Abstract
Objective
The quantitative interpretation of hip cartilage MRI has been limited by the difficulty of identifying and delineating the cartilage in a 3D dataset, thereby reducing its routine usage. In this paper a solution is suggested by unfolding the cartilage to planar 2D maps on which both morphology and biochemical degeneration patterns can be investigated across the entire hip joint.
Design
Morphological TrueFISP and biochemical dGEMRIC hip images were acquired isotropically for 15 symptomatic subjects with mild or no radiographic osteoarthritis. A multi-template based label fusion technique was used to automatically segment the cartilage tissue, followed by a geometric projection algorithm to generate the planar maps. The segmentation performance was investigated through a leave-one-out study, for two different fusion methods and as a function of the number of utilized templates.
Results
For each of the generated planar maps, various patterns could be seen, indicating areas of healthy and degenerated cartilage. Dice coefficients for cartilage segmentation varied from 0.76 with four templates to 0.82 with 14 templates. Regional analysis suggests even higher segmentation performance in the superior half of the cartilage.
Conclusions
The proposed technique is the first of its kind to provide planar maps that enable straightforward quantitative assessment of hip cartilage morphology and dGEMRIC values. This technique may have important clinical applications for patient selection for hip preservation surgery, as well as for epidemiological studies of cartilage degeneration patterns. It is also shown that 10-15 templates are sufficient for accurate segmentation in this application.
Keywords: Osteoarthritis, hip, dGEMRIC, MRI, segmentation, label fusion
Introduction
At early stages of osteoarthritis (OA) the glycosaminoglycan (GAG) content in the cartilage has been shown to decrease (1). The delayed gadolinium enhanced MRI of cartilage (dGEMRIC) method is a validated imaging technique for estimation of the GAG content, based on the principle that the intravenously injected contrast agent Gd(DTPA)2- (Magnevist®, Bayer Schering Pharma) distributes in the cartilage in an inverse relationship to the GAG content (1). A quantitative measurement of T1 in the cartilage is then performed, which correlates to the distributed amount of Gd(DTPA)2-.
Although three-dimensional (3D) volumetric hip-dGEMRIC is often used, the complexity of manually segmenting and visualizing the cartilage typically restricts the routine evaluation to just a few select slices. One of the few innovative attempts to date at evaluating the full 3D volume in a clinically useful way is a scheme presented by Domayer et al (2), where an isotropic 3D T1 dataset is manually reformatted to evaluate the cartilage quality at multiple locations around the femoral head.
This paper describes a new method for straightforward quantitative whole-joint assessment of both morphology and dGEMRIC values in hip cartilage, using novel two-dimensional (2D) planar maps. This work also includes the implementation and verification of an automated segmentation technique that has never previously been utilized with hip cartilage.
Method
A multi-template based label fusion technique is used to automatically segment the cartilage from the acquired 3D data. The segmented cartilage is then unfolded using a projection algorithm, resulting in two planar maps showing the T1 variation and cartilage thickness across the joint. In addition, a leave-one-out cross validation study was performed to verify the segmentation technique and its dependence on the included number of templates.
Image acquisition
A retrospective analysis was performed on data from 15 hip dGEMRIC patients (2 males and 13 females, 13–45 years old). The cohort consisted of patients who were sent for a dGEMRIC examination by an orthopedic specialist, either as a part of a pre-surgical examination or as part of a clinical workup for hip pain. All subjects were symptomatic with mild or no radiographic OA. Hence, sufficient cartilage tissue was present for assessment using MRI and the patients were deemed representative of the typical dGEMRIC population. The study was approved by the local Institutional Review Board.
All imaging was performed on a 1.5T Siemens Avanto scanner at 45 minutes after intravenous injection of a double dose (0.4 mL/kg) Gd(DTPA)2-. T1 maps were generated using the 3D-Variable Flip Angle (3D-VFA) method (3), acquired in the oblique sagittal plane along the axis of the femoral neck using isotropic 0.83 mm3 voxels and an acquisition matrix of 192x192x100. Excitation pulse flip angles were 5° and 28° (4). TR was 15 ms and TE was 4.8 ms. Scan time was 7 min and 26 s. The subsequent TrueFISP sequence was acquired in the same plane using 0.63 mm3 voxels and an acquisition matrix of 256×256×144, covering the same region as the 3D-VFA acquisition. TR was 12.6 ms and TE was 5.5 ms. Scan time was 7 min and 47 s. Parallel imaging acceleration factor was 2, for both sequences.
The bulk cartilage inside of the acetabular rim (i.e. the combined femoral and acetabular cartilage, excluding the area around fovea), the femur and the acetabulum were segmented manually in all 15 TrueFISP datasets in order to serve as a library of template segmentations to guide the automated segmentation algorithm.
Automated segmentation
Automated segmentation was performed using a method referred to as multi-template based label fusion (5). First, all template images are registered to a target image (i.e. a TrueFISP image on which automated segmentation is to be performed), using a combined seven-parameter (rigid plus global scaling) and non-rigid block matching registration (6). By then applying the deformation field retrieved from these registrations to the associated template segmentations, a set of segmentation candidates for the target image are constructed. Finally, the segmentation candidates are fused into a consensus segmentation estimate using the Local MAP STAPLE (LS) algorithm (7). For comparison, fusion was also performed using standard majority voting (MV) (5). Total processing time was typically around three hours using 20 Intel Xeon cores.
In this work, each of the 15 cases were automatically segmented through a leave-one-out scheme, using all or a subset of the remaining cases as templates, thus keeping target and template volumes completely disjoint.
Unfolding to planar maps
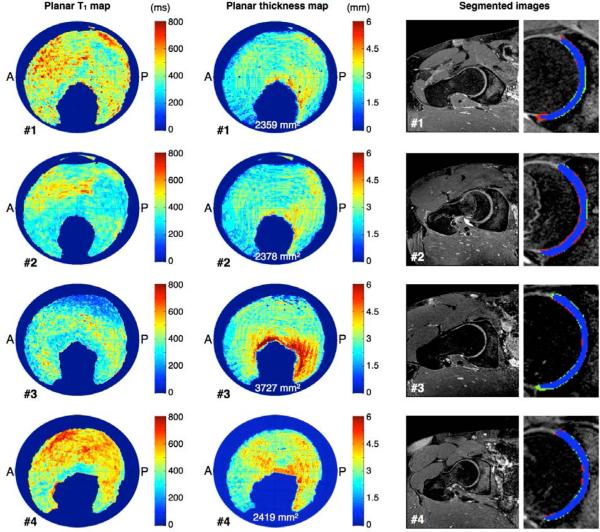
The automatically generated segmentations were used to mask out the cartilage from the T1 volume. This was followed by a sphere fitting routine (8) to find the center of the spherical cartilage structure. Finally, the Lambert azimuthal equal area projection algorithm (9) was applied to unfold the radially averaged acetabular cartilage to a T1 planar map (Fig. 1). The bulk cartilage morphological planar map and the acetabular surface area were also retrieved.
Fig 1.
Left and center column show automatically generated planar T1 maps and cartilage morphology maps for four representative cases. Anterior and posterior sides are indicated in the maps by letters A and P. Total acetabular cartilage areas are also shown within the morphology maps. Healthy hip cartilage is typically indicated by T1 values around 500 ms or above, whereas OA is typically indicated by lower T1 values. Right column show segmentation results through the weight-bearing cartilage for the same cases. Blue label represent tissue segmented in common by both manual and automatic methods, red is manual method only and green is automatic method only.
Leave-one-out cross validation
The impact of the number of templates on segmentation performance was investigated by repeatedly selecting n random cases from the 14 possible (n = 4, 6, 8, 10, 12, 14) and using them as templates for the segmentation pipeline. By repeating this 50 times, for each case and for each n < 14, average Dice's similarity coefficients (5) were calculated, for which the effect of individual template selection was averaged out. From these average Dice coefficients, the influence of the number of templates on segmentation performance can be determined. As an additional measurement to investigate regional variations in segmentation accuracy, the average Dice coefficient was also calculated only involving cartilage segmented superior to the fovea (i.e. the superior hemisphere of the femoral head cartilage).
Results
Automated segmentation
From the leave-one-out studies it was shown that LS performs better than MV for any number of templates and for all of the segmented structures (Fig. 2). In cartilage segmentation using four templates, average Dice coefficients (± SD between subjects) were 0.788 (± 0.053) and 0.761 (± 0.069) for LS and MV, respectively. Corresponding values using 14 templates were 0.824 (± 0.052) and 0.82 (± 0.056) (Fig 2a). Similar tendencies were seen in segmentations of femur and acetabulum, with a Dice coefficient in the approximate range of 0.94 to 0.96 in femur and 0.86 to 0.91 in acetabulum, as the number of templates was increased from four to 14 (Figs 2b and 2c). The separately calculated average Dice coefficient, only involving cartilage superior to the fovea, was 0.87 (± 0.039) when segmented using LS and 14 templates.
Fig 2.
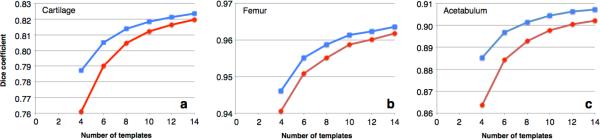
Segmentation accuracy measured by average Dice coefficient as a function of the number of templates used for Local MAP STAPLE fusion (blue curve, square markers) and Majority Voting fusion (red curve, circular markers). a) segmenting cartilage b) segmenting femur c) segmenting acetabulum
Planar maps
Various patterns of cartilage degeneration and morphology are readily demonstrated by the different color patterns on the planar maps (Fig. 1). T1 values range between 200-800 ms and bulk cartilage thickness mostly ranges between 1.0-3.5 mm. Cartilage surface area varies between 2359–3727 mm2 for the different cases.
Discussion
The primary purpose of this work is to introduce a method for assessing cartilage quality throughout the entire hip joint that is readily useful in clinical applications. As an important step, this work also verifies the performance of the segmentation method for use in this application. Although there are prior work done on automated segmentation of hip cartilage (12), to our best knowledge, this is the first work to use multi-template based label fusion for such segmentation.
From the generated planar maps (Fig. 1) the patterns of degeneration, as represented by the variations in T1 values, vary substantially between cases. Previous works have shown healthy hip cartilage to be indicated by T1 values around 500 ms or above, whereas OA at any stage is indicated by lower T1 values (10). Since the margin between the acetabular and femoral cartilages is almost indistinguishable with MRI, both cartilages were segmented together and subsequently divided. For this reason any gap between the femoral and acetabular cartilage will be included in the measurements, occasionally resulting in an overestimated bulk cartilage thickness. This is especially apparent in the area around fovea for case #3 (Fig. 1). Due to the retrospective nature of this work, the pulse sequences were originally optimized primarily for visual inspection. For this reason, bulk cartilage thickness is typically only 3-6 voxels at this current T1-resolution, which may be improved with a future protocol, optimized specifically for generating planar maps.
Since dGEMRIC investigations are normally performed on subjects with only mild OA, extensive morphological changes are not expected in the typical dGEMRIC population. The performance of the segmentation technique in severe OA is therefore not investigated in this work. Additionally, the performance effect of gender bias in the current templates is assumed to be low, as the non-rigid registration algorithm effectively accounts for morphological differences between templates and target. However, this assumption will be further investigated in a future work.
By using the multi-template based label fusion approach for automated segmentation, the effect of mis-registration of individual templates is limited (5). The demonstrated effect on the overall segmentation performance, when adjusting the number of templates, agrees well with previously shown segmentation results for neurological applications (5). The superior performance of LS compared to MV also conforms well to previously published findings (7). Given the limited segmentation improvement as more than ten templates are used, it is indicated that 10-15 templates are sufficient for use in this application. Since the total processing time increases linearly with the number of templates, keeping this number as low as possible is in the interest of making the method feasible in clinical routine.
The large difference in average Dice coefficients between the three types of structures is mostly explained by their difference in size and shape. The measured Dice coefficient for cartilage segmentation in this work is roughly equal to previously shown Dice coefficients in knee cartilage segmentation (13,14). However, since knee cartilage is known to be thicker than hip cartilage, the similar Dice coefficients actually imply superior performance of the segmentation in this work.
The lower segmentation accuracy in the anterior part of the cartilage is an effect of inconsistencies between template segmentations regarding which material is segmented in this region. In a future work, segmentations by additional raters will be included, which will reduce this type of inconsistencies.
It should be noted that as a consequence of the choice of projection algorithm, angles and distances within the planar map do not necessarily conform to true angles and distances. Instead, the chosen algorithm is optimized to preserve areas within the image, which was determined to be of highest priority in this application in order to obtain an intuitive perception of the planar maps. In this work, planar maps were generated from dGEMRIC T1 data. However, the technique can equally well be applied to any type of relevant hip cartilage data (e.g. T2 mapping or gagCEST (15)) acquired in 3D. Also, if only morphological maps are of interest the method will work well using TrueFISP images alone.
Potential and anticipated future applications for this technique include utilizing the maps to select appropriate candidates for hip arthroscopy and hip preservation surgery, or for planning tissue-engineering procedures. The overall integrity of the patient's cartilage at various locations within the hip joint plays an important, but not well understood, role on the ultimate success of the procedure. Cartilage planar maps may also help predict whether patients are likely to have pain after a surgical procedure. If patient outcome can be predicted based on pre-surgical cartilage maps, this would have enormous implications for determining which patients would benefit most from these procedures. Other applications include epidemiological studies where the patterns of degeneration may provide insight into the development of cartilage disease.
In conclusion, this work introduces a novel way of visualizing and assessing hip cartilage, by segmenting 3D volumetric data and subsequently unfolding it to a planar 2D map. The primary application of this technique is dGEMRIC T1 maps, although the technique can be applied to other types of 3D hip images as well. As part of this work it has also been shown that automated segmentation of hip cartilage using a multi-template based label fusion technique is both feasible and robust, with highly accurate segmentation results.
Acknowledgements
Portions of this research were conducted on the Orchestra High Performance Compute Cluster at Harvard Medical School, which is partially provided through NIH grant NCRR 1S10RR028832-01.
Role of the funding source
This research was supported by the Boston Children's Hospital Translational Research Program, Siemens Healthcare and The Swedish Agency for Innovation Systems. The funding sources had no involvement in the research or publishing of this work.
Footnotes
Contributions
The main drafting of the article was performed by CS, whereas all authors have reviewed and revised the manuscript critically for intellectual content. All authors have given their final approval of the submitted version of the manuscript. The conception and design of the work was made by authors CS, YJK and SKW. Software implementations were made by CS, AAA and SKW. Image acquisition and analysis was performed by CS, SB and YJK. Manual segmentation work was performed by CS and YJK. Authors CS, YJK and SKW take responsibility of the integrity of the work as whole.
Competing interest statement
CS, AAA, SB and SKW have no conflicts of interest to declare. YJK receives research support from Siemens Healthcare, which had no influence on this work.
References
- 1.Bashir A, Gray ML, Hartke J, Burstein D. Nondestructive imaging of human cartilage glycosaminoglycan concentration by MRI. Magn Reson Med. 1999;41(5):857–865. doi: 10.1002/(sici)1522-2594(199905)41:5<857::aid-mrm1>3.0.co;2-e. [DOI] [PubMed] [Google Scholar]
- 2.Domayer SE, Mamisch TC, Kress I, Chan J, Kim YJ. Radial dGEMRIC in developmental dysplasia of the hip and in femoroacetabular impingement: preliminary results. Osteoarthritis Cartilage. 2010;18(11):1421–1428. doi: 10.1016/j.joca.2010.08.010. [DOI] [PubMed] [Google Scholar]
- 3.Mamisch TC, Dudda M, Hughes T, Burstein D, Kim YJ. Comparison of delayed gadolinium enhanced MRI of cartilage (dGEMRIC) using inversion recovery and fast T1 mapping sequences. Magn Reson Med. 2008;60(4):768–773. doi: 10.1002/mrm.21726. [DOI] [PubMed] [Google Scholar]
- 4.Siversson C, Chan J, Tiderius CJ, Mamisch TC, Jellus V, Svensson J, Kim YJ. Effects of B(1) inhomogeneity correction for three-dimensional variable flip angle T(1) measurements in hip dGEMRIC at 3 T and 1.5 T. Magn Reson Med. 2012;67(6):1776–1781. doi: 10.1002/mrm.23150. [DOI] [PubMed] [Google Scholar]
- 5.Aljabar P, Heckemann RA, Hammers A, Hajnal JV, Rueckert D. Multi-atlas based segmentation of brain images: atlas selection and its effect on accuracy. Neuroimage. 2009;46(3):726–738. doi: 10.1016/j.neuroimage.2009.02.018. [DOI] [PubMed] [Google Scholar]
- 6.Suarez RO, Commowick O, Prabhu SP, Warfield SK. Automated delineation of white matter fiber tracts with a multiple region-of-interest approach. Neuroimage. 2012;59(4):3690–3700. doi: 10.1016/j.neuroimage.2011.11.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Commowick O, Akhondi-Asl A, Warfield SK. Estimating a reference standard segmentation with spatially varying performance parameters: local MAP STAPLE. IEEE Trans Med Imaging. 2012;31(8):1593–1606. doi: 10.1109/TMI.2012.2197406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Späth H. Least-Square Fitting with Spheres. J Optim Theory Appl. 1998;96(1):191–199. [Google Scholar]
- 9.Snyder JP. Map Projections: A Working Manual: USGS Professional Paper. 1987:1395. [Google Scholar]
- 10.Tiderius CJ, Jessel R, Kim YJ, Burstein D. Hip dGEMRIC in asymptomatic volunteers and patients with early osteoarthritis: the influence of timing after contrast injection. Magn Reson Med. 2007;57(4):803–805. doi: 10.1002/mrm.21190. [DOI] [PubMed] [Google Scholar]
- 11.Sur S, Mamisch TC, Hughes T, Kim YJ. High resolution fast T1 mapping technique for dGEMRIC. J Magn Reson Imaging. 2009;30(4):896–900. doi: 10.1002/jmri.21869. [DOI] [PubMed] [Google Scholar]
- 12.Nishii T, Sugano N, Sato Y, Tanaka H, Miki H, Yoshikawa H. Three-dimensional distribution of acetabular cartilage thickness in patients with hip dysplasia: a fully automated computational analysis of MR imaging. Osteoarthritis Cartilage. 2004;12(8):650–657. doi: 10.1016/j.joca.2004.04.009. [DOI] [PubMed] [Google Scholar]
- 13.Fripp J, Crozier S, Warfield SK, Ourselin S. Automatic segmentation and quantitative analysis of the articular cartilages from magnetic resonance images of the knee. IEEE Trans Med Imaging. 2010;29(1):55–64. doi: 10.1109/TMI.2009.2024743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Folkesson J, Dam EB, Olsen OF, Pettersen PC, Christiansen C. Segmenting articular cartilage automatically using a voxel classificationf approach. IEEE Trans Med Imaging. 2007;26(1):106–115. doi: 10.1109/TMI.2006.886808. [DOI] [PubMed] [Google Scholar]
- 15.Matzat SJ, van Tiel J, Gold GE, Oei EH. Quantitative MRI techniques of cartilage composition. Quant Imaging Med Surg. 2013;3(3):162–174. doi: 10.3978/j.issn.2223-4292.2013.06.04. [DOI] [PMC free article] [PubMed] [Google Scholar]