Abstract
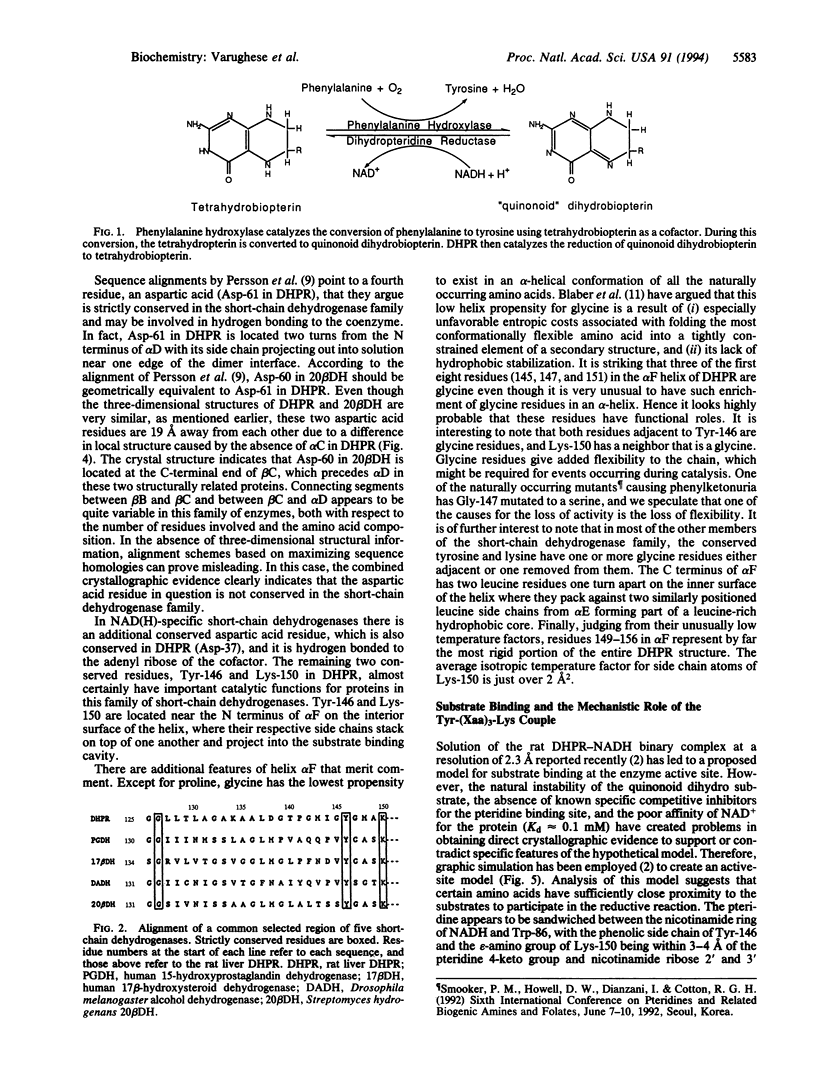
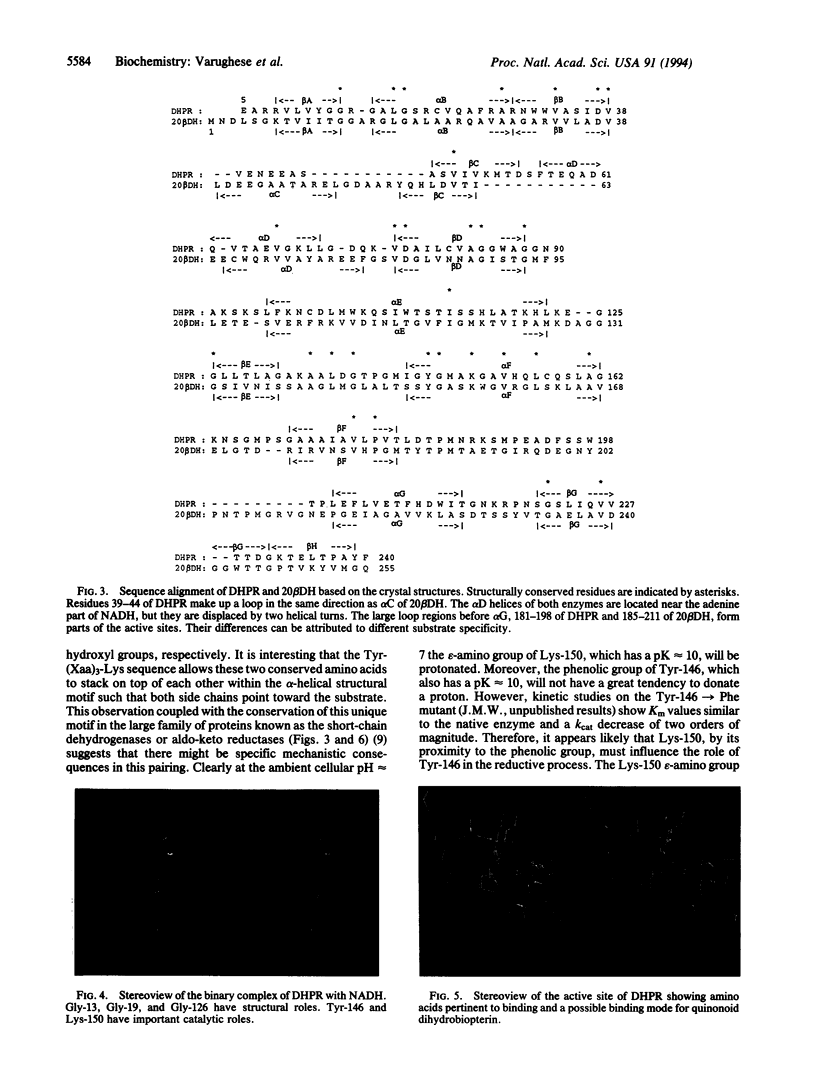
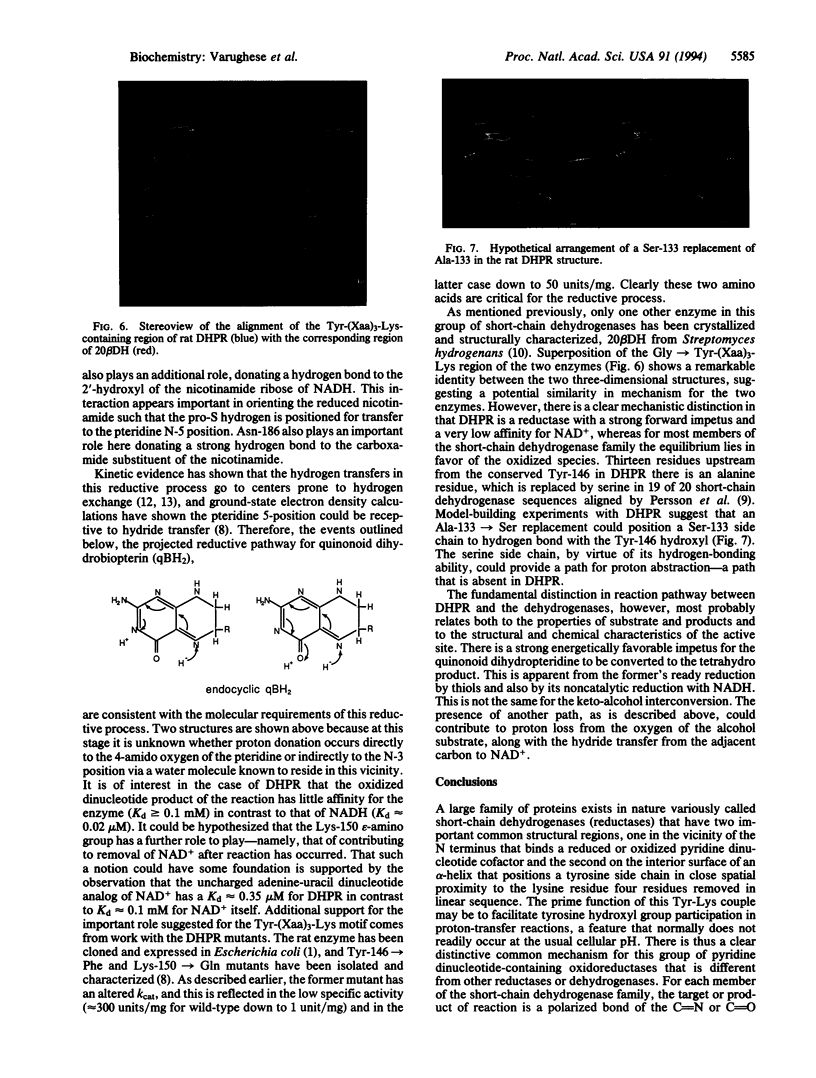
Dihydropteridine reductase (EC 1.6.99.7) is a member of the recently identified family of proteins known as short-chain dehydrogenases. When the x-ray structure of dihydropteridine reductase is correlated with conserved amino acid sequences characteristic of this enzyme class, two important common structural regions can be identified. One is close to the protein N terminus and serves as the cofactor binding site, while a second conserved feature makes up the inner surface of an alpha-helix in which a tyrosine side chain is positioned in close proximity to a lysine residue four residues downstream in the sequence. The main function of this Tyr-Lys couple may be to facilitate tyrosine hydroxyl group participation in proton transfer. Thus, it appears that there is a distinctive common mechanism for this group of short-chain or pyridine dinucleotide-dependent oxidoreductases that is different from their higher molecular weight counterparts.
Full text
PDF

Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Armarego W. L. Hydrogen transfer from 4-R and 4-S (4-3H) NADH in the reduction of d,l-cis-6,7-dimethyl-6,7 (8H) dihydropterin with dihydropteridine reductase from human liver and sheep liver. Biochem Biophys Res Commun. 1979 Jul 12;89(1):246–249. doi: 10.1016/0006-291x(79)90970-7. [DOI] [PubMed] [Google Scholar]
- Armarego W. L., Randles D., Waring P. Dihydropteridine reductase (DHPR), its cofactors, and its mode of action. Med Res Rev. 1984 Jul-Sep;4(3):267–321. doi: 10.1002/med.2610040302. [DOI] [PubMed] [Google Scholar]
- Blaber M., Zhang X. J., Matthews B. W. Structural basis of amino acid alpha helix propensity. Science. 1993 Jun 11;260(5114):1637–1640. doi: 10.1126/science.8503008. [DOI] [PubMed] [Google Scholar]
- Ghosh D., Weeks C. M., Grochulski P., Duax W. L., Erman M., Rimsay R. L., Orr J. C. Three-dimensional structure of holo 3 alpha,20 beta-hydroxysteroid dehydrogenase: a member of a short-chain dehydrogenase family. Proc Natl Acad Sci U S A. 1991 Nov 15;88(22):10064–10068. doi: 10.1073/pnas.88.22.10064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- KAUFMAN S. STUDIES ON THE STRUCTURE OF THE PRIMARY OXIDATION PRODUCT FORMED FROM TETRAHYDROPTERIDINES DURING PHENYLALAMINE HYDROXYLATION. J Biol Chem. 1964 Jan;239:332–338. [PubMed] [Google Scholar]
- Kaufman S., Holtzman N. A., Milstien S., Butler L. J., Krumholz A. Phenylketonuria due to a deficiency of dihydropteridine reductase. N Engl J Med. 1975 Oct 16;293(16):785–790. doi: 10.1056/NEJM197510162931601. [DOI] [PubMed] [Google Scholar]
- Persson B., Krook M., Jörnvall H. Characteristics of short-chain alcohol dehydrogenases and related enzymes. Eur J Biochem. 1991 Sep 1;200(2):537–543. doi: 10.1111/j.1432-1033.1991.tb16215.x. [DOI] [PubMed] [Google Scholar]
- Smith I., Clayton B. E., Wolff O. H. Letter: A variant of phenylketonuria. Lancet. 1975 Feb 8;1(7902):328–329. doi: 10.1016/s0140-6736(75)91230-1. [DOI] [PubMed] [Google Scholar]
- Varughese K. I., Skinner M. M., Whiteley J. M., Matthews D. A., Xuong N. H. Crystal structure of rat liver dihydropteridine reductase. Proc Natl Acad Sci U S A. 1992 Jul 1;89(13):6080–6084. doi: 10.1073/pnas.89.13.6080. [DOI] [PMC free article] [PubMed] [Google Scholar]