Abstract
N2O is a powerful greenhouse gas contributing both to global warming and ozone depletion. While fungi have been identified as a putative source of N2O, little is known about their production of this greenhouse gas. Here we investigated the N2O-producing ability of a collection of 207 fungal isolates. Seventy strains producing N2O in pure culture were identified. They were mostly species from the order Hypocreales order—particularly Fusarium oxysporum and Trichoderma spp.—and to a lesser extent species from the orders Eurotiales, Sordariales, and Chaetosphaeriales. The N2O 15N site preference (SP) values of the fungal strains ranged from 15.8‰ to 36.7‰, and we observed a significant taxa effect, with Penicillium strains displaying lower SP values than the other fungal genera. Inoculation of 15 N2O-producing strains into pre-sterilized arable, forest and grassland soils confirmed the ability of the strains to produce N2O in soil with a significant strain-by-soil effect. The copper-containing nitrite reductase gene (nirK) was amplified from 45 N2O-producing strains, and its genetic variability showed a strong congruence with the ITS phylogeny, indicating vertical inheritance of this trait. Taken together, this comprehensive set of findings should enhance our knowledge of fungi as a source of N2O in the environment.
Terrestrial ecosystems are a major source of nitrous oxide (N2O)1,2, a so-called greenhouse gas also commonly known as laughing gas. Although it has received much less attention than CO2, the 100 year global warming potential of N2O is 298 times greater than that of CO2 due to the much longer half life of N2O3. There is also growing concern over nitrous oxide concentrations because, following the reduction of chlorine- and bromine-containing halocarbons by the Montreal Protocol, N2O has become the main ozone-depleting substance emitted to the stratosphere4.
Nitrous oxide emissions are mostly due to two microbial processes: nitrification and denitrification. Nitrous oxide is a by-product of the first step of nitrification, the oxidation of ammonia to nitrite5. In contrast, N2O is either an intermediate or the end product of the denitrification cascade, which consists in the reduction of nitrate or nitrite into nitric oxide, nitrous oxide and dinitrogen6. Sixty-two percent of the total global N2O emissions are from natural and agricultural soils (6 and 4.2 Tg N yr−1, respectively7), and denitrification is traditionally considered as the main source of these emissions8.
It is well known that denitrification is widespread among prokaryotes—indeed, the ability to denitrify has been observed in more than 60 bacterial and archaeal genera9. Moreover, eukaryotes such as fungi in soils10 or foraminifers in aquatic environments11,12 are also capable of denitrification. Characterization of the fungal denitrification ability in Fusarium oxysporum and Cylindrocarpon tonkinense has shown that this reductive process was performed via a copper-containing nitrite reductase (NirK) and cytochrome P450 nitric oxide reductase10. However, no nitrous oxide reductase has been identified in fungi and N2O is the end product of denitrification in the few characterized fungal strains13,14. By using fungal or bacterial inhibitors to distinguish the microbial origin of N2O, previous studies have reported that fungi could contribute up to 18% of potential denitrification15 and be significant N2O producers in some terrestrial systems16,17. Despite the importance of fungi in several soil functions, such as organic matter decomposition18 and primary production through symbiotic or pathogenic relationships with plants19, the production of N2O by fungi has only been studied in a limited number of strains14,20. To what extent this trait is conserved amongst fungi remains unknown, but understanding the microbial sources of this greenhouse gas will be crucial for selecting mitigation strategies. Here, we screened a collection of 207 fungal strains belonging to 9 classes and 23 orders to determine the prevalence of the N2O-producing capacity among fungi. We further characterized the initial and end-products of denitrification of the N2O production-positive strains in pure culture and determined their N2O isotopic signature. Positive fungal strains were also inoculated into pre-sterilized arable, forest and grassland soils in order to verify their ability to produce this greenhouse gas in soil. Finally, we studied the phylogeny of the nirK gene, which encodes the copper-containing nitrite reductase using newly developed primers, and investigated the relationships between the nuclear ribosomal internal transcribed spacer (ITS) region and nirK phylogeny, N2O production rates and N2O isotopic signatures.
Results and Discussion
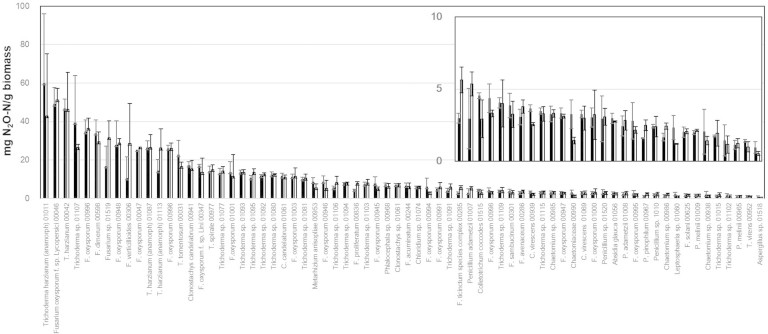
To assess how the N2O producing ability is distributed within fungi, 207 fungal strains comprising 23 orders and 54 genera were screened by incubating the strains in liquid culture under conditions that were previously reported to favour fungal denitrification21. The strains were selected to cover the largest possible fungal diversity within the Microorganisms of Interest for Agriculture and Environment collection (MIAE) (INRA, Dijon, France), which is dedicated to soil microbial diversity. At the end of the incubation, differences in the pH of the media were observed between strains. Since N2O can also be produced by chemical denitrification at low pH22, abiotic N2O production from nitrite was evaluated in sterile media with a pH gradient, and strains were scored positive when the N2O concentrations in the headspace were higher than those in the sterile flasks at the same pH (Fig. S1). When nitrite was used as an electron acceptor, more than a third of the strains were capable of producing N2O, with activities ranging from 0.5 ± 0.1 to 60.0 ± 36.0 mg N2O-N g−1 dry fungal biomass (Fig. 1). The N2O-producing activities were much lower when nitrate was used as an electron acceptor (<0.1 mg N2O-N g−1 fungal biomass; F = 108.55, P < 0.0001), supporting previous studies showing that nitrite rather than nitrate is preferable for fungal denitrification23. No difference was observed when incubating the positive strains with and without acetylene, indicating that the fungi does not reduce N2O, which was also in accordance with previous studies24. Accordingly, amplification of the nosZ gene using various primer sets25,26 was not successful (data not shown). The high proportion of N2O-producing fungal strains observed in our study contrasts with previous studies in which only 1% to 10% of examined bacteria were capable of denitrification based either on culture-based, direct-molecular approaches or genome analyses27,28,29. However, the maximum percentage of nitrogen recovered as N2O from nitrite in our work was about 38%, and most of the fungal strains reduced between 3% and 10% of the nitrite, while denitrifying bacteria are capable of reducing at least 80% of soluble nitrogen into gas30. Nonetheless, lower percentages were also reported for denitrifying bacteria such as Bacillus species, with ranges between 3.5% and 13.2% of the nitrate reduced to gaseous nitrogen after 48 h of growth31. Although we cannot rule out that growing the strains in different media or conditions may have resulted in different rates of nitrogen reduction, our incubation experiment using the standard media and conditions showed that N2O production is common within the fungal kingdom.
Figure 1. N2O production by the positive fungi strains with (white) and without (black) 10% C2H2 in the headspace.
Error bars indicate the standard deviation (n = 3). Both strain names and MIAE numbers are indicated.
Fifty out of the 70 positive strains belonged to the Hypocreales order, and Fusarium and Trichoderma were the main Hypocreales genera observed. Interestingly, many of the Fusarium strains identified as N2O-producers were Fusarium oxysporum. Even within this species, the production rate was highly variable, ranging from 2.8 to 34.7 mg N2O-N g−1 dry fungal biomass. This species was reported to be one of the dominant fungal taxa in several studies32, and accounted for up to 43% of the ITS pyrosequencing dataset retrieved from Mediterranean soils33. F. oxysporum includes non-pathogenic and pathogenic strains, with the latter causing disease to a broad range of host plants, but no plant-based bioassay has been conducted on the tested strains to discriminate pathogenic and non-pathogenic F. oxysporum. In any case, the high number of N2O-producing F. oxysporum individuals suggests this species is involved in greenhouse gas emissions, and therefore are potentially detrimental in terms of both primary production and climate regulation. In both Trichderma and Fusarium species capable of living in plant tissue, respiration of nitrogen oxides is likely due to the adaptation to hypoxic conditions. Indeed, the oxygen concentration near or within plant tissue is low (<1%)34, and several studies have reported that it is critical to the fungal infection of plants that the infecting fungus possess strategies to overcome hypoxia35. The other positive strains identified in this work belong to the Eurotiales (8), Sordariales (5), Chaetosphaeriales (3), Mucorales (1), Pleosporales (1), Glomerellales (1) and Ophiostomatales orders (1). Sordariales has also been reported as a dominant order in agricultural soil, while the order Pleosporales is frequently detected in that environment32. Collectively, our results indicate that N2O production is a common trait in fungal taxa that are frequently abundant in soils, although this feature seems to be more a strain-specific than a species-specific trait. It also underlines the importance of quantifying the fungal contribution to terrestrial N2O emissions.
Analysis of the N2O isotopomer ratios (relative abundance of 14N15N16O or 15N14N16O to that of 14N14N16O) has been proposed as a powerful method to obtain more information on the sources of this greenhouse gas36. However, the site preference (SP) values due to fungal N2O production have only been determined in 2 strains (F. oxysporum and C. tonkinense)37, and a more comprehensive analysis of the variability of the fungal 15N site preference is required for a robust distinction between the bacterial and fungal contributions to N2O emissions. The SP values of the 67 fungal strains tested herein varied from 15.8–36.7‰ with an average value of 30.0 ± 4.8‰. Our results are partly consistent with the previous study reporting a positive SP of 36.9–37.1‰37. However, we found a larger range of variation, with SP values as low as 15.8 ± 2.6‰ for Penicillium melinii and as high as 36.7 ± 2.2‰ for F. sambucinum. These strains with low SP values often had more acidic conditions at the end of the incubation (pH 3–5; Fig. S1). Therefore, we cannot rule out that these lower SP values resulted from the higher contribution of abiotic N2O production under more acidic conditions38. Interestingly, a significant taxa effect (P < 0.01 by Student's t-test) with lower SP values was observed for the Penicillium strains compared to the other fungal genera. We thus confirmed that the N2O isotopic signature can be used to determine which organisms, i.e., fungi or bacteria, are producing N2O by denitrification because of their distinct SP (about 0–10‰ for bacterial denitrification39). However, it will be difficult to distinguish N2O emissions from nitrification and fungal denitrification in the environment, since SP values for the N2O produced by both bacteria and archaea during ammonia oxidization ranged between 13.1 and 30.2‰40,41. Since fungi are microaerophilic denitrifiers, our results suggest that a stable isotopic approach alone is not enough to decipher whether nitrification or fungal denitrification is contributing to N2O emissions in environments where fungi are abundant.
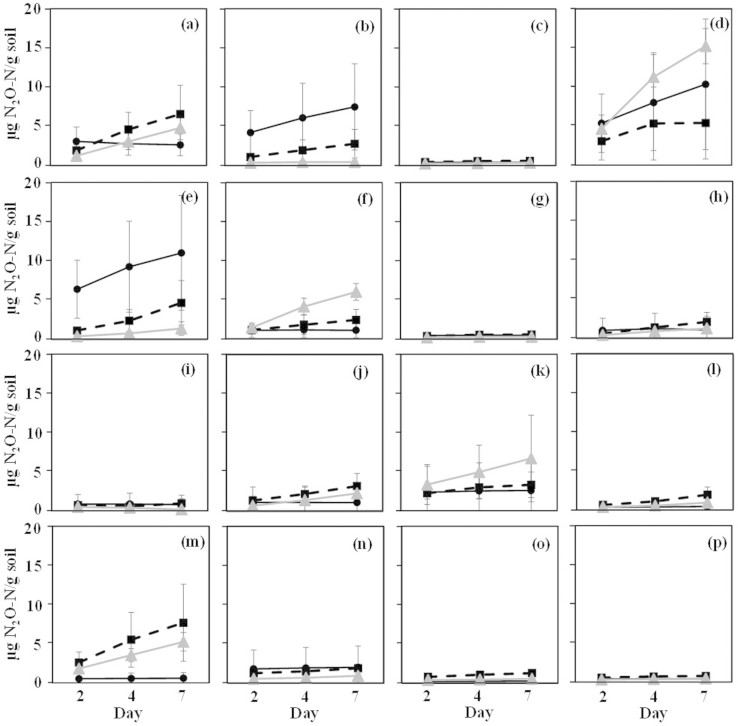
To further confirm that fungi could actually produce N2O in soil and not only in liquid culture, 15 of the positive strains were selected based on their high activity in pure culture and/or their taxonomic affiliation for inoculation into an arable, a grassland and a forest pre-sterilized soils. After allowing soil colonization by the inoculated fungal strains for a month, nitrite was added to induce denitrification and the concentration of N2O in the headspace was measured after 2, 4 and 7 days. Seven strains belonging to the Trichoderma, Fusarium, Penicillium and Phialocephala genera produced N2O in at least one of the soils, up to a maximum amount of 82.1 ng N2O-N g−1 h−1 (Fig. 2), which was significantly higher (F = 15590, P < 0.0001) than the amount produced by chemical denitrification in the non-inoculated sterile soils. This is comparable to previous studies focusing on fungal contribution15,16,17,42 or net N2O production from soil43,44,45. In our study, the highest amount of N2O produced was observed in the forest soil for Fusarium verticillioides and F. dimerium, while the grassland and the arable soils were the soils with the highest emissions for Trichoderma harzianum and Phialocephala spp. on one hand, and F. oxysporum f. sp. lini, Metarhizium anisopliae on the other. These significant strain-by-soil effects (F = 4.69, P < 0.0001) likely reflect differences in fungal nutrient requirements and/or preferences for different soil physico-chemical characteristics. Indeed, previous studies demonstrated that several soil physico-chemical parameters, such as extractable P concentrations, C/N ratio46, pH, sand content or litter cover47, can affect the soil fungal communities or fungal biomass48. The fungal N2O-producing capacity observed in our study could also be influenced by biotic interactions occurring in the natural ecosystems between fungi or with bacteria and other organisms.
Figure 2. N2O production by 15 selected fungi strains inoculated in 3 different sterile soils.
NO2− was used as the electron acceptor. The strains were incubated for 7 days, and headspace N2O concentration were measured 3 times (2, 4 and 7 days after NO2− addition). Code for the soils: Black circle: Sweden (Slogaryd), forest soil; grey triangle: France, arable soil; black rectangle: The Netherlands, grassland soil. Code for the fungi: a: Trichoderma harzianum (MIAE00042); b: Fusarium verticillioides (MIAE00306); c: Penicillium adametzii (MIAE01008); d: F. oxysporum f. sp. lini (MIAE00347); e: F. dimerum (MIAE00598); f: Metarhizium anisopliae (MIAE00953); g: Chaetomium sp. (MIAE00985); h: T. harzianum (anamorph) (MIAE01011); i: Leptosphaeria sp. (MIAE01060); j: T. tomentosum (MIAE00031); k: Fusarium sp. (MIAE01519); l: Clonostachys candelabrum (MIAE00941); m: Phialocephala sp. (MIAE00968); n: Colletotrichum coccodes (MIAE01515); o: Aspergillus sp. (MIAE01518);); p: No fungi control. The error bar represents the standard deviation (n = 3).
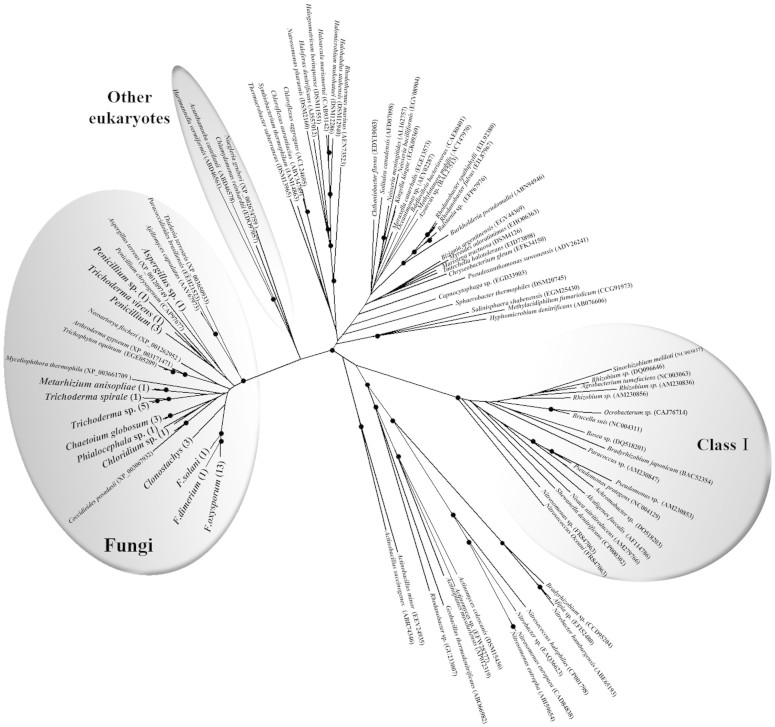
Both copper-containing nitrite reductase (encoded by the nirK gene) and P450nor (nitric oxide reductase) are key enzymes involved in fungal denitrification. However, P450nor belongs to a superfamily of proteins that are widely distributed among fungi and known to be involved in a wide variety of physiological reactions10, which prevents the use of the corresponding genes as molecular markers to target denitrifying fungi. To date, several primers targeting the nirK denitrification gene have been described in the literature, but none of them was designed to amplify denitrifying eukaryotes25,49. Despite the low number of fungal nirK sequences available in the databases (less than 30) and the high diversity of the tested fungal strains, the amplification of the fungal nirK denitrification gene was successful in 45 out of 70 strains using our newly designed primer set EunirK-F1 and EunirK-R2. This supports our findings that these fungal strains are capable of denitrification and that N2O was not produced by other processes. Notably, when used to amplify DNA extracted from soil, our primer set also amplified the bacterial nirK in part due to the lower proportion of fungi in soil compared to bacteria (data not shown). A phylogenetic tree was constructed using these fungi nirK sequences and bacterial nirK sequences from available databases (Fig. 3). Our fungal nirK sequences clustered with the other fungal nirK homologues retrieved from the database, and were distinct from the bacterial nirK sequences. The phylogeny also shows that fungal nirK sequences are closer to other eukaryotic sequences (amoeba, protozoa or green alga) than to bacterial ones, indicating that fungal nirK sequences branched from bacterial nirK sequences at a very early stage of their evolutional history, as suggested by Kim et al.50. In addition, we also found a strong congruence between the ITS and the nirK phylogenies (Fig. S2), indicating a vertical inheritance of nirK genes. Interestingly, we found no correlation between the genetic distance of the nirK genes and the N2O production rates. Similarly, weak or no relationships were observed between the bacterial genotypes and denitrification phenotypes in previous studies31,51. Phenotypic convergence within similar ecological niches of distantly related organisms can lead to such a discrepancy between genetic and phenotypic distances. In depth investigation of the ecology of denitrifying fungi would undoubtedly help clarify which environmental factors lead to a convergent denitrification phenotype.
Figure 3. Neighbour-joining phylogenetic tree of nirK amino acid sequences constructed by Clustal W with 1000 bootstrap samplings.
Strain names in bold indicate the sequences obtained in this study. The numbers in parentheses indicate the number of the strains. Bootstrap values greater than 75% are indicated as black circles.
In conclusion, the analysis of a vast collection of fungi showed that N2O production is a common and widespread trait in fungi. Nitrite instead of nitrate was the preferred substrate, while N2O was always the end-product of denitrification. We showed that the range of variation of the N2O isotopic signature was taxa-dependent and larger than previously reported, with values as low as 15%. Inoculation of 15 strains into previously sterilized arable, forest and grassland soils demonstrated the ability of fungi to contribute to soil N2O emissions with fluxes potentially as high as those reported in natural soils. Further studies are clearly warranted to elucidate the significance of denitrification in fungi and its consequences for N2O emissions.
Methods
Fungal strains and in vitro incubation experiments
The fungal strains tested in this study were previously isolated from agricultural soils or plant roots. The details of the isolating procedures have been described previously52. The strains were purified by single-spore isolation and preserved in the MIAE collection (INRA, Dijon, France). Fungal strains were first cultured under an aerobic condition in a 147 ml plasma flask containing 50 ml of liquid malt medium (pH 7.5). The flasks were incubated with a rotator shaker at 25°C, 120 rpm. After 3 days to 2 weeks depending on their growth, 50 ml of liquid malt medium with 10 mM NaNO2 (pH 7.5) was added to the plasma flasks, which were capped with a butyl rubber stopper. The headspace gas was then replaced by pure N2 gas and 1 ml of pure O2 to obtain microaerobic conditions. The flasks were incubated again at 25°C, 120 rpm for one additional week. The incubations were performed in triplicate flasks for each fungal strain. After the second incubation round, 0.5 ml of headspace gas was sampled and N2O concentrations were determined by gas chromatography. Three previously characterized strains24,53,54, F. oxysporum MT811 (JCM11502), Cylindrocarpon lichenicola (NBRC30561) and Aspergillus oryzae RIB40 (NBRC100959), were used as positive controls. At the end of the incubation, mycelia were collected and dried for 24 h at 105°C for fungal biomass determination, and the pH of the medium was determined by using a commercial electrode.
Strains capable of producing N2O were also incubated as described above, but with NO3− as electron acceptor (NaNO3 at a final concentration of 5 mM), and biomass and pH determination were performed as described above. Gas measurements were done with and without 10% C2H2 gas in the headspace55 to verify whether or not they were capable of reducing N2O into N2.
Fungal N2O production in sterile soil
The physical and chemical parameters of the three soils used in this study are described in Table 1. Triplicate samples were collected from the top 10 cm of three different soils, sieved to <2 mm and sterilized by γ -radiation (35 kGy; Conservatome, Dagneux, France).
Table 1. Soil characteristics and properties across a range of ecosystem types.
| Country | Description | Clay | Loam | Sand | Water content | WHC | Total-C | Total-N | OM | ||
|---|---|---|---|---|---|---|---|---|---|---|---|
| % | % | % | g g−1 soil DW | g g−1 soil DW | pH | g kg−1 soil DW | g kg−1 soil DW | C:N ratio | g kg−1 soil DW | ||
| France | Non-irrigated arable land | 18.4 | 66.4 | 15.1 | 0.11 | 0.50 | 6.6 | 10.2 | 0.97 | 10.6 | 17.7 |
| The Netherlands | Permanent Grassland | 3.4 | 2.7 | 93.8 | 0.17 | 0.44 | 5.1 | 20.3 | 1.25 | 16.3 | 35.1 |
| Sweden | Coniferous forest | 21.4 | 15.9 | 62.7 | 1.51 | 2.54 | 4.3 | 253.0 | 14.2 | 17.8 | 438.0 |
WHC: water holding capacity; OM: organic matter; DW: dry weight.
Denitrifying strains were incubated with liquid malt medium (400 mL) in 1 L flasks at 25°C, 120 rpm for 7 to 14 days so that enough biomass was obtained. Liquid cultures were centrifuged (12,000 rpm for 10 min) in 50 mL tubes and washed twice with 30 mL of sterile physiological water (0.9% NaCl). For each soil, fungal pellets were resuspended with sterile water and inoculated into two series of triplicate microcosms containing the γ-ray sterilized soil (5 g in 147 mL sterile bottles). Soil moisture was adjusted in order to obtain a water holding capacity (WHC) of 60% after the inoculation, and all the inoculated soil microcosms were incubated for 3–4 weeks at room temperature to allow soil colonization by the fungi. After this pre-incubation, 2 mL of NaNO2 solution (10 mM) was added and the soil moisture was adjusted at a WHC of 90%. In half of the replicated microcosms (three for each soil), 10 mL of the ambient air in the headspace was replaced with 10 mL C2H2, and all bottles were incubated at 25°C for one additional week. Production of N2O was also monitored in all three sterile soils without inoculation of fungi (negative controls).
Nitrous oxide emission measurements
Nitrous oxide production by fungal pure culture was determined by analysing the gas samples collected in the headspace gas using a gas chromatograph (TRACE GC Ultra; Thermo) equipped with an electron capture detector (GC-ECD).
In the inoculated soil microcosms, gas samples were collected 3 times (2, 4, and 7 days after NO2 addition) from the headspace and were analysed with GC-ECD to determine the N2O concentrations. The N2O concentrations were analysed by ANOVA using the GLM procedure in SAS56. For the liquid medium culture experiment, strain and substrate (NO2 or NO3) were used for the fixed effect. For the sterilized soil incubation experiment, strain and soil were used for the fixed effect and strain*soil was included in the model. Tukey's multiple comparisons test was used to separate the means. The significance level was 0.05.
Nitrous oxide isotopic signature measurements
Ten ml of each headspace gas sample was taken at the end of the incubation experiments and stored in a pre-evacuated vial. The samples were then introduced into a gas chromatograph-isotope ratio mass spectrometer (GC-IRMS) (MAT 252; Thermo Fisher Scientific K.K., Yokohama, Japan) system as described elsewhere to measure the N2O isotopomer ratios57. Site-specific N isotope analysis in N2O was conducted using ion detectors that had been modified for the mass analysis of fragment ions of N2O (NO+) containing N atoms in the centre positions of N2O molecules, whereas the bulk (average) N and oxygen isotope ratios were determined from molecular ions58. Pure N2O (purity > 99.999%; Showa Denko K.K., Japan) was calibrated with international standards and used as a working standard for the isotopomer ratios. The notation of the isotopomer ratios is shown below. The measurement precision was typically better than 0.1‰ for δ15Nbulk and δ18O, and better than 0.5‰ for δ15Nα and δ15Nβ.
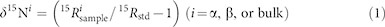 |
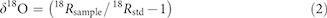 |
Here, 15Rα and 15Rβ respectively represent the 15N/14N ratios at the centre and end sites of the nitrogen atoms; 15Rbulk and 18R respectively indicate the average isotope ratios for 15N/14N and 18O/16O. The subscripts “sample” and “std” respectively indicate the isotope ratios for the sample and the standard, atmospheric N2 for N and Vienna Standard Mean Ocean Water (V-SMOW) for O. We also define the 15N site preference (hereinafter SP) as an illustrative parameter of the intramolecular distribution of 15N:
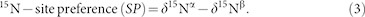 |
The N2O concentration was measured simultaneously with the isotopomer ratios by comparing the peak area of the major ion (mass 44 and 30 in molecular ion analysis and fragment ion analysis, respectively) obtained with the sample gas and with a reference gas (349 nL/L N2O in Air; Japan Fine Products Co., Ltd.)57.
Primer design
Full-length nirK nucleotide sequences of fungal genomes were obtained from the Functional Gene Pipeline public database (http://fungene.cme.msu.edu//index.spr). These sequences were aligned using MEGA in order to design fungal specific −nirK primers. The primer sequences were as follows: EunirK-F1 (5′-GGB AAY CCI CAY AAY ATC GA-3′) and EunirK-R2 (5′-GGI CCI GCR TTS CCR AAG AA-3′).
DNA extraction, and PCR amplification of the nirK gene and ITS from positive strains
DNA extraction from the denitrifying fungal cultures was performed using a commercially available DNA extraction kit, DNeasy® Plant Maxi (QIAGEN). The extraction was performed according to the manufacturer's instructions. The purified DNA samples were stored at −20°C until further analysis.
The PCR protocol for the nirK gene was as follows: 10 min at 94°C and 40 cycles consisting of 1 min at 94°C, 30 s at 53°C, and 1 m at 72°C. For amplification of the fungi Internal Transcribed Spacer (ITS) region, the primer sets ITS5 (5′-TCC TCC GCT TAT TGA TAT GC-3′) and ITS4 (5′-GGA AGT AAA AGT CGT AAC AAG G-3′) were used59. The PCR protocol was as follows: 10 min at 95°C and 35 cycles consisting of 95°C for 15 s, 30 s at 52°C, and 72°C for 1.5 s. These amplicons were purified and sequenced with the BigDye Terminator version 3.1 cycle sequencing kit (Applied Biosystems) and the ABI Prism 3100 genetic analyser. The nucleotide sequence of the ITS regions of other fungi and bacterial nirK (both class 1 and 2) amino acid sequences were also obtained from the database described above, and the phylogenetic tree was constructed based on the maximum likelihood method using CLUSTALW. Congruence between the ITS and nirK phylogenies was graphically illustrated using the cophyloplot function from the “ape” R package60.
Author Contributions
L.P. and C.S. designed the experiments. K.M., A.S. and L.P. wrote the paper. K.M., C.H., M.C.B. and F.B. screened the fungal isolates. V.E.H. verified the fungal isolates. K.M., S.T. and N.Y. worked on the isotopic experiments.
Additional Information
Accession codes: All sequence data were submitted to DDBJ under accession numbers AB904794-AB904830.
Supplementary Material
Supplementary Information
Acknowledgments
The authors are grateful to Nadine Gautheron for providing technical assistance on culturing fungal strains of the MIAE collection (Microorganisms of Interest for Agriculture and Environment). ST and NY were supported by KAKENHI (23224013) of the Ministry of Education, Culture, Sports, Science and Technology, Japan.
References
- Davidson E. A., Rogers J. & Whitman W. Fluxes of nitrous oxide and nitric oxide from terrestrial ecosystems. In: Microbial production and consumption of greenhouse gases: methane, nitrogen oxides, and halomethanes. (eds. Rogers J. E., , Whitman W. B., eds. ) 219–235. (American Society for Microbiology, 1991). [Google Scholar]
- Davidson E. The contribution of manure and fertilizer nitrogen to atmospheric nitrous oxide since 1860. Nat. Geosci. 2, 659–662 (2009). [Google Scholar]
- Forster P. et al. Changes in Atmospheric Constituents and in Radiative Forcing. In: Climate Change 2007: The Physical Science Basis. (eds. Solomon S., et al.) Ch.2, 130–234. (Cambridge University Press, 2007). [Google Scholar]
- Ravishankara A., Daniel J. & Portmann R. Nitrous oxide (N2O): the dominant ozone-depleting substance emitted in the 21st century. Science 326, 123–125 (2009). [DOI] [PubMed] [Google Scholar]
- Hooper A. B. & Terry K. Hydroxylamine oxidoreductase of Nitrosomonas: Production of nitric oxide from hydroxylamine. BBA-Enzymol. 571, 12–20 (1979). [DOI] [PubMed] [Google Scholar]
- Zumft W. Cell biology and molecular basis of denitrification. Microbiol. Mol. Biol. Rev. 61, 533–616 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomson A. J., Giannopoulos G., Pretty J., Baggs E. M. & Richardson D. J. Biological sources and sinks of nitrous oxide and strategies to mitigate emissions. Phil. Trans. R. Soc. B: Biol. Sci. 367, 1157–1168 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ostrom N. et al. Isotopologue data reveal bacterial denitrification as the primary source of N2O during a high flux event following cultivation of a native temperate grassland. Soil Biol. Biochem. 42, 499–506 (2010). [Google Scholar]
- Philippot L., Hallin S. & Schloter M. Ecology of denitrifying prokaryotes in agricultural soil. Adv. Agron. 96, 249–305 (2007). [Google Scholar]
- Shoun H., Fushinobu S., Jiang L., Kim S. W. & Wakagi T. Fungal denitrification and nitric oxide reductase cytochrome P450nor. Phil. Trans. R. Soc. B: Biol. Sci. 367, 1186–1194 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Risgaard-Petersen N. et al. Evidence for complete denitrification in a benthic foraminifer. Nature 443, 93–96 (2006). [DOI] [PubMed] [Google Scholar]
- Piña-Ochoa E. et al. Widespread occurrence of nitrate storage and denitrification among Foraminifera and Gromiida. Proc. Natl. Acad. Sci. 107, 1148–1153 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayatsu M., Tago K. & Saito M. Various players in the nitrogen cycle: diversity and functions of the microorganisms involved in nitrification and denitrification. Soil Sci. Plant Nutr. 54, 33–45 (2008). [Google Scholar]
- Shoun H., Kim D. H., Uchiyama H. & Sugiyama J. Denitrification by fungi. FEMS Microbiol. Lett. 94, 277–281 (1992). [DOI] [PubMed] [Google Scholar]
- Herold M. B., Baggs E. M. & Daniell T. J. Fungal and bacterial denitrification are differently affected by long-term pH amendment and cultivation of arable soil. Soil Biol. Biochem. 54, 25–35 (2012). [Google Scholar]
- Laughlin R. J., Rütting T., Müller C., Watson C. J. & Stevens R. J. Effect of acetate on soil respiration, N2O emissions and gross N transformations related to fungi and bacteria in a grassland soil. Appl. Soil Ecol. 42, 25–30 (2009). [Google Scholar]
- Marusenko Y., Huber D. P. & Hall S. J. Fungi mediate nitrous oxide production but not ammonia oxidation in aridland soils of the southwestern US. Soil Biol. Biochem. 63, 24–36 (2013). [Google Scholar]
- Lundell T. K., Mäkelä M. R. & Hildén K. Lignin‐modifying enzymes in filamentous basidiomycetes–ecological, functional and phylogenetic review. J. Bas. Microbiol. 50, 5–20 (2010). [DOI] [PubMed] [Google Scholar]
- Alabouvette C., Olivain C., Migheli Q. & Steinberg C. Microbiological control of soil‐borne phytopathogenic fungi with special emphasis on wilt‐inducing Fusarium oxysporum. New Phytol. 184, 529–544 (2009). [DOI] [PubMed] [Google Scholar]
- Tsuruta S. et al. Denitrification by yeasts and occurrence of cytochrome P450nor in Trichosporon cutaneum. FEMS Microbiol. Lett. 168, 105–110 (1998). [DOI] [PubMed] [Google Scholar]
- Zhou Z., Takaya N., Sakairi M. A. C. & Shoun H. Oxygen requirement for denitrification by the fungus Fusarium oxysporum. Arch. Microbiol. 175, 19–25 (2001). [DOI] [PubMed] [Google Scholar]
- Daum D. & Schenk M. K. Influence of nutrient solution pH on N2O and N2 emissions from a soilless culture system. Plant Soil 203, 279–288 (1998). [Google Scholar]
- Takaya N. Dissimilatory nitrate reduction metabolisms and their control in fungi. J. Biosci. Bioeng. 94, 506–510 (2002). [DOI] [PubMed] [Google Scholar]
- Shoun H. & Tanimoto T. Denitrification by the fungus Fusarium oxysporum and involvement of cytochrome P-450 in the respiratory nitrite reduction. J. Biol. Chem. 266, 11078–11082 (1991). [PubMed] [Google Scholar]
- Throback I. N., Enwall K., Jarvis A. & Hallin S. Reassessing PCR primers targeting nirS, nirK and nosZ genes for community surveys of denitrifying bacteria with DGGE. FEMS Microbiol. Ecol. 49, 401–417 (2004). [DOI] [PubMed] [Google Scholar]
- Jones C. M., Graf D. R. H., Bru D., Philippot L. & Hallin S. The unaccounted yet abundant nitrous oxide-reducing microbial community: a potential nitrous oxide sink. ISME J. 7, 417–426 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheneby D. et al. Denitrifying bacteria in bulk and maize-rhizospheric soil: diversity and N2O-reducing abilities. Canad. J. Microbiol. 50, 469–474 (2004). [DOI] [PubMed] [Google Scholar]
- Bru D. et al. Determinants of the distribution of nitrogen-cycling microbial communities at the landscape scale. ISME J. 5, 532–542 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones C. M., Stres B., Rosenquist M. & Hallin S. Phylogenetic analysis of nitrite, nitric oxide, and nitrous oxide respiratory enzymes reveal a complex evolutionary history for denitrification. Mol. Biol. Evol. 25, 1955–1966 (2008). [DOI] [PubMed] [Google Scholar]
- Mahne I. & Tiedje J. M. Criteria and methodology for identifying respiratory denitrifiers. Appl. Environ. Microbiol. 61, 1110–1115 (1995). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones C. M. et al. Phenotypic and genotypic heterogeneity among closely related soil‐borne N2- and N2O- producing Bacillus isolates harboring the nosZ gene. FEMS Microbiol. Ecol. 76, 541–552 (2011). [DOI] [PubMed] [Google Scholar]
- Klaubauf S. et al. Molecular diversity of fungal communities in agricultural soils from Lower Austria. Fung. Div. 44, 65–75 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orgiazzi A. et al. Unravelling soil fungal communities from different Mediterranean land-use backgrounds. PLoS One 7, e34847 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudolph N. et al. Dynamic oxygen mapping in the root zone by fluorescence dye imaging combined with neutron radiography. J. Soils Sed.12, 63–74 (2012). [Google Scholar]
- Corrales Escobosa A. R. et al. Fusarium oxysporum Adh1 has dual fermentative and oxidative functions and is involved in fungal virulence in tomato plants. Fung. Genet. Biol. 48, 886–895 (2011). [DOI] [PubMed] [Google Scholar]
- Yoshida N. & Toyoda S. Constraining the atmospheric N2O budget from intramolecular site preference in N2O isotopomers. Nature 405, 330–334 (2000). [DOI] [PubMed] [Google Scholar]
- Sutka R., Adams G., Ostrom N. & Ostrom P. Isotopologue fractionation during N2O production by fungal denitrification. Rapid Commun. Mass Spectrom. 22, 3989–3996 (2008). [DOI] [PubMed] [Google Scholar]
- Samarkin V. et al. Abiotic nitrous oxide emission from the hypersaline Don Juan Pond in Antarctica. Nat. Geosci. 3, 341–344 (2010). [Google Scholar]
- Sutka R. L. et al. Distinguishing nitrous oxide production from nitrification and denitrification on the basis of isotopomer abundances. Appl. Environ. Microbiol. 72, 638–644 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung M.-Y. et al. Isotopic signatures of N2O produced by ammonia-oxidizing archaea from soils. ISME J. 8, 1115–1125 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santoro A. E., Buchwald C., McIlvin M. R. & Casciotti K. L. Isotopic signature of N2O produced by marine ammonia-oxidizing archaea. Science 333, 1282–1285 (2011). [DOI] [PubMed] [Google Scholar]
- Wei W. et al. N2O emission from cropland field soil through fungal denitrification after surface applications of organic fertilizer. Soil Biol. Biochem. 69, 157–167 (2014). [Google Scholar]
- Enwall K., Philippot L. & Hallin S. Activity and composition of the denitrifying bacterial community respond differently to long-term fertilization. Appl. Environ. Microbiol. 71, 8335–8343 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braker G., Schwarz J. & Conrad R. Influence of temperature on the composition and activity of denitrifying soil communities. FEMS Microbiol. Ecol. 73, 134–148 (2010). [DOI] [PubMed] [Google Scholar]
- Orlando J., Carú M., Pommerenke B. & Braker G. Diversity and activity of denitrifiers of Chilean arid soil ecosystems. Front. Microbiol. 3, 101 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lauber C. L., Strickland M. S., Bradford M. A. & Fierer N. The influence of soil properties on the structure of bacterial and fungal communities across land-use types. Soil Biol. Biochem. 40, 2407–2415 (2008). [Google Scholar]
- Wubet T. et al. Differences in soil fungal communities between European beech (Fagus sylvatica L.) dominated forests are related to soil and understory vegetation. PloS One 7, e47500 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joergensen R. G. & Wichern F. Quantitative assessment of the fungal contribution to microbial tissue in soil. Soil Biol. Biochem. 40, 2977–2991 (2008). [Google Scholar]
- Braker G., Zhou J., Wu L., Devol A. H. & Tiedje J. M. Nitrite reductase genes (nirK and nirS) as functional markers to investigate diversity of denitrifying bacteria in pacific northwest marine sediment communities. Appl. Environ. Microbiol. 66, 2096–2104 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S.-W., Fushinobu S., Zhou S., Wakagi T. & Shoun H. Eukaryotic nirK genes encoding copper-containing nitrite reductase: originating from the protomitochondrion? Appl. Environ. Microbiol. 75, 2652–2658 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu B., Mao Y., Bergaust L., Bakken L. R. & Frostegård Å. Strains in the genus Thauera exhibit remarkably different denitrification regulatory phenotypes. Environ. Microbiol. 15, 2816–2828 (2013). [DOI] [PubMed] [Google Scholar]
- Edel V., Steinberg C., Gautheron N., Recorbet G. & Alabouvette C. Genetic diversity of Fusarium oxysporum populations isolated from different soils in France. FEMS Microbiol. Ecol. 36, 61–71 (2001). [DOI] [PubMed] [Google Scholar]
- Nakanishi Y. et al. A eukaryotic copper-containing nitrite reductase derived from a NirK homolog gene of Aspergillus oryzae. Biosci. Biotechnol. Biochem. 74, 984–991 (2010). [DOI] [PubMed] [Google Scholar]
- Usuda K., Toritsuka N., Matsuo Y., Kim D. H. & Shoun H. Denitrification by the fungus Cylindrocarpon tonkinense: anaerobic cell growth and two isozyme forms of cytochrome P-450nor. Appl. Environ. Microbiol. 61, 883–889 (1995). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshinari T. & Knowles R. Acetylene inhibition of nitrous oxide reduction by denitrifying bacteria. Biochem. Biophys. Res. Commun. 69, 705–710 (1976). [DOI] [PubMed] [Google Scholar]
- SAS Institute Inc. . SAS/STAT 9.1 user's guide. (SAS Institute Inc., 2004). [Google Scholar]
- Toyoda S., Mutobe H., Yamagishi H., Yoshida N. & Tanji Y. Fractionation of N2O isotopomers during production by denitrifier. Soil Biol. Biochem. 37, 1535–1545 (2005). [Google Scholar]
- Toyoda S. & Yoshida N. Determination of nitrogen isotopomers of nitrous oxide on a modified isotope ratio mass spectrometer. Anal. Chem. 71, 4711–4718 (1999). [Google Scholar]
- Schoch C. L. et al. Nuclear ribosomal internal transcribed spacer (ITS) region as a universal DNA barcode marker for Fungi. Proc. Natl. Acad. Sci. 109, 6241–6246 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paradis E., Claude J. & Strimmer K. APE: analyses of phylogenetics and evolution in R language. Bioinformatics 20, 289–290 (2004). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Information