its homologs harbor P113 and A118 instead (12). All these sensors are activated by +1 metal ions but do not respond to +2 ions, such as Zn(II), Cd(II), Hg(II), or Pb(II), that act as inducers of other members of the metal-responsive MerR family (2, 6, 14). Indeed, sequence alignments of CueR and other metal sensors of the family allow discrimination of sensors recognizing metal ions with a +1 charge from those activated by divalent metal ions (Fig. 1). ZntR and other sensors known to recognize divalent metal ions, such as the Hg(II) sensor MerR, the Cd(II) sensor CadR, and the Pb(II) sensor PbrR, have a conserved cysteine residue at the beginning of the α5 helix and a C-X8/9-C (where X is any amino acid residue) motif at the MBL. CueR and other monovalent metal ion sensors, such as the Au(I) sensors GolS and CupR, share a conserved R-X-S-A-D-V-K signature at the beginning of the α5 helix and the C-X-G-D-X3-D-C sequence at the MBL, followed by a conserved proline residue. Most of these sensors also respond to metals with similar charges and coordination chemistries, but as mentioned above, they are responsive to either +1 or +2 metal ions, not both types.
FIG 1.
Amino acid sequence alignment of the regions involved in metal ion recognition in mono- and divalent MerR metal ion sensors. The sensors studied were as follows: Salmonella Typhimurium CueR (accession no. NP_459494), Escherichia coli CueR (accession no. NP_415020), Pseudomonas putida CueR (accession no. NP_742748.1), Rhizobium leguminosarum HmrR (accession no. YP_771363), Sinorhizobium meliloti HmrR (accession no. NP_437559), Rhodobacter sphaeroides RSP_2889 (accession no. YP_352951), Salmonella Typhimurium GolS (accession no. NP_459349), Cupriavidus metallidurans CupR (accession no. YP_585664), Salmonella Typhimurium ZntR (accession no. NP_462316.1), Escherichia coli ZntR (accession no. NP_417751.1), Bordetella pertussis ZccR (accession no. NP_881331.1), Pseudomonas aeruginosa CadR (accession no. WP_003117472.1), Cupriavidus metallidurans PbrR (accession no. YP_581755.1), Cupriavidus metallidurans PbrR691 (accession no. YP_584450.1), Tn21 MerR (accession no. ADC80844.1), Tn501 MerR (accession no. CAA77320.1), and Serratia marcescens MerR (accession no. NP_941195.1). Conserved cysteine, serine, and histidine residues are highlighted. The MBL sequence and the residue at the beginning of the dimerization helix (arrowheads) are also highlighted.
The available structures of CueR interacting with Cu(I), Au(I), or Ag(I) showed that at least two residues from the conserved R-X-S-A-D-V-K sequence approach the metal coordination sphere, establishing hydrogen-bonding interactions with several residues of the MBL (7). The R75 residue interacts with D119, which is conserved in all sensors that recognize +1 metal ions, while the conserved S77 residue interacts with the metal-coordinating C112 residue and with D115. In fact, the S77 residue has been proposed to play an important role in maintaining the conformation of the shielded metal-binding site, providing hydrophobic and steric restrictions that privilege the binding of monovalent metal ions (7). In the present study, different GolS and CueR mutants at position 77 were analyzed to disclose the importance of this residue in metal discrimination. The presence of either the conserved serine residue or an alanine (present only in an uncharacterized MerR-like regulator in Shewanella) restricts activation of these metal sensors to +1 metal ions. In contrast, the sole replacement of S77 by cysteine in both GolS and CueR generates sensors that maintain wild-type sensitivity to +1 ions but become responsive to +2 metal ions. These sensors detect Hg(II) with high sensitivities, and in a mutant deleted of the Zn(II) exporter ZntA, they can also respond to either Zn(II), Cd(II), Pb(II), or Co(II). To our knowledge, these are the first characterized pan-metal ion sensors, with the ability to detect both +1 and +2 metal ions.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
Bacterial strains and plasmids used in this study are listed in Table 1. Cells were grown at 37°C either in Luria-Bertani (LB) broth, on LB-agar plates, or in minimal M9 medium, as indicated. Kanamycin was used at a final concentration of 25 μg ml−1, tetracycline at 15 μg ml−1, chloramphenicol at 10 μg ml−1, and ampicillin at 100 μg ml−1. Isopropyl-β-d-thiogalactopyranoside (IPTG) was added to a final concentration of 100 or 500 μM to induce the expression of the CueR or GolS variants, respectively, from plasmids. Stocks of bacterial strains containing 15% glycerol were stored frozen at −70°C.
TABLE 1.
S. enterica serovar Typhimurium strains and plasmids used in this study
| Strain or plasmid | Relevant genotype and/or description | Reference or source |
|---|---|---|
| Strains | ||
| 14028s | Wild type | ATCC 14028 |
| PB6699 | golSC111S-cat | Laboratory stock |
| PB6791 | golSC112S-cat | Laboratory stock |
| PB5449 | ΔcueR | 18 |
| PB7116 | golS golB::lacZY+-kan | 12 |
| PB8261 | golSsL golB::lacZY+-kan | 12 |
| PB7177 | golS-Q69–77 golB::lacZY+-kan | This work |
| PB7871 | golS-MBL(+1) golB::lacZY+-kan | This work |
| PB7873 | golS-MBL(−1) golB::lacZY+-kan | This work |
| PB7875 | golS-α5(+1) golB::lacZY+-kan | This work |
| PB7877 | golS-α5(−1) golB::lacZY+-kan | This work |
| PB7863 | golSS77C golB::lacZY+-kan | This work |
| PB7865 | golSS77E golB::lacZY+-kan | This work |
| PB8001 | golSS77A golB::lacZY+-kan | This work |
| PB8540 | golSS77T golB::lacZY+-kan | This work |
| PB7705 | golSS77P golB::lacZY+-kan | This work |
| PB7706 | golSS77K golB::lacZY+-kan | This work |
| PB9295 | cueRS77C-cat | This work |
| PB9297 | cueRS77A-cat | This work |
| PB9311 | cueR-cat | This work |
| PB8090 | zntA::cat | This work |
| PB8631 | golSS77C-C111S golB::lacZY+-kan | This work |
| PB9464 | golSS77C-C112S golB::lacZY+-kan | This work |
| Plasmids | ||
| pPB1225 | pMC1871 derivative; PcopA::lacZ tet | 13 |
| pUH21-2 lacIq | oripMB1 bla lacIq | 19 |
| pPB1205 | pUH::golS | 10 |
| pPB1358 | pUH::golSS77C | This work |
| pPB1359 | pUH::golSS77A | This work |
| pPB1360 | pUH::golSS77T | This work |
| pPB1361 | pUH::golSS77P | This work |
| pPB1362 | pUH::cueRS77C | This work |
| pPB1351 | pUH::cueRC120S | This work |
All reagents and chemicals were purchased from Sigma. LB culture media were obtained from Difco, and oligonucleotides were provided by Life Technology.
Genetic and molecular biology techniques.
GolS or CueR mutants were constructed basically as previously described (12). To generate the different golS alleles, a megaprimer was produced using the golS-F forward primer (5′-ATGAGGAGGAGCGTCATGAACATCG-3′) and reverse primers carrying each mutation (primer sequences are available on request). The megaprimer was extended using RvP1-golB-R (5′-GTGAACTCCTTTTGTGTGGGAACTG-3′). For the introduction of the mutation at position 77 in cueR, a megaprimer was amplified using the primer cueR-F (5′-GAGGATCCATATGAATATTAGCG-3′) and the corresponding reverse primer for each mutant version (primer sequences are available on request). Extension of the megaprimer was done using the oligonucleotide RvP1-cueR-R (5′-CCGCCGGTTTATGCTTGATGCCGCGTTAGT-3′). Chromosomal DNA from the ATCC 14028s strain was used as the template in all the amplifications, except for the generation of golSS77C-C111S or golSS77C-C112S, for which DNA from strain PB6699 (golSC111S) or PB6791 (golSC112S) was used. The PCR products were individually fused to a chloramphenicol resistance cassette (amplified from the plasmid pKD3 by use of the appropriate primer pairs) by splicing by overhang extension PCR (SOE PCR) (15). Final overlap extension PCR products were introduced into the chromosome of Salmonella LB5010 Δ(golS-golB)::kan and LB5010 ΔcueR::kan derivatives (Table 1) by one-step linear transformation followed by Lambda Red-mediated recombination (16). Deletion of the zntA gene was done as described previously (10). The mutations were transferred to the wild-type ATCC 14028s strain or between strains by using P22 transduction (17).
To generate the golB::lacZ transcriptional fusion, the lac fusion plasmid pKG136 (Table 1) was inserted at the remaining FLP recombination target (FRT) site generated after removing the chloramphenicol resistance cassette as previously described (12). To generate GolS or CueR variant expression plasmids, each golS or cueR mutant allele was PCR amplified from the chromosome of the mutant Salmonella strain, using the golS-ORF-F/golS-ORF-R or cueR-F/cueR-R primer pair (10, 18), and cloned into BamHI-HindIII-digested pUH21-2 laqIq (19).
All plasmids and linear DNAs were introduced into the bacterial strains by electroporation using a Bio-Rad device following the manufacturer's recommendations. All constructs were verified by DNA sequencing.
Metal induction assays.
β-Galactosidase assays of golB::lacZ and copA::lacZ transcriptional fusions were used to analyze the metal-mediated activation of GolS and CueR, respectively, essentially as described previously (13). The cells were grown overnight in LB medium supplemented with the addition of sublethal concentrations of different metal salts. For each metal, final curves for the optical density at 600 nm (OD600) were previously performed to establish the highest concentration of each metal salt that did not affect cell growth. Stock solutions (1 or 0.5 M) of AuHCl4 · 3H2O, CuSO4 · 5H2O, ZnCl2, Pb(NO3)2, CdCl2, HgCl2, and CoCl2 · 6H2O were prepared using sterile distilled water and stored at 4°C. All heavy metal salts were at least of analytical grade (>95% pure).
Protein purification and Western blot analysis.
GolS, CueR, and the corresponding S77C and S77T variants were purified from E. coli XL1-Blue strains transformed with the corresponding expression plasmids essentially as described previously (10, 18). All procedures were carried out at 4°C. Standard protein determinations were done using the Bradford assay, with bovine serum albumin as a standard. The concentrations of the purified proteins were determined by measuring the absorbance at 280 nm, considering a theoretical extinction coefficient (obtained using the free ProtParam tool) of 11,460 M−1 cm−1 or 4,470 M−1 cm−1 for the GolS or CueR variants, respectively. Detection of GolS or its variants in soluble cell extracts by Western blotting was done essentially as described previously (10), using rabbit polyclonal anti-GolS antibodies.
Protein-DNA interaction and spectroscopy assays.
Electrophoretic gel mobility shift assays were performed essentially as previously described (10). Approximately 6 fmol of labeled DNA fragment containing the golB promoter region was incubated at room temperature for 20 min with purified GolS, GolSS77C, or GolSS77T.
UV-visible (UV-Vis) absorption determinations for CueRS77C were carried out in a Jasco UV-Vis-NIR V-670 spectrometer. Prior to determinations, the purified protein was loaded into a Sephadex G50 gel filtration column (HiTrap desalting column; Amersham Pharmacia Biotech) equilibrated with 20 mM Bis-Tris, pH 7, 500 mM NaNO3, 1 mM tris(2-carboxyethyl)phosphine (TCEP), and 5% glycerol in order to eliminate all Cl− ions. The protein sample was then dialyzed in buffer containing 20 mM Bis-Tris, pH 7, 500 mM NaNO3, 1 mM TCEP, 5% glycerol, and 500 μM Pb(NO3)2, using a 10,000-molecular-weight-cutoff membrane for 3 h. The excess of Pb2+ was removed by incubation in the same buffer lacking Pb(NO3)2 for 2 h. This step was repeated once, and then the sample was left overnight under the same conditions. The absorptions of two Pb(II)-loaded CueRS77C samples and the corresponding apo-CueRS77C proteins at similar final concentrations were recorded between 200 and 500 nm. The extinction coefficient for the Pb(II)-CueRS77C complex at 300 nm was calculated as the ratio between the absorbance value measured at 300 nm and the protein concentration.
The Pb(II)-loaded CueRS77C samples were subjected to atomic absorption spectroscopy to determine the amount of total lead bound to the purified protein. The absorbance at 283.3 nm of the sample was determined in a Metrolab 250AA flame atomic absorption spectrometer. Calibration curves were done using a Pb(II) standard. The total amount of lead was normalized against the total amount of purified protein.
Bioinformatics.
Sequence alignment was performed using MEGA, version 3.0, software (20). Different point mutations of CueR were modeling in silico by using the data from the crystallographic structure of the CueR-Cu(I) complex (Protein Data Bank code 1Q05) as a template and the free PyMOL, version 1.5, tool (http://www.pymol.org/).
RESULTS
Residues from the α4-α5 loop of GolS do not affect metal selectivity.
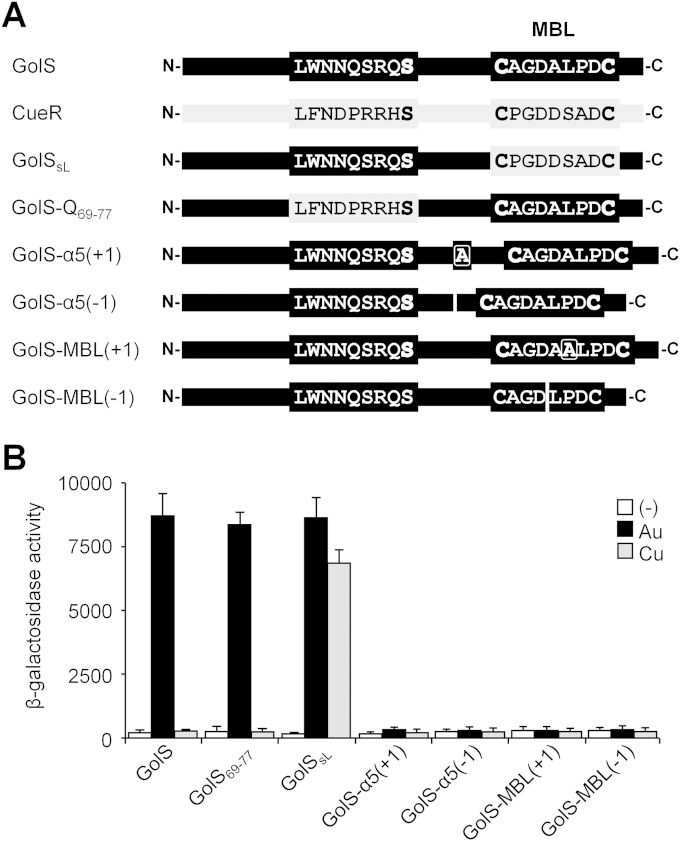
The crystallographic structure of the CueR-Cu(I) complex (7) reveals that besides residues from the MBL, a number of residues from the loop between the α4 and α5 helices or the N terminus of the α5 helix of the other monomer approach the first metal coordination sphere and contribute to neutralizing the negative charge of the buried S-Cu-S center. Because the sequences of the N-terminal region of the α5 helix (from S77 to K81) are identical in CueR and GolS, only the contribution to Au(I) selectivity of the α4-α5 loop, from L69 to S77, was analyzed. A chimeric golS allele coding for a sensor with the L69-to-S77 region of CueR (GolS69–77) was generated (Fig. 2A) and introduced into the chromosome, replacing the wild-type copy of golS, in an S. enterica serovar Typhimurium 14028s strain carrying the golB::lacZ reporter. The ability of the mutant sensor to activate transcription was analyzed in cells grown in Luria-Bertani (LB) medium in the absence or presence of either 40 μM AuHCl4 or 500 μM CuSO4, i.e., concentrations required to attain the maximal induction of the reporter gene (10, 21). The GolS69–77 sensor showed wild-type sensitivity to Au ions but not to Cu (Fig. 2B). In contrast, the nonselective GolSsL sensor, a chimeric sensor with a replacement of the MBL region (between A113 and P118) of GolS with the same region of CueR (12), was responsive to both Au and Cu, indicating that the α4-α5 loop of GolS does not contribute to Au(I) selectivity.
FIG 2.
The MBL size and distance from the GolS N terminus are optimized for metal detection. (A) Schematic representations of GolS, CueR, and the GolS variants GolSsL, GolS69–77, GolS-α5(+1), GolS-α5(−1), GolS-MBL(+1), and GolS-MBL(−1) (see the text for details on the construction of each golS mutant allele). The relevant sequences are shown. (B) β-Galactosidase activities (Miller units) from a golB::lacZ transcriptional fusion expressed by cells carrying either golS (GolS) or the mutant alleles coding for the above-mentioned GolS variants. The cells were grown overnight in LB broth without (−) or with the addition of 40 μM AuHCl4 (Au) or 1 mM CuSO4 (Cu). Extracellular Au(III) is reduced to Au(I) once inside the bacterial cytoplasm, where it is detected by the GolS sensor (14). The data correspond to mean values for four independent experiments performed in duplicate. Error bars correspond to standard deviations.
The GolS MBL size and distance from the N terminus are optimized for metal detection.
In the structures of the active CueR and ZntR dimers (7), the antiparallel coiled-coil interaction of the α5 helices from both monomers is proposed to have essential roles in signal transduction and in structuring the metal-binding site at the dimer interface. All characterized metal sensors of the MerR family have a 34-residue dimerization helix, but MBL regions vary in length from 7 amino acids in monovalent metal ion sensors to 9 residues in all characterized Zn(II) sensors (2) (Fig. 1). Besides these observations, to our knowledge, no studies have been done on the importance of these features for the functionality of these metal sensors. Four different GolS variants, each having either a single alanine residue inserted or deleted at position 107 of the α5 helix [GolS-α5(+1) and GolS-α5(−1)] or at position 116 within the MBL region [GolS-MBL(+1) and GolS-MBL(−1)], were generated (Fig. 2A). Increasing or decreasing the length of the α5 helix or the MBL in GolS modifies the relative position of the coordinating C112 residue for one monomer to the Ser77 residue of the other monomer at the dimer interface or the relative position of the selectivity-directing residues A113 and P118 (12) at the MBL, respectively. Each mutant allele was inserted into the chromosome, replacing the wild-type copy of golS, in order to test the ability of the sensor to activate transcription in response to Au and Cu ions, as described above. Affecting either the length of the dimerization helix or the MBL rendered an inactive sensor (Fig. 2B). These results indicate that the metal-binding site of GolS is precisely structured to achieve proper recognition of monovalent metal ions.
The residue at position 77 in either GolS or CueR affects metal sensing.
In the CueR metal-binding site (7), the S77 residue from one monomer approaches the S-Cu-S center formed by C112 and C120 of the other monomer but does not coordinate the metal (see Fig. 4A). Instead, it establishes hydrogen bonding with several residues of the MBL, including the metal-coordinating C112, D115, and A118 residues. These interactions were proposed to maintain the conformation of the buried metal-binding site and to stabilize the quaternary interactions at the dimer interface. The S77 residue is conserved in all characterized monovalent metal ion sensors of the family but not in homologs that respond to +2 metal ions, such as Hg(II) or Zn(II), which have a conserved cysteine residue at this position (Fig. 1). Different GolS S77-substituted variants were generated (Fig. 3). The serine was replaced by alanine, a residue present in an uncharacterized Shewanella CueR-like regulator (10–13); cysteine, present in the divalent metal ion sensors; or threonine, an amino acid residue with a polar uncharged side chain similar to that of serine or cysteine. It was also replaced by residues that are expected to severely affect the environment of the metal-binding site, such as proline, which can affect the local conformation and movement freedom in the folded protein, or charged residues, i.e., glutamic acid and lysine. Each mutant allele was introduced into the chromosome, replacing the wild-type golS copy, and evaluated for the ability to activate transcription of the golB::lacZ reporter fusion in response to Au or Cu ions (Fig. 3).
FIG 3.
The residue at position 77 is relevant to the metal-induced activation of GolS. β-Galactosidase activities (Miller units) were determined for cultures from golB::lacZ strains expressing the wild-type sensor or the indicated chromosomally encoded (A and B) or plasmid-expressed (D) GolS variants. For panels A and D, cells were grown as described in the legend to Fig. 2; for panel B, cells were grown overnight in LB broth without (−) or with the addition of 0.1, 1, or 10 μM AuHCl4. The data correspond to mean values for four independent experiments performed in duplicate. Error bars correspond to standard deviations. (C) Electrophoretic gel mobility shift assay using 6 fmol of 32P-3′-end-labeled PCR fragment from the golB promoter region and purified GolS, GolSS77C, or GolSS77T, at a final concentration of 5 or 20 nM. (E) Detection of the different GolS variants in whole-cell extracts from cultures grown overnight in LB without metals. The extracts were resolved by SDS-PAGE, followed by transfer to nitrocellulose and development using rabbit polyclonal anti-GolS antibodies. IPTG was added to a final concentration of 500 μM to activate expression of the plasmid-expressed GolS variants.
First, as expected, the S77A substitution did not modify the response to monovalent metal ions (Fig. 3A). Surprisingly, the S77C substitution, far from impairing monovalent metal detection, resulted in a pattern of metal activation similar to that of the native sensor or the GolSS77A mutant (Fig. 3A). In fact, no difference in the response to different Au concentrations was detected between these mutant variants and wild-type GolS, in both rich and minimal media (Fig. 3B and data not shown), indicating that both mutant sensors respond to metal ions with approximately the same sensitivity as that of GolS. Furthermore, the GolSS77C sensor could not be distinguished from the wild-type Au sensor in its ability to bind DNA (Fig. 3C). In contrast, replacement of S77 by threonine, proline, glutamic acid, or lysine rendered inactive sensors unable to trigger expression of the reporter gene (Fig. 3A).
The above-described experiments were done with strains in which the GolS variants were expressed from the chromosomal GolS-controlled promoter, located upstream of the preceding gene in the operon, i.e., golT. Therefore, their expression depended on self-activation derived from allosteric changes as a consequence of metal sensing (10). To separate sensor expression from metal detection, the golSS77T, golSS77P, golSS77A, and golSS77C variants were cloned under the control of the lac promoter in multicopy plasmids. A Salmonella Typhimurium strain carrying the golB::lacZ reporter was transformed with each of these plasmids, and LacZ induction was analyzed in cultures grown in LB and 500 μM IPTG, with or without addition of 40 μM AuHCl4. Under these conditions, a 9-fold Au induction of the reporter fusion was observed in the strain expressing GolSS77T, suggesting that the S77T substitution did not completely inactivate the sensor. Note that a >50-fold induction by Au was observed in cells expressing GolSS77A, GolSS77C, or the native Au sensor (Fig. 3D). On the other hand, GolSS77P was unable to induce the reporter gene in the presence of Au, even though this variant accumulated to levels similar to those of the fully active sensors or the GolSS77T variant in the cell (Fig. 3E).
The above results indicate that S77A and S77C substitutions do not affect the metal activation ability of GolS. Similarly, CueRS77A and CueRS77C exhibited patterns of metal induction of the copA::lacZ reporter fusion similar to that of the wild-type CueR sensor, even in a mutant deleted in golS (Fig. 4). This coincides with the modeling of different mutations over the crystallographic structure of CueR bound to Cu(I), which suggests that the S77A or S77C substitution provokes little or no disturbance in the MBL environment (Fig. 4A). All the putative hydrogen-bonding interactions of the wild-type sensor were conserved in the simulations for the CueRS77A variant, while the S77C replacement generated an additional putative interaction between the Cu(I) ion and the C77 residue.
GolSS77C and CueRS77C respond to both monovalent and divalent metal ions.
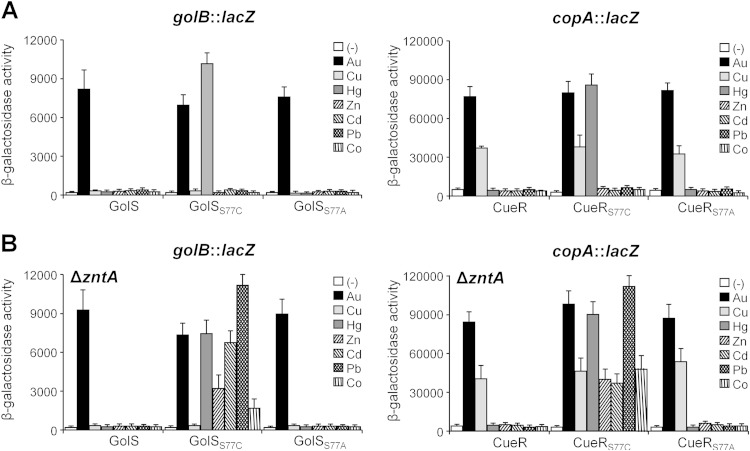
In view of the above results and the strict conservation of the cysteine residue at the N terminus of the dimerization helix in all members of the MerR family that detect divalent metal ions (Fig. 1), we tested if GolSS77C and CueRS77C also respond to divalent metal ions. HgCl2 triggered the expression of either golB::lacZ or copA::lacZ in the golSS77C or cueRS77C mutant strains, respectively, in both minimal and rich media (Fig. 5A and data not shown). This induction was not observed in the wild type or in the golSS77A or cueRS77A strain (Fig. 5A). Under these conditions, no activation of expression of the reporter genes was observed by addition of sublethal concentrations of ZnCl2, CdCl2, CoCl2, or Pb(NO3)2 for all tested strains, suggesting that GolS or CueR mutants with the S77C substitution acquired the ability to recognize Hg(II) without affecting their capability to sense either Au(I) or Cu(I) ions.
FIG 5.
GolSS77C and CueRS77C recognize +2 metal ions. β-Galactosidase activities (Miller units) from the chromosomal golB::lacZ fusion or from a plasmid-carried copA::lacZ transcriptional fusion were measured in cultures of otherwise wild-type (A) or ΔzntA (B) cells expressing the indicated chromosomal GolS or CueR variants. Cells were grown overnight in minimal M9 medium without (−) or with the addition of 10 μM AuHCl4 (Au), 10 μM CuSO4 (Cu), 1 μM HgCl2 (Hg), 100 μM ZnCl2 (Zn), 20 μM CdCl2 (Cd), 50 μM Pb(NO3)2 (Pb), or 100 μM CoCl2. The data correspond to mean values for four independent experiments performed in duplicate. Error bars correspond to the standard deviations.
The inability of GolSS77C or CueRS77C to respond to Zn(II), Cd(II), Co(II), or Pb(II) may be inherent to the low availability of these metal ions in the cytoplasmic environment. S. enterica codes for a Zn(II)-P-type ATPase homologous to E. coli ZntA, whose expression is triggered by Zn(II) but also by Cd(II) or Pb(II). ZntA is predicted to transport these metal ions out of the E. coli cytoplasm (22). Therefore, the unresponsiveness of GolSS77C or CueRS77C to Zn(II), Pb(II), or Cd(II) may be a consequence of the presence of ZntA. GolSS77C- or CueRS77C-controlled expression of golB::lacZ or copA::lacZ was analyzed in strains carrying a deletion of zntA (Fig. 5B). In contrast to the metal response observed for these mutant sensors in the otherwise wild-type background, in the ΔzntA strains, addition of Pb(II), Cd(II), Zn(II), or Co(II) triggered the expression of the reporter genes. The reporters' responsiveness to these metal ions is attributed to the S77C substitution in GolS or CueR, because no induction by these metal ions was observed in strains harboring either the wild-type sensors or the golSS77A or cueRS77A allele.
The in vitro ability of CueRS77C to bind Pb(II) was verified by spectroscopic studies. CueRS77C was able to bind up to 0.93 ± 0.23 equivalent of Pb per monomer. Neither CueR nor CueRC120S was able to bind the metal ion under the same conditions. The Pb(II)-loaded CueRS77C protein exhibited a peak at 300 nm (Fig. 6), with a calculated extinction coefficient of 4,774 M−1 cm−1. This peak probably represents the thiolate-Pb(II) charge-transfer band, reflecting the involvement of Cys residues as ligands of the Pb(II) atom.
FIG 6.
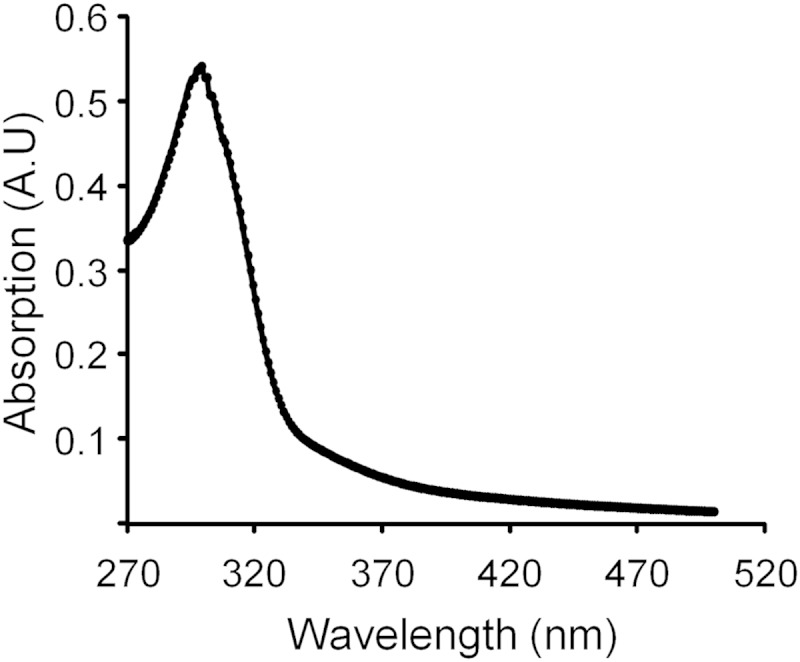
UV-Vis differential spectrum of Pb(II)-loaded CueRS77C. See Materials and Methods for details. A.U, instrument's arbitrary units.
The GolS C111 residue is not required for detection of divalent metal ions.
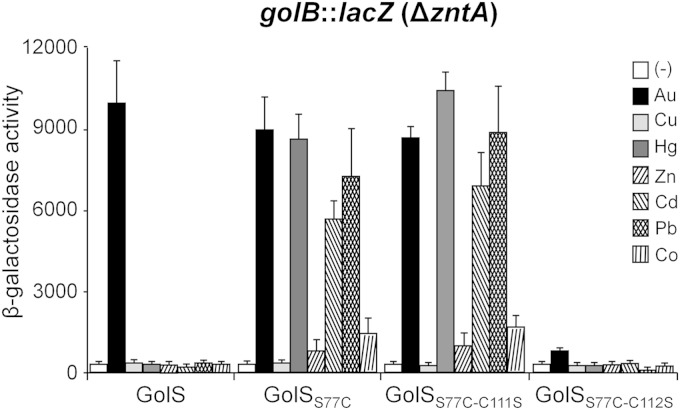
The interaction of two Zn(II) ions at the ZntR metal-binding site requires two consecutive cysteine residues, C114 and C115, besides C124 and H119 from the MBL and C79, equivalent to Cys77 in GolSS77C and CueRS77C (7). GolSS77C has a similar array of consecutive cysteine residues, i.e., C111 and C112, as well as C120 and C77, which can act as ligands of the divalent metal ion, but lacks the histidine residue equivalent to ZntR H119. CueRS77C lacks the extra cysteine residue at the beginning of the MBL but has a “CCHH” motif in the C-terminal portion that can serve as a ligand of the divalent metal ion. Neither GolS C111 nor the CueR CCHH motif is required for the detection of monovalent metal ions (10, 12). Thus, the role of the C111 residue of the GolSS77C sensor in Zn(II) detection was analyzed, considering that this ion requires a larger number of available ligands for coordination. ΔzntA golSS77C-C111S and ΔzntA golSS77C-C112S strains were used to test the expression of golB::lacZ in the presence of monovalent or divalent metal ions. As expected, expression of the reporter gene was similarly induced by Au(I), Hg(II), Zn(II), Cd(II), Pb(II), and Co(II) in either the ΔzntA golSS77C-C111S or ΔzntA golSS77C strain (Fig. 7), while no metal-induced expression was observed in the ΔzntA golSS77C-C112S strain. These results clearly indicate that C111 is not involved in metal sensing even in the absence of the C112 residue, which is necessary for metal-induced activation of the reporter gene.
FIG 7.
C111 of GolSS77C does not participate in metal recognition. The graph shows golB::lacZ β-galactosidase activities (Miller units) measured in ΔzntA cells expressing GolS, GolSS77C, GolSS77C-C111S, or GolSS77C-C112S. The cells were grown as described in the legend to Fig. 5. The data correspond to mean values for four independent experiments performed in duplicate. Error bars correspond to the standard deviations.
DISCUSSION
Specificity in metal recognition by MerR metalloregulators is directed mainly by the number and geometry of available ligands provided by the sensor protein for interaction with the metal (23, 24). In addition, other residues located in the metal-coordinated environment can indirectly influence metal recognition by providing charge neutralization as well as hydrophobic or steric restrictions that privilege interaction with some metals over others (7, 25). We previously reported that two residues from the MBL, A113 and P118, determine the ability of GolS to distinguish Au(I) from Cu(I) (10, 12). In the present study, we investigated the roles of residues from the N termini of the dimerization helices of GolS and CueR in metal ion discrimination. The crystallographic structure of the CueR-Cu(I) complex (7) revealed that a protomer's residues from this region approach and interact with residues from the metal-binding site of the other protomer at the dimer interface. Our results indicated that these residues in GolS are not involved in monovalent metal ion discrimination. However, they may be important for productively propagating the detected signal to the DNA-binding region to activate transcription (Fig. 2). In this sense, just increasing or decreasing the distance of the residues at the N terminus of the α5 helix from the coordinating cysteine residues of the MBL completely abolishes the functionality of the metal sensor.
Among the residues present at the beginning of the dimerization helix, the S77 residue was proposed—though not demonstrated—to be the main contributor to excluding +2 metal ions, such as Zn(II), from the metal-binding cavity of CueR, favoring the selective recognition of group 1B monovalent metal ions (7). In fact, the presence of either serine or cysteine at this position in uncharacterized MerR homologs is used as a marker to predict mono- or divalent metal ion preferences, respectively (2, 4). Replacement of C79 in ZntR and PbrR or C82 in MerR with alanine or serine rendered inactive mutant sensors unable to respond to their inducer +2 metals (26–28). As shown in Fig. 3 and 4, the replacement of the S77 residue by cysteine in both GolS and CueR not only did not modify the response to Cu(I), Ag(I), or Au(I) but also increased the spectrum of metal ions detected by the sensors. Contrasting with wild-type sensors, both GolSS77C and CueRS77C activated transcription of their target genes with Hg(II), and when the Zn(II) exporter ZntA was deleted, also with Zn(II), Pb(II), Cd(II), or Co(II) (Fig. 5 and 7). The ability of the CueRS77C sensor to interact with Pb(II) ions was also confirmed by spectroscopy (Fig. 6). Our results indicate that the S77C substitution provides a third S ligand to coordinate divalent metal ions, such as Pb(II) and Zn(II), which usually require three or four ligands for binding (29). The unique ability of GolSS77C and CueRS77C to respond to both +1 and +2 metal ions is rather an exception among the MerR metal sensors, since all characterized homologs detect one or a group of cognate metal ions with the same charge and similar coordination chemistries (23). Also, the ability of these mutant sensors to accommodate such a variety of metal ions that differ in ionic ratio, charge, and softness highlights the extraordinary flexibility of the metal-binding cavities of CueR and GolS.
Remarkably, all characterized MerR-like sensors responding to Cu(I)/Au(I)/Ag(I) or Hg(II) share a conserved Pro residue immediately following the MBL, although the MBL is larger in the latter group. Interestingly, sensors detecting Zn(II), Pb(II), Cd(II), or Co(II) lack this residue and have even longer MBL regions. Monovalent metal ion sensors, such as CueR and GolS, may have arisen from a promiscuous sensor after a single C-G transversion generating the C77S substitution, while divalent metal sensors fit to accommodate the preferred metals by modifying the sequence and length of the MBL region. It would be interesting to identify residues determining the exclusion of +1 metals from the metal-binding sites of divalent metal ion sensors, as well as those determining selectivity against different +2 metals.
Overall, this study contributes to the understanding of the strategies that CueR and GolS use to selectively recognize monovalent metal ions and, in particular, highlights the importance of the S77 residue in excluding +2 metals from their metal-binding site. Decoding the molecular bases that allow MerR sensors to distinguish between different metal ions would also be valuable for the rational design of improved metal-specific sensors with biotechnological applications.
ACKNOWLEDGMENTS
We thank Guillermo Bahr for his helpful advice with in silico modeling.
This work was supported by grants from the Agencia Nacional de Promoción Científica y Tecnológica, the Secretary of Science, Technology and Innovation of Santa Fe, and the National Scientific and Technical Research Council (CONICET) to S.K.C. and F.C.S. M.M.I. is a fellow of CONICET. S.K.C. and F.C.S. are career investigators of CONICET. F.C.S. is also a career investigator of the Rosario National University Research Council (CIUNR).
REFERENCES
- 1.Finney LA, O'Halloran TV. 2003. Transition metal speciation in the cell: insights from the chemistry of metal ion receptors. Science 300:931–936. doi: 10.1126/science.1085049. [DOI] [PubMed] [Google Scholar]
- 2.Brown NL, Stoyanov JV, Kidd SP, Hobman JL. 2003. The MerR family of transcriptional regulators. FEMS Microbiol Rev 27:145–163. doi: 10.1016/S0168-6445(03)00051-2. [DOI] [PubMed] [Google Scholar]
- 3.Helmann JD, Soonsanga S, Gabriel S. 2007. Metalloregulators: arbiters of metal sufficiency, p 37–71. In Nies DH, Silver S (ed), Molecular microbiology of heavy metals. Springer-Verlag, Berlin, Germany. [Google Scholar]
- 4.Hobman JL, Wilkie J, Brown NL. 2005. A design for life: prokaryotic metal-binding MerR family regulators. Biometals 18:429–436. doi: 10.1007/s10534-005-3717-7. [DOI] [PubMed] [Google Scholar]
- 5.Summers AO. 2009. Damage control: regulating defenses against toxic metals and metalloids. Curr Opin Microbiol 12:138–144. doi: 10.1016/j.mib.2009.02.003. [DOI] [PubMed] [Google Scholar]
- 6.Ma Z, Jacobsen FE, Giedroc DP. 2009. Coordination chemistry of bacterial metal transport and sensing. Chem Rev 109:4644–4681. doi: 10.1021/cr900077w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Changela A, Chen K, Xue Y, Holschen J, Outten CE, O'Halloran TV, Mondragon A. 2003. Molecular basis of metal-ion selectivity and zeptomolar sensitivity by CueR. Science 301:1383–1387. doi: 10.1126/science.1085950. [DOI] [PubMed] [Google Scholar]
- 8.Chen PR, He C. 2008. Selective recognition of metal ions by metalloregulatory proteins. Curr Opin Chem Biol 12:214–221. doi: 10.1016/j.cbpa.2007.12.010. [DOI] [PubMed] [Google Scholar]
- 9.Stoyanov JV, Brown NL. 2003. The Escherichia coli copper-responsive copA promoter is activated by gold. J Biol Chem 278:1407–1410. doi: 10.1074/jbc.C200580200. [DOI] [PubMed] [Google Scholar]
- 10.Checa SK, Espariz M, Pérez Audero ME, Botta PE, Spinelli SV, Soncini FC. 2007. Bacterial sensing of and resistance to gold salts. Mol Microbiol 63:1307–1318. doi: 10.1111/j.1365-2958.2007.05590.x. [DOI] [PubMed] [Google Scholar]
- 11.Humbert MV, Rasia RM, Checa SK, Soncini FC. 2013. Protein signatures that promote operator selectivity among paralog MerR monovalent metal ion regulators. J Biol Chem 288:20510–20519. doi: 10.1074/jbc.M113.452797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ibañez MM, Cerminati S, Checa SK, Soncini FC. 2013. Dissecting the metal selectivity of MerR monovalent metal ion sensors in Salmonella. J Bacteriol 195:3084–3092. doi: 10.1128/JB.00153-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pérez Audero ME, Podoroska BM, Ibáñez MM, Cauerhff A, Checa SK, Soncini FC. 2010. Target transcription binding sites differentiate two groups of MerR-monovalent metal ion sensors. Mol Microbiol 78:853–865. doi: 10.1111/j.1365-2958.2010.07370.x. [DOI] [PubMed] [Google Scholar]
- 14.Checa SK, Soncini FC. 2011. Bacterial gold sensing and resistance. Biometals 24:419–427. doi: 10.1007/s10534-010-9393-2. [DOI] [PubMed] [Google Scholar]
- 15.Aiyar A, Xiang Y, Leis J. 1996. Site-directed mutagenesis using overlap extension PCR. Methods Mol Biol 57:177–191. [DOI] [PubMed] [Google Scholar]
- 16.Datsenko KA, Wanner BL. 2000. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc Natl Acad Sci U S A 97:6640–6645. doi: 10.1073/pnas.120163297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Davis RW, Bolstein D, Roth JR. 1980. Advanced bacterial genetics. Cold Spring Harbor Press, Cold Spring Harbor, NY. [Google Scholar]
- 18.Espariz M, Checa SK, Pérez Audero ME, Pontel LB, Soncini FC. 2007. Dissecting the Salmonella response to copper. Microbiology 153:2989–2997. doi: 10.1099/mic.0.2007/006536-0. [DOI] [PubMed] [Google Scholar]
- 19.Soncini FC, García Véscovi E, Groisman EA. 1995. Transcriptional autoregulation of the Salmonella typhimurium phoPQ operon. J Bacteriol 177:4364–4371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kumar S, Tamura K, Nei M. 2004. MEGA3: integrated software for molecular evolutionary genetics analysis and sequence alignment. Brief Bioinform 5:150–163. doi: 10.1093/bib/5.2.150. [DOI] [PubMed] [Google Scholar]
- 21.Pontel LB, Pérez Audero ME, Espariz M, Checa SK, Soncini FC. 2007. GolS controls the response to gold by the hierarchical induction of Salmonella-specific genes that include a CBA efflux-coding operon. Mol Microbiol 66:814–825. doi: 10.1111/j.1365-2958.2007.05963.x. [DOI] [PubMed] [Google Scholar]
- 22.Sharma R, Rensing C, Rosen BP, Mitra B. 2000. The ATP hydrolytic activity of purified ZntA, a Pb(II)/Cd(II)/Zn(II)-translocating ATPase from Escherichia coli. J Biol Chem 275:3873–3878. doi: 10.1074/jbc.275.6.3873. [DOI] [PubMed] [Google Scholar]
- 23.Guerra AJ, Giedroc DP. 2012. Metal site occupancy and allosteric switching in bacterial metal sensor proteins. Arch Biochem Biophys 519:210–222. doi: 10.1016/j.abb.2011.11.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Waldron KJ, Rutherford JC, Ford D, Robinson NJ. 2009. Metalloproteins and metal sensing. Nature 460:823–830. doi: 10.1038/nature08300. [DOI] [PubMed] [Google Scholar]
- 25.Rao L, Cui Q, Xu X. 2010. Electronic properties and desolvation penalties of metal ions plus protein electrostatics dictate the metal binding affinity and selectivity in the copper efflux regulator. J Am Chem Soc 132:18092–18102. doi: 10.1021/ja103742k. [DOI] [PubMed] [Google Scholar]
- 26.Hobman J, Julian D, Brown N. 2012. Cysteine coordination of Pb(II) is involved in the PbrR-dependent activation of the lead-resistance promoter, PpbrA, from Cupriavidus metallidurans CH34. BMC Microbiol 12:109. doi: 10.1186/1471-2180-12-109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Khan S, Brocklehurst KR, Jones GW, Morby AP. 2002. The functional analysis of directed amino-acid alterations in ZntR from Escherichia coli. Biochem Biophys Res Commun 299:438–445. doi: 10.1016/S0006-291X(02)02660-8. [DOI] [PubMed] [Google Scholar]
- 28.Shewchuk LM, Helmann JD, Ross W, Park SJ, Summers AO, Walsh CT. 1989. Transcriptional switching by the MerR protein: activation and repression mutants implicate distinct DNA and mercury(II) binding domains. Biochemistry 28:2340–2344. doi: 10.1021/bi00431a053. [DOI] [PubMed] [Google Scholar]
- 29.Parr RG, Yang W. 1989. Density-functional theory of atoms and molecules, vol 16 Oxford University Press, New York, NY. [Google Scholar]