Abstract
Lipooligosaccharides are glycolipids found in the cell wall of many mycobacterial species including the opportunistic pathogen Mycobacterium kansasii. The genome of M. kansasii ATCC12478 contains a cluster with genes orthologous to Mycobacterium marinum LOS biosynthesis genes. To initiate a genetic dissection of this cluster and demonstrate its role in LOS biosynthesis in M. kansasii, we chose MKAN27435, a gene encoding a putative glycosyltransferase. Using Specialized Transduction, a phage-based gene knockout tool previously used to generate null mutants in other mycobacteria, we generated a MKAN27435 null mutant. The mutant strain was found to be defective in the biosynthesis of higher LOS subspecies, viz LOS-IV, LOS-V, LOS-VI and LOS-VII. Additionally, a range of low abundance species were detected in the mutant strain and mass spectroscopic analysis indicated that these were shunt products generated from LOS-III by the addition of up to six molecules of a pentose.
Introduction
Unique lipids found in the distinct cell wall of mycobacteria are important for integrity and some play a vital role in virulence [1,2]. Lipooligosaccharides (LOS’s) are cell wall associated glycolipids produced by many mycobacterial species including Mycobacterium canetti, the ‘smooth’ Mycobacterium tuberculosis complex (MTBC) strain [3]. Interestingly, Mycobacterium tuberculosis, also a MTBC strain, does not produce LOSs but the genome of M. tuberculosis H37Rv does contain a ‘reduced’ LOS biosynthetic cluster representative of approximately a third of genes found in M. canetti [4]. Other LOS producers include the opportunistic pathogen Mycobacterium kansasii and poikilotherm pathogen Mycobacterium marinum [5,6].
M. kansasii causes chronic pulmonary and disseminated infections in humans and is the second most common cause of non-tuberculosis mycobacterial (NTM) infections after Mycobacterium avium complex (MAC) [7–9]. There are subtle differences in lung lesions caused by M. kansasii and M. tuberculosis with regards to the location of lesions and the tendency to form cavitary lesions, and unlike M. tuberculosis, M. kansasii infections are not contagious. M. kansasii is also commonly found in HIV patients; genomic analysis of M. kansasii strains identified five subtypes associated with HIV patients [10,11]. Mouse infection studies have shown that the rough variant of M. kansasii, which is deficient in LOS biosynthesis, survives longer causing chronic pulmonary disease compared to smooth, LOS-producing strains [12,13]. Similarly, the LOS-producing MTBC strain M. canetti is an opportunistic human pathogen, unlike M. tuberculosis which is virulent in humans. These findings indicate that a loss of LOSs may be one, if not the only, defining factor in the increased virulence of M. tuberculosis in humans. Furthermore, the role of individual subclasses of LOSs in antigenicity and immunomodulation is poorly understood in context of co-relation between LOS production and severity of disease. Recently, LOS-IV, but not the other LOS-subclasses from M. marinum, was shown to inhibit TNF-α release in LPS-activated macrophages [14]. Similarly, a LOS-IV-deficient mutant showed increased virulence in infected zebrafish embryos [15]. Furthermore, Alibaud et al. [16] observed that M. marinum strains deficient in LOS production were phagocytized more efficiently than those accumulating only early LOS intermediates, while wild type strains and LOS-IV-lacking mutants were phagocytized with the least efficiency. A fine dissection of role of different LOS-subclasses and their co-relation to the ability to cause disease in models such as M.kansasii and M.marinum can eventually help us better understand how M. tuberculosis, which has lost the ability to make LOS’s through reductive evolution [4], became a more virulent human pathogen than other LOS producing members of the MTB complex like M. canetti.
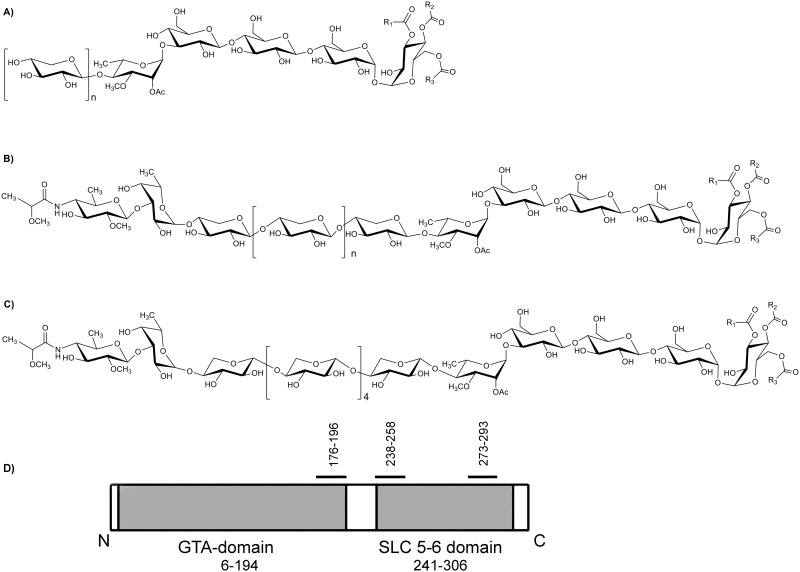
M. kansasii produces seven subclasses of LOS’s, all of which contain a common triacylated, tetra glucose core (Fig 1). In addition all the subclasses of LOSs contain 3-O-Me-rhamnose and varying amounts of xylose. Further, LOS subclasses LOS-IV, LOS-V, LOS-VI and LOS-VII contain a terminal fucose and unique sugar N-acyl kansosamine [17,18]. While chemical structures of LOS’s from various mycobacteria have been elucidated, far less is known about LOS biosynthesis pathways [17,18]. Most of our understanding of enzymes involved in LOS biosynthesis comes from genetic studies in M. marinum [4, 19, 20] in which mutant strains accumulating intermediates were used to identify glycosyl transferases involved in LOS biosynthesis. The LOS biosynthesis cluster in M. marinum includes at least 38 ORFs. By comparing the LOS biosynthesis gene cluster from M. marinum with the M. kansasii ATCC12478 genome sequence, we identified a potential LOS biosynthesis cluster in M. kansasii containing ORFs encoding putative proteins with domains found in glycosyl transferases (GTFs). Targeted deletion of LOS genes will allow us to generate strains that produce intermediate patterns of LOSs, which can be then tested in laboratory models of infection to study the impact of loss of all LOSs, and of individual LOS species, in virulence and immunomodulation. M. kansasii, with its ability cause opportunistic pulmonary infections in humans, and co-relation of the absence of LOSs to its ability to cause disease, make it a suitable model system to conduct the above mentioned studies. Here, we have generated and characterized a null mutant of the ORF MKAN27435 which encodes a putative glycosyltransferase, to probe its role in LOS biosynthesis.
Fig 1. Structures of different LOS subclasses from M. kansasii .
(A) LOS-I-III (n = 1–3), (B) LOS-IV-VII (n = 2–4), (C) LOS-VII; R1, R2 and R3 represents the acyl chain attached to tetra glucose core of LOS. (D) Schematic showing predicted domains and topology of MKAN27435. The amino and carboxy terminals are indicated as ‘N’ and ‘C’ respectively, and numbers represent predicted borders of domains which are depicted as grey areas. The regions corresponding to the predicted transmembrane domains are indicated with black bars.
Materials and Methods
Generation of MKAN27435 null mutant
The M. kansasii null mutant ΔMKAN27435 was generated by Specialized Transduction, a phage mediated gene knockout methodology used previously for other mycobacterial species [21,22]. Approximately 1kb regions upstream and downstream of MKAN27435 were PCR amplified and cloned on either side of a hygromycin resistance cassette (hyg) in the plasmid p0004S [23] to generate the allelic exchange substrate plasmid pΔMKAN27435. pΔMKAN27435 was then packaged into the temperature sensitive mycobacteriophage phAE159 [21] to generate the recombinant knockout phage phΔMKAN27435. The wild type (WT) strain M. kansasii ATCC12478 was transduced with phΔMKAN27435 using the protocols described by Larsen et al. [23] and the transductants were selected on 7H10 agar plate containing 100 μg/ml hygromycin. Replacement of MKAN27435 with the hyg cassette in the transductants was confirmed by Southern blot (S1 Fig). One such strain was designated ΔMKAN27435 and was used for all subsequent analysis.
Complementation of the ΔMKAN27435 strain
The MKAN27435 ORF was amplified from M. kansasii ATCC12478 genomic DNA using the primer pair F27435 (5’-ATGCGAATTCGTGGATTCGGTCAGCGTTG-3’) and R27435 (5’-AT GCAAGCTTTCAGTCCGCCGGATTGTCGAAG-3’). The PCR product was cloned into the E. coli-mycobacterial replicative shuttle plasmid pMV261 [24] using the primer-incorporated EcoRI and HindIII sites (underlined in primer sequence). The cloned PCR product was verified by sequencing and subsequently electroporated into the ΔMKAN27435 strain [24]. Transformants were selected on 7H10 plates containing 100 μg/ml hygromycin and 25 μg/ml kanamycin. One such transformant was designated ΔMKAN27435-C and was used for subsequent analysis.
Lipid analysis of M. kansasii strains
Wild type, mutant and complemented strains M. kansasii were grown in 10 ml 7H9 broth and lipids were labeled by adding 1μCi/ml [14C] acetate (57mCi/mmol) at midlog phase. Apolar and polar lipids were then extracted from the cell pellets obtained from these cultures using methods described by Dobson et al. [25]. The extracted lipids were re-constituted in chloroform: methanol (2:1 v/v). The polar lipids were separated by 2-dimensional (2D) TLC using solvent system E [25]. Polar lipids were visualized by autoradiography by exposing the TLC plates to a Kodak BioMax MR film for 24–48 h.
Purification of LOS’s from M. kansasii wild type and ΔMKAN27435
Cells of M.kansasii WT and ΔMKAN27435 strains were harvested from a 6 L culture, and fractions of polar and apolar lipids were extracted using a scaled up version of the protocol of Dobson et al. mentioned in the above section [25]. LOS’s were purified from the polar lipid fraction by using DEAE-cellulose an-ion exchange chromatography. Briefly, DEAE-cellulose is equilibrated in chloroform: methanol (2:1 v/v). The polar lipid fraction was applied to the column and eluted with 0, 10mM, 25mM, 100mM and 500mM ammonium acetate in chloroform: methanol (2:1 v/v). The fractions were collected and dried on a rotary evaporator. The dried fractions were reconstituted in chloroform: methanol (2:1 v/v) and separated by 2D TLC using solvent system E [25]. Purified LOS’s on the TLC plates were visualized by α-naphthol staining.
Mass spectroscopy analysis of LOS subclasses from M. kansasii WT and mutant strains
The LOS subclasses from M. kansasii WT and ΔMKAN27435 strains were permethylated using sodium hydroxide: for each sample, about 5 NaOH pellets were ground to fine powder in a dry mortar with a pestle. About 3 ml of anhydrous DMSO was added to form a slurry. About 1 ml of the resulting slurry was added to the sample before 0.5 ml of methyl iodide was added. The reaction mixture was mixed on a vortex mixer and then agitated on an automatic shaker for 30 min at room temperature. The reaction was quenched with water, while constantly shaking the tube. Permethylated samples were extracted with 1 ml of chloroform and washed several times with 3 ml of water. The chloroform was dried down under a gentle stream of nitrogen gas before sample purification using a Sep-Pak C18 cartridge. The permethylated LOS were then dissolved in methanol before an aliquot was mixed at a 1:1 ratio (v/v) with 10 mg/ml 3,4-diaminobenzophenone in 75% acetonitrile. The glycan-matrix mixture was spotted on a target plate and dried. MALDI-TOF MS data were obtained using a 4800 MALDI-TOF/TOF mass spectrometer (AB Sciex UK Limited) in the positive ion mode. The obtained MS data were viewed and processed using Data Explorer 4.9 (AB Sciex UK Limited).
Results
MKAN27435 encodes a putative glycosyl transferase
The LOS biosynthesis cluster in M. marinum contains at least 38 genes. In addition to numerous glycosyl transferases, the region also contains a polyketide synthase gene, pks5 responsible for the biosynthesis of the acyl chains found in LOSs. We used the M. marinum pks5 sequence as a query for a BLAST search to locate a putative LOS biosynthesis cluster in the genome of M. kansasii ATCC12478 (Table 1). Compared to five LOS-associated glycosyl transferase (GTF) genes found in M. marinum, the M. kansasii LOS biosynthetic cluster contains eight genes encoding putative GTFs. Only three orthologues of M. marinum LOS-associated GTFs are present in M. kansasii, suggesting that the other GTF genes found in the latter were involved in the addition of the unique sugars found in M. kansasii LOSs, viz fucose and N-acyl kansosamine. We then selected MKAN27435, a putative GTF-encoding gene that has no orthologues in M. marinum, for further genetic analysis. BLASTP analysis using the MKAN27435 sequence revealed matches to LosA (MMAR2313) and WcaA (MMAR2333) from M. marinum, but with relatively low scores (26% identity and 42% similarity with LosA and 23% identity and 43% similarity with WcaA). A preliminary amino acid sequence analysis of MKAN27435 revealed a domain characteristic of members of the GT-2 family of glycosyltransferases that are similar to eukaryotic dolichol phosphatemannose (DPM) synthases (Fig 1). These GTFs, that typically contain a GT-A fold [26], catalyze the transfer of sugars to dolichol phosphate by using nucleotide sugars as substrates. Topology predictions also indicated the presence of three transmembrane helices, indicating a membrane-associated function. Additionally, the C-terminal region of MKAN27435 contains a domain similar to the solute-binding domain of SLC5 proteins which are Na+/sugar co-transporters with affinity for D-galactose, D-glucose, and D-fucose. Given the chemical composition of M. kansasii LOSs (Fig 1), it seemed likely that MKAN27435 was involved in the addition of either a glucose, or a fucose residue during LOS biogenesis in M. kansasii. The role could be either direct, involving addition of one of these sugars to a growing LOS intermediate, or indirect, involving the formation of a polyprenyl-linked glucose or fucose moiety that was a substrate for another GTF involved in LOS biosynthesis (S2 Fig).
Table 1. Tabulated comparison of genes from LOS biosynthetic clusters of M. kansasii and M. marinum.
| M. kansasii | M.marinum | Putative function |
|---|---|---|
| MKAN27390 | MMAR2311 | Glycosyl transferase, GT-A type |
| - | MMAR2313 | Glycosyl transferase, GT-A type |
| - | MMAR2333 | Glycosyl transferase, GT-A type |
| MKAN27425 | - | Glycosyl transferase, GT-A type |
| MKAN27430c | - | NAD dependent epimerase/dehydratase family |
| MKAN27435 | - | Glycosyltransferase, GT-A type |
| MKAN27440 | - | Glucose-1-phosphate cytidiltransferase |
| MKAN27445 | - | NAD dependent epimerase/dehydratase family |
| MKAN27450 | - | Rhamnose epimerase |
| MKAN27475 | - | Methyltransferase |
| MKAN27485 | MMAR2340 | polyketide synthase |
| MKAN27490 | MMAR2341 | Fatty acyl co-A synthatase |
| MKAN27530 | MMAR2342 | MmpL family transport protein |
| MKAN27540 | MMAR2344 | polyketide synthase |
| MKAN27575 | MMAR2349 | Rhamnosyltransferase |
| MKAN27580 | MMAR2351 | Glycosyl transferase, GT-A type |
| MKAN27600 | MMAR2353 | Glycosyl transferase, GT-B type |
| MKAN27610 | MMAR2355 | Acyl transferase |
| MKAN27675 | MMAR2366 | Fatty acyl co-A reductase |
| MKAN27680 | MMAR2367 | keto acyl reductase |
| MKAN27695 | - | Glycosyl transferase, GT-A type |
Where genes are unique to each species, the absence of an ortholog in the other species is indicated by a hyphen (-).
Loss of MKAN27435 alters LOS biosynthesis
Given the presence of MKAN27435 in a putative LOS biosynthesis cluster, and the outcomes of the bioinformatics analysis described above, we sought to probe the role of MKAN27435 as a GTF involved in the biosynthesis of LOS’s in M. kansasii. First, a null mutant of MKAN27435 was generated in the parental M. kansasii ATCC12478 strain (referred to as M. kansasii WT henceforth) using Specialized Transduction. Next, cultures of the strains were grown in the presence of [14C]-acetate to label lipids, and LOS’s were isolated as part of the polar lipid fraction from the labeled cultures. Separation of the polar lipids by 2D TLC [25] revealed an altered pattern of LOS species in the mutant strain; while M. kansasii WT produced all the subclasses of LOS’s (LOS-I, LOS-II, LOS-III, LOS-IV, LOS-V, LOS-VI and LOS-VII) the mutant produced LOS-I, LOS-II and LOS-III, but LOS-IV, LOS-V, LOS-VI and LOS-VII were missing in the strain (Fig 2). Furthermore, a new set of less abundant polar lipid species were detected in the mutant strain (Fig 2). Synthesis of all LOS species was restored in the mutant strain on introduction of a plasmid-encoded copy of MKAN27435 (Fig 2), indicating that the observed alteration in LOS patterns in the mutant was solely due to the loss of MKAN27435 function. In addition to the above phenotypes, we did observe a faint spot migrating closest to the origin in the 2D-TLC plate for the M. kansasii WT strain, which was absent in the mutant strain. However, as the spot did not reappear in the complemented strain, we presumed that this was an artifact and was unrelated to MKAN27435 function.
Fig 2. Autoradiograph of 2-D TLC analysis of polar lipids extracted from M. kansasii WT, ΔMKAN27435, ΔMKAN27435-C strains.
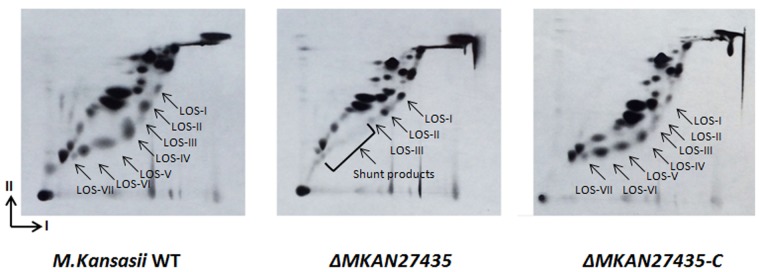
Arrows indicate the different LOS species. I and II indicate system E dimension 1 and 2 respectively. Dimension I: Chloroform: Methanol: H2O (60:30:6); Dimension II: Chloroform: Acetic acid: Methanol: H2O (40:25:3:6).
Mass spectroscopic analysis of LOS subclasses
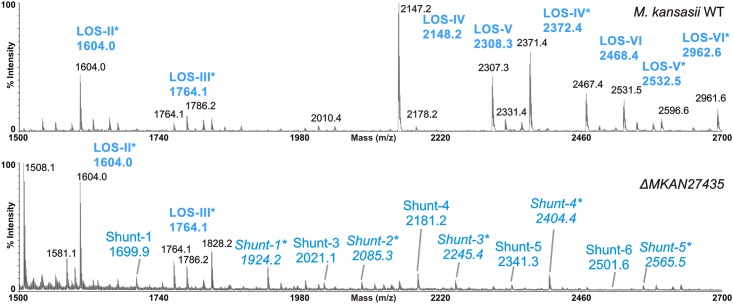
To further characterize the set of lower abundance polar lipid species detected in the mutant strain, fractionated polar lipids from the wild type and mutant strains, were subjected to MALDI-TOF mass spectroscopy (Tables 2 and 3; Fig 3). The sample preparation process included a permethylation step, which variably removes acyl groups and replaces them with a methyl group, and also methylates free hydroxyl groups. We were able to thus detect acylated and non-acylated variants. Acyl group retention is indicated by increments of 221Da, for example the species m/z 2148.2 corresponds to LOS-IV in the WT strain and m/z 2372.4 represents LOS-IV with acyl intact (represented as LOS-IV*). Consistent with the TLC data, higher LOSs were missing in the mutant strain. We also detected a range of low abundance species present only in the mutant strain with sizes that corresponded to incremental additions of m/z 160 to LOS-III (Table 3, Fig 3). Each increment corresponded to a per-O-methylated pentose and the size ranges observed indicated that the species were likely LOS-III elaborated further with up to six pentose residues.
Table 2. MALDI-TOF mass spectroscopy analyses tabulated to list LOS species with corresponding m/z values in M. kansasii WT and ΔMKAN27435 strains.
| m/z | LOS species | M.kansasii WT | ΔMKAN27435 |
|---|---|---|---|
| 1443.91 | LOS-I* | + | + |
| 1604 | LOS-II* | + | + |
| 1539.9 | LOS-III | + | + |
| 1764.1 | LOS-III* | + | + |
| 2148.2 | LOS-IV | + | - |
| 2372.4 | LOS-IV* | + | - |
| 2308.3 | LOS-V | + | - |
| 2532.5 | LOS-V* | + | - |
| 2468.4 | LOS-VI | + | - |
| 2692.6 | LOS-VI* | + | - |
All molecular ions are [M+Na]+and an asterisk (*) indicates species with an intact acyl group. Presence (+) or absence (-) of each species is indicated for each strain.
Table 3. MALDI-TOF mass spectroscopy analysis tabulated to list shunt products and corresponding m/z values in M. kansasii ΔMKAN27435.
| m/z | Product | Putative structure |
|---|---|---|
| 1699.9 | Shunt product-1 | LOS-III + 1 X pentose |
| 1860.1 | Shunt product-2 | LOS-III + 2 X pentose |
| 2021.1 | Shunt product-3 | LOS-III + 3 X pentose |
| 2181.2 | Shunt product-4 | LOS-III + 4 X pentose |
| 2341.3 | Shunt product-5 | LOS-III + 5 X pentose |
| 2501.6 | Shunt product-6 | LOS-III + 6 X pentose |
| 1924.2 | Shunt product-1* | LOS-III + 1X pentose |
| 2085.3 | Shunt product-2* | LOS-III + 2X pentose |
| 2245.3 | Shunt product-3* | LOS-III + 3X pentose |
| 2404.4 | Shunt product-4* | LOS-III + 4X pentose |
| 2565.5 | Shunt product-5* | LOS-III + 5X pentose |
All molecular ions are [M+Na]+ and products with an asterisk (*) correspond to m/z values with an intact acyl group.
Fig 3. MALDI-TOF mass spectroscopy analysis of purified LOS’s from M.kansasii WT and ΔMKAN27435 strains in positive ion mode.
All molecular ions are [M+Na]+ and species indicated with an asterisk (*) correspond to m/z values with an intact acyl group.
Discussion
Using Specialized Transduction, we were able to generate the first targeted gene knockout of M. kansasii, demonstrating the utility of the phage-based method as a genetic tool for this mycobacterial species. The MKAN27435 null mutant was defective in making four of the seven subclasses of LOS’s indicating that the mutant strain’s inability to extend LOS-III to subsequent LOS subclasses via the addition of a fucose residue. Given the presence of domains from DPM-like GTFs in MKAN27435, it was likely that MKAN27435 catalyzes the transfer of nucleotide-bound fucose to a polyprenol carrier. Polyprenol-bound fucose could then be ‘flipped’ across the membrane and subsequently utilized as substrate by an extracellular fucosyltransferase to add fucose to LOS-III (S2 Fig). There are a total of two DPM-like GTFs in the M. kansasii LOS cluster indicating that in part, sugar addition to the LOS core likely occurs via polyprenol-linked sugar donors and may thus occur extracellularly. However, there remains an alternative possibility that DPM-like GTFs encoded by LOS clusters directly utilize membrane attached LOS intermediates as substrate.
In addition to the loss of LOS-IV, LOS-V, LOS-VI and LOS-VII, we also observed a number of low abundance polar lipid species in the mutant strain; m/z values suggested that these were likely shunt products derived from LOS-III containing up to six additional pentose residues. Though the low abundance of these polar lipid species limited the detailed analysis we could conduct on these species, it could be likely that the pentose residues were additional xyloses transferred to LOS-III in the mutant strain. Surprisingly, we did not observe any difference in the colony morphology of the mutant suggesting that a complete loss of all LOS subclasses was essential for the transition to a rough colony phenotype. The generation and characterization of this mutant in M. kansasii opens up opportunities for the generation of further targeted mutants in M. kansasii not limited to the study of LOS biosynthesis.
Supporting Information
(A) Schematic representation of the MKAN27435 region in the M. kansasii ATCC12478 (WT) genome and its corresponding region in the ΔMKAN27435 mutant; HYG, hygromycin resistance gene from Streptomyces hygroscopicus, sacB, sucrose counterselectable gene from Bacillus subtilis. (B) Individual lanes from a Southern blot of KpnI digested genomic DNA from M. kansasii WT and ΔMKAN27435 mutant strains. Probes were the left and right flanking sequences originally PCR amplified to generate the allelic exchange substrate and were labelled using Roche DIG-High Prime DNA labelling and detection kit. Expected restriction bands that bind to the probe are indicated for each strain. Bands were visualised by exposing an X-ray film to chemiluminescence generated as apart of the Southern blotting protocol.
(DOCX)
Proposed pathway of LOS-IV biosynthesis in M. kansasii showing the involvement of MKAN27435 in transferring a nucleotide sugar (fucose) to a polyprenol unit. The polyprenol bound fucose may serve as a sugar donor for the synthesis of LOS-IV by a yet unknown glycosyl transferase. Glu, glucose; Me-Rha, 3-O-Me-Rhamnose; Xyl, Xylose; Fuc, Fucose and N-acyl Kan, N-acyl kansosamine.
(DOCX)
Data Availability
All relevant data are within the paper and its Supporting Information files.
Funding Statement
This work was funded by grants from the Medical Research Council (UK). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Brennan PJ, Nikaido H (1995) The envelope of mycobacteria. Annu Rev Biochem 64: 29–63. [DOI] [PubMed] [Google Scholar]
- 2. Daffe M, Draper P (1998) The envelope layers of mycobacteria with reference to their pathogenicity. AdvMicrobPhysiol 39: 131–203. [DOI] [PubMed] [Google Scholar]
- 3. Daffe M, McNeil M, Brennan PJ (1991) Novel type-specific lipooligosaccharides from Mycobacterium tuberculosis . Biochemistry 30: 378–388. [DOI] [PubMed] [Google Scholar]
- 4. Ren H, Dover LG, Islam ST, Alexander DC, Chen JM, Besra GS, et al. (2007) Identification of the lipooligosaccharide biosynthetic gene cluster from Mycobacterium marinum . Mol Microbiol 63: 1345–1359. [DOI] [PubMed] [Google Scholar]
- 5. Hunter SW, Jardine I, Yanagihara DL, Brennan PJ (1985) Trehalose-containing lipooligosaccharides from mycobacteria: structures of the oligosaccharide segments and recognition of a unique N-acylkanosamine-containing epitope. Biochemistry 24: 2798–2805. [DOI] [PubMed] [Google Scholar]
- 6. Minnikin DE, Ridell M, Wallerström G, Besra GS, Parlett JH, Bolton RC, et al. (1989) Comparative studies of antigenic glycolipids of mycobacteria related to the leprosy bacillus. Acta Leprol 7: 51–54. [PubMed] [Google Scholar]
- 7. Rombouts Y, Elass E, Biot C, Maes E, Coddeville B, Burguière A, et al. (2010) Structural analysis of an unusual bioactive N-acylated lipo-oligosaccharide LOS-IV in Mycobacterium marinum . J Am Chem Soc 132: 16073–16084. 10.1021/ja105807s [DOI] [PubMed] [Google Scholar]
- 8. Lillo M, Orengo S, Cernoch P, Harris RL (1990) Pulmonary and disseminated infection due to Mycobacterium kansasii: a decade of experience. Rev Infect Dis 12: 760–767. [DOI] [PubMed] [Google Scholar]
- 9. Tompkins JC, Witzig RS (2007) Mycobacterium kansasii in HIV patients: clarithromycin and antiretroviral effects. Int Tuberc Lung Dis 11: 331–337. [PubMed] [Google Scholar]
- 10. Picardeau M, Prod’Hom G, Raskine L, LePennec MP, Vincent V (1997) Genotypic characterization of five subspecies of Mycobacterium kansasii . J Clin Microbiol 35: 25–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Evans SA, Colville A, Evans AJ, Crisp AJ, Johnston ID (1996) Pulmonary Mycobacterium kansasii infection: comparison of the clinical features, treatment and outcome with pulmonary tuberculosis. Thorax 51: 1248–1252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Belisle JT, Brennan PJ (1989) Chemical basis of rough and smooth variation in mycobacteria. J Bacteriol 171: 3465–3470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Collins FM, Cunningham DS (1981) Systemic Mycobacterium kansasii infection and regulation of the alloantigenic response. Infect Immun 32: 614–624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Rombouts Y, Burguière A, Maes E, Coddeville B, Elass E, Guérardel Y, et al. (2009) Mycobacterium marinum lipooligosaccharides are unique caryophyllose-containing cell wall glycolipids that inhibit tumor necrosis factor-alpha secretion in macrophages. J Biol Chem 284: 20975–20988. 10.1074/jbc.M109.011429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Van der Woude AD, Sarkar D, Bhatt A, Sparrius M, Raadsen SA, Boon L, et al. (2012) Unexpected link between lipooligosaccharide biosynthesis and surface protein release in Mycobacterium marinum . J Biol Chem 287: 20417–20429. 10.1074/jbc.M111.336461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Alibaud L, Pawelczyk J, Gannoun-Zaki L, Singh VK, Rombouts Y, Drancourt M, et al. (2014) Increased phagocytosis of Mycobacterium marinum mutants defective in lipooligosaccharide production: a structure-activity relationship study. J Biol Chem 289: 215–228. 10.1074/jbc.M113.525550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Hunter SW, Murphy RC, Clay K, Gorent MB, Brennan PJ (1983) Trehalose-containing Lipooligosaccharides. J Biol Chem 258: 10481–10487. [PubMed] [Google Scholar]
- 18. Hunter SW, Fujiwara T, Murphy RC, Brennan PJ (1984) N-acylkanosamine. A novel N-acylamino sugar from the trehalose-containing lipooligosaccharide antigens of Mycobacterium kansasii . J Biol Chem 259: 9729–9734. [PubMed] [Google Scholar]
- 19. Burguière A, Hitchen PG, Dover LG, Kremer L, Ridell M, et al. (2005) LosA, a key glycosyltransferase involved in the biosynthesis of a novel family of glycosylated acyltrehalose lipooligosaccharides from Mycobacterium marinum . J Biol Chem 280: 42124–42133. [DOI] [PubMed] [Google Scholar]
- 20. Sarkar D, Sidhu M, Singh A, Chen J, Lammas D A, van der Sar AM, et al. (2011) Identification of a glycosyltransferase from Mycobacterium marinum involved in addition of a caryophyllose moiety in lipooligosaccharides. J Bacteriol 193: 2336–2340. 10.1128/JB.00065-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Bardarov S, Kriakov J, Carriere C, Yu S, Vaamonde C, McAdam RA, et al. (1997) Conditionally replicating mycobacteriophages: a system for transposon delivery to Mycobacterium tuberculosis . Proc Natl Acad Sci U S A 94: 10961–10966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Bardarov S, Bardarov S Jr, Pavelka MS Jr, Sambandamurthy V, Larsen M, Tufariello J, et al. (2002) Specialized transduction: an efficient method for generating marked and unmarked targeted gene disruptions in Mycobacterium tuberculosis, M. bovis BCG and M. smegmatis . Microbiology 148: 3007–3017. [DOI] [PubMed] [Google Scholar]
- 23. Larsen MH, Biermann K, Tandberg S, Hsu T, Jacobs WR (2007) Genetic Manipulation of Mycobacterium tuberculosis . Curr Protoc Microbiol. Vol. Chapter 10 pp. A2.1–A2.21. [DOI] [PubMed] [Google Scholar]
- 24. Stover CK, de La Cruz VF, Fuerst TR, Burlein JE, Benson LA, Bennett LT, et al. (1991) New use of BCG for recombinant vaccines. Nature 351: 456–460. [DOI] [PubMed] [Google Scholar]
- 25. Dobson G, Minnikin DE, Minnikin SM, Parlett M, Goodfellow M, Ridell M, et al. (1985) Systematic analysis of complex mycobacterial lipids In: Goodfellow M, Minnikin D., editors. In Chemical methods in bacterial systematics. London: Academic Press; pp. 237–265. [Google Scholar]
- 26. Breton C, Snajdrová L, Jeanneau C, Koca J, Imberty A (2006) Structures and mechanisms of glycosyltransferases. Glycobiology 16: 29R–37R. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(A) Schematic representation of the MKAN27435 region in the M. kansasii ATCC12478 (WT) genome and its corresponding region in the ΔMKAN27435 mutant; HYG, hygromycin resistance gene from Streptomyces hygroscopicus, sacB, sucrose counterselectable gene from Bacillus subtilis. (B) Individual lanes from a Southern blot of KpnI digested genomic DNA from M. kansasii WT and ΔMKAN27435 mutant strains. Probes were the left and right flanking sequences originally PCR amplified to generate the allelic exchange substrate and were labelled using Roche DIG-High Prime DNA labelling and detection kit. Expected restriction bands that bind to the probe are indicated for each strain. Bands were visualised by exposing an X-ray film to chemiluminescence generated as apart of the Southern blotting protocol.
(DOCX)
Proposed pathway of LOS-IV biosynthesis in M. kansasii showing the involvement of MKAN27435 in transferring a nucleotide sugar (fucose) to a polyprenol unit. The polyprenol bound fucose may serve as a sugar donor for the synthesis of LOS-IV by a yet unknown glycosyl transferase. Glu, glucose; Me-Rha, 3-O-Me-Rhamnose; Xyl, Xylose; Fuc, Fucose and N-acyl Kan, N-acyl kansosamine.
(DOCX)
Data Availability Statement
All relevant data are within the paper and its Supporting Information files.