Abstract
The conserved transcription factor HSF-1 is essential to cellular stress resistance and organismal lifespan determination. The canonical function of HSF-1 is to regulate a network of molecular chaperones that maintain protein homeostasis during extrinsic environmental stresses or intrinsic age related deterioration. In the metazoan C. elegans, we engineered a modified HSF-1 strain that increases stress resistance and longevity without enhancing chaperone induction. This HSF-1 dependent health assurance acts through the regulation of pat-10. Upon heat stress pat-10 upregulation maintains a functional actin cytoskeleton and endocytic network. Loss of pat-10 causes a collapse of organismal health and failure of stress resistance. Furthermore, overexpression of pat-10 is sufficient to increase both thermotolerance and longevity by mechanisms that affect actin stability. Our findings indicate that in addition to chaperone induction, HSF-1 plays a prominent role in cytoskeletal integrity to ensure proper cellular function during times of stress and aging.
The survival of an organism is intricately linked to its ability to maintain cellular quality control, including organelle integrity, lipid homeostasis, proper protein folding and cellular communication. The organismal response to unpredictable or extreme environmental changes is critical to mitigate damages caused by cellular stress. The heat shock protein (HSP) family of molecular chaperones is the most highly induced class of genes in response to thermal stress, suggesting these proteins are part of a fundamental defense against proteotoxic stress. Consistent with this hypothesis, ectopic expression of the master transcriptional regulator of HSPs, heat shock factor-1 (HSF-1), is sufficient to confer resistance to thermal stress and increase lifespan in the nematode C. elegans (1). Furthermore, overexpression of HSF-1 can alleviate the toxicity associated with diseases caused by misfolding or aggregating proteins (2).
Interestingly, recent studies have begun to reveal the potential dispensability of chaperone induction in thermotolerance and longevity. For example, neither a hypomorphic mutation of hsf-1, nor a block in the transcriptional upregulation of HSPs using α-amanitin affects the thermotolerance of C. elegans under conditions of heat stress (3, 4). However, other studies using the same mutant strain of hsf-1 find a significant decrease in heat stress resistance (5). The conflicting results may be due to experimental design differences, but taken together suggest a complexity in HSF-1 regulation and function that is not fully captured in traditional chaperone induction models of stress resistance and aging.
To test potential HSF-1 mediated mechanisms of cellular protection that are independent of enhancing chaperone induction, integrated transgenic nematode strains were generated that overexpressed either the full-length hsf-1 gene (hsf-1(FL)) or a variant of hsf-1 with a C-terminal truncation (hsf-1(CT)) of 84 amino acids, removing a transcriptional activation domain responsible for the upregulation of HSPs (6). Importantly, the hsf-1(CT) variant was designed to mimic the C-terminal missense mutation found in the hsf-1(sy441) mutant strain, a widely used allele that decreases stress induced HSP transcription (6). hsf-1(FL) was overexpressed in the N2 wild-type (WT) background and hsf-1(CT) was overexpressed in the hsf-1(sy441) mutant background. Therefore the hsf-1(CT) strain mirrored the overexpression of hsf-1(FL) but contained no endogenous copies of full length, wild-type hsf-1 (Fig. 1A). Both transgenes used the ubiquitous sur-5 promoter, resulting in approximately 3-fold higher transcriptional expression than endogenous hsf-1 expression (Fig. 1B).
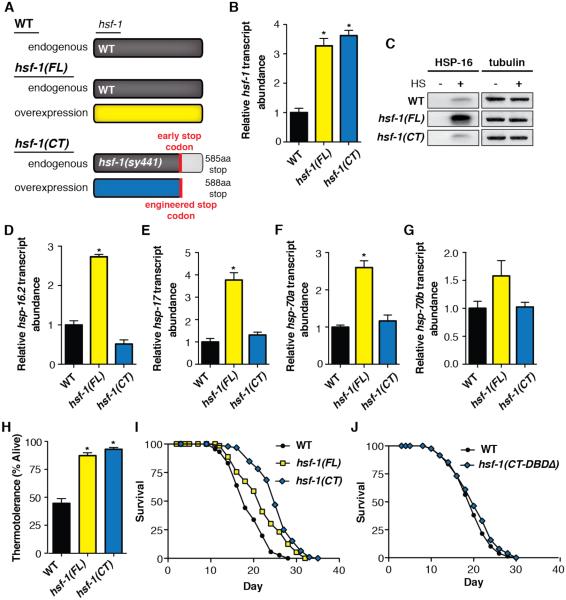
Fig. 1.
hsf-1(CT) increases lifespan and thermotolerance without enhancing the inducible chaperone network. (A) Diagram of hsf-1 genotypes in wild-type (WT), full-length (hsf-1(FL)), and truncated (hsf-1(CT)) overexpression strains. (B) hsf-1(FL) and hsf-1(CT) equally overexpress hsf-1, as determined by quantitative PCR (qPCR). (C) Chaperone induction in hsf-1(FL) is enhanced, as determined by western blot of HSP-16 before and after heat shock. (D to G) qPCR of hsp-16.2 (D), hsp-17 (E), hsp-70a (C12C8.1) (F), and hsp-70b (F44E5.4) (G) show enhanced chaperone induction in hsf-1(FL). (H) Thermotolerance assay of worms shifted from 20° to 34° for 13 hours. hsf-1(FL) and hsf-1(CT) survival is significantly increased. (I) Lifespan analysis of hsf-1(FL) and hsf-1(CT) strains show increased longevity. (J) Lifespan extension of the hsf-1(CT) strain is lost when the DNA binding domain is removed from the overexpression plasmid. *P < 0.005; error bars indicate SEM.
Analysis of protein and transcript abundance confirmed that overexpression of hsf-1(FL) enhanced hsp-16 heat inducible expression, whereas hsf-1(CT) overexpression showed no difference to the wild-type control strain (Fig. 1, C and D). While hsf-1(CT) overexpression had no effect, hsf-1(FL) worms also showed enhanced transcriptional upregulation of all HSF-1 regulated HSPs tested (Fig. 1, E to G). Furthermore, we performed transcriptome sequencing analysis of these strains and confirmed hsf-1(FL) enhanced transcription of all known heat inducible HSPs, whereas hsf-1(CT) did not (fig. S1).
Interestingly, both hsf-1(FL) and hsf-1(CT) transgenic worms had increased thermotolerance (Fig. 1H). Furthermore, both strains lived significantly longer than wild-type (Fig. 1I). Because the lifespan extension of hsf-1(CT) was unexpected, we tested if this phenotype was dependent on a functional DNA binding domain. We found that the increased longevity was abolished when the DNA binding domain was removed (hsf-1(CT-DBDΔ) (Fig. 1J). Taken together, increased lifespan and thermotolerance did not correlate with the induction of HSPs. These findings support a hypothesis in which thermotolerance and longevity of an organism mediated by overexpression of hsf-1 is independent of increased induction of chaperones.
Intrigued by the findings that hsf-1(CT) can regulate thermotolerance without enhanced HSP induction, we sought to find factors that were responsible for HSF-1 mediated thermotolerance. To determine which cellular networks are required in these long-lived, thermo-protected worms, we completed quantitative transcriptomic and proteomic analyses comparing hsf-1(FL) and hsf-1(CT) strains to wild-type and hsf-1(sy441) strains. We filtered for significantly upregulated transcripts or proteins, at either basal or heat stress conditions, unique to our thermotolerant strains (Fig. 2A). This filtering method only considered candidates that were similarly upregulated in the hsf-1(FL) strain, avoiding potential neomorphic effects of the hsf-1(CT) strain.
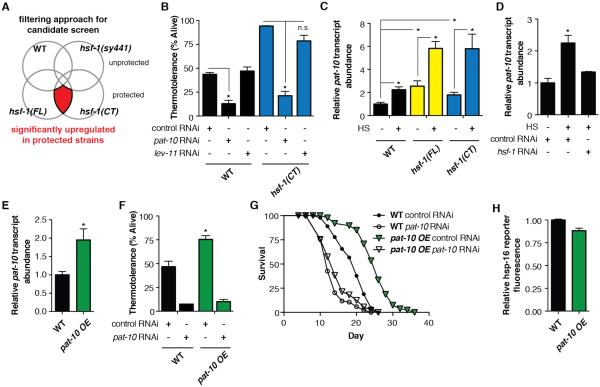
Fig. 2.
pat-10 is necessary and sufficient for thermotolerance and longevity. (A) Filtering selection method for RNAi based thermotolerance screen selected proteins or transcripts that were upregulated in our protected strains (hsf-1(FL) and hsf-1(CT)) but not in unprotected strains (WT and hsf-1(sy441)), before or after heat shock. (B) RNAi of pat-10 significantly reduces thermotolerance in WT and hsf-1(CT) strains, whereas control lev-11 RNAi has no effect. (C) qPCR shows pat-10 is upregulated by heat shock in all strains. Transcript abundance is further increased in hsf-1 overexpression strains hsf-1(FL) and hsf-1(CT). (D) Heat shock induced pat-10 upregulation is reduced by hsf-1 RNAi, as determined by qPCR. (E) qPCR of pat-10 overexpression in the pat-10 OE strain. This level of overexpression is similar to the increase in pat-10 expression after heat shock in WT worms. (F) pat-10 overexpression significantly increases thermotolerance, whereas pat-10 RNAi significantly impairs thermotolerance in WT and pat-10 OE strains. (G) pat-10 overexpression significantly increases lifespan. pat-10 RNAi significantly reduces longevity in WT and pat-10 OE strains. (H) qPCR shows pat-10 OE does not induce the hsp-16.2p::GFP reporter strain. *P < 0.05; error bars indicate SEM.
Fifty-two gene products passed our filters using the proteomic data and 46 genes met these criteria from the transcript data. The corresponding 98 unique genes had significant gene ontology enrichments for development, cytoskeleton organization, complex assembly, and immune defense response (fig. S2). Significantly, molecular chaperones were absent from the enriched gene ontologies.
Reducing expression of genes essential to thermotolerance should lower survival under heat stress. Therefore, we performed an RNAi based screen in the hsf-1(CT) strain of the 98 genes that passed our filtering criteria. Eighty-four genes were scored for their effect on thermotolerance (fig. S3). RNAi of the remaining 14 genes induced developmental arrest and were not further characterized. From the screen we identified a calcium binding protein, pat-10, as essential for thermotolerance (Fig. 2B).
Significantly, pat-10 transcription was heat inducible in all strains, including wild-type (Fig. 2C). Furthermore, hsf-1 overexpression strains showed a basal increase in pat-10 transcripts, as well as an enhanced induction upon thermal stress (Fig. 2C). Using the consensus heat shock element (HSE) binding site sequence for HSF-1 (7, 8) we examined the upstream promoter region of pat-10. We identified a strong putative HSE binding site less than 500bp from the transcription start site of pat-10 (fig. S4). Additionally, RNAi of hsf-1 blocked the upregulation of pat-10 upon heat shock (Fig. 2D). Therefore, pat-10 appears to be a direct target of HSF-1 transcriptional regulation.
Loss of pat-10 expression resulted in reduced thermotolerance of all strains tested; we therefore tested whether ectopic overexpression of pat-10 could be sufficient to render otherwise wild-type animals thermotolerant. Indeed, two-fold, overexpression of pat-10 (Fig. 2E) was sufficient to significantly increase thermotolerance (Fig. 2F) and extend lifespan (Fig. 2G). Furthermore, RNAi of pat-10 eliminated the increased thermotolerance (Fig. 2F) and lifespan (Fig. 2G) of the pat-10 overexpression strain. Taken together, pat-10 is necessary and sufficient for increased thermotolerance and longevity. Additionally, the beneficial effects of pat-10 overexpression were not due to an increase in basal HSP transcription (Fig. 2H) as assayed by a GFP reporter driven by the hsp-16.2 promoter (9).
One characterized function of pat-10 is its role in the troponin complex (10–12), which is necessary for body wall muscle contraction. However, RNAi towards the worm homologue of tropomyosin, lev-11, a partner component with pat-10 in the troponin complex, did not affect thermotolerance (Fig. 2B), while RNAi to pat-10 or lev-11 each result in adult onset paralysis (11, 13). This suggests that the role of pat-10 in muscle contraction is not responsible for thermotolerance.
In addition to playing a critical role in muscle contraction, loss of pat-10 has been shown to disrupt actin cytoskeleton dynamics and endocytosis (11, 12, 14, 15). To address these potential mechanisms of protection we first used GFP labeled muscle filaments to asses actin organization (16). Upon heat shock, muscle filaments became unorganized and damaged, leading to impaired motility (Fig. 3, A and B). However, overexpression of pat-10 was sufficient to prevent muscle and motility deterioration upon thermal stress (Fig. 3, A and B). Furthermore, we found that heat shock dramatically decreased the ratio of the filamentous (F) actin to globular (G) actin in wild-type worms, whereas the protected pat-10 overexpression animals maintained F actin when heat stressed (Fig. 3C, fig. S5). With regard to aging, we found that the ratio of F to G actin also decreases with age and that pat-10 overexpression lessened this decline (Fig. 3D, fig. S5). Collectively, these results suggest that under conditions of acute cellular stress or gradual age-related deterioration, the integrity of the actin cytoskeleton is fundamental to the survival of the organism and that overexpression of pat-10 can abrogate the collapse of actin filaments.
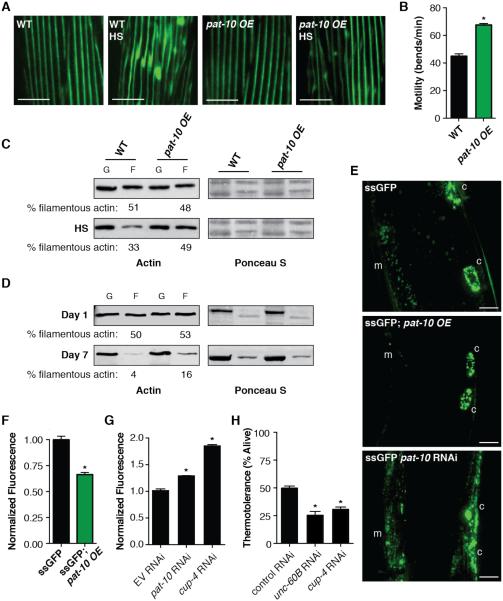
Fig. 3.
pat-10 overexpression increases actin cytoskeletal integrity and improves cellular trafficking. (A) GFP tagged to myosin heavy chain in muscle shows a breakdown of actin organization after heat shock in WT worms, whereas overexpressing pat-10 maintains actin organization after heat shock. (B) Worm thrashes per minute in liquid were used to monitor motility. Loss of motility after heat shock is lessened in the pat-10 OE strain. (C) Heat stress causes a breakdown of filamentous (F) actin into globular (G) actin. This breakdown is prevented by overexpression of pat-10. Ponceau S staining shown as a loading control. (D) Aging causes a breakdown of F actin that is lessened by overexpression of pat-10. Ponceau S staining shown as a loading control. (E) Reporter strain that secretes GFP (ssGFP) from muscle cells (m), which is then endocytosed by coelomocytes (c) to be degraded. pat-10 OE strain shows increased efficiency of secretion and endocytosis, whereas RNAi of pat-10 impairs secretion and uptake. (F) Normalized GFP fluorescence is significantly lower in ssGFP reporter strain overexpressing pat-10. (G) Blocking coelomocytic endocytosis increases GFP fluorescence in the ssGFP reporter strain. (H) RNAi of genes that block coelomocytic endocytosis decrease thermotolerance. *P < 0.05; error bars indicate SEM; scale bars indicate 10 μm.
Loss of pat-10 via RNAi has been found in several screens to disrupt endocytosis in multiple tissues, a process dependent upon a functional actin cytoskeleton (14, 15, 17). To visualize apical endocytosis within the intestine, we soaked animals in a solution containing the fluorescent FM 4-64FX dye (18), which acts as a passive, late endocytic pathway marker. As expected, pat-10 RNAi disrupted endocytosis in C. elegans (fig. S6). Next, to assay basolateral endocytosis, as well as quantitatively analyze the role of pat-10, we used a secretion and endocytosis reporter designed to actively secrete GFP (ssGFP) from muscle cells into the pseudocoelomic fluid, where it is then endocytosed by the coelomocyte cells and degraded (19) (fig. S7A). Therefore, the ssGFP reports upon effective secretion from the muscle cells and endocytosis by the coelomocyte; so that low GFP levels would correspond to effective secretion and uptake. Fitting with the hypothesis that overexpression of pat-10 improves transport and cellular processing via improved subcellular scaffolding, the pat-10 OE strain had a decrease in overall ssGFP fluorescence (Fig. 3, E and F). The decrease in ssGFP was due to improved secretion and uptake, as seen by the absence of fluorescence in the muscle and pseudocoelomic fluid (Fig 3E). This decrease was not due to an overall decrease in expression of GFP (fig. S7). RNAi of pat-10 increased overall ssGFP fluorescence and specifically decreased muscle secretion and coelomocytic endocytosis (Fig. 3, E and G). As a further control, we found that cup-4 RNAi, which blocks coelomocytic endocytosis (20), showed an even higher increase of fluorescence by blocking the uptake and degradation of ssGFP by the coelomocytes (Fig. 3G). Collectively, these data suggest that PAT-10 plays an active role in the maintenance of the cytoskeleton, which is in turn critical to cellular transport, specifically active secretion and endocytosis.
To further validate the importance of cytoskeletal integrity and endocytosis in thermotolerance, we tested whether key components in these pathways were required for survival at non-permissive temperatures. RNAi targeting a C. elegans homologue of cofilin, unc-60B, or a key regulator of coelomocytic endocytosis, cup-4, both reduced thermotolerance (Fig. 3H). Hence, it is clear that actin cytoskeletal maintenance and endocytosis are crucial to thermotolerance in C. elegans, however, we wanted to test if this mechanism is conserved.
To disrupt the actin cytoskeleton in human HEK293T cells, we used Cytochalasin D, which blocks the addition of actin monomers to filaments (21), or Latrunculin A, which binds monomers of actin, preventing polymerization of filaments (22) (Fig. 4A). Inhibiting filamentous actin formation by either Cytochalasin D or Latrunculin A significantly reduced thermotolerance in human cell culture (Fig. 4B). Importantly, these drugs did not cause cell death when cells were unstressed (fig. S8). Similar to our C. elegans data, these findings reiterate the importance of the actin cytoskeleton during times of cellular stress.
Fig. 4.
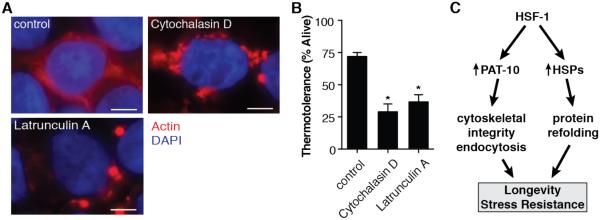
Impairing actin dynamics decreases thermotolerance in mammalian cell culture. (A) HEK293T cells treated with Cytochalasin D or Latrunculin A show disrupted filamentous actin formation in mammalian cell culture. (B) Both Cytochalasin D and Latrunculin A treatment reduce the thermotolerance of HEK293T cells after a heat shock of 45°C for 2 hours. (C) Proposed model of the duel pathways of HSF-1 mediated health assurance. *P < 0.05; error bars indicate SEM; scale bars indicate 5 μm.
Elevated levels of hsf-1 have been shown to benefit multiple organisms, yet its oncogenic properties are a major therapeutic drawback (23, 24). Because the inducible chaperone network promotes survival and proliferation of metastasizing cells (25), the ability to harness protective, non-chaperone components within the HSF-1 signal transduction cascade appears essential for future drug development. Our identification of pat-10 as a modifier of thermotolerance and longevity may apply to mammalian systems without the typical oncogenic dangers associated with hsf-1 overexpression that results in chaperone induction. An unbiased screen has already identified pat-10 as a positive modifier of toxicity for the neurodegenerative disease protein α– synuclein (15). This study further demonstrated that endocytosis played a major role in neuronal dysfunction caused by α–synuclein. We suggest a similar model by which HSF-1 activity maintains cellular scaffolding and transport in times of heat stress or aging through increased pat-10.
The hsf-1(CT) strain can mount a transcriptional response to heat shock, albeit reduced in complexity to that of hsf-1(FL). The molecular mechanism by which hsf-1(CT) regulates transcription without the C-terminal activation domain remains unclear, but possible explanations include HSF-1 containing additional activation domains. Alternatively, the hsf-1(CT) modification may cause affinity changes to HSE binding sites or different cofactors and coregulators, which could alter the transcriptional profile.
There is extensive evidence that molecular chaperones represent a useful defense against proteotoxic stress. Despite observations that we can enhance thermotolerance without further increasing the chaperone network, HSPs still play a pivotal role in stress resistance. In fact, small HSPs are known to interact with actin, promoting filament stability (26–28). Furthermore, our findings underscore the importance of maintaining filamentous actin, as opposed to the overall levels of total or globular actin. We propose a model in which HSF-1 regulates chaperones and actin cytoskeletal genes to act synergistically in a shared mechanism to promote thermotolerance and longevity (Fig. 4C). In the absence of chaperone induction, stabilization of the actin cytoskeleton is sufficient to promote survival under conditions of cellular stress and aging. Furthermore, a possible role for the massive induction of chaperones during heat stress is to manage the increasing pool of destabilized actin filaments.
Supplementary Material
Acknowledgments
Bioinformatic analysis is included in the Supplementary Materials. The following funding sources supported this research: Howard Hughes Medical Institute, National Center for Research Resources (5P41RR011823-17), National Institute of General Medical Sciences (8 P41 GM103533-17), National Institute on Aging (R01AG027463-04), NAB funded by a postdoctoral fellowship to the Salk Center for Nutritional Genomics from the Leona M. & Harry B. Helmsley Charitable Trust, PMD funded by George E. Hewitt Foundation for Medical Research and National Institute of Aging (1K99AG042495-01A1).
We thank the lab of Dr. G. Lithgow for generously sharing the HSP-16 antibody and Dr. J. Durieux for helping design figure illustrations. Some of the nematode strains used in this work were provided by the Caenorhabditis Genetics Center (University of Minnesota), which is supported by the NIH – Office of Research Infrastructure Programs (P40 OD010440).
References and Notes
- 1.Hsu A-L. Regulation of Aging and Age-Related Disease by DAF-16 and Heat-Shock Factor. Science. 2003;300:1142–1145. doi: 10.1126/science.1083701. [DOI] [PubMed] [Google Scholar]
- 2.Fujimoto M. Active HSF1 Significantly Suppresses Polyglutamine Aggregate Formation in Cellular and Mouse Models. J. Biol. Chem. 2005;280:34908–34916. doi: 10.1074/jbc.M506288200. [DOI] [PubMed] [Google Scholar]
- 3.McColl G, et al. Insulin-like Signaling Determines Survival during Stress via Posttranscriptional Mechanisms in C. elegans. Cell Metab. 2010;12:260–272. doi: 10.1016/j.cmet.2010.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kourtis N, Nikoletopoulou V, Tavernarakis N. Small heat-shock proteins protect from heat-stroke-associated neurodegeneration. Nature. 2012;490:213–218. doi: 10.1038/nature11417. [DOI] [PubMed] [Google Scholar]
- 5.Prahlad V, Cornelius T, Morimoto RI. Regulation of the Cellular Heat Shock Response in Caenorhabditis elegans by Thermosensory Neurons. Science. 2008;320:811–814. doi: 10.1126/science.1156093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hajdu-Cronin YM. The L-Type Cyclin CYL-1 and the Heat-Shock-Factor HSF-1 Are Required for Heat-Shock-Induced Protein Expression in Caenorhabditis elegans. Genetics. 2004;168:1937–1949. doi: 10.1534/genetics.104.028423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kroeger PE, Morimoto RI. Selection of new HSF1 and HSF2 DNA-binding sites reveals difference in trimer cooperativity. Mol. Cell. Biol. 1994;14:7592–7603. doi: 10.1128/mcb.14.11.7592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Trinklein ND, Murray JI, Hartman SJ, Botstein D, Myers RM. The role of heat shock transcription factor 1 in the genome-wide regulation of the mammalian heat shock response. Mol. Biol. Cell. 2004;15:1254–1261. doi: 10.1091/mbc.E03-10-0738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Link CD, Cypser JR, Johnson CJ, Johnson TE. Direct observation of stress response in Caenorhabditis elegans using a reporter transgene. Cell Stress Chaperones. 1999;4:235–242. doi: 10.1379/1466-1268(1999)004<0235:doosri>2.3.co;2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Terami H, et al. Genomic organization, expression, and analysis of the troponin C gene pat-10 of Caenorhabditis elegans. J. Cell Biol. 1999;146:193–202. doi: 10.1083/jcb.146.1.193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ono K, Ono S. Tropomyosin and Troponin Are Required for Ovarian Contraction in the Caenorhabditis elegans Reproductive System. Mol. Biol. Cell. 2004;15:2782–2793. doi: 10.1091/mbc.E04-03-0179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Obinata T, Ono K, Ono S. Troponin I controls ovulatory contraction of non-striated actomyosin networks in the C. elegans somatic gonad. J. Cell Sci. 2010;123:1557–1566. doi: 10.1242/jcs.065060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yu R, Ono S. Dual roles of tropomyosin as an F-actin stabilizer and a regulator of muscle contraction in Caenorhabditis elegans body wall muscle. Cell Motil. Cytoskeleton. 2006;63:659–672. doi: 10.1002/cm.20152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Balklava Z, Pant S, Fares H, Grant BD. Genome-wide analysis identifies a general requirement for polarity proteins in endocytic traffic. Nat. Cell Biol. 2007;9:1066–1073. doi: 10.1038/ncb1627. [DOI] [PubMed] [Google Scholar]
- 15.Kuwahara T, et al. A systematic RNAi screen reveals involvement of endocytic pathway in neuronal dysfunction in α-synuclein transgenic C. elegans. Hum. Mol. Genet. 2008;17:2997–3009. doi: 10.1093/hmg/ddn198. [DOI] [PubMed] [Google Scholar]
- 16.Herndon LA, et al. Stochastic and genetic factors influence tissue-specific decline in ageing C. elegans. Nature. 2002;419:808–814. doi: 10.1038/nature01135. [DOI] [PubMed] [Google Scholar]
- 17.Greene W, Gao S-J. Actin Dynamics Regulate Multiple Endosomal Steps during Kaposi’s Sarcoma-Associated Herpesvirus Entry and Trafficking in Endothelial Cells. PLoS Pathog. 2009;5:e1000512. doi: 10.1371/journal.ppat.1000512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Grant B, et al. Evidence that RME-1, a conserved C. elegans EH-domain protein, functions in endocytic recycling. Nat. Cell Biol. 2001;3:573–579. doi: 10.1038/35078549. [DOI] [PubMed] [Google Scholar]
- 19.Fares H, Greenwald I. Regulation of endocytosis by CUP-5, the Caenorhabditis elegans mucolipin-1 homolog. Nat. Genet. 2001;28:64–68. doi: 10.1038/ng0501-64. [DOI] [PubMed] [Google Scholar]
- 20.Patton A, et al. Endocytosis Function of a Ligand-Gated Ion Channel Homolog in Caenorhabditis elegans. Curr. Biol. 2005;15:1045–1050. doi: 10.1016/j.cub.2005.04.057. [DOI] [PubMed] [Google Scholar]
- 21.Casella JF, Flanagan MD, Lin S. Cytochalasin D inhibits actin polymerization and induces depolymerization of actin filaments formed during platelet shape change. Nature. 1981;293:302–305. doi: 10.1038/293302a0. [DOI] [PubMed] [Google Scholar]
- 22.Spector I, Shochet NR, Kashman Y, Groweiss A. Latrunculins: novel marine toxins that disrupt microfilament organization in cultured cells. Science. 1983;219:493–495. doi: 10.1126/science.6681676. [DOI] [PubMed] [Google Scholar]
- 23.Whitesell L, Lindquist S. Inhibiting the transcription factor HSF1 as an anticancer strategy. Expert Opin. Ther. Targets. 2009;13:469–478. doi: 10.1517/14728220902832697. [DOI] [PubMed] [Google Scholar]
- 24.Nguyen CH, et al. Heat-shock factor 1 both positively and negatively affects cellular clonogenic growth depending on p53 status. Biochem. J. 2013;452:321–329. doi: 10.1042/BJ20130098. [DOI] [PubMed] [Google Scholar]
- 25.Ciocca DR, Arrigo AP, Calderwood SK. Heat shock proteins and heat shock factor 1 in carcinogenesis and tumor development: an update. Arch. Toxicol. 2013;87:19–48. doi: 10.1007/s00204-012-0918-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lavoie JN, Gingras-Breton G, Tanguay RM, Landry J. Induction of Chinese hamster HSP27 gene expression in mouse cells confers resistance to heat shock. HSP27 stabilization of the microfilament organization. J. Biol. Chem. 1993;268:3420–3429. [PubMed] [Google Scholar]
- 27.Mounier N, Arrigo A-P. Actin cytoskeleton and small heat shock proteins: how do they interact? Cell Stress Chaperones. 2002;7:167–176. doi: 10.1379/1466-1268(2002)007<0167:acashs>2.0.co;2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Doshi BM, Hightower LE, Lee J. The role of Hsp27 and actin in the regulation of movement in human cancer cells responding to heat shock. Cell Stress Chaperones. 2009;14:445–457. doi: 10.1007/s12192-008-0098-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Brenner S. The Genetics of Caenorhabditis Elegans. Genetics. 1974;77:71–94. doi: 10.1093/genetics/77.1.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hoogewijs D, Houthoofd K, Matthijssens F, Vandesompele J, Vanfleteren J. Selection and validation of a set of reliable reference genes for quantitative sod gene expression analysis in C. elegans. BMC Mol. Biol. 2008;9:9. doi: 10.1186/1471-2199-9-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Dillin A. Timing Requirements for Insulin/IGF-1 Signaling in C. elegans. Science. 2002;298:830–834. doi: 10.1126/science.1074240. [DOI] [PubMed] [Google Scholar]
- 32.Gentleman RC, et al. Bioconductor: open software development for computational biology and bioinformatics. Genome Biol. 2004;5:R80. doi: 10.1186/gb-2004-5-10-r80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wu Z, Irizarry RA, Gentleman R, Martinez-Murillo F, Spencer F. A Model-Based Background Adjustment for Oligonucleotide Expression Arrays. J. Am. Stat. Assoc. 2004;99:909. [Google Scholar]
- 34.Smyth GK. Linear models and empirical bayes methods for assessing differential expression in microarray experiments. Stat. Appl. Genet. Mol. Biol. 2004;3 doi: 10.2202/1544-6115.1027. Article3. [DOI] [PubMed] [Google Scholar]
- 35.Benjamini Y, Hochberg Y. Controlling the False Discovery Rate: A Practical and Powerful Approach to Multiple Testing. J. R. Stat. Soc. Ser. B Methodol. 1995;57:289–300. [Google Scholar]
- 36.Wolters DA, Washburn MP, Yates JR., 3rd An automated multidimensional protein identification technology for shotgun proteomics. Anal. Chem. 2001;73:5683–5690. doi: 10.1021/ac010617e. [DOI] [PubMed] [Google Scholar]
- 37.McDonald WH, et al. MS1, MS2, and SQT-three unified, compact, and easily parsed file formats for the storage of shotgun proteomic spectra and identifications. Rapid Commun. Mass Spectrom. RCM. 2004;18:2162–2168. doi: 10.1002/rcm.1603. [DOI] [PubMed] [Google Scholar]
- 38.Peng J, Elias JE, Thoreen CC, Licklider LJ, Gygi SP. Evaluation of multidimensional chromatography coupled with tandem mass spectrometry (LC/LC-MS/MS) for large-scale protein analysis: the yeast proteome. J. Proteome Res. 2003;2:43–50. doi: 10.1021/pr025556v. [DOI] [PubMed] [Google Scholar]
- 39.Xu T, et al. ProLuCID, a Fast and Sensitive Tandem Mass Spectra-based Protein Identification Program. Mol. Cell. Proteomics. 2006;5:S174. [Google Scholar]
- 40.Tabb DL, McDonald WH, Yates JR., 3rd DTASelect and Contrast: tools for assembling and comparing protein identifications from shotgun proteomics. J. Proteome Res. 2002;1:21–26. doi: 10.1021/pr015504q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Park SK, Venable JD, Xu T, Yates JR. A quantitative analysis software tool for mass spectrometry–based proteomics. Nat. Methods. 2008;5:319–322. doi: 10.1038/nmeth.1195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ashburner M, et al. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat. Genet. 2000;25:25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Huang DW, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 44.Cartharius K, et al. MatInspector and beyond: promoter analysis based on transcription factor binding sites. Bioinforma. Oxf. Engl. 2005;21:2933–2942. doi: 10.1093/bioinformatics/bti473. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.