Abstract
This study assesses the nasal occurrence of β-lactamase-producing Enterobacteriaceae both in patients in a hospital department of infectious diseases at admission and in healthy Madagascan students and health care workers.
Nasal swabs from 681 students, 824 health care workers, and 169 patients were obtained in Antananarivo, Madagascar, and transferred to Germany. Screening for β-lactamase (ESBL, ampC) producing Enterobacteriaceae was performed by cultural and molecular approaches, comprising Brilliance ESBL agar, E-testing, ABCD-testing, and commercial hyplex ESBL and SuperBug ID PCR.
Regarding ESBL-positive strains and strains with resistance against at least three out of the four tested bactericidal antibiotic drugs, 0.3% (five out of 1541) of the students and health care workers group showed nasal colonization, whereas colonization was observed in 7.1% (12 out of 169) of the hospitalized patients at admission. No appreciably reduced detection rates after sample storage and intercontinental transport were observed. A considerable proportion of nasal colonization with cephalosporin-resistant Enterobacteriaceae was demonstrated in Madagascan hospital patients at admission, posing a risk of developing future endogenous infections. The nasal colonization of healthy individuals was negligible. Good storage and transport stability of Enterobacteriaceae will allow for future studies even in areas difficult to access.
Keywords: colonization, Enterobacteriaceae, extended-spectrum β-lactamase, Madagascar, resistance
Introduction
The increasing resistance of bacterial pathogens to antimicrobial drugs is a major public-health menace facing this century that does not spare tropical countries. In particular, extended-spectrum β-lactamase (ESBL)-positive Enterobacteriaceae are known to be prevalent in Madagascan hospital patients [1–4], including populations at particular risks such as newborns [1]. ESBL expression causes increased resistance to penicillins and cephalosporins, driven by a variety of molecular mechanisms [5, 6]. Among known mechanisms, blaCTX-M expression (CTX = resistance to the WHO-listed antibiotic drug ceftriaxone, M = Munich, Germany, as the site of first description) is the most prevalent one in Madagascar, being detected in three out of four Madagascan ESBL-positive Enterobacteriaceae [4].
So far, little is known about the occurrence of ESBL- positive strains as colonizers in the healthy Madagascan population. We therefore performed a screening for ESBL strains as colonizers of the nasal vestibulum in relation to the total nasal colonization with Gram-negative rod-shaped bacteria. In the authors’ own experience, nasal colonization with Enterobacteriaceae is frequent in resource-limited tropical countries, presumably due to limited access to facilities with adequate sanitary hygiene [7], although the gut is the major site of enterobacterial colonization. In an Israeli study, ESBL colonization of the upper airways was lower than colonization of the gut by a factor of 3–4 [8]. However, stool samples are much more difficult to obtain, particularly from healthy volunteers. In a previous Madagascan study using fresh stool samples, only patients from outpatient departments were included [3]. For these reasons, we chose to analyze the lower-yielding but more easily obtained nasal swabs to make adherence to the study protocol more likely. In a population with frequent nasal colonization with Gram-negative rod-shaped bacteria, even analyses from such atypical localizations might provide hints regarding the dimension of ESBL colonization if sampling from the gut or the inguinal region is difficult for logistic and socio-cultural reasons.
The primary focus of the analysis was nasal colonization with ESBL-positive strains both in apparently healthy Madagascan students and health care workers and in a small group of hospital patients at admission to hospital without risk of nosocomial transmission. The study’s secondary objective was to assess the stability of ESBL-positive Enterobacteriaceae during storage and transport in the tropical setting. Such information is of importance in estimating the reliability of culture-based diagnostic results if an immediate cultural assessment is not possible and samples have to be transported over large distances to an appropriate laboratory facility.
Material and methods
Study population
Healthy volunteers
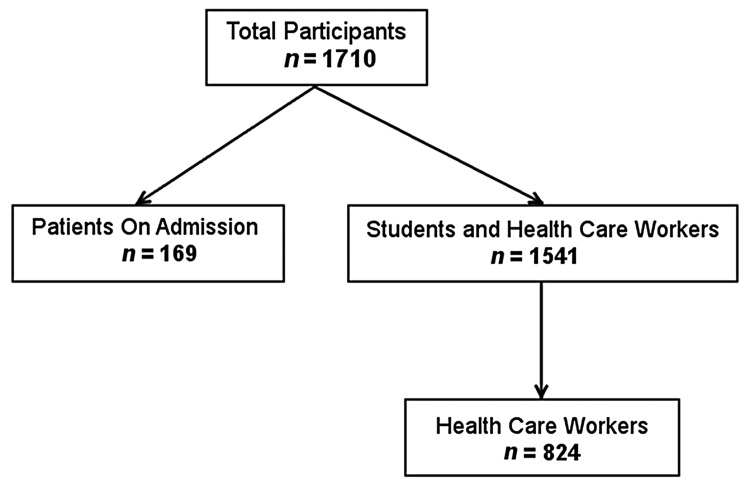
Nasal swabs (Amies w/o Ch, Copan Italia SpA, Brescia, Italy) from the nasal vestibulum were obtained from a group of 1541 healthy volunteers comprising students and health care workers from Antananarivo, Madagascar, and the nearby surroundings. There were no exclusion criteria. A total of 824 participants were health care workers (including students working in a hospital) (Fig. 1). All volunteers were asked to complete questionnaires to provide information on age, gender, residence, accommodation in a student hostel, study subject for students, job details for hospital workers, current or chronic diseases, recent hospital stays, and intake of antibiotics, as well as contacts with diseased persons or animals. Completed questionnaires were returned by 1505 of the volunteers. Age information was included in 1493 questionnaires; the median age was 23 years, ranging from 13 to 67 years. Gender information was provided in 1504 questionnaires; 66% of the participants were female.
Fig. 1.
Study populations. Two groups were analyzed, one comprising students and health care workers, the other comprising hospital patients at admission
Hospital patients
Nasal swabs were also collected from 169 patients from the Department of Infectious Diseases of the University Hospital Joseph Raseta de Befelatanana, Antananarivo, Madagascar directly at admission (Fig. 1). During the 6-month-sampling period, only patients with recent stays in intensive care units were excluded from the study. All other patients admitted were included. No samples were obtained from outpatient departments. All included patients completed a questionnaire to provide information on age, gender, residence, hospitalization, and intake of antibiotics during the previous 6 months, types of antibiotics used, chronic diseases, and professional contact with animals (Fig. 1). The median age was 34 years, ranging from 15 to 84 years; 41% of the patients were female.
Laboratory procedures
Screening for ESBL-positive Enterobacteriaceae
After sampling, the swabs were transferred to the laboratory, where they were stored at 4 °C. The storage and transport time ranged from 0 to 841 h (median: 53 h), depending on the geographical site of sampling. For nine samples, the transport time exceeded 300 h for logistical reasons. For 43 samples, the storage and transport time could not be documented.
After storage intervals ranging from 2 weeks to 4 months, several batches of 20 to 400 samples were transported by air to Hamburg, Germany. After arrival, all swabs were incubated in unselective thioglycolate enrichment broth (Heipha, Eppelheim, Germany) at 37 °C for 16–24 h to obtain maximum yields. Broth enrichment is known to double the yield of ESBL-expressing bacteria after swabbing in upper respiratory tract samples [9]. After broth enrichment, 10 µl of the incubated broths were cultured on nonselective Columbia agar enriched with 5% sheep blood (Oxoid, Basingstoke, UK), on MacConkey II agar (Becton Dickinson, Franklin Lakes, New Jersey, USA), which is selective for Gram-negative rod-shaped bacteria, and on Brilliance ESBL selective agar (Oxoid, Basingstoke, UK), which is made for selective growth of ESBL-positive Enterobacteriaceae. The sensitivity of the ESBL agar is 94.9–97.9%, and the specificity, 95.7–100% [10, 11]. Agar plates were incubated at 37 °C for 40–48 h.
For all samples, growth of Gram-negative rod-shaped bacteria on MacConkey II agar was assessed without further differentiation. From Brilliance ESBL selective agar, all colonies that looked suspicious for Enterobacteriaceae (blue, green, brown colonies) were isolated for further investigations. From colonies that looked suspicious for Gram-negative nonfermentative rod-shaped bacteria (i.e., yellow or yellowish-brown or greenish-brown colonies), a subset of 14 out of 194 strains was representatively analyzed. All isolated colonies were stored at −80 °C in Microbank™ tubes (Pro-Lab Diagnostics, Bromborough, UK).
Testing of storage and transport stability of ESBL strains
For logistic reasons, two different approaches were chosen to assess the storage and transport stability of ESBL strains from sampling in Madagascar to analysis in Germany.
For the patient subgroup at the University Hospital Joseph Raseta de Befelatanana, Antananarivo, swabs were immediately smeared on Brilliance ESBL agar after sampling and prior to storage and transport. The agar plates were incubated at 37 °C for 48 h in the University Hospital Laboratory. Colonies on Brilliance ESBL agar that looked suspicious for Enterobacteriaceae were isolated and shipped to Germany in addition to the swabs for further investigations.
For the subgroup of healthy students and health care workers, immediate analysis on Brilliance ESBL agar in Madagascar was impossible for logistic reasons. Therefore, commercial hyplex ESBL ID PCR (amPLEX, Giessen, Germany) targeting blaCTX-M as the most frequent ESBL resistance mechanism in Madagascar [4] was applied to the swabs of a subset of 251 samples in Hamburg according to the manufacturer’s instructions. As well as blaCTX-M [6], the hyplex ESBL kit also targets β-lactamases of the blaTEM- and blaSHV-types [5] as well as the blaOXA-1 carbapenemase in a consensus approach, though without specificity for the expression of an ESBL phenotype. Positive signals of the blaCTX-M-PCR were correlated with cultural results on Brilliance ESBL agar, and positive signals of the β-lactamase consensus PCR, with the general growth of Enterobacteriaceae on MacConkey II agar.
Assessment of isolated strains
The strains isolated from Brilliance ESBL agar were identified by 16S rRNA gene sequencing and matrix-assisted laser-desorption–ionization time-of-flight mass spectrometry (MALDI–TOF–MS). A previously described 16S rRNA gene PCR targeting an 817-base-pair fragment was used [12–14]. Sequencing results were interpreted using the CLSI (Clinical and Laboratory Standards Institute) guideline MM18-A “Interpretive Criteria for Identification of Bacteria and Fungi by DNA Target Sequencing; Approved Guideline” [15] as detailed [16]. MALDI–TOF–MS analysis was performed using a Shimadzu/Kratos “AXIMA Assurance” MALDI–TOF mass spectrometer (Shimadzu Germany Ltd., Duisburg, Germany) as described [14] with a minor modification. The databases Myla (version 3.2.0–6) and Saramis (version A2012/10 161150-219) were used for automated identification of the strains.
As well as Brilliance ESBL agar screening, the presence of ESBL- or AmpC-type resistance was confirmed or excluded by the commercial ABCD test kit Mast ID D68C (Mast Diagnostic, Amlens, France) as described by the manufacturer and others [17]. In addition, resistance testing was performed via E-tests (BioMerieux, Marcy-l’Étoile, France) for piperacillin, ceftazidime, ciprofloxacin, and meropenem as representatives of the four important bactericidal antibiotic substance groups of penicillins, cephalosporins, fluoroquinolones, and carbapenems, respectively. E-test results were interpreted as sensitive, intermediate sensitive, and resistant in accordance with the EUCAST guideline (version 4.0, 2014, http://www.eucast.org/fileadmin/src/media/PDFs/EUCAST_files/Breakpoint_tables/Breakpoint_table_v_4.0.pdf). Multidrug resistance was accepted if ≤1 of the four substances tested sensitive in accordance with German national guidelines [18].
All strains isolated from the ESBL agar were analyzed using hyplex ESBL ID PCR (amPLEX, Giessen, Germany) as described by the manufacturer. Strains that tested intermediate sensitive or resistant for meropenem were additionally assessed using hypex SuperBug ID PCR (amPLEX) [19] targeting the carbapenemase genes blaVIM, blaIMP, blaKPC, blaOXA-48, and blaNDM-1 according to the manufacturer’s instructions.
Analysis
Absolute occurrence as well as occurrence as a percentage of potential risk factors from the questionnaires was descriptively compared in study participants with and without proof of ESBL-positive strains and multidrug-resistant Enterobacteriaceae. Relative risks were assessed for nonnumeric parameters. The age of the study participant groups with and without proof of ESBL-positive strains and multidrug-resistant Enterobacteriaceae was compared using nonparametric Mann-Whitney testing.
To reduce the risk of a bias due to die-off of bacteria from inappropriately handled swabs, only samples showing Gram-negative growth on MacConkey II agar were included in this assessment.
Ethical clearance
The study complied with the principles of the Helsinki Declaration of 1975 and with all subsequent amendments by the World Medical Assembly. All study participants provided written informed consent for the sampling. If minors/children were enrolled in the study, written informed consent of the next to kin, caretakers, or guardians were obtained. Ethical clearance was obtained from the Ethical Committee of the Ministry of Health of the Republic of Madagascar.
Results
Screening results after broth enrichment
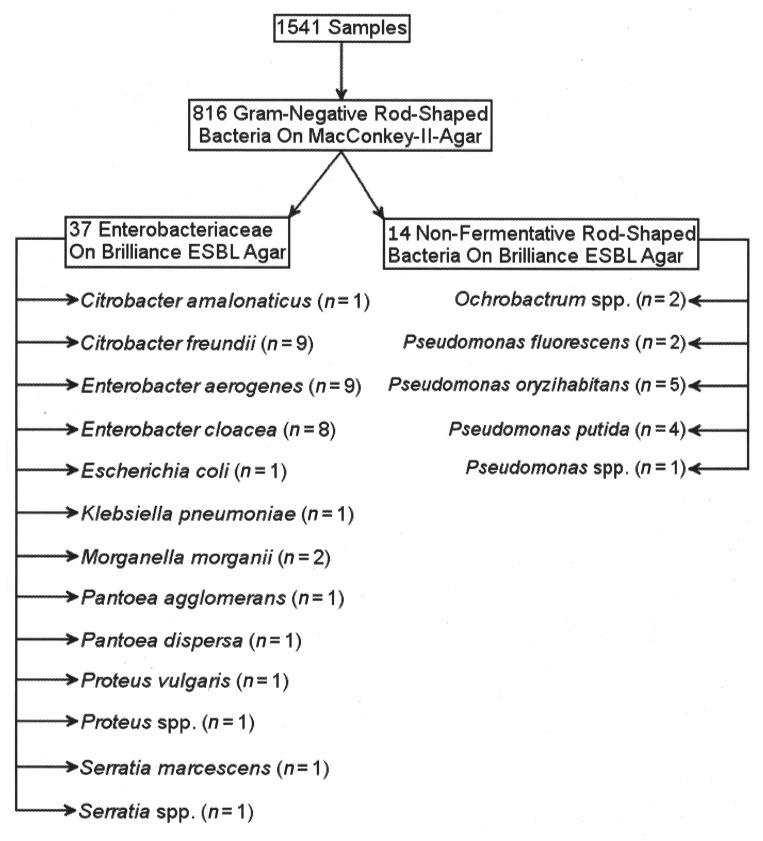
Of 1541 samples obtained from students and health care workers, Gram-negative rod-shaped bacteria grew on MacConkey II agar from 816 samples and enterobacterial growth was observed on Brilliance ESBL agar for 37 study participants. The Enterobacteriaceae detected on Brilliance ESBL agar comprised Citrobacter amalonaticus (n = 1), Citrobacter freundii (n = 9), Enterobacter aerogenes (n = 9), Enterobacter cloacae (n = 8), Escherichia coli (n = 1), Serratia marcescens (n = 1), Serratia spp. (n = 1), Klebsiella pneumoniae (n = 1), Morganella morganii (n = 2), Pantoea agglomerans (n = 1), Pantoea dispersa (n = 1), Proteus vulgaris (n = 1), and Proteus sp. (n = 1) as identified by 16S rRNA gene sequencing and MALDI–TOF–MS analysis. In addition, 14 out of 194 nonfermentative Gram-negative rod-shaped bacteria were representatively isolated and identified as Ochrobactrum spp. (n = 2), Pseudomonas fluorescens (n = 2), Pseudomonas oryzihabitans (n = 5), Pseudomonas putida (n = 4), and Pseudomonas sp. (n = 1) (Fig. 2).
Fig. 2.
Screening results of the students and health care workers group after broth enrichment and cultural growth on Brilliance ESBL agar
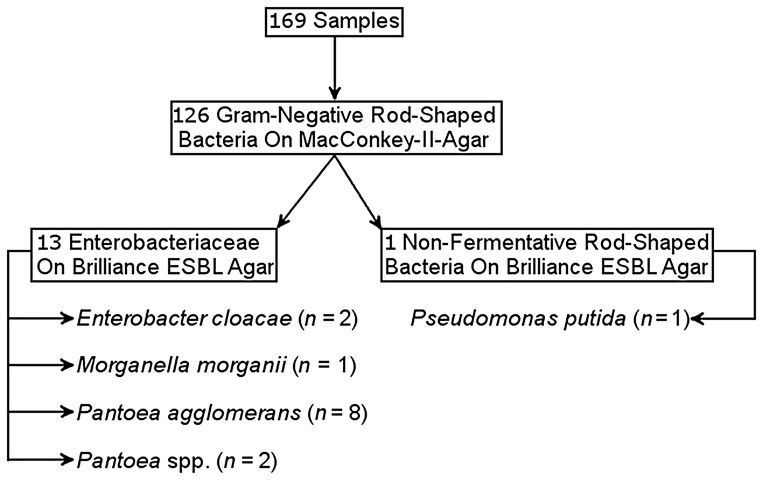
Of 169 samples from hospital patients obtained at admission, 126 showed Gram-negative rod-shaped bacteria growing on MacConkey II agar and 13 showed Enterobacteriaceae as well as 38 nonfermentative rod-shaped bacteria on Brilliance ESBL agar. The Enterobacteriaceae from Brilliance ESBL agar comprised Enterobacter cloacae (n = 2), Morganella morganii (n = 1), Pantoea agglomerans (n = 8), and Pantoea spp. (n = 2, further discrimination failed). Of the 38 Gram-negative nonfermentative rod-shaped bacteria on Brilliance ESBL, 1 was representatively isolated and identified as P. putida (Fig. 3).
Fig. 3.
Screening results of the patients group after broth enrichment and cultural growth on Brilliance ESBL agar
ESBL confirmation testing and resistance testing
ABCD testing by Mast ID D68C confirmed an ESBL phenotype in 3 out of 37 isolated Enterobacteriaceae from the students and health care workers group and in 11 out of 13 isolates from the patients group. In addition, an AmpC phenotype was identified in 21 Enterobacteriaceae from the students and health care workers group and in 1 isolate from the patients group.
BlaCTX-M expression was identified by commercial PCR in all 3 Enterobacteriaceae from the students and health care workers group for which ABCD testing had indicated an ESBL phenotype. For the patients group, blaCTX-M expression was confirmed in four out of 11 isolates with ESBL phenotype in ABCD testing. The consensus PCR, targeting blaTEM, blaSHV, blaCTX-M, and blaOXA-1, was positive in 6 out of 37 isolates from the students and health care workers group and in 12 out of 13 enterobacterial isolates from the patients group (Table 1).
Table 1.
Results of ABCD-testing and ESBL-PCR
| Species | Number of isolates | Results
of ABCD testing |
Results
of hyplex CTX-M-type β-lactamase PCR and consensus PCR
(targeting β-lactamase genes blaTEM,
blaSHV, blaCTX-M and
carbapenemase gene blaOXA-1) |
||
|---|---|---|---|---|---|
| Number of ESBL positive isolates | Number of AMPC positive isolates | Number of CTX-M positive Isolates | Number of consensus positive isolates | ||
| Students and health
care workers group | |||||
| Citrobacter amalonaticus | 1 | 0 | 0 | 0 | 0 |
| Citrobacter freundii | 9 | 0 | 5 | 0 | 3 |
| Enterobacter aerogenes | 9 | 0 | 7 | 0 | 0 |
| Enterobacter cloacae | 8 | 1 | 6 | 1 | 1 |
| Escherichia coli | 1 | 1 | 0 | 1 | 1 |
| Klebsiella pneumonia | 1 | 1 | 0 | 1 | 1 |
| Morganella morganii | 2 | 0 | 2 | 0 | 0 |
| Pantoea agglomerans | 1 | 0 | 0 | 0 | 0 |
| Pantoea dispersa | 1 | 0 | 0 | 0 | 0 |
| Proteus vulgaris | 1 | 0 | 0 | 0 | 0 |
| Proteus spp. | 1 | 0 | 0 | 0 | 0 |
| Serratia marcescens | 1 | 0 | 0 | 0 | 0 |
| Serratia spp. | 1 | 0 | 1 | 0 | 0 |
| Patients
group | |||||
| Enterobacter cloacae | 2 | 2 | 0 | 2 | 2 |
| Morganella morganii | 1 | 0 | 1 | 0 | 0 |
| Pantoea agglomerans | 8 | 7 | 0 | 2 | 8 |
| Pantoea spp. | 2 | 2 | 0 | 1 | 2 |
Multidrug-resistant Enterobacteriaceae were observed in four participants of the students and health care workers group and in 11 participants of the patients group, comprising two and ten ESBL-positive strains, respectively (Table 2). Multidrug-resistant strains were not observed among the 14 isolated nonfermentative Gram-negative rod-shaped bacteria from the students and health care workers group and the single isolate from the patients group.
Table 2.
Results of E-test-based resistance testing of multidrug-resistant isolates (n = 15) according to EUCAST interpretation guidelines
| Species | Piperacillin | Ceftazidim | Meropenem | Ciprofloxacin |
|---|---|---|---|---|
| Students and health
care workers group | ||||
| Citrobacter freundii | R | R | S | R |
| Enterobacter cloacae | R | R | S | R |
| Escherichia coli | R | R | S | R |
| Serratia marcescens | R | R | S | R |
| Patients
group | ||||
| Enterobacter cloacae | R | R | S | R |
| Enterobacter cloacae | R | R | S | R |
| Pantoea agglomerans | R | R | R | R |
| Pantoea agglomerans | R | R | S | R |
| Pantoea agglomerans | R | R | S | R |
| Pantoea agglomerans | R | R | S | R |
| Pantoea agglomerans | R | R | S | R |
| Pantoea agglomerans | R | R | S | R |
| Pantoea agglomerans | R | R | S | R |
| Pantoea agglomerans | R | R | S | R |
| Pantoea sp. | R | R | S | R |
| R, resistant; S, sensitive | ||||
A total of four isolates were tested intermediate sensitive or resistant to meropenem, comprising one ESBL-positive Pantoea agglomerans from the patients group and three nonfermentative Gram-negative rod-shaped bacteria. The latter comprised an intermediate sensitive Pseudomonas putida from the patients group and two intermediate sensitive Pseudomonas putida from the students and health care workers group. Commercial hyplex SuperBug ID PCR identified blaOXA-48 as a genetic determinant of carbapenem resistance in both intermediate sensitive Pseudomonas putida from the students and health care workers group.
Storage and transport stability
Among the samples from students and health care workers, 251 swabs were analyzed for blaCTX-M and other β-lactamase genes by PCR as well as cultural assessment. Both PCR approaches led to positive signals in just one sample, from which a blaCTX-M-positive Enterobacter cloacae could be grown on Brilliance ESBL agar and MacConkey II agar.
Among the 169 samples from patients, concordant culture results on Brilliant ESBL agar were observed in three instances. From all three swabs showing enterobacterial growth on Brilliance ESBL agar in Antananarivo, identical isolates were obtained by broth enrichment after transport of the swabs to Germany. Further, broth enrichment in Hamburg led to a total of ten additional Enterobacteriaceae on Brilliance ESBL agar that had not initially identified in Madagascar (Table 3).
Table 3.
Culture results on Brilliance ESBL agar directly in Madagascar and after broth enrichment in Germany
| Species | Growth
on ESBL agar in Antananarivo without broth-enrichment |
Growth
on ESBL agar in Germany after broth-enrichment |
|---|---|---|
| Enterobacter cloacae | 2 | 2 |
| Morganella morganii | 0 | 1 |
| Pantoea agglomerans | 1 | 8 |
| Pantoea spp. | 0 | 2 |
Risk factor analysis
The low number of ESBL-positive or multidrug resistant Enterobacteriaceae in the students and health care workers group (5 [0.3%]) did not allow for a sound statistical risk factor analysis. In the hospital patients group, study participants carrying ESBL-positive or multidrug-resistant Enterobacteriaceae were compared with participants who did not but who were nasal carriers of Gram-negative rod-shaped bacteria. Study participants resident in Antananarivo were less frequently colonized than patients from the surrounding rural areas; otherwise, there was no detectable difference between the groups (Table 4). Colonized patients were significantly younger (P = 0.0081, Mann–Whitney test; mean age ± standard deviation [SD] 26 ± 8.3 years) than noncolonized patients (mean age ± SD 37.9 ± 15.6 years).
Table 4.
Distribution of epidemiological risk factors among carriers and noncarriers of ESBL-positive strains and/or multidrug-resistant strains from the patients group in absolute numbers/total numbers of the group (and percent)
| Risk factors | Risk factor present | ESBL-positive or multidrug-resistant colonizing bacteria | RR (95% CI) |
|---|---|---|---|
| Number of patients with colonization | 12 | ||
| Female sex | Positive | 10.1% (7/69) | 2.0 |
| Negative | 5.3% (5/95) | (0.7–6.1) | |
| Beta-lactam antibiotic drugs during the last 6 months* | Positive | 4.9% (4/82) | 0.7 |
| Negative | 7.5% (5/67) | (0.2–2.3) | |
| Any antibiotic drugs during the last 6 months* | Positive | 6.6% (7/106) | 1.4 |
| Negative | 4.7% (2/43) | (0.3–6.6) | |
| Contact with animals | Positive | 9.5% (4/42) | 1.5 |
| Negative | 6.3% (8/127) | (0.5–4.8) | |
| Known chronic disease (not further specified) | Positive | 10.6% (5/47) | 1.9 |
| Negative | 5.7% (7/122) | (0.6–5.6) | |
| Residence in the capital Antananarivo | Positive | 3.5% (4/115) | 0.2 |
| Negative | 14.8% (8/54) | (0.1–0.8) | |
| Hospitalization within the last 14 days | Positive | 10.7% (3/28) | 1.7 |
| Negative | 6.4% (9/141) | (0.5–5.8) | |
| *Data were
extractable from 149 out of 169 questionnaires only RR, relative risk | |||
Discussion
The occurrence of ESBL-positive Enterobacteriaceae in Madagascan hospitals has been described repeatedly [1–4]. Here, we screened healthy volunteers and patients at admission for ESBL-positive or multidrug resistant enterobacterial colonization in the nasal vestibulum. The numbers of ESBL-positive or multidrug-resistant Enterobacteriaceae detected were respectively three and four out of 1541 samples from students and health care workers and 11 and 11 out of 169 patients. Cephalosporin-resistant Enterobacteriaceae were isolated from 37 students and health care workers and 13 hospital patients from ESBL selective agar.
Our data suggest that the colonization of healthy Madagascans with ESBL-positive or multidrug-resistant Enterobacteriaceae is low. Although the nose is not the primary site of human ESBL colonization, the observed high nasal colonization rate of 53.0% (816 out of 1541 samples) with Gram-negative rod-shaped bacteria is indicative of a low percentage of ESBL among colonizing Enterobacteriaceae in Madagascar. Of note, analysis of nasal ESBL colonization only makes sense if high rates of nasal carriage of Gram-negative rod-shaped bacteria are guaranteed. This has been described for countries with restricted sanitary hygiene [7]. The low number of carriers with ESBL-positive or multidrug-resistant colonization did not allow for a risk factor analysis in the students and health care workers group.
In contrast, a considerably higher rate of cephalosporin-resistant Gram-negative colonization was detected in hospital patients at admission. The quantitatively dominant Pantoea spp. are phylogenetically closely related to Enterobacter spp. [20] which are on rank eight of nosocomially transmitted patient isolates [21]. Pantoea spp. have been infrequently isolated in Madagascar [3] and can be associated with human disease and nosocomial spread [22–24].
Residence outside the capital Antananarivo represented a risk factor that was frequently observed in hospitalized carriers in association with ESBL-positive or multidrug-resistant colonization, and colonized patients were younger than noncolonized ones in this descriptive, hypothesis-forming study. The reasons remain speculative.
Our data from newly admitted patients suggest that a considerable proportion of cephalosporin-resistant colonization in Madagascan hospitals is not caused by nosocomial transmission. Rather, endogenous colonization is already present at the time of admission, reducing the effect of hygiene precautions for the prevention of infections due to resistant bacteria. Potential further spread of these agents should be kept in mind and monitored in future national surveillance programs.
Among the ESBL-positive Enterobacteriaceae, blaCTX-M was frequently observed in ESBL-positive strains. All ESBL-positive isolates from the students and health care workers group were blaCTX-M-positive; four out of 11 ESBL-positive strains were blaCTX-M-positive among the patients. These data are in line with previous investigations [4], which reported blaCTX-M as accounting for 75.5% of ESBL-positive Enterobacteriaceae in Antananarivo. Interestingly, the AmpC-resistance type accounted for a considerable proportion of observed cephalosporin resistance as well. These preliminary findings are of clinical importance regarding ceftriaxon being the antibiotic drug of choice for initial empirical antibiotic treatment in cases of suspected bacteremia or sepsis at the university hospital of Antananarivo. Furthermore, cultural demonstration of these life-threatening infections is not available for the majority of patients because of limited resources.
A considerable proportion (30% (15 out of 50)) of cephalosporin-resistant Enterobacteriaceae were multidrug-resistant, reducing the number of therapeutic options to fewer than two bactericidal antibiotic substance groups. While fluoroquinolone resistance was more frequently observed, lack of sensitivity to carbapenems was observed in only one enterobacterial isolate, a Pantoea agglomerans strain. Observed ESBL positivity of the respective strain and negative results in hypex SuperBug ID PCR might be compatible with porin loss or deficiency in this instance. Diagnostic assays to confirm this hypothesis are not established at our institutes. However, the absence of frequent carbapenemase genes in hypex SuperBug ID PCR makes it likely. In contrast, molecular characterization allowed the detection of carbapenem resistance in three out of 14 colonizing Gram-negative nonfermentative rod-shaped bacteria, two of them being Pseudomonas spp. harboring blaOxa-48.
As an interesting side effect, the study demonstrated a high degree of storage and transport stability of Gram-negative pathogens, facilitating future studies in resource-limited areas with transport of samples to well-equipped laboratories for further sample assessment, at least if broth enrichment can be provided.
Conclusions
The study showed a considerable proportion of nasal colonization with cephalosporin-resistant Enterobacteriaceae in Madagascan hospital patients at admission. Accordingly, the risk of endogenous infections due to such agents has to be considered. In contrast, the nasal colonization with cephalosporin-resistant or multidrug-resistant Enterobacteriaceae in the healthy Madagascan population is negligibly low.
Acknowledgements
The authors are grateful to Annett Michel and Steffen Lohr for excellent technical assistance. We also thank all participating health care workers and students as well as patients from the Befelatanana university hospital for taking part in the study.
Footnotes
Declaration of interest: The authors declare that there are no conflicts of interest. PCR analyses and bacterial culture were funded by the German Ministry of Defence (MoD), scientific project number 13K2-S-451215 “Development/evaluation of diagnostic molecular procedures for the diagnosis of infectious agents and symptom-based diagnostic procedures for tropical infectious diseases”. The funding source had no involvement in study design; in the collection, analysis, and interpretation of data; in the writing of the report; and in the decision to submit the article for publication.
Contributor Information
Volker Micheel, 1Department of Tropical Medicine at the Bernhard Nocht Institute, German Armed Forces Hospital of Hamburg, Hamburg, Germany.
Benedikt Hogan, 2Infectious Disease Epidemiology Department, Bernhard Nocht Institute for Tropical Medicine Hamburg, Hamburg, Germany.
Rivo Andry Rakotoarivelo, 3Infectious Disease Department, University Hospital Joseph Raseta de Befelatanana, Antananarivo, Madagascar.
Raphael Rakotozandrindrainy, 4Department of Microbiology and Parasitology, University of Antananarivo, Antananarivo, Madagascar.
Fetra Razafimanatsoa, 5Laboratory Department, University Hospital Joseph Raseta de Befelatanana, Antananarivo, Madagascar.
Tsiriniaina Razafindrabe, 4Department of Microbiology and Parasitology, University of Antananarivo, Antananarivo, Madagascar.
Jean Philibert Rakotondrainiarivelo, 4Department of Microbiology and Parasitology, University of Antananarivo, Antananarivo, Madagascar.
Sabine Crusius, 6Institute for Microbiology, Virology and Hygiene, University Hospital Rostock, Rostock, Germany.
Sven Poppert, 7Institute for Medical Microbiology, Justus-Liebig-University Giessen, Giessen, Germany.
Norbert Georg Schwarz, 2Infectious Disease Epidemiology Department, Bernhard Nocht Institute for Tropical Medicine Hamburg, Hamburg, Germany.
Jürgen May, 2Infectious Disease Epidemiology Department, Bernhard Nocht Institute for Tropical Medicine Hamburg, Hamburg, Germany.
Hagen Frickmann, 1Department of Tropical Medicine at the Bernhard Nocht Institute, German Armed Forces Hospital of Hamburg, Hamburg, Germany; 6Institute for Microbiology, Virology and Hygiene, University Hospital Rostock, Rostock, Germany.
Ralf Matthias Hagen, 1Department of Tropical Medicine at the Bernhard Nocht Institute, German Armed Forces Hospital of Hamburg, Hamburg, Germany.
References
- 1.Andriatahina T, Randrianirina F, Hariniana ER, Talarmin A, Raobijaona H, Buisson Y, Richard V. High prevalence of fecal carriage of extended-spectrum beta-lactamaseproducing Escherichia coli and Klebsiella pneumoniae in a pediatric unit in Madagascar. BMC Infect Dis. 2010;10:204. doi: 10.1186/1471-2334-10-204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Randrianirina F, Vaillant L, Ramarokoto CE, Adriamanarivo ML, Razafimahandry Hc, Randiranomenjanahary J, Raveloson JR, Hariniana ER, Carod JF, Talarmin A, Richard V. Antimicrobial resistance in pathogens causing nosocomial infections in surgery and intensive care units of two hospitals in Antananarivo, Madagascar. J Infect Dev Ctries. 2010;4:74–82. doi: 10.3855/jidc.454. [DOI] [PubMed] [Google Scholar]
- 3.Herindrainy P, Randrianirina F, Ratovoson R, Ratsima Hariniana E, Buisson Y, Genel N, Decré D, Arlet G, Talarmin A, Richard V. Rectal carriage of extended-spectrum beta-lactamase-producing gram-negative bacilli in community settings in Madagascar. PLoS One. 2011;6:e22738. doi: 10.1371/journal.pone.0022738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rakotonirina HC, Garin B, Randrianirina F, Richard V, Talarmin A, Arlet G. Molecular characterization of multid-rug-resistant extended-spectrum β-lactamase-producing Enterobacteriaceae isolated in Antananarivo, Madagascar. BMC Microbiol. 2013;13:85. doi: 10.1186/1471-2180-13-85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Moland ES, Hanson ND, Black JA, Hossain A, Song W, Thomson KS. Prevalence of newer beta-lactamases in Gram-negative clinical isolates collected in the United States from 2001 to 2002. J Clin Microbiol. 2006;44:3318–3324. doi: 10.1128/JCM.00756-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pitout JDD, Hossain A, Hanson ND. Phenotypic and molecular detection of CTX-M-beta-lactamases produced by Escherichia coli and Klebsiella spp. J Clin Microbiol. 2004;42:5715–5721. doi: 10.1128/JCM.42.12.5715-5721.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Farida H, Severin JA, Gasem MH, Keuter M, van den Broek P, Hermans PW, Wahyono H, Verbrugh HA. Nasopharyngeal carriage of Klebsiella pneumoniae and other gram-negative bacilli in pneumonia-prone age groups in Semarang, Indonesia. J Clin Microbiol. 2013;51:1614–1616. doi: 10.1128/JCM.00589-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Friedmann R, Raveh D, Zartzer E, Rudensky B, Broide E, Attias D, Yinnon AM. Prospective evaluation of colonization with extended-spectrum beta-lactamase (ESBL)-producing Enterobacteriaceae among patients at hospital admission and of subsequent colonization with ESBL-pro ducing Enterobacteriaceae among patients during hospitalization. Infect Control Hosp Epidemiol. 2009;30:534–543. doi: 10.1086/597505. [DOI] [PubMed] [Google Scholar]
- 9.Murk JL, Heddema ER, Hess DLJ, Boogards JA, Vandenbroucke-Grauls CM, Debets-Ossenkopp YJ. Enrichment broth improved detection of extended-spectrumbeta-lactamase-producing bacteria in throat and rectal surveillance cultures of samples from patients in intensive care units. J Clin Microbiol. 2009;47:1885–1887. doi: 10.1128/JCM.01406-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Huang TD, Bogaerts P, Berhin C, Guisset A, Glupczinsky Y. Evaluation of Brilliance ESBL agar, a novel chromogenic medium for detection of extended-spectrum-beta-lactamase-producing Enterobacteriaceae. J Clin Microbiol. 2010;48:2091–2096. doi: 10.1128/JCM.02342-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ongut G, Daloglu AE, Bayson BO, Daglar D, Ogunc D, Sekercioglu AO, Colak D, Gunseren F. Evaluation of a chromogenic medium for detection of extended-spectrumbeta-lactamase-producing Escherichia coli and Klebsiella pneumoniae strains. Clin Lab. 2014;60:1213–1215. doi: 10.7754/clin.lab.2013.130812. [DOI] [PubMed] [Google Scholar]
- 12.Cilia V, Lafay B, Christen R. Sequence heterogeneities among 16S ribosomal RNA sequences and their effect on phylogenetic analyses at the species level. Mol Biol Evol. 1996;13:451–461. doi: 10.1093/oxfordjournals.molbev.a025606. [DOI] [PubMed] [Google Scholar]
- 13.Hagen RM, Frickmann H, Elschner M, Melzer F, Neubauer H, Gauthier YP, Racz P, Poppert S. Rapid identification of Burkholderia pseudomallei and Burkholderia mallei by fluorescence in situ hybridization (FISH) from culture and paraffin-embedded tissue samples. Int J Med Microbiol. 2011;301:585–590. doi: 10.1016/j.ijmm.2011.04.017. [DOI] [PubMed] [Google Scholar]
- 14.Frickmann H, Christner M, Donat M, Berger A, Essig A, Podbielski A, Hagen RM, Poppert S. Rapid discrimination of Haemophilus influenzae, H. parainfluenzae, and H. haemolyticus by fluorescence in situ hybridization (FISH) and two matrix-assisted laser-desorption-ionization time-of-flight mass spectrometry (MALDI–TOF–MS) platforms. PLoS One. 2013;8:e63222. doi: 10.1371/journal.pone.0063222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Clinical and Laboratory Standards Institute. Interpretive criteria for identification of bacteria and fungi by DNA target sequencing. 1. Wayne, PA: Clinical and Laboratory Standards Institute; 2008. pp. 30–34. [Google Scholar]
- 16.Justesen US, Skov MN, Knudsen E, Holt HM, Sogaard P, Justesen T. 16S rRNA gene sequencing in routine identification of anaerobic bacteria isolated from blood cultures. J Clin Microbiol. 2010;48:946–948. doi: 10.1128/JCM.02075-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ingram PR, Inglis TJ, Vanzetti TR, Henderson BA, Harnett GB, Murray RJ. Comparison of methods for AmpC betalactamase detection in Enterobacteriaceae. J Med Microbiol. 2011;60:715–721. doi: 10.1099/jmm.0.029140-0. [DOI] [PubMed] [Google Scholar]
- 18.Mattner F, Bange FC, Meyer E, Seifert H, Wichelhaus TA, Chaberny IF. Preventing the spread of multidrug-resistant gram-negative pathogens: recommendations of an expert panel of the German Society for Hygiene and Microbiology. Dtsch Arztebl Int. 2012;109:39–45. doi: 10.3238/arztebl.2012.0039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kaase M, Szabados F, Wassill L, Gatermann SG. Detection of carbapenemases in Enterobacteriaceae by a commercial multiplex PCR. J Clin Microbiol. 2012;50:3115–3118. doi: 10.1128/JCM.00991-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Delétoile A, Decré D, Courant S, Passet V, Audo J, Grimont P, Arlet G, Brisse S. Phylogeny and identification of Pantoea species and typing of Pantoea agglomerans strains by multilocus gene sequencing. J Clin Microbiol. 2009;47:300–310. doi: 10.1128/JCM.01916-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sievert DM, Ricks P, Edwards JR, Schneider A, Patel J, Srinivasan A, Kallen A, Limbago B, Fridkin S National Healthcare Safety Network NHSN Team and Participating NHSN Facilities. Antimicrobial-resistant pathogens associated with healthcare-associated infections: summary of data reported to the National Healthcare Safety Network at the Centers for Disease Control and Prevention, 2009–2010. Infect Control Hosp Epidemiol. 2013;34:1–14. doi: 10.1086/668770. [DOI] [PubMed] [Google Scholar]
- 22.Bicudo EL, Macedo VO, Carrara MA, Castro FF, Rage RI. Nosocomial outbreak of Pantoea agglomerans in a pediatric urgent care center. Braz J Infect Dis. 2007;11:281–284. doi: 10.1590/s1413-86702007000200023. [DOI] [PubMed] [Google Scholar]
- 23.Cruz AT, Cazacu AC, Allen CH. Pantoea agglomerans, a plant pathogen causing human disease. J Clin Microbiol. 2007;45:1989–1992. doi: 10.1128/JCM.00632-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Liberto MC, Matera G, Puccio R, Lo Russo T, Colosimo E, Focà E. Six cases of sepsis caused by Pantoea agglomerans in a teaching hospital. New Microbiol. 2009;32:119–123. [PubMed] [Google Scholar]