SUMMARY
Histone mRNAs are rapidly degraded when DNA replication is inhibited during S-phase with degradation initiating with oligouridylation of the stemloop at the 3′ end. We developed a customized RNA-Seq strategy to identify the 3′ termini of degradation intermediates of histone mRNAs. Using this strategy, we identified two types of oligouridylated degradation intermediates: RNAs ending at different sites of the 3′ side of the stemloop that resulted from initial degradation by 3′hExo and intermediates near the stop codon and within the coding region. Sequencing of polyribosomal histone mRNAs revealed that degradation initiates and proceeds 3′ to 5′ on translating mRNA and many intermediates are capped. Knockdown of the exosome-associated exonuclease Pml/Scl-100, but not the Dis3L2 exonuclease, slows histone mRNA degradation, consistent with 3′ to 5′ degradation by the exosome containing PM/Scl-100. Knockdown of No-go decay factors also slowed histone mRNA degradation, suggesting a role in removing ribosomes from partially degraded mRNAs.
INTRODUCTION
The half-life of an mRNA is an important component in determining its steady-state levels, and regulation of degradation is an efficient way to rapidly down-regulate those levels. mRNAs can be potentially degraded 5′ to 3′ after decapping, 3′ to 5′, or by both mechanisms simultaneously. In mammalian cells, the precise intermediates that arise during degradation of a specific mRNA are not known. Degradation of most mRNAs in mammalian cells is initiated by deadenylation resulting in an oligo(A) tail that binds Lsm1-7 (Garneau et al., 2007); the relative importance of the 5′ to 3′ and 3′ to 5′ pathways is not known.
Replication-dependent histone mRNAs are the only known metazoan mRNAs that are not polyadenylated, ending instead in a conserved stemloop (SL) that plays a critical role in histone mRNA regulation (Marzluff et al., 2008). The stemloop binding protein (SLBP) binds the 5′ side of the stem (Tan et al., 2013) and is required for all steps in histone mRNA metabolism. The half-life of histone mRNA is tightly regulated to balance histone and DNA synthesis, and inhibition of DNA replication during S-phase reduces the histone mRNA half-life to ~10–15 min (Graves and Marzluff, 1984; Harris et al., 1991).
The coordinate expression of histone mRNAs coupled with the ability to induce histone mRNA degradation provides an opportunity to study the dynamics of degradation. Recently, we showed that histone mRNA degradation is initiated by oligouridylation of the 3′ end (Mullen and Marzluff, 2008; Su et al., 2013), resulting in a binding site for Lsm1-7 (Lyons et al., 2014). In vivo knockdown of the 5′ to 3′ exonuclease Xrn1, the decapping enzyme Dcp2 or the 3′ to ′5 exosome complex all partially stabilize histone mRNA, with the exosome knockdown having a larger stabilizing effect (Mullen and Marzluff, 2008), consistent with a major role for 3′ to 5′ degradation.
Ross and coworkers previously suggested that histone mRNA is degraded 3′ to 5′ after inhibition of DNA replication with initial intermediates resulting from partial degradation of the SL by a polyribosome associated 3′ to 5′ exonuclease (Ross et al., 1986; Ross et al., 1987; Caruccio and Ross, 1994). This exonuclease is clearly 3′hExo (Eri-1), a protein that specifically binds the histone SL. 3′hExo and SLBP form a complex on the 3′ end of histone mRNA (Yang et al., 2006; Tan et al., 2013), and 3′hExo was recently shown to be essential for the initial steps of degradation of histone mRNA (Hoefig et al., 2013).
Here, we report the development of a high-throughput sequencing strategy specifically targeting the 3′ terminus of histone mRNAs that allows us to detect and analyze the full range of degradation intermediates, including non-templated oligouridylated species. We find that initial oligouridylation occurs while the histone mRNA is on polyribosomes and degradation initially proceeds 3′ to 5′ without decapping while the mRNA is associated with ribosomes. Components of the No-go decay pathway likely play a role in removing ribosomes from stalled degradation complexes.
RESULTS
Histone mRNAs end in a conserved SL formed by an endonucleolytic cleavage event 5 nts 3′ of the SL (Scharl and Steitz, 1994). Following cleavage, the mRNA is trimmed by 3′hExo (Hoefig et al., 2013), resulting in a mature mRNA ending in a SL and a 2–3-nt tail (Fig. 1A). Mammalian histone mRNAs have a relatively short and tightly regulated half-life. When HeLa cells in S-phase are treated with inhibitors of DNA replication, histone mRNA is rapidly degraded (Mullen and Marzluff, 2008), providing a system for studying its degradation pathway.
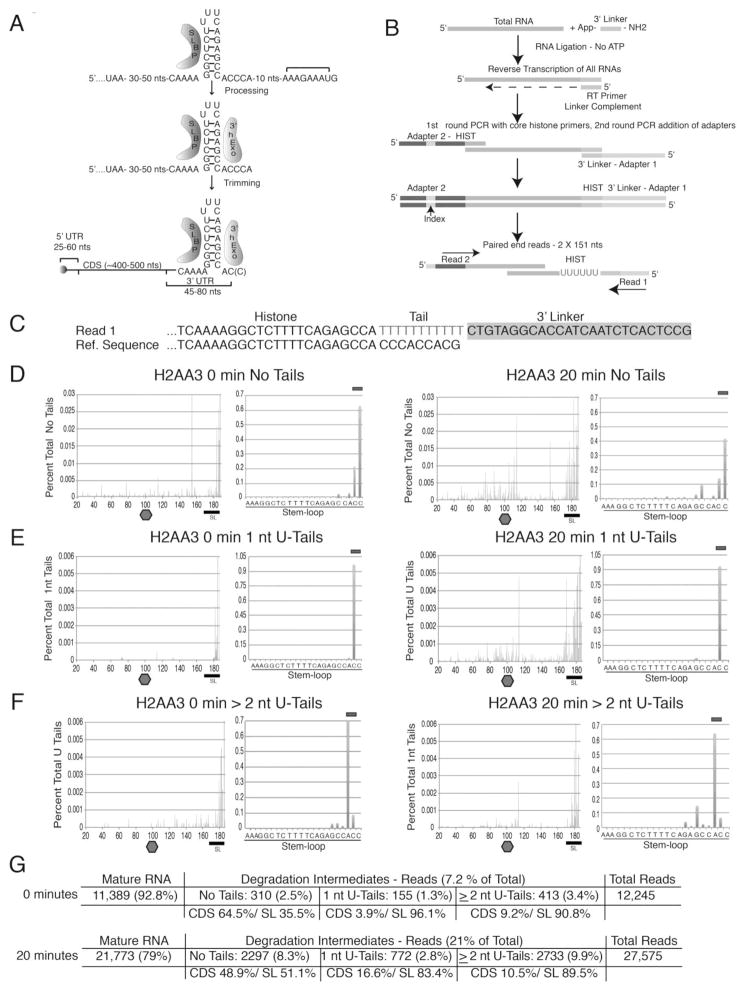
Figure 1. Strategy to detect histone mRNA degradation intermediates.
(A) Histone pre-mRNA processing steps resulting in the mature transcript bound by SLBP and 3′hExo.
(B) Diagram of the high-throughput sequencing strategy for 3′-end analysis of histone mRNA degradation intermediates (see the Supplemental Experimental Procedures). RNA ligated to a preadenylated linker is reverse transcribed with the linker complement (LC), followed by limited PCR with a histone-specfiic primer. A second round of limited PCR adds Illumina specific adaptors.
(C) Alignment of a sequence of a U-tailed histone mRNA ligated to the linker (top) with a histone reference sequence (bottom).
(D–F) The distribution of the 3′ ends of reads from the HIST2H2AA3 mRNA before (left) or after (right) 20 min HU treatment. The 3′ ends of the untailed RNAs (D), RNAs with 1-nt U-tail (E), and RNAs with U tails of ≥ 2 nt (F) were plotted. The stop codon was set at position 100, and the top bar indicates the mature mRNAs ending in AC or ACC (D), ACU (E) or AUU or ACUU (F) after the stem. On the right are the tails in the SL on an expanded scale.
(G) Table summarizing the number of mature and tailed RNAs.
We initially detected histone mRNA degradation intermediates using a circular RT-PCR assay (Mullen and Marzluff, 2008). Because these intermediates were isolated by circularization, they must have been decapped. We identified other putative oligouridylated degradation intermediates near the 3′ end (Mullen and Marzluff, 2008) and throughout the mRNA using d(A) priming and ligation-mediated RT-PCR (Supp. Fig. 1). However, the low number of isolated intermediates did not permit full analysis of this degradation pathway.
A high-throughput sequencing strategy for histone mRNA degradation intermediates
To determine the spectrum of 3′ ends of degradation intermediates, we used an approach similar to that of Newman and Hammond for pre-miRNAs (Newman et al., 2011; Fig. 1B). After ligation of a preadenylated linker, we primed cDNA with the linker complement to define the 3′ ends of all RNA molecules. Following reverse transcription, libraries were prepared using two rounds of limited PCR. The first round of PCR used gene-specific primers to the 15–20 distinct H2a and H2b replication-dependent histone mRNAs (Marzluff et al., 2002). Because the ORFs in each histone class are highly conserved, these primers simultaneously target most of the H2a and H2b genes. Illumina adapter sequences and bar codes were added in a subsequent, limited PCR reaction, and the libraries were sequenced using an Illumina MiSeq platform to detect mature mRNAs and degradation intermediates with non-templated 3′-end additions.
This method allows the detection of non-templated tails as short as 1 nt regardless of their composition. It also detects truncated histone mRNAs that are not tailed and are potential degradation intermediates. This non-biased approach allowed us to analyze the 3′ end of all histone mRNA molecules. Control experiments demonstrated that this method did not discriminate against RNAs ending in multiple U’s or full-length RNAs (Supp. Fig. 1).
Locating 3′ Transcript Ends and Detecting Non-Templated 3′ Additions
Our sequencing strategy produces paired-end reads: the first read contains the 3′ end of a histone transcript (reverse complement) and the second starts in the ORF (Fig. 1B). Because we are interested primarily in locating transcript 3′ ends, the second read serves to provide further constraints to aid in aligning the first read. To analyze these data, we developed a bioinformatics pipeline to determine the position and sequence of the 3′ ends of histone mRNAs, including any non-templated nucleotide additions (Fig. 1B; see supp. methods).
We determined the location and composition of non-templated additions by aligning the paired-end reads to the human genome (hg19) using the STAR aligner (Dobin et al., 2013) and locating the portions of the reads that do not align to the reference sequence. We then used a global alignment algorithm to align these non-templated bases to the preadenylated linker sequence. The unmatched portion of the read that does not align to the linker or the genome contains non-templated, 3′ transcript additions and indicates the precise 3′ end of the transcript (Fig. 1C). We analyzed the distribution of non-templated additions by plotting the percentages of histone mRNAs ending at each nucleotide (Fig. 1E) and generated similar plots for potential degradation intermediates without 3′ additions (Fig. 1D; see sup. methods for a detailed description).
Multiple sites of oligouridylation are found on histone mRNAs undergoing degradation
To test our custom RNA-seq and bioinformatics strategies, we treated S-phase HeLa cells with hydroxyurea (HU) to activate histone mRNA degradation and sequenced RNA from untreated and 20-min HU-treated cells. The results for histone HIST2H2AA3 (H2AA3) mRNA, an abundant histone H2a mRNA, are shown in Figure 1D–F. We divided our data into three categories: mRNA molecules with no tails, 1-nt or ≥2-nt U-tails. For each category, we plotted the reads as a percent of the reads in that category (y-axis) versus the position of the last encoded nt within the mRNA (x-axis). Due to the large number of reads in the SL, these are shown on an expanded scale to the right.
The mature-length H2AA3 mRNA normally ends with ACC or AC due to trimming by 3′hExo (Fig. 1D). Surprisingly, nearly all of the RNAs with a 1-nt U-tail (99%) ended with an ACU after the SL (Fig. 1E), and the majority of the RNAs with 2-nt U-tails ended with AUU or ACUU after the SL (Fig. 1F). Identical results were obtained for all other tested histone mRNAs. We consider these RNAs to be functional, full-length histone mRNAs that had 1–2 nts trimmed off followed by addition of 1–2 non-templated uridines that restored their full length. Sequence reads corresponding to these ‘mature’ RNAs are indicated by a bar over the final 3 nts in the expanded SL degradation profile. In the untreated (0 min) sample, 92.8% of the transcripts were mature mRNAs.
In addition to mature H2AA3 mRNA, shorter molecules with no tails or U-tails, were present. At 0 min, these totaled 7.2% of all reads (Fig. 1G), and 20 min after HU treatment, when histone mRNA levels had been reduced by 15% as judged by Northern blotting (not shown), these reads increased ~3-fold, consistent with them being degradation intermediates. The tails ranged from 2–15 nts and were composed primarily of uridines, although A and C were occasionally present (see Fig. 2B). A large number of tails were added to the final 4 nts of the 3′ side of the stem; the others were dispersed throughout the ORF and the 5′ portion of the 3′ UTR near the stop codon. This same distribution was observed for the no-tail category (Fig. 1D). A small percentage of 1-U tails that were not full-length mRNAs were found largely within the ORF (Fig. 1E).
Figure 2. High-throughput sequencing of histone mRNA degradation intermediates.
(A) S-phase HeLa cells were treated with HU for the indicated times (min), and whole cell RNA was analyzed by Northern blotting for histone H2a and 7SK RNA as a loading control.
(B) The distribution of reads at the stemloop for the HIST2H2AA3 gene is shown. Sequences of U-tails ≥ 6 nts at the indicated position (of 1042 tailed mRNAs) are shown.
(C–E) Libraries were prepared from the 0- and 20-min time points (panel A) as in Fig. 1B. The distribution of reads containing ≥2 nt U-tails are shown for the HIST2H2AA3 (C); HIST1H2AC (D) and HIST1H2BC genes (E), with the stop codon at 100 for H2a genes or 200 for the H2BC gene. Note the gap with few reads between the stemloop and 15 nts 3′ of the stop codon 20 min after HU treatment. On the right is an expanded scale for reads at the stemloop, and below is a summary of the percentage of reads in each class. Additional data on the reads with no tails and 1 nt tails are in Supp. Fig. 2.
(F) Total RNA from S-phase cells treated with HU for 20 min was incubated with an anti-m7G antibody. The bound and unbound RNA was recovered and analyzed by semi-quantitative RT-PCR for histone H2AA3, GAPDH or 7SK mRNA.
(G–H). The capped mRNA from panel F was sequenced, and the distribution of no tails (left) and ≥2 nt U-tailed reads (bottom) from HIST2H2AA3 mRNA are shown.
I. Quantification of histone mRNA transcripts from panels C–E.
J. Quantification of histone mRNA transcripts from panels G and H
Degradation does not proceed uniformly through the histone mRNA
We treated S-phase HeLa cells with HU for 1 hr and sequenced histone mRNAs from untreated and 20-min HU-treated cells when ~50% of the histone mRNA had been degraded (Fig. 2A). We show analyses of three histone mRNAs, HIST2H2AA3 (H2AA3), HIST1H2AC (H2AC) and HIST1H2BC (H2BC), in Fig. 2C–E and Supp. Fig. 2. The distribution of transcripts in the untreated and HU-treated cells that have ≥2-nt U-tails are shown in Fig. 2C–E, and the transcripts with no tails and 1-nt U-tails in Fig. 2I and Supp. Fig. 2C. For all three histone mRNAs, the pattern of oligouridylation was similar to Fig. 1. In untreated cells, 84–92% of the mRNAs were mature, and a large reduction to 23–48% took place after HU treatment, consistent with more extensive degradation compared to Fig. 1.
All three histone mRNAs show an accumulation of tailed reads 2–4 nts into the distal side of the SL (Fig. 2C–E; expanded graphs to the right), suggesting these represent oligouridylated intermediates accumulating at a rate-limiting step in degradation. The distribution of tails in the SL and the composition of tails that were ≥6 nts at the G 3-nts into the SL is shown in Fig. 2B. The majority of the tails were uridine homopolymers, but a C or A was occasionally present. The loop contains 4 consecutive U residues, making it impossible to assign tail locations in this region. The loop and the 5′ side of the stem contained very few tailed intermediates, suggesting that SLBP might still be bound to the uridylated RNA, blocking further degradation.
Oligo(U) tails of all three genes were dispersed throughout the ORF and present up to 15 nts after the stop codon; few tails were located beyond this point until the 3′ side of the SL (Fig 2C–E). We observed the same pattern in the non-tailed degradation intermediates (Fig. 1C; Sup. Fig. 2). The distance between the stop codon and start of the SL differs among these mRNAs: H2AA3, 65 nts; H2AC, 55 nts; and H2BC, 34 nts. The length of the gap correlates with the distance between the stop codon and SL, with accumulation of intermediates starting 15 nts 3′ of the stop codon. Statistical analysis with the Wilcoxon signed-rank test indicated the gap was significant (p< 3 × 10−7 (H2AA3); p<10−4 (H2BC); p<3 × 10−6 (H2AC). We propose this gap results from rapid degradation of the mRNA following removal of the SLBP/3′hExo complex from the 3′ end of histone mRNA, which proceeds until the exonuclease reaches a ribosome at the termination codon, causing it to stall. The ribosome must then be removed and the RNA reprimed by uridylation for further degradation of the mRNA.
Many histone mRNA degradation intermediates are capped
We sequenced capped histone mRNAs isolated by immunoprecipitation with anti-m7G with an anti-m7G antibody (Munns et al., 1977) from cells treated with HU for 20 min. Most of the histone mRNA and GAPDH mRNA were precipitated, as determined by RT-PCR (Fig. 2F). The capped histone mRNAs were oligouridylated on the 3′ side of the SL and within the ORF, similar to the distribution found in the total mRNA from HU-treated cells (Fig. 2H). A similar distribution of non-tailed intermediates in the ORF that were capped and truncated was also observed (Fig. 2G, J). Thus, a substantial number of histone mRNA molecules are degraded 3′ to 5′ without decapping for at least the first 150 nts from the 3′ end.
Oligo(U) tails are added to histone mRNAs on polyribosomes
Since translation of histone mRNA is necessary for its degradation (Kaygun and Marzluff, 2005; Graves et al., 1987), we tested whether the initial stages of histone mRNA oligouridylation and degradation occurred on polyribosomes. We isolated polyribosomes from Jurkat cells after HU treatment for 20 min (Fig. 3A). Pactamycin, a translation initiation inhibitor that prevents 60S joining and blocks a step immediately after synthesis of the first peptide bond (Kappen and Goldberg, 1976), was added for 5 min, 15 min after the treatment with HU. As a control, cycloheximide, which blocks elongation and maintains polyribosome association, was added for 5 min to a parallel culture 15 min after HU treatment (Fig. 3A). Because inhibiting protein synthesis stabilizes histone mRNA (Stimac et al., 1984; Graves and Marzluff, 1984), we restricted pactamycin treatment to 5 min, the minimal time that allowed many polyribosomes to run off and for mRNA to accumulate in 80S initiation complexes. This allowed us to 1) identify histone mRNAs that were being translated and 2) identify degradation intermediates that were still present after inhibiting protein synthesis.
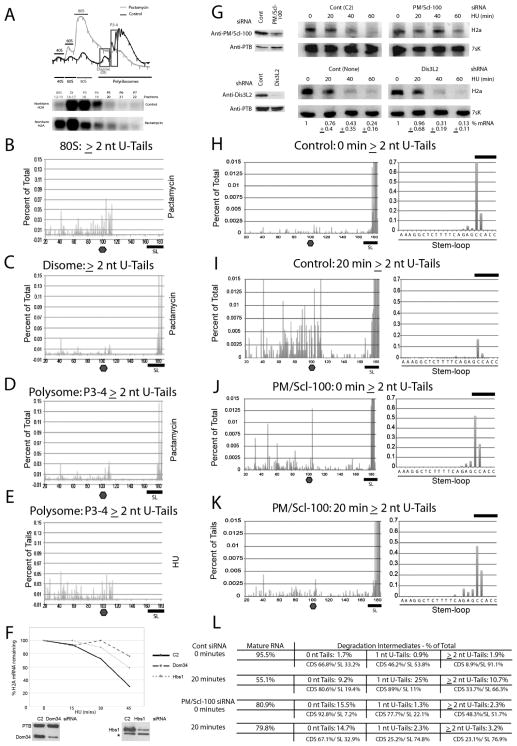
Figure 3. Histone mRNAs are degraded 3′ to 5′ on polyribosomes.
(A) Exponentially growing Jurkat cells were treated with HU for 20 min followed by a 5-min treatment with cycloheximide (black) or pactamycin (gray). Fractions were analyzed by Northern blotting, and libraries were prepared from each fraction and sequenced. Analysis of the complete gradient is shown in Supp. Fig. 3A.
(B–E) Distribution of reads for HIST2H2AA3 RNA with ≥2 nt U-tails (top) from the monosome (B), disome (C) or fractions with 3–5 ribosomes (D). The control fraction with 3–5 ribosomes from cells treated with HU-CHX is shown in panel E.
(F) HeLa cells were treated with siRNAs against Dom34 or Hsb1 for 48 hrs. Total cell protein was analyzed by western blotting for Dom 34 and PTB (right) and Hbs1 (left). The band with an * is a cross-reacting band. Parallel cultures were treated with HU for the indicated times (min) and analyzed for histone H2a and 7SK RNA by Northern blotting (Supp. Fig. 3G). Results were quantified on a PhosphorImager.
(G) Cells were treated with control or Pm/Scl100-targeting siRNAs or a shRNA targeting Dis3L2, and extracts were analyzed by Western blotting (top). Total RNA was prepared from cells treated with HU at the indicated times (bottom) and analyzed as in panel 2A.
(H–K) RNA was sequenced from the 0- (H, J) and 20-min (I, K) samples of the control (I. J) and PM/Scl-100 siRNA-treated cells (K, L) and sequenced. The percent of reads with ≥2 nt U-tails at each position is plotted. On the right is an expansion of the reads in the stemloop.
L. Quantification of the histone transcripts in panels H–K.
Northern analysis showed that most of the histone H2a mRNA in the control gradient (Fig. 3A, and sup. Fig. 3A) was in fractions 14–19, which corresponded to 2–4 ribosomes bound to the histone mRNA, consistent with the 375-nt ORF. There was a shift of histone mRNA to slower-sedimenting fractions after pactamycin treatment, and a portion of the histone mRNA shifted to 80S complexes. Based on the Northern and polyribosome profile, we prepared libraries from the pactamycin and control gradients by pooling fractions from three regions of the gradient: monosome (80S), disome (DI) and 3–4 ribosomes (P3 and P4; Fig. 3A). After pactamycin treatment, histone mRNAs containing ≥2-nt U-tails were distributed throughout the ORF in the 80S fraction (Fig. 3B), indicating that these mRNAs had been polyribosome associated prior to treatment. In contrast, many of the histone mRNAs associated with the disome and small polyribosomes had U-tails that were added at the 3′ end of the SL (Fig. 3C–D), suggesting that they were modified during the 5-min pactamycin treatment (Fig. 3C). In the control gradient, uridylated histone mRNA degradation intermediates were present in the fractions that contained small polyribosomes (Fig. 3E), and many histone mRNAs were uridylated within the ORF, suggesting that degradation that had initiated before CHX treatment continued on the CHX-stalled polyribosomes.
These results suggest that histone mRNA degradation intermediates still have associated ribosomes after removal of the 3′ UTR. These ribosomes will be stalled on the mRNA and not readily removed. A possible mechanism to remove the ribosomes is No-go decay (Passos et al., 2009), which, in yeast, requires two proteins, Dom34p and Hbs1p. Mammalian homologues with a similar function have been described (Saito et al., 2013). Knocking down either Dom34 or Hbs1 by RNAi slowed histone mRNA degradation (Fig. 3F, supp. Fig. 3E, F), consistent with an important role in removing ribosomes from partially degraded mRNA.
Knockdown of the exosome alters the distribution of degradation intermediates
We previously showed that knockdown of Rrp41, a core component of the exosome, or the distributive 3′ to 5′ exonuclease PM/Scl-100 (Rrp6) slowed histone mRNA degradation (Mullen and Marzluff, 2008). In addition to PM/Scl-100, mammalian cells express three processive 3′ to 5′ exonucleases: Dis3 (Rrp44) and Dis3L1 are associated with the nuclear and cytoplasmic exosome, respectively (Tomecki et al., 2010), while Dis3L2 functions in an exosome-independent manner in the cytoplasm. Dis3L2 has recently been implicated in the degradation of uridylated RNAs (Malecki et al., 2013; Chang et al., 2013). In contrast to PM/Scl-100 (Fig. 3G, top), knockdown of Dis3L2 had no effect on histone mRNA degradation (Fig. 3G, bottom).
We sequenced the histone mRNAs from untreated and HU-treated PM/Scl-100 knockdown cells and compared them to cells transfected with control siRNA (Fig. 3H–K). Control siRNA-treated cells showed similar profiles to untreated cells (Fig. 3H–I; Figs. 1,2). In contrast, prior to HU treatment, PM/Scl-100 knockdown cells contained a larger percentage of tailed and untailed degradation intermediates (20%) in the ORF and stemloop (Fig. 3J, L). The accumulation of degradation intermediates before HU treatment likely resulted from impaired histone mRNA degradation that normally occurs at a low rate in exponentially growing cells.
After treating PM/Scl-100 knockdown cells with HU for 20 min, we observed a large increase in the percentage of SL-associated, ≥2-nt U-tailed reads (76.9%), indicating that the initial step in histone mRNA degradation was activated (Fig. 3K). Fewer intermediates within the ORF and around the stop codon were observed, consistent with the slow conversion of partially degraded and uridylated SL intermediates to the second step in degradation that requires the PM/Scl-100-associated exosome.
The gap of intermediates between the stop codon and SL that was observed in our control experiments was reduced in PM/Scl-100-knockdown cells (Fig. 3K). This finding is consistent with the distributive PM/Scl-100 exosome degrading histone mRNA after removal of the SL and supports the idea that the gap results from rapid progression of the exosome followed by stalling near the stop codon due to encountering a terminating ribosome.
We conclude that many histone mRNAs are degraded 3′ to 5′ in two phases: degradation into the SL by the 3′hExo, then degradation by the PM/Scl-100 exosome. If the nuclease stalls during either phase of degradation, further degradation is primed by reuridylation.
Exogenous capped, non-polyadenylated RNAs are degraded by a similar pathway
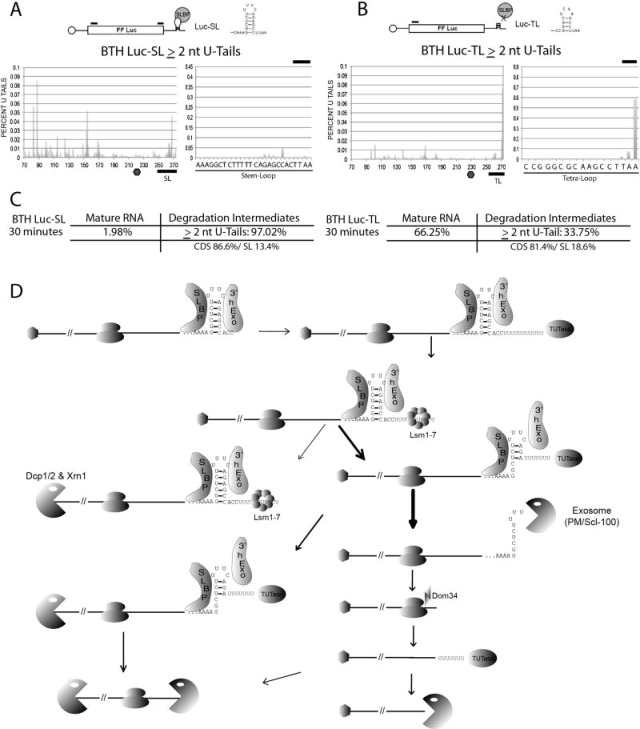
A chimeric, capped luciferase mRNA ending in a canonical histone SL (Luc-SL) is translated in HeLa cells after nucleoporation and undergoes a reduction in half-life upon HU treatment similar to that of endogenous histone mRNA (Su et al., 2013). A similar luciferase mRNA that ended in a tetraloop sequence (Luc-TL) is not translated. We sequenced libraries of luciferase-specific RNAs from cells that were nucleoporated with each mRNA. Both Sl and TL chimeric mRNAs had a substantial fraction of intermediates that were uridylated (Fig. 4A, B) located throughout the mRNA. Similar intermediates were found regardless of whether the RNA had an “uncleavable” BTH (Fig. 4) or conventional (ARCA) cap (sup. Fig. 4) or if it was actively translated. Thus, a pathway exists in mammalian cells that degrades capped, non-polyadenylated RNAs by oligouridylation followed by 3′ to 5′ exonuclease digestion.
Figure 4. Oligouridylation occurs on transfected, capped, non-polyadenylated RNAs.
(A–B) BTH-capped luciferase mRNA ending in a histone stemloop (LUC-SL, panel A) or tetraloop (LUC-TL, panel B) was nucleoporated into synchronized HeLa cells that were then treated with HU, and RNA was isolated 30 min after HU treatment. These are the same RNA samples that were previously analyzed (Su et al., 2013). Results with a set of ARCA capped mRNAs is shown in Supp. Fig. 4.
(C) Possible pathways of histone mRNA degradation initiated at the 3′ end of histone mRNA.
DISCUSSION
Regulation of mRNA half-life is critical for determining the steady-state levels of a transcript and allowing rapid changes in mRNA levels in response to cellular changes. The mechanism of poly(A) mRNA degradation in yeast (S. cerevisiae) is well understood (Parker and Song, 2004). The major pathway for degradation of most mRNAs in rapidly growing yeast is 5′ to 3′ with deadenylation followed by binding of Lsm1-7, which leads to decapping and subsequent degradation 5′ to 3′ by Xrn1. This pathway has been confirmed by genetic experiments and by insertion of secondary structures into mRNAs that arrest either Xrn1 or the exosome (Decker and Parker, 1993; Caponigro and Parker, 1996). A minor pathway in yeast is degradation 3′ to 5′ by the exosome (Anderson and Parker, 1998), and this pathway is dependent on deadenylation (Mitchell and Tollervey, 2003; Anderson and Parker, 1998). In contrast the relative importance of the 3′ to 5′ and 5′ to 3′ pathways for specific mRNAs is not known in mammals.
Pathway of histone mRNA degradation: requirement for two 3′ to 5′ exonucleases
We have elucidated a pathway of histone mRNA degradation that is activated when DNA synthesis is inhibited (Fig. 4C). The initial step is oligouridylation of the 3′ end of histone mRNA (Mullen and Marzluff, 2008; Su et al., 2013). This initial degradation intermediate may be decapped and degraded 5′ to 3′ by Xrn1, degraded 3′ to 5′ by the exosome or degraded from both ends by both enzymes (Mullen and Marzluff, 2008). 3′ to 5′ degradation is initiated by 3′hExo, which is essential for histone mRNA degradation (Hoefig et al., 2013) and forms a complex with SLBP on the 3′ end of the histone mRNA (Tan et al., 2013; Yang et al., 2006). 3′hExo degrades the mRNA into the SL, resulting in a degradation intermediate with 2–4 nts removed from the 3′ side of the stem that is reuridylated before further degradation (Fig 4C). We postulate that this intermediate still has SLBP bound to the 3′ end, blocking further degradation. After removal of SLBP, histone mRNAs are degraded by the exosome containing PM/Scl-100. When 3′ to 5′ degradation stalls, likely by the exosome contacting a ribosome near the termination codon or secondary structure in the mRNA, histone mRNA is reuridylated to resume degradation. Knockdown of the mammalian homologues of the Hbs1-Dom34 complex slows histone mRNA decay, suggesting this complex may remove stalled ribosomes to allow degradation to proceed (Fig. 3F).
Is decapping involved in histone mRNA degradation?
The previous degradation intermediates we isolated by circular RT-PCR were not capped (Mullen and Marzluff, 2008). In addition, when the Luc-SL RNA was introduced into HeLa cells by nucleoporation, 5′ to 3′ degradation clearly occurred based on stabilization by the uncleavable cap, although HU-treatment resulted in an increased rate of the 3′ to 5′ pathway and accelerated degradation of the RNA with an uncleavable cap (Su et al., 2013). Our results indicate that decapping does not occur efficiently after oligouridylation. The reason for this failure is not clear since Lsm1-7 can bind to the oligo(U) tail (Lyons et al., 2014) and is found in a complex containing SLBP on the 3′ end of histone mRNA after initiation of histone mRNA degradation (Mullen and Marzluff, 2008). It is consistent with the observation that in 3′hExo knockdown cells, histone mRNAs are not degraded although oligouridylated mRNAs accumulate (Hoefig et al., 2013). Binding of Lsm1-7 to oligo(U) tails has been reported to stimulate decapping in vitro (Song and Kiledjian, 2007); some feature of this reaction must be relatively inactive in vivo. It is possible that binding of Lsm1-7 to oligo(U) differs in its ability to activate decapping than when binding to oligo(A) after deadenylation.
Note that during 3′ to 5′ degradation, multiple opportunities exist for decapping to occur after stalling of the 3′ to 5′ exonuclease (Fig. 4C), and it is possible that most of the histone mRNAs are ultimately decapped prior to complete 3′ to 5′ degradation. Decapped RNAs may be degraded rapidly; therefore, they may not be a large fraction of the degradation intermediates. The critical role of Lsm1-7 in histone mRNA degradation is likely not simply activation of decapping.
How common is uridylation in triggering RNA degradation in mammals?
Oligouridylation has been previously implicated in the degradation of miRNA and pre-miRNA in plants (Zhao et al., 2012; Li et al., 2005) and animals (Heo et al., 2009; Thornton et al., 2012). In addition, the 5′ product of siRNA-directed mRNA cleavage has been reported to be oligouridylated (Shen and Goodman, 2004). The detailed pathway of degradation of mRNA following siRNA-mediated cleavage is not known but could occur by a similar mechanism that occurs for histone mRNA degradation intermediates. A hybrid GFP mRNA ending in the 3′ end of MALAT1 is oligouridylated, apparently as part of an RNA degradation pathway (Wilusz et al., 2012). Cells transfected with a luciferase RNA containing a tetraloop are degraded at the same rate regardless of whether they are decapped (Su et al., 2013), and we found a substantial number of oligouridylated intermediates in both samples. These results raise the possibility that oligouridylation plays a role in degradation of a large number of RNAs in mammalian cells. The high-throughput sequencing approaches described here can likely be adapted to the study of degradation intermediates of any mRNA.
EXPERIMENTAL PROCEDURES
Preparation and analysis of RNA
Total cell RNA was prepared from S-phase HeLa cells and analyzed by Northern blotting for histone mRNA as described previously (Mullen and Marzluff, 2008). Libraries containing the 3′ ends of histone mRNAs were prepared as described in Supplemental Procedures and sequenced on a MiSeq (Illumina). Cells were treated with PM/Scl-100 siRNAs as previously described (Mullen and Marzluff, 2008), or stable knockdowns were obtained with a lentivirus-encoded shRNA for Dis3L2.
Polyribosome analysis
Exponentially growing Jurkat cells in suspension culture were treated with HU for 15 min and then with pactamycin or cycloheximide for 5 minutes. Cell lysates were prepared and polyribosomes resolved by sucrose gradient centrifugation. RNA was prepared from each fraction and analyzed by Northern blotting. Libraries were prepared from the fractions.
Details of all procedures, including the bioinformatics analysis, are in Supplemental Methods. All graphs were made in Excel by plotting the ratio of reads at each position to the total reads of that sample (Y-axis) versus the position on the mRNA (X-axis).
Supplementary Material
Acknowledgments
Supported by NIH grants GM29832 to W.F.M; NIH grant HG06272 and NSF grant ABI/EF0850237 to J.F.P. and NIH grant GM20818 to R.E.R. M.K.S. and S.M. were supported by NIH grants F32GM87059 and F32GM80007, respectively. J.W. was supported by NSF Graduate Research Fellowship DGE-1144081. W.S. was supported by an Ike Muslow postdoctoral research fellowship from the LSUHSC-S Research Council. We thank Drs. Scott Hammond, Martin Newman, Will Jeck, Greg Matera, Gabrielle Phillips and Chris Nicchitta for helpful discussions; Hemant Kelkar, Piotr Mieczkowski, Kevin Weeks, Dave Mauger for assistance with HTS, and Dr. Rebecca Hartley for comments on the manuscript.
Footnotes
ACCESSION NUMBER
The GEO accession number for the data reported in this paper is GSE54922.
References
- Anderson JS, Parker R. The 3′ to 5′ degradation of yeast mRNAs is a general mechanism for mRNA turnover that requires the SKI2 DEVH box protein and 3′ to 5′ exonucleases of the exosome complex. EMBO J. 1998;17:1497–1506. doi: 10.1093/emboj/17.5.1497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caponigro G, Parker R. Mechanisms and control of mRNA turnover in Saccharomyces cerevisiae. Microbiol Rev. 1996;60:233–249. doi: 10.1128/mr.60.1.233-249.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caruccio N, Ross J. Purification of a human polyribosome-associated 3′ to 5′ exonuclease. J Biol Chem. 1994;269:31814–31821. [PubMed] [Google Scholar]
- Chang HM, Triboulet R, Thornton JE, Gregory RI. A role for the Perlman syndrome exonuclease Dis3l2 in the Lin28-let-7 pathway. Nature. 2013;497:244–248. doi: 10.1038/nature12119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Decker CJ, Parker R. A turnover pathway for both stable and unstable mRNAs in yeast: Evidence for a requirement for deadenylation. Genes Dev. 1993;7:1632–1643. doi: 10.1101/gad.7.8.1632. [DOI] [PubMed] [Google Scholar]
- Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, Batut P, Chaisson M, Gingeras TR. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29:15–21. doi: 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garneau NL, Wilusz J, Wilusz CJ. The highways and byways of mRNA decay. Nat Rev Mol Cell Biol. 2007;8:113–126. doi: 10.1038/nrm2104. [DOI] [PubMed] [Google Scholar]
- Graves RA, Marzluff WF. Rapid, reversible alterations in histone gene transcription and histone mRNA levels in mouse myeloma cells. Mol Cell Biol. 1984;4:351–357. doi: 10.1128/mcb.4.2.351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graves RA, Pandey NB, Chodchoy N, Marzluff WF. Translation is required for regulation of histone mRNA degradation. Cell. 1987;48:615–626. doi: 10.1016/0092-8674(87)90240-6. [DOI] [PubMed] [Google Scholar]
- Harris ME, Böhni R, Schneiderman MH, Ramamurthy L, Schümperli D, Marzluff WF. Regulation of histone mRNA in the unperturbed cell cycle: Evidence suggesting control at two posttranscriptional steps. Mol Cell Biol. 1991;11:2416–2424. doi: 10.1128/mcb.11.5.2416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heo I, Joo C, Kim YK, Ha M, Yoon MJ, Cho J, Yeom KH, Han J, Kim VN. TUT4 in concert with Lin28 suppresses microRNA biogenesis through pre-microRNA uridylation. Cell. 2009;138:696–708. doi: 10.1016/j.cell.2009.08.002. [DOI] [PubMed] [Google Scholar]
- Hoefig KP, Rath N, Heinz GA, Wolf C, Dameris J, Schepers A, Kremmer E, Ansel KM, Heissmeyer V. Eri1 degrades the stem-loop of oligouridylated histone mRNAs to induce replication-dependent decay. Nat Struct Mol Biol. 2013;20:73–81. doi: 10.1038/nsmb.2450. [DOI] [PubMed] [Google Scholar]
- Kappen LS, Goldberg IH. Analysis of the two steps in polypeptide chain initiation inhibited by pactamycin. Biochemistry. 1976;15:811–818. doi: 10.1021/bi00649a013. [DOI] [PubMed] [Google Scholar]
- Kaygun H, Marzluff WF. Translation termination is involved in histone mRNA degradation when DNA replication is inhibited. Mol Cell Biol. 2005;25:6879–6888. doi: 10.1128/MCB.25.16.6879-6888.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Yang Z, Yu B, Liu J, Chen X. Methylation protects miRNAs and siRNAs from a 3′-end uridylation activity in Arabidopsis. Curr Biol. 2005;15:1501–1507. doi: 10.1016/j.cub.2005.07.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyons SM, Ricciardi A, Guo AY, Kambach C, Marzluff WF. The C-terminal tail of Lsm4 interacts directly with the 3′ end of the histone mRNP and is required for efficient histone mRNA degradation. RNA. 2014;20:88–102. doi: 10.1261/rna.042531.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malecki M, Viegas SC, Carneiro T, Golik P, Dressaire C, Ferreira MG, Arraiano CM. The exoribonuclease Dis3L2 defines a novel eukaryotic RNA degradation pathway. EMBO J. 2013;32:1842–1854. doi: 10.1038/emboj.2013.63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marzluff WF, Gongidi P, Woods KR, Jin JP, Maltais L. The human and mouse replication-dependent histone genes. Genomics. 2002;80:487–498. [PubMed] [Google Scholar]
- Marzluff WF, Wagner EJ, Duronio RJ. Metabolism and regulation of canonical histone mRNAs: life without a poly(A) tail. Nat Rev Genet. 2008;9:843–854. doi: 10.1038/nrg2438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell P, Tollervey D. An NMD pathway in yeast involving accelerated deadenylation and exosome-mediated 3′-->5′ degradation. Mol Cell. 2003;11:1405–1413. doi: 10.1016/s1097-2765(03)00190-4. [DOI] [PubMed] [Google Scholar]
- Mullen TE, Marzluff WF. Degradation of histone mRNA requires oligouridylation followed by decapping and simultaneous degradation of the mRNA both 5′ to 3′ and 3′ to 5′. Genes Dev. 2008;22:50–65. doi: 10.1101/gad.1622708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munns TW, Liszewski MK, Sims HF. Characterization of antibodies specific for N6-methyladenosine and for 7-methylguanosine. Biochemistry. 1977;16:2163–2168. doi: 10.1021/bi00629a019. [DOI] [PubMed] [Google Scholar]
- Newman MA, Mani V, Hammond SM. Deep sequencing of microRNA precursors reveals extensive 3′ end modification. RNA. 2011;17:1795–1803. doi: 10.1261/rna.2713611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parker R, Song H. The enzymes and control of eukaryotic mRNA turnover. Nat Struct Mol Biol. 2004;11:121–127. doi: 10.1038/nsmb724. [DOI] [PubMed] [Google Scholar]
- Passos DO, Doma MK, Shoemaker CJ, Muhlrad D, Green R, Weissman J, Hollien J, Parker R. Analysis of Dom34 and its function in no-go decay. Mol Biol Cell. 2009;20:3025–3032. doi: 10.1091/mbc.E09-01-0028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross J, Kobs G, Brewer G, Peltz SW. Properties of the exonuclease activity that degrades H4 histone mRNA. J Biol Chem. 1987;262:9374–9381. [PubMed] [Google Scholar]
- Ross J, Peltz SW, Kobs G, Brewer G. Histone mRNA degradation in vivo: the first detectable step occurs at or near the 3′ terminus. Mol Cell Biol. 1986;6:4362–4371. doi: 10.1128/mcb.6.12.4362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saito S, Hosoda N, Hoshino S. The Hbs1-Dom34 Protein Complex Functions in Non-stop mRNA Decay in Mammalian Cells. J Biol Chem. 2013;288:17832–17843. doi: 10.1074/jbc.M112.448977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scharl EC, Steitz JA. The site of 3′ end formation of histone messenger RNA is a fixed distance from the downstream element recognized by the U7 snRNP. EMBO J. 1994;13:2432–2440. doi: 10.1002/j.1460-2075.1994.tb06528.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen B, Goodman HM. Uridine addition after microRNA-directed cleavage. Science. 2004;306:997. doi: 10.1126/science.1103521. [DOI] [PubMed] [Google Scholar]
- Song MG, Kiledjian M. 3′ Terminal oligo U-tract-mediated stimulation of decapping. RNA. 2007;13:2356–2365. doi: 10.1261/rna.765807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stimac E, Groppi VE, Jr, Coffino P. Inhibition of protein synthesis stabilizes histone mRNA. Mol Cell Biol. 1984;4:2082–2087. doi: 10.1128/mcb.4.10.2082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su W, Slepenkov SV, Slevin MK, Lyons SM, Ziemniak M, Kowalska J, Darzynkiewicz E, Jemielity J, Marzluff WF, Rhoads RE. mRNAs containing the histone 3′ stem-loop are degraded primarily by decapping mediated by oligouridylation of the 3′ end. RNA. 2013;19:1–16. doi: 10.1261/rna.034470.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan D, Marzluff WF, Dominski Z, Tong L. Structure of histone mRNA stem-loop, human stem-loop binding protein, and 3′hExo ternary complex. Science. 2013;339:318–321. doi: 10.1126/science.1228705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thornton JE, Chang HM, Piskounova E, Gregory RI. Lin28-mediated control of let-7 microRNA expression by alternative TUTases Zcchc11 (TUT4) and Zcchc6 (TUT7) RNA. 2012;18:1875–1885. doi: 10.1261/rna.034538.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomecki R, Kristiansen MS, Lykke-Andersen S, Chlebowski A, Larsen KM, Szczesny RJ, Drazkowska K, Pastula A, Andersen JS, Stepien PP, Dziembowski A, Jensen TH. The human core exosome interacts with differentially localized processive RNases: hDIS3 and hDIS3L. EMBO J. 2010;29:2342–2357. doi: 10.1038/emboj.2010.121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilusz JE, JnBaptiste CK, Lu LY, Kuhn CD, Joshua-Tor L, Sharp PA. A triple helix stabilizes the 3′ end of long non-coding RNAs that lack poly(A) tails. Genes and Development. 2012;26:2392–2407. doi: 10.1101/gad.204438.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang XC, Purdy M, Marzluff WF, Dominski Z. Characterization of 3′hExo, a 3′ exonuclease specifically interacting with the 3′ end of histone mRNA. J Biol Chem. 2006;281:30447–30454. doi: 10.1074/jbc.M602947200. [DOI] [PubMed] [Google Scholar]
- Zhao Y, Mo B, Chen X. Mechanisms that impact microRNA stability in plants. RNA Biol. 2012;9:1218–1223. doi: 10.4161/rna.22034. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.