Abstract
Background
The ability to measure the expression of pro-inflammatory cytokines from intestinal biopsies in patients with Crohn’s disease in an accurate and reproducible way is critical for proof-of-concept and mechanism-of-action trials; however, the number of biopsies from a segment of the ileum or colon required to yield reproducible results has not been rigorously evaluated. We examined intestinal biopsies from patients with Crohn’s disease to validate methods for detecting changes in inflammatory gene expression.
Methods
To evaluate the reproducibility of gene expression measurements, intestinal biopsies were obtained from designated segmentsfrom6 healthy controls, 6 patients with active Crohn’s disease, and 6 patients with inactive Crohn’s disease. Disease activity was based on the simple endoscopic score for Crohn’s disease (SES-CD). Expression of 7 pro-inflammatory genes was measured from each biopsy using quantitative PCR. Using a linear mixed effects model, the power to detect transcriptional changes corresponding to active and inactive Crohn’s disease was calculated.
Results
Total SES-CD score corresponds with expression of most inflammatory biomarkers. For most genes, 2 – 5biopsies are needed to reduce sampling error to <25% for most genes. To measure changes in mRNA expression corresponding to active versus inactive Crohn’s disease, one to two intestinal biopsies from 3 patients before and after treatment are needed to yield power of at least 80%.
Conclusion
Measuring pro-inflammatory gene expression from mucosal biopsies from patients with Crohn’s disease is practicable and provides objective biomarkers that can be utilized in proof-of-concept and mechanism-of-action trials to assess response to therapy.
Keywords: Crohn’s disease, gene expression, biomarker, coefficient of variation, sampling error, proof-of-concept, power
INTRODUCTION
Crohn’s disease (CD) is a chronic relapsing inflammatory disease affecting any portion of the gastrointestinal tract in a genetically susceptible individual.1 The treatment options for CD have expanded over the past twenty years with the emergence of anti-TNF and anti-integrin antibodies;2–6 however, a significant proportion of patients with CD will not respond to existing medications.7, 8 There remains an unmet need for new medications for Crohn’s disease with unique mechanisms of action. With significant advances in the understanding of the pathogenesis of Crohn’s disease, increasing numbers of candidate drugs have emerged. Evaluation of novel therapies relies upon the availability of relatively large numbers of patients for phase 1 and phase 2 studies. This existing system, however, is inefficient and slows the translation of research innovations into therapies for patients.
Measuring the expression of inflammatory genes in an accurate and reproducible manner from intestinal biopsies of patients with CD is critical for implementation of proof-of-concept and mechanism-of-action studies. Current clinical endpoints utilize clinical scores that rely heavily upon subjective symptoms reported by patients but may not reflect active inflammation as confirmed by endoscopy or inflammatory biomarkers, making it difficult to interpret negative clinical trials evaluating new medications in CD.9–11 In the absence of predictive inflammatory biomarkers from the blood, intestinal inflammatory gene expression is an appealing method for measuring disease activity in a quantitative manner.
Prior studies using intestinal biopsies have shown that expression of inflammatory cytokines from intestinal biopsies of patients with CD was increased not only in active areas of disease but also in endoscopic ally unaffected mucosa.12–14 Furthermore, inflammatory genes that were over expressed in the setting of active CD normalize after successful treatment with infliximab.15, 16 Since gene expression in different regions of the colon is not uniform in a healthy colon,17 and CD frequently leads to discontinuous inflammation at a macroscopic level, it is not clear whether one could draw reliable conclusions from a single intestinal biopsy from an individual with Crohn’s disease. To date, the accuracy and reproducibility of measuring inflammatory biomarkers from different regions of the intestine in CD have not been rigorously evaluated to validate methods for longitudinal comparisons of gene expression over time. We applied methodology that was developed for gene expression assays using quantitative PCR with cell-based standards in patients with rheumatoid arthritis.18 In an effort to develop methods pertinent to early proof-of-concept and mechanism-of-action trials, we aimed to evaluate the reproducibility of measuring expression of inflammatory genes from intestinal biopsies from a small cohort of patients with CD.
MATERIALS AND METHODS
Patient Selection
Patients with Crohn’s disease and healthy controls undergoing colonoscopy as part of routine medical care at the University of California, San Diego were enrolled in the study. The participants recruited had a diagnosis of Crohn’s disease, had not undergone colectomy, and were over 18 years of age. Healthy controls were undergoing colonoscopy as part of routine colon cancer screening. Based on the endoscopic appearance, each participant was classified as having active or inactive disease by a single experienced endoscopist (WJS). Simple endoscopic score for Crohn’s disease (SES-CD) was determined.19, 20 Healthy controls were not included if they reported gastrointestinal symptoms. The institutional review board at UCSD approved the study, and all participants provided written informed consent.
Intestinal Biopsies
Three biopsies were obtained from each of 5 segments of the ileum and colon (ileum, and ascending, transverse, descending, and recto sigmoid colon) from patients with CD. Biopsies were immediately placed into the RNA stat-60 (Tel-Test, Friendswood, TX, USA) and stored at −80 C. Assays were performed within 6 months of acquiring samples.
RNA Isolation
Biopsy samples were homogenized manually on ice in RNA Stat-60 using chilled Kontes-Duall tissue grinders. Following addition of chloroform and centrifugation, 70% ethanol was mixed with the aqueous phase, then run on RNeasy Lipid Tissue Mini columns following manufacturer protocol (Qiagen, Germantown, MD). RNA was eluted in 40 ul of TE buffer. Ribo-green reagent (Life Technologies, Carlsbad, CA) was used to quantify the RNA. For the reverse transcription reaction, 250 ng RNA was used (TaqMan Reverse Transcription Reagents, Life Technologies, Carlsbad, CA). Quantitative PCR (QPCR) reagents and primers for glyceraldehyde 3-phosphate dehydrogenase (GAPDH), interleukin (IL)-1β, IL-6, IL-8, interferon gamma-induced protein (IP)-10, matrix metalloproteinase (MMP)-3, S100 calcium binding protein A8 (S100A8), and TNFα were obtained from Life Technologies (Carlsbad, CA). The qPCR methods, generation of standards, and GAPDH normalization, were performed as described by Boyle et al.18 The inter-assay reproducibility based on the coefficient of variation (CV) was 4 – 6%. Though batch effect has increasingly been recognized as a potential confounder when performing multiplex or microarray systems, batch effect should not affect our methodology using quantitative PCR with a dynamic range and a 7-point standardized curve. As a consequence, sample randomization was not performed, nor is batch effect a confounding factor.
We intentionally measured gene expression from a limited number of inflammatory genes, rather than using a multiplex system, in order to maximize our ability to sample the colon, using 15 intestinal biopsies per patient. The seven genes measured have been implicated in the pathogenesis of inflammatory bowel disease. Upon activation by nuclear factor-κB (NF-κB) and mitogen activated protein (MAP) kinase, IL-1β, IL-6, and TNFα initiate pro-inflammatory signaling cascades that have been associated with multiple autoimmune conditions, including Crohn’s disease.21–24 IL-8 is a chemo attractant that induces neutrophil chemo taxis and phagocytosis, and IP-10 is a chemokine that attracts inflammatory cells.25–29 MMP-3 degrades extracellular matrix under inflammatory conditions,14 and S100-A8 is a subunit of cal protect in, a biomarker for fecal inflammation, that is used to monitor disease activity in inflammatory bowel disease.16, 30, 31
Data Analysis
The relationship between gene expression and SES-CD score by intestinal segment or in total as well as C-reactive protein (CRP) was evaluated using linear mixed effects models that control for repeated biopsies obtained within a single region for an individual subject. The estimated regression coefficient associated with the linear effect of the total SES-CD and segmental SES-CD was reported, and an approximate F-test with the Kenward-Roger approach was used to evaluate significance.32 Gene expression in each segment from individuals with active versus inactive CD, as well as from individuals within active CD versus healthy controls, was compared using a mixed model fit to control for repeated biopsies in a segment. The coefficient of variation for each inflammatory gene was determined using repeated measures.33 Power analyses were performed using a simulation-based approach derived from a linear mixed model for gene expression with repeat measures. Power was maintained at 80% and 90%, and p-values of <0.05 were considered statistically significant. All analyses were performed using the open source statistical language R34 with ‘lme4’35 and ‘pbkrtest’36 packages.
RESULTS
The protocol for collecting biopsies was intended to quantify variability within an intestinal segment and between segments. Pinch biopsies were obtained with cold forceps from 6 patients with active Crohn’s disease, 6 with inactive Crohn’s disease, and 6 healthy controls. As a result of a change in the study protocol after enrollment and biopsy collection from 3 patients, ileal biopsies were not obtained from 2 individuals with active CD and 1 with inactive CD; however, the ileum was assessed to calculate the SES-CD in those patients.
The mean age of patients with active and inactive Crohn’s disease was 39 and 44, respectively (Table 1). In contrast, the average age of healthy controls was 61 years old as these individuals were undergoing colonoscopy for colon cancer screening. On average, the healthy controls were older than the patients with Crohn’s disease as a result of the indication for the colonoscopy. While age may have an effect on inflammatory gene expression, only limited comparisons of inflammatory gene expression in patients with CD versus healthy controls were performed. Of the patients with CD, thirty-three percent of patients with active disease were men as compared to 67% with active disease. The average duration of disease was 11 as compared to 17 years in participants with active versus inactive CD. Thirty-three percent of patients with active CD were taking steroids as compared to none with inactive CD, and 67% with active CD were taking TNFα antagonists as compared to 50% with inactive disease. Thirty-three percent of patients with active disease as compared with 50% with inactive disease were taking immunosuppressive medications, such as a zathioprine, 6-mercaptopurine, or methotrexate. Of the patients with active disease, 67% of patients had ileocolonic disease and 33% had only colonic involvement. 33% of patients with active CD, and none with inactive disease, had a history of perianal disease. Of patients with active and inactive CD, 17% had a stricturing phenotype, and only 17% of participants with active CD had a penetrating disease phenotype. Prior bowel resections, including ileal and ileocolonic resections, had been performed in 17% patients with active disease and 50% of patients with inactive disease. The mean CRP (mg/dL) in active CD was 1.14 as compared with 0.17 in inactive disease, but this difference was not significant. All of the patients with active CD had some degree of endoscopic rectal inflammation. The average SES-CD was significantly higher in patients with active CD (10.3, SD 4.4) as compared to inactive CD (0.7, SD 0.8, p<0.05), and these scores were consistent with moderately active and inactive CD, respectively. All of the patients with active CD had evidence of active disease based on histology.
Table 1.
Baseline Characteristics of Patients with Crohn’s Disease
| Active (n= 6) |
Inactive (n=6) |
P-value | |
|---|---|---|---|
| Age (years +/− S.D.) | 39.3 +/− 18.3 | 43.5 +/− 26.1 | NS |
| % Male | 33% | 66% | NS |
| TNFα antagonists use, % | 66% | 50% | NS |
| Steroid use, % | 33% | 0% | NS |
| Immunosuppressive use, % | 33% | 50% | NS |
| Disease duration (years +/− S.D.) | 11.2 +/− 8.7 | 17.0 +/− 16.4 | NS |
| History of Perianal Disease, % | 33% | 0% | NS |
| Prior Bowel Resection, % | 17% | 50% | NS |
| Disease Location | |||
| Ileal (L1) | 0% | 33% | NS |
| Colonic (L2) | 33% | 0% | |
| Ileocolonic (L3) | 67% | 67% | |
| Disease Behavior | |||
| Non-stricturing, non-penetrating (B1) | 67% | 83% | NS |
| Stricturing (B2) | 17% | 17% | |
| Penetrating (B3) | 17% | 0% | |
| SES-CD (mean +/− S.D.) | 10.3 +/− 4.4 | 0.7 +/− 0.8 | <0.05 |
| Active Histologic Disease, % | 100% | 0% | <0.05 |
| CRP (mg/dL) (mean +/− S.D.) | 1.14 +/− 0.93 | 0.17 +/− 0.12 | NS |
S.D.: standard deviation; CDAI: Crohn’s Disease Activity Index; SES-CD: Simple endoscopic score for Crohn’s Disease; CRP: C-reactive protein; mg: milligram; dL: deciliter
Correlation between Simple Endoscopic Score for Crohn’s Disease and Gene Expression
The relationship between the total SES-CD score and gene expression was evaluated using a linear mixed model to validate the use of inflammatory gene biomarkers. Expression of IL-6, IL-8, TNFα, IP-10, MMP-3, and S100-A8 was associated with the total SES-CD score; however, IL-1β expression did not correlate with the total SES-CD score (Table 2). Specifically, the total SES-CD had the greatest relative effect on S100-A8 and IP-10. Using a similar linear mixed effects model, the association between simple endoscopic score from a segment of the intestine and its corresponding gene expression was evaluated independent of total SES-CD. Interestingly, IL-1β, IL-6, IL-8, MMP-3, and S100-A8 expression were significantly associated with the segmental SES-CD score; however, TNFα and IP-10 were not significantly associated (Table 2). The differences in the two analyses suggest that expression of certain genes, like TNFα and IP-10, are more closely associated with the overall endoscopic disease activity as represented by total SES-CD regardless of whether the segment of the intestinal sample appears to be actively inflamed or ulcerated, raising the possibility that these genes may be useful biomarkers for global disease activity. In contrast, expression of other genes, such as IL-1β, maybe more closely related to local inflammation and disease activity in a given portion of the intestine rather than to overall disease severity. Thus, expression of the majority of inflammatory genes is associated with total endoscopic disease activity and may be useful as biomarkers that can potentially predict global disease activity using biopsies from a single part of the intestine.
Table 2.
Association Between Inflammatory Gene Expression and Simple Endoscopic Scores for Crohn’s Disease
| Inflammatory Gene |
Estimated Effect of SES-CD from Segment |
Estimated Effect of Total SES-CD |
|---|---|---|
| IL-1β | 0.167* | 0.109 |
| IL-6 | 0.232* | 0.196* |
| IL-8 | 0.198* | 0.268* |
| TNFα | 0.040 | 0.192* |
| IP-10 | 0.040 | 0.310* |
| MMP-3 | 0.397* | 0.286* |
| S100-A8 | 0.326* | 0.348* |
SES-CD: Simple Endoscopic Score for Crohn’s Disease
The regression coefficient associated with the effect of simple endoscopic score for Crohn’s disease was calculated using a linear regression with a correction for ileal or colonic segment. Kenward-Roger F was used to determine significance. P-values less than 0.05 were considered significant and are indicated by *.
The estimated effect represents the increase in the log of gene expression for one unit increase in the simple endoscopic score for Crohn’s disease.
Correlation between C-reactive Protein and Gene Expression
To further validate gene expression as an indicator of disease, the relationship between CRP and gene expression was examined with a similar linear mixed model. Expression of IL-6, IP-10, and MMP3 was significantly associated with CRP (p-value <0.05); however, there was no significant association between CRP and IL-8, TNFα, and IL-1β when analyzing all patients with CD. One patient with active endoscopic CD had a normal CRP, which has been described in the literature37 and may be a consequence of genetic polymorphisms in the CRP promoter region.38 Excluding the patient with active Crohn’s disease without an elevated CRP, expression of IL-6, IL-8, TNFα, IP-10, MMP-3, and S100-A8 was significantly associated with the CRP concentration (p-value <0.05). Although a single outlier has a significant effect on this relationship, the association between CRP and expression of the majority of inflammatory genes provides further support for the validity of using intestinal biopsies to measure disease activity.
Gene Expression in Active versus Inactive Crohn’s Disease
Intestinal inflammatory gene expression from patients with active versus inactive Crohn’s disease was compared using a cell means mixed model. Expression of mRNA from the ileum was significantly greater in the setting of active Crohn’s disease as compared with inactive disease for all 7 genes that were examined. With the exception of IL-1β, expression of IL-6, IL-8, TNFα, IP-10, MMP-3, and S100-A8 was also significantly greater in patients with active as compared to inactive CD in the ascending, transverse, and descending colon (Figure 1, Figure 2, Figure S1). In the rectum, IL-8, TNFα, IP-10, MMP-3, and S100-A8 expression was significantly greater in active compared to inactive CD. There was a trend towards greater expression of IL-1β and IL-6 in active versus inactive CD that did not reach statistical significance.
Figure 1.
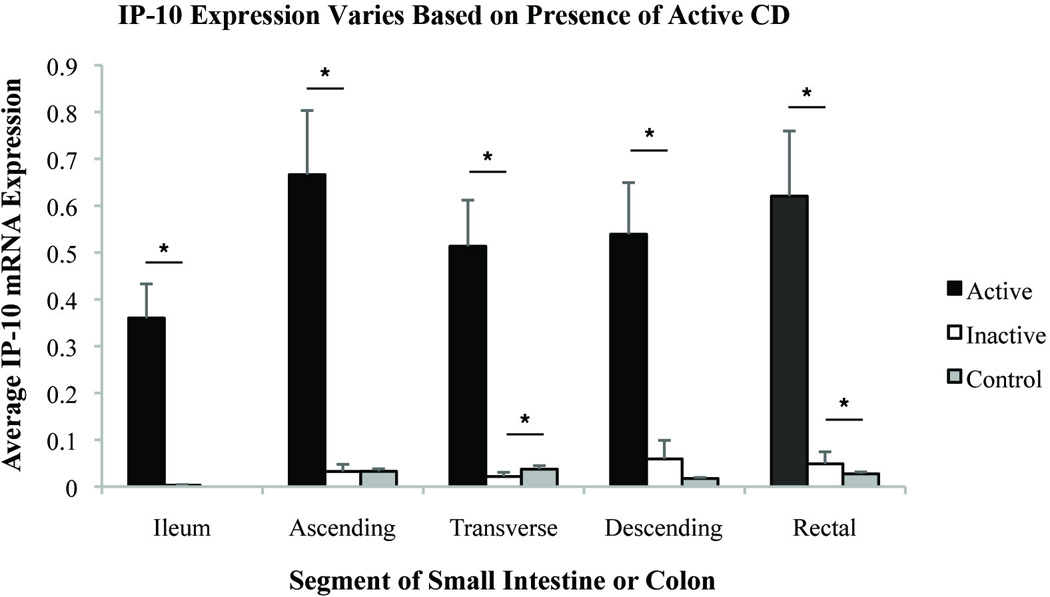
Comparison of relative IP-10 expression by segment of small intestine or colon in patients with active CD, inactive CD, and healthy controls. Relative gene expression units were averaged. Standard error shown. * indicates significance with p-value <0.05.
Figure 2.
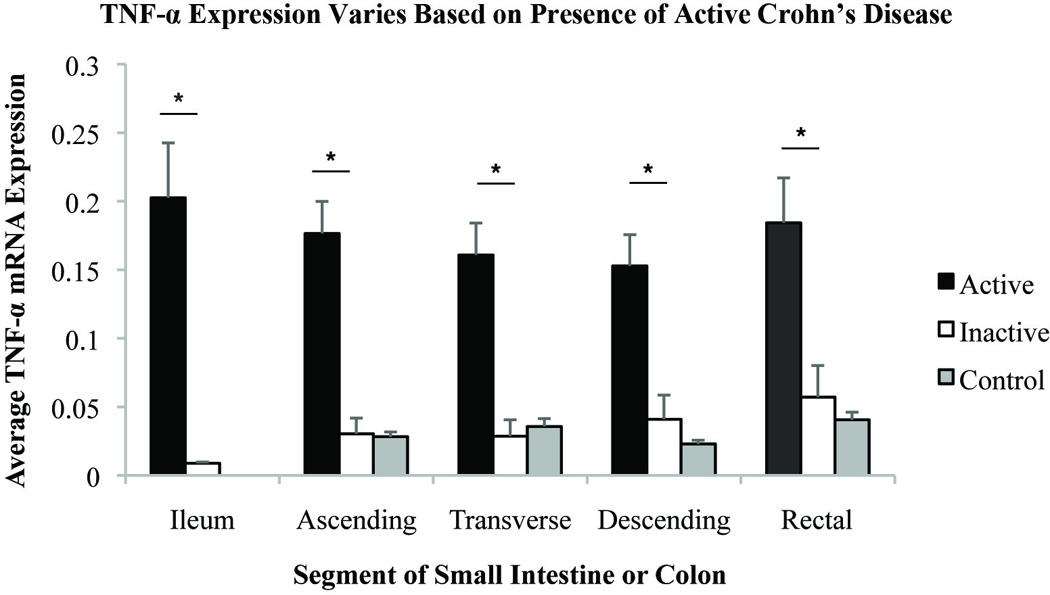
Comparison of relative TNFα expression by segment of small intestine or colon in patients with active CD, inactive CD, and healthy controls. Relative gene expression units were averaged. Standard error shown. * indicates significance with p-value <0.05.
The relative differences in gene expression between patients with active and inactive CD were greatest in the ileal samples where there was a significant effect for each gene that was measured (Table S1). In the colon, the magnitude of differences in gene expression in active and inactive CD was similar between segments and appeared to be dependent on the gene measured, rather than the segment of the intestine sampled. Overall, the differences in gene expression between active and inactive CD were greatest forIP-10, MMP-3, and IL-8. Even in areas that appeared to uninvolved endoscopically, inflammatory gene expression was greater if the individual had active CD. These results raise the possibility that measuring inflammatory genes from an unaffected distal portion of the colon may provide diagnostic information about the presence of proximal disease activity.
Coefficient of Variation for Inflammatory Genes in Crohn’s Disease
The coefficient of variation (CV) for intestinal biopsies obtained from patients with Crohn’s disease was measured, revealing significant variability with differences based on the region sampled and gene measured (Table 3). The coefficient of variation was lowest for IL-1β and TNFα with CVs ranging from 27% to 40% and 25% to 40%, respectively, depending upon the region of the intestine sampled. In contrast, the variability in MMP-3 expression was the highest with the CV ranging from 48% in the ascending colon to 68% in the ileum. Based on FDA guidelines for industry bio analytical method validation, a standard error of 25% or less was selected a priori as a benchmark for quality in sampling error.39 The number of intestinal biopsies needed from an individual patient to reduce sampling error to <25% was determined, and for TNFα, 2 biopsies would be needed for any segment of the colon, whereas 3 ileal biopsies would be needed (Table 4). With the exception of MMP-3, which had higher CV values, 2 – 5 intestinal biopsies was generally required to reduce standard error to <25%, and this was relatively consistent across different intestinal segments.
Table 3.
Coefficient of Variation for Inflammatory Genes in by Intestinal Segment For Patients with Crohn’s Disease
| Inflammatory Gene |
Ileum | Ascending Colon |
Transverse Colon |
Descending Colon |
Rectum |
|---|---|---|---|---|---|
| TNF-α | 40% | 27% | 29% | 25% | 28% |
| IL-1β | 27% | 40% | 40% | 29% | 35% |
| IP-10 | 44% | 44% | 45% | 41% | 33% |
| S100-A8 | 42% | 43% | 44% | 42% | 40% |
| IL-6 | 48% | 56% | 61% | 35% | 48% |
| IL-8 | 53% | 40% | 43% | 37% | 51% |
| MMP-3 | 68% | 48% | 56% | 57% | 52% |
The coefficient of variation was calculated for each gene by segment from intestinal and colonic biopsies obtained from patients with CD. The coefficient of variation differed by gene and by segment of the intestine sampled.
Table 4.
Number of Intestinal Biopsies from Patients with Crohn’s Disease to Reduce Standard Error to <25%
| Inflammatory Gene |
Ileum | Ascending Colon |
Transverse Colon |
Descending Colon |
Rectum |
|---|---|---|---|---|---|
| TNF-α | 3 | 2 | 2 | 1 | 2 |
| IL-1β | 2 | 3 | 3 | 2 | 2 |
| IP-10 | 4 | 4 | 4 | 3 | 2 |
| S100-A8 | 3 | 3 | 4 | 3 | 3 |
| IL-6 | 4 | 6 | 6 | 2 | 4 |
| IL-8 | 5 | 3 | 4 | 3 | 5 |
| MMP-3 | 8 | 4 | 6 | 6 | 5 |
The coefficient of variation was calculated for each gene for each segment. The number of biopsies required to reduce the standard error to less than 25% was determined.
The CV was also measured for inflammatory gene expression in healthy controls (Table S2). As in patients with CD, the CV varied based on gene measured and region examined. Overall, the values were slightly higher than in the CD patients, and this may in part be explained by the low absolute expression of inflammatory genes in a healthy colon.
Power Analyses with Mixed Models
We subsequently assessed whether we could apply our results to a hypothetical interventional clinical trial where intestinal biopsies would be obtained before and after therapy. We used two methods to evaluate whether we could reliably detect differences or changes in gene expression in response to a given treatment. In the first approach, we theorized that a clinical response to therapy may result in a hypothetical two-fold reduction in gene expression, and we calculated how many intestinal biopsies would be needed before and after treatment from a given number of patients in order to detect such a two-fold reduction with 80% power and alpha of 0.05. For the majority of the inflammatory genes that we measured, including IL-1β, IL-6, IL-8, TNFα, IP-10, and S100-A8, 3 – 6 biopsies from 3 patients before and after therapy would be sufficient to detect a two-fold change in expression (Table 5). If additional power were desired to reduce the risk of a type II error, 1–2 additional intestinal biopsies would increase the power to 90% (Table S3). However, more than 6 biopsies would need to be obtained from 6 patients to detect such changes in MMP-3, making it a poor biomarker candidate for evaluating response to a therapy. This model was applied to each segment and yielded fairly consistent results across different segments and appeared to vary predominantly based on the gene measured in spite of the discontinuous nature of inflammation in CD. The incremental information obtained from proximal biopsies, as compared to distal ones, was limited.
Table 5.
Number of Biopsies from 3 Patients Required Detect a Two-Fold Reduction in Gene Expression
| Inflammatory Gene |
Ileum | Ascending Colon |
Transverse Colon |
Descending Colon |
Rectum |
|---|---|---|---|---|---|
| TNF-α | 2 | 3 | 3 | 3 | 3 |
| IL-1β | 2 | 3 | 3 | 3 | 3 |
| IL-6 | 4 | 6 | 6 | 6 | 6 |
| IL-8 | 4 | 6 | 6 | 6 | 6 |
| S100-A8 | 4 | 5 | 5 | 5 | 6 |
| IP-10 | 4 | 6 | 6 | 6 | 6 |
| MMP-3 | 8 | >9 | >9 | >9 | >9 |
With a linear mixed effect model using repeated measures, the power to detect two-fold reduction in the expression of inflammatory genes from intestinal or colonic biopsies was determined. The minimum number of biopsies prior to and following treatment required from 3 patients to obtain a power of 80% is reported.
Using a more clinically relevant model, we hypothesized that a meaningful clinical response to a therapy in a patient with CD might lead to a reduction in expression of inflammatory genes. With a second linear mixed effect model, we quantified the hypothetical number of biopsies that would be needed before and after treatment to detect differences in gene expression comparable to active versus inactive CD. Interestingly, a total of one to two intestinal biopsies from three patients before and after treatment could reliably distinguish active from inactive Crohn’s disease for all 7 genes with a power of 80% (Table 6). Given the importance of minimizing a type II error in a proof-of-concept study, we repeated the analysis using power of 90% and found that with two biopsies from three patients, one could detect differences in mRNA gene expression between active and inactive CD from any region of the intestine (Table S4). These findings were consistent across different regions in the intestine and suggest that distal colonic biopsies might be sufficient to measuring disease activity in the setting of colonic Crohn’s disease.
Table 6.
Number of Intestinal Biopsies from 3 Patients to Detect Differences in Gene Expression Comparable to Active versus Inactive Disease
| Segment of Small Intestine or Colon | |||||
|---|---|---|---|---|---|
| Inflammatory Gene |
Ileum | Ascending Colon |
Transverse Colon |
Descending Colon |
Rectum |
| TNF-α | 1 | 1 | 1 | 1 | 1 |
| IL-1β | 1 | 1 | 1 | 1 | 1 |
| IL-6 | 1 | 2 | 1 | 1 | 1 |
| IL-8 | 1 | 1 | 1 | 1 | 1 |
| S100-A8 | 1 | 1 | 1 | 1 | 1 |
| IP-10 | 1 | 1 | 1 | 1 | 1 |
| MMP-3 | 1 | 1 | 1 | 1 | 1 |
With a linear mixed effect model using repeated measures, the power to detect changes in expression of inflammatory genes comparable to active as compared to inactive disease was determined in patients with CD. The minimum number of intestinal or colonic biopsies prior to and following treatment required from 3 patients to obtain a power of 80% is reported.
A previously published analysis performed using colonic biopsies from patients with ulcerative colitis (UC) showed consistent results.40 Two to 4 rectal biopsies in UC, as compared to 2–5 intestinal or colonic biopsies in CD, were needed to reduce sampling error to <25%. In the study of patients with UC, the mixed model for power calculations used a fixed number of colonic biopsies to determine the number of patients required to obtain a power of 80%, and 3 rectal biopsies before and after treatment from 2 patients would yield power of at least 80% or greater.40 When we applied this model using three intestinal or colonic biopsies before and after treatment to gene expression in CD, we found that 2 patients with CD were also needed to obtain a power of 80% or greater (Table S5). Together these two sets of data suggest that similar strategies may be applied to UC and CD to detect statistically significant differences between active and inactive disease.
DISCUSSION
Using a systematic approach, we assessed the variability in quantifying inflammatory gene expression and the sensitivity to detect changes in mRNA from intestinal biopsies in patients with CD. Though we measured a select number of genes from a small cohort of patients with Crohn’s disease, we maximized the number of intestinal biopsies obtained from each individual to examine sampling of different segments of the intestine. Specifically, the discontinuous nature of inflammation in CD makes validation studies in this population necessary so that appropriate, reproducible gene expression assays can be included in future proof-of-concept and mechanism-of-action studies.
There are no current standards to guide the sampling of the intestine for gene expression studies in patients with Crohn’s disease. Prior gene expression studies have obtained a single biopsy from an endoscopic ally active and/or inactive portion of colonic mucosa and assumed that the intestinal gene expression was representative of the intestine; however, studies have not been specifically performed to assess replicate sampling in CD where intestinal tissue is heterogeneously inflamed. The technical replicate CV for qPCR ranged from 4–6%, values within the FDA guidelines for assays. In our analysis of the variation in intestinal tissue gene expression, we determined the number of intestinal biopsies needed to reduce standard error to <25%, and this a priori assumption was based on the least restrictive variation for a single sample assay from FDA recommendations.39 Though proposed specifically for ligand binding assays, the guidelines served as a reference for sample size and power calculations. Though our cohort of patients was relatively small, we maximized the number of intestinal biopsies obtained from each individual. In effect, our results effectively establish the number of intestinal biopsies required in order to provide sampling reliability comparable to a single biopsy. We previously showed40 in UC that 2 – 5 intestinal biopsies are generally needed to reduce standard error to <25%, and we now extend this finding to CD.
Overall, the variability was not significantly different based on the intestinal segment sampled. The consistent variability and robust differences in gene expression suggest that distal biopsies could potentially used as a proxy to evaluate for proximal disease. Using gene expression from descending colonic or rectal biopsies, it is possible that one may be able to assess for proximal disease and avoid performing a complete colonoscopy to evaluate disease activity in the proximal colon or ileum. Distal colonic gene expression may act as an accurate biomarker, reflecting proximal disease activity. A caveat, however, is that the sample size from our study was small. All of the patients with active CD had some degree of rectal inflammation and consequently may have had higher distal inflammatory gene expression than one would expect in the setting of isolated ileal disease. Our results should be applicable to Crohn’s colitis, particularly with any degree of rectal involvement; however, further validation in patients with isolated ileal disease will be needed to confirm whether sampling an endoscopically uninvolved colon is an accurate biomarker for proximal disease.
Our methodical evaluation of variability in sampling the intestine provides the foundation for hypothesis testing at the site of disease in CD using therapeutic agents. A similar approach for proof-of-concept and mechanistic clinical trials using serial synovial biopsies has been successfully in rheumatoid arthritis,41, 42 and an analogous biopsy-based study from patients with UC provided a framework for performing serial biopsy studies.40 Further validity for our approach comes from the association between SES-CD and inflammatory gene expression, indicating that inflammatory biomarkers correlate with endoscopic disease activity. Ultimately, using the variability from the intestinal sampling, we determined the minimum number of patients and the number of biopsies prior to and following treatment needed to power a study to detect a reduction in gene expression corresponding to a transition from active to inactive disease.
In conclusion, molecular biomarker analysis from intestinal biopsies in Crohn’s disease is feasible, offering a reproducible method for measuring site-specific inflammation with a highly sensitive and high dynamic range assays. For early clinical trials, two intestinal biopsies from three patients before and after therapy should provide over 90% power to evaluate a new therapeutic agent. Using this approach, early proof-of-concept and mechanism-of-action clinical trials could enroll fewer patients than studies based on clinical, and even endoscopic, endpoints. Thus, gene expression studies using serial intestinal biopsies are an appealing and quantitative approach to measure response to therapy and facilitate mechanism-of-action studies focusing on the effect at the site of disease.
Supplementary Material
Comparison of relative IL-8, IL-1β, IL-6, MMP-3, and S100-A8 expression by segment of small intestine or colon in patients with active CD, inactive CD, and healthy controls. Relative gene expression units were averaged. Standard error shown. * indicates significance with p-value <0.05.
Acknowledgements
We would like to thank Elisabeth Evans, Marianne Fahmy, Suresh Pola, PreetBagi, and the endoscopy staff at UCSD Thornton Hospital for their assistance in collecting the intestinal samples, and the UCSD Clinical Translational Research Institute for their assistance with this project.
Source of Funding:
This study was supported by grants from the US National Institutes of Health (DK093507 to J. Chang) and the UCSD Digestive Disease Research Development Center Grant DK080506. The study was partially supported by the National Institutes of Health Grant UL1TR000100. B. Boland was supported by NIH grant T32 DK 007202. B. Zhang was supported by grant 1TL1RR03197 from the NIH National Center for Research Resources.
Footnotes
Conflicts of Interest: None.
REFERENCES
- 1.Abraham C, Cho JH. Inflammatory bowel disease. N Engl J Med. 2009;361:2066–2078. doi: 10.1056/NEJMra0804647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Baumgart DC, Sandborn WJ. Crohn's disease. Lancet. 2012;380:1590–1605. doi: 10.1016/S0140-6736(12)60026-9. [DOI] [PubMed] [Google Scholar]
- 3.Sandborn WJ, Feagan BG, Rutgeerts P, et al. Vedolizumab as induction and maintenance therapy for Crohn's disease. N Engl J Med. 2013;369:711–721. doi: 10.1056/NEJMoa1215739. [DOI] [PubMed] [Google Scholar]
- 4.Dignass A, Van Assche G, Lindsay JO, et al. The second European evidence-based Consensus on the diagnosis and management of Crohn's disease: Current management. J Crohns Colitis. 2010;4:28–62. doi: 10.1016/j.crohns.2009.12.002. [DOI] [PubMed] [Google Scholar]
- 5.Lichtenstein GR, Hanauer SB, Sandborn WJ. Management of Crohn's disease in adults. Am J Gastroenterol. 2009;104:465–483. doi: 10.1038/ajg.2008.168. quiz 464, 484. [DOI] [PubMed] [Google Scholar]
- 6.Sandborn WJ, Colombel JF, Enns R, et al. Natalizumab induction and maintenance therapy for Crohn's disease. N Engl J Med. 2005;353:1912–1925. doi: 10.1056/NEJMoa043335. [DOI] [PubMed] [Google Scholar]
- 7.D'Haens GR, Panaccione R, Higgins PD, et al. The London Position Statement of the World Congress of Gastroenterology on Biological Therapy for IBD with the European Crohn's and Colitis Organization: when to start, when to stop, which drug to choose, and how to predict response? Am J Gastroenterol. 2011;106:199–212. doi: 10.1038/ajg.2010.392. quiz 213. [DOI] [PubMed] [Google Scholar]
- 8.Allez M, Karmiris K, Louis E, et al. Report of the ECCO pathogenesis workshop on anti-TNF therapy failures in inflammatory bowel diseases: definitions, frequency and pharmacological aspects. J Crohns Colitis. 2010;4:355–366. doi: 10.1016/j.crohns.2010.04.004. [DOI] [PubMed] [Google Scholar]
- 9.Gomes P, du Boulay C, Smith CL, et al. Relationship between disease activity indices and colonoscopic findings in patients with colonic inflammatory bowel disease. Gut. 1986;27:92–95. doi: 10.1136/gut.27.1.92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Peyrin-Biroulet L, Reinisch W, Colombel JF, et al. Clinical disease activity, C-reactive protein normalisation and mucosal healing in Crohn's disease in the SONIC trial. Gut. 2014;63:88–95. doi: 10.1136/gutjnl-2013-304984. [DOI] [PubMed] [Google Scholar]
- 11.Lahiff C, Safaie P, Awais A, et al. The Crohn's disease activity index (CDAI) is similarly elevated in patients with Crohn's disease and in patients with irritable bowel syndrome. Aliment Pharmacol Ther. 2013;37:786–794. doi: 10.1111/apt.12262. [DOI] [PubMed] [Google Scholar]
- 12.Reimund JM, Wittersheim C, Dumont S, et al. Increased production of tumour necrosis factor-alpha interleukin-1 beta, and interleukin-6 by morphologically normal intestinal biopsies from patients with Crohn's disease. Gut. 1996;39:684–689. doi: 10.1136/gut.39.5.684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Leon AJ, Gomez E, Garrote JA, et al. High levels of proinflammatory cytokines, but not markers of tissue injury, in unaffected intestinal areas from patients with IBD. Mediators Inflamm. 2009;2009:580450. doi: 10.1155/2009/580450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Noble CL, Abbas AR, Lees CW, et al. Characterization of intestinal gene expression profiles in Crohn's disease by genome-wide microarray analysis. Inflamm Bowel Dis. 2010;16:1717–1728. doi: 10.1002/ibd.21263. [DOI] [PubMed] [Google Scholar]
- 15.Arijs I, De Hertogh G, Machiels K, et al. Mucosal gene expression of cell adhesion molecules, chemokines, and chemokine receptors in patients with inflammatory bowel disease before and after infliximab treatment. Am J Gastroenterol. 2011;106:748–761. doi: 10.1038/ajg.2011.27. [DOI] [PubMed] [Google Scholar]
- 16.Leal RF, Planell N, Kajekar R, et al. Identification of inflammatory mediators in patients with Crohn's disease unresponsive to anti-TNFalpha therapy. Gut. 2014 doi: 10.1136/gutjnl-2013-306518. [DOI] [PubMed] [Google Scholar]
- 17.Noble CL, Abbas AR, Cornelius J, et al. Regional variation in gene expression in the healthy colon is dysregulated in ulcerative colitis. Gut. 2008;57:1398–1405. doi: 10.1136/gut.2008.148395. [DOI] [PubMed] [Google Scholar]
- 18.Boyle DL, Rosengren S, Bugbee W, et al. Quantitative biomarker analysis of synovial gene expression by real-time PCR. Arthritis Res Ther. 2003;5:R352–R360. doi: 10.1186/ar1004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Daperno M, D'Haens G, Van Assche G, et al. Development and validation of a new, simplified endoscopic activity score for Crohn's disease: the SES-CD. Gastrointest Endosc. 2004;60:505–512. doi: 10.1016/s0016-5107(04)01878-4. [DOI] [PubMed] [Google Scholar]
- 20.Ferrante M, Colombel JF, Sandborn WJ, et al. Validation of endoscopic activity scores in patients with Crohn's disease based on a post hoc analysis of data from SONIC. Gastroenterology. 2013;145:978–986. e5. doi: 10.1053/j.gastro.2013.08.010. [DOI] [PubMed] [Google Scholar]
- 21.Chang M, Chow SC. Analysis strategies for adaptive designs with multiple endpoints. J Biopharm Stat. 2007;17:1189–1200. doi: 10.1080/10543400701645348. [DOI] [PubMed] [Google Scholar]
- 22.Lapunzina P, Monk D. The consequences of uniparental disomy and copy number neutral loss-of-heterozygosity during human development and cancer. Biol Cell. 2011;103:303–317. doi: 10.1042/BC20110013. [DOI] [PubMed] [Google Scholar]
- 23.Romanelli V, Nevado J, Fraga M, et al. Constitutional mosaic genome-wide uniparental disomy due to diploidisation: an unusual cancer-predisposing mechanism. J Med Genet. 2011;48:212–216. doi: 10.1136/jmg.2010.081919. [DOI] [PubMed] [Google Scholar]
- 24.Raddatz D, Bockemuhl M, Ramadori G. Quantitative measurement of cytokine mRNA in inflammatory bowel disease: relation to clinical and endoscopic activity and outcome. Eur J Gastroenterol Hepatol. 2005;17:547–557. doi: 10.1097/00042737-200505000-00012. [DOI] [PubMed] [Google Scholar]
- 25.Wang SL, Chang CH, Yang HC, et al. Performance evaluation of coherence-based adaptive imaging using clinical breast data. IEEE Trans Ultrason Ferroelectr Freq Control. 2007;54:1669–1679. doi: 10.1109/tuffc.2007.438. [DOI] [PubMed] [Google Scholar]
- 26.Lian NX, Chang L, Tan YP, et al. Adaptive filtering for color filter array demosaicking. IEEE Trans Image Process. 2007;16:2515–2525. doi: 10.1109/tip.2007.904459. [DOI] [PubMed] [Google Scholar]
- 27.Wu CC, Chen JS, Chen SJ, et al. Kinetics of adaptive immunity to cationic bovine serum albumin-induced membranous nephropathy. Kidney Int. 2007;72:831–840. doi: 10.1038/sj.ki.5002426. [DOI] [PubMed] [Google Scholar]
- 28.Grip O, Janciauskiene S. A torvastatin reduces plasma levels of chemokine (CXCL10) in patients with Crohn's disease. PLoS One. 2009;4:e5263. doi: 10.1371/journal.pone.0005263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ostvik AE, Granlund AV, Bugge M, et al. Enhanced expression of CXCL10 in inflammatory bowel disease: potential role of mucosal Toll-like receptor 3 stimulation. Inflamm Bowel Dis. 2013;19:265–274. doi: 10.1002/ibd.23034. [DOI] [PubMed] [Google Scholar]
- 30.Cayatte C, Joyce-Shaikh B, Vega F, et al. Biomarkers of Therapeutic Response in the IL-23 Pathway in Inflammatory Bowel Disease. Clin Transl Gastroenterol. 2012;3:e10. doi: 10.1038/ctg.2012.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sipponen T, Karkkainen P, Savilahti E, et al. Correlation of faecal cal protect in and lactoferrin with an endoscopic score for Crohn's disease and histological findings. Aliment Pharmacol Ther. 2008;28:1221–1229. doi: 10.1111/j.1365-2036.2008.03835.x. [DOI] [PubMed] [Google Scholar]
- 32.Kenward MG, Roger JH. Small sample inference for fixed effects from restricted maximum likelihood. Biometrics. 1997;53:983–997. [PubMed] [Google Scholar]
- 33.Rosengren S, Firestein GS, Boyle DL. Measurement of inflammatory biomarkers in synovial tissue extracts by enzyme-linked immunosorbent assay. Clin Diagn Lab Immunol. 2003;10:1002–1010. doi: 10.1128/CDLI.10.6.1002-1010.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Team RC. R: A language and environment for statistical computing. Vienna, Austria: R Foundation for Statistical Computing; 2013. [Google Scholar]
- 35.Bates D, Maechler M, Bolker B, et al. lme4: Linear mixed-effects models using Eigen and S4. R package version 1.0–4. 2013 http://CRNA.R-project.org/package=lme4. [Google Scholar]
- 36.Halekoh U, Hojsgaard S. pbkrtest: Parametric bootstrap and Kenward Roger based methods for mixed model comparison. R package version 0.3–7. 2013 http://CRAN.R-project.org/package=pbkrtest. [Google Scholar]
- 37.Florin TH, Paterson EW, Fowler EV, et al. Clinically active Crohn's disease in the presence of a low C-reactive protein. Scand J Gastroenterol. 2006;41:306–311. doi: 10.1080/00365520500217118. [DOI] [PubMed] [Google Scholar]
- 38.Carlson CS, Aldred SF, Lee PK, et al. Polymorphisms within the C-reactive protein (CRP) promoter region are associated with plasma CRP levels. Am J Hum Genet. 2005;77:64–77. doi: 10.1086/431366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.U.S. Department of Health and Human Services FaDA, Center for Drug Evaluation and Research. Guidance for Industry: Bioanalytical Method Validation. Volume Revision 1. 2013 Sep; ed, 2013. [Google Scholar]
- 40.Boland BS, Boyle DL, Sandborn WJ, et al. Validated gene expression biomarker analysis for biopsy-based clinical trials in ulcerative colitis. Aliment Pharmacol Ther. 2014 doi: 10.1111/apt.12862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Buch MH, Boyle DL, Rosengren S, et al. Mode of action of a batacept in rheumatoid arthritis patients having failed tumour necrosis factor blockade: a histological, gene expression and dynamic magnetic resonance imaging pilot study. Ann Rheum Dis. 2009;68:1220–1227. doi: 10.1136/ard.2008.091876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kavanaugh A, Rosengren S, Lee SJ, et al. Assessment of rituximab's immunomodulatory synovial effects (ARISE trial). 1: clinical and synovial biomarker results. Ann Rheum Dis. 2008;67:402–408. doi: 10.1136/ard.2007.074229. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Comparison of relative IL-8, IL-1β, IL-6, MMP-3, and S100-A8 expression by segment of small intestine or colon in patients with active CD, inactive CD, and healthy controls. Relative gene expression units were averaged. Standard error shown. * indicates significance with p-value <0.05.


