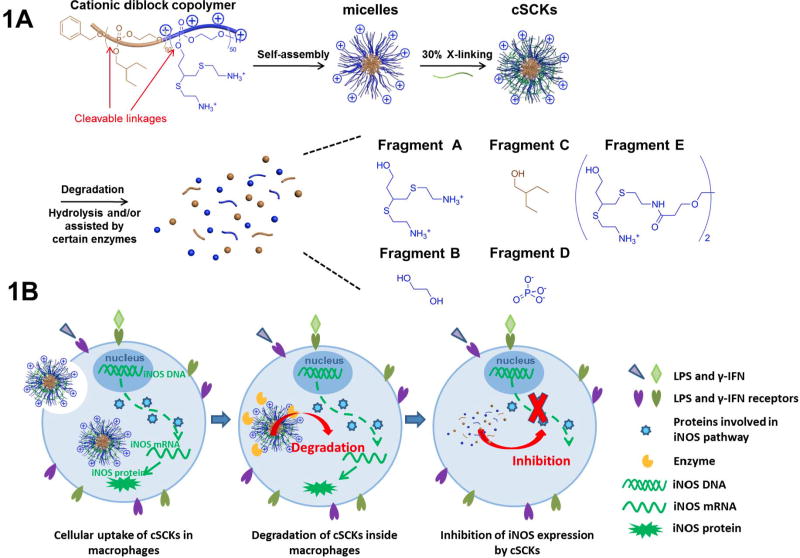
Degradable polymeric nanoparticles have attracted great attention for various biomedical applications due to their extended biodistribution and more favorable pharmacokinetics than small molecules.[1] Of particular interest has been to use polymeric nanoparticles to deliver therapeutic payloads, either by direct chemical conjugation or by physical encapsulation, accounting for more than half of the research activity and nanomedicine market.[2-4] More often than not, degradable nanoparticles used in the treatment of diseases have shown cytotoxic effects or off-target effects,[5] and rarely, if ever, have there been reports of beneficial properties of the nanoparticles.[6,7] Here, we report that a polyphosphoester (PPE)-based cationic degradable nanoparticle without loading of any external drugs, is able to efficiently inhibit inducible nitric oxide synthase (iNOS) transcription, which eventually results in the inhibition of nitric oxide (NO) over-production, and may have use for the treatment of acute lung injury (ALI), among other diseases that involve inflammatory responses.
ALI, physiologically characterized by pulmonary edema and arterial hypoxemia, is a clinical syndrome of acute respiratory failure with high morbidity and mortality, which requires improved treatment approaches.[8,9] iNOS, an enzyme with abundant mediators such as LPS, γ-IFN, and IL-1 is found in activated lung macrophages, lung epithelial and endothelial cells, and results in over-production of nitric oxide (NO), which with unregulated fashion leads to tissue damage.[10,11] The inhibition of iNOS has been shown to reduce the accumulation of NO, enhance cell survival and, therefore, improve the outcome of ALI.[12] Most therapeutic agents for the inhibition of iNOS mRNA and protein, are small molecules,[13,14] though siRNAs and antibodies have been used.[15-17]
We intended to take advantage of a recently-developed approach that involves the rapid and efficient preparation of a versatile class of functional nanoparticles,[18,19] and advance the chemistry toward the production of polyphosphoester (PPE)-based cationic shell crosslinked nanoparticles (cSCKs) for the complexation and delivery of therapeutic siRNAs to treat ALI, however, it was observed that these bioinspired nanomaterials possessed biological activity themselves. Our interest was in the translational aspects of these materials and in detailed examination of their hydrolytic degradation characteristics, as they are produced on multi-gram scales within 2 days starting from small compounds. The cationic micelles and cSCKs were constructed by the direct dissolution of amphiphilic cationic diblock PPE cationic and amphiphilic diblock PPE into ultrapure water and followed by crosslinking the shell region of these micelles with 3,3′-(ethane-1,2-diylbis(oxy))dipropanoic acid (Bis-dPEG®2-acid) via amidation chemistry (Figure 1A). As shown in Figure 1B, the nanometer size (about 15 nm) and cationic surface of cSCKs facilitated the cellular uptake in RAW 264.7 macrophages, where iNOS is over-expressed by the activation of LPS and γ-IFN.[20] It was expected that the cSCKs would undergo rapid degradation under the hydrolytic and possibly enzymatic conditions inside macrophages to generate small fragments, which would also release any therapeutic cargo that the cSCKs would carry. Interestingly, however, one or more of the degradation products served as an inhibitor of certain steps along the iNOS transcription process, leading to the inhibition of iNOS protein expression. Therefore, without further carrying any therapeutic payloads, the cSCK itself, as an inherent degradable drug device, demonstrated the ability of both efficient cellular internalization and significant inhibition of iNOS protein over-expression.
Figure 1.
(A) Schematic illustration of the preparation of cSCKs from cationic and amphiphilic diblock PPE, and the degradation of cSCKs into small fragments. (B) Schematic illustration of cSCKs cellular uptake, degradation, and degradation products mediated inhibition of iNOS expression.
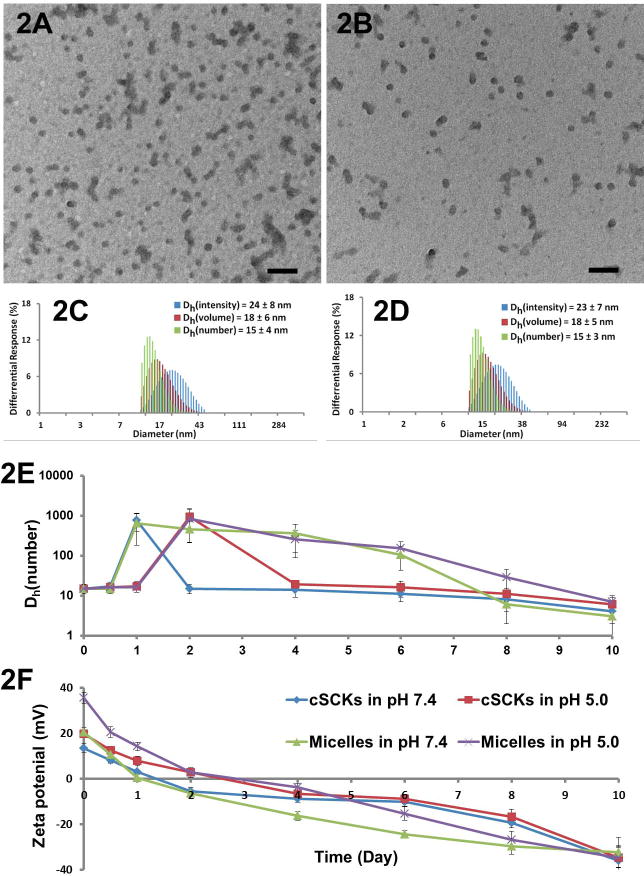
After the construction of cSCKs by self-assembly of amphiphilic diblock PPE and shell-cross-linking, the self-assembled micelles and chemically-crosslinked cSCKs were characterized by transmission electron microscopy (TEM), dynamic light scattering (DLS) and zeta potential measurements (Figure 2 A-D). The hydrodynamic diameters (by number) of both micelles and cSCKs were ca. 15 nm. The crosslinking process resulted in no discernible change in DLS and TEM sizes and size distributions. Cationic micelles exhibited positive surface charge with average zeta potential values of 20.3 mV at pH 7.4 and 35.6 mV at pH 5.0, while cationic cSCKs had less positive average zeta potentials of 16.9 mV at pH 7.4 and 30.8 mV at pH 5.0, which may result from the partial consumption of amino groups during the cross-linking process.
Figure 2.
(A) TEM image of micelles, average diameter is 17 ± 4 nm. (B) TEM image of cSCKs, average diameter is 16 ± 4 nm. (C) DLS results of micelles: Dh(intensity) = 24 ± 8 nm, Dh(volume) = 18 ± 6 nm, Dh(number) = 15 ± 4 nm. (D) DLS results of cSCKs: Dh(intensity) = 23 ± 7 nm, Dh(volume) = 18 ± 5 nm, Dh(number) = 15 ± 3 nm. (E) Changes in hydrodynamic diameters by DLS for micelles and cSCKs as a function of hydrolytic degradation times. (F) Changes in zeta potentials for micelles and cSCKs as a function of hydrolytic degradation times.
The phosphoester linkages of the PPEs are susceptible to cleavage[21] either by spontaneous hydrolysis[22-26] or by the action of enzymes.[27-30] To study the hydrolytic degradation of PPE-based cationic nanoparticles, the nanoparticles were incubated in pH 5.0 and pH 7.4 aqueous PBS buffers at 37 °C for ten days, and the degradation profiles were examined by measuring changes in particle size and surface charge (Figure 2E, F). After ten days of degradation, at both pH 5.0 and pH 7.4, no particles could be observed by TEM, and could barely be detected by DLS. The zeta potentials of both micelles and cSCKs changed from positive to neutral, and finally reached around -35 mV. This change from positive to negative values was due to the loss of cationic side chains (ammonium groups, Fragment A) after hydrolysis (Figure S1). Both nanoparticles reached neutrality in pH 7.4 one day earlier than that in pH 5.0 solution. The increased rate at higher pH, may be due in part to attack by the deprotonated amino groups on the phosphoester bonds. Hydrodynamic diameters of cSCKs had a tendency to become increasingly smaller with time, until macromolecules or particles were undetectable by DLS, except for the formation of large aggregates at the isoelectric point, where zeta potentials were almost zero. As degradation continued and zeta potential became negative, the large aggregates were dispersed in the cSCK samples. Large aggregates were also observed during the degradation of the micelles at the isoelectric point, but those species didn't disappear until a later stage. This difference may be explained by reassembly of the micelles during the degradation,[31] whereas the cSCKs maintained the micellar structures because they were crosslinked.[32,33] The degradation study demonstrated that PPE-based cationic nanoparticles spontaneously degraded with a half-life of days at neutral to slightly acidic pH and lost their cationic feature rapidly.[34] It was expected that degradation within cells may be more greatly accelerated by enzymes.
The major degradation fragments of PPE-based cSCKs would be, after the complete breakdown of the phosphoester linkages along the backbone and connecting to the side chain functionalities, 3,4-bis(2-aminoethylthio)butan-1-ol (Fragment A), 2-ethylbutan-1-ol (Fragment B), ethylene glycol (Fragment C), phosphate ion (Fragment D), and two Fragment A's linked together by a cross-linker (Fragment E).[22,35,36] Independently synthesized Fragment A, 3,4-bis(2-aminoethylthio)butan-1-ol (see supporting information), shared structural similarity with reported iNOS inhibitors, most of which have a positively-charged group or a thioether bond.[37] Due to the synthetic difficulty, Fragment E and other less possible fragments, such as phosphate esters of Fragment A and E, which might also have the iNOS inhibition effects, were not synthesized.
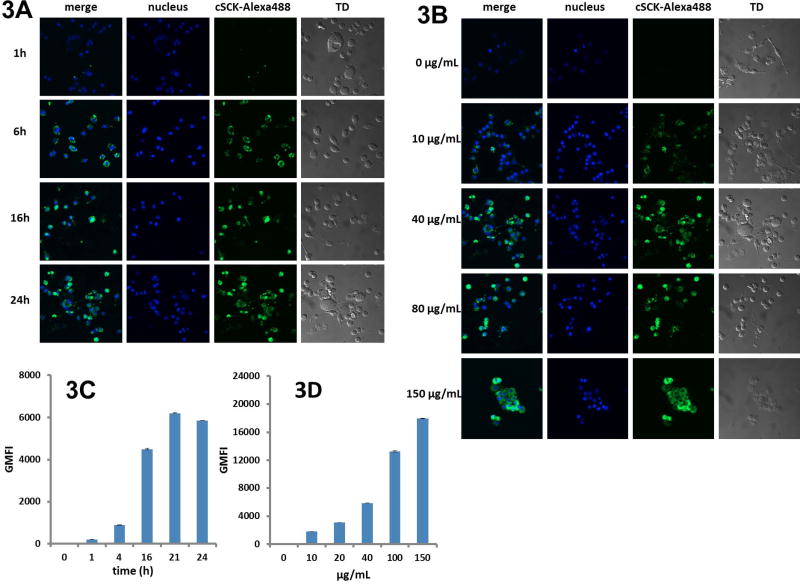
AlexaFluor 488 dye was conjugated to the cSCK at a ratio of 0.5 dye molecules per polymer chain to examine cellular uptake of cSCKs in LPS- and γ-IFN-induced RAW 264.7 macrophages, as measured by confocal microscopy and flow cytometry. By confocal microscopy, the uptake of AlexaFluor 488-cSCKs dramatically increased with an increase of incubation times (Figure 3A), and with an increase of particle concentrations after a 24 h incubation (Figure 3B). Of note, AlexaFluor488-cSCK or its degradation products appeared to be located diffusely in the cytoplasm, rather than in a punctate pattern typical of endosomal-like vesicles, as occurs with non-degradable cationic SCKs we have reported previously.[17,38] This difference might be explained by more rapid release of the cSCKs and their degradation products as a result of the fast degradation of the particles into small fragments, which increased the osmotic pressure within the endosomes. Alternatively, the degradable cSCKs could have entered cells through a different mechanism. The cellular uptake of AlexaFluor 488-cSCKs in activated RAW cells was also quantitatively studied by flow cytometry (Figures 3C and 3D), which also confirmed the dose-dependent and time-dependent pattern. In contrast, the partially-degraded cSCKs had limited cellular uptake, due to the loss of the cationic feature during the degradation (Figure S2).
Figure 3.
Cellular uptake of cSCKs in LPS- and γ-IFN-activated RAW 264.7 cells. Confocal microscopic images of (A) cSCKs (40 μg/mL) at different incubation times and (B) 24 h incubation with different concentrations. Flow cytometry studies of (C) cSCKs (40 μg/mL) at different incubation times and (D) 24 h incubation with different concentrations. GMFI (Geometric mean fluorescence intensity).
We next evaluated the toxicity of cSCKs and their degradation products in LPS- and γ-IFN-activated RAW 264.7 cells by an MTT assay. cSCKs demonstrated dose dependent cytotoxicity only above 150 μg/mL at 24 h (Figure 4A). Degraded cSCKs showed less toxicity, however, the four degradation products showed no significant cytotoxicity at concentrations up to 600 μg/mL.
Figure 4.

(A) Cytotoxicity of cSCKs and fragments in LPS- and γ-IFN-activated RAW 264.7 cells. (B) Inhibition of NO production by cSCKs and fragments by the measurement of nitrite levels. (C) Inhibition of iNOS mRNA. Relative iNOS mRNA level was measured by qRT-PCR.
To test the ability of cSCKs and its degradation products to inhibit iNOS activity, we incubated them with activated RAW 264.7 cells, and monitored the nitrite levels by the Griess Assay. Nitrite is one of the products of nitric oxide that is produced by the iNOS enzyme. As shown in Figure 4B, cSCKs demonstrated dose-dependent nitrite inhibition at 24 h, with relative nitrite levels of 52% at a particle concentration of 10 μg/mL, and decreasing dramatically to 5% at 160 μg/mL, a point at which the nanoparticles do not show significant cytotoxicity. Degraded cSCKs also inhibited nitrite production, decreasing the relative nitrite level of 94% at 10 μg/mL to 36% at 150 μg/mL at 24 h. Of four degradation products investigated, only fragment A demonstrated dose-dependent iNOS inhibition with the relative nitrite level decreasing from 85% at 10 μg/mL to 8% at 400 μg/mL, while no nitrite inhibition was observed for the other fragments (Fragment B, C and D).
The observation that fragment A inhibits nitrite production supports the concept that nitrite inhibition caused by cSCKs results from degradation of the cSCKs inside cells, thereby, generating Fragment A. Taking into consideration that 100 μg/mL of cSCKs generates only 48 μg/mL of Fragment A after complete degradation (48 wt% loading of Fragment A), the increased nitrite inhibition ability of cSCK over Fragment A is much larger than it appears in Figure 4B. This enhanced performance by the cSCK could be explained by poor cellular permeability of Fragment A alone, due to its small size. In comparison, the cSCK is capable of entering cells rapidly by endocytosis, due to its nanometer size and highly positively-charged surface, and then degrading and releasing Fragment A inside the cells. Inhibition of nitrite production by Fragment A could then either occur by directly inhibiting the iNOS enzyme, or by inhibiting the transcription or translation of the iNOS mRNA.
To determine whether the cSCK or fragment A inhibits nitrite production by inhibiting the iNOS transcription, iNOS mRNA levels were determined by qRT-PCR (quantitative Reverse Transcription Polymerase Chain Reaction). As shown in Figure 4C, cSCKs led to significant iNOS mRNA knockdown from an mRNA level of 41% at 20 μg/mL particle concentration to 3% at 160 μg/mL, which correlates with the nitrite inhibition levels. Degraded cSCKs also showed iNOS mRNA inhibition, but not as efficient as with fresh cSCKs. Fragment A also decreased iNOS mRNA levels from 95% at 20 μg/mL to 50% at a high concentration of 200 μg/mL, while none of the other fragments showed significant iNOS mRNA inhibition. These results suggested that cSCK and Fragment A were inhibiting some upstream regulatory steps in the iNOS transcription process, rather than serving as a direct inhibitor to iNOS protein. Unlike other reported cases,[39] in which known biologically-active compounds were incorporated through degradable linkages into polymer nanoparticles and then released, this paper presents the integrated construction of polymer nanoparticles by biologically-active compounds, and generation of those novel biologically-active compounds after the degradation.
In conclusion, a degradable PPE-based cationic nanoparticle (cSCK) has been developed, which serendipitously demonstrated efficient inhibition of iNOS transcription and eventually led to the inhibition of NO over-production. We were able to ascribe this inhibition to rapid spontaneous hydrolytic degradation of the nanoparticle within cells to release the degradation Fragment A, which was shown to inhibit iNOS transcription. The higher specific activity of the cSCK than the identified Fragment A could be attributed to higher cellular uptake of the cationic nanoparticle by endocytosis followed by rapid degradation to release high quantities of Fragment A inside the cell. As such, this nanoparticle may have the potential to serve as a promising anti-iNOS-based anti-inflammatory agent for the treatment of acute lung injury. This nanoparticle might also serve as a paradigm for the general design of diagnostic and therapeutic nanoparticles with built-in inflammatory-reducing properties, as integral components of their frameworks. Future work will focus on mechanistic studies aimed at identifying the mechanism by which cSCK or Fragment A inhibits iNOS transcription. This study also suggests the necessity of examining the biological activities of all degradation products of polymer nanoparticles to identify beneficial ones.
Experimental Section
Materials
The amphiphilic and cationic diblock copolymer consisting of poly(2-ethylbutyl phospholane) (PEBP) as a hydrophobic segment and poly(3,4-bis((2-aminoethyl)thio)butylphospholane)(PAETBP) as a hydrophilic segment was synthesized according to the literature.[1] 3,3′-(Ethane-1,2-diylbis(oxy))dipropanoic acid (commercial name: Bis-dPEG®2-acid) was used as received from Quanta BioDesign, Ltd. (Powell, Ohio). Alexa Fluor® 488 Sulfodichlorophenol Ester (Alexa Fluor® 488 5-SDP Ester) was used as received from Life Technologies Corp. (Carlsbad, CA). 2,2-Dimethoxy-2-phenylacetophenone (DMPA) and 1-[3′-(dimethylamino)propyl]-3-ethylcarbodiimide methiodide (EDCI) were used as received from Sigma-Aldrich Company (St. Louis, MO). Nanopure water (18 MΩ·cm) was acquired by means of a Milli-Q water filtration system, Millipore Corp. (St. Charles, MO). Ultrapure water (Molecular Biology Grade, Fisher BioReagents) was used as received from Thermo Fisher Scientific Inc (Pittsburgh, PA). The RAW 264.7 cell line was obtained from the American Type Culture Collection. CellTiter 96 non-radioactive cell proliferation assay and Griess assay were from Promega Co. All cell culture media, TRIzol Reagent, Lipofectamine™2000 were purchased from Invitrogen, Inc. TURBO DNA-free kit, high-capacity cDNA reverse transcription kit and Power SYBR green master mix were from Ambion. LPS (lipopolysaccharides from Escherichia coli 055:B5) was from Sigma and IFN-gamma (γ-IFN, mouse, recombinant, E. coli) was from Peprotech.
Characterization Techniques
1H NMR, 31P NMR and 13C NMR spectra were recorded on an Inova 300 or Mercury 300 spectrometer interfaced to a UNIX computer using VnmrJ software. Transmission electron microscopy (TEM) was conducted on a Hitachi H-7500 microscope, operating at 100 kV. Samples for TEM measurements were prepared as follows: 4 μL of the dilute solution (with a polymer concentration of 0.1 mg/mL) was deposited onto a carbon-coated copper grid, and after 2 min, the excess of the solution was quickly wicked away with a piece of filter paper. The samples were then negatively stained with 1 wt% phosphotungstic acid (PTA) aqueous solution. After 1 min, the excess staining solution was quickly wicked away with a piece of filter paper and the samples were left to dry under ambient conditions overnight. The average diameter of nanoparticles on the TEM grid was obtained by measuring the core domain of 200 particles at different areas of the TEM specimen and the standard deviation was calculated. Dynamic light scattering (DLS) measurements were conducted using a Delsa Nano C from Beckman Coulter, Inc. (Fullerton, CA) equipped with a laser diode operating at 658 nm. Scattered light was detected at a 165° angle and analyzed using a log correlator over 70 accumulations for a 0.5 mL of sample in a glass size cell (0.9 mL capacity). The photomultiplier aperture and the attenuator were automatically adjusted to obtain a photon counting rate of ca. 10 kcps. The calculations of the particle size distribution and distribution averages were performed using CONTIN particle size distribution analysis routines using Delsa Nano 2.31 software. The peak averages of histograms from intensity, volume and number distributions out of 70 accumulations were reported as the average diameter of the particles. All determinations were repeated 10 times. The zeta potential values of the nanoparticles were determined by Delsa Nano C particle analyzer (Beckman Coulter. Fullerton, CA) equipped with a 30 mW dual laser diode (658 nm). The zeta potential of the particles in suspension was obtained by measuring the electrophoretic movement of charged particles under an applied electric field. Scattered light was detected at a 30° angle at 25 °C. The zeta potential was measured at five regions in the flow cell and a weighted mean was calculated. These five measurements were used to correct for electroosmotic flow that was induced in the cell due to the surface charge of the cell wall. All determinations were repeated 6 times.
Formation of cationic micelles
The cationic diblock copolymers (10.0 mg) were suspended into ultrapure water (2.0 mL) and sonicated for 10 min. After being stirred for 2 h, a clear solution containing the cationic micelles for the cationic SCKs was obtained.
Formation of cationic SCKs (cSCKs)
The cationic diblock copolymers (10.0 mg, 33.3 μmol of amino groups) were suspended into ultrapure water (2.0 mL) and sonicated for 10 min. After being stirred for 1 h, a clear solution containing the cationic micelles was obtained. To a stirred solution of micelle in a 5-mL vial was added a solution of the diacidcrosslinker (Bis-dPEG®2-acid, 1.0 mg, 5.0 μmol) dropwise in ultrapure water (0.2 mL). The mixture solution was allowed to stir for 1 h at room temperature. To this reaction mixture was added a solution of EDCI (4.3 mg, 15 μmol) dissolved in ultrapure water (0.5 mL) dropwise. The reaction mixture was allowed to stir overnight at room temperature and was then transferred to presoaked dialysis membrane tubing (MWCO ca. 6-8 kDa), and dialyzed against ultrapure water for 36 h in the cold room (4-8 °C) to remove small molecules. The final concentration of cSCKs was adjusted to 2.5 mg/mL by ultrapure water. The cSCK solution was able to be lyophilized into powder and kept in the freezer (-20 °C) for long-term storage and transportation, before being tested.
Formation of dye-labeled cSCKs
The cationic diblock copolymers (10.0 mg, 33.3 μmol of amino groups) were suspended into ultrapure water (2.0 mL) and sonicated for 10 min. After being stirred for 1 h, a clear solution containing the cationic micelles was obtained. To a stirred solution of micelle in a 5-mL vial was added a solution of the diacidcrosslinker (Bis-dPEG®2-acid, 1.0 mg, 5.0 μmol) in ultrapure water (0.2 mL) dropwise, followed by a solution of Alexa Fluor® 488 5-SDP ester (0.13 mg, 0.17 μmol) in ultrapure water (0.1 mL) dropwise. The mixture solution was allowed to stir for 1 h at room temperature. To this reaction mixture was added a solution of EDCI (4.3 mg, 15 μmol) dissolved in ultrapure water (0.5 mL) dropwise. The reaction mixture was allowed to stir overnight at room temperature and was then transferred to presoaked dialysis membrane tubing (MWCO ca. 6-8 kDa), and dialyzed against ultrapure water for 36 h in the cold room (4-8 °C) to remove small molecules. The final concentration of dye-labeled cSCKs was adjusted to 2.5 mg/mL by the addition of ultrapure water. The dye-labeled cSCK solution was able to be lyophilized into powder and kept in the freezer (-20 °C) for the long term storage and transportation, before being tested
General procedure of degradation studies
In a typical degradation experiment, a solution of cSCKs was adjusted to pH 5.0 or pH 7.4 by 150 mM PBS buffer, 150 mM acetic buffer (for 31P NMR studies) or 150 MOPS buffer (for 31P NMR studies). The mixture solution was incubated in a 37 °C shaker. The size, size distribution, zeta potential and 31P chemical shifts were measured by Delsa Nano C particle analyzer and Inova 300 spectrometer, respectively, during the degradation.
Preparation of degraded cSCKs
A solution of cationic SCKs (2.5 mg/mL, 8 mL) was adjusted to pH 7.4 by PBS buffer (12 mM, 2 mL, without NaCl or KCl) and incubated in the 37 °C shaker. After full degradation of nanoparticles (12 days), the solution was lyophilized into powder before being tested.
Synthesis of 3,4-bis(2-aminoethylthio)butan-1-ol hydrochloride salt (Fragment A)
A solution of 3-butyn-1-ol (0.20 g, 2.86mmol), cysteamine hydrochloride (3.24 g, 28.6 mmol), and DMPA (14.6 mg, 0.57 mmol) in 10.0 mL of methanol was sparged with nitrogen for 5 min and then irradiated under UV irradiation (365 nm) for 2 h. The reaction mixtures were precipitated from methanol into acetone four times to remove excess thiols and photoinitiator by-products to give 3,4-bis(2-aminoethylthio)butan-1-ol hydrochloride salt as a yellow liquid at a yield of 20%. 1H NMR (D2O, ppm): δ 3.74 (d, J = 5.7 Hz, HOCH2CH2), 3.34 (s, HOCH2CH2), 3.21 (t, J = 6.9 Hz, CH2CH2NH3), 2.95 (m, SCH2CH2NH3, SCH2CHS), 2.01, 1.75 (dt, J =5.7, 6.0 Hz, HOCH2CH2CH). 13C NMR (CD3OD, ppm): δ 58.75, 42.11, 38.85, 38.42, 36.87, 35.46, 29.13, 27.15. ESI MS: calculated [M-H]- for C8H21Cl2N2OS2: 295.0472, found: 295.0594. IR: 3600-2400, 2000, 1693, 1599, 1379, 1322, 1141, 1036, 939, 887 cm‐1.
Cell culture
RAW 264.7 cells (mouse monocyte-macrophage cell line) were maintained in DMEM containing 10% FBS, streptomycin (100 μg/mL), and penicillin (100 units/mL) at 37 ܰC in a humidified atmosphere with 5% CO2.
Confocal microscopy of cSCKs cellular uptake
RAW 264.7 cells were plated in 35 mm MatTek glass bottom microwell dishes (MatTek) at a density of 2×104 cells per well and cultured in 100 μL DMEM containing 10% FBS. 24 hours later, AlexaFluor 488 labeled degradable cSCKs were incubated with the RAW 264.7 cells, and then cells were activated by LPS (1 μg/mL) and γ-IFN (20 ng/mL) after 6 h. At the time of confocal microscopy, 1 μL of Hoechst 33342 (10 μg/mL) was added to each dish to stain nuclei for one hour and then each dish was washed three times with PBS buffer and examined with a Nikon A1 Confocal Microscope (Nikon). In one experiment, different concentrations of AlexaFluor 488-cSCKs (0, 10, 40, 80, 150 μg/mL) were added to each dish and images were collected by confocal microscopy after 24 h incubation. In another experiment, degradable AlexaFluor 488-cSCKs (40 μg/mL) were added and images were collected at different time points (1 h, 6 h, 16 h, 24 h).
Flow cytometry of cSCKs cellular uptake
RAW 264.7 cells were plated in a 6 well plate at a density of 3×105 cells per well and cultured in 1 mL DMEM containing 10% FBS. After 24 h, AlexaFluor 488-cSCKs were added to RAW 264.7 cells, and cells were activated by LPS (1 μg/mL) and γ-IFN (20 ng/mL) 6 h later. At the time of flow cytometry, cells were washed 3 times with PBS, collected by trypsinization, pelleted, and resuspended in 0.5 mL PBS for flow cytometric analysis on an FACS-calibur instrument (Becton Dickinson) equipped with a 488 nm Ar laser. For each sample, 10,000 events were collected and the fluorescence from two channels was detected on a logarithmic scale, respectively. The data were processed by FlowJo software. In one experiment, different concentrations of AlexaFluor 488-cSCKs (0, 10, 20, 40, 100, 150 μg/mL) were added to each dish and data were collected after 24 h incubation. In another experiment, degradable cSCK-AlexaFluor 488 (40 μg/mL) were added and data were collected at different time points (0, 1 h, 4 h, 16 h, 21 h, 24 h).
Cytotoxicity assay of cSCKs in RAW 264.7 cells
The cytotoxicties of cSCKs were examined by CellTiter 96 non-radioactive cell proliferation assay (Promega). RAW 264.7 cells were seeded in a 96 well plate at a density of 2×104 cells per well and cultured in 100 μL DMEM containing 10% FBS. After 24 h, the medium was replaced with 100 μL fresh culture medium containing cSCKs (fresh), cSCKs (degraded), fragment A, fragment B, fragment C and fragment D with different concentrations. 6 h later, cells were activated with LPS (1 μg/mL) and γ-IFN (20 ng/mL) to induce iNOS expression. After another 24 h, 15 μL of dye solution was added to each well and incubated at 37 °C in a humidified CO2 incubator. 4 h later, 100 μL of stop solution was added to each well and incubated overnight. The absorbance of colored formazan product was recorded at 570 nm using a microplate reader (Molecular Devices) and normalized to the control group with no treatment. An average of three determinations was made.
Inhibition of iNOS expression by cSCKs determined by Griess assay
RAW 264.7 cells were seeded in a 96 well plate at a density of 2×104 cells per well and cultured in 100 μL DMEM containing 10% FBS. After 24 h, the medium was replaced with 100 μL fresh culture medium containing cSCKs (fresh), cSCKs (degraded), fragment A, fragment B, fragment C and fragment D with different concentrations. 6 h later, cells were activated with LPS (1 μg/mL) and γ-IFN (20 ng/mL). After another 24 h, nitrite level was measured by Griess assay (Promega) at 540 nm using a microplate reader (Molecular Devices) and normalized to the control group with no treatment. An average of three determinations was made.
Inhibition of iNOS mRNA by cSCKs determined by qRT-PCR
RAW 264.7 cells were seeded in 6 well plate at a density of 3×105 cells per well and cultured in 1000 μL DMEM containing 10% FBS. 24 h later, cells were transfected with cSCKs (fresh), cSCKs (degraded), fragment A, fragment B, fragment C and fragment D with different concentrations. Cells were then activated with LPS (1 μg/mL) and γ-IFN (20 ng/mL) after 6 h. After another 24 h, cells were washed twice with PBS and total RNA was extracted using TRIzol Reagent (Invitrogen) according to the manufacturer's instruction and quantified spectrophotometrically by Nanophotometer (Implen). Approximately 500 ng of total RNA was treated with TURBO DNA-free kit (Ambion) and reverse transcribed with high-capacity cDNA reverse transcription kit (Ambion) according to standard protocols supplied by the manufacturer. cDNA synthesis was performed for 10 min at 25 °C and 2 h at 37 °C, followed by heat inactivation for 5 min at 85 °C. The real-time PCR assay was performed using the Applied Biosystems StepOne plus Real-Time PCR System (Applied Biosystems) with 40 cycles of 95 °C for 15 sec and 60 °C for 1 min. PCR reactions were carried out in a 25 μL of reaction mixture consisting of 12.5 μL Power SYBR Green Master Mix (2*) (Ambion), 200 nM primers, cDNA and water. The primers used to amplify iNOS were d(TGGTGGTGACAAGCACATTT) and d(AAGGCCAAACACAGCATACC), and to amplify GAPDH were d(TGGAGAAACCTGCCAAGTATG) and d(GTTGAAGTCGCAGGAGACAAC). The threshold cycle (Ct) was calculated by the instrument's software (StepOne). The expression of each mRNA was calculated by the comparative Ct method (ΔΔCt). The average of 3 determinations was calculated.
Supplementary Material
Figure S1. A) 1H NMR (in d6-DMSO) of cationic amphiphilic diblock PPE. B) 1H NMR (in D2O) of collected precipitated polymer/nanoparticle (around isoelectric point) after 1-2 days degradation. The disapperance of signals between 1.7 ppm and 3.2 ppm (on the Fragment A) verfied that the loss of cationic feature was due to the release of Fragment A after the cleavage of phosphoester linkages.
Figure S2. A) Cellular uptake of degraded cSCKs. Images were obtained under confocal microscopy. B) Degraded cSCKs with different concentrations at 24 h incubation. GMFI was quantitatively measured by Flow Cytometry. C) Cellular uptake of non-degradable cSCKs[2] (10 μg/mL) at 24 h incubation.
Acknowledgments
We gratefully acknowledge financial support from the National Heart Lung and Blood Institute of the National Institutes of Health as a Program of Excellence in Nanotechnology (HHSN268201000046C) (J.-S. A. T. and K. L. W.) and the National Science Foundation under grant number DMR-1105304 (K. L. W.). The Welch Foundation is gratefully acknowledged for support through the W. T. Doherty-Welch Chair in Chemistry, Grant No. A-0001 (K. L. W.). The transmission electron microscopy facilities at Washington University in St. Louis, Department of Otolaryngology, Research Center for Auditory and Visual Studies funded by NIH P30 DC004665 are gratefully acknowledged.
Footnotes
Supporting Information: Supporting Information is available online from the Wiley Online Library or from the author.
Contributor Information
Dr. Yuefei Shen, Department of Chemistry, Washington University in St. Louis, St. Louis, Missouri, 63130, USA.
Dr. Shiyi Zhang, Department of Chemistry, Washington University in St. Louis, St. Louis, Missouri, 63130, USA, Department of Chemistry, Department of Chemical Engineering, Laboratory for Synthetic-Biologic Interactions, Texas A&M University, P.O. BOX 30012, 3255 TAMU, College Station, Texas, 77842, USA.
Fuwu Zhang, Department of Chemistry, Department of Chemical Engineering, Laboratory for Synthetic-Biologic Interactions, Texas A&M University, P.O. BOX 30012, 3255 TAMU, College Station, Texas, 77842, USA.
Alexander Loftis, Department of Chemistry, Washington University in St. Louis, St. Louis, Missouri, 63130, USA.
Adriana Pavía-Sanders, Department of Chemistry, Department of Chemical Engineering, Laboratory for Synthetic-Biologic Interactions, Texas A&M University, P.O. BOX 30012, 3255 TAMU, College Station, Texas, 77842, USA.
Dr. Jiong Zou, Department of Chemistry, Department of Chemical Engineering, Laboratory for Synthetic-Biologic Interactions, Texas A&M University, P.O. BOX 30012, 3255 TAMU, College Station, Texas, 77842, USA
Jingwei Fan, Department of Chemistry, Department of Chemical Engineering, Laboratory for Synthetic-Biologic Interactions, Texas A&M University, P.O. BOX 30012, 3255 TAMU, College Station, Texas, 77842, USA.
Prof. John-Stephen A. Taylor, Email: taylor@wuchem.wustl.edu, Department of Chemistry, Washington University in St. Louis, St. Louis, Missouri, 63130, USA.
Prof. Karen L. Wooley, Email: wooley@chem.tamu.edu, Department of Chemistry, Department of Chemical Engineering, Laboratory for Synthetic-Biologic Interactions, Texas A&M University, P.O. BOX 30012, 3255 TAMU, College Station, Texas, 77842, USA.
References
- 1.Nair LS, Laurencin CT. Prog Polym Sci. 2007;32:762. [Google Scholar]
- 2.Wagner V, Dullaart A, Bock AK, Zweck A. Nat Biotechnol. 2006;24:1211. doi: 10.1038/nbt1006-1211. [DOI] [PubMed] [Google Scholar]
- 3.Langer R. Nature. 1998;392:5. [PubMed] [Google Scholar]
- 4.Elsabahy M, Wooley KL. J Polym Sci, Part A: Polym Chem. 2012;50:1869. doi: 10.1002/pola.25955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Becker AL, Orlotti NI, Folini M, Cavalieri F, Zelikin AN, Johnston APR, Zaffaroni N, Caruso F. ACS Nano. 2011;5:1335. doi: 10.1021/nn103044z. [DOI] [PubMed] [Google Scholar]
- 6.Mitragotri S, Lahann J. Nature Materials. 2009;8:15. doi: 10.1038/nmat2344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.(a) Wang Z, Liu S, Ma J, Qu G, Wang X, Yu S, He J, Liu J, Xia T, Jiang GB. ACS Nano. 2013;7:4171. doi: 10.1021/nn400594s. [DOI] [PubMed] [Google Scholar]; (b) Fuhrmann K, Schulz JD, Gauthier MA, Leroux JC. ACS Nano. 2012;6:1667. doi: 10.1021/nn2046554. [DOI] [PubMed] [Google Scholar]
- 8.Johnson ER, Matthay MA. J Aerosol Med Pulm D. 2010;23:243. doi: 10.1089/jamp.2009.0775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Matthay MA, Ware LB, Zimmerman GA. J Clin Invest. 2012;122:2731. doi: 10.1172/JCI60331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mehta S. Vascul Pharmacol. 2005;43:390. doi: 10.1016/j.vph.2005.08.013. [DOI] [PubMed] [Google Scholar]
- 11.Guo FH, De Raeve HR, Rice TW, Stuehr DJ, Thunnissen FB, Erzurum SC. Proc Natl Acad Sci USA. 1995;92:7809. doi: 10.1073/pnas.92.17.7809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hosogi S, Iwasaki Y, Yamada T, Komatani-Tamiya N, Hiramatsu A, Kohno Y, Ueda M, Arimoto T, Marunaka Y. Biomed Res. 2008;29:257. doi: 10.2220/biomedres.29.257. [DOI] [PubMed] [Google Scholar]
- 13.Hesslinger C, Strub A, Boer R, Ulrich WR, Lehner MD, Braun C. Biochem Soc Trans. 2009;37:886. doi: 10.1042/BST0370886. [DOI] [PubMed] [Google Scholar]
- 14.Bonnefous C, Payne JE, Roppe J, Zhuang H, Chen X, Symons KT, Nguyen PM, Sablad M, Rozenkrants N, Zhang Y, Wang L, Severance D, Walsh JP, Yazdani N, Shiau AK, Noble SA, Rix P, Rao TS, Hassig CA, Smith ND. J Med Chem. 2009;52:3047. doi: 10.1021/jm900173b. [DOI] [PubMed] [Google Scholar]
- 15.Huang H, Pierstorff E, Osawa E, Ho D. ACS Nano. 2008;2:203. doi: 10.1021/nn7000867. [DOI] [PubMed] [Google Scholar]
- 16.Koh WC, Chandra P, Kim DM, Shim YB. Anal Chem. 2011;83:6177. doi: 10.1021/ac2006558. [DOI] [PubMed] [Google Scholar]
- 17.Shen YF, Fang HF, Zhang K, Shrestha R, Wooley KL, Taylor JSA. Nucleic Acid Ther. 2013;23:95. doi: 10.1089/nat.2012.0390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.(a) Zhang S, Li A, Zou J, Lin LY, Wooley KL. ACS Macro Lett. 2012;1:328. doi: 10.1021/mz200226m. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Zhang S, Wang H, Shen Y, Zhang F, Seetho K, Zou J, Taylor JSA, Dove AP, Wooley KL. Macromolecules. 2013;46:5141. doi: 10.1021/ma400675m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhang SY, Zou J, Zhang FW, Elsabahy M, Felder SE, Zhu JH, Pochan DJ, Wooley KL. J Am Chem Soc. 2012;134:18467. doi: 10.1021/ja309037m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Noda T, Amano F. J Biochem. 1997;121:38. doi: 10.1093/oxfordjournals.jbchem.a021566. [DOI] [PubMed] [Google Scholar]
- 21.Baran J, Penczek S. Macromolecules. 1995;28:5167. [Google Scholar]
- 22.Wang YC, Tang LY, Li Y, Wang J. Biomacromolecules. 2009;10:66. doi: 10.1021/bm800808q. [DOI] [PubMed] [Google Scholar]
- 23.Liu JY, Huang W, Pang Y, Zhu XY, Zhou YF, Yan DY. Biomacromolecules. 2010;11:1564. doi: 10.1021/bm100188h. [DOI] [PubMed] [Google Scholar]
- 24.Iwasaki Y, Nakagawa C, Ohtomi M, Ishihara K, Akiyoshi K. Biomacromolecules. 2004;5:1110. doi: 10.1021/bm049961m. [DOI] [PubMed] [Google Scholar]
- 25.Zhang S, Zou J, Elsabahy M, Karwa A, Li A, Moore DA, Dorshow RB, Wooley KL. Chem Sci. 2013;4:2122. doi: 10.1039/C3SC50252J. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Xiong MH, Li YJ, Bao Y, Yang XZ, Hu B, Wang J. Adv Mater. 2012;24:6175. doi: 10.1002/adma.201202847. [DOI] [PubMed] [Google Scholar]
- 27.Wang DA, Williams CG, Yang F, Cher N, Lee H, Elisseeff JH. Tissue Engineering. 2005;11:201. doi: 10.1089/ten.2005.11.201. [DOI] [PubMed] [Google Scholar]
- 28.Wang YC, Tang LY, Sun TM, Li CH, Xiong MH, Wang J. Biomacromolecules. 2008;9:388. doi: 10.1021/bm700732g. [DOI] [PubMed] [Google Scholar]
- 29.Du JZ, Chen DP, Wang YC, Xiao CS, Lu YJ, Wang J, Zhang GZ. Biomacromolecules. 2006;7:1898. doi: 10.1021/bm051003c. [DOI] [PubMed] [Google Scholar]
- 30.Wachiralarpphaithoon C, Iwasaki Y, Akiyoshi K. Biomaterials. 2007;28:984. doi: 10.1016/j.biomaterials.2006.10.024. [DOI] [PubMed] [Google Scholar]
- 31.Zhu J, Zhang S, Zhang F, Wooley KL, Pochan DJ. Adv Func Mater. 2013;23:1767. [Google Scholar]
- 32.Samarajeewa S, Shrestha R, Li YL, Wooley KL. J Am Chem Soc. 2012;134:1235. doi: 10.1021/ja2095602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhang S, Li Z, Samarajeewa S, Sun G, Yang C, Wooley KL. J Am Chem Soc. 2011;133:11046. doi: 10.1021/ja203133h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wang J, Mao HQ, Leong KW. J Am Chem Soc. 2001;123:9480. doi: 10.1021/ja016062m. [DOI] [PubMed] [Google Scholar]
- 35.Yang XZ, Wang YC, Tang LY, Xia H, Wang J. J Polym Sci, Part A: Polym Chem. 2008;46:6425. [Google Scholar]
- 36.Liu X, Ni PH, He JL, Zhang MZ. Macromolecules. 2010;43:4771. [Google Scholar]
- 37.Alderton WK, Cooper CE, Knowles RG. Biochem J. 2001;357:593. doi: 10.1042/0264-6021:3570593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Shen Y, Shrestha R, Ibricevic A, Gunsten SP, Welch MJ, Wooley KL, Brody SL, Taylor JSA, Liu Y. Interface Focus. 2013;3:3. doi: 10.1098/rsfs.2012.0059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Cho S, Hwang O, Lee I, Lee G, Yoo D, Khang G, Kang PM, Lee D. Adv Func Mater. 2012;22:4038. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. A) 1H NMR (in d6-DMSO) of cationic amphiphilic diblock PPE. B) 1H NMR (in D2O) of collected precipitated polymer/nanoparticle (around isoelectric point) after 1-2 days degradation. The disapperance of signals between 1.7 ppm and 3.2 ppm (on the Fragment A) verfied that the loss of cationic feature was due to the release of Fragment A after the cleavage of phosphoester linkages.
Figure S2. A) Cellular uptake of degraded cSCKs. Images were obtained under confocal microscopy. B) Degraded cSCKs with different concentrations at 24 h incubation. GMFI was quantitatively measured by Flow Cytometry. C) Cellular uptake of non-degradable cSCKs[2] (10 μg/mL) at 24 h incubation.