Abstract
Background
Brachiopods undergo radial cleavage, which is distinct from the stereotyped development of closely related spiralian taxa. The mesoderm has been inferred to derive from the archenteron walls following gastrulation, and the primary mesoderm derivative in the larva is a complex musculature. To investigate the specification and differentiation of the mesoderm in the articulate brachiopod Terebratalia transversa, we have identified orthologs of genes involved in mesoderm development in other taxa and investigated their spatial and temporal expression during the embryonic and larval development of T. transversa.
Results
Orthologs of 17 developmental regulatory genes with roles in the development of the mesoderm in other bilaterian animals were found to be expressed in the developing mesoderm of T. transversa. Five genes, Tt.twist, Tt.GATA456, Tt.dachshund, Tt.mPrx, and Tt.NK1, were found to have expression throughout the archenteron wall at the radial gastrula stage, shortly after the initiation of gastrulation. Three additional genes, Tt.Pax1/9, Tt.MyoD, and Tt.Six1/2, showed expression at this stage in only a portion of the archenteron wall. Tt.eya, Tt.FoxC, Tt.FoxF, Tt.Mox, Tt.paraxis, Tt.Limpet, and Tt.Mef2 all showed initial mesodermal expression during later gastrula or early larval stages. At the late larval stage, Tt.dachshund, Tt.Limpet, and Tt.Mef2 showed expression in nearly all mesoderm cells, while all other genes were localized to specific regions of the mesoderm. Tt.FoxD and Tt.noggin both showed expression in the ventral mesoderm at the larval stages, with gastrula expression patterns in the archenteron roof and blastopore lip, respectively.
Conclusions
Expression analyses support conserved roles for developmental regulators in the specification and differentiation of the mesoderm during the development of T. transversa. Expression of multiple mesodermal factors in the archenteron wall during gastrulation supports previous morphological observations that this region gives rise to larval mesoderm. Localized expression domains during gastrulation and larval development evidence early regionalization of the mesoderm and provide a basis for hypotheses regarding the molecular regulation underlying the complex system of musculature observed in the larva.
Electronic supplementary material
The online version of this article (doi:10.1186/s13227-015-0004-8) contains supplementary material, which is available to authorized users.
Keywords: Brachiopod, Terebratalia transversa, Mesoderm, Spiralia, Ectomesoderm
Background
The mesoderm is frequently considered the ‘third germ layer’ in metazoans. As its name suggests, the mesoderm is a ‘middle’ layer, located between the ectoderm and endoderm. The mesoderm gives rise to tissues including muscle, parenchyma, cartilage, hemolymph, and somatic gonads, as well as forms the lining of coelomic cavities [1]. The mesoderm is considered to be a unique feature of bilaterian animals, and it is hypothesized that it evolved from the endoderm [2]. The homology of the mesoderm among bilaterians is supported by the deployment of a conserved set of transcription factors in the specification and differentiation of the mesoderm in the classical model systems of the mouse, Drosophila, and Caenorhabditis elegans, where the molecular basis for mesoderm development has been investigated in the greatest detail [3-6]. Orthologs of the transcription factors Eya [7-9], MyoD [10-12], and Mef2 [13-15] have all been shown to have important roles in mesoderm development and myogenesis in each of these taxa. However, the specific gene regulatory network architecture underlying mesoderm development differs in each of these species, reflecting the significant differences between them in modes of gastrulation, which forms the endomesoderm, and the character and organization of mesodermal derivatives [6,16].
Although mesoderm formation has been studied in detail in these and other model systems, comparatively little is known about mesoderm specification and differentiation in members of the large protostome clade Spiralia [17,18]. This clade is defined as including all descendents from the last common ancestor of animals with quartet spiral cleavage (that is, mollusks, annelids, nemerteans, and platyhelminths) and is likely more inclusive than, or a senior synonym of, the clade termed Lophotrochozoa [19,20]. In spiral cleavage, stereotyped cell divisions result in an invariant developmental program, with tissues and organs in the larva traceable to individual, homologous, blastomeres in the early embryo [21,22]. Almost all taxa that display spiral cleavage form visceral mesoderm from a homologous blastomere in the early cleavage stages termed micromere 4d or the mesentoblast [23]. In addition to this mesendodermal component of the mesoderm, many spiral cleaving embryos also develop mesoderm from ectodermal sources [24,25].
While the internal evolutionary relationships of the Spiralia remain a subject of investigation, nearly all recent phylogenies resolve brachiopods (a.k.a. lamp shells) as having evolved from within the clade for which spiral cleavage was plesiomorphic [20,26-29]. This suggests that brachiopods most likely evolved from an ancestor with spiral cleavage; however, extant brachiopods show no trace of this stereotyped cleavage program [30-33].
Brachiopods develop through a form of radial cleavage, and endomesoderm is formed through invagination of cells at the vegetal pole during gastrulation [34]. Morphological analyses have shown that during early gastrulation, the embryo remains radially symmetrical (Figure 1A,E) and invaginating tissue forming the archenteron extends towards the animal pole [34]. The archenteron consists of two domains, the presumptive endoderm (located on the ‘roof’) and mesoderm located in a ring at the boundary of the ectoderm and endoderm (Figure 1A,E) [33-35]. As gastrulation proceeds, the gastrula becomes asymmetric as the animal and vegetal poles shift positions relative to one another, establishing the anterior-posterior and dorsal-ventral axes (Figure 1B,F). After the blastopore elongates along the ventral side of the embryo, the dorsal surface of the archenteron expands asymmetrically at the boundary of the roof and walls, extending a curtain of cells down towards the ventral side of the embryo [34]. This process generates the mesoderm as a distinct tissue layer, surrounding the endoderm (Figure 1C,G). As the blastopore closes from posterior to anterior, the endoderm seals dorsally to form a sac open to the environment through the remnant of the blastopore anteriorly on the ventral surface of the embryo and closed in a blind ending posteriorly. In the late larval stage, the mesoderm is present in all three main regions of the larva, termed the apical, mantle, and pedicle lobes (Figure 1D,H). Mesodermal differentiation is first indicated by expression of the actin-binding gene Tt.tropomyosin, which is expressed in the anterior and lateral regions of the archenteron wall in the asymmetric gastrula and early larval stages (Figure 1I,J). In the late larval stage, Tt.tropomyosin is expressed in the mesoderm of the mantle lobe, including the chaetal sacs (black arrowheads in Figure 1K), and in the lateral mesoderm of the pedicle lobe (black arrows in Figure 1K). In the final competent larval form, the predominant mesodermal derivative is a complex system of musculature, which includes prominent longitudinal muscles in the pedicle lobe and two pairs of chaetal sacs in the mantle lobe (Figure 1L,M) [36,37].
Figure 1.
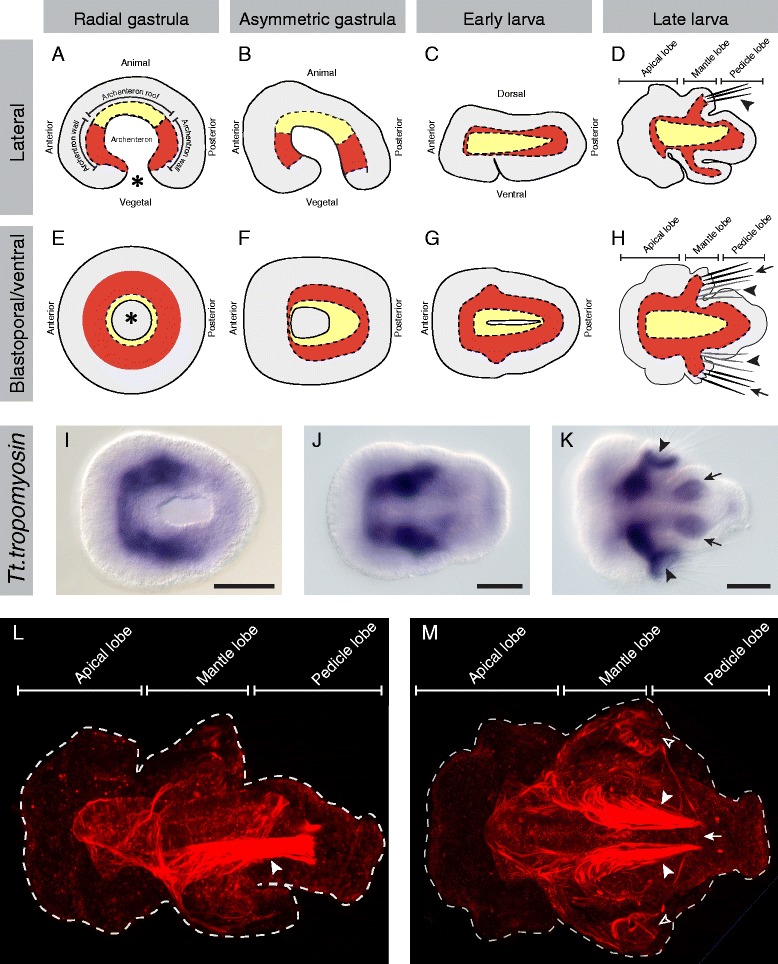
Diagrams of T. transversa development and distribution of musculature in the competent larva. All images are oriented with anterior to the left. Panels (A-D) and (I) are lateral views. Panels (E, F) and (J) are blastoporal/ventral views. Panels (G, H) are ventral views. (A-H) Diagrammatic views of T. transversa gastrula and larval stages. The mesoderm is shaded red, the endoderm is shaded yellow, and the ectoderm is shaded gray. (A, E) The location of the blastopore in the radial gastrula stage is denoted by an asterisk. (D) Lateral view of the late larval stage. One set of dorsal chaetae (black arrowhead) is shown emerging from the mesodermal chaetal sac in the mantle lobe. (H) Ventral view of the late larval stage. Lateral chaetae (black arrows) are shown emerging from the mesodermal chaetal sacs in the mantle lobe. Dorsal chaetae are behind the plane of the cross section (black arrowheads). (I-K) Expression of Tt.tropomyosin during T. transversa development. (I, J) Tt.tropomyosin is expressed in the anterior and lateral portions of the archenteron wall during late gastrula and early larval stages. (K) Tt.tropomyosin is expressed in the mesoderm of the mantle lobe, including the chaetal sacs (black arrowheads) and the lateral regions of the pedicle lobe (black arrows) during the late larval stage. Scale bars are 50 μm in length. (L, M) Phalloidin staining of filamentous actin in the musculature of the competent larval stage. Images are projections of confocal z-series through half of the larva. (L) Lateral view showing complex musculature in the apical, mantle, and pedicle lobes. The prominent pedicle muscles can be observed the ventral region of the larva (white arrowhead). (M) Ventral view of larval musculature. Bundles of muscles are present in the lateral chaetal sacs of the mantle lobe (open arrowheads). Relatively little staining is detected medially in the pedicle lobe (white arrow) between the paired pedicle muscles (white arrowheads).
The available data from Terebratalia suggests that all mesodermal derivatives in the larva are of an endomesodermal origin, derived from cells invaginated at the vegetal blastopore during the radial gastrula stage [34,35,38]. A second source of mesoderm, the ectomesoderm, is present in many other taxa in the clade Spiralia [21,22,39], including in phoronids [40], which are closely related to [20,41-48], or derived from [49-52], brachiopods. To date, no evidence has been presented for an ectodermal source of mesoderm in brachiopods, although detailed lineage analysis of blastomere fates has not been conducted for any member of the group.
In this study, we have analyzed mesoderm development in the articulate brachiopod Terebratalia transversa by cloning and examining the spatiotemporal patterns of genes orthologous to ones that have been shown to have roles in mesoderm specification and differentiation in a variety of bilaterian taxa (Additional file 1). These included orthologs of the transcription factor genes FoxC, FoxD, FoxF, GATA4/5/6, MEF2, Mox, mesoPrx, MyoD, NK1, paraxis, Pax1/9, Six1/2, and twist, the nuclear protein genes dachshund, eyes absent, and Limpet, and the BMP inhibitor noggin. Expression data on these genes provides insight into the molecular basis of mesoderm formation and differentiation in brachiopods and enhances our understanding of potential conservation of mesoderm patterning mechanisms across bilaterian taxa.
Methods
Gene cloning and orthology assignment
Genes of interest were chosen based on literature searches for developmental regulators expressed in the mesoderm of diverse bilaterian taxa. Putative homologs of these genes were identified from a T. transversa transcriptome using TBLASTN search, followed by reciprocal BLASTX searches against NCBI GenBank. Oligonucleotide primers were designed from recovered contigs for RT-PCR or RACE amplification of genes of interest. PCR amplification was performed on a cDNA library synthesized from mixed-stage embryonic RNA with the Advantage RT-for-PCR Kit (Clontech Laboratories, Inc., Mountain View, CA, USA). RACE amplification was performed on cDNA libraries synthesized from mixed-stage embryonic RNA with the SMARTer RACE Kit (Clontech Laboratories, Inc., Mountain View, CA, USA). Amplified fragments were cloned into pGEM-T vector (Promega, Madison, WI, USA) and verified by Sanger sequencing. Sequences for cloned genes are available in GenBank (accession numbers in Additional file 2). Gene orthology was determined by phylogenetic reconstruction. FASTA-formatted files were generated with the inferred amino acid sequences for cloned genes and representative homologs from other metazoan taxa. Sequence alignment was performed with MUSCLE [53], and resultant alignments were trimmed and corrected by eye to remove non-conserved regions and correct obvious errors. The best-fit likelihood model for each alignment was determined using ProtTest [54]. Phylogenetic reconstruction was performed with MrBayes 3.2 with 4 independent runs of 4 chains and 10,000,000 generations each [55].
Fertilization and fixation
Adult T. transversa (Sowerby 1846) were collected by dredging in San Juan Channel, between San Juan Island and Shaw Island, WA, USA, between October and January in 2008, 2010, and 2012, and were maintained in flow-through seawater aquaria at Friday Harbor Laboratories. In vitro fertilization was performed by manual dissection and maceration of gonads. Prior to fertilization, oocytes were maintained in clean seawater until germinal vesicle breakdown and shedding of follicle cells were observed (between 4 and 8 h after stripping of gonads). Sperm were activated with seawater buffered to pH 9.8 with Tris. Following fertilization, embryos were reared in 1-L glass beakers with daily water changes. Embryos were fixed with 4% paraformaldehyde in filtered seawater for 1 h, washed four times in phosphate-buffered saline with 0.1% Tween-20, rinsed with distilled water, and subsequently dehydrated and stored in 100% methanol until in situ hybridization.
In situ hybridization
In situ hybridization of transcripts for cloned genes was performed using protocols established for chromogenic detection in the cnidarian Nematostella vectensis [56]. A detailed protocol is presented in Additional file 3. Hybridization was performed at 62°C for 48 h with DIG-UTP-labeled probes at a concentration of 1 ng/μL. Detection of hybridized probes was performed by staining with NBT and BCIP, after labeling with alkaline phosphatase-conjugated anti-DIG antibody. At least 20 embryos were processed per stage for each gene, and development of staining was checked by a stereomicroscope prior to completion of the in situ protocol and mounting for imaging. In all cases, staining was highly consistent within stages. Embryos were cleared and mounted in 80% glycerol, and imaging was performed on a Zeiss AxioSkop microscope equipped with Plan-Apochromat 20×/08 N.A. objective and differential interference contrast optics (Carl Zeiss, Jena, Germany). Images were acquired with a Zeiss AxioCam HRc digital camera and Zeiss AxioVision v4.8 software (Carl Zeiss, Jena, Germany).
Results
Phylogenetic analysis and orthology assignment
Full-length or partial cDNA sequences were isolated from T. transversa for putative homologs of the transcription factor genes Forkhead C (FoxC), Forkhead D (FoxD), Forkhead F (FoxF), GATA4/5/6, MEF2, Mox, mesoPrx (mPrx), MyoD, NK1, paraxis, Pax1/9, Six1/2, and twist, the nuclear protein genes dachshund, eyes absent, and Limpet, and the BMP inhibitor gene noggin. Orthology assignments were verified by Bayesian analysis of phylogenetics using inferred amino acid sequences of the cloned transcripts and representative sequences from other metazoan taxa (Additional files 4, 5, 6, 7, 8, 9, 10, and 11). T. transversa genes are subsequently referred to as Tt.dachshund, Tt.eya, Tt.FoxC, Tt.FoxD, Tt.FoxF, Tt.GATA4/5/6, Tt.Limpet, Tt.MEF2, Tt.Mox, Tt.mPrx, Tt.MyoD, Tt.NK1, Tt.noggin, Tt.paraxis, Tt.Pax1/9, Tt.Six1/2, and Tt.twist.
Whole-mount in situ expression patterns
Mesodermal expression was observed for all 17 genes listed above, and representative photomicrographs of the radial gastrula, asymmetric gastrula, early larval, and late larval stages are presented (Figures 2, 3, 4, 5, 6, 7, 8, and 9). Photomicrographs and descriptions of gene expression patterns are organized in order of the earliest developmental stage when mesodermal expression was detected for each gene. Two other genes with conserved mesodermal expression in other bilaterian taxa, NK3/bagpipe and NK4/tinman, were investigated but were not found to have mesodermal expression in the embryonic stages evaluated here (data not shown).
Figure 2.
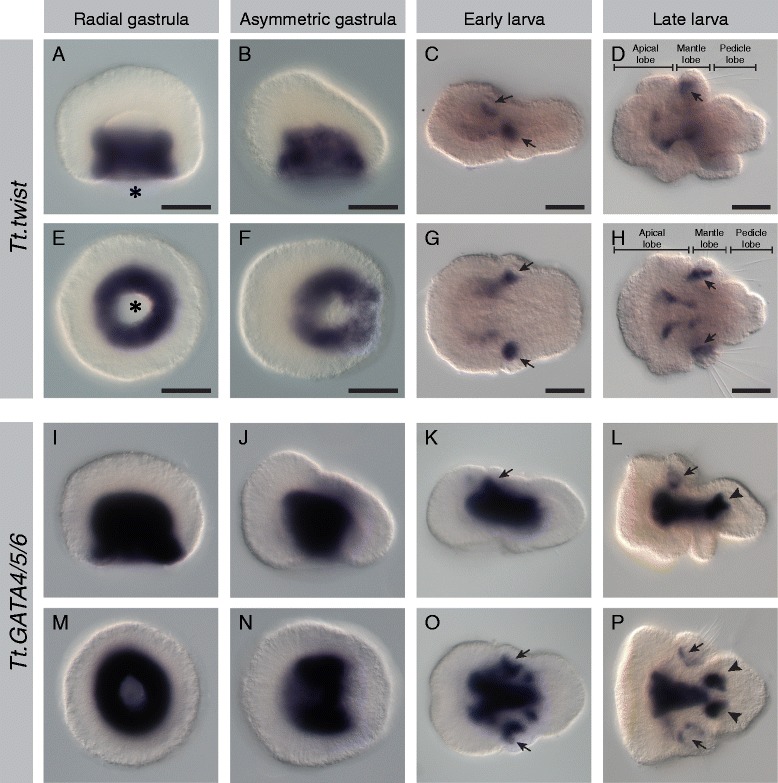
Expression patterns of Tt.twist and Tt.GATA456. All images are oriented with anterior to the left. Panels (A-D) and (I-L) are lateral views. Panels (E, F) and (M, N) are blastoporal views. Panels (G, H) and (O, P) are ventral views. For detailed descriptions of expression patterns, see text. (A-H) Tt.twist expression throughout the archenteron wall at gastrula stages and in the anterior and chaetal sac mesoderm (black arrows) in larval stages. (A, E) The location of the blastopore in the radial gastrula stage is denoted by an asterisk. (I-P) Tt.GATA456 expression in the archenteron roof and walls at gastrula stages. Expression in the endoderm, pedicle mesoderm (black arrowheads), and chaetal sac mesoderm (black arrows) in larval stages. Scale bars are 50 μm in length.
Figure 3.
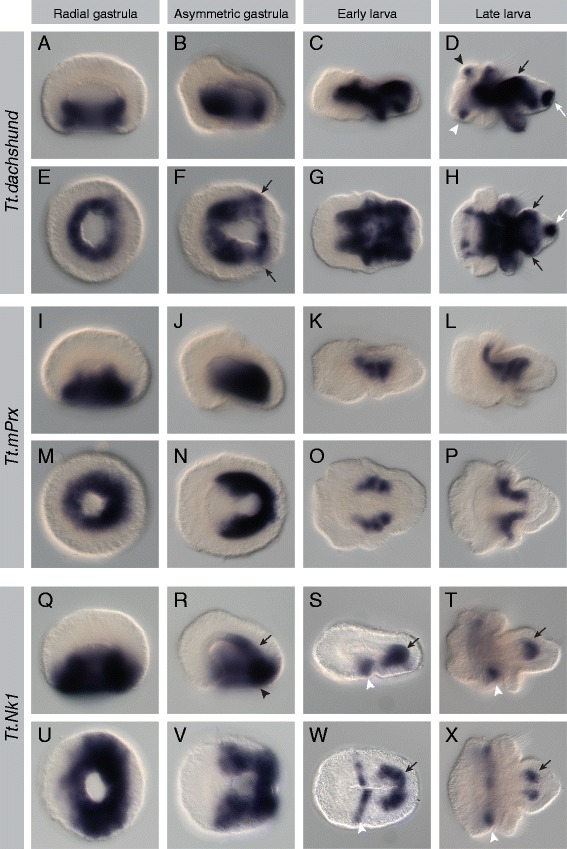
Expression patterns of Tt.dachshund, Tt.mPrx, and Tt.NK1. All images are oriented with anterior to the left. Panels (A-D) and (I-L) are lateral views. Panels (E, F) and (M, N) are blastoporal views. Panels (G, H) and (O, P) are ventral views. For detailed descriptions of expression patterns, see text. (A-H) Tt.dachshund expression in the archenteron walls at gastrula stages. Broad mesodermal expression at larval stages, along with additional domains in the pedicle lobe ectoderm (black and white arrows) and the dorsal eyespot (black arrowhead) and ventral ganglion (white arrowhead) regions of the apical lobe. (I-P) Tt. mPrx expression in the archenteron walls at gastrula stages. Expression in the lateral mesoderm at the boundary of the mantle and pedicle lobes in larval stages. (Q, R, U, V) Tt.NK1 expression in the archenteron walls (black arrow) and blastopore lip (black arrowhead) at gastrula stages. (S, T, W, X) Expression of Tt.NK1 in the pedicle mesoderm (black arrow) and posterior apical ectoderm (white arrowhead) at larval stages.
Figure 4.
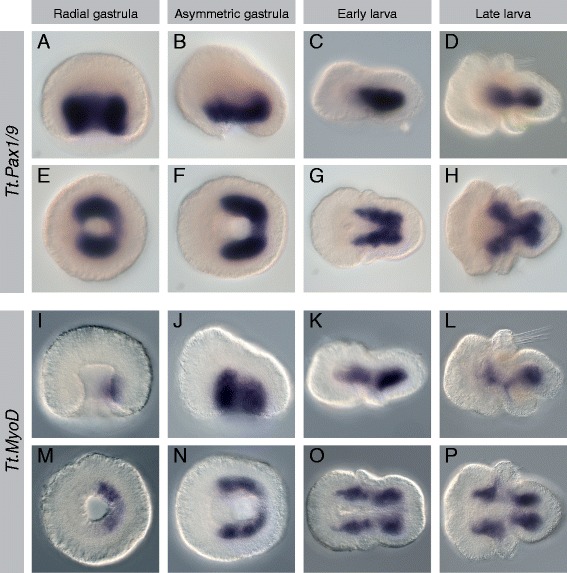
Expression patterns of Tt.Pax1/9 and Tt.MyoD. All images are oriented with anterior to the left. Panels (A-D) and (I-L) are lateral views. Panels (E, F) and (M, N) are blastoporal views. Panels (G, H) and (O, P) are ventral views. For detailed descriptions of expression patterns, see text. (A-H) Tt.Pax1/9 expression in the lateral and posterior archenteron walls at gastrula stages. Expression in the mesoderm of the mantle and pedicle lobes in larval stages. (I-P) Tt.MyoD expression in the lateral and posterior archenteron walls at gastrula stages. Expression in the lateral mesoderm of the apical, mantle, and pedicle lobes at larval stages.
Figure 5.
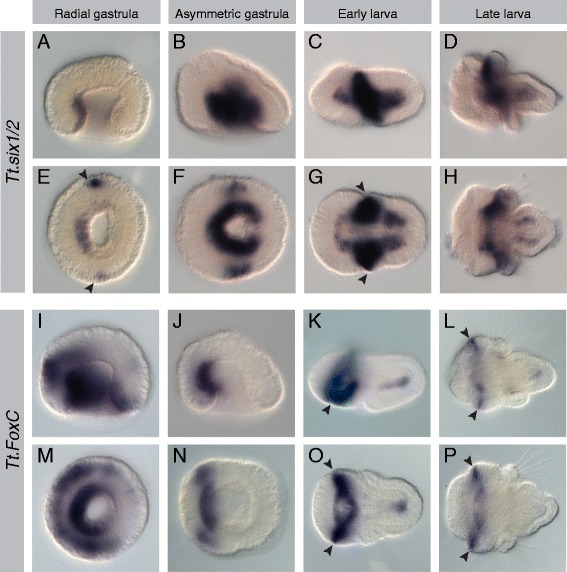
Expression patterns of Tt.Six1/2 and Tt.FoxC. All images are oriented with anterior to the left. Panels (A-D) and (I-L) are lateral views. Panels (E, F) and (M, N) are blastoporal views. Panels (G, H) and (O, P) are ventral views. For detailed descriptions of expression patterns, see text. (A-H) Tt.Six1/2 is expressed in the archenteron walls and lateral ectoderm (black arrowheads) in the late gastrula stage and in the apical, mantle, and pedicle lobe mesoderm and mantle lobe ectoderm (black arrowheads) in the larval stages. (I-P) Tt.FoxC expression is first detected in the radial gastrula in the anterior of the archenteron wall and broadly in the adjacent anterior ectoderm. In the asymmetric gastrula, expression persists in the anterior archenteron wall and anterior ectodermal expression has resolved into two lateral bands that extend along the animal-vegetal axis. Ectodermal expression is out of the plane of focus in the lateral view. In larval stages, expression is in the ventral anterior and posterior mesoderm. In the early larval stage, a ventral band of apical ectoderm is positioned near the anterior of the blastopore, and in the late gastrula stage, ectodermal expression is expanded dorsally to form a circumferential ring just anterior of the ciliary band (black arrowheads).
Figure 6.
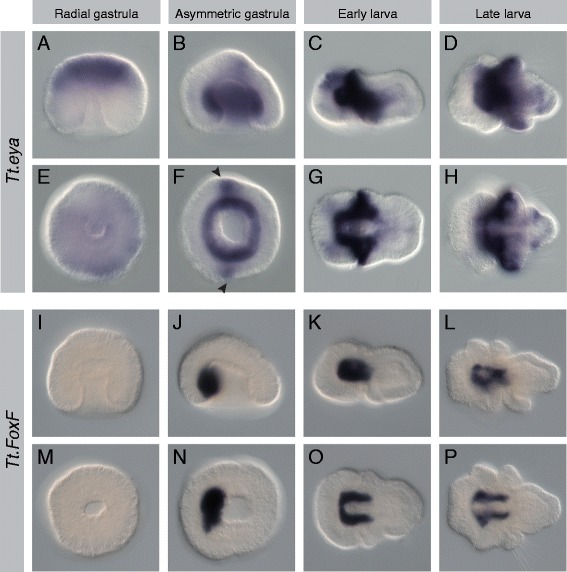
Expression patterns of Tt.eya and Tt.FoxF. All images are oriented with anterior to the left. Panels (A-D) and (I-L) are lateral views. Panels (E, F) and (M, N) are blastoporal views. Panels (G, H) and (O, P) are ventral views. For detailed descriptions of expression patterns, see text. (A-H) Tt.eya is expressed in the animal cap at the radial gastrula stage and in the archenteron walls and lateral ectoderm (black arrowheads) at the asymmetric gastrula stage. Tt.eya is expressed in the apical and mantle lobe mesoderm and anterior mantle ectoderm at larval stages. (I-P) Tt.FoxF expression is first detected in the asymmetric gastrula in the anterior of the archenteron wall. Expression is in the mesoderm laterally and anteriorly flanking the endoderm in larval stages.
Figure 7.
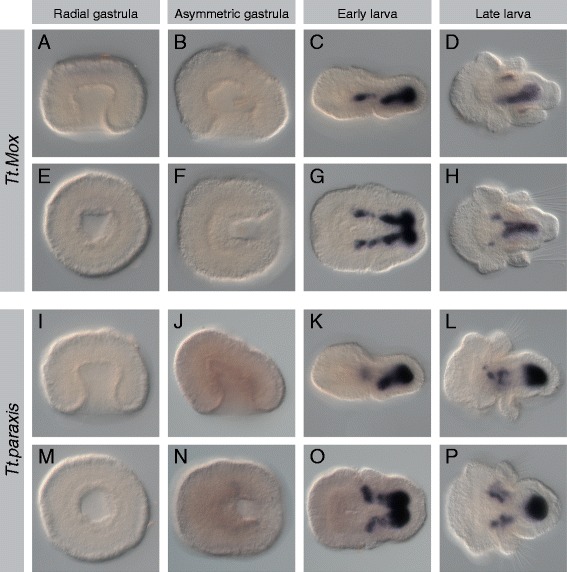
Expression patterns of Tt.Mox and Tt.paraxis. All images are oriented with anterior to the left. Panels (A-D) and (I-L) are lateral views. Panels (E, F) and (M, N) are blastoporal views. Panels (G, H) and (O, P) are ventral views. For detailed descriptions of expression patterns, see text. (A-H) Tt.Mox expression is first detected in the early larva in lateral mesodermal bands flanking the endoderm. Expression in the late larva is in the ventromedial mesoderm. (I-P) Expression of Tt.paraxis is first detected in the early larva, in the mesoderm of the mantle and pedicle lobes. In the late larva, there are distinct domains of expression underlying the mantle lobe and in the posterior mesoderm of the pedicle lobe.
Figure 8.
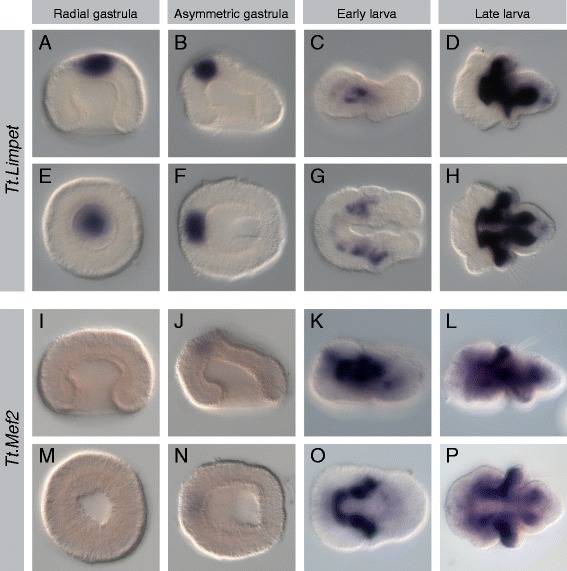
Expression patterns of Tt.Limpet and Tt.Mef2. All images are oriented with anterior to the left. Panels (A-D) and (I-L) are lateral views. Panels (E, F) and (M, N) are blastoporal views. Panels (G, H) and (O, P) are ventral views. For detailed descriptions of expression patterns, see text. (A-H) Tt.Limpet is expressed in the apical ectoderm at the gastrula stages. Mesodermal expression is first observed in the early larva in irregular bands in the developing apical and mantle lobes. In the late larva, strong expression is observed in all but the most posterior region of the mesoderm. (I-P) Weak expression of Tt.Mef2 is observed in the apical ectoderm at the late gastrula stage. In the early larva, a strong continuous band of mesodermal expression flanks the anterior portion of the endoderm and extends laterally into the developing mantle lobe. In the late larva, strong expression is observed flanking the endoderm in the apical lobe and extending into the mantle lobe, including the chaetal sacs. Expression to Tt.Mef2 is also observed in the pedicle lobe mesoderm.
Figure 9.
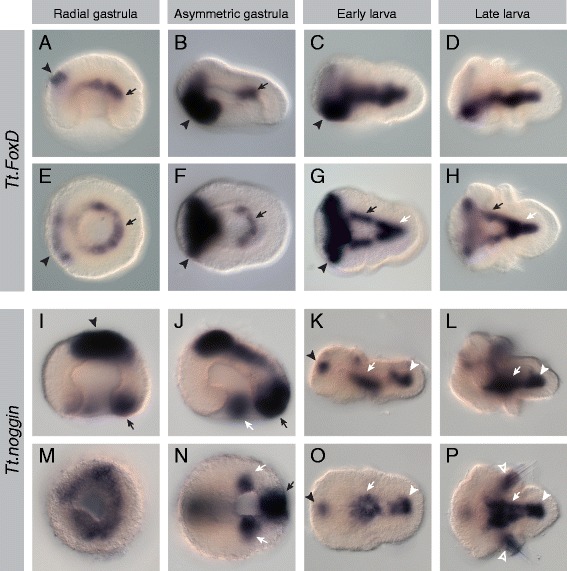
Expression patterns of Tt.FoxD and Tt.noggin. All images are oriented with anterior to the left. Panels (A-D) and (I-L) are lateral views. Panels (E, F) and (M, N) are blastoporal views. Panels (G, H) and (O, P) are ventral views. For detailed descriptions of expression patterns, see text. (A-H) Tt.FoxD is expressed in a narrow band of cells at the border of the archenteron wall and roof in the radial gastrula (black arrows). A second band of expression is present in ectodermal cells at the anterior of the animal half of the embryo (black arrowheads). In the larval stages, Tt.FoxD is expressed in two bands of the mesoderm in the mantle lobe (black arrows), which converge ventromedially in the pedicle lobe (white arrows). In the asymmetric gastrula and larval stages, a broad band of ectodermal expression is present on the ventral side just anterior of the blastopore. (I-P) Tt.noggin is expressed in the blastopore lip (black arrow) and the ectodermal animal cap (black arrowhead) of the radial gastrula. Distinct domains of expression are present in the lateral (white arrows) and posterior (black arrows) regions of the blastopore lip in the asymmetric gastrula. Expression of Tt.noggin in the ventral mesoderm of the forming mantle (white arrows) and pedicle lobes (white arrowheads) in the early larva expands to form a single broad domain of expression in the ventromedial mesoderm of the late larva. Additional domains of expression also appear in the ectodermal portions of the chaetal sacs (open white arrowheads) in the late larva.
Tt.twist
In the radial gastrula, Tt.twist is expressed symmetrically throughout the archenteron wall, which is fated to form the mesoderm, but is absent from the archenteron roof, which will form the endoderm (Figure 2A,E). Tt.twist continues to be expressed in the archenteron wall at the asymmetric gastrula stage, as the animal pole begins to shift relative to the vegetal pole and the anterior-posterior axis is established (Figure 2B,F). In the early larval stage, Tt.twist becomes localized to a horseshoe-shaped domain of the mesoderm surrounding the anterior endoderm in the developing apical lobe (Figure 2C,G). Four additional spots of mesodermal Tt.twist expression also appear in the developing mantle lobe of the early larva (black arrows in Figure 2C,G). These paired dorsal and lateral domains correspond to the positions of chaetal sac formation. In the late larva, Tt.twist expression remains in the anterior mesoderm and the chaetal sacs (Figure 2D,H).
Tt.GATA4/5/6
In the radial gastrula, Tt.GATA4/5/6 is expressed in both the mesodermal archenteron wall and the endodermal archenteron roof (Figure 2I,M). In the asymmetric gastrula, Tt.GATA4/5/6 expression persists in the archenteron roof and the lateral domains of the archenteron wall, but is absent from the anterior and posterior regions of the archenteron wall (Figure 2J,N). In the early larva, Tt.GATA4/5/6 expression in the endoderm becomes localized to the developing midgut, and mesodermal expression is in two paired domains, laterally flanking the anterior and posterior ends of the endoderm (Figure 2K,O). Four additional spots of mesoderm expression form laterally and dorsally in the developing chaetal sacs in the mantle lobe (black arrows in Figure 2K,O). In the late larva, Tt.GATA4/5/6 expression persists in the midgut, in two paired mesodermal domains adjacent to the anterior and posterior endoderm (black arrowheads in Figure 2L,P), and in the chaetal sacs (black arrows in Figure 2L,P).
Tt.dachshund
In the radial gastrula, Tt.dachshund is expressed throughout the mesodermal region of the archenteron wall (Figure 3A,E). In the asymmetric gastrula, Tt.dachshund is expressed in the archenteron wall, as well as in the lateral ectoderm adjacent to the posterior end of the archenteron (black arrows in Figure 3F). In the early larva, Tt.dachshund is expressed throughout the mesoderm, with the exception of the most anterior and posterior regions of the tissue (Figure 3C,G). A broad contiguous band of expression is also present in the dorsal and lateral ectoderm of the developing pedicle lobe (Figure 3C,G). In the late larva, Tt.dachshund shows continued expression in nearly all the mesoderm, including the chaetal sacs and the belt of mesoderm in the extended mantle lobe (Figure 3D,H). Four ectodermal domains of Tt.dachshund expression are present in the late larva: in punctate spots in the region of the eyespots on the dorsal side of the apical lobe (black arrowhead in Figure 3D), in punctate spots in the region of the ganglion on the ventral side of the apical lobe (white arrowhead in Figure 3D), in a dorsolateral saddle on the anterior half of the pedicle lobe (black arrows in Figure 3D,H), and in the posterior end of the pedicle lobe (white arrow Figure 3D,H).
Tt.mPrx
In the radial gastrula, Tt.mPrx is expressed throughout the archenteron wall, with slightly stronger expression in the posterior compared to the anterior (Figure 3I,M). In the asymmetric gastrula, Tt.mPrx is expressed in a horseshoe-shaped band of cells covering the lateral and posterior sides of the archenteron wall and is absent from the anterior archenteron wall (Figure 3J,N). In the early larva, Tt.mPrx is expressed in two lateral mesodermal bands, flanking the endoderm in the region of the developing apical lobe (Figure 3K,O). In the late larva, Tt.mPrx expression laterally flanks the endoderm in the anterior region of the pedicle lobe and extends dorsolaterally into the pedicle lobe, contacting the chaetal sacs (Figure 3L,P).
Tt.NK1
In the radial gastrula, Tt.NK1 is expressed throughout the archenteron walls and blastopore lip and extends laterally into the vegetal ectoderm (Figure 3Q,U). In the asymmetric gastrula, Tt.NK1 is expressed in the lateral and posterior archenteron walls (black arrow in Figure 3R) and blastopore lip (black arrowhead in Figure 3R) and extends laterally into the vegetal ectoderm adjacent to the blastopore lip (Figure 3R,V). Expression of Tt.NK1 is absent from the anterior archenteron wall and blastopore lip at this stage. In the early larval stage, Tt.NK1 is expressed in a crescent of mesoderm around the posterior of the endoderm in the developing pedicle lobe (black arrow in Figure 3S,W) and in a chevron in the ventral ectoderm just anterior of the furrow forming between the apical and mantle lobes (white arrowhead in Figure 3S,W). In the late larva, Tt.NK1 is expressed in bilateral regions of ectoderm laterally flanking the posterior endoderm in the pedicle lobe (black arrow in Figure 3T,X). A band ectodermal expression is present in the ventral ectoderm at the posterior margin of the apical lobe (white arrowhead in Figure 3T,X).
Tt.Pax1/9
In the radial gastrula, Tt.Pax1/9 is expressed strongly in the lateral sides of the archenteron wall (Figure 4A,E). It is expressed more weakly in the posterior of the archenteron wall and is absent from the anterior of the archenteron wall. In the asymmetric gastrula, Tt.Pax1/9 is expressed in the ventral portion of the lateral and posterior archenteron wall (Figure 4B,F). In the early larvae, Tt.Pax1/9 is expressed in a broad V-shaped mesodermal domain, lateral and ventral to the endoderm (Figure 4C,G). In the late larva, Tt.Pax1/9 expression expands into a Y-shaped domain, with two large regions lateral and ventral to the posterior end of the endoderm, a broad band of expression ventral to the midgut, and two broad domains extending laterally in the mantle lobe (Figure 4D,H).
Tt.MyoD
At the radial gastrula stage, Tt.MyoD expression is restricted to the posterior wall of the archenteron (Figure 4I,M). By the asymmetric gastrula stage, expression has expanded to a horseshoe shape and includes the lateral archenteron walls (Figure 4J,N). In the early larva, Tt.MyoD extends in two lateral bands, extending from the apical lobe to the pedicle lobe and flanking the endoderm (Figure 4K,O). Expression is strongest at the boundary of the apical and mantle lobes and the anterior of the pedicle lobe and is absent from the most posterior medial portion of the mesoderm. The anterior and posterior expression domains persist in the late larvae and are connected by a weaker band of expression that extends ventrally into the mantle lobe (Figure 4L,P).
Tt.Six1/2
Expression of Tt.Six1/2 is first detected at the radial gastrula stage in a narrow band in the anterior mesodermal region of the archenteron wall (Figure 5A,E). At this stage, two additional spots of expression are also observed in the lateral ectoderm (black arrowheads in Figure 5E). By the asymmetric gastrula stage, TtSix1/2 expression has expanded to all but the most posterior portion of the archenteron wall (Figure 5B,F). In the early larvae, Tt.Six1/2 is strongly expressed in the mesoderm and ectoderm at the anterior border of the forming mantle lobe (black arrowheads in Figure 5G) and more weakly expressed in the mesoderm of the apical and pedicle lobes (Figure 5C,G). In the late larvae, Tt.Six1/2 expression remains strong in the mesoderm and ectoderm at the anterior of the mantle lobe and weaker in the apical and pedicle mesoderm (Figure 5D,H).
Tt.FoxC
Expression of Tt.FoxC is first detected at the radial gastrula stage in the anterior of the archenteron wall and in a broad band in the anterior ectoderm (Figure 5I,M). At the asymmetric gastrula stage, mesodermal expression of Tt.FoxC remains localized to the anterior archenteron wall, and ectodermal expression forms two lateral anterior bands (Figure 5J,N). Two bands of expression are also observed in the adjacent anterior lateral ectoderm. In the early larva, there are two mesodermal domains of Tt.FoxC expression. In the apical lobe, a dorsal crescent of expression extends laterally, just anterior of the endoderm (Figure 5K,O). In the pedicle lobe, a medial band of expression extends ventrally, below the endoderm. The ventral ectodermal domains of expression are expanded and converge medially at the mouth (black arrowhead in Figure 5K). In the late larva, two dorsolateral domains of mesodermal expression remain at the anterior edge of the endoderm (Figure 5L,P). Ectodermal expression is circumferential at the anterior edge of the ciliary band in the apical lobe.
Tt.eya
In the radial gastrula, Tt.eya is weakly expressed in the animal cap (Figure 6A,E). Mesodermal expression of Tt.eya is first detected at the asymmetric gastrula stage throughout the archenteron wall, as well as in the lateral bands of the ectoderm (black arrowheads in Figure 6F). In the early larva, Tt.eya is strongly expressed in a U-shaped domain surrounding the anterior endoderm and in the dorsolateral anterior ectoderm of the developing mantle lobe (Figure 6C,G). Weaker ectodermal bands of expression are also observed dorsolaterally in the apical lobe. Strong expression in the anterior mesoderm and the dorsal anterior mantle ectoderm persists in the late larva (Figure 6D,H). Weaker ectodermal expression is also observed in the ventral half of the mantle lobe and laterally in the pedicle lobe.
Tt.FoxF
Expression of Tt.FoxF is first observed at the asymmetric gastrula stage in the anterior archenteron wall (Figure 6J,N). In the early larva, a U-shaped domain of mesodermal expression surrounds the anterior endoderm (Figure 6K,O). In the late larva, two lateral bands of mesodermal expression laterally flank the endoderm in the apical and pedicle lobes, with a weak band of expression connecting the anterior of the endoderm (Figure 4L,P).
Tt.Mox
Expression of Tt.Mox is first observed at the early larval stage (Figure 7C,G). Two domains of strong expression are observed in the posterior mesoderm, with weaker bands of expression laterally flanking the endoderm and extending anteriorly to the apical lobe. In the late larva, a medial band of expression in the posterior mesoderm extends ventrally below the posterior endoderm (Figure 7D,H). Two small domains of expression flank the endoderm at the boundary of the apical and mantle lobes.
Tt.paraxis
Expression of Tt.paraxis is first observed at the early larval stage (Figure 7K,O). As for Tt.Mox, two domains of strong expression are observed in the posterior mesoderm, with weaker expression extending anteriorly, terminating with two lateral mesodermal bands at the boundary of the apical and mantle lobes. In the late larva, a strong domain of expression persists at the posterior mesoderm (Figure 7L,P). Two disjunct and weaker domains of expression are also observed laterally at the boundary of the apical and mantle lobes.
Tt.Limpet
In the radial and asymmetric gastrula stages, Tt.Limpet expression is exclusively ectodermal, in the central region of the animal cap, where the ciliary apical tuft is located (Figure 8A,B,E,F). In the early larva, expression of Tt.Limpet is absent from the ectoderm, and weak expression is detected in the anterior and lateral mesoderm of the developing apical and mantle lobes (Figure 8C,G). In the late larva, expression of Tt.Limpet is expressed in nearly all regions of the mesoderm, surrounding the endoderm, extending into the mantle lobe, and forming two large lateral domains in the pedicle lobe (Figure 8D,H). The only region of the late larva mesoderm lacking Tt.Limpet expression is in the posterior region of the pedicle lobe, where Tt.paraxis is expressed.
Tt.Mef2
Tt.Mef2 is first detected in the asymmetric gastrula, when it is weakly expressed in the apical ectoderm (Figure 8J,N). At the early larval stage, strong expression is detected in the mesoderm of the developing apical and mantle lobes (Figure 8K,O). In the late larva, strong expression persists in the lateral mesoderm of the apical and mantle lobes, and lateral mesoderm expression is also detected in the pedicle lobe (Figure 8L,P).
Tt.FoxD
Tt.FoxD is expressed at the radial gastrula stage in a narrow band of cells at the border of the archenteron wall and roof in the radial gastrula (black arrows in Figure 9A,E). A second band of expression is present in ectodermal cells at the anterior of the animal half of the embryo at this stage (black arrowheads in Figure 9A,E). In the asymmetric gastrula, endomesodermal expression of Tt.FoxD remains in the posterior boundary of the archenteron roof and wall (black arrows in Figure 9B,F). Strong ectodermal expression is observed in a band on the ventral side, just anterior of the blastopore (black arrowheads in Figure 9B,F). In the later bilateral gastrula stage, during which the blastopore becomes elongate and then closes from posterior to anterior, the Tt.FoxD expression domain in the archenteron becomes positioned more ventrally and extends anteriorly (black arrows in Additional file 12B,C,F,G). An additional domain of expression also develops ventromedially in the posterior of the archenteron (white arrows in Additional file 12B,C,F,G). In the larval stages, expression is observed in two bands of ventral mesoderm in the mantle lobe (black arrows in Figure 9G,H), which connect to a ventromedial domain of expression in the pedicle lobe (white arrows in Figure 9G,H). A band of ectodermal expression in the ventral portion of the apical lobe persists through larval development (Figure 9C,D,G,H).
Tt.noggin
Tt.noggin is expressed in the radial gastrula in the blastopore lip (Figure 9M), with prominent expression in the posterior region (black arrow in Figure 9I). Strong ectodermal expression is also observed in the animal cap (black arrowhead in Figure 9I). By the asymmetric gastrula stage, expression in the blastopore lip has resolved into three distinct domains, a region of strong expression in the posterior of the blastopore lip (black arrows in Figure 9J,N) and two smaller domains in the lateral regions of the blastopore lip (white arrows in Figure 9J,N). In the later bilateral gastrula stage, lateral domains of Tt.noggin expression in the blastopore lip converge along the midline and shift in from the ventral surface of the embryo as the blastopore elongates and closes (white arrows in Additional file 12 J, K, N, and O). Similarly to Tt.FoxD, an additional domain of expression also develops medially in the posterior of the archenteron (white arrowheads in Additional file 12 J, K, N, and O). During the bilateral gastrula stage, the Tt.noggin expression domain shifts to the dorsal ectodermal surface of the embryo and decreases in intensity (black arrows in Additional file 12 J and K). In the early larval stage, two domains of Tt.noggin expression are observed in the ventral mesoderm of the developing mantle and pedicle lobes (white arrows and white arrowheads, respectively, in Figure 9K,O). A small domain of ectodermal expression is observed in the anterior of the apical lobe (black arrowheads in Figure 9K,O). In the late larva, a single domain of expression in the ventral mesoderm extends from the pedicle lobe to the posterior edge of the apical lobe (white arrowheads and white arrows in Figure 9L,P). Additional expression of Tt.noggin is observed in the ectodermal portion of the chaetal sacs (open white arrowheads in Figure 9L,P).
Discussion
In the present study, we have detected mesodermal expression for 17 developmental regulator genes during the embryonic and larval stages of development in the articulate brachiopod T. transversa. Each of these genes shows a unique pattern of expression with regard to both their spatial and temporal deployment, suggesting the dynamic mechanisms underlying the development of the complex larval musculature.
Expression in the radial gastrula
Five transcription factor genes, twist, GATA456, dachshund, NK1, and mPrx, showed expression in the whole archenteron wall in the radial gastrula stage. An additional four genes, Pax1/9, MyoD, Six1/2, and FoxC, showed localized expression in a portion of the archenteron wall at this stage. Expression in the archenteron wall is consistent with previous morphological observations that this region gives rise to the mesoderm in the larva [34,35]. These genes are therefore all expressed mesodermally during gastrulation and likely play roles in later aspects of mesoderm specification and determination. For twist and GATA456, comparison with expression and functional data from other bilaterian taxa suggests that these genes are widely utilized in mesoderm specification and differentiation (Additional file 1). Both genes are expressed in the larval mesoderm of the annelids [57-61] and the mesodermal parenchyma of planarian embryos [62,63]. Interestingly, while a twist ortholog is expressed in the developing mesoderm of the mollusk Patella, it is localized to the ectomesoderm [64], rather than the endomesoderm as it is in Terebratalia. Both genes are also involved in the development of mesoderm in ecdysozoans [65-69], although the role of GATA456 in the arthropod Drosophila is limited to the development of the heart [68], a structure which in brachiopods forms only in juveniles after metamorphosis. Twist orthologs are also involved in the multiple aspects of mesoderm development in deuterostomes [70-73]. The expression of both twist and GATA456 orthologs in the acoel Isodiametra [74] suggests that these genes may have ancestral roles in mesoderm development among bilaterian animals.
The available data is less conclusive for the other three early panmesodermal genes, dachshund, mPrx, and NK1 (Additional file 1), although each of these genes is expressed in the developing mesoderm of some taxa. Expression of Tt.NK1 in the developing pedicle musculature shows similarities to segmental expression in the annelid Platynereis [75] and a subset of the somatic musculature in Drosophila [76,77]. These results support a role for NK1 in myogenesis among protostomes, derived from a more ancestral role in ectodermal or neural patterning, which is shared between protostomes and deuterostomes [75,76,78-80]. In the case of mPrx, expression patterns have not been described for other protostome taxa. However, broad expression in the developing mesoderm of a hemichordate [81] and mouse [82-84] suggests that greater taxonomic sampling may reveal a previously unrecognized conservation of this gene’s participation in the formation of mesoderm. The case of Tt.dachshund is intriguing because of its mesodermal broad expression not only during the gastrula stages but also throughout the larval development. In the annelid Neanthes, expression of dachshund is also observed in the mesoderm but only during the initial formation of new segments at the posterior growth zone [85]. While mesodermal expression of dachshund is also reported from deuterostome taxa [86-89], this seems to be derived from a conserved ancestral role in neural development [90,91]. It therefore appears that dachshund has been recruited to play a novel role in mesoderm formation in among brachiopods and annelids and may play an additional role in maintenance of mesodermal identity or mesodermal differentiation in Terebratalia.
For those genes that first show regionalized expression in the archenteron wall, there is good evidence that both MyoD and FoxC have widespread roles in mesoderm development among bilaterians (Additional file 1). FoxC is particularly intriguing, as it is expressed in the anterior and posterior mesoderm of annelids [92], mollusks [92], and arthropods [93]. This may be indicative of an evolutionarily conserved role for FoxC in patterning mesoderm at the anterior and posterior extremities. Six1/2 is broadly expressed in the mesoderm in several deuterostomes [81,94,95] and the acoel Isodiametra [74], but among protostomes, mesodermal expression has only been reported from C. elegans, where it is restricted to the non-muscle coelomocyte lineage [96]. The restriction of Six1/2 expression to the eyes of an annelid [97], platyhelminth [91], and arthropod [98] suggests that its expression in the mesoderm of Terebratalia may be an independent evolutionary acquisition. In the case of Pax1/9, mesodermal expression in Drosophila [99] and mouse [100,101] is likely acquired independently in the two lineages, given its restriction to the pharyngeal endoderm of more basally divergent deuterostomes [102-104]. However, taxonomic sampling is insufficient to infer a potentially conserved role in protostome mesoderm.
Expression in the asymmetric gastrula
Mesodermal expression of two genes, Tt.eya and Tt.FoxF, was first detected in the asymmetric gastrula stage. This later onset of expression suggests that these two genes likely function in a later stage of specification or differentiation than the genes discussed above and may be downstream of them in the mesodermal gene regulatory network (GRN).
Although Tt.eya is strongly expressed throughout the archenteron wall in the asymmetric gastrula and in the anterior mesoderm of the larval stages, there is limited evidence for mesodermal expression in other members of the Spiralia. Expression of an eya ortholog has been reported from the mesodermally derived photophore (or light organ) of the bobtail squid Euprymna scolopes [105]. However, this structure is an evolutionary novelty within cephalopod mollusks, and expression of eya appears to be part of a redeployment of the Pax-Six-Eya-Dachshund network from eye specification to facilitate acquisition of light sensitivity in the photophore [105]. In the platyhelminth Schmidtea, expression of eya is restricted to the eyes during embryonic development and regeneration [91,106]. A mesodermal function for eya is more widely reported in ecdysozoans and deuterostomes, with orthologs playing key roles in myogenesis in both Drosophila [107] and vertebrates [108] and showing expression in the invaginating endomesoderm in the cephalochordate Branchiostoma [94].
The restriction of Tt.FoxF expression to the anterior mesoderm of the asymmetric gastrula and larval stages of Terebratalia may be comparable to expression in the anterior mesoderm of the mollusk Patella [92]. However, in both Patella and the annelid Capitella, there was also expression in the posterior mesoderm [92], for which no equivalent was observed in Terebratalia. More broadly, the expression of Tt.FoxF surrounds the larval endoderm, which may share an evolutionary origin with expression in the visceral mesoderm of Drosophila [109], the hemichordate Saccoglossus [110], and the mouse [111].
Expression in the larval stages
Tt.Mox and Tt.paraxis show very similar expression patterns in the mantle and pedicle lobes of the larval stages. Mox expression in the ventral mesoderm appears to be a conserved feature in Spiralia, given that orthologs in the annelid Platynereis [112] and the mollusk Haliotis [113] show comparable expression. Conserved expression of Mox is further supported by expression of orthologs in the ventral mesoderm of Drosophila [114] and the hemichordate Saccoglossus [115]. In chordates, Mox also shows mesodermal expression, although primarily in the paraxial mesoderm [116-118]. Less taxa have been sampled for expression of paraxis; however, most available data support a conserved role in mesoderm development. Comparable to the expression of Tt.paraxis, in the annelid Platynereis, paraxis is in the ventrolateral mesoderm [112], and in the cephalochordate Branchiostoma, paraxis is in developing somites as they form at the posterior of the embryo [119]. In mouse, there are two paralogs, paraxis, which is required in somite formation [120], and scleraxis, which is required for the initial specification of mesoderm as well as for subsequent chondrogenesis in tissues derived from the somites [121]. The one exception to this trend of mesodermal expression and function for paraxis orthologs is in Drosophila, where expression of the ortholog CG33557 (previously CG12648) is restricted to neural cells [122].
Mesodermal expression of Tt.Limpet and Tt.Mef2 is very similar with both first detected anteriorly in the early larva and then expanding throughout nearly all mesodermal tissues by the late larval stage. These patterns of expression suggest that both genes likely have roles in myogenic differentiation, as their localization closely matches that of musculature labeled by phalloidin in slightly older competent larvae. This included the notable absence of expression in the medial and posterior mesoderm where there is no apparent muscle formation. Expression has not been described of either of these genes on taxa in the Spiralia, but the myogenic function of Mef2 has been well characterized in both Drosophila [13,123]. Myogenic roles have also been described for paralogs in mouse [15], and an ancestral function in myogenesis is further supported by expression in the musculature of the acoel Isodiametra [74]. Two exceptions to this trend of a myogenic role for Mef2 are its apparent lack of a developmental function in C. elegans [14] and its restriction to non-myogenic mesoderm in the developing sea urchin larva [124]. Data is not available on expression of Limpet in other taxa in the Spiralia, but orthologs in Drosophila and C. elegans are expressed in subsets of the mesoderm [125-127]. There are no direct orthologs of Limpet genes in deuterostomes, but the FHL family in vertebrates appear to be the most closely related, having lost the PET domain characteristic of Limpet genes but sharing an organization of four-and-a-half LIM domains with them [128]. Members of the FHL family show expression in musculature and heart [129], suggesting that the ancestral role for Limpet/FHL may have been in the mesoderm.
Morphogenesis of ventral mesoderm and a possible source of ectomesoderm
Two genes with larval expression in the ventral mesoderm, Tt.FoxD and Tt.noggin, have patterns of early expression distinct from those of the genes discussed above, which show expression in the archenteron wall. Orthologs of Tt.FoxD and Tt.noggin show comparable expression in the ventromedial mesoderm of the annelid Platynereis [112], a region which has been termed the ‘axochord’. In contrast to Platynereis, where the ventromedial region is described as being contractile [112], there is no evidence for ventromedial musculature in T. transversa, based on reconstructions of phalloidin-stained larvae (Figure 1 and [37]). Vertebrate noggin orthologs are well known for their expression in the developing mesoderm [130,131] and their role in dorsal morphogenesis [131,132]; however, additional taxonomic sampling will aid in resolving whether mesodermal noggin expression is conserved between deuterostomes and protostomes. Consistent with the expression of Tt.FoxD in both mesodermal and ectodermal tissues, FoxD orthologs in other taxa show expression in a range of tissues, including in the developing mesoderm of several species. In ecdysozoans, the C. elegans ortholog unc-130 is expressed in the ventral mesoderm [133], while the Drosophila ortholog fd59A is restricted only to ectodermal neural tissues [134]. In the hemichordate Saccoglossus, the FoxD ortholog is expressed in the ventral mesoderm [110]; however, in echinoderms, FoxD expression has only been reported in ectodermal tissues [78,135]. Among chordates, FoxD is expressed in the notochord and somites of Branchiostoma [136], and the paralog FoxD2 is expressed in the paraxial mesoderm of Xenopus [137] and the mouse [138]. In nearly all investigated taxa, FoxD orthologs also show ectodermal expression, suggesting that both mesodermal and ectodermal expression of FoxD may have been an ancestral trait of bilaterians, but mesodermal expression appears to have been subject to loss in multiple lineages.
Changes in the position of Tt.noggin and Tt.FoxD expression over the course of gastrulation and larval development provide fascinating clues about the morphogenesis of the mesoderm. Expression of Tt.FoxD at the radial gastrula stage is in the region of the archenteron wall closest to the animal pole, at the boundary with the archenteron roof. This has previously been described as the site at which endomesodermal tissue enfolds downwards to partition the endoderm and mesoderm and form a tubular gut [34]. This downward movement repositions the portion of the archenteron wall closest to the animal pole of gastrula and displaces it to the ventral side of the larva. The transition of Tt.FoxD expression from the animal pole boundary of the archenteron wall in the gastrula to the ventral mesoderm in the larva is consistent with these morphogenetic movements and suggests that the ventral mesoderm is specified early in development of the boundary of the endoderm and mesoderm in the archenteron. Tt.noggin is expressed in the lateral and posterior regions of the blastopore lip in gastrula stages but in the ventral mesoderm of larval stages. The position of these expression domains suggests that the same population of cells may express Tt.noggin in gastrula and larval stages. While the majority of the mesoderm is formed from the endomesoderm, which invaginates during early gastrulation to form the archenteron, the expression of Tt.noggin presents the possibility that cells in the lateral dorsal lip contribute to the ventral mesoderm. For both Tt.FoxD and Tt.noggin, it appears that expression in the most posterior mesodermal cells is upregulated in the early larva independent of the morphogenetic movements that place cells in the ventral mesoderm and that expression in the two distinct populations of cells coalesces by the late larval stage. A third source of tissue in the ventral mesoderm appears to be cell originally situated in the region of the archenteron wall closest to the blastopore at the vegetal pole, based on the fact that Tt.Pax1/9 becomes localized there in the asymmetric gastrula and subsequently is expressed in the ventral mesoderm. It therefore appears that three populations of cells contribute to the ventral mesoderm of the brachiopod larva. Two endomesodermal sources, at the animal and vegetal limits of the archenteron wall, are brought together through the folding of the archenteron roof that creates the gut, while a third ectomesodermal source invaginates from the blastopore lip during closure of the blastopore.
The presence of ectomesoderm fits with current phylogenetic hypotheses which suggest that although all extant brachiopods display radial or bilateral cleavage [139], they are descended from ancestors that had spiral cleavage [19,22]. Given that the spiral cleaving taxa which are closely related to brachiopods (that is, annelids [140], nemerteans [141,142], and mollusks [143]) all develop ectomesoderm, it appears that this is plesiomorphic at least for the group Trochozoa [22] and possibly more broadly for the Spiralia in general [21], given the formation of ectomesoderm in platyhelminths [24]. Indeed, the formation of ectomesoderm in phoronids [40], which are sister to [45], or nested within [51], the brachiopod lineage, evidences that potential for an evolutionary decoupling of the specification of ectomesoderm from the stereotyped spiral cleavage program. In the future, cell lineage studies in brachiopods will help resolve whether cells expressing Tt.noggin in the lateral blastopore lips do indeed invaginate to form mesoderm, as our in situ hybridization results suggest.
Conservation and variation in the mesodermal gene regulatory network
Previous comparisons of the genes underlying specification and differentiation of the mesoderm (and in particular musculature) have shown commonalities across bilaterian taxa, in particular between mouse and Drosophila, in which the most extensive developmental genetic studies have been conducted [4,6,144]. Although numerous transcription factors play roles in mesoderm specification and myogenesis in both mouse and Drosophila, the details of their connections and interactions in the mesodermal/myogenic gene regulatory network are divergent in these two systems [6]. Our results from expression analyses on embryonic stages in the brachiopod T. transversa support that a conserved set of transcription factors and nuclear proteins have roles in mesoderm specification and differentiation across the major bilaterian clades. As discussed above, the early expression of Tt.twist may suggest a role in the initial specification of mesoderm comparable to its function in Drosophila [65], despite the fact that expression of paralogs in the more closely related annelid Capitella is not detected until well after gastrulation [57], and in the mollusk Patella, the ortholog is only expressed in the ectomesoderm [64]. Later in development, broad expression of Tt.MyoD and Tt.Mef2 in portions of the mesoderm fated to form muscle is consistent with conserved roles for each of these genes in myogenic specification and differentiation. On the other hand, a number of the genes described in this study, including Tt.dachshund and Tt.Six1/2, show mesodermal expression that is quite distinct from the predominantly ectodermal expression of orthologs in most other protostome taxa described to date. It may be that this represents novel aspects of mesoderm formation associated with the transition from spiral cleavage to radial cleavage in the stem lineage of brachiopods. Looking forward, sampling from a broader range of protostome taxa will likely help to distinguish how many of the genes presented here have conserved roles in mesoderm formation, versus independent recruitments with brachiopods and other taxa.
Conclusions
The expression patterns observed suggest that several transcription factors, including Tt.twist, Tt.GATA456, Tt.dachshund, Tt.NK1, and Tt.mPrx, likely all play roles in specification of the mesoderm as a whole, given their expression throughout the archenteron wall during the radial gastrula stage. At the same time, localized expression of Tt.Pax1/9, Tt.MyoD, and Tt.Six1/2 in specific regions of the archenteron wall suggests that the mesoderm is being regionalized even during the early phases of its specification. This regionalization is further reflected as larval development progresses and the expression of most genes is restricted to a subset of the larval mesoderm. The diversity of expression patterns for mesodermal genes during the development of T. transversa likely forms the basis for the complex musculature observed in the larva. While the majority of gene expression patterns are consistent with an endomesodermal source of mesoderm, dynamic expression of Tt.noggin at the blastopore suggests a previously unrecognized contribution of the ectomesoderm. Expression patterns of many genes, including Tt.twist, Tt.MyoD, and Tt.Mef2, are consistent with conserved roles in mesoderm differentiation and specification. Widespread mesodermal expression of Tt.dachshund and Tt.Six1/2 may be an evolutionary novelty within brachiopods associated with their secondarily derived mode of radial cleavage.
Acknowledgements
This work was supported in part by a grant from NASA to MQM and by funding from the Sars International Centre for Marine Molecular Biology to AH. The authors wish to thank Dennis Willows, David Duggins, the crew of the R/V Centennial, and the staff of Friday Harbor Laboratories, University of Washington, for the assistance in collection of adult T. transversa specimens for this study. We thank Scott Santaga, José M. (Chema) Martín-Durán, and Bruno Vellutini for the assistance with rearing and collecting of embryos and for the valuable discussions on brachiopods and the evolution of the mesoderm. We also thank three anonymous reviewers, whose comments improved the manuscript.
Abbreviations
- BCIP
5-bromo-4-chloro-3-indolyl phosphate
- BLAST
Basic Local Alignment Search Tool
- BLASTX
translated nucleotide BLAST against protein database
- cDNA
complementary DNA
- DIG
digoxigenin
- DIG-UTP
digoxigenin-conjugated uracil triphosphate
- NBT
nitro blue tetrazolium chloride
- NCBI
National Center for Biotechnology Information
- PCR
polymerase chain reaction
- RACE
rapid amplification of cDNA ends
- RT-PCR
reverse translation polymerase chain reaction
- TBLASTN
protein BLAST against translated nucleotide database
Additional files
Summary table of mesodermal gene expression. Compilation of mesodermal gene expression data for T. transversa and other bilaterian taxa, for all genes investigated in this study. Genes are organized by the developmental stage at which mesodermal expression was first detected in T. transversa.
GenBank accession numbers. GenBank accession numbers are listed for T. transversa genes used in this study.
In situ hybridization protocol. A detailed protocol is presented for in situ hybridization of riboprobes in whole-mount T. transversa embryos, as performed for all gene expression data presented in this study.
Bayesian phylogenetic analysis of bHLH transcription factors. Bayesian phylogenetic analysis supports orthology assignments for Tt.MyoD, Tt.paraxis, and Tt.twist.
Bayesian phylogenetic analysis of dachshund nuclear proteins. Bayesian phylogenetic analysis supports orthology assignment for Tt.dachshund, based on placement with orthologs from the annelid Platynereis and the mollusk Crassostrea.
Bayesian phylogenetic analysis of Forkhead (Fox) transcription factors. Bayesian phylogenetic analysis supports orthology assignments for Tt.FoxC, Tt.FoxD, and Tt.FoxF.
Bayesian phylogenetic analysis of MADS-box transcription factors. Bayesian phylogenetic analysis supports orthology assignment for Tt.MEF2.
Bayesian phylogenetic analysis of ANTP-class homeobox transcription factors. Bayesian phylogenetic analysis supports orthology assignment for Tt.Mox.
Bayesian phylogenetic analysis of NK-class homeobox transcription factors. Bayesian phylogenetic analysis supports orthology assignment for Tt.NK1.
Bayesian phylogenetic analysis of Paired box (Pax) transcription factors. Bayesian phylogenetic analysis supports orthology assignment for Tt.Pax1/9.
Bayesian phylogenetic analysis of sine oculis (Six) class homeobox transcription factors. Bayesian phylogenetic analysis supports orthology assignment for Tt.Six1/2.
Expression patterns of Tt.FoxD and Tt.noggin during the transition for gastrula to larval stages. All images are oriented with anterior to the left. Panels A-D and I-L are lateral views. Panels E-F and M-N are blastoporal views. Panels G-H and O-P are ventral views. For detailed descriptions of expression patterns, see text. (A-H) Tt.FoxD is expressed in a narrow band of cells at the border of the archenteron wall and roof in the asymmetric gastrula and transitions ventrally in the bilateral gastrula (black arrows). A second region of mesodermal expression develops in two ventrolateral posterior bands, which converge medially as the blastopore closes (white arrows). (I-P) Domains of Tt.noggin in the lateral regions of the blastopore lip of the asymmetric gastrula invaginate to contribute to the ventromedial mesoderm as the blastopore closes in the bilateral gastrula (white arrows). Expression in the posterior of the blastopore lip shifts to the dorsal ectoderm (black arrows), while a second domain of posterior mesodermal expression forms medially in the region that will form the pedicle lobe (white arrowheads).
Footnotes
Competing interests
The authors declare that they have no competing interests.
Authors’ contributions
YJP helped conceptualize and design the study, isolated and cloned the genes, performed the expression analyses, analyzed and interpreted the data, and drafted the manuscript. AH helped conceptualize and design the study, isolated and cloned the genes, performed the expression analyses, analyzed and interpreted the data, and aided in the manuscript preparation. MQM helped conceptualize and design the study and aided in the manuscript preparation. All authors read and approved the final manuscript.
Contributor Information
Yale J Passamaneck, Email: yale@hawaii.edu.
Andreas Hejnol, Email: andreas.hejnol@uib.no.
Mark Q Martindale, Email: mqmartin@whitney.ufl.edu.
References
- 1.Gilbert SF. Developmental biology. 10. Sunderland: Sinauer Associates Incorporated; 2013. [Google Scholar]
- 2.Martindale MQ, Pang K, Finnerty JR. Investigating the origins of triploblasty: “mesodermal” gene expression in a diploblastic animal, the sea anemone Nematostella vectensis (phylum, Cnidaria; class, Anthozoa) Development. 2004;131:2463–74. doi: 10.1242/dev.01119. [DOI] [PubMed] [Google Scholar]
- 3.Olson EN, Perry M, Schulz RA. Regulation of muscle differentiation by the MEF2 family of MADS box transcription factors. Dev Biol. 1995;172:2–14. doi: 10.1006/dbio.1995.0002. [DOI] [PubMed] [Google Scholar]
- 4.Black BL, Olson EN. Transcriptional control of muscle development by myocyte enhancer factor-2 (MEF2) proteins. Annu Rev Cell Dev Biol. 1998;14:167–96. doi: 10.1146/annurev.cellbio.14.1.167. [DOI] [PubMed] [Google Scholar]
- 5.Amin NM, Shi H, Liu J. The FoxF/FoxC factor LET-381 directly regulates both cell fate specification and cell differentiation in C. elegans mesoderm development. Development. 2010;137:1451–60. doi: 10.1242/dev.048496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ciglar L, Furlong EE. Conservation and divergence in developmental networks: a view from Drosophila myogenesis. Curr Opin Cell Biol. 2009;21:754–60. doi: 10.1016/j.ceb.2009.10.001. [DOI] [PubMed] [Google Scholar]
- 7.Clark IBN, Boyd J, Hamilton G, Finnegan DJ, Jarman AP. D-six4 plays a key role in patterning cell identities deriving from the Drosophila mesoderm. Dev Biol. 2006;294:220–31. doi: 10.1016/j.ydbio.2006.02.044. [DOI] [PubMed] [Google Scholar]
- 8.Li X, Oghi KA, Zhang J, Krones A, Bush KT, Glass CK, et al. Eya protein phosphatase activity regulates Six1-Dach-Eya transcriptional effects in mammalian organogenesis. Nature. 2003;426:247–54. doi: 10.1038/nature02083. [DOI] [PubMed] [Google Scholar]
- 9.Furuya M, Qadota H, Chisholm AD, Sugimoto A. The C. elegans eyes absent ortholog EYA-1 is required for tissue differentiation and plays partially redundant roles with PAX-6. Dev Biol. 2005;286:452–63. doi: 10.1016/j.ydbio.2005.08.011. [DOI] [PubMed] [Google Scholar]
- 10.Balagopalan L, Keller CA, Abmayr SM. Loss-of-function mutations reveal that the Drosophila nautilus gene is not essential for embryonic myogenesis or viability. Dev Biol. 2001;231:374–82. doi: 10.1006/dbio.2001.0162. [DOI] [PubMed] [Google Scholar]
- 11.Chen L, Krause M, Sepanski M, Fire A. The Caenorhabditis elegans MYOD homologue HLH-1 is essential for proper muscle function and complete morphogenesis. Development. 1994;120:1631–41. doi: 10.1242/dev.120.6.1631. [DOI] [PubMed] [Google Scholar]
- 12.Rudnicki MA, Schnegelsberg P, Stead RH, Braun T. MyoD or Myf-5 is required for the formation of skeletal muscle. Cell. 1993;75:1351–9. doi: 10.1016/0092-8674(93)90621-v. [DOI] [PubMed] [Google Scholar]
- 13.Bour BA, O‘Brien MA, Lockwood WL, Goldstein ES, Bodmer R, Taghert PH, et al. Drosophila MEF2, a transcription factor that is essential for myogenesis. Genes Dev. 1995;9:730–41. doi: 10.1101/gad.9.6.730. [DOI] [PubMed] [Google Scholar]
- 14.Dichoso D, Brodigan T, Chwoe KY, Lee JS, Llacer R, Park M, et al. The MADS-Box factor CeMEF2 is not essential for Caenorhabditis elegans myogenesis and development. Dev Biol. 2000;223:431–40. doi: 10.1006/dbio.2000.9758. [DOI] [PubMed] [Google Scholar]
- 15.Potthoff MJ, Olson EN. MEF2: a central regulator of diverse developmental programs. Development. 2007;134:4131–40. doi: 10.1242/dev.008367. [DOI] [PubMed] [Google Scholar]
- 16.Technau U, Scholz CB. Origin and evolution of endoderm and mesoderm. Int J Dev Biol. 2003;47:531–9. [PubMed] [Google Scholar]
- 17.Schleip W. Die Determination Der Primitiventwicklung. Leipzig: Akademische Verlags; 1929. [Google Scholar]
- 18.Giribet G, Dunn CW, Edgecombe GD, Hejnol A, Martindale MQ, Rouse GW. Assembling the spiralian tree of life. In: Telford MJ, Littlewood DTJ, editors. Animal evolution: genomes, fossils, and trees. Oxford: Oxford University Press; 2009. pp. 52–64. [Google Scholar]
- 19.Dunn CW, Giribet G, Edgecombe GD, Hejnol A. Animal phylogeny and its evolutionary implications. Annu Rev Ecol Evol Syst. 2014;45:371–95. [Google Scholar]
- 20.Halanych KM, Bacheller JD, Aguinaldo AM, Liva SM, Hillis DM, Lake JA. Evidence from 18S ribosomal DNA that the lophophorates are protostome animals. Science. 1995;267:1641–3. doi: 10.1126/science.7886451. [DOI] [PubMed] [Google Scholar]
- 21.Henry JJ, Martindale MQ. Conservation and innovation in spiralian development. Hydrobiologia. 1999;402:255–65. [Google Scholar]
- 22.Hejnol A. A twist in time–the evolution of spiral cleavage in the light of animal phylogeny. Integr Comp Biol. 2010;50:695–706. doi: 10.1093/icb/icq103. [DOI] [PubMed] [Google Scholar]
- 23.Lambert JD. Mesoderm in spiralians: the organizer and the 4d cell. J Exp Zool B Mol Dev Evol. 2007;310B:15–23. doi: 10.1002/jez.b.21176. [DOI] [PubMed] [Google Scholar]
- 24.Boyer BC, Henry JQ, Martindale MQ. Dual origins of mesoderm in a basal spiralian: cell lineage analyses in the polyclad turbellarian Hoploplana inquilina. Dev Biol. 1996;179:329–38. doi: 10.1006/dbio.1996.0264. [DOI] [PubMed] [Google Scholar]
- 25.Boyer BC, Henry JQ. Evolutionary modifications of the spiralian developmental program. Integr Comp Biol. 1998;38:621–33. [Google Scholar]
- 26.Hausdorf B, Helmkampf M, Meyer A, Witek A, Herlyn H, Bruchhaus I, et al. Spiralian phylogenomics supports the resurrection of bryozoa comprising Ectoprocta and Entoprocta. Mol Biol Evol. 2007;24:2723. doi: 10.1093/molbev/msm214. [DOI] [PubMed] [Google Scholar]
- 27.Dunn CW, Hejnol A, Matus DQ, Pang K, Browne WE, Smith SA, et al. Broad phylogenomic sampling improves resolution of the animal tree of life. Nature. 2008;452:745–9. doi: 10.1038/nature06614. [DOI] [PubMed] [Google Scholar]
- 28.Hejnol A, Obst M, Stamatakis A, Ott M, Rouse GW, Edgecombe GD, et al. Assessing the root of bilaterian animals with scalable phylogenomic methods. Proc R Soc Lond B Biol. 2009;276:4261–70. doi: 10.1098/rspb.2009.0896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Struck TH, Wey-Fabrizius AR, Golombek A, Hering L, Weigert A, Bleidorn C, et al. Platyzoan paraphyly based on phylogenomic data supports a noncoelomate ancestry of Spiralia. Mol Biol Evol. 2014;31:1833–49. doi: 10.1093/molbev/msu143. [DOI] [PubMed] [Google Scholar]
- 30.Freeman G. Regional specification during embryogenesis in the inarticulate brachiopod Glottidia. Dev Biol. 1995;172:15–36. doi: 10.1006/dbio.1995.0003. [DOI] [PubMed] [Google Scholar]
- 31.Freeman G. Regional specification during embryogenesis in the inarticulate brachiopod Discinisca. Dev Biol. 1999;209:321–39. doi: 10.1006/dbio.1999.9251. [DOI] [PubMed] [Google Scholar]
- 32.Freeman G. Regional specification during embryogenesis in the craniiform brachiopod Crania anomala. Dev Biol. 2000;227:219–38. doi: 10.1006/dbio.2000.9857. [DOI] [PubMed] [Google Scholar]
- 33.Freeman G. Regional specification during embryogenesis in Rhynchonelliform brachiopods. Dev Biol. 2003;261:268–87. doi: 10.1016/s0012-1606(03)00307-5. [DOI] [PubMed] [Google Scholar]
- 34.Long JA, Stricker S. Brachiopoda. In: Geise AC, Pearse JS, Pearse V, editors. Reproduction of marine invertebrates vol VI: echinoderms and lophophorares. Pacific Grove, CA: Boxwood Pr; 1991. pp. 47–85. [Google Scholar]
- 35.Freeman G. Regional specification during embryogenesis in the articulate brachiopod Terebratalia. Dev Biol. 1993;160:196–213. doi: 10.1006/dbio.1993.1298. [DOI] [PubMed] [Google Scholar]
- 36.Altenburger A, Wanninger A. Comparative larval myogenesis and adult myoanatomy of the rhynchonelliform (articulate) brachiopods Argyrotheca cordata, A. cistellula, and Terebratalia transversa. Front Zool. 2009;6:3. doi: 10.1186/1742-9994-6-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Santagata S. Evaluating neurophylogenetic patterns in the larval nervous systems of brachiopods and their evolutionary significance to other bilaterian phyla. J Morphol. 2011;272:1153–69. doi: 10.1002/jmor.10975. [DOI] [PubMed] [Google Scholar]
- 38.Long JA. The embryology of three species representing three superfamilies of articulate Brachiopoda. University of Washington PhD Dissertation. 1964:1–241
- 39.Lambert JD. Developmental patterns in spiralian embryos. Curr Biol. 2010;20:R72–7. doi: 10.1016/j.cub.2009.11.041. [DOI] [PubMed] [Google Scholar]
- 40.Freeman G, Martindale MQ. The origin of mesoderm in phoronids. Dev Biol. 2002;252:301–11. doi: 10.1006/dbio.2002.0854. [DOI] [PubMed] [Google Scholar]
- 41.Hyman LH. The invertebrates: smaller coelomate groups. New York: McGraw-Hill; 1959. [Google Scholar]
- 42.Helmkampf M, Bruchhaus I, Hausdorf B. Phylogenomic analyses of lophophorates (brachiopods, phoronids and bryozoans) confirm the Lophotrochozoa concept. Proc R Soc Lond B Biol. 2008;275:1927. doi: 10.1098/rspb.2008.0372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Paps J, Baguñà J, Riutort M. Lophotrochozoa internal phylogeny: new insights from an up-to-date analysis of nuclear ribosomal genes. Proc R Soc Lond B Biol. 2009;276:1245–54. doi: 10.1098/rspb.2008.1574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hausdorf B, Helmkampf M, Nesnidal MP, Bruchhaus I. Phylogenetic relationships within the lophophorate lineages (Ectoprocta, Brachiopoda and Phoronida) Mol Phylogenet Evol. 2010;55:1121–7. doi: 10.1016/j.ympev.2009.12.022. [DOI] [PubMed] [Google Scholar]
- 45.Sperling EA, Pisani D, Peterson KJ. Molecular paleobiological insights into the origin of the Brachiopoda. Evol Dev. 2011;13:290–303. doi: 10.1111/j.1525-142X.2011.00480.x. [DOI] [PubMed] [Google Scholar]
- 46.Nesnidal MP, Helmkampf M, Meyer A, Witek A, Bruchhaus I, Ebersberger I, et al. New phylogenomic data support the monophyly of Lophophorata and an Ectoproct-Phoronid clade and indicate that Polyzoa and Kryptrochozoa are caused by systematic bias. BMC Evol Biol. 2013;13:253. doi: 10.1186/1471-2148-13-253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Murdock DJE, Bengtson S, Marone F, Greenwood JM, Donoghue PCJ. Evaluating scenarios for the evolutionary assembly of the brachiopod body plan. Evol Dev. 2014;16:13–24. doi: 10.1111/ede.12059. [DOI] [PubMed] [Google Scholar]
- 48.Skovsted CB, Brock GA, Topper TP, Paterson JR, Holmer LE. Scleritome construction, biofacies, biostratigraphy and systematics of the tommotiid Eccentrotheca helenia sp. nov. from the Early Cambrian of South Australia. Palaeontology. 2011;54:253–86. [Google Scholar]
- 49.Cohen B, Weydmann A. Molecular evidence that phoronids are a subtaxon of brachiopods (Brachiopoda: Phoronata) and that genetic divergence of metazoan phyla began long before the early Cambrian. Org Divers Evol. 2005;5:253–73. [Google Scholar]
- 50.Santagata S, Cohen BL. Phoronid phylogenetics (Brachiopoda; Phoronata): evidence from morphological cladistics, small and large subunit rDNA sequences, and mitochondrial cox1. Zool J Linnean Soc. 2009;157:34–50. [Google Scholar]
- 51.Cohen BL. Rerooting the rDNA gene tree reveals phoronids to be ‘brachiopods without shells’; dangers of wide taxon samples in metazoan phylogenetics (Phoronida; Brachiopoda) Zool J Linnean Soc. 2012;167:82–92. [Google Scholar]
- 52.Balthasar U, Butterfield NJ. Early cambrian “soft-shelled” brachiopods as possible stem-group phoronids. Acta Palaeontol Pol. 2009;54:307–14. [Google Scholar]
- 53.Edgar RC. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32:1792–7. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Abascal F, Zardoya R, Posada D. ProtTest: selection of best-fit models of protein evolution. Bioinformatics. 2005;21:2104–5. doi: 10.1093/bioinformatics/bti263. [DOI] [PubMed] [Google Scholar]
- 55.Ronquist F, Huelsenbeck JP. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics. 2003;19:1572–4. doi: 10.1093/bioinformatics/btg180. [DOI] [PubMed] [Google Scholar]
- 56.Wolenski FS, Layden MJ, Martindale MQ, Gilmore TD, Finnerty JR. Characterizing the spatiotemporal expression of RNAs and proteins in the starlet sea anemone. Nematostella vectensis. Nat Protoc. 2013;8:900–15. doi: 10.1038/nprot.2013.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Dill KK, Thamm K, Seaver EC. Characterization of twist and snail gene expression during mesoderm and nervous system development in the polychaete annelid Capitella sp. I Dev Genes Evol. 2007;217:435–47. doi: 10.1007/s00427-007-0153-4. [DOI] [PubMed] [Google Scholar]
- 58.Pfeifer K, Schaub C, Wolfstetter G, Dorresteijn A. Identification and characterization of a twist ortholog in the polychaete annelid Platynereis dumerilii reveals mesodermal expression of Pdu-twist. Dev Genes Evol. 2013;223:319–28. doi: 10.1007/s00427-013-0448-6. [DOI] [PubMed] [Google Scholar]
- 59.Gillis WJ, Bowerman B, Schneider SQ. Ectoderm and endomesoderm specific GATA transcription factors in the marine annelid Platynereis dumerilli. Evol Dev. 2007;9:39–50. doi: 10.1111/j.1525-142X.2006.00136.x. [DOI] [PubMed] [Google Scholar]
- 60.Boyle MJ, Seaver EC. Developmental expression of foxA and gata genes during gut formation in the polychaete annelid. Capitella sp. I. Evol Dev. 2008;10:89–105. doi: 10.1111/j.1525-142X.2007.00216.x. [DOI] [PubMed] [Google Scholar]
- 61.Boyle MJ, Seaver EC. Expression of FoxA and GATA transcription factors correlates with regionalized gut development in two lophotrochozoan marine worms: Chaetopterus (Annelida) and Themiste lageniformis (Sipuncula) Evodevo. 2010;1:2. doi: 10.1186/2041-9139-1-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Martín-Durán JM, Amaya E, Romero R. Germ layer specification and axial patterning in the embryonic development of the freshwater planarian Schmidtea polychroa. Dev Biol. 2010;340:145–58. doi: 10.1016/j.ydbio.2010.01.018. [DOI] [PubMed] [Google Scholar]
- 63.Martín-Durán JM, Romero R. Evolutionary implications of morphogenesis and molecular patterning of the blind gut in the planarian Schmidtea polychroa. Dev Biol. 2011;352:164–76. doi: 10.1016/j.ydbio.2011.01.032. [DOI] [PubMed] [Google Scholar]
- 64.Nederbragt AJ, Lespinet O, van Wageningen S, van Loon AE, Adoutte A, Dictus WJAG. A lophotrochozoan twist gene is expressed in the ectomesoderm of the gastropod mollusk Patella vulgata. Evol Dev. 2002;4:334–43. doi: 10.1046/j.1525-142x.2002.02020.x. [DOI] [PubMed] [Google Scholar]
- 65.Leptin M. twist and snail as positive and negative regulators during Drosophila mesoderm development. Genes Dev. 1991;5:1568–76. [DOI] [PubMed]
- 66.Baylies MK, Bate M. twist: a myogenic switch in Drosophila. Science. 1996;272:1481–4. [DOI] [PubMed]
- 67.Harfe BD, Gomes AV, Kenyon C, Liu J, Krause M, Fire A. Analysis of a Caenorhabditis elegans Twist homolog identifies conserved and divergent aspects of mesodermal patterning. Genes Dev. 1998;12:2623–35. doi: 10.1101/gad.12.16.2623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Gajewski K, Fossett N, Molkentin JD, Schulz RA. The zinc finger proteins Pannier and GATA4 function as cardiogenic factors in Drosophila. Development. 1999;126:5679–88. doi: 10.1242/dev.126.24.5679. [DOI] [PubMed] [Google Scholar]
- 69.Maduro MF, Meneghini MD, Bowerman B, Broitman-Maduro G, Rothman JH. Restriction of mesendoderm to a single blastomere by the combined action of SKN-1 and a GSK-3beta homolog is mediated by MED-1 and −2 in C. elegans. Mol Cell. 2001;7:475–85. doi: 10.1016/s1097-2765(01)00195-2. [DOI] [PubMed] [Google Scholar]
- 70.Wu S-Y, Yang Y-P, McClay DR. Twist is an essential regulator of the skeletogenic gene regulatory network in the sea urchin embryo. Dev Biol. 2008;319:406–15. doi: 10.1016/j.ydbio.2008.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Yasui K, Zhang SC, Uemura M, Aizawa S, Ueki T. Expression of a twist-related gene, Bbtwist, during the development of a lancelet species and its relation to cephalochordate anterior structures. Dev Biol. 1998;195:49–59. doi: 10.1006/dbio.1997.8834. [DOI] [PubMed] [Google Scholar]
- 72.Tokuoka M, Satoh N, Satou Y. A bHLH transcription factor gene, Twist-like1, is essential for the formation of mesodermal tissues of Ciona juveniles. Dev Biol. 2005;288:387–96. doi: 10.1016/j.ydbio.2005.09.018. [DOI] [PubMed] [Google Scholar]
- 73.Chen ZF, Behringer RR. twist is required in head mesenchyme for cranial neural tube morphogenesis. Genes Dev. 1995;9:686–99. [DOI] [PubMed]
- 74.Chiodin M, Børve A, Berezikov E, Ladurner P, Martinez P, Hejnol A. Mesodermal gene expression in the acoel Isodiametra pulchra indicates a low number of mesodermal cell types and the endomesodermal origin of the gonads. PLoS One. 2013;8:e55499. doi: 10.1371/journal.pone.0055499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Saudemont A, Dray N, Hudry B, Le Gouar M, Vervoort M, Balavoine G. Complementary striped expression patterns of NK homeobox genes during segment formation in the annelid Platynereis. Dev Biol. 2008;317:430–43. doi: 10.1016/j.ydbio.2008.02.013. [DOI] [PubMed] [Google Scholar]
- 76.Dohrmann C, Azpiazu N, Frasch M. A new Drosophila homeo box gene is expressed in mesodermal precursor cells of distinct muscles during embryogenesis. Genes Dev. 1990;4:2098–111. doi: 10.1101/gad.4.12a.2098. [DOI] [PubMed] [Google Scholar]
- 77.Knirr S, Azpiazu N, Frasch M. The role of the NK-homeobox gene slouch (S59) in somatic muscle patterning. Development. 1999;126:4525–35. doi: 10.1242/dev.126.20.4525. [DOI] [PubMed] [Google Scholar]
- 78.Yankura KA, Martik ML, Jennings CK, Hinman VF. Uncoupling of complex regulatory patterning during evolution of larval development in echinoderms. BMC Biol. 2010;8:143. doi: 10.1186/1741-7007-8-143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Schubert FR, Fainsod A, Gruenbaum Y, Gruss P. Expression of the novel murine homeobox gene Sax-1 in the developing nervous system. Mech Dev. 1995;51:99–114. doi: 10.1016/0925-4773(95)00358-8. [DOI] [PubMed] [Google Scholar]
- 80.Simon R, Lufkin T. Postnatal lethality in mice lacking the Sax2 homeobox gene homologous to Drosophila S59/slouch: evidence for positive and negative autoregulation. Mol Cell Biol. 2003;23:9046–60. doi: 10.1128/MCB.23.24.9046-9060.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Green SA, Norris RP, Terasaki M, Lowe CJ. FGF signaling induces mesoderm in the hemichordate Saccoglossus kowalevskii. Development. 2013;140:1024–33. doi: 10.1242/dev.083790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Opstelten D, Vogels R, Robert B, Kalkhoven E. The mouse homeobox gene, S8, is expressed during embryogenesis predominantly in mesenchyme. Mech Dev. 1991;34:29–41. doi: 10.1016/0925-4773(91)90089-o. [DOI] [PubMed] [Google Scholar]
- 83.Cserjesi P, Lilly B, Bryson L, Wang Y, Sassoon DA, Olson EN. MHox: a mesodermally restricted homeodomain protein that binds an essential site in the muscle creatine kinase enhancer. Development. 1992;115:1087–101. doi: 10.1242/dev.115.4.1087. [DOI] [PubMed] [Google Scholar]
- 84.Leussink B, Brouwer A, El Khattabi M, Poelmann RE, Gittenberger-de Groot AC, Meijlink F. Expression patterns of the paired-related homeobox genes MHox/Prx1 and S8/Prx2 suggest roles in development of the heart and the forebrain. Mech Dev. 1995;52:51–64. doi: 10.1016/0925-4773(95)00389-i. [DOI] [PubMed] [Google Scholar]
- 85.Winchell CJ, Valencia JE, Jacobs DK. Expression of Distal-less, dachshund, and optomotor blind in Neanthes arenaceodentata (Annelida, Nereididae) does not support homology of appendage-forming mechanisms across the Bilateria. Dev Genes Evol. 2010;220:275–95. doi: 10.1007/s00427-010-0346-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Candiani S, Kreslova J, Benes V, Oliveri D, Castagnola P, Pestarino M, et al. Cloning and developmental expression of amphioxus Dachschund. Gene Expr Patterns. 2003;3:65–9. doi: 10.1016/s1567-133x(02)00070-4. [DOI] [PubMed] [Google Scholar]
- 87.Mazet F, Hutt JA, Milloz J, Millard J, Graham A, Shimeld SM. Molecular evidence from Ciona intestinalis for the evolutionary origin of vertebrate sensory placodes. Dev Biol. 2005;282:494–508. doi: 10.1016/j.ydbio.2005.02.021. [DOI] [PubMed] [Google Scholar]
- 88.Caubit X, Thangarajah R, Theil T, Wirth J, Nothwang HG, Rüther U, et al. Mouse Dac, a novel nuclear factor with homology to Drosophila dachshund shows a dynamic expression in the neural crest, the eye, the neocortex, and the limb bud. Dev Dyn. 1999;214:66–80. doi: 10.1002/(SICI)1097-0177(199901)214:1<66::AID-DVDY7>3.0.CO;2-7. [DOI] [PubMed] [Google Scholar]
- 89.Davis RJ, Shen W, Heanue TA, Mardon G. Mouse Dach, a homologue of Drosophila dachshund, is expressed in the developing retina, brain and limbs. Dev Genes Evol. 1999;209:526–36. doi: 10.1007/s004270050285. [DOI] [PubMed] [Google Scholar]
- 90.Lemons D, Fritzenwanker JH, Gerhart J, Lowe CJ, McGinnis W. Co-option of an anteroposterior head axis patterning system for proximodistal patterning of appendages in early bilaterian evolution. Dev Biol. 2010;344:358–62. doi: 10.1016/j.ydbio.2010.04.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Martín-Durán JM, Monjo F, Romero R. Morphological and molecular development of the eyes during embryogenesis of the freshwater planarian Schmidtea polychroa. Dev Genes Evol. 2012;222:45–54. doi: 10.1007/s00427-012-0389-5. [DOI] [PubMed] [Google Scholar]
- 92.Shimeld SM, Boyle MJ, Brunet T, Luke GN, Seaver EC. Clustered Fox genes in lophotrochozoans and the evolution of the bilaterian Fox gene cluster. Dev Biol. 2010;340:234–48. doi: 10.1016/j.ydbio.2010.01.015. [DOI] [PubMed] [Google Scholar]
- 93.Häcker U, Kaufmann E, Hartmann C, Jürgens G, Knöchel W, Jäckle H. The Drosophila fork head domain protein crocodile is required for the establishment of head structures. EMBO J. 1995;14:5306–17. doi: 10.1002/j.1460-2075.1995.tb00215.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Kozmik Z, Holland ND, Kreslova J, Oliveri D. Pax–Six–Eya–Dach network during amphioxus development: conservation in vitro but context specificity in vivo. Dev Biol. 2007;306:143–59. doi: 10.1016/j.ydbio.2007.03.009. [DOI] [PubMed] [Google Scholar]
- 95.Oliver G, Wehr R, Jenkins NA, Copeland NG, Cheyette BN, Hartenstein V, et al. Homeobox genes and connective tissue patterning. Development. 1995;121:693–705. doi: 10.1242/dev.121.3.693. [DOI] [PubMed] [Google Scholar]
- 96.Amin NM, Lim SE, Shi H, Chan TL, Liu J. A conserved Six–Eya cassette acts downstream of Wnt signaling to direct non-myogenic versus myogenic fates in the C. elegans postembryonic mesoderm. Dev Biol. 2009;331:350–60. doi: 10.1016/j.ydbio.2009.05.538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Arendt D, Tessmar K, de Campos-Baptista M-IM, Dorresteijn A, Wittbrodt J. Development of pigment-cup eyes in the polychaete Platynereis dumerilii and evolutionary conservation of larval eyes in Bilateria. Development. 2002;129:1143–54. doi: 10.1242/dev.129.5.1143. [DOI] [PubMed] [Google Scholar]
- 98.Cheyette BN, Green PJ, Martin K, Garren H, Hartenstein V, Zipursky SL. The Drosophila sine oculis locus encodes a homeodomain-containing protein required for the development of the entire visual system. Neuron. 1994;12:977–96. doi: 10.1016/0896-6273(94)90308-5. [DOI] [PubMed] [Google Scholar]
- 99.Duan H, Zhang C, Chen J, Sink H, Frei E, Noll M. A key role of Pox meso in somatic myogenesis of Drosophila. Development. 2007;134:3985–97. doi: 10.1242/dev.008821. [DOI] [PubMed] [Google Scholar]
- 100.Wallin J, Wilting J, Koseki H, Fritsch R, Christ B, Balling R. The role of Pax-1 in axial skeleton development. Development. 1994;120:1109–21. doi: 10.1242/dev.120.5.1109. [DOI] [PubMed] [Google Scholar]
- 101.Peters H, Wilm B, Sakai N, Imai K, Maas R, Balling R. Pax1 and Pax9 synergistically regulate vertebral column development. Development. 1999;126:5399–408. doi: 10.1242/dev.126.23.5399. [DOI] [PubMed] [Google Scholar]
- 102.Ogasawara M, Wada H, Peters H, Satoh N. Developmental expression of Pax1/9 genes in urochordate and hemichordate gills: insight into function and evolution of the pharyngeal epithelium. Development. 1999;126:2539–50. doi: 10.1242/dev.126.11.2539. [DOI] [PubMed] [Google Scholar]
- 103.Lowe CJ, Wu M, Salic A, Evans L, Lander E, Stange-Thomann N, et al. Anteroposterior patterning in hemichordates and the origins of the chordate nervous system. Cell. 2003;113:853–65. doi: 10.1016/s0092-8674(03)00469-0. [DOI] [PubMed] [Google Scholar]
- 104.Holland ND, Holland LZ, Kozmik Z. An amphioxus Pax gene, AmphiPax-1, expressed in embryonic endoderm, but not in mesoderm: implications for the evolution of class I paired box genes. Mol Mar Biol Biotechnol. 1995;4:206–14. [PubMed] [Google Scholar]
- 105.Peyer SM, Pankey MS, Oakley TH, McFall-Ngai MJ. Eye-specification genes in the bacterial light organ of the bobtail squid Euprymna scolopes, and their expression in response to symbiont cues. Mech Dev. 2014;131:111–26. doi: 10.1016/j.mod.2013.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Mannini L, Rossi L, Deri P, Gremigni V, Salvetti A. Djeyes absent (Djeya) controls prototypic planarian eye regeneration by cooperating with the transcription factor Djsix-1. Dev Biol. 2004;269:346–59. doi: 10.1016/j.ydbio.2004.01.042. [DOI] [PubMed] [Google Scholar]
- 107.Liu Y-H, Jakobsen JS, Valentin G, Amarantos I, Gilmour DT, Furlong EEM. A systematic analysis of Tinman function reveals Eya and JAK-STAT signaling as essential regulators of muscle development. Dev Cell. 2009;16:280–91. doi: 10.1016/j.devcel.2009.01.006. [DOI] [PubMed] [Google Scholar]
- 108.Heanue TA, Reshef R, Davis RJ, Mardon G, Oliver G, Tomarev S, et al. Synergistic regulation of vertebrate muscle development by Dach2, Eya2, and Six1, homologs of genes required for Drosophila eye formation. Genes Dev. 1999;13:3231–43. doi: 10.1101/gad.13.24.3231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Zaffran S, Küchler A, Lee HH, Frasch M. biniou (FoxF), a central component in a regulatory network controlling visceral mesoderm development and midgut morphogenesis in Drosophila. Genes Dev. 2001;15:2900–15. doi: 10.1101/gad.917101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Fritzenwanker JH, Gerhart J, Freeman RM, Lowe CJ. The Fox/Forkhead transcription factor family of the hemichordate Saccoglossus kowalevskii. Evodevo. 2014;5:17. doi: 10.1186/2041-9139-5-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Ormestad M, Astorga J, Carlsson P. Differences in the embryonic expression patterns of mouse Foxf1 and −2 match their distinct mutant phenotypes. Dev Dyn. 2004;229:328–33. doi: 10.1002/dvdy.10426. [DOI] [PubMed] [Google Scholar]
- 112.Lauri A, Brunet T, Handberg-Thorsager M, Fischer AHL, Simakov O, Steinmetz PRH, et al. Development of the annelid axochord: insights into notochord evolution. Science. 2014;345:1365–8. doi: 10.1126/science.1253396. [DOI] [PubMed] [Google Scholar]
- 113.Hinman VF, Degnan BM. Mox homeobox expression in muscle lineage of the gastropod Haliotis asinina: evidence for a conserved role in bilaterian myogenesis. Dev Genes Evol. 2002;212:141–4. doi: 10.1007/s00427-002-0223-6. [DOI] [PubMed] [Google Scholar]
- 114.Chiang C, Patel NH, Young KE, Beachy PA. The novel homeodomain gene buttonless specifies differentiation and axonal guidance functions of Drosophila dorsal median cells. Development. 1994;120:3581–93. doi: 10.1242/dev.120.12.3581. [DOI] [PubMed] [Google Scholar]
- 115.Lowe CJ, Terasaki M, Wu M, Freeman RM, Runft L, Kwan K, et al. Dorsoventral patterning in hemichordates: insights into early chordate evolution. PLoS Biol. 2006;4 doi: 10.1371/journal.pbio.0040291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Minguillón C, Garcia-Fernàndez J. The single amphioxus Mox gene: insights into the functional evolution of mox genes, somites, and the asymmetry of amphioxus somitogenesis. Dev Biol. 2002;246:455–65. doi: 10.1006/dbio.2002.0660. [DOI] [PubMed] [Google Scholar]
- 117.Imai KS, Hino K, Yagi K, Satoh N, Satou Y. Gene expression profiles of transcription factors and signaling molecules in the ascidian embryo: towards a comprehensive understanding of gene networks. Development. 2004;131:4047–58. doi: 10.1242/dev.01270. [DOI] [PubMed] [Google Scholar]
- 118.Mankoo BS, Collins NS, Ashby P, Grigorieva E, Pevny LH, Candia A, et al. Mox2 is a component of the genetic hierarchy controlling limb muscle development. Nature. 1999;400:69–73. doi: 10.1038/21892. [DOI] [PubMed] [Google Scholar]
- 119.Beaster-Jones L, Kaltenbach SL, Koop D, Yuan S, Chastain R, Holland LZ. Expression of somite segmentation genes in amphioxus: a clock without a wavefront? Dev Genes Evol. 2008;218:599–611. doi: 10.1007/s00427-008-0257-5. [DOI] [PubMed] [Google Scholar]
- 120.Burgess R, Rawls A, Brown D, Bradley A, Olson EN. Requirement of the paraxis gene for somite formation and musculoskeletal patterning. Nature. 1996;384:570–3. doi: 10.1038/384570a0. [DOI] [PubMed] [Google Scholar]
- 121.Brown D, Wagner D, Li X, Richardson JA, Olson EN. Dual role of the basic helix-loop-helix transcription factor scleraxis in mesoderm formation and chondrogenesis during mouse embryogenesis. Development. 1999;126:4317–29. doi: 10.1242/dev.126.19.4317. [DOI] [PubMed] [Google Scholar]
- 122.Peyrefitte S, Kahn D, Haenlin M. New members of the Drosophila Myc transcription factor subfamily revealed by a genome-wide examination for basic helix-loop-helix genes. Mech Dev. 2001;104:99–104. doi: 10.1016/s0925-4773(01)00360-4. [DOI] [PubMed] [Google Scholar]
- 123.Ranganayakulu G, Zhao B, Dokidis A, Molkentin JD. A series of mutations in the D-MEF2 transcription factor reveal multiple functions in larval and adult myogenesis in Drosophila. Dev Biol. 1995;171:169–81. doi: 10.1006/dbio.1995.1269. [DOI] [PubMed] [Google Scholar]
- 124.Andrikou C, Iovene E, Rizzo F, Oliveri P, Arnone MI. Myogenesis in the sea urchin embryo: the molecular fingerprint of the myoblast precursors. Evodevo. 2013;4:33. doi: 10.1186/2041-9139-4-33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Tomancak P, Berman BP, Beaton A, Weiszmann R, Kwan E, Hartenstein V, et al. Global analysis of patterns of gene expression during Drosophila embryogenesis. Genome Biol. 2007;8:R145. doi: 10.1186/gb-2007-8-7-r145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Qadota H, Mercer KB, Miller RK, Kaibuchi K, Benian GM. Two LIM domain proteins and UNC-96 link UNC-97/PINCH to myosin thick filaments in Caenorhabditis elegans muscle. Mol Biol Cell. 2007;18:4317–26. doi: 10.1091/mbc.E07-03-0278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Xiong G, Qadota H, Mercer KB, McGaha LA. A LIM-9 (FHL) / SCPL-1 (SCP) complex interacts with the C-terminal protein kinase regions of UNC-89 (Obscurin) in Caenorhabditis elegans muscle. J Mol Biol. 2009;386:976–88. doi: 10.1016/j.jmb.2009.01.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Koch BJ, Ryan JF, Baxevanis AD. The diversification of the LIM superclass at the base of the Metazoa increased subcellular complexity and promoted multicellular specialization. PLoS One. 2012;7 doi: 10.1371/journal.pone.0033261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Chu P-H, Ruiz-Lozano P, Zhou Q, Cai C, Chen J. Expression patterns of FHL/SLIM family members suggest important functional roles in skeletal muscle and cardiovascular system. Mech Dev. 2000;95:259–65. doi: 10.1016/s0925-4773(00)00341-5. [DOI] [PubMed] [Google Scholar]
- 130.Smith WC, Harland RM. Expression cloning of noggin, a new dorsalizing factor localized to the Spemann organizer in Xenopus embryos. Cell. 1992;70:829–40. doi: 10.1016/0092-8674(92)90316-5. [DOI] [PubMed] [Google Scholar]
- 131.McMahon JA, Takada S, Zimmerman LB, Fan CM, Harland RM, McMahon AP. Noggin-mediated antagonism of BMP signaling is required for growth and patterning of the neural tube and somite. Genes Dev. 1998;12:1438–52. doi: 10.1101/gad.12.10.1438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Smith WC, Knecht AK, Wu M, Harland RM. Secreted noggin protein mimics the Spemann organizer in dorsalizing Xenopus mesoderm. Nature. 1993;361:547–9. doi: 10.1038/361547a0. [DOI] [PubMed] [Google Scholar]
- 133.Nash B, Colavita A, Zheng H, Roy PJ, Culotti JG. The forkhead transcription factor UNC-130 is required for the graded spatial expression of the UNC-129 TGF-beta guidance factor in C. elegans. Genes Dev. 2000;14:2486–500. doi: 10.1101/gad.831500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Lee H-H, Frasch M. Survey of forkhead domain encoding genes in the Drosophila genome: classification and embryonic expression patterns. Dev Dyn. 2004;229:357–66. doi: 10.1002/dvdy.10443. [DOI] [PubMed] [Google Scholar]
- 135.Tu Q, Brown CT, Davidson EH, Oliveri P. Sea urchin forkhead gene family: phylogeny and embryonic expression. Dev Biol. 2006;300:49–62. doi: 10.1016/j.ydbio.2006.09.031. [DOI] [PubMed] [Google Scholar]
- 136.Yu JK, Holland ND, Holland LZ. An amphioxus winged helix/forkhead gene, AmphiFoxD: insights into vertebrate neural crest evolution. Dev Dyn. 2002;225:289–97. doi: 10.1002/dvdy.10173. [DOI] [PubMed] [Google Scholar]
- 137.Pohl BS, Knöchel W. Temporal and spatial expression patterns of FoxD2 during the early development of Xenopus laevis. Mech Dev. 2002;111:181–4. doi: 10.1016/s0925-4773(01)00617-7. [DOI] [PubMed] [Google Scholar]
- 138.Sasaki H, Hogan BL. Differential expression of multiple fork head related genes during gastrulation and axial pattern formation in the mouse embryo. Development. 1993;118:47–59. doi: 10.1242/dev.118.1.47. [DOI] [PubMed] [Google Scholar]
- 139.Freeman G. A developmental basis for the Cambrian radiation. Zool Sci. 2007;24:113–22. doi: 10.2108/zsj.24.113. [DOI] [PubMed] [Google Scholar]
- 140.Meyer NP, Boyle MJ, Martindale MQ, Seaver EC. A comprehensive fate map by intracellular injection of identified blastomeres in the marine polychaete Capitella teleta. EvoDevo. 2010;1:8. doi: 10.1186/2041-9139-1-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Henry JQ, Martindale MQ. The establishment of embryonic axial properties in the nemertean. Cerebratulus lacteus. Dev Biol. 1996;180:713–21. doi: 10.1006/dbio.1996.0340. [DOI] [PubMed] [Google Scholar]
- 142.Henry JJ, Martindale MQ. Conservation of the spiralian developmental program: cell lineage of the nemertean. Cerebratulus lacteus. Dev Biol. 1998;201:253–69. doi: 10.1006/dbio.1998.8966. [DOI] [PubMed] [Google Scholar]
- 143.Render J. Cell fate maps in the Ilyanassa obsoleta embryo beyond the third division. Dev Biol. 1997;189:301–10. doi: 10.1006/dbio.1997.8654. [DOI] [PubMed] [Google Scholar]
- 144.Barnes RM, Firulli AB. A twist of insight - the role of Twist-family bHLH factors in development. Int J Dev Biol. 2009;53:909–24. doi: 10.1387/ijdb.082747rb. [DOI] [PMC free article] [PubMed] [Google Scholar]


