Abstract
Background
Cytidine and uridine are produced commercially by Bacillus subtilis. The production strains of cytidine and uridine were both derivatives from mutagenesis. However, the exact metabolic and genetic factors affecting the productivity remain unknown. Genetic engineering may be a promising approach to identify and confirm these factors.
Results
With the deletion of the cdd and hom genes, and the deregulation of the pyr operon in Bacillus subtilis168, the engineered strain produced 200.9 mg/L cytidine, 14.9 mg/L uridine and 960.1 mg/L uracil. Then, the overexpressed prs gene led to a dramatic increase of uridine by 25.9 times along with a modest increase of cytidine. Furthermore, the overexpressed pyrG gene improved the production of cytidine, uridine and uracil by 259.5%, 11.2% and 68.8%, respectively. Moreover, the overexpression of the pyrH gene increasesd the yield of cytidine by 40%, along with a modest augments of uridine and uracil. Lastly, the deletion of the nupC-pdp gene resulted in a doubled production of uridine up to 1684.6 mg/L, a 14.4% increase of cytidine to 1423 mg/L, and a 99% decrease of uracil to only 14.2 mg/L.
Conclusions
The deregulation of the pyr operon and the overexpression of the prs, pyrG and pyrH genes all contribute to the accumulation of pyrimidine nucleoside compounds in the medium. Among these factors, the overexpression of the pyrG and pyrH genes can particularly facilitate the production of cytidine. Meanwhile, the deletion of the nupC-pdp gene can obviously reduce the production of uracil and simultaneously improve the production of uridine.
Electronic supplementary material
The online version of this article (doi:10.1186/s12934-015-0237-1) contains supplementary material, which is available to authorized users.
Keywords: Bacillus subtilis, Cytidine, Uridine, Gene deletion, Gene expression, Pyrimidine nucleotide biosynthesis
Introduction
Bacillus subtilis is able to synthesize the uridine 5'-monophosphate (UMP) de novo. The excess UMP can be further converted to terminal metabolites (cytidine, uridine and uracil), which could then be secreted out of the cell (Figure 1). The pyrimidine nucleotide biosynthetic (pyr) operon of B. subtilis contains 10 genes. The first gene of the pyr operon encodes a bifunctional protein PyrR which is the regulator protein for pyr operon and a uracil phosphoribosyl transferase [1]. The second gene, pyrP, encodes a uracil permease. The remaining eight genes encode the six enzymes involved in the de novo biosynthesis of UMP [2]. The expression of the pyr operon is regulated by transcriptional attenuation mechanism involving PyrR. The PyrR protein is mainly activated by UMP and the regulating elements are three specific binding loops (BL1, BL2 and BL3) on the nascent pyr mRNA. The combination of the PyrR protein and BLs disrupts the antiterminator, permitting the formation of terminator hairpin and leading to the reduced expression of the downstream genes [3,4]. The resulting high intracellular concentration of UMP would strongly inhibit the transcription of the pyr operon.
Figure 1.

The biosynthetic pathway of pyrimidine nucleotide in B. subtilis.
In B. subtilis, L-aspartate is the precursor of both amino acids (lysine, methionine, threonine, and isoleucine) and pyrimidine nucleotide biosynthesis (Figure 1). The deficiency of homoserine dehydrogenase (encoded by the hom gene) can prevent the aspartate entering to the methionine and threonine biosynthesis and improve the supplement to the de novo biosynthesis of UMP [5]. Phosphoribosyl pyrophosphate (PRPP) is another important precursor of pyrimidine nucleotide biosynthesis (Figure 1). Phosphoribosyl pyrophosphate synthetase (PRS) is encoded by the prs gene of the gcaD-prs-ctc operon whose transcription regulation mechanism has been unknown yet [6]. Improving the expression level of the prs gene can significantly increase the intracellular pool of PRPP [7,8].
In B. subtilis, UMP kinase, which is encoded by the pyrH gene, catalyzes the phosphorylation of UMP by ATP to yield UDP and ADP (Figure 1). The UMP kinase activity is regulated allosterically by GTP (activator) and UTP (inhibitor) [9,10]. The pyrG gene encodes CTP synthetase which aminates UTP to form CTP (Figure 1). The transcription of the pyrG gene is tightly regulated by a CTP-sensitive reiterative transcription attenuation control mechanism [11-13]. The inherent regulation of the pyrG gene transcription and CTP synthetase activity limit the excessive synthesis of CTP in B. subtilis.
Catalyzed by non-specific 5′-phosphatase, the excessive intracellular CMP and UMP can be dephosphorylated to form cytidine and uridine, respectively. The cytidine can be further deaminized to form uridine by cytidine deaminase (encoded by the cdd gene) (Figure 1) [14]. In the dra-nupC-pdp operon, the pdp gene encodes pyrimidine nucleoside phosphorylase which catalyzes the degradation of uridine to form uracil and D-ribose-1-phosphate [15]. Then, uracil will be secreted out of the cell. Therefore, the terminal metabolites of pyrimidine nucleotide are generally uracil instead of cytidine and uridine in the wild-type B. subtilis. The dra gene encodes deoxyriboaldolase and the nupC gene encodes the transporter responsible for pyrimidine nucleoside uptake. The expression of the dra-nupC-pdp operon is repressed by glucose and induced by deoxyribonucleosides and deoxyribose [16,17]. When the carbon source is poor, by the physiological function of the dra-nupC-pdp operon, the pyrimidine nucleoside accumulated in the medium could be recycled as carbon source. Obviously, the physiological functions of nupC and pdp genes make no contribution to the accumulation of pyrimidine nucleoside in the medium.
B. subtilis strains with defect and pyrimidine analogue resistant could accumulate large amounts of cytidine or uridine [18-21]. Nevertheless, the exact genetic and metabolic mechanisms resulting in pyrimidine nucleoside over-production have not been fully identified and confirmed.
In this study, by using genetic manipulation method, we modified some key genes and operons related to the pyrimidine nucleotide biosynthesis in B. subtilis 168 and investigated the influence of these modifications on the production of pyrimidine nucleoside compounds.
Results
Deletion of the cdd and hom genes
In order to observe the effects of the related genetic modification on cytidine and uridine synthesis separately, we blocked the reaction from cytidine to uridine by deleting 151 bp coding sequences of the cdd gene in B. subtilis 168 N and obtained the cdd gene deficient strain B. subtilis TD01 (Additional file 1: Figure S1). Subsequently, in order to improve the supplement of aspartate for the pyrimidine nucleotide biosynthesis, we deleted 827 bp coding sequences of the hom gene in the strain TD01 and obtained the strain B. subtilis TD02 (Additional file 1: Figure S2). The shake-flask culture experiments demonstrated that the strain TD02 could accumulate cytidine and uracil in detectable level in medium while no uridine was detected (Figure 2).
Figure 2.

Pyrimidine and pyrimidine nucleoside accumulation by B. subtilis strains after 72 h fermentation. Results are the average of three replicates with error bars indicating standard error from the mean (TD02, TD12, TD13, TD231, TD232 and TD33).
Deregulation of the pyr operon
As described before, the transcription regulation mechanism of the pyr operon restricted the over-synthesis of UMP and its derivatives. We deleted 738 bp coding sequences of the pyrR gene in the strain TD02 and constructed the recombinant B. subtilis TD12 (Additional file 1: Figure S3). The pyr operon mRNA transcription level in the ΔpyrR strain TD12 was compared with parent strain TD02 through RT-qPCR analysis. We chose the sequences which lay in the middle of the pyr operon as the detective point. The transcript abundance increased 6.28-fold in TD12 (Figure 3), which indicated that the pyr operon was deregulated in ΔpyrR strain. The shake-flask culture experiments showed that the strain TD12 could accumulate 200.9 ± 8.3 mg/L cytidine, 14.9 ± 0.8 mg/L uridine and 960.1 ± 39.1 mg/L uracil, respectively (Figure 2). The deregulation of the pyr operon expression could significantly increase the accumulation of both cytidine and uracil, and have little effect on the accumulation of uridine in the medium.
Figure 3.

Relative transcriptional levels of mRNA in the recombinant strains. The mRNA expression levels of WT strain is regarded as 1(blank). The relative mRNA expression levels of recombinant strains (filled) are compared with WT strain. (A) Relative transcriptional levels of the pyrR gene. ΔpyrR: the pyrR gene was deleted. (B) Relative transcriptional levels of the prs gene. prs +: the prs gene was overexpressed. (C) Relative transcriptional levels of the pyrG gene. pyrG +: the pyrG gene was constitutively expressed. pyrG +*: the pyrG gene was overexpressed. (D) Relative transcriptional levels of the pyrH gene. pyrH +: the pyrH gene was overexpressed. The ccpA gene was used as the internal control gene to normalize the results. Results are the average of three replications with error bars indicating standard error from the mean.
Overexpression of the prs gene
PRPP is not only a precursor for UMP de novo biosynthesis but also locates in the crossing point in multiple metabolic pathways. To illustrate the impact of intracellular PRPP level on the biosynthesis of UMP and its derivatives, we overexpressed the prs gene in TD12 and constructed recombinant strain B. subtilis TD13. The RT-qPCR analyses showed that a 72.86-fold improvement of the prs mRNA transcription level occurred in recombinant TD13 compared with parental strain TD12, which indicated that the prs gene was successful overexpressed (Figure 3). The flask culture revealed that the recombinant strain TD13 accumulated 247.2 ± 9.6 mg/L cytidine, 440.7 ± 17.1 mg/L uridine and 935.7 ± 31.9 mg/L uricil in medium, respectively (Figure 2). Compared with strain TD12, the accumulation of cytidine and uracil of strain TD13 slightly rose or dropped, but the accumulation of uridine significantly increased by 25.9-fold. These results demonstrated that pyrimidine nucleotide precursor can be further increased by overexpressing the prs gene, and intracellular PRPP level was an important rate-limiting factor in biosynthesis of UMP and its derivatives. In other words, the improvement of PRPP supplement could increase the synthesis of pyrimidine nucleoside which was mainly reflected in the improvement of uridine.
Overexpression of the pyrG gene
The CTP synthetase (encoded by the pyrG gene) catalyzes the reaction from UTP to CTP. In order to investigate the effects of CTP synthetase level on pyrimidine nucleoside biosynthesis, we constructed the recombinant B. subtilis TD231 by introducing 4 extra G residues at the 5′ ends of the pyrG transcripts based on the strain TD13 [13]. Meanwhile, on the basis of strain TD13, we constructed recombinant B. subtilis TD232 by repleacing the ITR of the pyrG gene with the gsiB mRNA stabilizer and made the promoter of the pyrG gene consensus (Figure 4). The RT-qPCR analysis showed that the intracellular pyrG mRNA level of recombinant TD231 and TD232 were about 325-fold and 675-fold, respectively, in comparison to the parent strain TD13 containing the pyrG gene (Figure 3). In the flask fermentation medium, the TD231 and TD232 could accumulate 301.3 ± 13.7 mg/L and 888.7 ± 25.7 mg/L cytidine, which achieved a 21.9% and a 259.5% improvement compared to the pyrG gene WT strain TD13, respectively (Figure 2). Meanwhile, the uracil production increased about 33% (1244.9 ± 10.9 mg/L) and 68.8% (1579 ± 49.8 mg/L), respectively, while the uridine production only increased ~7.9% (474.6 ± 11.8 mg/L) and ~11.2% (490.1 ± 19.1 mg/L), respectively (Figure 2). These results revealed that the improvement of CTP synthetase level could not only contribute to the production of the downstream metabolites of CTP (known as cytidine), but also those of UMP (known as uracil & uridine). This may be explained by that the improvement of metabolic flux to cytidine led to the reduction of the intracellular UMP pool and subsequently the UMP synthesis was released from UMP-sensitive feedback inhibition to some extent, so that UMP synthesis increased in actually and a part of the increased UMP contributed to the elevation of uridine/uracil production.
Figure 4.

Nucleotide sequence of PSB expression cassette. The sequence of the nontemplate strand is shown. The −10 and −35 regions of the promoter, Shine–Dalgarno sequence (SD) and initiation codon are bold and underlined. The RNA-stabilizing elements are shown in italics.
Overexpression of the pyrH gene
Among the reactions from UMP to cytidine, UMP kinase (encoded by the pyrH gene) might serve as a rate-limiting factor. In order to confirm this, we overexpressed the pyrH gene in strain TD232 and obtained the recombinant B. subtilis TD33. The RT-qPCR analyses showed that the pyrH mRNA level of TD33 was about 3.75-fold higher than that of strain TD232 of which the pyrH gene was wild-type (Figure 3). The shake flask fermentation experiments showed that the recombinant TD33 accumulated 1244.2 ± 53.9 mg/L cytidine, 558.6 ± 24.2 mg/L uridine and 1696.7 ± 39.7 mg/L uracil in the culture broth and increased by 40%, 14% and 7.5%, respectively, compared with strain TD232 (Figure 2). The production of cytidine, uridine and uracil all increased with the improvement of UMP kinase express level. These results proved that UMP kinase express level was also a rate-determining factor of pyrimidine nucleoside compounds production.
Disruption of the nupC-pdp gene
To ascertain the effect of the nupC-pdp gene on pyrimidine nucleoside accumulation in B. subtilis, the nupC-pdp gene was deleted from the chromosome of strains TD12, TD13 and TD33, respectively (Additional file 1: Figure S4). The corresponding derivative strains were named as B. subtilis TD12np, TD13np and TD33np, respectively. The culture broth analyses of the six strains were shown in Table 1. In the strains (TD13np and TD33np) with the prs gene overexpressed, the knockout of the nupC-pdp gene led to a decrease of the uracil yield by 99% on average, while an increase of cytidine and uridine yield by 23% and 230% on average, respectively. In strain TD12 (the prs gene is wild-type), the knockout of the nupC-pdp gene led to a decreased accumulation of uracil by about 50%, while an increased accumulation of cytidine and uridine by about 0.3-fold and 79-fold, respectively. These results indicated that the accumulated uracil in medium was mainly derived from the degradation of uridine catalyzed by pyrimidine nucleoside phosphorylase. Meanwhile, there may be another metabolic reaction remains unknown, which could generate uracil in B. subtlis, and the rate of the unknown reaction may be controlled by intracellular PRPP pool level.
Table 1.
Pyrimidine and pyrimidine nucleoside produced by modifying different genes of the pyrimidine nucleotide biosynthesis pathway in B. subtilis after 72 h fermentation
| Strain | Productivity (mg/L) | Ratio (%) | Productivity (mg/L) | Ratio (%) | Productivity (mg/L) | Ratio (%) | Molarity (mmol/L) |
|---|---|---|---|---|---|---|---|
| Cytidine | Uridine | Uracil | Total | ||||
| TD12 | 200.9 ± 8.3 | 14.9 ± 0.8 | 960.1 ± 39.1 | 9.5 | |||
| TD12np | 260.3 ± 11.3 | 129.6 | 1187.5 ± 49.9 | 7970 | 508.2 ± 18.5 | 52.9 | 10.5 |
| TD13 | 247.2 ± 9.6 | 440.7 ± 17.1 | 935.7 ± 31.9 | 11.2 | |||
| TD13np | 326.9 ± 7.0 | 132.2 | 1571.4 ± 38.9 | 356.6 | 16.9 ± 0.1 | 1.8 | 7.9 |
| TD33 | 1244.2 ± 53.9 | 558.6 ± 24.2 | 1696.7 ± 39.6 | 22.6 | |||
| TD33np | 1423.0 ± 47.5 | 114.4 | 1684.6 ± 62.4 | 301.6 | 14.2 ± 0.2 | 0.8 | 22.9 |
Data shown are the average of three independent experiments.
Together with the inactivation of the cdd, hom and pyrR genes, the deletion of nupC-pdp gene led to a slight increase of the total amount of pyrimidine compounds (i.e., cytidine, uridine, and uracil) accumulated in the culture. However, together with the inactivation of the cdd, hom and pyrR genes and the overexpression of the prs gene, the deletion of the nupC-pdp gene led to a modest decrease of the total amount of pyrimidine compounds in the culture. Nevertheless, if the pyrH and pyrG genes were both overexpressed subsequently, the total amount of pyrimidine compounds in the culture would be doubled to about 22 mmol/L (Table 1). These results revealed that the overexpression of the pyrH and pyrG genes caused a metabolic flux enlargement from UMP to cytidine, thereby, reducing the intracellular UMP level which may relieve feedback inhibition for UMP to carbamyl phosphate synthetase and lead to more UMP synthesis.
The growth of recombinant strains
In shaking flask fermentation, we measured the biomass of the recombinant strains mentioned above. The data showed that the cell growth of recombinant strains were similar to that of the parental strains (Figure 5). Therefore, the above modification of relevant genes of pyrimidine nucleotide biosynthesis had no detectable effect on cell growth.
Figure 5.
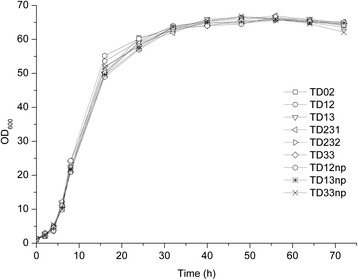
The biomass of recombinant strains in 72 h fermentation. Data obtained are the result of three independent fermentations.
Discussion
As already shown in the “introduction”, the pyrimidine nucleotide biosynthesis of B. subtilis was strictly regulated so that no excess pyrimidine nucleoside would be synthesized and secreted to the medium. In order to well illuminate the rate-determining factors affecting pyrimidine nucleoside excess synthesis, we chose genetic engineering as a desired strategy.
The inactivation of the cdd gene abolished the reaction from cytidine to uridine and resulted in the accumulation of cytidine in the medium, indicating that the deficiency of the cdd gene is a key factor of the accumulation of cytidine. The result coincided with the experimental findings of previous studies [22].
Based on the hom and cdd genes deficiency, the deregulation of the pyr operon by deleting the pyrR gene doubled the yield of both cytidine and uracil, while mutagenesis was the mere prevailing method to achieve the same genetic effect in the past [21]. However, though one determining factor of UMP biosynthesis have been revealed with deregulation of the pyr operon, the carbamoyl phosphate synthetase (encoded by the pyrAA/pyrAB gene), which catalyzes the first reaction of UMP de novo biosynthesis, is still subjected to feedback inhibition by UMP [23,24]. Hence, the excess synthesis of pyrimidine nucleoside compounds is still considered to be limited.
The overexpression of the prs gene resulted in an increase of uridine yield by 25.9-fold in this work. Analogous experimental results also existed in the study of biosynthesis of purine nucleoside and its derivatives in B. subtilis [8,25]. Thus, PRPP pool level is one important restrictive factor affecting the overproduction of nucleotide and its derivatives. However, PRS is an allosteric enzyme and its activity is intensively feedback inhibited by purine nucleotide [26]. By using site-directed mutation to release the feedback inhibition of phosphoribosyl pyrophosphate synthetase (PRS), satisfactory results may be obtained.
The overexpression of the pyrG and pyrH genes resulted in a significantly increased amount of cytidine in the medium, proving that the activity of both UMP kinase and CTP synthetase were rate-determining factors for cytidine over-synthesis. Since both UMP kinase and CTP synthetase are allosteric enzymes, it is believed that by releasing their feedback inhibition, the cytidine production could be further improved. The overexpression of the pyrG and pyrH genes also led to an increase in the total pyrimidine compounds accumulation in the medium. We interpret that the augmented metabolic flux from UMP to CTP could reduce the intracellular UMP level and thereby release the feedback inhibition on UMP biosynthesis, especially the activity of carbamyl phosphate synthetase.
The pyrimidine nucleoside phosphorylase (encoded by the pdp gene) involves in the reaction from uridine to uracil and the loss of the pdp gene should make cell accumulate only nucleosides and no uracil in theory [27,28]. However, according to the experimental results, by deleting the nupC-pdp gene, we obtained that the uracil accumulation reduced by 99% rather than 100%. The small amount of uracil remaining in the medium likely resulted from some unspecific reactions. Since the decrease of uracil yield was always accompanied with a remarkable increase of uridine yield, the uracil accumulated in the medium was mainly derived from uridine or UMP. Under the precondition of the pdp gene deficiency, the amount of residual uracil in the medium seems to be negatively correlated with the amount of the available PRPP in the cell (TD12np). Hence, the unspecific reactions which result in the residual uracil are likely subject to the level of intercellular PRPP pool.
Conclusions
We deleted/overexpressed the genes which were closely related to pyrimidine nucleoside biosynthesis by using genetic manipulation method in B. subtilis168 and constructed a series of recombinant strains. The results of shaking flask fermentation demonstrated that the deregulation of pyr operon, the overexpression of the prs, pyrG and pyrH genes, and the deletion of the nupC-pdp gene all facilitated the over-synthesis of pyrimidine nucleoside compounds. The production of uridine and cytidine were up to 1684.6 mg/L and 1423 mg/L in the result strain TD33np, respectively. (i) The UMP synthesis operon (pyr operon) and the PRPP synthesis (encoded by the prs gene) are both rate-determining factors for the UMP biosynthesis. If the feedback inhibitions of PRPP synthetase and carbamoyl phosphate synthetaserelease are released, the production of pyrimidine nucleoside compounds may be improved further. (ii) The overexpression of the pyrH and pyrG genes can improve the proportion of cytidine in the pyrimidine nucleoside products, while reduce the proportion of uridine. If the feedback inhibition of the CTP synthetase and UMP kinase are released, the proportion of cytidine in the pyrimidine nucleoside products may be improved further. (iii) The pyrimidine nucleoside phosphorylase (encoded by the pdp gene) activity is of close correlation with the accumulation of uracil in the medium, and the deletion of the nupC-pdp gene can reduce the accumulation of uracil to a very low level (1%) in the medium. (iv) The cdd gene is a key factor of the accumulation of cytidine, and if cytidine deaminase activity is restored, the proportion of uridine in the accumulated pyrimidine compounds will increase sharply.
Materials and methods
The bacterial strains and general culture conditions
The strains used in this study are listed in Table 2. All organisms were cultured in Luria-Bertani (LB) medium or on 1.5% (w/v) agar plates supplemented with the appropriate antibiotics when required. Chloramphenicols (Cm), 6 μg ml−1and neomycin (Nm), 15 μg ml−1, were used to select resistant B. subtilis cells. For liquid cultures, B. subtilis were incubated at 37°C and were shaken at 220 rpm.
Table 2.
Strains
| Strain | Relevant genotype | Reference |
|---|---|---|
| B.subtilis 168 (BGSC 1A1) | trpC2 | Laboratory stock |
| B.subtilis 168 N | trpC2, ΔaraR::neo R | Laboratory stock |
| B.subtilis TD01 | trpC2, ΔaraR::neo R ,Δcdd | This study |
| B.subtilis TD02 | trpC2, ΔaraR::neo R , Δcdd, Δhom | This study |
| B.subtilis TD12 | trpC2,ΔaraR::neo R , Δcdd, Δhom, ΔpyrR | This study |
| B.subtilis TD12np | trpC2,ΔaraR::neo R , Δcdd, Δhom, ΔpyrR, ΔnupC-pdp | This study |
| B.subtilis TD13 | trpC2,ΔaraR::neo R , Δcdd, Δhom,ΔpyrR, ΔxylR::prs | This study |
| B.subtilis TD13np | trpC2,ΔaraR::neo R , Δcdd, Δhom,ΔpyrR, ΔxylR::prs, ΔnupC-pdp | This study |
| B.subtilis TD231 | trpC2,ΔaraR::neo R , Δcdd, Δhom, ΔpyrR, ΔxylR::prs, pyrG + | This study |
| B.subtilis TD232 | trpC2,ΔaraR::neo R , Δcdd, Δhom, ΔpyrR,ΔxylR::prs, pyrG +* | This study |
| B.subtilis TD33 | trpC2,ΔaraR::neo R , Δcdd, Δhom, ΔpyrR, ΔxylR::prs,pyrG +* ,pyrH + | This study |
| B.subtilis TD33np | trpC2,ΔaraR::neo R , Δcdd, Δhom, ΔpyrR,ΔxylR::prs,pyrG +* ,pyrH + , ΔnupC-pdp | This study |
pyrG +: the pyrG gene was constitutively expressed. pyrG +*: the pyrG gene was overexpressed.
DNA manipulation techniques and PCR
The isolation and manipulation of DNA were conducted according to standard procedures [29]. All chromosomal DNA were extracted from B. subtilis and isolated by the protocol of Sangon Biotech (Shanghai, China). PCR was performed with DNA Ploymerase HiFi or Taq DNA polymerase (TransGen, Beijing, China) in a DNA thermal cycler (DNAEngine, BIO-RED, Hercules, CA, USA) through the procedure recommended by the manufacturer. Overlapped extension PCR (SOE-PCR) was carried out as decribed [30,31]. PCR products were purified with a PCR Purification kit (Biomed, Beijing, China) and analyzed by electrophoresis in 1% (w/v) agarose gels.
Transformation and transformants’ seletion
Transformation of B. subtilis was performed by using competent cells as described by Anagnostopoulos and Spizizen [32]. Competent transformation used liner DNA frangments. The transformants seletion relied on the method described by Liu et al. [33,34].
Gene deletion
The method of marker-free gene deletion was as described by Liu et al. [33,34]. We took pyrR gene as an example. The fragment using for deleting the pyrR gene was constructed as follows. The 0.9 kb cat (C) fragment was amplified from the pC194 plasmid using the primers Cat1qr and Cat2r (Table 3) [35]. The 1.2 kb araR (R) fragment, including the whole coding region of the araR gene, was amplified from the B. subtilis 168 genome using the primers araR1qr and araR2qr. The 1.3 kb UPpyrR (U), 0.9 kb DNpyrR (D) and 0.5 kb GpyrR (G) fragments were amplified from the B. subtilis 168 genome using the primers pyrUP1 and pyrUP2, pyrDN1q and pyrDN2, and pyrG1and pyrG2, respectively. These five PCR fragments were then ligated in the order U-D-C-R-G by splicing by overlapped extension PCR (SOE-PCR) using the primers pyrUP1 and pyrG2 and then were used to transform. The deletions of the cdd, hom and nupC-pdp genes were similar to the pyrR gene.
Table 3.
Primers and synthetized fragments
| Oligonucleotides for pyrR gene deletion | |
|---|---|
| Primer number | Sequence 5'- 3' |
| pyrUP1 | AAAAGTGAGCGGATTGA |
| pyrUP2 | TCCTGCCAGAGCATAGAG |
| pyrDN1q | CTCTATGCTCTGGCAGGAGGGGTTTTTTCTTCAACAATCAGGGGGAAAT |
| pyrDN2 | GGGCCCGGATCCCACTGTCACCCATAATAGAGC |
| pyrG1 | GCATTCTTGTCGGCATTA |
| pyrG2 | GCCACAGCAGGACTCATT |
| Cat1qr | CATAAAAGCAGGTCTTCATCGCTCTATTATGTCTTCAACTAAAGCACCCAT |
| Cat2pr | GGGCCCGGATCCTCTTCAACTAAAGCACCCAT |
| araR1qr | CTGCCCCGTTAGTTGAAGGCATTTTCTGTCAATGTTTTC |
| araR2qr | AATCCCTCTTGTCTTAATGCTTATTCATTCAGTTTTCGTG |
| Oligonucleotides for pyrH gene overexpression | |
| Primer number | Sequence 5'- 3' |
| pyrHUP1 | TACGGCATTCACATCAGG |
| pyrHUP2q | TCCACTTCATCCACTCCATCGCTTAACGCATTGATATGA |
| P1h | ATGGAGTGGATGAAGTGGA |
| P2h | CATTCTTTACCCTCTCCTTT |
| pyrH1q | AAAGGAGAGGGTAAAGAATGGAAAAACCAAAATACAAACG |
| pyrH2q | CCTTCTACACGATATGTGCTGCGTACAGTAGCCAATTCG |
| pyrHDN1 | AGCACATATCGTGTAGAAGG |
| pyrHDN2q | ATGGGTGCTTTAGTTGAAGAAATGGCTGTCGCTATTGTT |
| Cat2h | GCTGTAATATAAAAACCTTC |
| Cat1qh | AAGAAGAAGGCAATGACACGTCTTCAACTAAAGCACCCAT |
| araR1qh | CTGCCCCGTTAGTTGAAGGCATTTTCTGTCAATGTTTTC |
| araR2h | TTATTCATTCAGTTTTCGTG |
| pyrHG1q | CACGAAAACTGAATGAATAATCAGCCTAATGATGTCTTGT |
| pyrHG2 | ACTTCTTGAACGACTTCCA |
| Promoter-h | ATGGAGTGGATGAAGTGGAATCGTTTTAGAATGGGAGAATTAACTATTAATGTTTGACAACTATTACAGAGTATGCTATAATAAATTCACAGAATAGTCTTTTAAGTAAGTCTACTCTGAATTTTTTTAAAAGGAGAGGGTAAAGA |
| CX1h | ATGGAGTGGATGAAGTGGA |
| CX2h | GCGTACAGTAGCCAATTCG |
| Oligonucleotides for prs gene overexpression | |
| Primer number | Sequence 5'- 3' |
| prsUP1 | AATCCGCCGCTTCCAA |
| prsUP2q | TGTCAAACATTAATAGTTAATTCTCCCATTCTAAAACGATTCCACTTCATCCACTCCAT |
| p1p | CAAATGGAGTGGATGAAG |
| p2p | ACTTCCCCGTCACTAAAT |
| prs1 | TTAAAAGGAGAGGGTAAAGAATGTCTAATCAATACGGAGATAAG |
| prs2 | TTAGCTGAACAGATAGCTGACT |
| prsDN1q | AGTCAGCTATCTGTTCAGCTAATGTCCTCCATTGTGATTGAT |
| prsDN2 | ACGCATGATGAAGAACTTG |
| CR1qp | CCAAGTTCTTCATCATGCGTTCTTCAACTAAAGCACCCAT |
| CR2p | TTATTCATTCAGTTTTCGTG |
| prsG1q | CACGAAAACTGAATGAATAAATCAAGTGGCGGAAGAAG |
| prsG2 | AGGATGCGATTCAATTATGC |
| Promoter-p | CAAATGGAGTGGATGAAGTGGAATCGTTTTAGAATGGGAGAATTAACTATTAATGTTTGACAACTATTACAGAGTATGCTATAATAAATTCACAGAATAGTCTTTTAAGTAAGTCTACTCTGAATTTTTTTAAAAGGAGAGGGTAAAGAATGTCTAATCAATACGGAGATAAGAATTTAAAGATTTTTTCTTTGAATTCGAATCCAGAGCTTGCAAAAGAAATCGCATATATAGTTGGAGTTCAATTAGGGAAATGTTCTGTCACAAGATTTAGTGACGGGGAAGT |
| CX1p | TCCGCCGATTACTTCTTG |
| CX2p | CCATGTCACTATTGCTTCAG |
| Oligonucleotides for hom gene deletion | |
| Primer number | Sequence 5'- 3' |
| homUP1 | TGAACTGACATTTGAACAT |
| homUP2 | CCTTTCTTTTGATTGTCC |
| homDN1q | CCAAGAGGACAATCAAAAGACGCTTTCTGCTGTTCATAA |
| homDN2 | CGTGTCATTGCCTTCTTC |
| Cat1qm | AAGAAGAAGGCAATGACACGTCTTCAACTAAAGCACCCAT |
| Cat2m | GCTGTAATATAAAAACCTTC |
| araR1qm | CTGCCCCGTTAGTTGAAGGCATTTTCTGTCAATGTTTTC |
| araR2m | TTATTCATTCAGTTTTCGTG |
| homG1q | CACGAAAACTGAATGAATAAAAGCGATTCGTGTAGGG |
| homG2 | GATGGTCAGGAAGCAGT |
| Oligonucleotides for nupC-pdp gene deletion | |
| Primer number | Sequence 5'- 3' |
| pnUP1 | GAAGTGTGCGAAGGATGT |
| pnUP2 | GAGGAGAATGTAGCCAAGAA |
| pnDN1q | GCAATTTATTCTTGGCTACATTCATTGGACGAAGCGAGAG |
| pnDN2 | CTGCGGAGTTCCTTGTATC |
| CR1qpn | ATGATACAAGGAACTCCGCAGTCTTCAACTAAAGCACCCAT |
| CR2pn | TTATTCATTCAGTTTTCGTG |
| pnG1q | CACGAAAACTGAATGAATAAATCTGCGATGTCAACTGTAT |
| pnG2 | GGCTGCGTCTTCTTCTGTT |
| Oligonucleotides for cdd gene deletion | |
| Primer number | Sequence 5'- 3' |
| cddUP1 | GCTTCAGGACGGACAGTTCAGT |
| cddUP2q | AGATACTGGTCCAGGGGTGTCAGCACTTGGATTTGGAATACGGCG |
| cddDN1 | GCTGACACCCCTGGACCAGTATCT |
| cddDN2q | CGTTTGTTGAACTAATGGGTGCTTTCAGAACCGTTGTCCTGCCCTTT |
| Cat1c | CACCCATTAGTTCAACAAACG |
| Cat2c | TTCAACTAACGGGGCAG |
| araR1qc | CTGCCCCGTTAGTTGAAGGCATTTTCTGTCAATGTTTTC |
| araR2E | CGCGGATCCTGACACCCCTGGACCAGTATCT |
| cddG1E | CGCGGATCCTGACACCCCTGGACCAGTATCT |
| cddG2 | CCCACAATTCAAGGTAGACACG |
| Oligonucleotides for pyrG gene constitutive expression and overexpression | |
| Primer number | Sequence 5'- 3' |
| pyrGUP1 | GGATACGGCGATGAAGGT |
| pyrGUP2q | GTTCTCTCTTCGTTTTTGAAGAGCCCCCCAAAATACATACTACATAGTTCGAC |
| pyrGDN1 | TTCAAAAACGAAGAGAGAACATAG |
| pyrGDN2q | GTTTGTTGAACTAATGGGTGCTAATCGTTTGAGACAGGTTGC |
| pyrGG1 | GTCCGCACGAAAACTGAATGAATACCAGCACGGTGAAGTATT |
| pyrGG2 | CCAAGATCCTCAACATCCTT |
| Cat1g | CACCCATTAGTTCAACAAACG |
| Cat2g | TTCAACTAACGGGGCAG |
| araR1qg | CTGCCCCGTTAGTTGAAGGCATTTTCTGTCAATGTTTTC |
| araR2g | TTATTCATTCAGTTTTCGTG |
| pyrGUP1b | TACGGCGATGAAGGTAAC |
| pyrGUP2qb | AGCTCCCTTTCAATTTCTTGCGTACTATGTTCTCTCTTCGT |
| gsiB1 | CAAGAAATTGAAAGGGAGCT |
| gsiB2 | TTCCAAGTGAGGATACAACTCC |
| pyrGDCRG1 | GGAGTTGTATCCTCACTTGG |
| pyrGDCRG2 | CAATGATGCCGTCTGTTC |
| Promoter-g | CAAGAAATTGAAAGGGAGCTATGTTTTTCTCAAATTGTAAGTTTATTTCATTGACAACTTTAAAAAGGATCGCTATAATAACCAATAAGGACAAAAGGAGGAATTCAAAATGACGAAATATATTTTTGTAACCGGGGGAGTTGTATCCTCACTTGGAA |
| CX1g | ATGGAGTGGATGAAGTGGA |
| CX2g | GCGTACAGTAGCCAATTCG |
Gene overexpression
To overexpress the pyrH gene, the inherent ITR of the pyrH was replaced by a stronger promoter A1 of bacteriophage φ29 ligated to the mRNA stabilizer of the aprE gene (PAE expression cassette) (Figure 6). To overexpress the prs gene, another prs gene copy controlling by PAE expression cassette was integrated in chromosome the xylR gene locus. The method to overexpress the pyrG gene is that a standardized promoter along with the the gsiB mRNA stabilizer (PSB expression cassette) was inserted after the inherent pyrG promoter (Figure 4). The method to constitutively express the pyrG gene is that 4 extra G residues at the 5′ ends of the pyrG ITR. The method of marker-free gene modification was derived from Liu et al. [33,34]. The specific method of gene modification was described as below; for example, the modified the pyrH gene fragment was constructed as follows. The 0.9 kb cat (C) fragment was amplified from the pC194 plasmid using the primers Cat1qh and Cat2h (Table 3). The 1.2 kb araR (R) fragment was amplified from the B. subtilis 168 genome using the primers araR1qh and araR2. The 1 kb UPpyrH (U), 0.7 kb DNpyrH (D) and 0.6 kb GpyrH (G) fragments were amplified from the B. subtilis 168 genome using the primers pyrHUP1 and pyrHUP2q, pyrHDN1q and pyrHDN2, and pyrHG1q and pyrHG2, respectively. The 0.8 kb pyrH (H) fragment, including the whole coding region of the pyrH gene, was amplified from the B. subtilis 168 genome using the primers pyrH1q and pyrH2q. The 0.15 kb Promoter (P) fragment was whole sequence synthesized by AuGCT DNA-SYN Biotechnology Corporation (Beijing, China), including PAE expression cassette, and ligated to plasmid pGH-A0981Gn. P fragment was amplified from plasmid pGH-A0981Gn by using the primers p1h and p2h. These seven PCR fragments were then ligated in the order G-R-C-D-H-P-U by splicing by SOE-PCR using the primers pyrHUP1 and pyrHG2 and then be used to transform. DNA sequencing was done at AuGCT DNA-SYN Biotechnology Corporation (Beijing, China) by using primers CX1h and CX2h. The constitutive expression and overexpression of the pyrG gene, and the overexpressions of the prs gene were similar to the pyrH gene.
Figure 6.

Nucleotide sequence of PAE expression cassette. The sequence of the nontemplate strand is shown. The −10 and −35 regions of the promoter, transcription start site (+1), Shine–Dalgarno sequence (SD) and initiation codon are bold and underlined. The RNA-stabilizing elements are shown in italics.
Fermentation
A loop of cells grown on an agar plate of LB medium was inoculated into a 250 mL flask containing 30 mL of stock culture medium (2% glucose, 2% soybean meal hydrolysate, 1% yeast extract, 0.25% NaCl, 0.1% MgSO4.7H2O, 0.1% KH2PO4, 0.5% sodiumglutamate, pH = 7.0) and was then cultured for 16 h at 37°C, with shaking at 220 rpm. 1.5 mL of the culture was transferred to a 250 mL flask containing 30 mL of fermentation medium (8% glucose, 3% soybean meal hydrolysate, 2% cornsteepliquor, 1.5% yeast extract, 0.25% NaCl, 0.8% MgSO4.7H2O, 0.25% KH2PO4, 1.5% (NH4)2SO4, 1.5% sodiumglutamate, pH = 7.0) and was then cultured for 72 h at 37°C, with shaking at 220 rpm. Samples were drawn at various time-points during the fermentation and were analyzed for cell growth (OD600) and pyrimidine compounds content.
Fermentation compounds analysis
The qualitative analysis of pyrimidine nucleoside and pyrimidine compounds in the medium was conducted by massspectrograph (Q-Exactive, Thermo Scientific, Waltham, MA, USA).The quantitative analysis of cytidine, uridine and uracil were conducted by HPLC (Waters 2695, Waters, Milford, MA, USA) with an HYPERSIL ODS C18 column (Thermo Scientific, Waltham, MA, USA). Separation was performed at 40°C with 0.05 M KH2PO4 at a flowrate of 1.5 mL min−1. The detective wavelength was 270 nm.
Quantitative real-time reverse transcription (RT)-PCR analysis
Total RNA of B. subtilis was extracted with RNAprep pure Cell/Bacteria Kit (TIANGEN, Beijing, China) as recommended by the supplier and was reverse-transcribed in cDNA using the Maxima First Strand cDNA Synthesis Kit (Thermo Scientific, Waltham, MA, USA) according to the manufacturer’s instructions. The Real-time PCR was carried out by LightCycler 480 (Roche Diagnostics GmbH, Mannheim, Germany) using SYBR Green Ι Master (Roche Diagnostics GmbH, Mannheim, Germany). The ccpA gene was served as internal control of Real-time PCR. The quantification cycle (Cq) was determined according to the second derivative maximum method using the LightCycler software 4.1 (Roche Diagnostics GmbH, Mannheim, Germany). The relative expression ratio (RE) was calculated according to Pfaffl [36].
Acknowledgements
We gratefully acknowledge the generous support of Lei Guo and Dr. Xi-xian Xie from Tianjin University of Science and Technology in fermentation experiments. We also appreciate Thermo Fisher Scientific for its important contributions (Shanghai, China) to qualitative analyses of fermentation products with Mass Spectrometry.
Additional file
The identification of the gene deletion. Figure S1 The identification of the cdd gene deletion. (A) Lane 1: marker. Lanes 2–7: PCR products of the recombinants (lanes 2–6, 2004 bp) and the WT strain (lane 7, 2155 bp) by using the primers cddUP1 and cddG2. (B) Lane 1: marker. Lanes 2–7: PCR products of the recombinants (lanes 2–6, 512 bp, 604 bp and 888 bp) and the WT strain (lane 7, 512 bp, 604 bp and 1039 bp) digested by SacI and EcoRI. Figure S2 The identification of the hom gene deletion. Lane 1: marker. Lanes 2, 4: PCR products of the WT strain (lane 2, 2531 bp) and the recombinant (lane 4, 1700 bp) by using the primers homUP1 and homDN2. Lanes 3, 5: PCR products of the WT strain (lane 3, 619 bp and 1912 bp) and the recombinant (lane 5, 619 bp and 1081 bp) digested by SphI. Figure S3 The identification of the pyrR gene deletion. Lane 1: marker. Lanes 2, 4: PCR products of the WT strain (lane 2, 2642 bp) and the recombinant (lane 4, 1904 bp) by using the primers pyrUP1 and pyrDN2. Lanes 3, 5: PCR products of the WT strain (lane 3, 531 bp and 2111 bp) and the recombinant (lane 5, 531 bp and 1373 bp) digested by PstI. Figure S4 The identification of the nupC-pdp gene deletion. Lane 1: marker. Lanes 2, 5: PCR products of the WT strain (lane 2, 3433 bp) and the recombinant (lane 5, 1495 bp) by using the primers pnUP1 and pnDN2. Lanes 3, 6: PCR products of the WT strain (lane 3, 474 bp and 2959 bp) and the recombinant (lane 6, 474 bp and 1021 bp) digested by HindIII. Lanes 4, 7: PCR products of the WT strain (lane 4, 174 bp, 339 bp, 1452 bp and 1468 bp) and the recombinant (lane 7, 174 bp, 339 bp and 982 bp) digested by EcoRV.
Footnotes
Competing interests
The authors declare that they have no competing interests.
Authors’ contributions
HZ designed the experiments; HZ, ZMY and SMY performed the experiments; HZ and RB wrote the manuscript; RB supervised the work; and all authors contributed to the discussion of the research. All authors read and approved the final manuscript.
Contributor Information
Hui Zhu, Email: zhuhui0505@hotmail.com.
Shao-Mei Yang, Email: shaomei@tju.edu.cn.
Zhao-Min Yuan, Email: yuanzhaomin@tju.edu.cn.
Rui Ban, Email: banrui@tju.edu.cn.
References
- 1.Turner RJ, Bonner ER, Grabner GK. Purification and characterization of bacillus subtilis PyrR, a bifunctional pyr mRNA-binding attenuation protein/uracil phosphoribosyl transferase. J Biol Chem. 1998;273(10):5932–8. doi: 10.1074/jbc.273.10.5932. [DOI] [PubMed] [Google Scholar]
- 2.Quinn CL, Stephenson BT, Switer RL. Functional organization and nucleotide sequence of the Bacillus subtilis pyrimidine biosynthetic operon. J Biol Chem. 1991;266:9113–27. [PubMed] [Google Scholar]
- 3.Turner RJ, Lu Y, Switzer RL. Regulation of the Bacillus subtilis pyrimidine biosynthetic (pyr) gene cluster by an atogenous transcriptional attenuation mechanism. J Bacterol. 1994;76(12):3708–22. doi: 10.1128/jb.176.12.3708-3722.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hobl B, Mack M. The regulator protein PyrR of Bacillus subtilis specifically interacts in vivo with three untranslated regions within pyr mRNA of pyrimidine biosynthesis. Microbiology. 2007;153(3):693–700. doi: 10.1099/mic.0.2006/003772-0. [DOI] [PubMed] [Google Scholar]
- 5.Parsot C, Cohen GN. Cloning and nucleotide sequence of the Bacillus subtilis hom gene coding for homoserine dehydrogense. Structural and evolutionary relationships with Escherichia coli aspartokinases–homoserine dehydrogenases I and II. J Biol Chem. 1988;263:14654–60. [PubMed] [Google Scholar]
- 6.Hilden I, Krath BN, Hove-jensen B. Tricistronic operon expression of the genes gcaD (tms), which encodes N-acetylglucosamine 1-phosphate uridyltransferase, prs, which encodes phosphoribosyl diphosphate synthetase, and ctc in vegetative cells of Bacillus subtilis. J Bacteriol. 1995;177(24):7280–4. doi: 10.1128/jb.177.24.7280-7284.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Shimaoka M, Takenaka Y, Kurahashi O, Kawasaki H, Matsui H. Effect of amplification of desensitized purF and prs on inosine accumulation in Escherichia coli. J Biosci Bioeng. 2007;103(3):255–61. doi: 10.1263/jbb.103.255. [DOI] [PubMed] [Google Scholar]
- 8.Zakataeva NP, Romanenkov DV, Skripnikova VS, Vitushkina MV, Livshits VA, Kivero AD, et al. Wild-type and feedback-resistant phosphoribosyl pyrophosphate synthetases from Bacillus amyloliquefaciens: purification, characterization, and application to increase purine nucleoside production. Appl Microbiol Biotechnol. 2012;93:2023–33. doi: 10.1007/s00253-011-3687-3. [DOI] [PubMed] [Google Scholar]
- 9.Gagyi C, Ionescu M, Gounon P, Sakamoto H, Rousslle JC, Laurent-Winter C. Identification and immunochemical location of UMP kinase from Bacillus subtilis. Curr Microbiol. 2004;48:62–7. doi: 10.1007/s00284-003-4117-2. [DOI] [PubMed] [Google Scholar]
- 10.Serina L, Blondin C, Krin E, Sismeiro O, Danchin A, Sakamoto H, et al. Escherichia coli UMP-kinase, a member of the aspartokinase family, is a hexamer regulated by guanine nucleotides and UTP. J Biol Chem. 1995;34:5066–74. doi: 10.1021/bi00015a018. [DOI] [PubMed] [Google Scholar]
- 11.Meng Q, Turnbough CL, Switzer RL. Attenuation control of pyrG expression in Bacillus subtilis is mediated by CTP-sensitive reiterative transcription. Proc Natl Acad Sci. 2004;101:10943–8. doi: 10.1073/pnas.0403755101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Meng Q, Turnbough CL, Switzer RL. Regulation of pyrG expression in Bacillus subtilis: CTP-regulated antitermination and reiterative transcription with pyrG templates in vitro. Mol Microbiol. 2007;63(5):1440–52. doi: 10.1111/j.1365-2958.2007.05595.x. [DOI] [PubMed] [Google Scholar]
- 13.Elsholz AKW, Jogenson CM, Switzer RL. The number of G residues in the Bacillus subtilis pyrG initially transcribed region governs reiterative transcription-mediated regulation. J Bacteriol. 2007;189(5):2176–80. doi: 10.1128/JB.01611-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Song B-H, Jan N. Chromosomal location, cloning and nucleotide sequence of the Bacillus subtilis cdd gene encoding cytidine/deoxycytidine deaminase. Mol Gen Genet. 1989;216:462–8. doi: 10.1007/BF00334391. [DOI] [PubMed] [Google Scholar]
- 15.Saxild HH, Andersen LN, Hammer K. Dra-nupC-pdp operon of Bacillus subtilis: nucleotide sequence, induction by deoxyribonucleosides, and transcriptional regulation by the deoR-encoded DeoR repressor protein. J Bacteriol. 1996;178:424–34. doi: 10.1128/jb.178.2.424-434.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zeng X, Saxild HH. Identication and characterization of a DeoR specic operator sequence essential for induction of the dra-nupC-pdp operon expression in Bacillus subtilis. J Bacteriol. 1999;181:1719–27. doi: 10.1128/jb.181.6.1719-1727.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zeng X, Saxild HH, Switzer RL. Purication and characterization of the DeoR repressor of Bacillus subtilis. J Bacteriol. 2000;182:1916–22. doi: 10.1128/JB.182.7.1916-1922.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Asahi S, Tsunemi Y, Lzawa M, Doi M. Cytidine production by Mutants of Bacillus subtilis. Biosci Biotech Biochem. 1994;58(8):1399–402. doi: 10.1271/bbb.58.1399. [DOI] [Google Scholar]
- 19.Asahi S, Tsunemi Y, Lzawa M, Doi M. A 3-deazauracil-resistant mutant of Bacillus subtilis with increased production of cytidine. Biosci Biotech Biochem. 1994;59(5):915–6. doi: 10.1271/bbb.59.915. [DOI] [PubMed] [Google Scholar]
- 20.Asahi S, Tsunemi Y, Doi M. Improvement of a cytidine-producing mutant of Bacillus subtilis introducing a mutantion by homologous recombination. Biosci Biotech Biochem. 1995;59(11):2123–6. doi: 10.1271/bbb.59.2123. [DOI] [Google Scholar]
- 21.Doi M, Asahi S, Tsunemi Y, Akiyama S. Mechanism of uridine production by Bacillus subtilis mutants. Appl Microbiol Biotechnol. 1989;30:234–8. doi: 10.1007/BF00256210. [DOI] [Google Scholar]
- 22.Su J, Huang J, Xie XX, Xu QY, Chen N. Knockout of the cdd gene in Bacillus subtilis and its influence on cytidine fermentation. Lett Biotechnol. 2010;21(1):39–42. [Google Scholar]
- 23.Paulus TJ, McGarry TJ, Shekelle PG, Rosenzweig S, Switzer RL. Coordinate synthesis of the enzymes of pyrimidine biosynthesis in Bacillus subtilis. J Bacteriol. 1982;149:775–8. doi: 10.1128/jb.149.2.775-778.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Potvin BW, Kelleher RJ. Pyrimidine biosynthetic pathway of Bacillus subtilis. J Bacteriol. 1975;123:604–15. doi: 10.1128/jb.123.2.604-615.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Asahara T, Mori Y, Zakataeva NP, Livshits VA, Yoshida K, Matsuno K. Accumulation of gene-targeted Bacillus subtilis mutationsthat enhance fermentative inosine production. Appl Microbiol Biotechnol. 2010;87:2195–207. doi: 10.1007/s00253-010-2646-8. [DOI] [PubMed] [Google Scholar]
- 26.Switzer RL, Sogin DC. Regulation and mechanism of phosphoribosyl pyrophosphate synthetase. V. Inhibition by end products and regulation by adenine diphosphate. J Biol Chem. 1973;248:1063–73. [PubMed] [Google Scholar]
- 27.Martinussen J, Glaser P, Andersen PS, Saxild HH. Two genes encoding uracil phosphoribosyltransferase are present in Bacillus subtilis. J Bacteriol. 1995;177(1):271–4. doi: 10.1128/jb.177.1.271-274.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Neuhard J. In: Utilization of preformed pyrimidine bases and nucleosides: Metabolism of nucleotides, nucleosides and nucleobases in microorganisms. Munch-Petersen, editor. London: Academic Press; 1983. pp. 95–148. [Google Scholar]
- 29.Sambrook J, Russell D. Molecular cloning: a laboratory manual. 3. Cold Spring Harbor: Cold Spring Harbor Laboratory Press; 2001. [Google Scholar]
- 30.Kuwayama H, Obara S, Morio T, Katoh M, Urushihara H, Tanaka Y. PCR-mediated generation of a gene disruption construct without the use of DNA ligase and plasmid vectors. Nucleic Acids Res. 2002;30(2):e2. doi: 10.1093/nar/30.2.e2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Shevchuk NA, Bryksin AV, Nusinovich YA, Cabello FC, Sutherland M, Ladisch S. Construction of long DNA molecules using long PCR-based fusion of several fragments simultaneously. Nucleic Acids Res. 2004;32(2):e19. doi: 10.1093/nar/gnh014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Anagnostopoulos C, Spizizen J. Requirements for transformation in Bacillus subtilis. J Bacteriol. 1961;81(5):741–6. doi: 10.1128/jb.81.5.741-746.1961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Liu S, Endo K, Ara K, Ozaki K, Naotake O. Introduction of marker-free deletions in Bacillus subtilis using the AraR repressor and the ara promoter. Microbiology. 2008;154(9):2562–70. doi: 10.1099/mic.0.2008/016881-0. [DOI] [PubMed] [Google Scholar]
- 34.Dong HN, Zhang DW. Current development in genetic engineering strategies of Bacillus species. Microb Cell Fact. 2014;13:63. doi: 10.1186/1475-2859-13-63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Horinouchi S, Weisblum B. Nucleotide sequence and functional map of pC194, a plasmid that specifies inducible chloramphenicol resistance. J Bacteriol. 1982;150(2):815–25. doi: 10.1128/jb.150.2.815-825.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pfaffl MV. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 2001;29(9):e45. doi: 10.1093/nar/29.9.e45. [DOI] [PMC free article] [PubMed] [Google Scholar]


